Significance
Studies of the adult Drosophila intestine have led to many insights in our understanding of cell-type diversity, stem cell regeneration, tissue homeostasis, and cell fate decision. Recent technological advances in RNA sequencing at the single-cell level allow profiling gene expression of individual cells, thereby shedding light on different cell types and their states. This approach reveals molecular markers for each cell type, including the roles of transcription factors in progenitors and mature cell types, such as enteroendocrine cells and enterocytes. Moreover, such a rich resource will pave ways for more efficient cross-species conservation, thus leading for a better understanding of the complex biology of the intestine.
Keywords: Drosophila, gut, stem cell, enteroblast, enteroendocrine cell
Abstract
Studies of the adult Drosophila midgut have led to many insights in our understanding of cell-type diversity, stem cell regeneration, tissue homeostasis, and cell fate decision. Advances in single-cell RNA sequencing provide opportunities to identify new cell types and molecular features. We used single-cell RNA sequencing to characterize the transcriptome of midgut epithelial cells and identified 22 distinct clusters representing intestinal stem cells, enteroblasts, enteroendocrine cells (EEs), and enterocytes. This unbiased approach recovered most of the known intestinal stem cells/enteroblast and EE markers, highlighting the high quality of the dataset, and led to insights on intestinal stem cell biology, cell type-specific organelle features, the roles of new transcription factors in progenitors and regional variation along the gut, 5 additional EE gut hormones, EE hormonal expression diversity, and paracrine function of EEs. To facilitate mining of this rich dataset, we provide a web-based resource for visualization of gene expression in single cells. Altogether, our study provides a comprehensive resource for addressing functions of genes in the midgut epithelium.
Like its mammalian counterpart, the adult Drosophila midgut is a complex tissue composed of various cell types performing diverse functions, such as digestion, absorption of nutrients, and hormone production. Enterocytes (ECs) secrete digestive enzymes, and absorb and transport nutrients, whereas enteroendocrine cells (EEs) secrete gut hormones that regulate gut mobility and function in response to external stimuli and bacteria. The fly midgut is a highly regenerative organ that has been used extensively in recent years as a model system to characterize the role of signaling pathways that coordinate stem cell proliferation and differentiation in the context of homeostasis and regeneration. For example, EGFR, JAK/STAT, and Hippo signaling control intestinal stem cell (ISC) growth and proliferation (1–5), while Notch signaling regulates ISC differentiation (6–9). To maintain homeostasis, ISC proliferates and gives rise to a transient progenitor, the enteroblast (EB), defined by the expression of Su(H)GBE-lacZ, a Notch pathway activity reporter (6, 7). In addition, both ISCs and EBs express the SNAIL family transcription factor escargot (esg). Polyploid ECs, characterized by the expression of Myosin31DF (Myo1A) and nubbin (also called pdm1), differentiate from EBs (6, 7). In contrast, EEs, marked by the expression of prospero (pros), are derived from ISCs through distinct progenitors, called pre-EEs, that express Piezo, a cation channel that senses mechanical tension (10, 11) (Fig. 1A). In addition, the midgut is surrounded by visceral muscles, which control midgut movements and secrete niche signals, such as Wingless (Wg), the EGFR ligand Vein (Vn), and the JAK-STAT ligand Unpaired1 (Upd1) to control ISC activities (1, 12, 13).
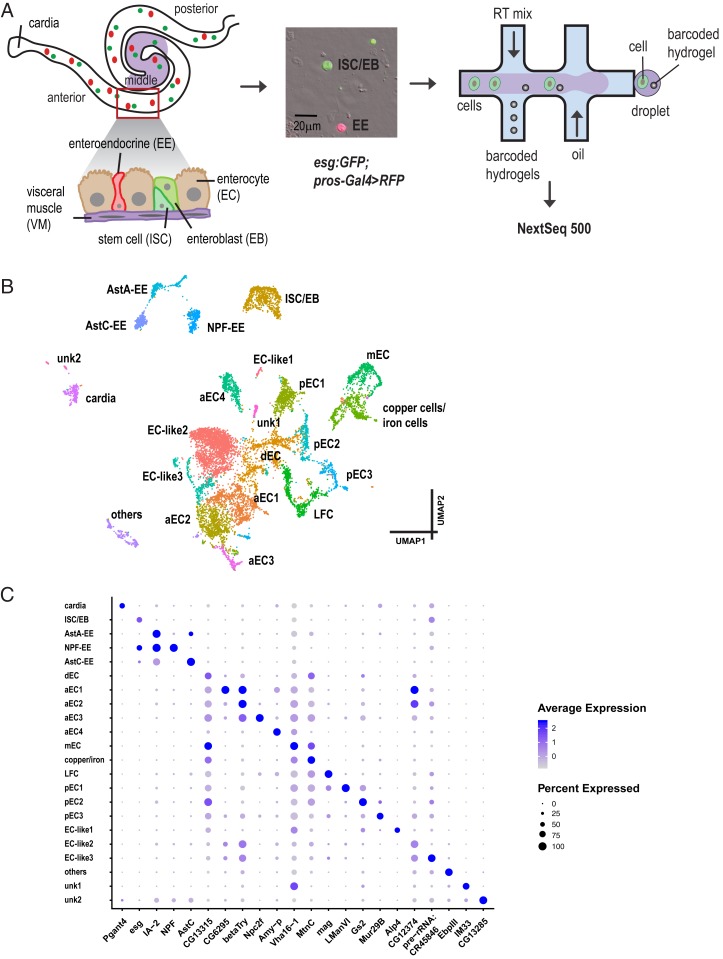
Fig. 1.
Single-cell expression survey of the adult intestinal gut. (A) Experimental design. Different regions of the midgut and different cell types are shown. For simplicity, we distinguish 3 regions of the midgut: Anterior, middle, and posterior. Green, ISC/EB; red, EE. Cells were isolated and encapsulated using inDrop and 10x Genomics approaches. (B) Annotated cell types visualized on the UMAP of 10,605 cells. (C) Expression levels and the percentage of cells expressing markers in each cluster are shown as a dot plot.
Similar to the compartmentalized mammalian digestive tract, the fly midgut can be divided into regions with distinct morphological, histological, and genetic properties (14, 15). For example, the middle region of the midgut, which contains a group of specialized copper cells, is acidic and resembles the mammalian stomach (16). In addition, EEs produce at least 10 different gut hormone peptides that are produced in specific regions: Allatostatins (AstA, AstB/Mip, AstC), Tachykinin (Tk), neuropeptide F (NPF), DH31, CCHa1, CCHa2, Orcokinin B, and Bursicon (Burs) (17–20). AstA-producing EEs are located in the posterior region of the gut, whereas EEs in the anterior, middle, and first half of the posterior midgut produce AstC (18). Moreover, individual EEs are able to produce 2 combinations of different hormones. In particular, some NPF-producing EEs also produce Tk (18, 21). The diversity and regional differences in EEs hinder our ability to comprehensively characterize subtypes of EEs using bulk RNA sequencing (RNA-seq).
To further characterize gene expression and cell types in the adult fly midgut, we used single-cell RNA sequencing (scRNA-seq), as it provides an unbiased approach to survey cell-type diversity, function, and define relationships between cell types (22, 23). Our study reveals molecular markers for each cell type, cell type-specific organelle features, regional differences among ECs, a transitional state of premature ECs, transcriptome differences between ISCs and EBs, 5 additional gut hormones, diverse hormone expression of EEs, paracrine function of EEs, a subset of EEs, and cell-type similarity between the fly and the mammalian gut. We demonstrate how the dataset can be used to characterize new genes involved in gut cell lineage and in particular, we demonstrate that the transcription factor klumpfuss suppresses EE formation. Finally, we built a web-based visualization resource (https://www.flyrnai.org/scRNA/) that allows users to browse scRNA-seq data, query the expression of any genes of interest in different cell types, and compare the expression of any 2 genes in individual cells. Altogether, our study provides a valuable resource for future studies of the Drosophila midgut.
