Abstract
Congenital scoliosis (CS) is a form of scoliosis caused by congenital vertebral malformations. Genetic predisposition has been demonstrated in CS. We previously reported that TBX6 loss-of-function causes CS in a compound heterozygous model; however, this model can explain only 10% of CS. Many monogenic and polygenic CS genes remain to be elucidated. In this study, we analyzed exome sequencing (ES) data of 615 Chinese CS from the Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) project. Cosegregation studies for 103 familial CS identified a novel heterozygous nonsense variant, c.2649G>A (p.Trp883Ter) in FBN1. The association between FBN1 and CS was then analyzed by extracting FBN1 variants from ES data of 574 sporadic CS and 828 controls; 30 novel variants were identified and prioritized for further analyses. A mutational burden test showed that the deleterious FBN1 variants were significantly enriched in CS subjects (OR = 3.9, P = 0.03 by Fisher’s exact test). One missense variant, c.2613A>C (p.Leu871Phe) was recurrent in two unrelated CS subjects, and in vitro functional experiments for the variant suggest that FBN1 may contribute to CS by upregulating the transforming growth factor beta (TGF-β) signaling. Our study expanded the phenotypic spectrum of FBN1, and provided nove insights into the genetic etiology of CS.
Subject terms: Medical genomics, Genetics research
Introduction
Congenital scoliosis (CS) is a form of scoliosis caused by congenital vertebral malformations potentially resulting from formation failure, segmentation defects, or a combination of both [1]. The prevalence of CS is ~0.5–1 per 1000 live births [2]. As a major cause to infantile and adolescent disability, CS perturbs patient lives and daily activities both physically and psychologically [3].
Genetic predisposition has been demonstrated in CS [4, 5]. CS is a consequence of single-gene mutations. We previously reported that a heterozygous 16p11.2 deletion or rare TBX6 loss-of-function (LoF) variants together with a common hypomorphic risk haplotype composed by three SNPs in trans cause CS [6]. We subsequently recapitulated this compound heterozygosity model in a gene-edited mouse [7], and defined the unique and clinically actionable phenotype of a monogenic form of CS, TBX6-associated CS (TACS) [8]. However, this model can explain only about 10% of CS [8, 9]. Many other contributor genes remain to be elucidated.
CS is also a consequence of multifactorial gene-environment mutual interaction [10]. The susceptibility of some polygenic defects has been illustrated in CS cases, accompanied with multiple birth defects [11]. Previous studies have reported several CS-associated genes, including PAX1 (MIM #167411) [12], PTK7 (MIM #601890) [13], DLL1 (MIM #606582) [14], DDR2 (MIM #191311) [15], T (MIM #601397) [16], and numerous other genes are thus far known to cause phenotypes involving scoliosis with vertebral malformations; however, their potential contribution to CS has been poorly investigated.
Here, as a part of the Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) project, we conducted exome sequencing (ES) for a CS cohort. Trio-based analyses on familial cases identified a novel nonsense variant in FBN1, the gene implicated in Marfan syndrome (MFS; MIM#154700) and many monogenic diseases with scoliosis and spinal dysplasia. We then examined the effect of rare FBN1 variants on sporadic CS and observed that deleterious missense variants were significantly enriched in CS. Functional analyses of a recurrent FBN1 missense variant revealed the potential association between upregulation of transforming growth factor beta (TGF-β) signaling and CS. The subjects carrying highly deleterious FBN1 variants had vertebral malformations, malformations of the ribs, and intraspinal defects.
Materials and methods
Participant recruitment
Initially, 615 Chinese CS subjects with complete clinical data and ES data were recruited at Peking Union Medical College Hospital (PUMCH) in China from 2010 to 2018, as a pivotal part of DISCO study (http://www.discostudy.org/). There were 103 familial cases with available samples for first-degree relatives. TBX6 mutations were examined and 41 cases molecularly diagnosed as TACS [6, 8] were removed. Consequently, 574 CS cases were retained for further study. For the control subjects, 828 unrelated Chinese iindividuals without apparent spinal deformity were ascertained. The ethical committees or institutional review boards at PUMCH that contributed to the patient samples approved the study. Written informed consent was obtained from each participant or their guardians (for those younger than 16 years old).
Exome Sequencing and Sanger sequencing
Genomic DNA from peripheral blood samples was extracted using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The ES data were processed and analyzed using the PUMCH Pipeline as previously described [17]. The following public available databases were used for mutation annotation: the 1000 Genomes Project (1KG; http://www.internationalgenome.org), the Exome Sequencing Project (ESP; http://evs.gs.washington.edu/EVS/), Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), and Genome Aggregation Database (gnomAD; http://gnomad-old.broadinstitute.org), Single Nucleotide Polymorphism database (dbSNP; https://www.ncbi.nlm.nih.gov/snp). In silico prediction tools, including Sorting Intolerant From Tolerant (SIFT) [18], Polymorphism Phenotyping v2 (Polyphen-2) [19], Mutation Taster [20], and Combined Annotation Dependent Depletion (CADD) [21] were utilized to predict deleterious properties of variants. We also annotated the detected variants using customized database based on Human Gene Mutation Database (HGMD; www.hgmd.cf.ac.uk/) and Online Mendelian Inheritance in Man (https://omim.org/). Empirically, we utilized the recommended filters of the Genome Analysis Toolkit (v2.2–3) prior to performing association analyses in order to guarantee that the risk of false negatives is minimized to select quality control passed variants [22]. Sanger sequencing by using an Applied Biosystem 3730xl DNA Analyzer (Life Technologies, CA, USA) was conducted to confirm the candidate variants identified by ES, and segregation analyses were performed in family members. The RefSeq accession numbers of the transcript and corresponding protein isoform of FBN1 we used for mutation nomenclature were NM_000138.4 and NP_000129.3, respectively.
