Significance
A previously unknown biomolecular binding interaction of metal complexes is uncovered. The periphery of coordinated ligands is activated by the electrophilic metal center to selectively form covalent adducts with cysteine thiols of cancer-associated proteins. This is exemplified by gold(III) mesoporphyrin IX, which we found to 1) form thiol adducts with cysteines of thioredoxin, peroxiredoxin, and deubiquitinases via the meso-carbon atom of the porphyrin ligand; 2) target cellular thiol proteins, as revealed by correlative nanoSIMS and EM imaging techniques as well as thermal proteome profiling; and 3) display effective in vitro and in vivo anticancer activity. It is envisaged that metal coordination complexes with ligand periphery active toward nucleophilic substitution–addition can serve as new platforms for anticancer molecular targeting.
Keywords: gold, porphyrin, ligand reactivity, cysteine thiol conjugation, antitumor agents
Abstract
Cysteine thiols of many cancer-associated proteins are attractive targets of anticancer agents. Herein, we unequivocally demonstrate a distinct thiol-targeting property of gold(III) mesoporphyrin IX dimethyl ester (AuMesoIX) and its anticancer activities. While the binding of cysteine thiols with metal complexes usually occurs via M–S bond formation, AuMesoIX is unique in that the meso-carbon atom of the porphyrin ring is activated by the gold(III) ion to undergo nucleophilic aromatic substitution with thiols. AuMesoIX was shown to modify reactive cysteine residues and inhibit the activities of anticancer protein targets including thioredoxin, peroxiredoxin, and deubiquitinases. Treatment of cancer cells with AuMesoIX resulted in the formation of gold-bound sulfur-rich protein aggregates, oxidative stress-mediated cytotoxicity, and accumulation of ubiquitinated proteins. Importantly, AuMesoIX exhibited effective antitumor activity in mice. Our study has uncovered a gold(III)-induced ligand scaffold reactivity for thiol targeting that can be exploited for anticancer applications.
Transition metal complexes can be developed to display effective anticancer therapeutic properties by the judicious choice/combination of metal ions and coordination ligands (1–8). The mechanisms of cytotoxic action of many anticancer metal complexes are usually attributable to 3 modes: 1) the coordination of metal ions to biomolecules via ligand exchange reactions, with notable examples being the formation of the platinum–DNA adducts of cisplatin (1) and gold–thiol/selenol adducts of gold(I) phosphine compounds such as auranofin (9). 2) Metal complexes with metal ions supported by strong donor ligands can also be used as stable scaffolds to noncovalently interact with biomolecules. Thus, substitutionally inert metal complexes of platinum (10), gold (11), rhodium (12, 13), ruthenium, and iridium (14) have been developed to target protein chaperones, transporters, cytoskeletons, protein kinases, and DNA as well as mismatched base pairs via supramolecular noncovalent interactions. 3) Redox-active organometallic complexes of ruthenium, iridium, and osmium have been developed to mediate anticancer cytotoxicity via intracellular catalytic oxidation/reduction processes (15). Nonetheless, metal complexes with coordinated ligands that covalently bind anticancer biomolecular targets and display favorable anticancer and antitumor properties have not yet been described.
We have a longstanding interest in anticancer gold compounds that have been historically known to display potent cytotoxicity in vitro and in vivo (16–23). The gold(III) ion is generally unstable in solution but can be stabilized by the use of strong σ-donor ligands, affording stable structures for specific biomolecular interactions and targeting (11, 24–26). We have developed antitumor gold(III) porphyrins, as exemplified by stable gold(III) meso-tetraphenylporphyrin that specifically targets a mitochondrial chaperone protein, likely through noncovalent interactions (24, 27–30). The initial objective of the use of a porphyrin ligand was to stabilize the gold(III) ion and to allow Au(III)–S interaction with anticancer biomolecular targets. Nonetheless, such interaction has not been observed. A less commonly known aspect is the activation of the reactivity of the porphyrin ligand of metalloporphyrins for nucleophilic attack/substitution reactions, as revealed in early works on the reactivity of rhodium(III) octaethylporphyrin (31) and antimony(V) porphyrins (octaethylporphyrin and tetraphenylporphyrin) (32) as well as the functionalization of gold(III) meso-tetraphenylporphyrin (33) via nucleophilic reactions on the meso-carbon atom of the porphyrin ligand. In a biological context, the heme moiety in natural or engineered hemoproteins is inherently reactive through the iron center to mediate the formation of covalent links of vinyl, methyl, or meso-carbons with nucleophilic amino acids (34). Given the strong inductive effect of the gold(III) ion, we are intrigued by the possibility that meso-unsubstituted gold(III) porphyrins can react with various nucleophilic substrates in biological systems, particularly those containing thiol groups.
Cysteine thiols are nucleophiles that are abundant in cells and play important roles in enzyme catalysis, signal transduction, and cellular redox regulation (35, 36). Cellular thiols from protein cysteines also represent a large redox-active pool directly involved in the defense against oxidative stress, which may be targeted to achieve cytotoxic effects (37, 38). Furthermore, the functional cysteine thiols of many cancer-driving proteins are appealing anticancer molecular targets (39). Herein, the interactions with cellular thiols and the anticancer activities of the gold(III) porphyrin complex AuMesoIX bearing a meso-unsubstituted mesoporphyrin IX dimethyl ester ligand are described. The electrophilic methine group of AuMesoIX is capable of forming covalent thiolato adducts with protein thiols through nucleophilic aromatic substitution. These findings uncover a biochemical feature of metal complexes via metal-induced ligand reactivity that could be harnessed for anticancer applications.
Results
Synthesis and Characterization.
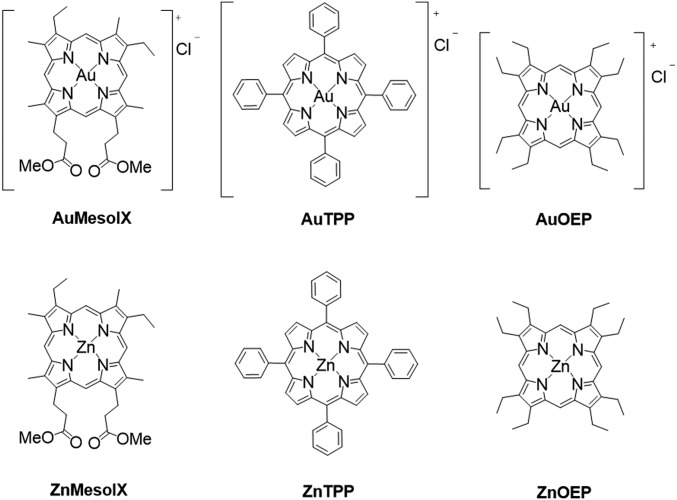
Three gold(III) porphyrin complexes [Au(Por)]Cl (Por = mesoporphyrin IX dimethyl ester [AuMesoIX], octaethylporphyrin [AuOEP], and meso-tetraphenylporphyrin [AuTPP]) were synthesized in 40 to 53% isolated yield (Fig. 1). Experimental procedures and characterization data of the complexes are detailed in SI Appendix. The crystal structures of AuMesoIX and AuOEP show the average Au–N distances being 2.029 and 2.025 Å, respectively (SI Appendix, Fig. S1 and Table S1). The porphyrin planes of both complexes do not show significant ruffling or saddling deformation. The UV–vis absorption spectra of AuMesoIX and AuOEP feature a hypsochromic shift of 10 to 30 nm for both the Soret and Q bands compared with those of the absorption maxima of AuTPP (SI Appendix, Fig. S2A). A similar trend was also observed for the zinc(II) complexes (SI Appendix, Fig. S2B), which can be attributed to the smaller degree of porphyrin macrocycle distortion in complexes with MesoIX and OEP ligands compared to that in AuTPP and ZnTPP, the porphyrin scaffolds of which have meso-phenyl substituents.