Results
Unbiased Single-Cell Transcriptomics Identifies 22 Distinct Clusters in the Adult Drosophila Midgut.
We used the inDrop (24) and 10x Genomics (25) platforms to profile the transcriptome of 10,605 midgut epithelial cells from 7-d-old females expressing GFP in progenitors (i.e., ISCs and EBs), and RFP in EEs (esg:GFP/+; pros-Gal4,UAS-RFP/+) (Fig. 1A). In total, 4 experiments were performed with 2 technical replicates for each experiment. Details on the number of cells and statistics are summarized in Dataset S1. Next, we used Seurat (26) to identify highly variable genes, performed linear dimensional reduction, determined statistically significant principal components, and generated unsupervised graph-based clustering. Strikingly, cells tend to cluster by cell type (ISCs, EBs, EEs, ECs, and cardia cells) but also by technology (SI Appendix, Fig. S1). Indeed, although replicates within a platform were consistent, we observed significant batch effects between technologies. To remove the technology effect, we used canonical correlation analysis in Seurat to align the datasets from the 2 technologies. The integrated data revealed 22 distinct clusters that can be visualized using a uniform manifold approximation and projection (UMAP) plot (27) (Fig. 1B). Each cluster was assigned to a specific cell type based on known specific markers (see markers listed in SI Appendix, Fig. S2 and Dataset S2). Finally, to facilitate mining of the datasets, we developed a visualization web portal (https://www.flyrnai.org/scRNA/) that allows users to query the expression of any genes of interest in different cell types and to compare the expression of any 2 genes in individual cells.
One cluster, ISC and EB progenitors (ISC/EB), is based on the expression of Dl and esg (SI Appendix, Fig. S2 B and C). Interestingly, based on the detection of downstream targets of Notch signaling, Enhancer of split complex E(spl)-C: E(spl)m3-HLH, E(spl)malpha-BFM, and E(spl)mbeta-HLH, we were not able to distinguish ISCs from EBs (28), suggesting that the transcriptome of ISCs and EBs are highly similar. EEs were assigned to 3 different clusters—AstA-EE, AstC-EE, and NPF-EE—based on the expression of pros (SI Appendix, Fig. S2D) and gut hormones. Fifteen different clusters mapped to ECs from distinct regions of the gut based on the expression of different Trypsin genes (14, 15). Specifically, 3 of the 15 EC clusters, anterior enterocytes 1 to 3 (aEC1-3), mapped to the anterior region of the midgut because they express alphaTry, betaTry, gammaTry, deltaTry, epsilonTry, and thetaTry (SI Appendix, Fig. S3 B–G). Another cluster, aEC4, most likely localizes to the anterior region due to the expression of Amy-p and Amy-d, and low levels of alphaTry, betaTry, and gammaTry, a characteristic of anterior ECs. Another cluster, middle ECs (mEC), mapped to the middle region of the midgut based on the regional expression of the lab transcription factor (SI Appendix, Fig. S2E) (14, 29–31), as well as Vha100-4, a component of Vacuolar H+ ATPase required for acid generation (32). Another cluster mapped to copper and iron cells based on the expression of PGRP-SC2, and a number of metal ion binding proteins, such as MtnA, MtnB, MtnC, and MtnD. Another cluster mapped to large flat cells (LFC) as they expressed PGRP-SC1a and PGRP-SC1b (14). Three clusters, posterior ECs 1 to 3 (pEC1-3), mapped to the posterior midgut based on kappaTry, lambdaTry, iotaTry, etaTry, and zetaTry expression (SI Appendix, Fig. S3 H–L). Another cluster, dEC, corresponds to differentiating ECs because cells have features of both ISCs and ECs. In addition, a small cluster of cells expressed alkaline phosphatase (Alp2, Alp4, and Alp13) and a number of genes involved in vesicle–membrane fusion and intercellular communication. Surprisingly, these cells also express 2 transporters, Smvt and Oatp58Dc, previously reported to be expressed in ISCs and EBs (33). However, as they do not express Dl and esg, we believe that they most likely correspond to ECs, and refer to them as “EC-like 1.” Further characterization of these cells will be required to address their identity. Two additional clusters, although they have relative low number of unique molecular identifiers compared to other clusters, still passed our quality control threshold and has the number of unique molecular identifiers similar to EEs. They express EC genes, such as Jon99Ciii, Bace, and betaTry (digestive enzymes), so we named them as EC-like 2 and EC-like 3, respectively. The last EC cluster mapped to cardia secretory cells, based on the expression of Pgant4 (34), which synthesizes and secretes the peritrophic membrane that lines and protects the gut (SI Appendix, Fig. S2F). In addition, one cluster of cells is enriched in genes involved in lipid metabolism and fatty acid biosynthesis but far away from the ECs based on the UMAP coordinates, suggesting that they may not be ECs, so we called it “others.” Finally, we could not assign an identity to 2 clusters, unknown cell type 1 (unk1) and unk2. Although unk1 may still relate to the ECs, unk2 could be related to the cardia cells based on the UMAP coordinates. The expression levels and the percentage of cells expressing markers in each cluster are shown as a dot plot (Fig. 1C). Overall, the number of cells we captured was not affected by the dissociation method and downstream analysis as EEs represent 8% of the cells [compared to 10% expected (35)], ISCs and EBs 8% of the cells [compared to 10% expected (7)], ECs 81% of the cells, and 4% for others (Dataset S2), which is consistent with the composition of each cell type in the fly gut.
Gene-Expression Signatures of Each Cluster.
Next, we analyzed the gene-expression signatures of each cluster and determined the enrichment of specific groups of genes involved in distinct cellular processes using the Gene List Annotation for Drosophila (GLAD) online resource (36). Genes categorized as major signaling pathways, transcription factors, cytoskeletal proteins, and RNA-binding are enriched in ISC/EB progenitor cells, whereas genes involved in metabolic processes, serine proteases, and transporters are enriched in ECs (SI Appendix, Fig. S4A; P value in Dataset S3). Interestingly, enriched genes of cells in the EC-like 1 cluster are involved in metabolic process, transporters, and phosphatases, which are more like the EC signature, although they express some ISC/EB markers. Detailed analyses of the expression of canonical genes of signaling pathways revealed that components of the Notch, EGFR and PVR RTK, Hippo, and insulin signaling pathways are enriched in the ISC/EB cluster, and components of the insulin and JNK signaling pathways are enriched in EEs. In addition, components of Imd and Toll immune pathways are enriched in the aEC1 and LFC clusters (SI Appendix, Fig. S4B). Further analyses showed that different EC clusters are enriched for different types of metabolic processes. For example, aECs (aEC1, -2, and -4) and cardia ECs are enriched in genes involved in carbohydrate metabolism, and posterior ECs (pEC1 and -2) are enriched in genes involved in lipid metabolism (SI Appendix, Fig. S4C), which is consistent with the observation that many lipid droplets are present in a subregion of the posterior gut (14). Among pECs, pEC1 are enriched in genes involved in lysosomal oligosaccharide catabolism (LManI, LManII, LManV, LManIII, LManIV); pEC2 are enriched in genes involved in glutamine and glutamate metabolism (Gs2, Gdh); pEC3 are enriched in mucin, sugar binding (lectin), and proteasomes. In addition, the ISC/EB cluster is enriched in integrin signaling, laminins (LanA, LanB1, LanB2, and wb), and their integrin receptors (mew, scb, and mys). The α-integrin subunits encoded by mew and scb and the β-integrin subunit encoded by mys have been shown to be highly expressed in ISCs and are required for stem cell maintenance (37). Cardia, aEC3, and unk2 clusters are enriched for chitin binding, a component of the peritrophic matrix secreted by cardia that functions as a permeability barrier between the food and the midgut epithelium that protects ECs from mechanical and bacteria damage. Finally, mEC and copper cells are enriched for ion channel transport genes.