Mutational burden analyses
Mutational burden analyses of FBN1 were implemented between 574 CS cases and 828 controls. To alleviate the biased factors attributable to differential sequencing coverage, we conducted harmonization analyses between case and control exomes. An individual RefSeq coding sequence site was excluded from the analysis if the absolute difference in percentages of cases compared with controls with adequate coverage of the site differed by >10%. This site-based pruning resulted in exclusion of 4.8% of the Refseq coding sequence sites. We also introduced a likely gene-disrupting (LGD) model [23] to prioritize the candidate variants. The LGD model is defined by clustering LoF variants (nonsense, splice-site, and insertion/deletion). A damaging missense (D-mis) model was also applied for variant prioritization. D-mis is defined by selection of D-mis variants with a predicted CADD score ≥ 20. In viewing of the dominant traits that FBN1 may have, inclusion criteria were strictly set to select the presumably LGD and D-mis variants to identify risk-conferring variants to CS. Variants that are not present at this time in 1KG, ESP, ExAC, dbSNP, the Universal Mutation Database for FBN1 (UMD-FBN1; http://www.umd.be/FBN1/) [24] were defined as “novel.” Only novel variants were subjected to the burden analysis. We applied a collapsing method [25] to detect the association of mutational burden. The CADD score of 20, which corresponds to the top 1% of damage when evaluating all known allelic variants [26], was set as the cutoff value for creating a stratified variants subgroup for collapsing.
Construction of expression plasmids
We constructed a plasmid expressing full-length FBN1 (GenBank: NM_000138.4) cDNA with enhanced green fluorescent protein (EGFP) fusion, pEGFP-FBN1. A full-length FBN1 cDNA having suitable restriction sites was PCR-amplified using KOD-Plus-Neo (Toyobo, Japan). The PCR amplicons were cloned into the NheI and SacII sites of the pEGFP-N1 expression vector (Clontech, Takara Bio, Japan). The construct for the Leu871Phe variant was generated by QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, CA, USA). All plasmid constructs were subjected to Sanger sequencing for verification.
Cell culture and transfection
HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Waltham, MA, USA) supplemented with 10% FCS (Biological Industries, CT, USA) and 1% penicillin/streptomycin (Gibco) at 37 °C with 5% CO2. Cells were seeded in a six-well plates and transfected with Lipofectamine™ 3000 Transfection Reagent (Invitrogen, CA, USA) according to the manufacturers’ instructions. The medium was replaced with fresh DMEM culture medium 6 h after transfection. The cells were further incubated for 48 h.
Real-time quantitative polymerase chain reaction
Total RNA was isolated using an RNeasy® Mini Kit (Qiagen) and treated with RNase-Free DNase Set (Qiagen). Total RNA was reversely transcribed to cDNA by using the PrimeScript™ RT reagent Kit (#RR037A, Clontech). The relative abundance of targeted mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers 5’-CAAGGGCATCCTGGGCTACACT-3′ and 5′- CTCTCTCTTCCTCTTGTGCTCTTGC-3′ were used for amplification of GAPDH; 5′-ACCTGGTTACTTCCGCATAG-3′ and 5′-GAGGCATCAGTTTCGTTTGT-3′ were used for amplification of EGFP-FBN1. Gene expression was determined using SYBR Green PCR Master Mix (Applied Biosystems, CA, USA) on the 7500Fast Real-Time PCR Systems (Applied Biosystems).
Western blot analysis
Cells were lysed with modified RIPA (50 mM Tris-HCL, 1% NP40, 0.25% Na-deoxycholate, 150 mM NaCl, and 1 mM EDTA; CompleteTM Protease Inhibitor Cocktail [Roche, Mannheim, Germany]), and protein concentrations were determined with the PierceTM BCA Protein Assay Kit (Pierce Biotechnology, Rockford, USA). A total of 5 mg protein was size separated on an 8% SDS polyacrylamide gel, and proteins were electrophoresed and transferred to nitrocellulose membranes. Membranes were blocked in powdered milk for 30 min at room temperature (RT, 25 °C), and primary antibodies (Phospho-Smad2 [Ser465/467] (138D4) Rabbit mAb #3108, Cell Signaling Technology, MA, USA; Smad2/3 [D7G7] XP® Rabbit mAb #8685, Cell Signaling Technology; Mouse Anti-β actin mAb, TA-09, ZSGB-bio, China) were incubated overnight at 4 °C. After washing, the corresponding horseradish-peroxidase-coupled IRDye® 800CW Goat anti-Mouse IgG Secondary Antibody (926-32210, LI-COR, NE, USA) were incubated for 1 h at RT. Bands were visualized with the WesternBright ECL HRP substrate chemiluminescent system (Advansta, CA, USA). Chemiluminescent signals were quantified using ImageJ [27].
Statistical analyses
A one-sided Fisher’s exact test or a one-sided Chi-square test was used for all mutational burden analyses by using SPSS Statistics 23.0 (SPSS Inc., Al Monk, NY, USA), and a Student's t test was used to compare the differences of qPCR and WB results. All cell experiments were independently repeated three times with different cell lysates for each single assay, and data were presented as mean ± S.E.M. A P value < 0.05 was considered statistically significant.
Results
Identification of a novel FBN1 null variant transmitted in a CS family
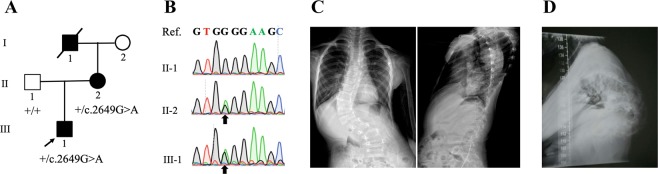
We applied ES to identify responsible variants with autosomal dominant inheritance in all 103 familial CS. We identified a maternally inherited LGD variant associated with the vertebral phenotypes in a family. The novel heterozygous LGD variant was c.2649G>A (p.Trp883Ter) in FBN1, which co-segregated with CS in a dominant family (Fig. 1a, b). The nonsense variant was considered to exert a significant LoF effect by triggering the nonsense-mediated mRNA decay mechanism and subsequently loss of FBN1 protein. The proband III-1 displayed CS with structural segmentation defect of the vertebrae (T10-L1) and the formation of vertebral bar on the left side (T10-L1) (Fig. 1c), whereas the mother II-2 exhibited severe congenital kyphosis with segmentation defects (Fig. 1d). Although the mitral valve prolapse with mild mitral insufficiency was found in the proband, no other apparent cardiovascular or ocular phenotypes featuring MFS were identified in the proband and his mother. The aortic root measurements of the proband still remained normal (Z score = 1.4) by echocardiographic measurements, and the diagnosis of MFS could not be implemented according to the Ghent Criteria [28]. As for now, no obvious “marfanoid” appearances are observed in the family members. i.e., III-1 and II-2 (Supplementary Table 1).