Fig. 1.
Chemical structures of gold(III) and zinc(II) porphyrin complexes.
Reactivity of Meso-Unsubstituted Gold(III) Porphyrin with Cysteine Thiols.
The 3 gold(III) porphyrin complexes were stable in biological buffer solutions as revealed by no change in the absorption spectra for a day (SI Appendix, Fig. S3). However, upon incubation with excess glutathione (GSH) for 24 h, a new absorption band at 430 nm was observed for AuMesoIX and AuOEP but not for AuTPP (SI Appendix, Fig. S4). Ultraperformance liquid chromatography (UPLC)–quadrupole time-of-flight (QTOF)–mass spectrometry (MS) analysis showed that incubation of AuMesoIX with GSH resulted in the formation of a new species with a distinct mass peak at m/z 1,094.3396 (z = +1) attributable to [AuMesoIX–GSH]+ (Fig. 2A and SI Appendix, Fig. S5). Likewise, a new species of m/z 1,034.3918 (z = +1) consistent with the [AuOEP–GSH]+ formulation was observed upon incubation of AuOEP with GSH (SI Appendix, Figs. S6 and S7). Formation of [AuMesoIX–GSH]+ and [AuOEP–GSH]+ adducts was also confirmed by matching the isotopic patterns with those of the simulated ones and by tandem-MS analysis of the fragment ions (SI Appendix, Figs. S5, S7–S9). No GSH-adduct was found for the AuTPP and MesoIX ligand (SI Appendix, Figs. S10 and S11). Individual amino acids (cysteine, glutamic acid, or glycine) of GSH were incubated with AuMesoIX or AuOEP and only the formation of adducts (m/z 908.2755 or 848.3202; z = +1; SI Appendix, Figs. S12 and S13) between the gold complexes and cysteine was observed in MS analysis.
Fig. 2.
Cysteine thiol conjugation by meso-unsubstituted gold(III) porphyrins. (A) UPLC-QTOF-MS analysis of the thiolato adduct for the parent ion of [AuMesoIX–GSH)]+ at a retention time of 4.13 min. (B) 1H and (C) 1H–13C HMBC NMR spectra of [AuOEP–GSH)]+. (D) MALDI-TOF-MS spectra of thioredoxin (Trx) before and after reaction with AuMesoIX. (E) Schematic reaction for the bioconjugation of meso-unsubstituted gold(III) porphyrins with GSH under physiological conditions.
The chemical structure of the gold(III) porphyrin-thiol adduct was also examined by NMR spectroscopy. Regarding the possible regioisomeric adducts that could be derived from AuMesoIX at different methine groups, symmetric AuOEP and GSH were used for the structural analysis (Fig. 2 B and C and SI Appendix, Figs. S16–S20). The 1H signals at δ 8.8 to 9.9 ppm are indicative of an asymmetric OEP porphyrin ring with 3 protons at different meso-carbon atoms (Fig. 2B) and the 1H signals of α-CH and β-CH2 of the cysteine residue were significantly shifted (SI Appendix, Fig. S16). Moreover, the 1H–13C heteronuclear multiple-bond correlation spectroscopy (HMBC) spectrum showed cross-peaks between the protons of β-CH2 of cysteine at 2.4 to 2.7 ppm and one meso-carbon atom of OEP at 114 ppm (Fig. 2C). Taken together, the findings reveal a covalent conjugation of the sulfhydryl group of the cysteine of GSH to the methine group of octaethylporphyrin (Fig. 2E). The UV–vis absorption spectrum of the [AuOEP–GSH]+ adduct shows a bathochromic shift (∼22 to 32 nm), weakening in intensity and band broadening of both the Soret and Q bands when compared with that of AuOEP alone (SI Appendix, Fig. S21). All of these spectral changes are indicative of an increase in the distortion of the OEP macrocyclic ring.
The interaction between AuMesoIX and a small thiol protein, thioredoxin (Trx), was studied by matrix-assisted laser desorption/ionization (MALDI)–time-of-flight (TOF)–MS. Incubation of Trx (molecular weight, 11,729.0 Da) with AuMesoIX resulted in the emergence of a new mass peak at 12,515.7 Da (Fig. 2D and SI Appendix, Fig. S22), consistent with formation of an adduct with a mass of ∼787 Da comprising one AuMesoIX and Trx. Pretreatment of Trx with the thiol-alkylating agent iodoacetamide prior to incubation with AuMesoIX resulted in no adduct formation (SI Appendix, Fig. S23). Purified and cellular Trx activities were inhibited by AuMesoIX in a dose-dependent manner (SI Appendix, Fig. S24).
The UV–vis absorption spectroscopy, NMR, and MS data allowed us to identify a nucleophilic aromatic substitution reaction between the meso-carbon atom of meso-unsubstituted gold(III) porphyrins and cysteine thiols (Fig. 2E).
In Vitro Anticancer Activity.
