Abstract
Streptococcus suis strains isolated from porcine endocarditis and tonsils in the Tokai area of Japan during 2004–2007 and 2014–2016 (n=114) were tested for antimicrobial susceptibility and distribution of selected resistance genes. No strains showed resistance to penicillin, ampicillin, cefotaxime, meropenem, vancomycin, and levofloxacin. High resistance to tetracycline (80.7%), clindamycin (65.8%), erythromycin (56.1%), and clarithromycin (56.1%) was observed. In chloramphenicol and sulfamethoxazole-trimethoprim, there was a trend towards increased resistance between the first (2004–2007) and second (2014–2016) periods. tet(O) and erm(B) genes were the most frequently detected, and tet(M) and mef(A/E) genes were only detected in strains isolated during 2014–2016. These results indicate that chloramphenicol and sulfamethoxazole-trimethoprim resistance, and tet(M) and mef(A/E) genes emerged in S. suis of this area after 2014.
Keywords: antimicrobial resistance, genotype, phenotype, pig, Streptococcus suis
Streptococcus suis causes a variety of diseases in pigs, including meningitis, septicemia, endocarditis, arthritis, and pneumonia [19]. S. suis is also a zoonotic pathogen related to the pork industry, which can cause meningitis and septicemia in humans [1, 27]. Although S. suis has been detected at high rates in porcine bacterial endocarditis lesions during meat hygiene inspection [9, 10, 24], it has also been found in the upper respiratory tract, such as the tonsils, of healthy pigs [10]. Thus, asymptomatic carriers might be a source of S. suis infection in pigs and humans [4, 10]. Of the approximately 30 known serotypes of S. suis, serotype 2 is the most virulent and is responsible for severe infections in both pigs and humans worldwide [21, 22, 27]. We have also reported the high rates of detection of the cps2J+ strains of S. suis in porcine bacterial endocarditis lesions [10].
Several studies have shown that S. suis strains isolated from both pigs and humans are highly resistant (92.0–99.6%) to at least one of the antimicrobial agents examined [5, 7, 28]. A study on Japanese S. suis strains isolated from pigs before 1996 documented that only 11.3% were sensitive to all antimicrobial agents examined [11]. Especially, high level of resistance to tetracycline (TC) and macrolides have been reported [2, 5, 7, 17, 25, 28]. The resistance genes, tet(O) and erm(B), are the most common in TC and macrolide-resistant S. suis, respectively [2, 7, 17]. Moreover, S. suis strains resistant to β-lactams, chloramphenicol (CP), and aminoglycosides have also been reported in several countries [5, 23]. Understanding the antimicrobial susceptibility of S. suis, especially strains of serotypes that are highly associated with disease, is important in the treatment and prevention of S. suis infection in animals and humans. However, there is limited information on the antimicrobial susceptibility of Japanese S. suis, particularly those recently isolated [11]. We performed antimicrobial susceptibility tests using chronologically diverse 114 S. suis isolates and investigated the relationship between their antimicrobial susceptibility and isolation period or cps types.
We have reported the cps types, putative multilocus sequence typing (MLST) complex, and virulence gene profiles of S. suis isolated from pigs brought into slaughterhouses in Nagoya City between 2004–2007, and between 2014–2016 [10, see in the Supplemental file]. In this study, of the 197 strains detected, 114 were selected. In principle, only one strain was selected from each farm, origin (endocarditis and tonsils) and isolation period (2004–2007 and 2014–2016). Multiple strains were selected only if the cps type, the putative MLST complex, or the virulence gene profile of strains were different. The 114 S. suis strains consisted of 50 strains from bacterial endocarditis of pigs obtained from 34 farms, and 64 strains from tonsils of healthy pigs obtained from 27 farms (Table 1).
Table 1. Streptococcsu suis strains tested in this study.
| Origin | Isolated period | Number of strains (Number of farms) |
||
|---|---|---|---|---|
|
cps type |
Total | |||
| cps2J+ | Othersa) | |||
| Endocarditis | 2004–2007 | 13 (12) | 3 (3) | 16 (12) |
| 2014–2016 | 25 (22) | 9 (7) | 34 (25) | |
| Subtotal | 38 (31) | 12 (10) | 50 (34) | |
| Tonsil | 2014–2016 | 11 (10) | 53 (25) | 64 (27) |
| Total | 49 (36) | 65 (30) | 114 (48) | |
a) cps types 3 (7strains), 4(6), 5(2), 6(2), 7(1), 8(1), 9(1), 10(1), 11(1), 15(3), 16(5), 21(1), 23(1), 25(2), 28(1), 30(1), 31(6), 1 or 14(4) and untypable (19).