Fig. 1.
The family with congenital scoliosis and an FBN1 nonsense variant, c.2649G>A (p.Trp883Ter). a Pedigree and segregation of the variant. b Electropherograms of Sanger sequencing showing the heterozygous c.2649G>A in the proband (III-1) and his mother (II-2). Ref., the reference sequence. Black arrows indicate the nucleotide substitution positions. c Anteroposterior and lateral X-ray film of the whole spine of the proband III-1 displaying congenital scoliosis with segmentation defects of thoracic 10-lumbar 1 spine with formation of vertebral bar on the left. d Lateral X-ray film of the whole spine of II-2 showing severe congenital kyphosis with segmentation defects.
In addition, we did not observe any LoF variants other than the FBN1 p.Trp883Ter in any of the family members.
Novel deleterious coding variants in FBN1 are enriched in CS patients
Concerning the potential association between FBN1 and CS, we further compared the difference of the frequency of FBN1 variants between sporadic CS cases and controls. We subsequently identified a total of 30 novel variants (28 missense, 1 frameshift, and 1 splicing variant) in 16 out of the 574 CS cases and 14 out of 828 in-house controls. When all 30 variants were evaluated by the collapsing method, no significant association with CS was observed (OR = 1.6, P = 0.15 by one-sided Chi-square test). Next, the variants were stratified based on their deleteriousness determined by the CADD score. In the group of CADD score < 20, a total of 19 variants were subjected to the collapsed analysis without mutation types stratification and no statistical difference was identified. Further stratification within this group based on mutation types did not show any statistical difference in any mutation types (Table 1). In contrast, in the group of CADD score ≥ 20, a total of 11 missense variants were identified; eight in CS and three in controls, respectively. The eight missense variants (including one recurrent variant) were significantly enriched in CS (OR = 3.9, P = 0.03 by one-sided Fisher’s exact test) (Table 1). Sanger sequencing results of the eight cases are shown in Supplementary Fig. 1. Intriguingly, we have also checked the FBN1 mutations in our study but did not identify any overlapping variants with the idiopathic scoliosis cohort reported by Buchan et al. [29].
Table 1.
Mutational burden analysis of FBN1 variants in congenital scoliosis.
| Variant | Total number of variants | Number of alternative alleles | OR (95% CI) | P value | |
|---|---|---|---|---|---|
| Congenital scoliosis (n = 574) | In-house control (n = 828) | ||||
| Total | 30 | 16 | 14 | 1.6 (0.8–3.2) | 0.15 |
| CADD score < 20 | 19 | 8 | 11 | 1.0 (0.4–2.6) | 0.55 |
| Frameshift | 1 | 0 | 1 | – | 0.60 |
| Splicing | 1 | 1 | 0 | – | 0.41 |
| Missense | 17 | 7 | 10 | 1.0 (0.4–2.7) | 0.60 |
| CADD score ≥ 20 | 11 | 8 | 3 | 3.9 (1.0–14.6) | 0.03 |
| Missense | 11 | 8 | 3 | 3.9 (1.0–14.6) | 0.03 |
Association in the allelic model was calculated using Fisher’s exact test. P value < 0.05 was considered statistically significant
OR odds ratio, CI confidence interval, CADD Combined Annotation Dependent Depletion
Genetic and phenotypic analyses of the CS individuals harboring candidate variants
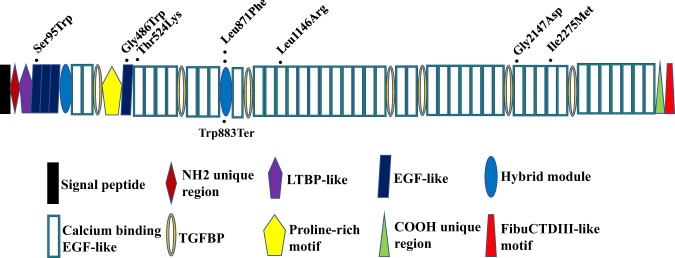
The genetic information and in silico deleteriousness predictions of the eight variants are presented in Table 2. One missense variant: p.Leu871Phe (CADD score = 23.9) was recurrent in two unrelated CS subjects. Leu871 was located within the hybrid 2 motif region of the protein (Fig. 2), which is an evolutionary ‘hybrid’ between TGF-β binding protein-like (TB) and calcium-binding epidermal growth factor-like (cbEGF-like) domains [30]. Previous studies indicated that mutations situated within this domain disrupted the ratio of an α-helix to β-sheet leading to a more compact conformation [31]. In addition, conservation analysis of amino acid residue of Leu871 in FBN1 indicated that it is highly conserved throughout evolution and across many selected species, suggesting it is required for the normal function of the protein (Fig. S2). The remaining six missense variants predicted with highly deleterious properties were located in epidermal growth factor-like (EGF-like) domains or cbEGF-like domains (Fig. 2), playing a seminal role in diseases [32].
Table 2.
