Significance
Steroids are mainly produced by animals, while bacteria are major steroid consumers in the biosphere. Anaerobic environments are major reservoirs for estrogens; however, prior to this study, biochemical mechanisms involved in anaerobic estrogen catabolism remained completely unknown. Here, we characterized anaerobic estrogen catabolic pathway in denitrifying Denitratisoma sp. strain DHT3, which includes the transformation of estrogens into androgens via a cobalamin-dependent methylation. Data presented here complete the central pathways for bacterial steroid catabolism and reveal an unprecedented role of cobalamin in microbial steroid metabolism. Sex steroids are involved in bidirectional metabolic interactions between bacteria and their eukaryotic hosts; the discovery of retroconversion of estrogens into androgens in bacteria portends unexplored microbe–host metabolic interdependencies via this cobalamin-dependent estrogen methylation reaction.
Keywords: estrogens, biocatalysis, cobalamin-dependent methyltransferase, microbial metabolism, steroids
Abstract
Steroid estrogens modulate physiology and development of vertebrates. Conversion of C19 androgens into C18 estrogens is thought to be an irreversible reaction. Here, we report a denitrifying Denitratisoma sp. strain DHT3 capable of catabolizing estrogens or androgens anaerobically. Strain DHT3 genome contains a polycistronic gene cluster, emtABCD, differentially transcribed under estrogen-fed conditions and predicted to encode a cobalamin-dependent methyltransferase system conserved among estrogen-utilizing anaerobes; an emtA-disrupted DHT3 derivative could catabolize androgens but not estrogens. These data, along with the observed androgen production in estrogen-fed strain DHT3 cultures, suggested the occurrence of a cobalamin-dependent estrogen methylation to form androgens. Consistently, the estrogen conversion into androgens in strain DHT3 cell extracts requires methylcobalamin and is inhibited by propyl iodide, a specific inhibitor of cobalamin-dependent enzymes. The identification of the cobalamin-dependent estrogen methylation thus represents an unprecedented metabolic link between cobalamin and steroid metabolism and suggests that retroconversion of estrogens into androgens occurs in the biosphere.
Sex steroids, namely androgens and estrogens, modulate physiology, development, and reproduction of animals (1–4). While required by animals, estrogens are also classified as group 1 carcinogens by the World Health Organization (https://monographs.iarc.fr/list-of-classifications/) and are often detected in surface waters of industrialized countries (5, 6). Long-term exposure to estrogen-contaminated water at nanomolar levels can disrupt the endocrine system and sexual development in animals (7, 8).
Biosynthesis of C18 estrogens from C19 androgens proceeds through the removal of the C-19 angular methyl group, resulting in the formation of an aromatic A-ring (9). This aromatization proceeds through 2 consecutive hydroxylations of the C-19 methyl group and subsequent oxidative bond cleavage between steroidal C-10 and C-19, which is catalyzed by an aromatase (namely P450arom or CYP19) at the cost of 3 NADPH and 3 O2 (SI Appendix, Fig. S1) (10). The reverse reaction (from estrogens to androgens) is thermodynamically challenging and has not been reported in any organisms.
Sex steroids are exclusively de novo synthesized by eukaryotes. However, bacteria are major consumers of steroids in the biosphere (11). Interestingly, recent studies suggested that sex steroids mediate bidirectional interactions between bacteria and their eukaryotic hosts (12, 13). Meanwhile, bacteria can also alter a host’s sex steroid profile (14). For example, intestinal Clostridium scindens is capable of converting glucocorticoids into androgens (15); Comamonas testosteroni, an opportunistic human pathogen, is capable of using a host’s androgens as the sole carbon source and electron donor (16). Furthermore, an earlier study of fecal microbiome suggested that the phylogenetic profile of gut microbiota likely affects endogenous estrogen metabolism in postmenopausal women (17).
Biochemical mechanisms involved in bacterial androgen catabolism have been studied extensively, which includes an O2-dependent 9,10-seco pathway and an O2-independent 2,3-seco pathway (Fig. 1) (18–22). In contrast, current knowledge of the mechanisms involved in estrogen catabolism is very limited. The low aqueous solubility of estrogens (∼1.5 mg/L at room temperature) (23) and the stable aromatic A-ring render estrogen a difficult substrate. Therefore, aerobic bacteria employ O2 as a cosubstrate of oxygenases to activate and to cleave the aromatic A-ring through the 4,5-seco pathway (Fig. 1) (24–26). In general, microorganisms degrade estrogens slowly under oxygen-limited or -fluctuating conditions (27). Thus, anaerobic environments, such as river sediments and marine sediments, are considered as the major reservoirs for estrogens (28). To date, only Denitratisoma oestradiolicum and Steroidobacter denitrificans have been reported to utilize estrogens under anaerobic conditions (29, 30). However, the biochemical mechanisms and catabolic genes involved in the anaerobic estrogen catabolism remain completely unknown.
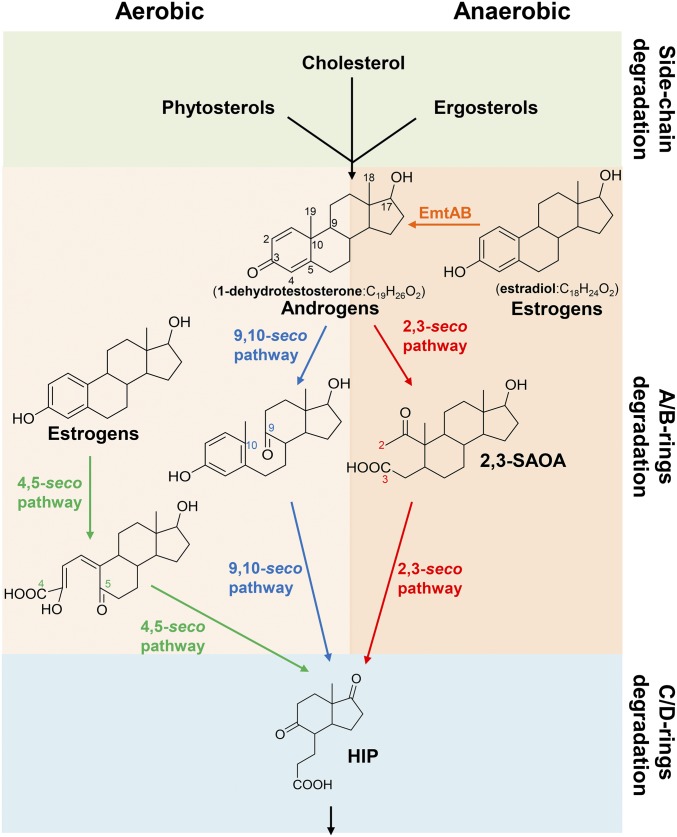
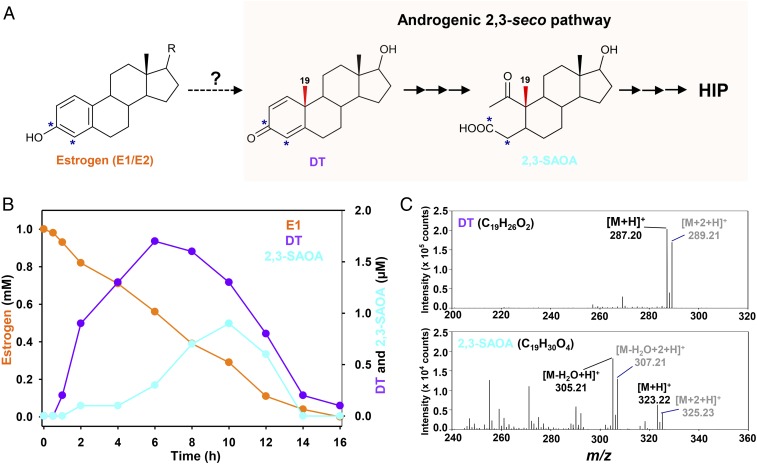
Fig. 1.