Antimicrobial susceptibility tests were performed by determining the minimum inhibitory concentration (MIC) of strains using the Dry plate Eiken broth microdilution method (Eiken Kagaku, Tochigi, Japan) according to the manufacturer’s instructions. Twelve antimicrobial agents were tested: penicillin (PCG), ampicillin (ABPC), cefotaxime (CTX), meropenem (MEPM), erythromycin (EM), clarithromycin (CAM), clindamycin (CLDM), TC, CP, vancomycin (VCM), levofloxacin (LVFX), and sulfamethoxazole-trimethoprim (ST, 19:1). The MIC breakpoints were taken from the Clinical and Laboratory Standard Institute (CLSI) criteria 2018 (M100-ED28) for the Streptococcus spp. viridans group [3]. Because the MIC distribution of ST that were not defined in the guideline showed bimodality, microbiological breakpoints were determined (19/1µg/ml) [13]. S. pneumoniae ATCC 49619 was used for quality control in all tests. The presence of the following resistance genes was examined by PCR assays: TC resistance genes-tet(O), tet(M), tet(L), and tet(K) [14]; macrolides resistance genes-erm(A), erm(B), erm(C), msr(A/B), ere(A), ere(B), mph(A), and mef(A/E) [20]. DNA templates were prepared by the boiling method, and PCR was performed in a total volume of 25 µl using Takara Ex Taq (TaKaRa Bio, Kusatsu, Japan). Statistical significance was determined by the chi-square test, Fisher’s exact test, and Yates corrections, depending on the number of samples. P values <0.05 were considered significant.
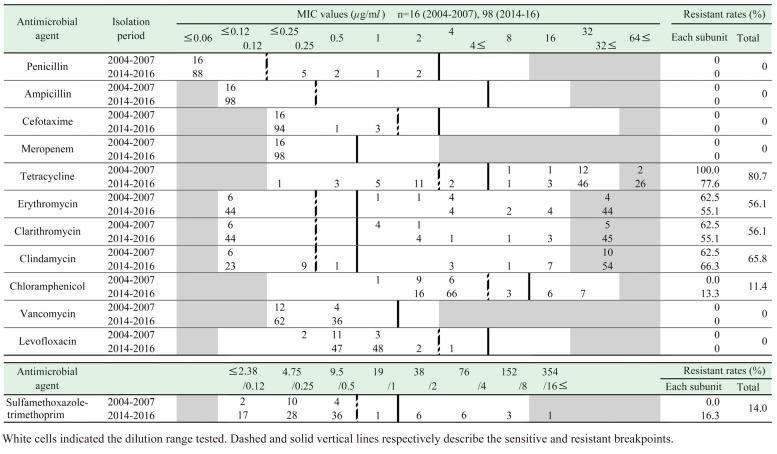
The highest resistance rate observed was against TC (80.7%, 92 strains), followed by CLDM (65.8%), EM (56.1%), and CAM (56.1%) (Table 2). 11.4% and 14.0% of the strains exhibited resistance to CP and ST respectively. All strains were susceptible to β-lactams (PCG, ABPC, CTX, and MEPM), VCM, and LVFX. Overall, 101 (88.6%) of the 114 strains were resistant to at least one of the antimicrobial agents examined (Table 4). Fifty-eight strains (50.9%) were resistant to both TC and macrolides.
Table 2. Minimum inhibitory concentration (MIC) distribution and resistance rates of all Streptococcus suis isolates.
Table 4. Antimicrobial resistance pattarns and detected resistance genes for each pattarns.