Candidate FBN1 variants in congenital scoliosis.
| Subject ID | Genomic positiona | Nucleotide changeb | Protein changec | Exon number | Family and domain | AF in gnomAD | In silico prediction toold | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | EAS | SIFT | Polyphen-2 | Mutation taster | CADD score | ||||||
| XH1162 | 48902987 | c.284C>G | p.Ser95Trp | 4 | EGF-likee #01 | 0 | 0 | D | D | D | 22.7 |
| XH810 | 48807596 | c.1456G>T | p.Gly486Trp | 12 | EGF-like #04 | 0 | 0 | D | D | D | 24.6 |
| XH152 | 48805763 | c.1571C>A | p.Thr524Lys | 13 | cbEGF-likef #03 | 0 | 0 | T | D | D | 32 |
| XH73 | 48787384 | c.2613A>C | p.Leu871Phe | 22 | Hybrid motif #02 | 0.00000406 | 0.000058 | D | D | D | 23.9 |
| XH579 | 48787384 | c.2613A>C | p.Leu871Phe | 22 | Hybrid motif #02 | 0.00000406 | 0.000058 | D | D | D | 23.9 |
| XH766 | 48779535 | c.3437T>G | p.Leu1146Arg | 28 | cbEGF-like #13 | 0 | 0 | D | P | D | 26.4 |
| XH902 | 48729214 | c.6440G>A | p.Gly2147Asp | 53 | cbEGF-like #32 | 0 | 0 | T | D | D | 34 |
| XH441 | 48722914 | c.6825C>G | p.Ile2275Met | 56 | cbEGF-like #35 | 0 | 0 | T | D | D | 20.6 |
gnomAD Genome Aggregation Database, AF allele frequency, ALL all populations, EAS east Asian, D damaging, P probably damaging, T tolerated
aChromosomal position based on GRCh37/hg19
b,cThe RefSeq transcript number of FBN1 is NM_000138.4
dIn silico prediction tools to evaluate a genetic variant for its deleterious potential
eEpidermal growth factor-like domain
fCalcium-binding epidermal growth factor-like domain
Fig. 2.
Schematic representation of the distribution of the candidate FBN1 variants identified in congenital scoliosis. Protein structure is redrawn from the UMD-FBN1 database. Black dots indicate the location of missense and nonsense variants.
The eight subjects harbored the FBN1 variants displayed different types of vertebral malformations (Table 3). Malformations of the ribs were also found in three subjects (Table 3). Notably, intraspinal defects (diastematomyelia, syringomyelia, and tethered cord) were identified in five subjects (Table 3). Although mild mitral insufficiency and pectus carinatum deformity presented in XH902, aortic root measurements still remained normal (Z score = 1.6), and ocular phenotypes supporting MFS were not present. This patient would be categorized as “potential MFS” until the aorta reaches threshold (Z score = 3) [28]. The remaining seven subjects had no cardiovascular or ocular manifestations (Table 3), and hence they were not diagnosed as MFS. We have also re-evaluated the other marfanoid musculoskeletal phenotypes of these eight cases, and observed that no apparent manifestations except Patient XH902 with pectus carinatum deformity (Supplementary Table 2).
Table 3.
Phenotypic manifestations of the congenital scoliosis subjects with deleterious FBN1 missense variants.
| Subject ID | Age (year) | Gender | MCA (degree) | Malformation involved | Other complication | ||||
|---|---|---|---|---|---|---|---|---|---|
| Vertebral | Costal | Intraspinal | Cardiovascular | Ocular | |||||
| XH1162 | 6 | M | 65 | SD, FL, LF, H | N | N | N | N | Short neck |
| XH810 | 7 | F | 48 | B | N | Tethered cord | N | N | N |
| XH152 | 11 | M | 72 | H, W, FL | N | N | N | N | Pulmonary dysfunction |
| XH73 | 12 | F | 40 | SD | Intercostal cohesion of bilateral 2–5th ribs | Diastematomyelia, hypoplastic spinal processes | N | N | N |
| XH579 | 2 | M | 104 | W, FF, SD | Bilateral 12th rib absence | syringomyelia, tethered cord | N | N | Joint contractures |
| XH766 | 1 | M | 80 | B, H | N | N | N | N | N |
| XH902 | 11 | M | 77 | SD, W | N | Syringomyelia | Mitral insufficiency | N | Pectus carinatum |
| XH441 | 1 | M | 50 | SD, FF, B | Bilateral 13th ribs | Diastematomyelia, syringomyelia | N | N | N |
MCA maximal Cobb angle, M male, F female, SD segmentation defect, FL fused lamina, LF lamina fissure, H hemivertebrae, B butterfly-vertebrae, W wedge-shaped vertebrae, FF formation failure of vertebrae, N not present
Functional study of the recurrent variant, Leu871Phe
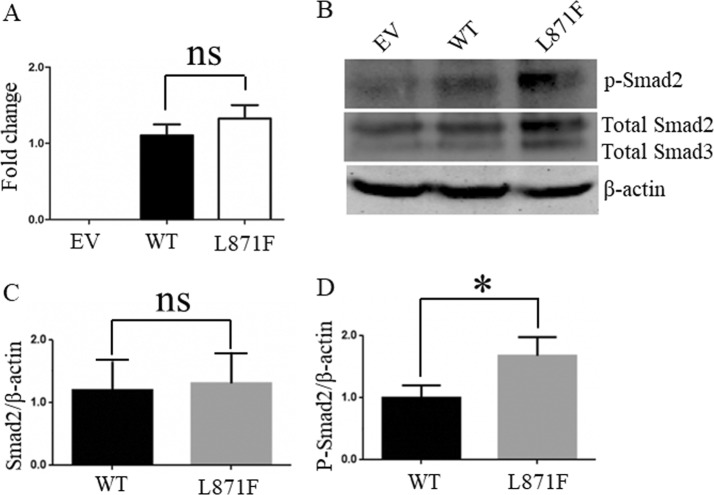
We overexpressed either human FBN1 wild-type or p.Leu871Phe in HEK293T cells. Total RNA was reversely transcribed to cDNA 48 h after transfection. The mRNA expression level of the p.Leu871Phe FBN1 showed no statistical difference to that of the WT plasmid (P = 0.38 by Student’s t test) (Fig. 3a).
Fig. 3.