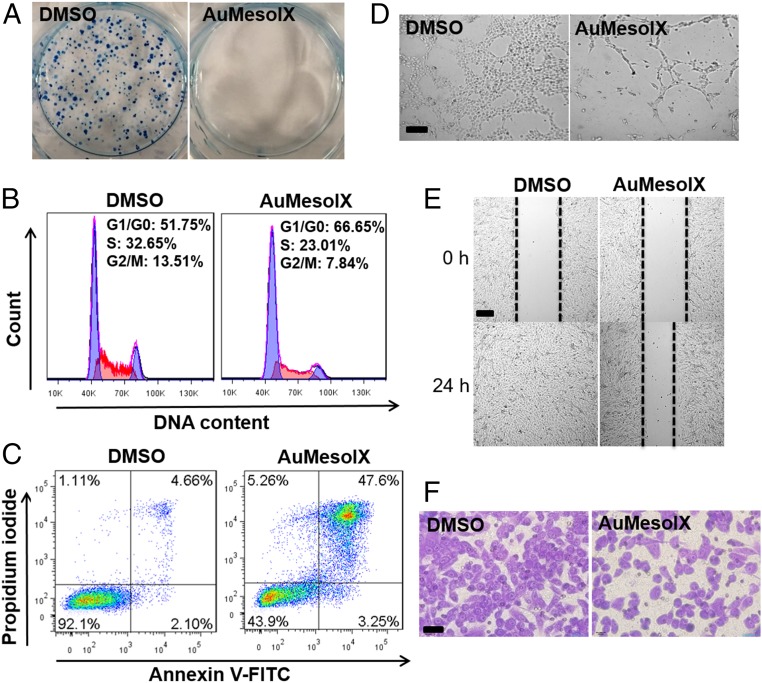
All of the gold(III) porphyrin complexes were remarkably cytotoxic in a panel of lung, colon, liver, breast, and ovarian cancer cell lines (half-maximal inhibitory concentration [IC50]: <1 μM; Table 1 and SI Appendix, Fig. S25). The corresponding zinc(II) porphyrins were almost inactive (IC50: >100 µM). AuMesoIX demonstrated a strong cytotoxic effect against both cisplatin-resistant A2780cis ovarian cancer cells (IC50: 0.12 μM) and the cisplatin-sensitive parental cells (IC50: 0.06 μM). AuMesoIX displayed a higher cytotoxicity in HCT116 colon cancer and NCI-H460 lung cancer cells compared to noncancerous colon epithelial cells (NCM460) and normal lung fibroblasts (CCD-19Lu) with 25- and 10-fold differences in IC50 values, respectively. AuMesoIX, but not the other 2 gold porphyrins, completely inhibited the clonogenic growth of A2780 ovarian cancer cells (Fig. 3A and SI Appendix, Fig. S26). AuMesoIX induced cell cycle arrest in the G0/G1 phase in A2780 cells (Fig. 3B) and triggered apoptosis (Fig. 3C). Moreover, AuMesoIX exhibited in vitro antiangiogenic activity (Fig. 3 D and E) and displayed antimetastatic activity in breast cancer cells (Fig. 3F).
Table 1.
In vitro cytotoxicity of gold(III) and zinc(II) porphyrin complexes and cisplatin on human cancerous and normal cell lines
| IC50 values, μM; 72 h | ||||||||
| Complex | A2780 (ovarian cancer) | A2780cis (ovarian cancer, cisplatin-resistant) | NCI-H460 (lung cancer) | MDA-MB-231 (breast cancer) | HepG2 (liver cancer) | HCT116 (colon cancer) | NCM460 (colon epithelial) | CCD-19Lu (lung fibroblast) |
| AuMesoIX | 0.06 ± 0.01 | 0.1 ± 0.01 | 0.07 ± 0.02 | 0.4 ± 0.02 | 0.8 ± 0.06 | 0.06 ± 0.01 | 1.5 ± 0.15 | 0.7 ± 0.04 |
| AuTPP | 0.2 ± 0.02 | 0.1 ± 0.01 | 0.2 ± 0.03 | 0.5 ± 0.03 | 0.3 ± 0.02 | 0.20 ± 0.02 | 0.4 ± 0.03 | 0.4 ± 0.04 |
| AuOEP | 0.3 ± 0.03 | 0.3 ± 0.03 | 0.2 ± 0.04 | 0.7 ± 0.05 | 0.5 ± 0.04 | 0.3 ± 0.02 | 1.4 ± 0.12 | 0.5 ± 0.03 |
| ZnMesoIX | 32.3 ± 6.9 | >100 | 15.5 ± 1.4 | 84.3 ± 9.6 | >100 | 10.2 ± 1.2 | >100 | >100 |
| ZnTPP | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| ZnOEP | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| Cisplatin | 0.7 ± 0.1 | 26.2 ± 1.9 | 3.7 ± 0.7 | 35.5 ± 1.5 | 22.3 ± 3.2 | 10.1 ± 2.2 | 93.8 ± 8.9 | 30.8 ± 4.9 |
Fig. 3.
In vitro anticancer activities of AuMesoIX. (A) Clonogenic assay of A2780 cells incubated with AuMesoIX (1 μM) for 2 h followed by incubation in fresh growth medium for 10 d. Flow-cytometric analyses of (B) the cell cycle and (C) apoptotic cell death of A2780 cells treated with 1 μM AuMesoIX for 24 h. (D) Tube formation assay of MS-1 endothelial cells treated with AuMesoIX (1 μM) for 3 h. (Scale bar, 100 μm.) (E) Wound-healing assay of MS-1 cells incubated with AuMesoIX (0.5 μM) for 24 h. (Scale bar, 100 μm.) (F) Matrigel invasion assay of MDA-MB-231 breast cancer cells treated with AuMesoIX (0.5 μM) for 24 h. (Scale bar, 40 μm.)
Cellular Uptake of AuMesoIX and In Cellulo Gold–Sulfur Interaction Revealed by Nanoscale Secondary Ion Mass Spectrometry.
AuMesoIX demonstrated rapid time- and dose-dependent cellular accumulation in A2780 cancer cells through determining the gold content by using inductively coupled plasma–MS (SI Appendix, Fig. S27 and Table S2). While AuTPP and AuOEP have similar lipophilicities as AuMesoIX (SI Appendix, Table S2), the uptake of gold for AuMesoIX-treated cells was markedly higher than that of the other 2 complexes. Such a difference was also observed in other cancer and noncancer cell lines (SI Appendix, Fig. S28). Notably, a lower gold content was found in AuMesoIX-treated noncancer cells (CCD-19Lu fibroblasts and NCM460 cells) than in cancer cells (NCI-H460 lung and HCT116 colon cancer). The uptake of gold for AuMesoIX was temperature dependent with a significant reduction at low temperature (4 °C) compared to that at 37 °C, but those of AuTPP and AuOEP displayed only minor temperature-dependent differences (SI Appendix, Fig. S29). Liquid chromatography–MS analysis of AuMesoIX-treated cells revealed the appearance of a species with m/z 775.2232 and an isotopic pattern matching that of hydrolyzed AuMesoIX with monopropionic acid (SI Appendix, Fig. S30), suggesting intracellular retention of AuMesoIX in the carboxylated form.
To examine the interaction of AuMesoIX with cellular components, nanoscale secondary ion mass spectrometry (nanoSIMS) was employed to image the intracellular elemental distribution (40, 41). NanoSIMS allows the simultaneous detection of multiple elements inside cells at high spatial resolution (50 nm). In combination with electron microscopy (EM), the correlative imaging allows the subcellular localization of added metal compounds to be visualized. As depicted in Fig. 4A and SI Appendix, Fig. S31A, the 197Au map of AuMesoIX-treated A2780 ovarian cancer cells clearly showed the presence of gold in the form of small aggregates in the cytosol. The region of 197Au aggregates was also colocalized with signals with high 32S/12C14N ratio, which could be attributed to sulfur-rich proteins. In contrast, the 197Au map of AuTPP-treated cells showed diffuse gold signals in the cytosol, and the association between the Au and S signals was not significant (SI Appendix, Fig. S31C). Scanning electron microscopy images of the same AuMesoIX-treated cells revealed the presence of small electron-dense aggregates in the mitochondria (Fig. 4B and SI Appendix, Fig. S31B). In particular, the mitochondria of AuMesoIX-treated cells displayed a disrupted morphology along with disintegrated cristae, compared to that of the untreated control (Fig. 4 B and C and SI Appendix, Fig. S32). For AuTPP-treated cells, electron-dense aggregates were hardly observed in the SEM image (SI Appendix, Fig. S31D). Taken together, the results of the nanoSIMS and EM analyses of AuMesoIX-treated cancer cells revealed a possible tight association between the gold and sulfur-rich proteins in the form of aggregates in the cytosol and disrupted mitochondria.