Central pathways for bacterial steroid catabolism. Bacteria adopt a convergent pathway (the 2,3-seco pathway) to catabolize different steroids under anaerobic conditions and adopt divergent pathways to catabolize estrogens (the 4,5-seco pathway) and other steroids (sterols and androgens; the 9,10-seco pathway) under aerobic conditions. All of the 3 steroid catabolic pathways converge at HIP. 2,3-SAOA, 17β-hydroxy-1-oxo-2,3-seco-androstan-3-oic acid (2,3-SAOA); HIP, 3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione.
In this study, we enriched an estrogen-degrading denitrifying bacterium Denitratisoma sp. strain DHT3 from a municipal wastewater treatment plant, which exhibits high efficiency in estrogen degradation under denitrifying conditions. We first characterized strain DHT3 and annotated its circular genome. Subsequently, we performed comparative transcriptomic analysis to identify the genes potentially involved in the anaerobic estrogen catabolism. The results along with bridging PCR analysis revealed a polycistronic gene cluster emtABCD (emt: estradiol methylation) that is differentially expressed in the strain DHT3 transcriptome under estrogen-fed conditions. Bioinformatic analysis predicted that the emtABCD gene cluster encodes a putative cobalamin-dependent methyltransferase, which is also present in D. oestradiolicum and S. denitrificans but not in other steroid-degrading anaerobes incapable of utilizing estrogens. Moreover, the emtA-disrupted strain DHT3, although incapable of growing on estrogens, is capable of growing on androgens. Finally, the 13C metabolite profile revealed estrogen consumption followed by androgen production in estradiol-fed strain DHT3 cultures. These data indicate the involvement of a cobalamin-mediated conversion of estrogens into androgens in strain DHT3.
Results and Discussion
Enrichment and Characterization of Denitratisoma sp. Strain DHT3.
The estrogen-degrading mixed culture was enriched from a denitrifying sludge that was collected from the Dihua Sewage Treatment Plant (Taipei, Taiwan). The estrogen-degrading denitrifier was highly enriched by repeating 10−8 dilution transfers in a chemically defined mineral medium containing estradiol as the sole substrate and nitrate as the terminal electron acceptor until a microscopically pure culture (vibrio-shaped cells) was obtained (SI Appendix, Fig. S2). Growth of the estradiol-degrading denitrifier on solid media (agar or gelrite plate) was not observed. The circular genome of strain DHT3 (3.66 Mb; 64.9% G + C; accession CP020914) has been sequenced and annotated (31). The phylogenetic analysis showed that the estradiol-degrading denitrifier shares a 97.5% 16S rRNA gene similarity with D. oestradiolicum DSM 16959, suggesting that it belongs to the genus Denitratisoma (31). Therefore, the estradiol-degrading denitrifier is named as Denitratisoma sp. strain DHT3 in this study.
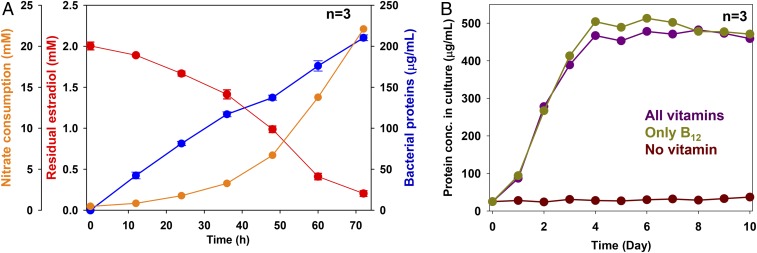
Stoichiometric analysis suggested that estradiol was mineralized to CO2 during the denitrifying growth of strain DHT3 (Fig. 2A). Strain DHT3’s growth (measured based on the increasing protein concentration in culture over time) was in parallel to the consumption of estradiol (electron donor) and nitrate (electron acceptor) in culture (1 L). After 72 h of incubation, ∼0.5 g estradiol (∼1.8 mmol) and 1.3 g of nitrate (∼21 mmol) were consumed, and ∼380 mg dry cell mass was produced in the 1-L bacterial culture. The complete oxidation of estradiol with nitrate follows the dissimilation equation (30): C18H24O2 + 23NO3− + 23H+→18CO2 + 11.5N2 + 23.5H2O. Hence, based on nitrate consumption, ∼0.25 g of estradiol (∼0.9 mmol) was completely oxidized to CO2, leaving ∼0.25 g estradiol (∼0.9 mmol) of estradiol to be assimilated into biomass. The amount of assimilated carbon from 0.9 mmol estradiol corresponds to ∼195 mg carbon. Assuming that carbon constitutes 50% of dry cell mass (32), the calculated dry cell mass produced from estradiol should be ∼390 mg. This value is close to the observed cell yield (380 mg).
Fig. 2.
Anaerobic growth of Denitratisoma sp. strain DHT3 with estradiol under denitrifying conditions and under different vitamin-supplementing conditions. (A) Anaerobic growth of strain DHT3 using estradiol as the sole substrate under denitrifying conditions. (B) Anaerobic growth of strain DHT3 on estradiol in the medium supplemented with different vitamins or without vitamins. Bacterial growth was measured based on the increasing total protein concentrations in the cultures. Results are representative of 3 individual experiments. Data shown are means ± SEM of 3 technical replicates.
Next, we characterized the substrate spectrum (SI Appendix, Table S1) and vitamin requirements of strain DHT3. Strain DHT3 is able to utilize estradiol, estrone, testosterone, acetate, fumarate, glycerol, and hexanoate as the sole substrate, but was not able to utilize other steroids, including synthetic estrogen 17α-ethynylestradiol, cholic acid, or cholesterol. The doubling time of strain DHT3 when it anaerobically grows on estrogens and testosterone ranged from 10 to 14 h and 8 to 10 h, respectively. No denitrifying growth with the following substrates was observed: yeast extract, peptone, formate, laureate, oleate, pimelate, 2-propanol, butanol, cyclohexanol, cyclopentanone, citrate, glutamate, glucose, fructose, sucrose, benzoate, toluene, or phenol. Moreover, strain DHT3 cannot utilize Fe3+, sulfate, O2, or perchlorate as the alternative electron acceptor to degrade estradiol. The vitamin requirements revealed by the growth experiments are consistent with the presence or absence of a complete set of biosynthetic genes for the given vitamin as annotated in the genome (31). Only cobalamin (namely cyanocobalamin; 20 μg/L) is required for strain DHT3’s growth (Fig. 2B). Addition of other vitamins did not facilitate the growth of strain DHT3 on estradiol (SI Appendix, Fig. S3), suggesting that strain DHT3 is cobalamin auxotrophic.
The genomes of 3 characterized anaerobic estrogen utilizers do not possess a complete set of biosynthetic genes to produce cobalamin for estrogen utilization (31). Interestingly, a previous study on microbial cholesterol catabolism also observed common cobalamin auxotrophy among cholesterol-utilizing anaerobes (33). These findings suggest that 1) the occurrence of interspecies cobalamin transfer between the cobalamin producers and the anaerobic steroid utilizers in natural habitats and 2) estrogens and other steroids are prone to accumulating in O2-limited ecosystems devoid of cobalamin or cobalamin-producing microbes. Therefore, cobalamin or cobalamin-producing anaerobes likely can be augmented into O2-limited ecosystems contaminated by sex steroids to boost in situ bioremediation.
Identification of Estrogen Catabolic Genes in Strain DHT3.