Functional analyses of the recurrent missense variant, c.2613A>C (p.Leu871Phe). a The expression of FBN1 mRNAs from HEK293T cells transfected with the empty vector (EV), wild type (WT), or mutant (p. Leu871Phe: L871F) FBN1 plasmids. ns not significant. b Western blotting for the protein lysates. c, d Quantification of the western blotting results. The ratio of phosphorylated Smad2 and total Smad2 to β-actin are shown normalized to WT plasmid. Data are expressed as arbitrary units indicating mean ± SEM of three independent experiments. Statistical significance was calculated by Student’s t test, *P < 0.05. ns not significant.
Previous studies have demonstrated that deleterious FBN1 mutations upregulate endogenous TGF-β signaling, which could be measured via phosphorylation of Smad2 (p-Smad2), a downstream target [33]. Transfected cells with the p.Leu871Phe FBN1 showed significantly elevated p-Smad2 level compared with the WT (P = 0.02 by Student’s t test) (Fig. 3b–d), while they showed no statistical difference in expression of total Smad2 compared with the WT (P = 0.96 by Student’s t test).
Discussion
In the present study, we conducted family-based ES analysis and allele-based mutational burden analysis, and provided evidence for the genetic association between FBN1 and CS. We then performed in vitro functional assay for a recurrent FBN1 variant (p.Leu871Phe) and revealed the upregulation of TGF-β signaling caused by the variant. Clinical features for the CS cases with FBN1 variants showed the expansion of the FBN1-related disease entity.
FBN1 encodes a connective tissue protein essential to extracellular microfibrils organization in dermal fibroblasts and skeletal muscle cells [34]. FBN1 is well known as a causal gene for MFS, which often has scoliosis without structural changes of the vertebrae and ribs. On the other hand, FBN1 mutations also cause various skeletal dysplasia, including Acromicric dysplasia (MIM #102370), Geleophysic dysplasia 2 (MIM #614185), Marfan lipodystrophy syndrome (MIM #616914), and Weill–Marchesani syndrome 2 (MIM #608328). These monogenic diseases have a broad range of vertebral phenotypes quite different from MFS. In a previous study, a novel FBN1 variant, p.Gly1796Glu co-segregated in a family with an autosomal dominant vertebral dysplasia featuring CS, in the absence of cardiac or ocular findings [35], indicating that FBN1 could be a disease gene for monogenic form of CS. Our case also confirmed that FBN1 causes CS as an autosomal dominant disease trait pattern.
The index case in our study was from an unusual three-generation CS family segregating FBN1 LoF variant, c.2649G>A (p.Trp883Ter), which has been previously reported to be a pathogenic nonsense mutation associated with MFS [36], and deposited in HGMD (accession ID: CM161831). This nonsense substitution truncates the protein at codon 883, which is 1989 amino acids from the end of the protein. According to the ACMG guidelines for classifying pathogenic variants [37], this nonsense mutation is a convincing pathogenic LoF allele based on the following evidences: (a) PVS1: this nonsense variant is interpreted at the internal part of FBN1 gene where LoF is a known mechanism [38]; (b) PP1: cosegregation with CS phenotypes in the affected family members; (c)PP3: multiple computational evidence supports highly deleterious properties (Mutation Taster prediction shows deleterious; conservation prediction score by GERP is 4.81; CADD score is 42); (d)PP5: Franken et al. recently reported this variant as a pathogenic LoF allele associated with MFS, but the detailed evaluations for spinal/vertebral phenotypes were unavailable [36]. Notably, it has been widely reported that the high degree of clinical variability associated with FBN1 variants. For instance, distinct genetic mechanisms, encompassing a second deleterious variant in another gene or a polygenic model involving modifier loci, are proposed to account for this clinical variability [39]. Thus, the occurrence of a pathogenic variant in FBN1 is not necessarily associated with or produces MFS. Consistently with our findings, some FBN1 nonsense mutations (c.284C>A, p.Ser95Ter; c.1347_1348dupTA, p.Thr450IlefsTer130) only caused isolated skeletal features, with no cardiac or ocular findings [40]. Pathogenic FBN1 variants cause MFS but can also be found in patients presenting with apparently isolated features. For instance, the c.1453C>T, p.(Arg485Cys) mutation in FBN1 has been identified in both autosomal dominant and recessive diseases characterized by a high extent of phenotypic variability [41, 42]. Hence, it is reasonable to postulate that the c.2649G>A allele, which has been associated with MFS, could also account for the isolated CS in our study.
FBN1 is implicated in vertebral growth and development [43]. Fibrillin microfibrils in the calf are ubiquitously distributed in the vertebral growth plate and involved in growth factor binding, playing a pivotal role in regulating bone growth [43]. Moreover, a murine model with an intragenic duplication of Fbn1 presented markedly enlarged lumbar vertebra and elongated costal cartilages [44]. Functional follow-up of Fbn1Tsk mouse unveiled the interactions of FBN1 with the Wnt signaling pathway [45] and its pivotal role for somitegenesis and vertebrate segmentation [4]. Since CS is induced by localized changes in vertebral body development [46], we proposed that factors distorting the normal localization process of the spine column may contribute to the pathogenesis of CS, such as FBN1.
TGF-β plays critical roles in development and maintenance of the skeleton [47]. The two unrelated patients (XH73 and XH579) sharing the p.Leu871Phe variant presented CS with vertebral segmentation defects, rib anomalies, and diastematomyelia. Our in vitro experimental results showed upregulation of the TGF-β signaling by the p.Leu871Phe variant, supporting the pathogenicity of the mutation. TGF-β signaling plays a critical role in cartilage and spine tissue development, upregulation of the TGF-β signaling pathway within the intervertebral disc could cause the occurrence of vertebral fusions [48]. The observations suggest that the upregulation of the TGF-β signaling pathway confer susceptibility to CS. In prior reports, the excess TGF-β activation and signaling have been frequently observed in FBN1-deficient humans and mice [29, 49].