Fig. 4.
NanoSIMS and EM analyses of AuMesoIX-treated cells. (A) NanoSIMS images of the 197Au and 32S/12C14N secondary ion signals of A2780 cells treated with AuMesoIX (3 μM) for 18 h. (Scale bar, 4 μm.) (B) SEM images of A2780 cells treated with AuMesoIX (Left). The yellow arrows show the intracellular electron-dense aggregates in degraded mitochondria (Right). (Scale bar, 4 μm.) (C) TEM image shows the morphology of intact mitochondria in cells treated with DMSO vehicle for 18 h. (Scale bar, 0.5 μm.)
Interactions with Cancer-Associated Thiol-Containing Proteins.
Peroxiredoxin III. Thermal proteome profiling (TPP) was performed to identify the proteins engaged by AuMesoIX in cancer cells (42). Protein targets become more resistant to heat-induced denaturation upon interaction with a small molecule compound. Measurement of the protein levels in the soluble fraction (nondenatured) allows the extent of target engagement by the compound to be determined. In conjunction with MS-based quantitative proteomics analysis, the thermal stability of the potential targets against a temperature gradient can be profiled (SI Appendix, Table S3). Using this approach, peroxiredoxin III (PRDX3) was identified as one of the proteins targeted by AuMesoIX; this was confirmed by an independent cellular thermal shift assay (CETSA) using immunoblotting, which showed an increase in the melting temperature (Tm) of PRDX3 by 2.6 °C following treatment of cells with AuMesoIX (Fig. 5A). PRDX3 is a mitochondrial antioxidant thiol enzyme that functions to decompose hydrogen peroxide via 2 catalytic cysteine residues (43, 44). In this work, AuMesoIX was found to react with PRDX3-derived peptides containing catalytic cysteines as revealed by electrospray ionization (ESI)-QTOF-MS analysis (Fig. 5B). Further tandem-MS analysis of the observed adducts (m/z 1,142.5017 and 1,143.4646) revealed that AuMesoIX was conjugated to the cysteine residues (SI Appendix, Fig. S33 and Tables S4 and S5). AuMesoIX reduced the peroxidase activity of purified PRDX3 in a dose-dependent manner in which ∼40% of the activity was inhibited by 10 μM AuMesoIX (Fig. 5C and SI Appendix, Fig. S34). In immunoblot analysis, both the monomer and disulfide-bonded dimer of mitochondrial PRDX3 were observed in untreated control cells (Fig. 5D and SI Appendix, Fig. S35). Intriguingly, expression levels of the PRDX3 monomer and dimer were down-regulated upon prolonged treatment with AuMesoIX. The level of the housekeeping cytochrome c oxidase (COXIV), which resides in the mitochondrial inner membrane, was unaffected. Immunofluorescence analysis showed that AuMesoIX treatment resulted in a significant reduction in overall PRDX3 staining, while the mitochondrial staining pattern and intensity of COXIV remained unaffected (Fig. 5E). These results are indicative of specific targeting of PRDX3 by AuMesoIX.
Fig. 5.
Peroxiredoxin III PRDX3 as a target of AuMesoIX. (A) Thermal stabilization of PRDX3 following treatment of A2780 ovarian cancer cells with AuMesoIX as determined by CETSA. (B) Mass spectra of the adducts formed between AuMesoIX and peptides of PRDX3 containing different active-site cysteines; red square indicates the modification site of AuMesoIX. (C) Dose-dependent effect of AuMesoIX on recombinant PRDX3 activity as measured by NADPH-dependent hydrogen peroxide reduction via a coupled Trx/Trx reductase (TrxR) system. (D) Immunoblotting analysis of PRDX3 expression in cells treated with indicated concentrations of AuMesoIX for 8 h. (E) Immunofluorescence staining of PRDX3 (green) and COXIV (red) in cells treated with AuMesoIX or DMSO vehicle. Nuclei were stained with DAPI (blue). (F and G) Flow cytometry analysis of intracellular oxidants of cells treated with AuMesoIX (3 μM) for 8 h as examined by (F) DCFH-DA or (G) MitoPY1 probe.
Inhibition of antioxidant enzymes such as thioredoxins and peroxiredoxins will lead to an imbalance in cellular redox status. AuMesoIX treatment augmented the intracellular and mitochondrial oxidant levels as measured with the fluorogenic probes DCFH-DA (Fig. 5F), MitoSOX (SI Appendix, Fig. S36), and MitoPY1 (45) (Fig. 5G), the last of which is specific for mitochondrial hydrogen peroxide. The increases in cellular and mitochondrial oxidant levels induced by AuMesoIX could be blocked by pretreatment of cells with the antioxidant N-acetylcysteine (NAC). Notably, pretreatment of cells with NAC also attenuated AuMesoIX-induced cytotoxicity, increasing the IC50 by 6.8-fold from 0.07 to 0.48 μM (SI Appendix, Fig. S37A and Table S6), whereas treatment with the GSH synthesis inhibitor l-buthionine sulfoximine (BSO) potentiated the cytotoxic effect of AuMesoIX, decreasing the IC50 by 3.5-fold to 0.02 μM (SI Appendix, Fig. S37B and Table S6). These results suggest that AuMesoIX-induced cytotoxicity was in part mediated by oxidative stress.
Protein Deubiquitinases.
Protein deubiquitinases (DUBs) catalyze the removal of ubiquitin from ubiquitinated proteins that participate in cancer growth (46, 47). Most of the DUBs are cysteine proteases containing catalytic thiols, which are potential drug targets for DUB inhibitors. Incidentally, our TPP analysis revealed that AuMesoIX treatment increased the cellular thermal stability of a DUB of the cysteine protease class, the ubiquitin carboxyl-terminal hydrolase isozyme L3 (UCHL3) (SI Appendix, Table S3); this was verified by CETSA in which the Tm increase was found to be 1.8 °C (Fig. 6A). The interaction of AuMesoIX with peptides derived from UCHL3 and several other DUBs containing catalytic cysteines was examined using ESI-QTOF-MS. A number of doubly charged adduct ions were found in the corresponding reaction mixtures and the isotopic patterns were consistent with the simulated AuMesoIX-peptide adducts (Fig. 6B and SI Appendix, Figs. S38–S41). No adducts with peptides containing other catalytic residues such as aspartic acid and histidine were found (SI Appendix, Figs. S42 and S43). AuMesoIX inhibited the activities of several purified DUBs as well as that in cell extracts (Fig. 6C). As anticipated, treatment of cells with AuMesoIX resulted in accumulation of high–molecular-weight, polyubiquitinated proteins (Fig. 6D). We also found that AuMesoIX targets the cancer chaperone heat shock protein 90 (HSP90) (SI Appendix, Fig. S44), which mediates folding of tumor-promoting client proteins and contains functional cysteine residues (48, 49). AuMesoIX could form adducts with HSP90-derived cysteines, inhibit both in vitro and cellular HSP90 activity, and down-regulate the expression of HSP90 client proteins including several signaling proteins known to drive cancer progression (SI Appendix, Figs. S45–S49).