Interestingly, several ribosomal proteins are enriched in the ISC/EB cluster cells, consistent with the high abundance of ribosomes in ISCs visualized by electron micrographs (7, 38) (Fig. 2 A and A″). The majority of ribosomes in ISCs are free ribosomes, consistent with the low level of expression of components of signal recognition particles (SRPs) in ISCs, as endoplasmic reticulum (ER)-bound ribosomes require binding to facilitate their docking onto the ER through SRP receptors. Specifically, 14% of ISCs express SRPs vs. 32% for ECs, further supporting our observation that ISCs contain more free ribosomes than ECs (Fig. 2 A and B″). Interestingly, a reduction in ribosome levels impairs lineage commitment in human hematopoiesis (39). Another characteristic of stem cells is that they often use glycolysis rather than oxidative phosphorylation as their energy source due to immature mitochondria, as evidenced by the presence of fewer cristae, whose shape is important for the assembly of respiratory chain complexes (40, 41; see and reviews in refs. 42 and 43). Consistent with this, we observe fewer mitochondrial cristae in ISCs than in ECs (Fig. 2 B′ and B″). During differentiation, ATP synthase (complex V) promotes the maturation of mitochondrial cristae through dimerization and up-regulation of the ATP synthase complex (40, 44). Interestingly, we observe an enrichment of ATP synthase complex V in clusters aEC4, dEC, and mEC (SI Appendix, Fig. S4C). In particular, cluster dEC retains some characteristics of ISCs that are attributable to ribosome enrichment, suggesting that dECs are relatively premature ECs in the process of differentiation (see results from the lineage inference analysis below). Finally, Golgi-associated vesicle biogenesis genes are enriched in EEs (Fig. 2 C and C′ and SI Appendix, Fig. S4C), consistent with their roles in gut hormone production and secretion.
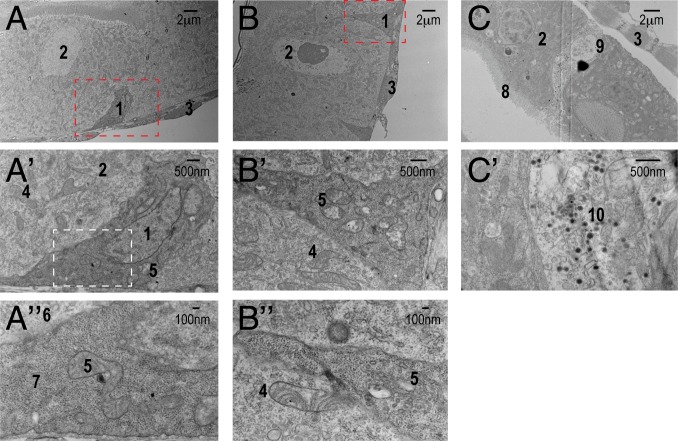
Fig. 2.
Electron micrographs of the ultrastructure of different cell types. (A) ISC (1) with darker density than its neighboring EC (2) resides on the basal side of the epithelium. They are often triangular shape with extensive basement membrane contact. (A′) Magnified view of the region shown in A outlined by the red dash box. (A″) Magnified view of A′ outline by the white dash box. The density of ribosomes (7) in ISCs is more pronounced than that of ribosomes (6) compared to ECs. B′ is a magnified view of B. Mitochondria in ISCs (5) contain less cristae than mitochondria of ECs (4), which can be visualized better in B″. Mitochondria in ISCs look “empty.” (C) EEs (9) contain many “loaded” secreted vesicles (10, dark vesicle), better visualized in C′. 1, ISC; 2, EC; 3, visceral muscle; 4, mitochondria in EC; 5, mitochondria in ISC; 6, ribosomes in EC; 7, ribosomes in ISC; 8, microvilli; 9, EE; 10, secretory vesicles.
Reconstructing Lineage Trajectories.
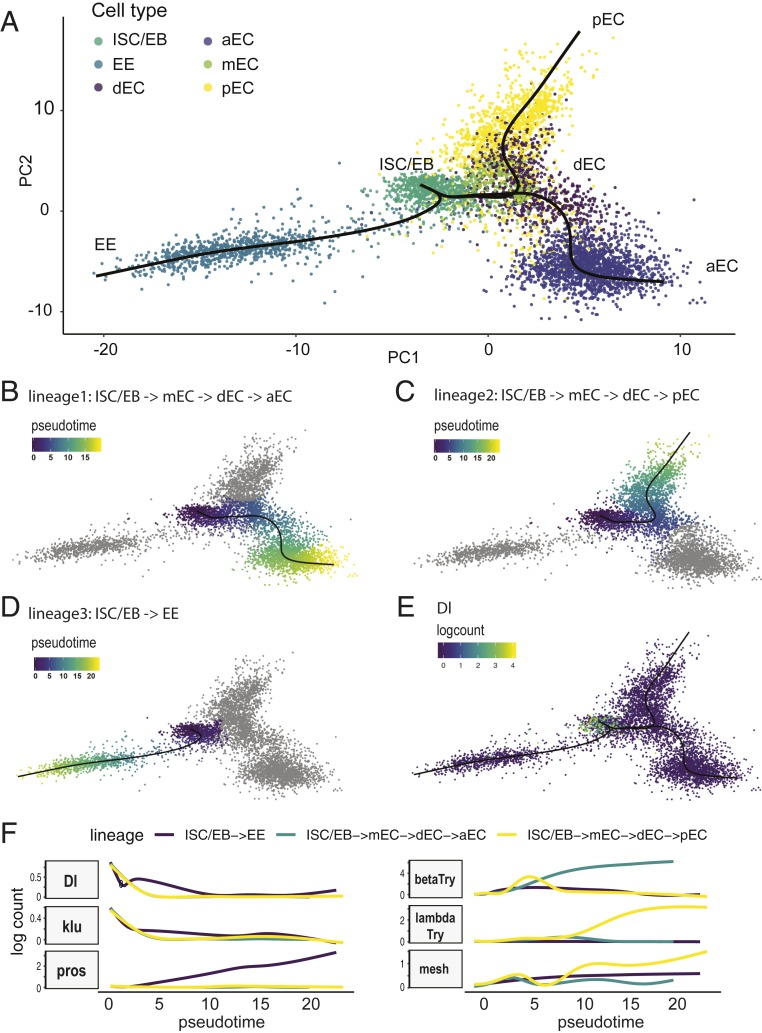
To investigate the relationship between different clusters and explore the cell differentiation process, we used Slingshot, a tool for inferring cell lineage and pseudotime from single-cell transcriptomics data (45). To infer cell lineage, we selected ISC/EB, dECs, ECs, and EEs and excluded cell types from cardia, others, and other unknown cell types, which are unlikely to have direct lineage relationships with ISC/EBs. Slingshot identified 3 lineages: 1) ISC/EBs → mECs → dECs → aECs; 2) ISC/EBs → mECs → dECs → pECs; and 3) ISC/EBs → EE (Fig. 3 A–D). The main start and end points are consistent with the current knowledge that EEs and ECs are derived from ISC/EBs. In addition, Dl, an ISC marker gene, is expressed at high levels at the start point and decreases over pseudotime in differentiating EEs and ECs (Fig. 3E), indicating that Slingshot has built accurate trajectories. Based on ribosome enrichment, we predicted that dECs are premature ECs that have undergone the transition from ISC/EBs to ECs. Indeed, Slingshot placed dECs in an intermediate state, along the path of ISC/EB differentiation into aECs or pECs, indicating that dECs represent premature ECs (Fig. 3A, purple). However, Slingshot also placed mECs in an intermediate state because they do not express many different digestive enzymes, such as trypsins, maltases, Jonah proteases, and mannosidase. We observed an increase in lambdaTry and mesh expression as ISCs differentiate to pECs, and an increase in betaTry expression as ISCs differentiate to aECs. We also discovered genes with patterns similar to Dl, such as klumpfuss (klu) (Fig. 3F). In addition, the expression of genes encoding septate junction protein complex components, mesh, ssk, and Tsp2A, gradually increased as ISCs differentiate to ECs or EEs, consistent with the observation that septate junction complex proteins are present in the apicolateral regions between ECs and EEs but not ISCs (Fig. 3F) (46–50).