Regarding a mutational burden test for CS with a complex disease trait, the most important thing is not to focus disproportionately on specific variants, but rather to integrate across all types of risk-associated variants. In some individuals, risk may be caused by an unusual combination of common variants and rare variants, like the TACS compound inheritance model [6–8], whereas in others it may be due to an accumulation of rare or ultra-rare variants. In this context, it seemed actionable to stratify the FBN1 variants by the mutation types and CADD C-score prior to detecting the association by the collapsing method [25]. The knowledge about the variants associated with CS and the variable genetic penetrance will blaze the trail for genetic counseling and evaluating of the relatives of the variant carriers.
In conclusion, our observations indicated that FBN1 may be a susceptibility gene for CS, expanding our knowledge to the existing list of predisposition and candidate genes in CS phenotypes. It remains unknown why distinct mutations in the FBN1 gene can convey a variable continuum of skeletal disorders, ranging from the overlapping features to discordant phenotypes. Future studies focusing on exploring the underlying mechanisms of pleiotropic effects of FBN1 may be warranted.
Supplementary information
Acknowledgements
We are grateful to the patients, their families, clinical surgeons, and genetic counselors for providing samples and clinical histories. This research was supported in part by the National Natural Science Foundation of China (81822030 to NW, 81772299 and 81930068 to ZHW, 81672123 and 81972037 to JGZ, 81772301 and 81972132 to GXQ, 81572097 and 81871746 to YPW, 81902271 to GL, 81902178 to SRW), Beijing Natural Science Foundation (7191007 to ZHW, 7172175 to NW, 7184232 to SL,), CAMS Initiative Fund for Medical Sciences (2016-I2M-3-003 to GXQ, NW and ZW, 2016-I2M-2-006 and 2017-I2M-2-001 to ZHW), the Central Level Public Interest Program for Scientific Research Institute (2018RC31003 to NW and Mao Lin), the International Program Associate grant from RIKEN of Japan (190038 to ML), grants from Japan Agency For Medical Research and Development (AMED) (18ek0109280, 18ek0109212 to SI), the National Key Research and Development Program of RIKEN & Ministry of Science and Technology of China (RIKEN-MOST) (2016YFE0128400, to SI and ZW), RIKEN Incentive Research Projects (201801062228 to ZW), and the National Key Research and Development Program of China (No. 2018YFC0910506 to NW and ZHW, 2016YFC0901501 to SYZ).
Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study
Guixing Qiu1,4,9, Zhihong Wu4,8,9, Jianguo Zhang1,4,9, Nan Wu1,4,9, Shengru Wang1, Jiaqi Liu4,5, Sen Liu1,4, Yuzhi Zuo1, Gang Liu1,4, Yuanqiang Zhang1,2, Chenxi Yu1,2, Sen Zhao1,4, Lianlei Wang1,2, Yanxue Zhao1, Zihui Yan1,2, Xinzhuang Yang8, Hengqiang Zhao10, Yuchen Niu8, Mao Lin1,2,3, Jiachen Lin1.
Author contributions
ML and NW performed the research, analyzed and interpreted the data. ML and NW drafted the manuscript. SZ, GL, YZH, CXY, YXZ, SL, JQL, YYY, YPC, XY, and BDT helped sample collection. LLW, YQZ, and SRW performed phenotyping of patients. ZHY and XZY helped analysis and interpretation of ES data. YCN and XXL provided technique support. YPW, SYZ, ZW, SI, GXQ, and JGZ offered professional discussions and instructions. JZS, JY, and HQZ helped bioinformatic analyses. ZHW and NW conceived and designed the study, revised the manuscript, and provided final approval of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Members of the Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study are listed below Acknowledgements.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhihong Wu, Email: wuzh3000@126.com.
Nan Wu, Email: dr.wunan@pumch.cn.
on behalf of the Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study:
Guixing Qiu, Zhihong Wu, Jianguo Zhang, Nan Wu, Shengru Wang, Jiaqi Liu, Sen Liu, Yuzhi Zuo, Gang Liu, Yuanqiang Zhang, Chenxi Yu, Sen Zhao, Lianlei Wang, Yanxue Zhao, Zihui Yan, Xinzhuang Yang, Hengqiang Zhao, Yuchen Niu, Xiaoxin Li, and Mao Lin
Supplementary information
The online version of this article (10.1038/s10038-019-0698-x) contains supplementary material, which is available to authorized users.