Fig. 6.
Protein deubiquitinase UCHL3 as a target of AuMesoIX. (A) Thermal stabilization of UCHL3 in AuMesoIX-treated A2780 cells as determined by CETSA. (B) List of the m/z of adducts formed between AuMesoIX and peptides derived from DUBs containing the catalytic amino acid residues (cysteine, aspartic acid, or histidine) as determined by ESI-QTOF-MS. (C) DUB inhibitory activity of AuMesoIX (0.5 μM) toward purified enzymes (UCHL1, 3 or 5; 0.4 μg) and A2780 cellular lysate (5 μg). (D) Immunoblot analysis of ubiquitinated protein accumulation in cells treated with AuMesoIX for 18 h. MG132 treatment (5 μM) was used as a positive control.
In Vivo Inhibition of Tumor Growth, Biodistribution, and Metabolism.
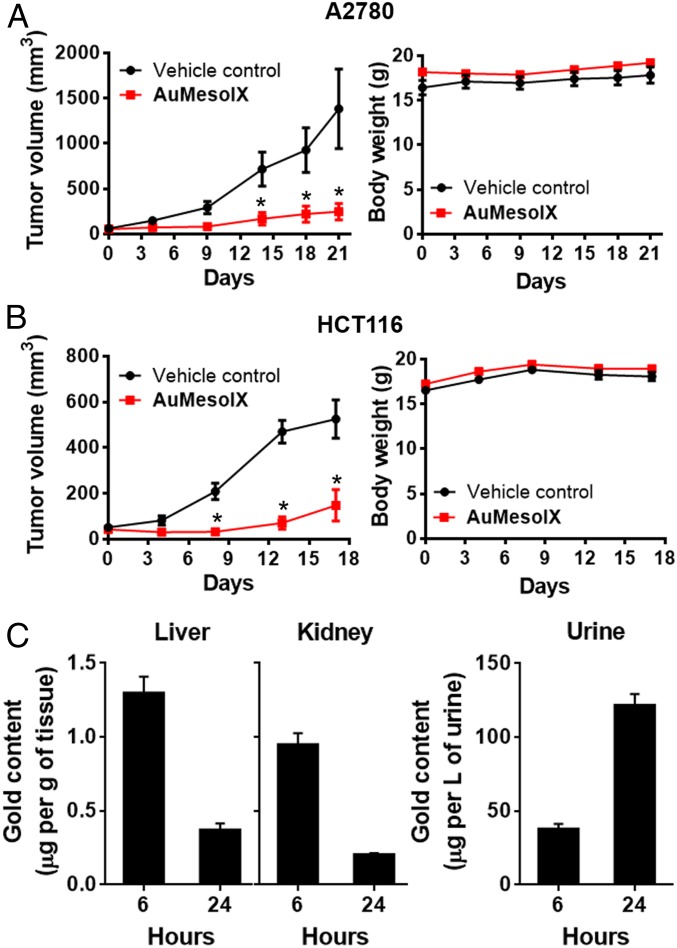
Treatment of nude mice bearing human ovarian cancer A2780 xenografts with AuMesoIX (2 mg/kg) by intravenous (i.v.) injection twice per week for 21 d resulted in suppression of tumor growth by 80% compared to those treated with vehicle control (Fig. 7A). Similar AuMesoIX treatment of mice bearing human colon cancer HCT116 xenografts was shown to reduce tumor growth by 72% (Fig. 7B). All mice survived, and no signs of toxicity or body weight loss were observed. Biodistribution of the gold(III) compounds in BALB/c mice following a single i.v. injection was studied. In AuMesoIX-administered mice, the gold content was found to be transient in the liver and kidney followed by urinary excretion (Fig. 7C). By contrast, in AuTPP-treated mice, the gold compounds accumulated in the liver and kidney with little urinary excretion (SI Appendix, Fig. S50). Possible metabolites of the gold(III) compounds were identified using the rat liver microsome as a source of biotransformation enzymes. Several metabolites of AuMesoIX including hydroxylated species and hydrolyzed products of the dimethyl ester (monopropionic and dipropionic acids) were found (SI Appendix, Fig. S51). However, metabolites of AuTPP were not detected under the same experimental conditions (SI Appendix, Fig. S52).
Fig. 7.
Antitumor activity of AuMesoIX and biodistribution in mice. Tumor volume and body weight of nude mice bearing (A) A2780 or (B) HCT116 xenografts following i.v. injection of AuMesoIX (2 mg/kg) or solvent control twice per week. Data are represented as means ± SEM (n = 5; Student’s t test; *P < 0.05, compared with vehicle control group). (C) Biodistribution of gold content in liver, kidney, and urine of BALB/C mice following i.v. injection of a single dose of AuMesoIX (2 mg/kg; n = 3).
Discussion
Conjugation of Coordinated Ligands to Biomolecules.
For decades, it has been well documented that the reactivity of coordinated organic ligands toward nucleophilic attack/addition is significantly enhanced attributable to the electron-withdrawing inductive effect and/or π-back-bonding interaction involving the metal ion. In gold porphyrin chemistry, it has also been reported that the reaction of gold(III) meso-tetraphenylporphyrin (AuTPP) with a strong base (hydroxide ion) gave gold(III) hydroxyphlorin with functionalization at the meso-carbon atom of the porphyrin scaffold (33). Nonetheless, addition/substitution reactions at the periphery of coordinated chelating ligands with biological nucleophiles have not been reported as a mechanism of action to account for the anticancer properties of metal complexes. In this work, we uncovered a previously unknown mode of interactions between metal compounds and biomolecules as illustrated by bioconjugation of cysteine thiol via the porphyrin scaffold of AuMesoIX with unsubstituted meso-carbons. The biological activities of many gold compounds are usually proposed to involve Au–S or Au–Se coordination via ligand substitution with protein thiols/selenols (18, 22). Herein, a direct Au–S interaction was not observed in the case of AuMesoIX; rather, thiol-specific bioconjugation to the porphyrin scaffold was demonstrated. This distinct thiol reactivity may account for the cytotoxic mechanisms of the gold(III) complexes and could be further exploited for anticancer applications.