The cps2J+ strains were significantly (P<0.01) more resistant to TC, EM, and CAM and significantly (P<0.01) less resistant to CP and ST than the other cps type strains (Table 3). CP resistance was found in strains with cps genes typed as 4, 15, 16, 25, 28 (1 strain each), 31 (3 strains) and untypable (4 strains), while ST resistance was found in strains with cps genes typed as 6 (1 strain), 3, 15 (2 strains each), 16, 31 (3 strains each), and untypable (4 strains). Additionally, all the CP-and the ST-resistant strains were isolated in 2014–2016. There was no difference in the antimicrobial resistance rates (except for ST) between the strains isolated from endocarditis lesions and those isolated from tonsils (Table 3).
Table 3. Antimicrobial resitance rates in Streptococcus suis strains tested by origins and cps types.
| Antimicrobial agents | Origins |
cps types |
||
|---|---|---|---|---|
| Endocarditis | Tonsils | cps2J+ | Others | |
| n=50 | n=64 | n=49 | n=65 | |
| Tetracycline | 86.0 | 76.6 | 91.8b) | 72.3 |
| Erythromycin | 66.0 | 48.4 | 75.5b) | 41.5 |
| Clarithromycin | 66.0 | 48.4 | 75.5b) | 41.5 |
| Clindamycin | 70.0 | 62.5 | 75.5 | 58.5 |
| Chloramphenicol | 6.0 | 15.6 | 2.0 | 18.5b,c) |
| Sulfamethoxazole-trimethoprim | 6.0 | 20.3a) | 2.0 | 23.1b,d) |
Only drugs with resistant strains were shown. a) P<0.05. b) P<0.01. c) cps types 4, 15, 16, 25, 28 (1 strain each), 31 (3), and untypable (4). d) cps types 6 (1 strain), 3, 15 (2 each), 16, 31 (3 each), and untypable (4).
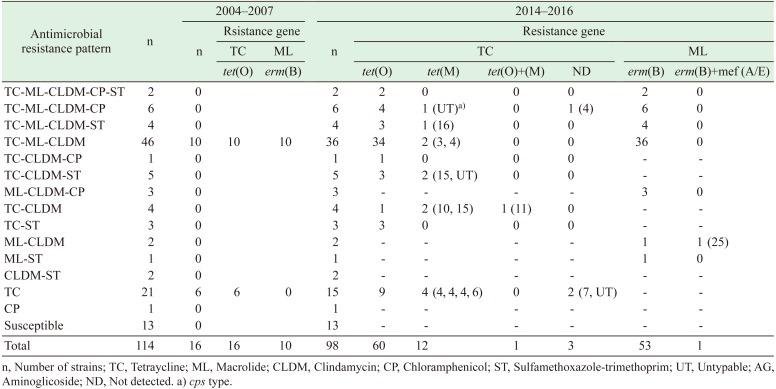
In this study, the tet gene was detected in 89 (96.7%) of the 92 TC-resistant strains. The most common tet gene identified was tet(O) (n=77, 83.7%), followed by tet(M) (n=13, 14.1%). One strain (1.1%) possessed both the tet(O) and tet(M) genes (Table 4). The erm(B) gene was detected in all 64 strains that were resistant to macrolides (CLDM, EM, or CAM), and one of the strains possessed both the erm(B) and mef(A/E) genes. All strains that possessed tet(M) and mef(A/E), as well as TC-resistant strains in which tet genes were negative, were isolated during 2014–2016. Moreover, these strains belonged to cps types other than the cps2J+.
High rates of TC resistance were observed during 2004–2007 (100%), 2014–2017 (77.6%), as well as during 1987–1996 (86.9%) [11]. Thus, TC resistance appears to be consistently prevalent in Japanese pig strains. High TC resistance rates in S. suis have been reported in pig strains (91% in the UK [6], 91.7% in China [28], 90% in Italy [16]) and also in humans (90.9% in Vietnam [7]). Because S. suis infections in humans are associated with exposure to pigs and contaminated pork, the high level of resistance rate among strains isolated from pigs is important, as it also affects the resistance rate in human strains. Our results indicate that the tet(O) gene is also a major determinant of TC resistance in S. suis in the Tokai area. Among the 59 tet genes, the tet(O) gene encoding a ribosome protective protein (RPP) appears to be the most common determinant of resistance in S. suis strains isolated from both pigs and humans globally [2, 5, 17, 23, 26]. On the other hand, although the tet(M) gene (also encoding a RPP) was detected only in 2.0% and 3.9% of Korean [5] and Italian [17] S. suis pig strains, respectively, it was detected in 36.4% of human strains in Hong Kong [2]. The tet(M) gene was the most commonly detected TC resistance gene in Enterococcus faecalis isolated from swine feces (20/22) [12]. The tet gene is often present on conjugative plasmids or transposons, and helps in transmission of resistance from one bacterium to another. In particular, since the tet(M) gene is thought to be related to the conjugative transposon Tn916, which has a very wide host range [15], it appears that tet(M) gene might increase, even in S. suis. Among all the antimicrobial agents that have been sold as veterinary medicines in Japan in 2016, TC and macrolides were the first (41%) and second (17%) most common agents, respectively [16]. Thus, the high frequency of resistance to TC and macrolides could be explained by the fact that they are the most widely used antimicrobial agents in veterinary medicine. Although, such in Salmonella and Escherichia coli, strains isolated from diseased pigs tend to have high-level resistance rates than those isolated from healthy pigs, no differences were found in this study in the resistance to TC and macrolides between bacterial endocarditis-derived and tonsils-derived strains.