References
- 1.Hedequist D, Emans J. Congenital scoliosis: a review and update. J Pediatr Orthop. 2007;27:106–16. doi: 10.1097/BPO.0b013e31802b4993. [DOI] [PubMed] [Google Scholar]
- 2.Wynne-Davies R. Congenital vertebral anomalies: aetiology and relationship to. spina bifida cystica. J Med Genet. 1975;12:280–8. doi: 10.1136/jmg.12.3.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen J, Wang Z, Liu J, Xue X, Qiu G. Abnormalities associated with congenital. scoliosis: a retrospective study of 226 Chinese surgical cases. Spine. 2013;38:814–8. doi: 10.1097/BRS.0b013e31827ed125. [DOI] [PubMed] [Google Scholar]
- 4.Pourquie O. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell. 2011;145:650–63. doi: 10.1016/j.cell.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, O'Reilly VC, et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306. doi: 10.1016/j.cell.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 6.Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–50. [DOI] [PMC free article] [PubMed]
- 7.Yang Nan, Wu Nan, Zhang Ling, Zhao Yanxue, Liu Jiaqi, Liang Xiangyu, Ren Xiaojun, Li Weiyu, Chen Weisheng, Dong Shuangshuang, Zhao Sen, Lin Jiachen, Xiang Hang, Xue Huadan, Chen Lu, Sun Hao, Zhang Jianguo, Shi Jiangang, Zhang Shuyang, Lu Daru, Wu Xiaohui, Jin Li, Ding Jiandong, Qiu Guixing, Wu Zhihong, Lupski James R, Zhang Feng. TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Human Molecular Genetics. 2018;28(4):539–547. doi: 10.1093/hmg/ddy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Jiaqi, Wu Nan, Yang Nan, Takeda Kazuki, Chen Weisheng, Li Weiyu, Du Renqian, Liu Sen, Zhou Yangzhong, Zhang Ling, Liu Zhenlei, Zuo Yuzhi, Zhao Sen, Blank Robert, Pehlivan Davut, Dong Shuangshuang, Zhang Jianguo, Shen Jianxiong, Si Nuo, Wang Yipeng, Liu Gang, Li Shugang, Zhao Yanxue, Zhao Hong, Chen Yixin, Zhao Yu, Song Xiaofei, Hu Jianhua, Lin Mao, Tian Ye, Yuan Bo, Yu Keyi, Niu Yuchen, Yu Bin, Li Xiaoxin, Chen Jia, Yan Zihui, Zhu Qiankun, Meng Xiaolu, Chen Xiaoli, Su Jianzhong, Zhao Xiuli, Wang Xiaoyue, Ming Yue, Li Xiao, Raggio Cathleen L., Zhang Baozhong, Weng Xisheng, Zhang Shuyang, Zhang Xue, Watanabe Kota, Matsumoto Morio, Jin Li, Shen Yiping, Sobreira Nara L., Posey Jennifer E., Giampietro Philip F., Valle David, Liu Pengfei, Wu Zhihong, Ikegawa Shiro, Lupski James R., Zhang Feng, Qiu Guixing. TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genetics in Medicine. 2019;21(7):1548–1558. doi: 10.1038/s41436-018-0377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otomo Nao, Takeda Kazuki, Kawai Shunsuke, Kou Ikuyo, Guo Long, Osawa Mitsujiro, Alev Cantas, Kawakami Noriaki, Miyake Noriko, Matsumoto Naomichi, Yasuhiko Yukuto, Kotani Toshiaki, Suzuki Teppei, Uno Koki, Sudo Hideki, Inami Satoshi, Taneichi Hiroshi, Shigematsu Hideki, Watanabe Kei, Yonezawa Ikuho, Sugawara Ryo, Taniguchi Yuki, Minami Shohei, Kaneko Kazuo, Nakamura Masaya, Matsumoto Morio, Toguchida Junya, Watanabe Kota, Ikegawa Shiro. Bi-allelic loss of function variants of TBX6 causes a spectrum of malformation of spine and rib including congenital scoliosis and spondylocostal dysostosis. Journal of Medical Genetics. 2019;56(9):622–628. doi: 10.1136/jmedgenet-2018-105920. [DOI] [PubMed] [Google Scholar]
- 10.Giampietro PF, Pourquie O, Raggio C, Ikegawa S, Turnpenny PD, Gray R, et al. Summary of the first inaugural joint meeting of the International Consortium for scoliosis genetics and the International Consortium for vertebral anomalies and scoliosis, March 16-18, 2017, Dallas. Tex Am J Med Genet A. 2018;176:253–6. doi: 10.1002/ajmg.a.38550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnei G, Gavriliu S, Vlad C, Georgescu I, Ghita RA, Dughila C, et al. Congenital scoliosis: an up-to-date. J Med Life. 2015;8:388–97. [PMC free article] [PubMed] [Google Scholar]
- 12.Giampietro PF, Raggio CL, Reynolds CE, Shukla SK, McPherson E, Ghebranious N, et al. An analysis of PAX1 in the development of vertebral malformations. Clin Genet. 2005;68:448–53. doi: 10.1111/j.1399-0004.2005.00520.x. [DOI] [PubMed] [Google Scholar]
- 13.Hayes M, Gao X, Yu LX, Paria N, Henkelman RM, Wise CA, et al. ptk7 mutant zebrafish models of congenital and idiopathic scoliosis implicate dysregulated Wnt signalling in disease. Nat Commun. 2014;5:4777. doi: 10.1038/ncomms5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barhoumi T, Nashabat M, Alghanem B, Alhallaj A, Boudjelal M, Umair M, et al. Delta like-1 gene mutation: a novel cause of congenital vertebral malformation. Front Genet. 2019;10:534. doi: 10.3389/fgene.2019.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bargal R, Cormier-Daire V, Ben-Neriah Z, Le Merrer M, Sosna J, Melki J, et al. Mutations in DDR2 gene cause SMED with short limbs and abnormal calcifications. Am J Hum Genet. 2009;84:80–4. doi: 10.1016/j.ajhg.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Liu J, Yuan D, Zuo Y, Liu Z, Liu S, et al. Progress and perspective of TBX6 gene in congenital vertebral malformations. Oncotarget. 2016;7:57430–41. doi: 10.18632/oncotarget.10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Zhao S, Liu B, Zhang Q, Li Y, Liu J, et al. Perturbations of BMP/TGF-beta and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM) J Med Genet. 2018;55:675–84. doi: 10.1136/jmedgenet-2017-105224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 19.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 21.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson AR, Smith EN, Matsui H, Braekkan SK, Jepsen K, Hansen JB, et al. Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinformatics. 2014;15:125. doi: 10.1186/1471-2105-15-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng H, Dharmadhikari AV, Varland S, Ma N, Domingo D, Kleyner R, et al. Truncating variants in NAA15 are associated with variable levels of Intellectual Disability, Autism Spectrum Disorder, and Congenital Anomalies. Am J Hum Genet. 2018;102:985–94. doi: 10.1016/j.ajhg.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collod-Beroud G, Le Bourdelles S, Ades L, Ala-Kokko L, Booms P, Boxer M, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat. 2003;22:199–208. doi: 10.1002/humu.10249. [DOI] [PubMed] [Google Scholar]