Fig. 3.
Results of cell lineage inference and pseudotime analysis using Slingshot. (A) Cell lineages were inferred using Slingshot. Three lineages were constructed: The first lineage is from ISC to aEC: ISC/EB → mEC → dEC → aEC (B). Pseudotime was inferred. The second lineage is from ISC to pEC: ISC/EB → mEC → dEC → pEC (C). The third lineage is from ISC to EE (D). (E) The expression level of Dl was plotted using logcount and its expression is restricted to the initial state (ISC/EB). (F) Plot of gene expression as a function of pseudotime for each lineage. The expression pattern of klu is similar to Dl (i.e., high in the beginning and gradually decreasing over pseudotime).
Identifying New Progenitor Markers.
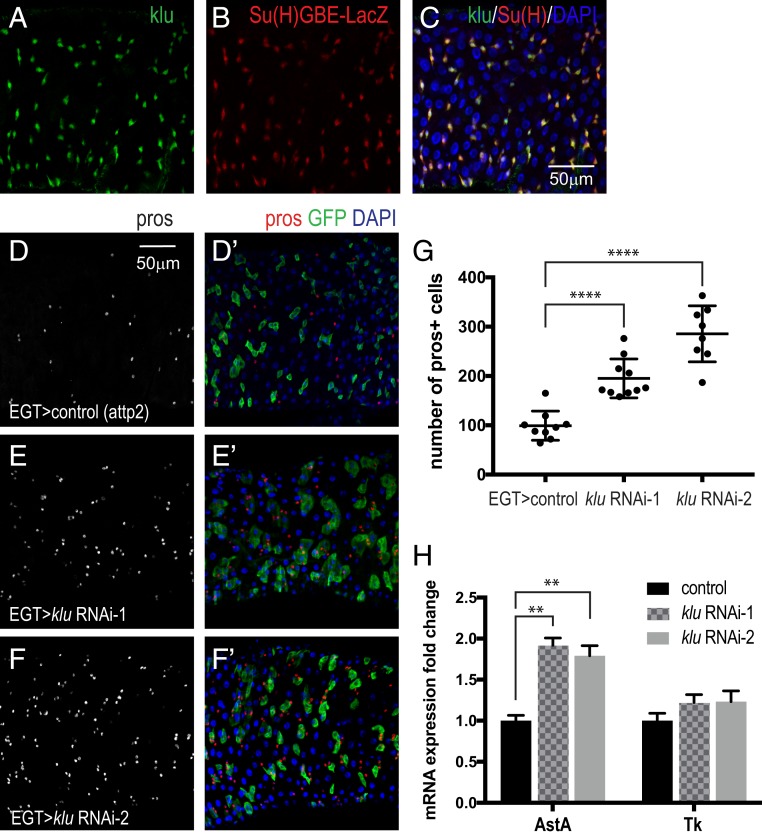
Our scRNA-seq analysis identified many previously known transcription factors (bun, esg, Myc, E(spl)mβ-HLH, peb, E(spl)mα-BFM, E(spl)m3-HLH, fkh, fru, CtBP, Rel, HmgD, Smr, Sox100B, jumu), polarity and adhesion proteins (mira, mew, shg, mys, scb, LanA, LanB1, LanB2, Nrg), and other molecules (robo2, hdc, Ets21C, and so forth) that are important for stem cell proliferation and maintenance (see Dataset S5 for top 32 transcription factors and Dataset S7 for the full list). We also identified additional transcription factors that are enriched in the ISC/EBs cluster: Df31, mamo, Unr, Eip75B, fs(1)h, Blimp-1, Trf, klu, Xrp1, lola, cwo, nej, elB, and so forth. Interestingly, although the expression of klu as a function of pseudotime is similar to Dl in the Slingshot analysis, we found an opposite expression pattern of Dl and klu in individual cells (inverse correlation). We confirmed that klu is expressed in EBs, as it is coexpressed with the EB reporter Su(H)GBE-LacZ (Fig. 4 A–C). klu is a WT1-like transcription factor, previously characterized as a regulator of self-renewal in Drosophila neuroblasts (51, 52) and during the preparation of this report it was shown to maintain lineage commitment of enterocyte progenitors in the gut (53). During hematopoiesis, klu acts downstream of Notch and Lozenge (Lz) to promote crystal differentiation (54). Since Notch signaling plays an important role in ISC differentiation, we speculated that klu also affects ISC differentiation. Indeed, knockdown of klu with 2 independent RNAi lines in ISCs and EBs with esgGal4-tubgal80ts in the adults for 7 d resulted in an increase of the EE population (Fig. 4 D–G), which is similar to the Notch loss of function phenotype (6, 7, 53). We observed an increase in the number of AstA-expressing EEs (inferred by mRNA level) when knocking down klu in ISCs and EBs (Fig. 4H), which is consistent with an increase of AstA-expressing EEs in Notch loss-of-function MARCM clones (35). Thus, klu is a new EB marker and downregulation of klu promotes ISC/EBs differentiation to EEs, in particular AstA-expressing EEs. We also confirmed that lola, a transcription repressor that antagonizes Notch (55), is expressed in esg+ cells, but only in the middle region of the midgut (SI Appendix, Fig. S5A).
Fig. 4.
klu expression in EBs and its loss-of-function phenotype. (A–C) Coexpression of klu and the EB reporter Su(H)GBE-LacZ. (D–F′). Knockdown of klu in ISCs/EBs results in an increase of EEs, marked by pros staining: red, pros; green, esg; blue, DAPI. (G) Quantification of EE number. Data are represented as mean ± SEM. Two-tailed t test, ****P < 0.0001. (H) qRT-PCR measurement of AstA and Tk expression from midguts expressing klu RNAi in adult ISCs/EBs for 7 d. rp49 was used for normalization. Data are represented as mean ± SEM. Two-tailed t test, **P = 0.0011 for control vs. RNAi-1; **P = 0.0047 for control vs. RNAi-2.
In addition to transcription factors, we also confirmed that leucine-rich-repeats and calponin homology domain protein (Lrch), Zipper (zip), and inscutable (insc) are expressed in esg+ progenitor cells (SI Appendix, Fig. S5 B–D) in different regions of the midgut. Interestingly, all of them are related to the cytoskeleton, suggesting that the cytoskeleton plays an important role in stem cell shape and biology, and consistent with the results of GLAD enrichment analyses (SI Appendix, Fig. S4A). Lrch, which is expressed in both anterior and posterior regions of the midgut, encodes a scaffold protein well conserved across animals that interacts with actin to stabilize the cell cortex and position of the mitotic spindle during cell division (56). Zip encodes cytoplasmic myosin II, which also binds to the actin cytoskeleton (57) and is coexpressed with esg+ in the middle of the midgut. Insc, which encodes a cytoskeletal adaptor protein required for apical–basal spindle orientation for asymmetric cell division in neuroblasts (58), is coexpressed with esg+ throughout the midgut, suggesting that it may be important for cell fate decisions.
Finally, the 2 amino acid transporters Pathetic (path) and minidiscs (mnd) are preferentially expressed in ISC/EBs. Path, a new type of PAT-related transporter that transports/uptakes alanine and glycine (59), genetically interacts with TOR and other InR signaling components to control growth. mnd is a leucine transporter that transports dietary leucine and induces insulin-like peptide release by the insulin-producing cells (60). We speculate that as EBs differentiate into ECs, they undergo extensive growth and DNA endoreplication, requiring uptake of a large amount of amino acids through these transporters to synthesize DNA.
Differences Between ISCs and EBs.