We first performed the comparative genomic analysis of the strain DHT3 genome to identify genes potentially involved in the anaerobic estrogen catabolism (Dataset S1). Consistent with the observed phenotype, the strain DHT3 genome contains a complete set of androgen catabolic genes in the established 2,3-seco pathway, including the genes involved in steroidal A/B-ring degradation (B9N43_01910 to 1920) and C/D-ring degradation (B9N43_4420 to 4465) (Fig. 3A). Moreover, the strain DHT3 genome lacks most genes for anaerobic cobalamin biosynthesis (34, 35), while it possesses the genes for cobalamin transport (B9N43_10265 to 10280) and utilization (B9N43_10425 to 10440) (Fig. 3A).
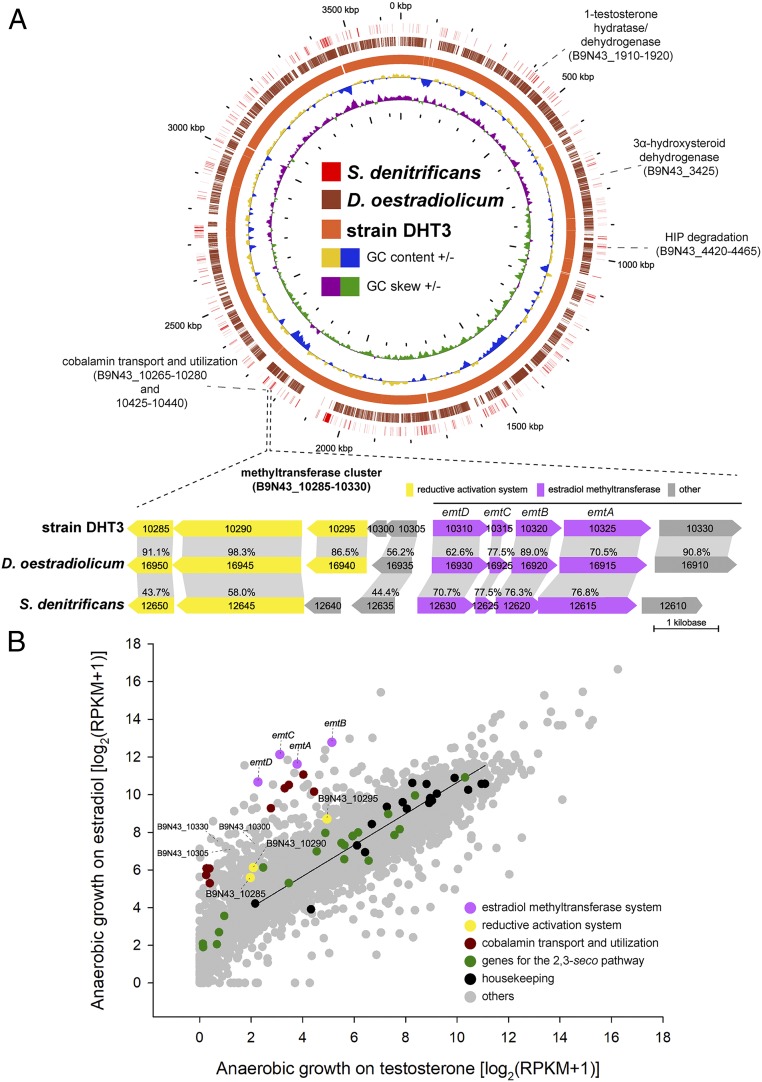
Fig. 3.
Comparative genomic analysis and comparative transcriptomic analysis of Denitratisoma sp. strain DHT3. (A) Steroid catabolic genes and the putative estrogen catabolic genes on circular genomes of strain DHT3, D. oestradiolicum DSM 16959, and S. denitrificans DSM 18526. The gene cluster emtABCD encoding putative estradiol methyltransferase is polycistronically transcribed in strain DHT3 and is present in these 3 estrogen-degrading anaerobes. Homologous open reading frames (ORFs) (colored arrows) between different bacterial genomes are connected with gray-colored blocks. Percentage (%) indicates the shared identity of the deduced amino acid sequences. (B) Global gene expression profiles (RNA-Seq) of strain DHT3 anaerobically grown on estradiol or testosterone. Each spot represents a gene. The linear regression line is based on the data points of the selected housekeeping genes (SI Appendix, Table S3). Relative gene expression values were estimated by calculating reads per kilobase transcript per million mapped reads (RPKM).
Subsequently, we performed a comparative transcriptomic analysis to detect the genes up-regulated in the estradiol-fed cultures but not in the testosterone-fed cultures. Our data suggested that genes involved in the 2,3-seco pathway are expressed at similar levels (<4-fold difference) in both estradiol-fed and testosterone-fed cultures (Fig. 3B). In contrast, the genes involved in transport, salvage, and reductive activation of cobalamin are differentially up-regulated (>5-fold difference) in the estradiol-fed cultures (Fig. 3B and Dataset S1). Among them, B9N43_10285 and _10290 encode a putative methyltransferase-activating protein (36) and a RamA-like ferredoxin (37), respectively. Additionally, a gene cluster putatively encoding a methyltransferase system (B9N43_10310 to 10325; denoted as emtABCD) was up-regulated (>5-fold difference) in the estradiol-fed culture. Notably, the emtABCD gene cluster is also present in estrogen-degrading anaerobes D. oestradiolicum and S. denitrificans but not in other steroid-degrading bacteria that cannot utilize estrogens (Fig. 3A).
We then characterized whether the mRNA products of the emtABCD cluster are polycistronically transcribed. Bridging PCR reactions were performed using primers spanning the intergenic regions of these genes (see SI Appendix, Table S2 for individual sequences). The results suggested that B9N43_10310 to 10330 are transcribed polycistronically, including emtABCD, and B9N43_10330 that encodes a putative serine hydroxymethyltransferase (SI Appendix, Fig. S4). However, the B9N43_10330-coding protein is less likely a necessary component of the putative cobalamin-dependent methyltransferase since 1) the B9N43_10330 homolog is not cooperonic with the emtABCD gene cluster in the genome of estrogen-utilizing S. denitrificans (Fig. 3A) and 2) the expression of B9N43_10330 in the transcriptome of the estradiol-fed culture is significantly lower than that of emtABCD (Fig. 3B and Dataset S1).
Functional Validation and Phylogenetic Analysis of emtA.
Our functional genomic analysis suggested that the polycistronic emtABCD is likely involved in the anaerobic estrogen catabolism. Thus, we disrupted the emtA gene in strain DHT3 using the TargeTron Gene Knockout System (with a group II intron and the kanamycin-resistant gene inserted) to validate the function of EmtABCD in the anaerobic estrogen catabolism in strain DHT3. We selected emtA for the gene disruption experiment since it is annotated as the catalytic subunit of EmtABCD. The emtA-disrupted mutant was isolated via 2 successive 10−8 dilution transfers in a defined mineral medium with testosterone as the sole substrate and kanamycin (20 μg/mL). PCR with primers flanking the emtA gene confirmed successful intragenic insertion of the group II intron into emtA in the mutant strain (Fig. 4A). The emtA-disrupted strain DHT3 can only utilize testosterone but not estradiol (Fig. 4B and SI Appendix, Fig. S5), revealing that emtA is involved in the anaerobic estrogen catabolism in strain DHT3.