The unsubstituted meso-positions of porphyrins are known to be the most reactive sites for both electrophilic and nucleophilic substitutions (50–52). In the literature, modification of the porphyrin scaffold at meso-positions has been reported on activated porphyrin systems with aromatic π-cation radical/π-dications on an oxidized porphyrin scaffold (51), electron-withdrawing substituents at meso-carbons (52), or high-valent metal centers (31–33). By varying the inductive effect on the conjugated π-electron system, the chemical reactivity of the porphyrin macrocycle can be modulated. For instance, the central metal ion of metalloporphyrin, such as Sb(V) (32) or Au(III) (33) of meso-tetraphenylporphyrin, with a high oxidation state and Lewis acidity strongly reduces the electron density at the porphyrin periphery and hence is susceptible to nucleophilic meso-hydroxylation. On the other hand, divalent metal ions, such as Zn(II) or Cd(II), is incapable to activate the peripheral atoms of porphyrin toward nucleophilic agents (53). In our case, the strong Lewis acidity of Au(III) rendered the porphyrin ring of AuMesoIX or AuOEP more reactive toward bioconjugation with the cysteine thiol of GSH compared with ZnMesoIX wherein only trace amount of adduct was observed (SI Appendix, Fig. S15). The MesoIX free ligand without the Au(III) ion also displayed no adduct formation under similar reaction conditions (SI Appendix, Fig. S11), indicating that thiol conjugation of the porphyrin scaffold of AuMesoIX is mediated by the electrophilic Au(III) center. In addition to the electronic effect, the meso-reactivity of porphyrin can be masked by the presence of meso- or β-pyrrolic substituents (50). For AuTPP with tetra-phenylated meso-substituents, no detectable adduct in the reactions with GSH/cysteine was found in MS analysis (SI Appendix, Figs. S10 and S14). Moreover, the lower abundance of adduct formed from the reaction between AuOEP and GSH compared to that with AuMesoIX (Fig. 2A and SI Appendix, Fig. S6A) can be explained by the lower meso-reactivity of AuOEP due to the steric hindrance of β-ethyl substituents.
Thiol-Selective Conjugation of Gold(III) Meso-Unsubstituted Porphyrins.
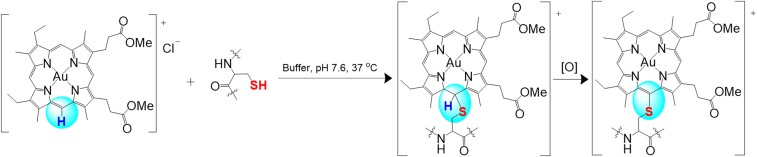
The porphyrin ligand bioconjugation of gold(III) meso-unsubstituted porphyrins was demonstrated to be cysteine-selective in experiments with individual amino acids, different peptides with or without cysteine, and protein with thiol blocking agent (Fig. 2D and SI Appendix, Figs. S12, S13, S22, S23, and S38–S43). This bioconjugation was confirmed to be nucleophilic aromatic substitution between the thiol group of cysteine and the methine H atom of meso-unsubstituted gold(III) porphyrin, as verified by the 1H and 1H–13C HMBC NMR spectroscopy data as well as the high-resolution MS measurements (Fig. 2 and SI Appendix, Figs. S5–S9, S12, S13, and S16). To date, only one report on gold(III) mesoporphyrin IX, which was limited to the study of electrochemical properties, has been published, in which the central Au(III) ion was demonstrated to be electrochemically inactive upon coordination by the mesoporphyrin IX ligand (54). By considering the steric hindrance and electron-withdrawing inductive effect of the gold(III) ion, we propose that the delta-meso-carbon of the porphyrin of AuMesoIX is activated for nucleophilic aromatic substitution with thiols of peptides and proteins (Scheme 1).
Scheme 1.
Proposed reaction of AuMesoIX with thiol-containing biomolecules under physiological conditions.
Distinct Anticancer Properties of AuMesoIX.
Targeting antioxidant and oncogenic signaling proteins by blocking functional cysteine thiols is an attractive anticancer strategy (37–39). Our studies show that thiol-modifying properties of AuMesoIX may in part contribute to its anticancer activities. In AuMesoIX-treated cells, a tight association of gold with sulfur-rich proteins in mitochondria was demonstrated by correlative nanoSIMS and EM imaging (Fig. 4), whereas in AuTPP-treated cells only weak correlation of gold and sulfur signals was observed (SI Appendix, Fig. S31C). From thermal proteome profiling and individual cellular experiments, we identified 2 possible protein targets of AuMesoIX, mitochondrial peroxiredoxin III (43, 44) and deubiquitinase UCHL3 (47), which are thiol-dependent enzymes essential for cancer cell survival and hence attractive anticancer molecular targets. AuMesoIX could conjugate with active cysteine residues on peptides of these thiol-based proteins and inhibit their biological activities, which may be linked to its ability to induce accumulation of intracellular reactive oxidants and polyubiquitinated proteins (Figs. 5 and 6). AuMesoIX also forms a covalent adduct with thioredoxin (Fig. 2D) and inhibits thioredoxin activity (SI Appendix, Fig. S24), which may further impose oxidative stress on cancer cells (55). The increases in cellular oxidant levels, as well as the cytotoxicity induced by AuMesoIX, could be attenuated by pretreatment of cells with the antioxidant NAC and potentiated by treatment with the GSH synthesis inhibitor BSO (Fig. 5 F and G and SI Appendix, Figs. S36 and S37 and Table S6). However, the cytotoxicity of AuTPP was not significantly affected by these redox modulators. These results suggest that AuMesoIX-induced cytotoxicity may be, at least in part, mediated by oxidative stress attributable to the thiol-targeting property of the compound. Importantly, AuMesoIX displayed effective in vivo antitumor activities with favorable metabolism, biodistribution, and clearance conferred by the quasiphysiological mesoporphyrin IX ligand (Fig. 7).
Implications of Metal-Induced Ligand Reactivity for Anticancer Compound Design.
Our findings demonstrate that anticancer transition metal complexes can target biological thiols through metal-induced ligand bioconjugation. In the literature, the activation of coordinative ligand reactivity by electron-deficient metal ions is fundamental to organometallic chemistry for synthetic applications. Different metal catalysts with electrophilic metal ions such as gold, platinum, and palladium have been reported for the activation of olefins toward nucleophilic reactions and C–H bond functionalization of small organic compounds via carbon–carbon and/or carbon–heteroatom bond formation (56, 57). In addition, transition-metal–mediated bioconjugation for the modification of peptides and proteins has recently been described (58). Although the high chemoselectivity of thiol conjugation can be achieved by these metal catalysts, their biocompatibility and pharmacological application remain to be determined.
Protein thiol conjugation is usually mediated by organic Michael addition-type thiol alkylators involving the α,β-unsaturated carbonyl group as a functional handle (59). Only the carbonyl group adjacent to the alkene allows polarization of the Michael acceptor for nucleophilic conjugation. By virtue of the versatility of metal–ligand coordination, it is conceivable that transition metal complexes can be a distinct platform for thiol conjugation via adjustment of the electronic properties and optimization of the ligand scaffold. We envision that by judicious selection of Lewis acidic metal ions and electrophile-containing ligands such as meso-unsubstituted porphyrin, specific targeting to cancer-associated proteins with tunable bioconjugation reactivity can be achieved.