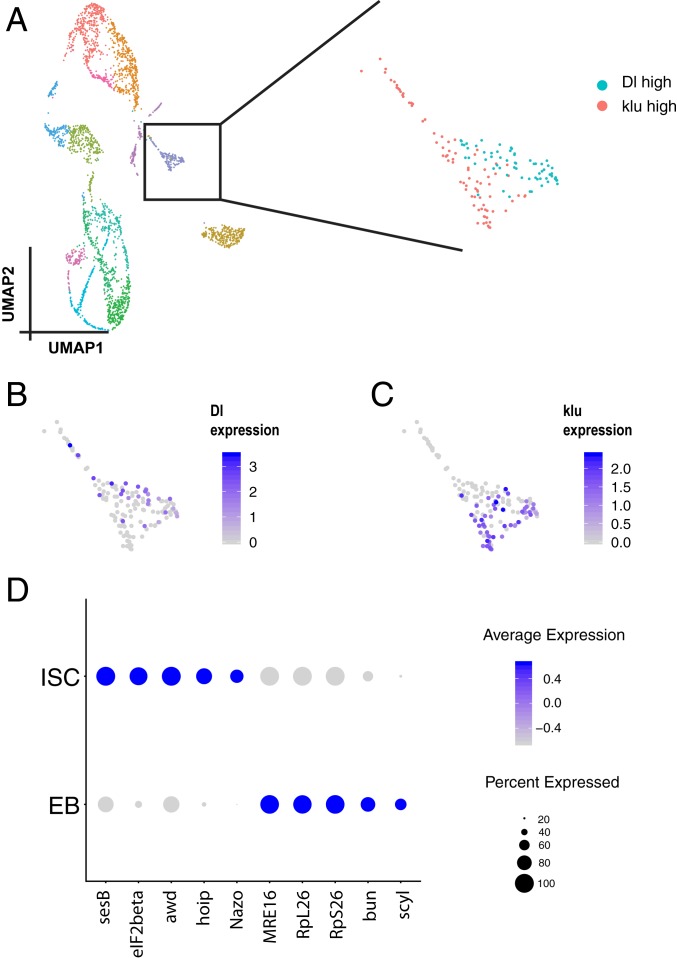
Initially, our inDrop data alone did not separate ISCs and EBs into 2 different clusters, and we wondered whether this could be resolved using another technology, 10x Genomics. Interestingly, analysis of 10x Genomics data alone (without integrating the data from inDrop) reveals that one subset of cells in the ISC/EB cluster is Dl+ klu− and another subset is Dl− klu+ (Fig. 5 A–C). Further analysis of these 2 clusters revealed that genes encoding translation elongation factors (eIF2beta, eEF1delta), small ribonucleoprotein particles (SmE, SmF, SNRPG), and chaperone and heat-shock proteins (Hsp60A, Hsp10, CCT1, CCT5) were expressed at higher levels in ISCs than in EBs (also see Dataset S6). Finally, EBs expressed MRE16, RpL26, RpS26, bun, and scyl at higher levels compared to ISCs (Fig. 5D). Thus, analysis of the 10x Genomics data alone allowed us to successfully distinguish ISC and EB cell populations.
Fig. 5.
Analysis of differences between ISCs and EBs. (A) UMAP of 2,979 cells from 10x Genomics. The ISC/EB cluster is outlined by a black box. There are 2 groups of cells in these clusters. One expresses high Dl (B) and the other expresses klu (C). (D) Dot plot of top 5 genes that express differentially in ISCs and EBs.
Identifying New EE Markers and Gut Hormones.
EEs are chemosensory cells that secrete regulatory hormones in response to luminal contents, such as nutrients and bacterial metabolites, to regulate gut physiology, food intake, and glucose homeostasis. We detected all 10 gut hormones in the EE cluster—AstA, Mip, AstC, Tk, NPF, Dh31, CCHa1, CCHa2, Burs, and Orcokinin (61)—as well as several key enzymes important for hormone biosynthesis, processing, and secretion—such as amon, Phm, Pal2, svr—that are also targets of the transcription factor dimmed (19, 62, 63). We also identified markers enriched in the EE population, IA-2, 7B2, nrv3, CG30183, and unc-13-4A, and genes involved in vesicle docking and secretion, such as Sytalpha, Sytbeta, Syt1, cac, nSyb, and Syx1A (Dataset S7). In addition, we found 5 neuropeptides—sNPF, ion transporter peptide (ITP), neuropeptide-like precursor 2 (Nplp2), crustacean cardioactive peptide (CCAP), and CNMa—previously reported to be expressed in neurons that are also expressed in a subset of EEs. ITP is a thirst-promoting hormone controlling the homeostasis of animal body fluids. Its expression is elevated when flies are dehydrated; however, the function of ITP in EEs is unknown (64). Nplp2 was long thought to be a neuropeptide and has recently been demonstrated to be a heat-inducible apolipoprotein required for thermal acclimation (65). Nplp2 facilitates dietary lipid extraction from the gut and fat storage in the fat body. CCAP has a cardio acceleratory effect when it is applied in vitro, but knockdown of CCAP in CCAP neurons did not compromise heartbeat (66). Finally, the functional role of CCAP as a gut hormone is not known. In summary, we identified a number of EEs markers and showed that 5 previously characterized neuropeptides are also expressed in gut EEs.
Diversity of EEs Revealed by Coexpression of Hormones.
Individual EEs are known to express a combination of 2 to 3 gut hormones (18, 20). Interestingly, our scRNA-seq data reveals that each individual EE expresses 2 to 5 different types of gut peptides. For example, the most frequent combination of gut hormones in our dataset are as follows: AstA-EEs are able to produce AstA, AstC, Orcokinin, CCHa1, and CCHa2; AstC-EEs tend to express AstC and Orcokinin; and NPF-EEs express NPF, Tk, Nplp2, and Orcokinin (SI Appendix, Fig. S6A). Next, we looked for common transcription factor binding sites upstream of these gut hormones from Chip-seq data, to identify transcription factors that could be regulating the coexpression of gut hormones (67). Interestingly, su(Hw) binding sites are present in the 5k window upstream of each of the 4 gut peptide genes (AstA, AstC, CCHa2, and CCHa2), suggesting that Su(Hw) may coregulate these hormones. Similarly, binding sites for Pdp1 and CG15515 are found upstream of NPF/Tk/Nplp2/Orcokinin and AstC/Orcokinin, respectively. A full list of the common transcription factor binding sites for coexpressed gut peptides can be found in Dataset S8.
Paracrine Function of EEs.
“Enteroendocrine” implies that cells in the gut secrete hormones and thereby exert effects in a distal organ. For example, CCHa2, secreted from the EEs in the gut, has been proposed to signal to the brain, where the CCHa2 receptor is detected (68). Interestingly, we found that a subset of AstC-EEs also express NPF receptors, suggesting that NPF hormone may regulate AstC-EEs in a paracrine manner (i.e., cell to cell short-range communication). Similarly, we also found that NPF-EEs express AstC receptors (AstC-R2), suggesting not only that NPF-EEs communicate to AstC-EEs but also that AstC-EEs signal back to NPF-EEs. NPF-EEs and AstC-EEs are both present in the middle region of the gut, supporting the idea that this feedback communication may occur in vivo. Interestingly, NPF-EEs also express AstA receptors (AstA-R2), suggesting that NPF-EEs in the middle region can receive signals from the posterior region (short-range communication) (SI Appendix, Fig. S6B). In addition, AstA-EEs also express the serotonin receptor (5-HT1A). Although serotonin is a neurotransmitter, it is predominantly produced and secreted in the mammalian gut and regulates peristaltic movement, fluid, and mucus secretion (69). Gut microbes can also modulate serotonin production, providing a molecular dialogue for microbe and host communication. Whether gut microbes in the fly interact with host cells in the same manner as observed for mammalian gut–microbe interactions is not known. The expression of serotonin receptor in AstA-EEs provides an interesting hypothesis that AstA-EEs may sense gut–microbe status to coordinate the host response.