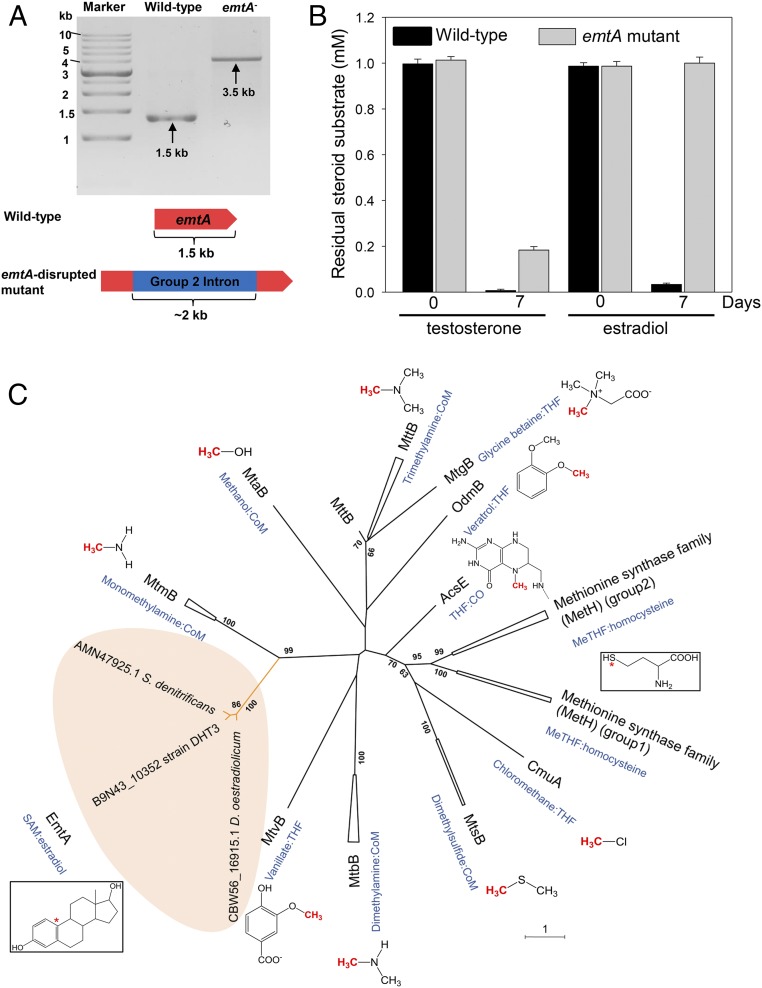
Fig. 4.
EmtA is involved in the anaerobic estrogen catabolism in Denitratisoma sp. strain DHT3. (A) Confirmation of intragenic insertion of the group II intron (∼2 kb) into the emtA of the emtA-disrupted mutant (emtA−) using PCR with the primers flanking this gene. (B) Testosterone and estradiol utilizations by the wild type or emtA-disrupted mutant of strain DHT3 (emtA−). (C) Phylogenetic relationship of EmtA and other cobamide-dependent methyltransferases. The phylogenetic tree was constructed using the neighbor-joining method with Jukes–Cantor parameter and a bootstrap value of 1,000. The asterisk (*) represents the terminal methyl acceptors for MetH and EmtA.
Subsequently, we elucidated the phylogenetic relationship of EmtA and other cobamide-dependent methyltransferases (SI Appendix, Table S4). Based on sequence homology, the most EmtA-similar protein in other organisms is MtmB (identity of protein sequence ∼30%), a catalytic subunit of the monomethylamine methyltransferase in methanogenic archaea (38). Moreover, pyrrolysine codons, a hallmark of archaeal methylamine:cobamide methyltransferases (39), are absent in emtA (SI Appendix, Fig. S6A). The phylogenetic tree showed that EmtA orthologs from the 3 estrogen-degrading anaerobes form a distinct lineage (Fig. 4C), separated from other experimentally characterized cobamide-dependent methyltransferases in prokaryotes. These EmtA orthologs were closely placed into the same clade with the MtmB in archaea Methanosarcina spp. (sequence accession nos. are provided in SI Appendix, Table S4), whereas other bacterial cobamide-dependent methyltransferases were phylogenetically distant from the EmtA orthologs (<30% sequence similarity) (Fig. 4C).
Initial Step of the Anaerobic Estrogen Catabolism in Strain DHT3 Proceeds through Estrogen Conversion into Androgens.
Next, we managed to identify the estrogen-derived metabolites by analyzing the 13C-labeled metabolite profile of the estrogen-fed strain DHT3 cultures (Fig. 5A). The strain DHT3 cultures were anaerobically incubated with a mixture of [3,4C-13C]estrone and unlabeled estrone in a 1:1 molar ratio (13C-labeled estradiol is not commercially available). The ultraperformance liquid chromatography–high resolution mass spectrometry (UPLC–HRMS) analysis revealed estrone consumption and sequential appearance of several 13C-labeled metabolites in the strain DHT3 cultures (Fig. 5B), including 6 androgenic metabolites (SI Appendix, Table S5). After 10 h of incubation, the amounts of the androgenic metabolites in the strain DHT3 cultures significantly decreased with a temporal spike of 17β-hydroxy-1-oxo-2,3-seco-androstan-3-oic acid (2,3-SAOA) (Fig. 5) and 3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione (HIP) (SI Appendix, Fig. S7), 2 characteristic ring-cleaved intermediates in the 2,3-seco pathway (20, 22), revealing that the anaerobic estrogen catabolism in strain DHT3 proceeds via C18 estrogen conversion into C19 androgens. The androgens were further degraded to HIP via the established 2,3-seco pathway. Consistently, genes in the 2,3-seco pathway were also expressed in the estradiol-fed strain DHT3 cells (Fig. 3B).
Fig. 5.
Anaerobic estrogen catabolism by Denitratisoma sp. strain DHT3 via estrogen conversion into androgens. (A) Schematic of anaerobic estrogen catabolism in strain DHT3 through a step of androgen production and subsequent degradation via the established 2,3-seco pathway. *, 13C-labeled carbon. (B) Time-dependent estrone (E1) consumption and intermediate production in the strain DHT3 cultures incubated with estrone (1 mM). Data are averages (deviations <5%) of 3 experimental measurements. (C) UPLC–HRMS-based identification of the androgenic metabolites in the estrone-fed strain DHT3 culture. The estrogen substrate contained unlabeled estrone and [3,4C-13C]estrone mixed in a 1:1 molar ratio.
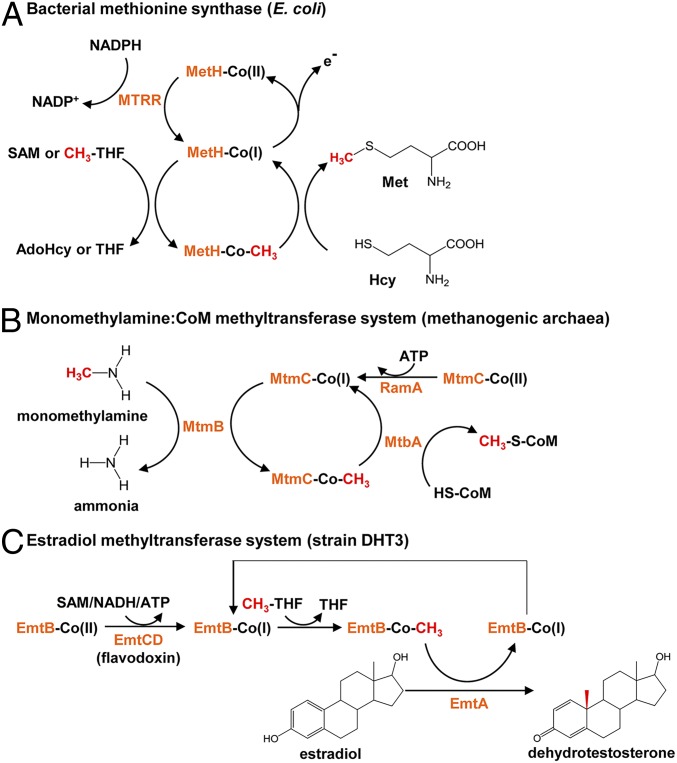
Our data suggested that the observed estrogen conversion into androgens in the strain DHT3 cultures is a methylation reaction likely catalyzed by the putative cobalamin-dependent methyltransferase EmtAB. Methionine synthase MetH, the best characterized cobalamin-dependent methyltransferase, catalyzes the methyl transfer from 5-methyl-tetrahydrofolate to homocysteine (40). In the primary catalytic cycle (Fig. 6A), the cob(I)alamin prosthetic group of MetH is methylated to form the methylcobalamin using a 5-methyl-tetrahydrofolate as the methyl donor. Subsequently, the methyl group of methylcobalamin is transferred to homocysteine to produce a methionine (41). However, the cob(I)alamin prosthetic group is prone to undergoing single-electron oxidation during the catalytic cycle, yielding the inactive cob(II)alamin even under anaerobic conditions (42); therefore, endergonic reductive activation of the cob(II)alamin prosthetic group is required for the reentry of MetH to the catalytic cycle (43), which consumes an NAD(P)H (electron donor) and a SAM (methyl donor) for the flavodoxin-mediated methylcobalamin salvage (41, 44–46) (Fig. 6A). On the other hand, reductive activation of the cob(II)amide (naturally occurring analogs of cobalamin) prosthetic group in methylamine methyltransferase in methanogenic archaea proceeds through an ATP-dependent mechanism catalyzed by RamA (37) (Fig. 6B). This ATP-dependent mechanism and RamA-like proteins also function in bacteria (47).