Conclusion
Our study showed a special mode of biomolecular interaction of metal complexes, as illustrated by cysteine thiol conjugation to the gold(III)-activated porphyrin scaffold of AuMesoIX through nucleophilic aromatic substitution. AuMesoIX can covalently modify the cysteine thiols of cancer-associated proteins and exhibit potent cancer cell cytotoxicity as well as effective in vivo antitumor activities, demonstrating an alternative modality for cysteine targeting and for the development of cancer therapeutics.
Materials and Methods
Synthesis.
The gold(III) porphyrin complexes were synthesized by reacting the corresponding porphyrin ligands with KAuCl4 in glacial acetic acid following Fleischer’s procedure (SI Appendix, Materials and Methods). The AuOEP-GSH adduct was prepared by the reaction of AuOEP with GSH in ammonium bicarbonate buffer solution (pH 7.4) at 37 °C for 18 h and purified by flash column chromatography (silica gel). Detailed procedures and characterization of all of the synthesized compounds and the thiolato adduct are described in SI Appendix.
Reactivity of Gold(III) Meso-Unsubstituted Porphyrins with Cysteine Thiols.
Gold(III) porphyrin complexes were incubated with respective freshly prepared GSH, cysteine-containing peptides derived from PRDX3, DUBs, and HSP90, or Trx protein in ammonium bicarbonate buffer solution (pH 7.4) at 37 °C for 18 h. For the peptide adducts, the reaction mixtures were diluted with methanol and subjected to ESI-QTOF-MS analysis. For the protein adduct, samples were diluted with saturated sinapinic acid solution and analyzed by MALDI-QTOF-MS.
Physical Measurements, MS, Proteomic Analysis, and Bioimaging.
1H, 13C, 1H–13C heteronuclear single-quantum coherence, and 1H–13C HMBC NMR spectra were recorded on a Bruker Avance DPX 400 or 500 spectrometer at 298 K with chemical shift relative to tetramethylsilane (for CDCl3) or residual nondeuterated solvent (for CD3OD). UV-vis spectra were recorded on a Hewlett-Packard 8452A diode array spectrophotometer. X-ray diffraction data were collected on a Bruker Proteum X8 diffractometer with monochromated Cu-Kα radiation (λ = 1.54178 Å). The gold contents in cells and mouse tissues were determined using an inductively coupled plasma mass spectrometer (Agilent 7500a). Mass spectra of metal compounds, peptides, and proteins were recorded on a Q-TOF Premier quadrupole time-of-flight tandem mass spectrometer (Waters Micromass) or MALDI-TOF-MS (Bruker ultrafleXtreme). Thermal proteome profiling was performed by bottom-up proteomic analysis of tandem mass tag (TMT)-labeled peptides from trypsin-digested proteins using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) (42). Bioimaging of the elemental distribution in cells was performed on a nanoscale secondary ion mass spectrometer (Cameca NanoSIMS 50) with correlative electron microscopic imaging (40).
Data Availability.
The X-ray crystallographic data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) (accession nos. 1919615 [AuMesoIX] and 1919616 [AuOEP]). Thermal proteome profiling datasets are available via ProteomeXchange Consortium with identifier PXD016875. Detailed experimental procedures on general chemical and biological analyses; analytical data including NMR and mass spectra, UPLC/MS and UV-vis absorption spectra, NanoSIMS and EM images, thermal proteome profiling data, and biological results are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Harry Gray for valuable comments. We also thank Dr. Clive Yik-Sham Chung and Dr. Ka-Pan Shing for helpful discussions. We acknowledge the Core Facility of Li Ka Shing Faculty of Medicine for confocal imaging and flow cytometry. This work was supported by the Innovation and Technology Fund (ITS/130/14FP and ITS/188/18) and Health@InnoHK administered by Innovation and Technology Commission; and General Research Fund (HKU 17300518 and 17160916) from the Research Grants Council.
Footnotes
The authors declare no competing interest.
Data deposition: The X-ray crystallographic data have been deposited in the Cambridge Crystallographic Data Centre (CCDC), https://www.ccdc.cam.ac.uk/ (accession nos. 1919615 [AuMesoIX] and 1919616 [AuOEP]). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, https://www.ebi.ac.uk/pride/archive/ (ID code PXD016875).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915202117/-/DCSupplemental.
References
- 1.Jung Y., Lippard S. J., Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 107, 1387–1407 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bruijnincx P. C., Sadler P. J., New trends for metal complexes with anticancer activity. Curr. Opin. Chem. Biol. 12, 197–206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berners-Price S. J., Filipovska A., Gold compounds as therapeutic agents for human diseases. Metallomics 3, 863–873 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Gasser G., Ott I., Metzler-Nolte N., Organometallic anticancer compounds. J. Med. Chem. 54, 3–25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suntharalingam K., et al. , Bidentate ligands on osmium(VI) nitrido complexes control intracellular targeting and cell death pathways. J. Am. Chem. Soc. 135, 14060–14063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mjos K. D., Orvig C., Metallodrugs in medicinal inorganic chemistry. Chem. Rev. 114, 4540–4563 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Muhammad N., Guo Z., Metal-based anticancer chemotherapeutic agents. Curr. Opin. Chem. Biol. 19, 144–153 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Miodragović Đ., et al. , Arsenoplatin-1 is a dual pharmacophore anticancer agent. J. Am. Chem. Soc. 141, 6453–6457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiskus W., et al. , Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res. 74, 2520–2532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun R. W.-Y., et al. , Luminescent cyclometalated platinum(II) complexes containing N-heterocyclic carbene ligands with potent in vitro and in vivo anti-cancer properties accumulate in cytoplasmic structures of cancer cells. Chem. Sci. 2, 728–736 (2011). [Google Scholar]
- 11.Fung S.-K., et al. , Cyclometalated gold(III) complexes containing N-heterocyclic carbene ligands engage multiple anti-cancer molecular targets. Angew. Chem. Int. Ed. Engl. 56, 3892–3896 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Boyle K. M., Barton J. K., A family of rhodium complexes with selective toxicity toward mismatch repair-deficient cancers. J. Am. Chem. Soc. 140, 5612–5624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierre V. C., Kaiser J. T., Barton J. K., Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl. Acad. Sci. U.S.A. 104, 429–434 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dörr M., Meggers E., Metal complexes as structural templates for targeting proteins. Curr. Opin. Chem. Biol. 19, 76–81 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Soldevila-Barreda J. J., Sadler P. J., Approaches to the design of catalytic metallodrugs. Curr. Opin. Chem. Biol. 25, 172–183 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Simon T. M., Kunishima D. H., Vibert G. J., Lorber A., Screening trial with the coordinated gold compound auranofin using mouse lymphocyte leukemia P388. Cancer Res. 41, 94–97 (1981). [PubMed] [Google Scholar]
- 17.Shaw C. F., III, Gold-based therapeutic agents. Chem. Rev. 99, 2589–2600 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Ott I., On the medicinal chemistry of gold complexes as anticancer drugs. Coord. Chem. Rev. 253, 1670–1681 (2009). [Google Scholar]