Characterization of a Subset of EEs.
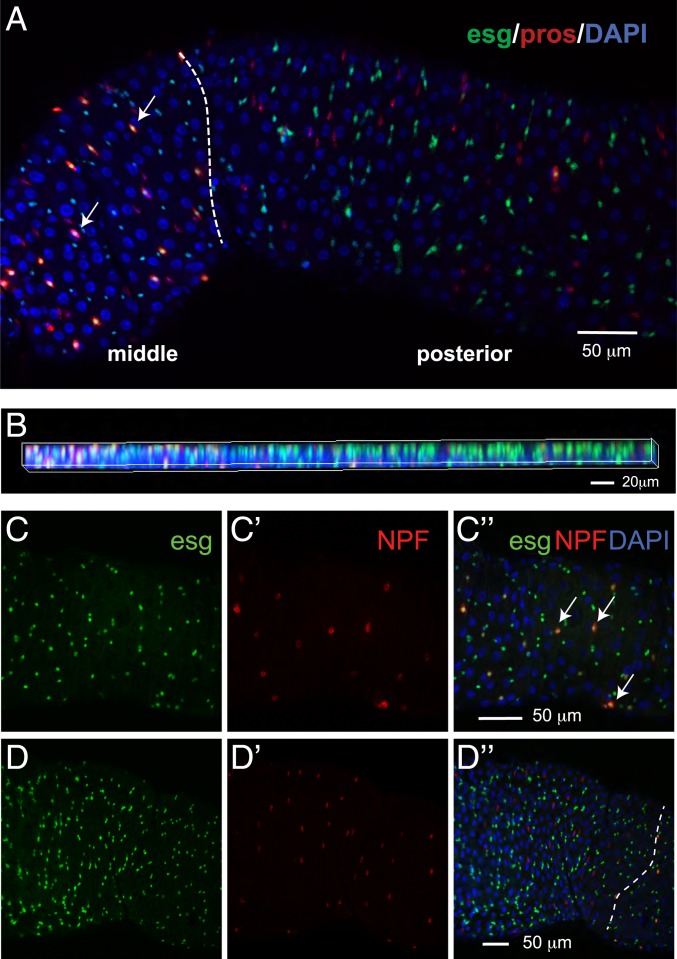
We noticed that esg, a marker for ISC/EBs, is not only expressed in ISCs and EBs but also in a subset of EEs (pros+) (SI Appendix, Fig. S2 C and D). To validate this observation, we examined the expression of a GFP insertion in the esg locus and pros-Gal4 driven RFP. In most cases, consistent with esg being a progenitor marker and pros a differentiation EE marker, esg and pros do not colocalize in individual cells. This mutually exclusive expression pattern is observed in the cells localized to the anterior and posterior regions of the midgut. However, esg+ pros+ double-positive cells are detectable in the middle region and a small portion of the posterior midgut (Fig. 6 A and B). To further characterize these esg+ pros+ cells, we analyzed the gene-expression profile in esg+ pros+ cells. Unexpectedly, esg+ pros+ cells do not express Dl but express Tk and NPF hormones, suggesting that esg+ pros+ cells are mature EEs rather than EE progenitors (Dl+) (SI Appendix, Fig. S6C). To further analyze this cell type, we examined the expression of esg and NPF and found that these markers colocalize in cells located in the middle region of the midgut, but not in the anterior region (Fig. 6 C and D″). Altogether, these analyses identify a subset of EEs with different molecular properties.
Fig. 6.
Characterization of a subset of EEs. (A and B) The majority of pros+ esg+ double-positive cells are located in the middle of the midgut, whereas very few double-positive cells are located in the posterior region. pros, an EE marker; esg, a progenitor marker. Dashed line indicates the boundary between the middle and posterior midgut regions. Arrow, pros+ esg+ double-positive cells. (B) Z-stacking 2D image showing the colocalization of pros and esg in the middle but not the posterior region. Genotype, esg-sfGFP/+, pros-Gal4; UAS-mCD8.RFP/+. (C and D) Image of middle (C and C″) and anterior (D and D″) midguts. Arrow, NPF+ esg+ double-positive cells. Dash line indicates the boundary between anterior and middle midgut regions. Genotype: esg-sfGFP/+, UAS-mcherryCAAX; NPF-Gal4/+. (Scale bars in C″ and D″ also apply to C, C′, D, and D′.)
Comparison of Fly Midgut and Mammalian Intestinal Cell Types.
Recent scRNA-seq studies have provided a cell atlas of the mammalian small intestine (70), thus providing a reference to evaluate whether gene signatures of gut cell types are evolutionary conserved. Cell types in these tissues are similar because they both contain progenitors (stem cells and transit amplifying cells [TAs] in mammals; ISC/EBs in flies), enterocytes and enteroendocrine (EE, tuft, goblet, and paneth cells in mammals; EEs in flies). We used DIOPT (DRSC Integrative Ortholog Prediction Tool) to map the mammalian marker genes of each cell types to Drosophila orthologs (71). Drosophila ISC/EBs are reminiscent of ISCs and TAs (in cell cycle G2 state) (SI Appendix, Fig. S7), whereas Drosophila EEs are similar to EEs, tuft, and goblet cells. Tuft cells are chemosensory cells. Drosophila EEs have also been reported to express various gustatory receptors, and potentially play a role in chemosensation (72). Drosophila cardia cells are similar to goblet cells, consistent with their role in secreting mucins. Paneth cells are similar to Drosophila aEC2 and aEC3, possibly due to the expression of lysozymes (function as bacteria defense) in both cell types. Overall, Drosophila ECs are similar to mammalian intestinal ECs. Drosophila posterior ECs show highest similarity to mammalian mature distal enterocytes; however, the Drosophila anterior ECs only show modest similarity with mammalian mature proximal ECs (SI Appendix, Fig. S7). P values of these comparisons are summarized in Dataset S9.
Discussion
The Drosophila intestinal epithelium is widely used to study stem cell regeneration and maintenance, as well as how signaling events from different cell types influence stem cell behaviors. Here, we surveyed the cell types of the adult intestinal epithelium using scRNA-seq and identified all known cell types, 1 cell type (esg+ pros+) in the middle region of the midgut, differentiating ECs, and 5 unknown cell types (unk1, unk2, EC-like 1 to 3). Our study recovered most previously known ISC/EBs and EEs markers, demonstrating the robustness of the scRNA-seq approach. Interestingly, gene expression analysis revealed that ISCs are enriched for free ribosomes and possess mitochondria with fewer cristae. We also identified transcription factors expressed differently along the guts, and cytoskeletal proteins and transcription factors preferentially expressed in the ISC/EB population. In particular, we validated that klu is specifically expressed in EBs and that knockdown of klu in ISC/EBs (with esg-Gal4) results in an increase of EEs, suggesting that klu inhibits EE differentiation. When we performed the scRNA-seq study using inDrop, we could not observe a clear separation between cells that expressed Dl and cells expressing Notch downstream targets, E(spl)m3-HLH, E(spl)malpha-BFM, E(spl)mbeta-HLH, and the EB marker, klu. Thus, we wondered whether this could be resolved using the 10x Genomics technology. Interestingly, using data from 10x Genomics we were able to detect one subset of cells in the ISC/EB cluster that expresses Dl+ klu− (ISC) and another subset expressing Dl− klu+ (EB). Therefore, using data from 10x Genomics alone allows us to demarcate the ISCs and EBs.
We started with a small number of cells in the first sample for inDrop and 10x Genomics technologies, which recovered 344 cells and 256 cells, respectively. This allowed us to test and compare the 2 technologies. Next, we increased the number of cells for each replicate (7,282 for inDrop and 2,723 for 10x Genomics). The 2 replicates allowed us to evaluate the consistency of cell-type discovery between the 2 platforms. Indeed, all of the major cell types (cardia, ISC/EB, EE, dEC, aEC, mEC, pEC, LFC, copper, and iron cells) were detected using both approaches (detailed comparison can be found in Dataset S2).