Fig. 6.
Proposed mechanisms involved in the catalytic cycles of EmtABCD based on the mechanisms of other characterized cobamide-dependent methyltransferases. (A) Methionine synthase MetH: In the catalytic cycle, the cobalamin prosthetic group is methylated by 5-methyl-tetrahydrofolate (CH3-THF), followed by the methyl transfer to homocysteine. For reductive activation of the cob(II)alamin prosthetic group, NAD(P)H and SAM serve as the electron donor and the methyl donor, respectively. AdoHcy, S-adenosylhomocysteine. (B) Monomethylamine:CoM methyltransferase MtmBC: the cobamide-binding subunit MtmC forms a heterotrimeric complex with MtbA (the CoM-binding subunit) and MtmB (the catalytic subunit). Reductive activation of the co(II)bamide prosthetic group proceeds through an ATP-dependent reduction catalyzed by RamA. (C) Proposed mechanism for the estradiol methylation to form 1-dehydrotestosterone by estradiol methyltransferase EmtABCD in strain DHT3. The cobalamin-binding subunit EmtB and the catalytic subunit EmtA are involved in the catalytic cycle of the estradiol methylation. Reductive activation of the co(II)balamin prosthetic group likely catalyzed by EmtCD (flavodoxin) at the cost of SAM, ATP, or NADH.
A Cobalamin-Mediated Estradiol Methylation to Form Androgens in Strain DHT3.
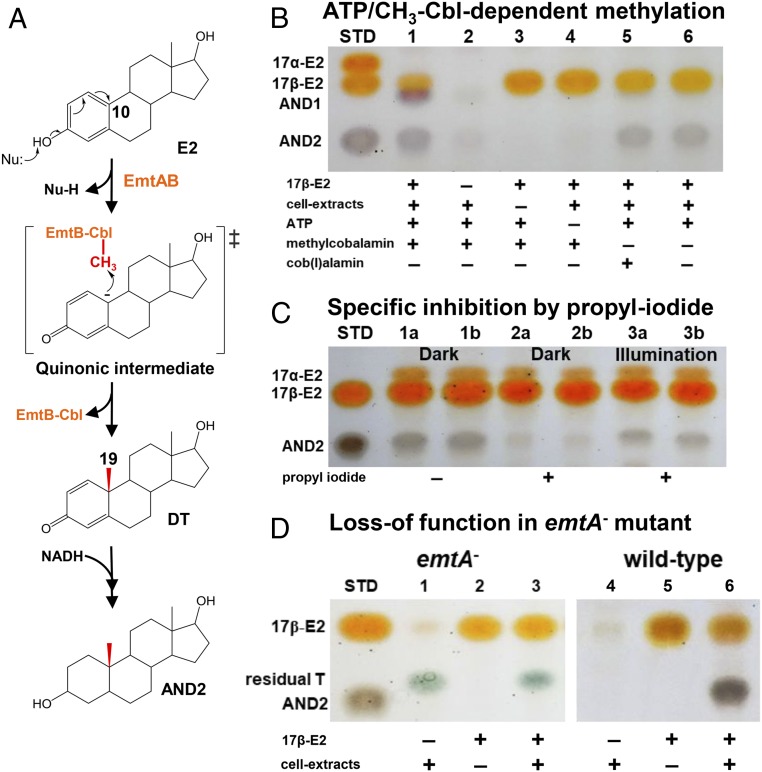
We then managed to validate the occurrence of the cobalamin-mediated estradiol methylation in strain DHT3. Thus, we monitored the methylation of estradiol (mixture of 2H-labeled estradiol and unlabeled estradiol in 1:1 molar ratio) in strain DHT3 cell extracts under anaerobic conditions, along with the addition of the reported methyl donors (SAM and methylcobalamin) and cofactors [cob(I)alamin, NAD(P)H, and ATP] required for the catalysis and reductive activation of cobalamin-dependent methyltransferases (37, 44, 45, 48, 49). After incubation, the estradiol-derived metabolites were extracted and analyzed using thin-layer chromatography (TLC) and UPLC–HRMS. We observed apparent production of 2 metabolites in the assay containing the strain DHT3 cell extracts, estradiol, ATP, and methylcobalamin, along with an obvious consumption of estradiol (lane 1 in Fig. 7B). The UPLC–HRMS analysis indicated that the 2 estradiol-derived metabolites AND1 and AND2 are both androgens (SI Appendix, Fig. S8). The 1H- and 13C-NMR spectra of AND2 (SI Appendix, Table S6) suggest the presence of 2 hydroxyl groups at C-3 and C-17 as well as 2 methyl groups (C-18 and C-19) in the steroidal structure, respectively. The fragment ion profile of the HRMS spectra (SI Appendix, Fig. S8) and the TLC characteristics of the 2 androgen metabolites were identical to those of the authentic standards of 17β-hydroxyandrostan-3-one ([M+H]+ = 291.23; C19H30O2; AND1) and 3β,17β-dihydroxyandrostane ([M+H]+ = 293.25; C19H32O2; AND2). The relative abundance of the M1 isotopomers of the 2 androgenic metabolites in the MS spectra is highly enriched (SI Appendix, Fig. S8), suggesting that they are downstream metabolites of the 2H-labeled estradiol mixture. The androgenic metabolites were not produced in the assays when estradiol, cell extracts, or ATP were excluded (lanes 2 through 4 in Fig. 7B). However, small amounts of AND2 were produced in the assays without the methylcobalamin addition (lanes 5 and 6), likely due to the presence of endogenous methylcobalamin in the strain DHT3 cell extracts. SAM addition significantly enhanced androgenic metabolite production in the strain DHT3 cell extracts in a dose-dependent manner (SI Appendix, Fig. S9). These results are analogous to the case of reductive activation of the cob(II)amide prosthetic group in MetH or in MtmC (Fig. 6).
Fig. 7.
Proposed mechanism for the Emt-catalyzed, cobalamin-mediated estradiol methylation. (A) Proposed mechanism involved in estrogenic A-ring activation and subsequent cobalamin-mediated C-10 methylation to form androgens. (B–D) TLC analysis of the cobalamin-mediated estradiol methylation in the strain DHT3 cell extracts. (B) ATP (lane 4) and methylcobalamin (lanes 5 and 6) are required for the estradiol (E2) methylation. (C) Specific inhibition of the E2 methylation in the strain DHT3 cell extracts by propyl iodide (lanes 2a/2b) in a reversible manner with daylight (lanes 3a/3b). Assays a and b are technical replicates in each treatment. All assays in C contain E2, cell extracts, ATP, NADH, and with or without propyl iodide. (D) Loss of E2 methylation activity in the cell extracts of the emtA-disrupted strain DHT3 mutant (lane 3). Assays 3 and 6 in D contain E2, cell extracts, ATP, NADH, and methylcobalamin. Abbreviations: AND1, 17β-hydroxyandrostan-3-one; AND2, 3β,17β-dihydroxyandrostane; DT, 1-dehydrotestosterone; Nu, nucleophile; STD, steroidal standards; and T, testosterone.