- 25.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen LL, Terczynska-Dyla E, Mork N, Scavenius C, Enghild JJ, Honing K, et al. Frequently used bioinformatics tools overestimate the damaging effect of allelic variants. Genes Immun. 2019;20:10–22. doi: 10.1038/s41435-017-0002-z. [DOI] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeys BL, Dietz HC, Braverman AC, Callewaert BL, De Backer J, Devereux RB, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet. 2010;47:476–85. doi: 10.1136/jmg.2009.072785. [DOI] [PubMed] [Google Scholar]
- 29.Buchan JG, Alvarado DM, Haller GE, Cruchaga C, Harms MB, Zhang T, et al. Rare variants in FBN1 and FBN2 are associated with severe adolescent idiopathic scoliosis. Hum Mol Genet. 2014;23:5271–82. doi: 10.1093/hmg/ddu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen SA, Iqbal S, Lowe ED, Redfield C, Handford PA. Structure and interdomain interactions of a hybrid domain: a disulphide-rich module of the fibrillin/LTBP superfamily of matrix proteins. Structure. 2009;17:759–68. doi: 10.1016/j.str.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellody KT, Freeman LJ, Baldock C, Jowitt TA, Siegler V, Raynal BD, et al. Marfan syndrome-causing mutations in fibrillin-1 result in gross morphological alterations and highlight the structural importance of the second hybrid domain. J Biol Chem. 2006;281:31854–62. doi: 10.1074/jbc.M602743200. [DOI] [PubMed] [Google Scholar]
- 32.Handford P, Downing AK, Rao Z, Hewett DR, Sykes BC, Kielty CM. The calcium binding properties and molecular organization of epidermal growth factor-like domains in human fibrillin-1. J Biol Chem. 1995;270:6751–6. doi: 10.1074/jbc.270.12.6751. [DOI] [PubMed] [Google Scholar]
- 33.Andelfinger G, Loeys B, Dietz H. A decade of discovery in the genetic understanding of thoracic aortic disease. Can J Cardiol. 2016;32:13–25. doi: 10.1016/j.cjca.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Park JW, Yan L, Stoddard C, Wang X, Yue Z, Crandall L, et al. Recapitulating and correcting Marfan Syndrome in a cellular model. Int J Biol Sci. 2017;13:588–603. doi: 10.7150/ijbs.19517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ades LC, Sreetharan D, Onikul E, Stockton V, Watson KC, Holman KJ. Segregation of a novel FBN1 gene mutation, G1796E, with kyphoscoliosis and radiographic evidence of vertebral dysplasia in three generations. Am J Med Genet. 2002;109:261–70. doi: 10.1002/ajmg.10333. [DOI] [PubMed] [Google Scholar]
- 36.Franken R, Groenink M, de Waard V, Feenstra HM, Scholte AJ, van den Berg MP, et al. Genotype impacts survival in Marfan syndrome. Eur Heart J. 2016;37:3285–90. doi: 10.1093/eurheartj/ehv739. [DOI] [PubMed] [Google Scholar]
- 37.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuguchi T, Matsumoto N. Recent progress in genetics of Marfan syndrome and Marfan-associated disorders. J Hum Genet. 2007;52:1–12. doi: 10.1007/s10038-006-0078-1. [DOI] [PubMed] [Google Scholar]
- 39.Aubart M, Gazal S, Arnaud P, Benarroch L, Gross MS, Buratti J, et al. Association of modifiers and other genetic factors explain Marfan syndrome clinical variability. Eur J Hum Genet. 2018;26:1759–72. doi: 10.1038/s41431-018-0164-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groth KA, Von Kodolitsch Y, Kutsche K, Gaustadnes M, Thorsen K, Andersen NH, et al. Evaluating the quality of Marfan genotype-phenotype correlations in existing FBN1 databases. Genet Med. 2017;19:772–7. doi: 10.1038/gim.2016.181. [DOI] [PubMed] [Google Scholar]
- 41.Overwater E, Efrat R, Barge-Schaapveld D, Lakeman P, Weiss MM, Maugeri A, et al. Autosomal dominant Marfan syndrome caused by a previously reported recessive FBN1 variant. Mol Genet Genom Med. 2019;7:e00518. doi: 10.1002/mgg3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries BB, Pals G, Odink R, Hamel BC. Homozygosity for a FBN1 missense mutation: clinical and molecular evidence for recessive Marfan syndrome. Eur J Hum Genet. 2007;15:930–5. doi: 10.1038/sj.ejhg.5201865. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Urban J. Immunolocalisation of fibrillin microfibrils in the calf metacarpal and vertebral growth plate. J Anat. 2013;223:641–50. doi: 10.1111/joa.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barisic-Dujmovic T, Boban I, Adams DJ, Clark SH. Marfan-like skeletal phenotype in the tight skin (Tsk) mouse. Calcif Tissue Int. 2007;81:305–15. doi: 10.1007/s00223-007-9059-4. [DOI] [PubMed] [Google Scholar]
- 45.Bayle J, Fitch J, Jacobsen K, Kumar R, Lafyatis R, Lemaire R. Increased expression of Wnt2 and SFRP4 in Tsk mouse skin: role of Wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J Investig Dermatol. 2008;128:871–81. doi: 10.1038/sj.jid.5701101. [DOI] [PubMed] [Google Scholar]
- 46.Giampietro PF, Blank RD, Raggio CL, Merchant S, Jacobsen FS, Faciszewski T, et al. Congenital and idiopathic scoliosis: clinical and genetic aspects. Clin Med Res. 2003;1:125–36. doi: 10.3121/cmr.1.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacFarlane EG, Haupt J, Dietz HC, Shore EM. TGF-beta family signaling in connective tissue and skeletal diseases. Cold Spring Harb Perspect Biol. 2017;9:a022269. doi: 10.1101/cshperspect.a022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zieba J, Forlenza KN, Khatra JS, Sarukhanov A, Duran I, Rigueur D, et al. TGFbeta and BMP dependent cell fate changes due to loss of filamin B produces disc degeneration and progressive vertebral fusions. PLoS Genet. 2016;12:e1005936. doi: 10.1371/journal.pgen.1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng CM, Cheng A, Myers LA, Martinez-Murillo F, Jie C, Bedja D, et al. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J Clin Investig. 2004;114:1586–92. doi: 10.1172/JCI22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.