Cell morphology and digestive functions are different along the length of the Drosophila midgut (14). For example, ECs in the middle midgut secrete acid and absorb metal ions, whereas ECs in the posterior midgut contain lipid droplets and uptake lipid nutrients. These characteristics reflect regionalized gene-expression differences as previously shown by bulk RNA-seq analyses (14, 15). We searched for differentially expressed transcription factors that could underlie regionalized gene expression and identified a number of potential candidates. In particular, vnd, odd, caup, and tup are preferentially expressed in the anterior region. We note only one discrepancy (odd) between our results and previously published bulk RNA-seq data (expressed in the posterior region from the Flygut-seq). Further studies will be required to resolve these differences. lab, Ptx1, CREG, apt, and dve are preferentially expressed in the middle midgut, consistent with the previous observation that the homeobox genes lab, Ptx1, and dve have been shown to be expressed in the adult middle midgut (15, 29, 33). Finally, bab2, ham, cad, Ets21C, Hnf4, and hth are preferentially expressed in the posterior midgut; the homeobox gene cad and Ets21C have been previously reported to be expressed in the posterior midgut (15, 73, 74). The expression pattern of these regionalized transcription factors from the Flygut-seq are listed in Dataset S4 to help compare these findings. Recently, scRNA-seq of the mouse embryo identified a group of 20 transcription factors that are expressed spatially along the anterior–posterior axis of the gut tube (75). Interestingly, 6 out of 20 transcription factors expressed in the mouse gut have fly orthologs that are also expressed differently along the anterior–posterior axis of the fly midgut. For example, mouse Irx3 and fly caup are expressed in the anterior region, mouse Hoxb1 and fly lab are expressed in the anterior–middle region, and mouse Cdx2 and fly cad are expressed in the posterior region (Dataset S4).
The regional expression of the transcription factors described above may also underlie the regionalization of EE populations. For example, cad, which is expressed in posterior ECs, is also highly expressed in AstA-EEs that are localized in the posterior midgut. We also identified another transcription factor expressed in posterior EEs, Poxn, that is homologous to mouse Pax8, which is expressed regionally in the mouse gut tube (75). Whether Poxn is expressed in posterior EEs has not yet been experimentally tested. Similarly, stem cell morphology and proliferation activity also differ along the anterior–posterior axis of the gut. However, although previous cell-specific RNA-seq studies revealed regional differences in stem cell transcriptomes (33), our scRNA-seq analysis was not able to identify subgroups or regional ISC/EB clusters, despite the fact that some stem cells express some regional markers, such as lab or Ptx1. It is possible that the regional differences in ISC transcriptomes are less prominent than the regional differences in EC transcriptomes (14, 15).
Regarding EEs, we identified candidate markers and 5 additional gut hormones: sNPF, ITP, Nplp2, CCAP, and CNMa. In addition, we found that individual EEs are able to express up to 5 different hormones, in contrast to the traditional view that these cells only produce 2 hormones (18, 21). Interestingly, a recent mammalian study showed that EEs express different hormones and that they can switch their hormonal repertoire depending on their tissue location (76). The most frequent combinations of gut hormones were AstA/AstC/Orcokinin/CCHa1/CCHa2 for AstA-EEs, AstC/Orcokinin for AstC-EEs, and NPF/Tk/Nplp2/Orcokinin for NPF-EEs. In addition, we found that EEs may also act in a paracrine manner because NPF-EEs expressed AstC-R2, which can receive signals from AstC-EEs. Finally, we showed that a subset of EEs expressing NPF and Tk in the middle of the midgut also expressed the esg progenitor marker.
Our study provides a rich resource to further characterize the molecular signature of each cell type and gene functions in different cell types in homeostatic conditions. Further scRNA-seq of the fly gut will allow a number of questions to be addressed. These include changes in cell states, cell-type composition, and transcriptomes in the context of regeneration, aging, infection, axenic condition, different diet, various mutant backgrounds, and disease models, such as the Yorkie-induced intestine tumor model (77). In addition to the higher ISC proliferation activity (78), the female midgut is larger and longer than the male midgut. Hence, it is highly warranted to use scRNA-seq to delineate the gut at physiological and functional levels based on sex differences. Furthermore, during aging, changes in ISC proliferation, regeneration capacity, innate immune and inflammatory response, and tissue integrity occurs, which can be analyzed using scRNA-seq. Taking these data together, we feel that future scRNA-seq will provide a fundamental understanding of the changes in cell states and interplay among cell types and disease.
Data Availability.
All data are available within this report and the associated SI Appendix.
Methods
Detailed methods and materials, including single-cell suspension preparation, high-throughput sequencing, dataset processing, gene-set enrichment analysis, cell-type conservation analysis between fly and mammalian intestine, fly genetics, staining and fluorescence imaging, and RT-PCR, are listed in SI Appendix. All of the analyses scripts were deposited in GitHub: https://github.com/hbc/drosophila-midgut-analysis. Sequencing files are available in the Gene Expression Omnibus, accession no. GSE120537, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120537.
Supplementary Material
Acknowledgments
The authors thank Drs. Mandovi Chatterjee and Sarah Boswell from the Single Cell Core at the Institute of Chemistry and Cell Biology–Longwood screening facility for inDrop encapsulation and library preparation; the Transgenic RNAi Project (TRiP) and the Bloomington Drosophila Stock Center or providing fly stocks; the Developmental Studies Hybridoma Bank for monoclonal antibodies; Luis Felipe Mirabella Escobedo and Henrique Camara for their help in gut dissection; and Drs. Li He, Stephanie Mohr, Qing Lan, and members of the N.P. laboratory for discussion and critical comments on the manuscript. This work was supported by the Jane Coffin Childs Foundation (R.-J.H.), TRiP R01GM084947, and Drosophila RNAi Sceening Center R01GM067761. N.P. is an investigator of the Howard Hughes Medical Institute. Work by R.K. and S.H.S. at the Harvard Chan Bioinformatics Core was funded by the Harvard Medical School Tools and Technology Committee and with the support of Harvard Catalyst, The Harvard Clinical and Translational Science Center (NIH Award UL1 RR 025758), and financial contributions from participating institutions. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health. Portions of this research were conducted on the Orchestra High Performance Compute Cluster at Harvard Medical School. This NIH-supported shared facility consists of thousands of processing cores and terabytes of associated storage and is partially provided through Grant NCRR 1S10RR028832-01. See http://rc.hms.harvard.edu for more information.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE120537). All the analyses scripts have been deposited in GitHub: https://github.com/hbc/drosophila-midgut-analysis.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916820117/-/DCSupplemental.
References
- 1.Jiang H., Grenley M. O., Bravo M. J., Blumhagen R. Z., Edgar B. A., EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang H., et al. , Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw R. L., et al. , The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147–4158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karpowicz P., Perez J., Perrimon N., The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137, 4135–4145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wisidagama D. R., Thummel C. S., Regulation of Drosophila intestinal stem cell proliferation by enterocyte mitochondrial pyruvate metabolism. G3: Genes, Genomes, Genet. 9, 3623–3630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Micchelli C. A., Perrimon N., Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Ohlstein B., Spradling A., The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Ohlstein B., Spradling A., Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–992 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Guo X., Huang H., Yang Z., Cai T., Xi R., Division of labor: Roles of groucho and CtBP in notch-mediated lateral inhibition that controls intestinal stem cell differentiation in Drosophila. Stem Cell Rep. 12, 1007–1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng X., Hou S. X., Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development 142, 644–653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L., Si G., Huang J., Samuel A. D. T., Perrimon N., Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature 555, 103–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin G., Xu N., Xi R., Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455, 1119–1123 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Biteau B., Jasper H., EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045–1055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marianes A., Spradling A. C., Physiological and stem cell compartmentalization within the Drosophila midgut. eLife 2, e00886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchon N., et al. , Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Dubreuil R. R., Copper cells and stomach acid secretion in the Drosophila midgut. Int. J. Biochem. Cell Biol. 36, 745–752 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Chen J., et al. , Isoform-specific expression of the neuropeptide orcokinin in Drosophila melanogaster. Peptides 68, 50–57 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Veenstra J. A., Agricola H. J., Sellami A., Regulatory peptides in fruit fly midgut. Cell Tissue Res. 334, 499–516 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Reiher W., et al. , Peptidomics and peptide hormone processing in the Drosophila midgut. J. Proteome Res. 10, 1881–1892 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Veenstra J. A., Ida T., More Drosophila enteroendocrine peptides: Orcokinin B and the CCHamides 1 and 2. Cell Tissue Res. 357, 607–621 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Song W., Veenstra J. A., Perrimon N., Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 9, 40–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner D. E., et al. , Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusi B. K., et al. , Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein A. M., et al. , Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng G. X., et al. , Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McInnes L. Healy J., Melville J., UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 (9 February 2018).