Next, we managed to validate the involvement of cobalamin-dependent methyltransferase in the estradiol methylation by adding propyl iodide, a specific inhibitor for cobalamin-dependent enzymes, to the assays. Propyl iodide inactivates cobalamin-dependent methyltransferases by propylating the cob(I)alamin prosthetic group in the dark (50, 51). Nevertheless, cobalamin-dependent methyltransferases can regain activity upon exposure to daylight (52). Consistently, propyl iodide addition significantly inhibited the production of the androgenic metabolites in the assays; the inhibition by propyl iodide was much less effective in the daylight-exposed assays (Fig. 7C). Furthermore, the addition of exogenous methylcobalamin, estradiol, NADH, and ATP to the cell extracts of the emtA-disrupted strain DHT3 cultures did not result in the androgen production (Fig. 7D), as opposed to the apparent production of AND2 in the assays added with wild-type strain DHT3 cell extracts. The emtA-disrupted mutant is unable to grow on estrogens; thus, both the wild-type and mutant were grown on testosterone (2 mM), with estradiol (50 μM) as an inducer of the emt genes. The emtA-disrupted mutant grew slower than the wild type (SI Appendix, Fig. S5A) and was not able to exhaust the fed testosterone (Fig. 7D), likely related to the toxicity of kanamycin in the growth medium.
Altogether, our data confirm that the anaerobic estradiol conversion into androgens in strain DHT3 is a cobalamin-mediated methylation reaction catalyzed by EmtA, the catalytic subunit of a putative cobalamin-dependent methyltransferase. Nonetheless, the specific roles of the products of emtBCD remain to be elucidated. The most EmtB-similar protein is MtbC (identity of protein sequence ∼42%), the cobamide-binding subunit of the dimethylamine methyltransferase in Methanosarcina spp (53). Pairwise alignment of the EmtB and MtbC sequences (SI Appendix, Fig. S6B) revealed that the EmtB homologs have the cobamide-binding motifs (D-x-H-x2-G-x41–42-S-x-L-x24–28-G-G) conserved in cobamide-dependent methyltransferases (54). EmtC is a hypothetical protein; the most similar protein to EmtD is F420/FMN-dependent oxidoreductase (flavodoxin) involved in reductive activation of cobamide-dependent methyltransferases (46).
For most methyl transfer reactions catalyzed by the cobamide-dependent methyltransferases (e.g., monomethylamine:coenzyme M [CoM] methyltransferase), 3 components are required to complete the methyl transfer cycle (Fig. 6B) (38, 45, 49, 55). First, the catalytic subunit MtmB (i.e., the MT1 component) transfers the methyl group from the monomethylamine to the highly reduced cob(I)amide prosthetic group in the cobamide-binding protein MtmC, followed by a second methyl transfer from methylcobamide to the final methyl acceptor CoM by the MtbA (i.e., the MT2 component). Accordingly, EmtA and EmtB are functionally analogous to MtmB and MtmC, respectively. However, the MT2 component-coding gene is missing from the strain DHT3 genome. The results of the cell-extract assays also suggested that SAM or methylcobalamin can serve as the direct methyl donor for the Emt-mediated estradiol methylation. Therefore, the Emt-mediated estradiol methylation resembles a reverse reaction of the MtmB-catalyzed methylamine demethylation and seems to employ a hybrid mechanism of RamA (ATP dependent) and MetH [NAD(P)H- and SAM-dependent] to reductively activate the cob(II)alamin prosthetic group (Fig. 6). Nevertheless, purification of the estradiol methyltransferase and further biochemical investigations are required to elucidate the specific functions of each Emt subunit in the estradiol methylation.
In this study, we found that strain DHT3 converts estrogens into androgens through a cobalamin-mediated methylation at C-10 on estrogens (Fig. 6C). However, activation of the phenolic A-ring of estrogens is a prerequisite for this methylation reaction. Given that the activated methyl group (CH3+) of methylcobalamin serves as an electrophile, the estradiol methylation in strain DHT3 likely includes the formation of a nucleophilic carboanion at C-10 on the phenolic A-ring (Fig. 7A). This catalytic strategy has been reported in many studies of anaerobic aromatic catabolism in denitrifying bacteria (56–59), which first proceeds through the deprotonation of the phenolic hydroxyl group to form a phenolate anion. Subsequently, the lone pair electrons on the deprotonated hydroxyl group are migrated to the para carbon atom through resonance of the pi system, forming a transient quinonic ring. Similarly, in the case of the estradiol methylation, the formation of a quinonic A-ring would come along with the formation of a C-10 nucleophilic carboanion, enabling the electrophilic attack by the methyl cation (CH3+) on methylcobalamin, yielding the androgenic products with a quinonic A-ring (Fig. 7A). Consistently, the 13C metabolite profile also showed that 1-dehydrotestosterone with a quinonic A-ring was produced in the strain DHT3 cultures following estradiol consumption (Fig. 5B). In the cell extracts, the produced 1-dehydrotestosterone was further converted into AND1 and AND2 via the NADH-dependent reduction of both the C-1 double bond and the C-3 keto group (Fig. 7A) by 3-ketosteroid Δ1-reductase and 3β-hydroxysteroid dehydrogenase that are constitutively expressed in strain DHT3 cells (Dataset S1). This claim is supported by the observed transformation of 1-dehydrotestosterone into AND2 and 2,3-SAOA in the strain DHT3 cell extracts with and without addition of NADH (2 mM), respectively (SI Appendix, Fig. S10).
Conclusion
In this study, we demonstrated that strain DHT3 converts estrogens into androgens via a cobalamin-mediated methylation and subsequently catabolizes the androgenic intermediates to HIP through the established 2,3-seco pathway. The discovery completes central pathways for bacterial steroid catabolism (Fig. 1). Briefly, anaerobic bacteria utilize a convergent catabolic pathway (the 2,3-seco pathway) to catabolize sterols, androgens, and estrogens, while aerobic bacteria adopt divergent pathways to catabolize 1) sterols and androgens (9,10-seco pathway) and 2) estrogens (4,5-seco pathway). Nevertheless, all 3 steroid catabolic pathways finally converge at HIP (Fig. 1) and HIP catabolic genes are conserved in the genomes of all characterized steroid-utilizing bacteria (SI Appendix, Fig. S11) (11, 60).
Cobamides such as cobalamin are a family of cobalt-containing tetrapyrrole biomolecules with essential biochemical functions in all 3 domains of life, serving as the prosthetic group for various methyltransferases, isomerases, and reductive dehalogenases (61). Before this study, the known methyl acceptors for cobalamin-dependent methyltransferases included tetrahydrofolate and homocysteine in most organisms (43, 48, 62) as well as CoM and tetrahydromethanopterin in methanogenic archaea (38, 53). Here, we demonstrated that estrogens are terminal methyl acceptors of a cobalamin-dependent methyltransferase in a denitrifying proteobacterium, revealing an unexpected role of cobalamin in steroid metabolism. Given that sex steroids are involved in bidirectional metabolic interactions between bacteria and their eukaryotic hosts, finding retroconversion of estrogens into androgens in bacteria portends unexplored microbe–host metabolic interdependencies via this Emt-mediated estrogen methylation reaction. Therefore, the emtABCD gene cluster can serve as a biomarker to elucidate the occurrence of retroconversion of estrogens in eukaryotic microbiota.