None of the strains were resistant to β-lactams. PCG resistance in S. suis was first reported in the UK in a serotype 2 strain isolated from a human patient in 1980 [18], and has since emerged in S. suis isolated from pigs globally [5, 6, 28]. However, China [28] and the UK [6] isolates exhibit low resistance rates (9.1% and 5%, respectively), and resistant strains have not been detected in Vietnam [7]. Moreover, in Japan, low (0–3.3%) resistance rates to β-lactams was observed in strains isolated from pigs during 1987–1996 [11]. Our results indicate that the high susceptibility to β-lactams has been maintained, and that β-lactams are still effective against S. suis infection in Japan.
Understanding the change in antimicrobial resistance profiles is important in the selection of antimicrobials against S. suis infections in pigs and humans. The emergence of CP and ST resistance is more important than that of tet(M) and mef(A/E) genes, which are involved in TC or macrolides resistance, as the former type of resistance is indicative of resistance against new antimicrobial agents. In particular, since ST is also used as a therapeutic agent for humans, the increase in ST-resistant bacteria is considered more important in the management of infections. The increase in resistance to CP, which is currently prohibited in livestock farming, might be due to the cross-resistance to thiamphenicol and florfenicol. Another possible cause is co-selection due to the use of other antimicrobial agents, because 11 of 13 CP-resistant strains were also resistant to macrolides and 9 strains were also resistant to TC. It has been reported that the CP resistance gene cat and the erm(B) and tet(O) genes are located within the same 40 kb DNA region of a conjugative mobile element in S. suis [8].
Differences in resistance rates might be involved in the virulence of S. suis. The prevalence of resistance to TC and macrolides was significantly higher in cps2J+ strains than in the other cps type strains. Macrolides and TC are likely to be used frequently for pigs in Japan. Therefore, it can be presumed that the disease-relevant cps2J + strain is likely to be exposed to these drugs. However, because there was no difference in the resistance rates between cps2J+ strains isolated from endocarditis lesions and those from tonsils (data not shown), TC- and macrolides-resistant cps2J+ strains have likely continued to be maintained on the farms. On the other hand, the resistance rates to CP and ST were significantly higher in other cps type strains compared to the cps2J+ strains, indicating that the resistance patterns can differ depending on the S. suis cps type. Notably, all strains that possessed tet(M) and mef(A/E), and those lacking the tet genes, were cps types other than cps2J+. However, no bias was found in the cps types among strains resistant to CP and ST and among strains that possessed these new genes. Therefore, the relationship between S. suis cps types and resistance genes should be studied further.
In conclusion, we showed the changes in antimicrobial susceptibilities and resistance genes of S. suis strains isolated from pigs between 2004–2007 and 2014–2017 in the Tokai area of Japan. Because understanding the antimicrobial susceptibility of S. suis is important for the treatment and prevention of S. suis infections in animals and humans, continuous surveillance of S. suis strains is needed.