- 19.Nobili S., et al. , Gold compounds as anticancer agents: Chemistry, cellular pharmacology, and preclinical studies. Med. Res. Rev. 30, 550–580 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Casini A., Messori L., Molecular mechanisms and proposed targets for selected anticancer gold compounds. Curr. Top. Med. Chem. 11, 2647–2660 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Berners-Price S. J., Barnard P. J., “Therapeutic gold compounds” in Ligand Design in Medicinal Inorganic Chemistry, Storr T., Ed. (Wiley, 2014), pp. 227–256. [Google Scholar]
- 22.Zou T., Lum C.-T., Lok C.-N., Zhang J. J., Che C.-M., Chemical biology of anticancer gold(III) and gold(I) complexes. Chem. Soc. Rev. 44, 8786–8801 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Bertrand B., Williams M. R. M., Bochmann M., Gold(III) complexes for antitumor applications: An overview. Chemistry 24, 11840–11851 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Hu D., et al. , Anticancer gold(III) porphyrins target mitochondrial chaperone hsp60. Angew. Chem. Int. Ed. Engl. 55, 1387–1391 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Yan J. J., et al. , Cyclometalated gold(III) complexes with N-heterocyclic carbene ligands as topoisomerase I poisons. Chem. Commun. (Camb.) 46, 3893–3895 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Teo R. D., et al. , A cytotoxic and cytostatic gold(III) corrole. Chem. Commun. 50, 13789–13792 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Che C.-M., et al. , Gold(III) porphyrins as a new class of anticancer drugs: Cytotoxicity, DNA binding and induction of apoptosis in human cervix epitheloid cancer cells. Chem. Commun. (Camb.) 1718–1719 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., He Q. Y., Sun R. W.-Y., Che C.-M., Chiu J.-F., GoldIII porphyrin 1a induced apoptosis by mitochondrial death pathways related to reactive oxygen species. Cancer Res. 65, 11553–11564 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Tu S., et al. , Gold (III) porphyrin complexes induce apoptosis and cell cycle arrest and inhibit tumor growth in colon cancer. Cancer 115, 4459–4469 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Lum C.-T., Sun R. W.-Y., Zou T.-T., Che C.-M., Gold(III) complexes inhibit growth of cisplatin-resistant ovarian cancer in association with upregulation of proapoptotic PMS2 gene. Chem. Sci. 5, 1579–1584 (2014). [Google Scholar]
- 31.Setsune J. I., Yazawa T., Ogoshi H., Yoshida Z. I., Meso-substitution reactions of rhodium(III)-octaethylporphyrins with organolithium reagents. J. Chem. Soc. Perkin Trans. 1 1980, 1641–1645 (1980). [Google Scholar]
- 32.Knör G., Spontaneous nucleophilic addition of hydroxide ions to the meso-position of high-valent antimony-oxo porphyrin complexes. J. Inorg. Biochem. 84, 297–299 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Segawa H., Azumi R., Shimidzu T., Direct hydroxylation at the meso position of gold(III) tetraphenylporphyrin by nucleophilic addition: Novel hydroxyphlorin derivatives. J. Am. Chem. Soc. 114, 7564–7565 (1992). [Google Scholar]
- 34.Lin Y. W., The broad diversity of heme-protein cross-links: An overview. Biochim. Biophys. Acta 1854, 844–859 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Pace N. J., Weerapana E., Diverse functional roles of reactive cysteines. ACS Chem. Biol. 8, 283–296 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Paulsen C. E., Carroll K. S., Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen R. E., Roth D., Winther J. R., Quantifying the global cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 106, 422–427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorrini C., Harris I. S., Mak T. W., Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Visscher M., Arkin M. R., Dansen T. B., Covalent targeting of acquired cysteines in cancer. Curr. Opin. Chem. Biol. 30, 61–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee R. F. S., et al. , Application of imaging mass spectrometry approaches to facilitate metal-based anticancer drug research. Metallomics 9, 365–381 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Legin A. A., et al. , Multi-scale imaging of anticancer platinum(IV) compounds in murine tumor and kidney. Chem. Sci. 7, 3052–3061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savitski M. M., et al. , Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Wood Z. A., Schröder E., Robin Harris J., Poole L. B., Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Chang T. S., et al. , Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J. Biol. Chem. 279, 41975–41984 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Dickinson B. C., Chang C. J., A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J. Am. Chem. Soc. 130, 9638–9639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pal A., Young M. A., Donato N. J., Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 74, 4955–4966 (2014). [DOI] [PubMed] [Google Scholar]
- 47.D’Arcy P., Wang X., Linder S., Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Whitesell L., Lindquist S. L., HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Taipale M., Jarosz D. F., Lindquist S., HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Fuhrhop J. H., “Irreversible reactions on the porphyrin periphery (excluding oxidations, reductions, and photochemical reactions)” in The Porphyrins, Dolphin D., Ed. (Academic Press, San Diego, 1978), pp. 131–159. [Google Scholar]
- 51.Smith K. M., Barnett G. H., Evans B., Martynenko Z., Novel meso-substitution reactions of metalloporphyrins. J. Am. Chem. Soc. 101, 5953–5961 (1979). [Google Scholar]
- 52.Jiang X., Nurco D. J., Smith K. M., Direct meso-alkylation of meso-formylporphyrins using Grignard reagents. Chem. Commun. 1996, 1759–1760 (1996). [Google Scholar]
- 53.Vicente M. G., “Reactivity and functionalization of β‐substituted porphyrins and chlorins” in The Porphyrin Handbook, Kadish K. M., Smith K. M., Guilard R., Eds. (Academic Press, Boston, 2003), pp. 149–199. [Google Scholar]
- 54.Jamin M. E., Iwamoto R. T., Gold porphyrin complexes. Evidence for electrochemically inert gold(III). Inorg. Chim. Acta 27, 135–143 (1978). [Google Scholar]
- 55.Arnér E. S., Holmgren A., The thioredoxin system in cancer. Semin. Cancer Biol. 16, 420–426 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Chianese A. R., Lee S. J., Gagné M. R., Electrophilic activation of alkenes by platinum(II): So much more than a slow version of palladium(II). Angew. Chem. Int. Ed. Engl. 46, 4042–4059 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Hartwig J. F., Carbon-heteroatom bond formation catalysed by organometallic complexes. Nature 455, 314–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C., Vinogradova E. V., Spokoyny A. M., Buchwald S. L., Pentelute B. L., Arylation chemistry for bioconjugation. Angew. Chem. Int. Ed. Engl. 58, 4810–4839 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boutureira O., Bernardes G. J., Advances in chemical protein modification. Chem. Rev. 115, 2174–2195 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray crystallographic data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) (accession nos. 1919615 [AuMesoIX] and 1919616 [AuOEP]). Thermal proteome profiling datasets are available via ProteomeXchange Consortium with identifier PXD016875. Detailed experimental procedures on general chemical and biological analyses; analytical data including NMR and mass spectra, UPLC/MS and UV-vis absorption spectra, NanoSIMS and EM images, thermal proteome profiling data, and biological results are provided in SI Appendix.