Materials and Methods
The estrogen-degrading denitrifying betaproteobacterium strain DHT3 was isolated from the estradiol-spiked anoxic sludge collected from the Dihua Sewage Treatment Plant (Taipei, Taiwan). Isolation and routine cultivation of strain DHT3 was performed at 28 °C in the dark with a headspace consisted of N2/CO2 (80:20, vol/vol). The resting cells of strain DHT3 were incubated with 13C-labeled estrone under denitrifying conditions. The cultural samples were withdrawn at different time intervals. The estrone-derived metabolites were extracted using ethyl acetate and analyzed through UPLC–atmospheric pressure chemical ionization (APCI)–HRMS. Cell extracts of strain DHT3 were prepared using a French pressure cell. After removing the membrane proteins through ultracentrifugation, the soluble proteins were anaerobically incubated with 2H-labeled estradiol (0.25 mM), ATP (5 mM), and methylcobalamin (1 mM) overnight. The resulting androgen metabolites were separated through liquid–liquid partition, TLC, and HPLC. The chemical structure of a HPLC-purified product was elucidated using NMR spectroscopy. Total RNA was extracted from strain DHT3 cells anaerobically grown with estradiol or testosterone. rRNA was removed from the total RNA samples, and cDNA samples were prepared using the RNA fragments as reverse-transcription templates. The constructed DNA libraries were sequenced using the Illumina HiSEq. 2000 system (Illumina). The emtA gene of strain DHT3 was disrupted using the TargeTron Gene Knockout System kit (Sigma-Aldrich, St. Louis, MO). Detailed materials and methods are described in SI Appendix, Supplemental Materials and Methods.
Data Availability.
Oligonucleotide primers used in this study are listed in SI Appendix, Table S2. The NMR spectral data of AND2 are provided in SI Appendix, Table S6. Nucleotide sequences of the 16S rRNA and emtABCD genes of Denitratisoma sp. strain DHT3 are shown in SI Appendix, Appendices S1–S5. Transcriptomic data of the strain DHT3 are available in SI Appendix, Dataset S1. Genome sequence of the strain DHT3 has been deposited in the National Center for Biotechnology Information (NCBI) Genome database, accession no. CP020914. The transcriptomes of the strain DHT3 have been deposited in the NCBI database [accession nos. SRR10362955 (testosterone-grown condition) and SRR10362956 (estradiol-grown condition)].
Supplementary Material
Acknowledgments
This research was funded by the Ministry of Science and Technology of Taiwan (MOST 107-2311-B-001 -021 -MY3). We gratefully appreciate Ms. Yu-Ching Wu at the Small Molecule Metabolomics Core Facility, Institute of Plant and Microbial Biology, Academia Sinica for the UPLC–HRMS analysis and Dr. Mei-Yeh Lu and her team at the High-Throughput Genomics Core Facility of the Biodiversity Research Center, Academia Sinica for genome sequencing. We also gratefully acknowledge Prof. Tony Z. Jia in Earth-Life Science Institute, Tokyo Institute of Technology for English editing. We also acknowledge Ms. Ching-Yen Tsai, Ms. I-Ting Lin, Mr. Yi-Shiang Huang, and Mr. Yu-Sheng Wang for their contributions in culture enrichments, RNA-Seq, and cell-extract activity assays.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.L.D. is a guest editor invited by the Editorial Board.
Data deposition: The genome sequence of the strain DHT3 has been deposited in the National Center for Biotechnology Information (NCBI) Genome database (accession no. CP020914). The transcriptomes of the strain DHT3 have been deposited in the NCBI database (accession nos. SRR10362955 and SRR10362956).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914380117/-/DCSupplemental.
References
- 1.Albrecht E. D., Pepe G. J., Steroid hormone regulation of angiogenesis in the primate endometrium. Front. Biosci. 8, d416–d429 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Ryan K. J., Biochemistry of aromatase: Significance to female reproductive physiology. Cancer Res. 42 (suppl. 8), 3342s–3344s (1982). [PubMed] [Google Scholar]
- 3.Mechoulam R., Brueggemeier R., Denlinger D., Estrogens in insects. Cell. Mol. Life Sci. 40, 942–944 (1984). [Google Scholar]
- 4.Tarrant A. M., Blomquist C. H., Lima P. H., Atkinson M. J., Atkinson S., Metabolism of estrogens and androgens by scleractinian corals. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136, 473–485 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Griffith D. R., Kido Soule M. C., Eglinton T. I., Kujawinski E. B., Gschwend P. M., Steroidal estrogen sources in a sewage-impacted coastal ocean. Environ. Sci. Process. Impacts 18, 981–991 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Wise A., O’Brien K., Woodruff T., Are oral contraceptives a significant contributor to the estrogenicity of drinking water? Environ. Sci. Technol. 45, 51–60 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Jobling S., et al. , Predicted exposures to steroid estrogens in U.K. rivers correlate with widespread sexual disruption in wild fish populations. Environ. Health Perspect. 114 (suppl. 1), 32–39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert M. R., Giller G. S., Barber L. B., Fitzgerald K. C., Skelly D. K., Suburbanization, estrogen contamination, and sex ratio in wild amphibian populations. Proc. Natl. Acad. Sci. U.S.A. 112, 11881–11886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyairi S., Fishman J., Radiometric analysis of oxidative reactions in aromatization by placental microsomes. Presence of differential isotope effects. J. Biol. Chem. 260, 320–325 (1985). [PubMed] [Google Scholar]
- 10.Praporski S., et al. , Organization of cytochrome P450 enzymes involved in sex steroid synthesis: PROTEIN-PROTEIN INTERACTIONS IN LIPID MEMBRANES. J. Biol. Chem. 284, 33224–33232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holert J., et al. , Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. MBio 9, e02345-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karavolos M. H., Winzer K., Williams P., Khan C. M., Pathogen espionage: Multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Vom Steeg L. G., Klein S. L., Sex steroids mediate bidirectional interactions between hosts and microbes. Horm. Behav. 88, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markle J. G., et al. , Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Ridlon J. M., et al. , Clostridium scindens: A human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 54, 2437–2449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horinouchi M., Hayashi T., Kudo T., Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 129, 4–14 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Fuhrman B. J., et al. , Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab. 99, 4632–4640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieslich K., Microbial side-chain degradation of sterols. J. Basic Microbiol. 25, 461–474 (1985). [DOI] [PubMed] [Google Scholar]
- 19.Horinouchi M., Koshino H., Malon M., Hirota H., Hayashi T., Steroid Degradation in Comamonas testosteroni TA441: Identification of metabolites and the genes involved in the reactions necessary before D-ring cleavage. Appl. Environ. Microbiol. 84, e01324-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P. H., et al. , Anaerobic and aerobic cleavage of the steroid core ring structure by Steroidobacter denitrificans. J. Lipid Res. 54, 1493–1504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang F. C., et al. , Integrated multi-omics analyses reveal the biochemical mechanisms and phylogenetic relevance of anaerobic androgen biodegradation in the environment. ISME J. 10, 1967–1983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warnke M., et al. , A patchwork pathway for oxygenase-independent degradation of side chain containing steroids. Environ. Microbiol. 19, 4684–4699 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Shareef A., Angove M. J., Wells J. D., Johnson B. B., Aqueous solubilities of estrone, 17β-estradiol, 17α-ethynylestradiol, and bisphenol A. J. Chem. Eng. Data 51, 879–881 (2006). [Google Scholar]
- 24.Coombre R. G., Tsong Y. Y., Hamilton P. B., Sih C. J., Mechanisms of steroid oxidation by microorganisms. X. Oxidative cleavage of estrone. J. Biol. Chem. 241, 1587–1595 (1966). [PubMed] [Google Scholar]
- 25.Chen Y. L., et al. , Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem. Biol. 24, 712–724.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Wu K., et al. , Identification of metabolites involved in the aerobic degradation of estrogen A/B-rings. Appl. Environ. Microbiol. 85, e02223-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czajka C. P., Londry K. L., Anaerobic biotransformation of estrogens. Sci. Total Environ. 367, 932–941 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Hanselman T. A., Graetz D. A., Wilkie A. C., Manure-borne estrogens as potential environmental contaminants: A review. Environ. Sci. Technol. 37, 5471–5478 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Fahrbach M., Kuever J., Meinke R., Kämpfer P., Hollender J., Denitratisoma oestradiolicum gen. nov., sp. nov., a 17β-oestradiol-degrading, denitrifying betaproteobacterium. Int. J. Syst. Evol. Microbiol. 56, 1547–1552 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Fahrbach M., et al. , Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 58, 2215–2223 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Chen Y. L., Wei S., Chiang Y. R., Genome analysis of the steroid-degrading denitrifying Denitratisoma oestradiolicum DSM 16959 and Denitratisoma sp. strain DHT3. bioRxiv:10.1101/710707 (22 July 2019).