Supplementary
REFERENCES
- 1.Chang B., Wada A., Ikebe T., Ohnishi M., Mita K., Endo M., Matsuo H., Asatuma Y., Kuramoto S., Sekiguchi H., Yamazaki M., Yoshikawa H., Watabe N., Yamada H., Kurita S., Imai Y., Watanabe H.2006. Characteristics of Streptococcus suis isolated from patients in Japan. Jpn. J. Infect. Dis. 59: 397–399. [PubMed] [Google Scholar]
- 2.Chu Y. W., Cheung T. K. M., Chu M. Y., Tsang V. Y. M., Fung J. T. L., Kam K. M., Lo J. Y. C.2009. Resistance to tetracycline, erythromycin and clindamycin in Streptococcus suis serotype 2 in Hong Kong. Int. J. Antimicrob. Agents 34: 181–182. doi: 10.1016/j.ijantimicag.2009.01.007 [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute (CLSI). 2018. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. Document M100-S28, Wayne. [Google Scholar]
- 4.Gottschalk M., Segura M., Xu J.2007. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim. Health Res. Rev. 8: 29–45. doi: 10.1017/S1466252307001247 [DOI] [PubMed] [Google Scholar]
- 5.Gurung M., Tamang M. D., Moon D. C., Kim S. R., Jeong J. H., Jang G. C., Jung S. C., Park Y. H., Lim S. K.2015. Molecular Basis of Resistance to Selected Antimicrobial Agents in the Emerging Zoonotic Pathogen Streptococcus suis. J. Clin. Microbiol. 53: 2332–2336. doi: 10.1128/JCM.00123-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez-Garcia J., Wang J., Restif O., Holmes M. A., Mather A. E., Weinert L. A., Wileman T. M., Thomson J. R., Langford P. R., Wren B. W., Rycroft A., Maskell D. J., Tucker A. W., BRADP1T Consortium. 2017. Patterns of antimicrobial resistance in Streptococcus suis isolates from pigs with or without streptococcal disease in England between 2009 and 2014. Vet. Microbiol. 207: 117–124. doi: 10.1016/j.vetmic.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoa N. T., Chieu T. T., Nghia H. D., Mai N. T., Anh P. H., Wolbers M., Baker S., Campbell J. I., Chau N. V., Hien T. T., Farrar J., Schultsz C.2011. The antimicrobial resistance patterns and associated determinants in Streptococcus suis isolated from humans in southern Vietnam, 1997–2008. BMC Infect. Dis. 11: 6. doi: 10.1186/1471-2334-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holden M. T., Hauser H., Sanders M., Ngo T. H., Cherevach I., Cronin A., Goodhead I., Mungall K., Quail M. A., Price C., Rabbinowitsch E., Sharp S., Croucher N. J., Chieu T. B., Mai N. T. H., Diep T. S., Chinh N. T., Kehoe M., Leigh J. A., Ward P. N., Dowson C. G., Whatmore A. M., Chanter N., Iversen P., Gottschalk M., Slater J. D., Smith H. E., Spratt B. G., Xu J., Ye C., Bentley S., Barrell B. G., Schultsz C., Maskell D. J., Parkhill J.2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4: e6072. doi: 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichikawa T., Kondo T., Yamahara A.1991. Streptococci isolated from pigs with bacterial endocarditis. Nippon Juishikai Zasshi 44: 153–157. [Google Scholar]
- 10.Ichikawa T., Niwa T., Nakanishi T.2018. Virulence-associated gene profiling of Streptococcus suis isolated from porcine bacterial endocarditis and tonsils. Nippon Juishikai Zasshi 71: 193–197. [Google Scholar]
- 11.Kataoka Y., Yoshida T., Sawada T.2000. A 10-year survey of antimicrobial susceptibility of streptococcus suis isolates from swine in Japan. J. Vet. Med. Sci. 62: 1053–1057. doi: 10.1292/jvms.62.1053 [DOI] [PubMed] [Google Scholar]
- 12.Kobashi Y., Hasebe A., Nishio M., Uchiyama H.2007. Diversity of tetracycline resistance genes in bacteria isolated from various agricultural environment. Microbes Environ. 22: 44–51. doi: 10.1264/jsme2.22.44 [DOI] [Google Scholar]
- 13.MacGowan A. P., Wise R.2001. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J. Antimicrob. Chemother. 48 Suppl 1: 17–28. doi: 10.1093/jac/48.suppl_1.17 [DOI] [PubMed] [Google Scholar]
- 14.Malhotra-Kumar S., Lammens C., Piessens J., Goossens H.2005. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob. Agents Chemother. 49: 4798–4800. doi: 10.1128/AAC.49.11.4798-4800.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marosevic D., Kaevska M., Jaglic Z.2017. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage - a review. Ann. Agric. Environ. Med. 24: 338–344. doi: 10.26444/aaem/74718 [DOI] [PubMed] [Google Scholar]
- 16.National Veterinary Assay Laboratory Ministry of Agriculture, Forestry and Fisheries. 2016. Sales amounts and sales volumes (active substance) of antibiotics, synthetic antibacterials, anthelmintics and antiprotozoals. National veterinary assay laboratory (online). http://www.maff.go.jp/nval/iyakutou/ hanbaidaka/pdf/h28hanbaikoukin20180205.pdf [accessed on October 1, 2019].