- 28.Bardin A. J., Perdigoto C. N., Southall T. D., Brand A. H., Schweisguth F., Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137, 705–714 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strand M., Micchelli C. A., Quiescent gastric stem cells maintain the adult Drosophila stomach. Proc. Natl. Acad. Sci. U.S.A. 108, 17696–17701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubreuil R. R., Grushko T., Baumann O., Differential effects of a labial mutation on the development, structure, and function of stomach acid-secreting cells in Drosophila melanogaster larvae and adults. Cell Tissue Res. 306, 167–178 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Hoppler S., Bienz M., Two different thresholds of wingless signalling with distinct developmental consequences in the Drosophila midgut. EMBO J. 14, 5016–5026 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overend G., et al. , Molecular mechanism and functional significance of acid generation in the Drosophila midgut. Sci. Rep. 6, 27242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutta D., Buchon N., Xiang J., Edgar B. A., Regional cell specific RNA expression profiling of FACS isolated Drosophila intestinal cell populations. Curr. Protoc. Stem Cell Biol. 34, 2F.2.1–2F.2.14 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zhang L., et al. , O-glycosylation regulates polarized secretion by modulating Tango1 stability. Proc. Natl. Acad. Sci. U.S.A. 111, 7296–7301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beehler-Evans R., Micchelli C. A., Generation of enteroendocrine cell diversity in midgut stem cell lineages. Development 142, 654–664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y., Comjean A., Perkins L. A., Perrimon N., Mohr S. E., GLAD: An online database of gene list annotation for Drosophila. J. Genomics 3, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin G., et al. , Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev. Biol. 377, 177–187 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Miller A., “The internal anatomy and histology of the imago of Drosophila melanogaster” in Biology of Drosophila, Demerec M., Ed. (Hafner, New York, 1950), p. 435. [Google Scholar]
- 39.Khajuria R. K., et al. , Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173, 90–103.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira F. K., et al. , ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat. Cell Biol. 17, 689–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cogliati S., et al. , Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu C., et al. , Energy metabolism plays a critical role in stem cell maintenance and differentiation. Int. J. Mol. Sci. 17, 253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo B. J., Yoon S. H., Do J. T., Mitochondrial dynamics in stem cells and differentiation. Int. J. Mol. Sci. 19, E3893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss M., Hofhaus G., Schröder R. R., Kühlbrandt W., Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27, 1154–1160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Street K., et al. , Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izumi Y., Motoishi M., Furuse K., Furuse M., A tetraspanin regulates septate junction formation in Drosophila midgut. J. Cell Sci. 129, 1155–1164 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Izumi Y., Yanagihashi Y., Furuse M., A novel protein complex, Mesh-Ssk, is required for septate junction formation in the Drosophila midgut. J. Cell Sci. 125, 4923–4933 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Yanagihashi Y., et al. , Snakeskin, a membrane protein associated with smooth septate junctions, is required for intestinal barrier function in Drosophila. J. Cell Sci. 125, 1980–1990 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Chiwei X., Ericsson M., Perrimon N., Understanding cellular signaling and systems biology with precision: A perspective from ultrastructure and organelle studies in the Drosophila midgut. Curr. Opin. Syst. Biol. 11, 24–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korzelius J., et al. , Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 33, 2967–2982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger C., et al. , FACS purification and transcriptome analysis of Drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Rep. 2, 407–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao Q., Komori H., Lee C. Y., Klumpfuss distinguishes stem cells from progenitor cells during asymmetric neuroblast division. Development 139, 2670–2680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korzelius J., et al. , The WT1-like transcription factor Klumpfuss maintains lineage commitment of enterocyte progenitors in the Drosophila intestine. Nat. Commun. 10, 4123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terriente-Felix A., et al. , Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development 140, 926–937 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng L., Carthew R. W., Lola regulates cell fate by antagonizing Notch induction in the Drosophila eye. Mech. Dev. 125, 18–29 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foussard H., et al. , LRCH proteins: A novel family of cytoskeletal regulators. PLoS One 5, e12257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franke J. D., Boury A. L., Gerald N. J., Kiehart D. P., Native nonmuscle myosin II stability and light chain binding in Drosophila melanogaster. Cell Motil. Cytoskeleton 63, 604–622 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Kraut R., Chia W., Jan L. Y., Jan Y. N., Knoblich J. A., Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, 50–55 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Goberdhan D. C., Meredith D., Boyd C. A., Wilson C., PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 132, 2365–2375 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Manière G., Ziegler A. B., Geillon F., Featherstone D. E., Grosjean Y., Direct sensing of nutrients via a LAT1-like transporter in Drosophila insulin-producing cells. Cell Rep. 17, 137–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J., Kim S. M., Kwon J. Y., A systematic analysis of Drosophila regulatory peptide expression in enteroendocrine cells. Mol. Cells 39, 358–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hadžić T., et al. , Genome-wide features of neuroendocrine regulation in Drosophila by the basic helix-loop-helix transcription factor DIMMED. Nucleic Acids Res. 43, 2199–2215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beebe K., Park D., Taghert P. H., Micchelli C. A., The Drosophila prosecretory transcription factor dimmed is dynamically regulated in adult enteroendocrine cells and protects against gram-negative infection. G3 (Bethesda) 5, 1517–1524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gáliková M., Dircksen H., Nässel D. R., The thirsty fly: Ion transport peptide (ITP) is a novel endocrine regulator of water homeostasis in Drosophila. PLoS Genet. 14, e1007618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rommelaere S., Boquete J. P., Piton J., Kondo S., Lemaitre B., The exchangeable apolipoprotein Nplp2 sustains lipid flow and heat acclimation in Drosophila. Cell Rep. 27, 886–899.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Dulcis D., Levine R. B., Ewer J., Role of the neuropeptide CCAP in Drosophila cardiac function. J. Neurobiol. 64, 259–274 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Kudron M. M., et al. , The ModERN resource: Genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics 208, 937–949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ren G. R., et al. , CCHamide-2 is an orexigenic brain-gut peptide in Drosophila. PLoS One 10, e0133017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banskota S., Ghia J. E., Khan W. I., Serotonin in the gut: Blessing or a curse. Biochimie 161, 56–64 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Haber A. L., et al. , A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu Y., et al. , An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park J. H., Kwon J. Y., Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS One 6, e29022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi Y. J., et al. , Age-related upregulation of Drosophila caudal gene via NF-kappaB in the adult posterior midgut. Biochim. Biophys. Acta 1780, 1093–1100 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Jin Y., et al. , EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via capicua-regulated genes. PLoS Genet. 11, e1005634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nowotschin S., et al. , The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beumer J., et al. , Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20, 909–916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon Y., et al. , Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 33, 36–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hudry B., Khadayate S., Miguel-Aliaga I., The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature 530, 344–348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within this report and the associated SI Appendix.