- 32.von Stockar U., Liu J., Does microbial life always feed on negative entropy? Thermodynamic analysis of microbial growth. Biochim. Biophys. Acta 1412, 191–211 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Wei S. T. S., et al. , Microbial functional responses to cholesterol catabolism in denitrifying sludge. mSystems 3, e00113-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore S. J., Warren M. J., The anaerobic biosynthesis of vitamin B12. Biochem. Soc. Trans. 40, 581–586 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Shelton A. N., et al. , Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 13, 789–804 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wassenaar R. W., Daas P. J., Geerts W. J., Keltjens J. T., van der Drift C., Involvement of methyltransferase-activating protein and methyltransferase 2 isoenzyme II in methylamine:coenzyme M methyltransferase reactions in Methanosarcina barkeri Fusaro. J. Bacteriol. 178, 6937–6944 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson T., Soares J. A., Lienard T., Gottschalk G., Krzycki J. A., RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic archaea. J. Biol. Chem. 284, 2285–2295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burke S. A., Lo S. L., Krzycki J. A., Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J. Bacteriol. 180, 3432–3440 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzycki J. A., Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Peariso K., Goulding C. W., Huang S., Matthews R. G., Penner-Hahn J. E., Characterization of the zinc binding site in methionine synthase enzymes of Escherichia coli: The role of zinc in the methylation of homocysteine. J. Am. Chem. Soc. 120, 8410–8416 (1998). [Google Scholar]
- 41.Bianchi V., et al. , Escherichia coli ferredoxin NADP+ reductase: Activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J. Bacteriol. 175, 1590–1595 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menon S., Ragsdale S. W., The role of an iron-sulfur cluster in an enzymatic methylation reaction. Methylation of CO dehydrogenase/acetyl-CoA synthase by the methylated corrinoid iron-sulfur protein. J. Biol. Chem. 274, 11513–11518 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Banerjee R. V., Frasca V., Ballou D. P., Matthews R. G., Participation of cob(I) alamin in the reaction catalyzed by methionine synthase from Escherichia coli: A steady-state and rapid reaction kinetic analysis. Biochemistry 29, 11101–11109 (1990). [DOI] [PubMed] [Google Scholar]
- 44.Banerjee R., Ragsdale S. W., The many faces of vitamin B12: Catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72, 209–247 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Ragsdale S. W., Enzymology of the wood-Ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci. 1125, 129–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall D. A., Jordan-Starck T. C., Loo R. O., Ludwig M. L., Matthews R. G., Interaction of flavodoxin with cobalamin-dependent methionine synthase. Biochemistry 39, 10711–10719 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Hennig S. E., Jeoung J. H., Goetzl S., Dobbek H., Redox-dependent complex formation by an ATP-dependent activator of the corrinoid/iron-sulfur protein. Proc. Natl. Acad. Sci. U.S.A. 109, 5235–5240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufmann F., Wohlfarth G., Diekert G., O-demethylase from Acetobacterium dehalogenans–Substrate specificity and function of the participating proteins. Eur. J. Biochem. 253, 706–711 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Ragsdale S. W., Catalysis of methyl group transfers involving tetrahydrofolate and B(12). Vitam. Horm. 79, 293–324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brot N., Weissbach H., Enzymatic synthesis of methionine. Chemical alkylation of the enzyme-bound cobamide. J. Biol. Chem. 240, 3064–3070 (1965). [PubMed] [Google Scholar]
- 51.Burke G. T., Mangum J. H., Brodie J. D., Mechanism of mammalian cobalamin-dependent methionine biosynthesis. Biochemistry 10, 3079–3085 (1971). [DOI] [PubMed] [Google Scholar]
- 52.Ankel-Fuchs D., Thauer R. K., Methane formation from methyl-coenzyme M in a system containing methyl-coenzyme M reductase, component B and reduced cobalamin. Eur. J. Biochem. 156, 171–177 (1986). [DOI] [PubMed] [Google Scholar]
- 53.Ferguson D. J. Jr, Gorlatova N., Grahame D. A., Krzycki J. A., Reconstitution of dimethylamine:coenzyme M methyl transfer with a discrete corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 275, 29053–29060 (2000). [DOI] [PubMed] [Google Scholar]
- 54.McAnulla C., et al. , Chloromethane utilization gene cluster from Hyphomicrobium chloromethanicum strain CM2(T) and development of functional gene probes to detect halomethane-degrading bacteria. Appl. Environ. Microbiol. 67, 307–316 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naidu D., Ragsdale S. W., Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J. Bacteriol. 183, 3276–3281 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lack A., Fuchs G., Carboxylation of phenylphosphate by phenol carboxylase, an enzyme system of anaerobic phenol metabolism. J. Bacteriol. 174, 3629–3636 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van den Heuvel R. H., Fraaije M. W., Laane C., van Berkel W. J., Regio- and stereospecific conversion of 4-alkylphenols by the covalent flavoprotein vanillyl-alcohol oxidase. J. Bacteriol. 180, 5646–5651 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheng X., Lind M. E., Himo F., Theoretical study of the reaction mechanism of phenolic acid decarboxylase. FEBS J. 282, 4703–4713 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Ewing T. A., et al. , Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase. J. Biol. Chem. 292, 14668–14679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergstrand L. H., Cardenas E., Holert J., Van Hamme J. D., Mohn W. W., Delineation of steroid-degrading microorganisms through comparative genomic analysis. MBio 7, e00166 (2016). Correction: MBio7, e00865-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan J., et al. , Purinyl-cobamide is a native prosthetic group of reductive dehalogenases. Nat. Chem. Biol. 14, 8–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doukov T., Seravalli J., Stezowski J. J., Ragsdale S. W., Crystal structure of a methyltetrahydrofolate- and corrinoid-dependent methyltransferase. Structure 8, 817–830 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Oligonucleotide primers used in this study are listed in SI Appendix, Table S2. The NMR spectral data of AND2 are provided in SI Appendix, Table S6. Nucleotide sequences of the 16S rRNA and emtABCD genes of Denitratisoma sp. strain DHT3 are shown in SI Appendix, Appendices S1–S5. Transcriptomic data of the strain DHT3 are available in SI Appendix, Dataset S1. Genome sequence of the strain DHT3 has been deposited in the National Center for Biotechnology Information (NCBI) Genome database, accession no. CP020914. The transcriptomes of the strain DHT3 have been deposited in the NCBI database [accession nos. SRR10362955 (testosterone-grown condition) and SRR10362956 (estradiol-grown condition)].