- 17.Princivalli M. S., Palmieri C., Magi G., Vignaroli C., Manzin A., Camporese A., Barocci S., Magistrali C., Facinelli B.2009. Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003–2007). Euro Surveill. 14: 19310. doi: 10.2807/ese.14.33.19310-en [DOI] [PubMed] [Google Scholar]
- 18.Shneerson J. M., Chattopadhyay B., Murphy M. F., Fawcett I. W.1980. Permanent perceptive deafness due to Streptococcus suis type II infection. J. Laryngol. Otol. 94: 425–427. doi: 10.1017/S0022215100089040 [DOI] [PubMed] [Google Scholar]
- 19.Staats J. J., Feder I., Okwumabua O., Chengappa M. M.1997. Streptococcus suis: past and present. Vet. Res. Commun. 21: 381–407. doi: 10.1023/A:1005870317757 [DOI] [PubMed] [Google Scholar]
- 20.Sutcliffe J., Grebe T., Tait-Kamradt A., Wondrack L.1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40: 2562–2566. doi: 10.1128/AAC.40.11.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takamatsu D., Wongsawan K., Osaki M., Nishino H., Ishiji T., Tharavichitkul P., Khantawa B., Fongcom A., Takai S., Sekizaki T.2008. Streptococcus suis in humans, Thailand. Emerg. Infect. Dis. 14: 181–183. doi: 10.3201/eid1401.070568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thravichitkul P., Wongsawan K., Takenami N., Pruksakorn S., Fongcom A., Gottschalk M., Khanthawa B., Supajatura V., Takai S.2014. Correlation between PFGE group and mrp/epf/sly genotypes of human Streptococcus suis serotype 2 in Northern Thailand. J. Pathogens 2014: Article ID 350416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Y., Aarestrup F. M., Lu C. P.2004. Characterization of Streptococcus suis serotype 7 isolates from diseased pigs in Denmark. Vet. Microbiol. 103: 55–62. doi: 10.1016/j.vetmic.2004.07.009 [DOI] [PubMed] [Google Scholar]
- 24.Tsuchiya Y., Sato S.2009. Epidemiological study of Streptococcus suis isolated from pigs brought to slaughterhouse. Nippon Juishikai Zasshi 62: 563–567. [Google Scholar]
- 25.Wisselink H. J., Veldman K. T., Van den Eede C., Salmon S. A., Mevius D. J.2006. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet. Microbiol. 113: 73–82. doi: 10.1016/j.vetmic.2005.10.035 [DOI] [PubMed] [Google Scholar]
- 26.Ye C., Bai X., Zhang J., Jing H., Zheng H., Du H., Cui Z., Zhang S., Jin D., Xu Y., Xiong Y., Zhao A., Luo X., Sun Q., Gottschalk M., Xu J.2008. Spread of Streptococcus suis sequence type 7, China. Emerg. Infect. Dis. 14: 787–791. doi: 10.3201/eid1405.070437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H., Jing H., Chen Z., Zheng H., Zhu X., Wang H., Wang S., Liu L., Zu R., Luo L., Xiang N., Liu H., Liu X., Shu Y., Lee S. S., Chuang S. K., Wang Y., Xu J., Yang W., Streptococcus suis study groups. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12: 914–920. doi: 10.3201/eid1206.051194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C., Ning Y., Zhang Z., Song L., Qiu H., Gao H.2008. In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet. Microbiol. 131: 386–392. doi: 10.1016/j.vetmic.2008.04.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.