Abstract
We performed a clonality analysis using polymerase chain reaction (PCR) for immunoglobulin heavy chain (IgH) gene rearrangement, specifically with regard to its utility as a method to diagnose bovine B-cell lymphoma. PCR for IgH gene rearrangement indicated monoclonal proliferation of B-cells in 24 of 35 cattle with B-cell lymphoma. In contrast, PCR for IgH gene rearrangement in lymph nodes and tumor tissues from 65 cattle diagnosed with tumors other than B-cell lymphoma and non-tumors revealed polyclonal population of B-cells. Sensitivity, specificity, positive predictive value, and negative predictive value for PCR for IgH gene rearrangement for bovine B-cell lymphoma were 68.6%, 100%, 100%, and 85.5%, respectively. Clonality analysis using PCR for IgH gene rearrangement may be useful for adjunctive diagnosis of bovine B-cell lymphoma.
Keywords: bovine B-cell lymphoma, clonality, polymerase chain reaction
Lymphoma is one of the most common neoplasms in cattle, and enlargement of superficial lymph nodes is an important clinical finding that can lead to the suspicion of bovine lymphoma [1, 8]. Cytology of enlarged superficial lymph nodes obtained by fine-needle aspiration (FNA) are used for definitive diagnosis of bovine lymphoma [10]. However, distinguishing between neoplastic and reactive lymphocytes is difficult and FNA of enlarged superficial lymph nodes is not always reliable [10]. Therefore, other methods offering objective results are required to determine whether lymphocytes are tumor cells or not.
The immunoglobulin heavy chain (IgH) gene in B-cells is produced through the recombination of V, D, and J genes, which collectively possess an extensive combinatorial repertoire [6]. Because all cells with malignancy share a common clonal origin, each B-cell lymphoma contains IgH variable regions of unique lengths. Therefore, evaluation of the clonal nature of a population of lymphocytes within tissue samples can facilitate the diagnosis of B-cell neoplasia. Assessment of clonality in suspected B-cell neoplasia is commonly used as an adjunctive diagnostic procedure in dogs, cats, and humans [2, 4, 7, 9].
We recently reported that clonal proliferation of B-cells was identified by polymerase chain reaction (PCR) for IgH gene rearrangement in cattle with B-cell lymphoma [3]. Others have reported on a B-cell clonality analysis using semi-nested PCR in cattle with lymphoma [5]. However, in that particular study, the B-cell clonality analysis was performed only in a few cattle without lymphoma. Therefore, the use of PCR for IgH gene clonality analysis as a diagnostic tool for bovine B-cell lymphoma has yet to be evaluated. In the present study, we evaluated the sensitivity, specificity, positive predictive value, and negative predictive value of PCR analysis for IgH gene clonality as a diagnostic tool for bovine B-cell lymphoma.
In total, 100 cattle were examined in the present study. These cattle were divided into 4 groups. Group 1 (B-cell lymphoma) comprised 35 cattle with B-cell lymphoma (14 diffuse large, 17 diffuse median, and 4 diffuse small B-cell lymphoma) (Fig. 1) and Group 2 (T-cell lymphoma) comprised 10 cattle with T-cell lymphoma. In these two groups, diagnoses were confirmed by immunohistochemical analysis using BLA-36 (Biogenex Laboratories, Fremont, CA, USA) and CD3 (Biogenex Laboratories) monoclonal antibodies. Bovine leukemia virus (BLV) infection in all cattle of Group 1 was confirmed by nested PCR for BLV 5′ LTR [3]. Group 3 (Other tumors) comprised 5 cattle with tumors other than lymphoma (2 osteosarcoma, 2 undifferentiated carcinoma and 1 mesothelioma); Group 4 (Non-tumor) comprised 50 cattle diagnosed with non-tumor diseases or healthy (30 cattle with inflammatory diseases (Inflammation) (9 cases of pneumonia, 6 with abscess formation in the mandible, pharynx, lung or liver, 5 with peritonitis, 3 with endocarditis, 3 with mastitis, 3 with arthritis and 1 with enteritis)); 10 cattle diagnosed with diseases other than tumors and inflammatory diseases (Other diseases) (3 with ventricular septal defects, 2 with diplomyelia, 2 with knuckling of the fetlock joint, 1 with pure red cell aplasia, 1 with meningocele and 1 with a persistent bovine viral diarrhea virus infection); and 10 clinically healthy cattle (Healthy). Cattle with tumors were definitively diagnosed by clinical, macroscopic, and histopathological examination, and non-tumor cattle were diagnosed based on clinical and macroscopic findings without histopathological examination at the Obihiro University of Agriculture and Veterinary Medicine, Hokkaido, Japan. Specimens were reviewed by board-certified veterinary pathologists. This study was approved by Obihiro University of Agriculture and Veterinary Medicine Committee for Experiments Using Animals.
Fig. 1.

Representative histopathological image of diffuse large (A), diffuse median (B) and diffuse small (C) B-cell lymphoma. Hematoxylin and eosin stain. Bar=20 µm.
Genomic DNA samples were extracted from lymph nodes of cattle in Groups 1, 2, and 4 (B-cell lymphoma, T-cell lymphoma, and Non-tumor), from tumor tissues in Group 3 (Other tumors) using QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany). Samples were stored at −30°C until analysis. Clonality analysis using PCR for IgH gene rearrangement was performed as described previously [3]. Briefly, PCR was conducted using a HotStarTaq DNA Polymerase (QIAGEN GmbH) with primer pair (BoVHF1: 5′-AGCCCTGAAATCCCGGCTCA-3′ and BoVHR1: 5′-TCCAGGAGTCCTTGGCCCCA-3′). The primer set was designed based on sequence information of the immunoglobulin heavy chain variable region (GenBank Accession No. DQ485764 to DQ485788). The amplification program was the same as previously reported [3]. PCR products were electrophoresed on a 3% agarose gel. A single band between 100 and 300 base pairs was considered monoclonal proliferation, while a smear or no visible band were considered polyclonal population of B-cells.
Results are shown in Table 1. In Group 1 (B-cell lymphoma), PCR yielded a single band in 24 of the 35 cattle (Fig. 2). In contrast, a smear or no visible bands were noted in cattle in Groups 2 (T-cell lymphoma), 3 (Other tumors), and 4 (Non-tumor) (Fig. 2). The sensitivity, specificity, positive predictive value, and negative predictive value of PCR analysis for IgH gene clonality were 68.6% (24/35), 100% (65/65), 100% (24/24), and 85.5% (65/76), respectively. In Group 1 (B-cell lymphoma), single PCR bands were observed in 12 (85.7%) diffuse large, 9 (52.9%) diffuse median, and 3 (75.0%) diffuse small B-cell lymphoma (Table 1). Results of the clonality analysis using PCR for IgH gene rearrangement in histological subclassification of B-cell lymphoma were not statistically different (P=0.141 by χ2 Independence Test).
Table 1. Polymerase chain reaction results for immunoglobulin heavy chain gene.
| Group | N | Band | Smear or no visible band | |
|---|---|---|---|---|
| B-cell lymphoma | 35 | 24 (68.6%) | 11 (31.4%) | |
| Diffuse large | 14 | 12 (85.7%) | 2 (14.3%) | |
| Diffuse median | 17 | 9 (52.9%) | 8 (47.1%) | |
| Diffuse small | 4 | 3 (75.0%) | 1 (25.0%) | |
| T-cell lymphoma | 10 | 0 (0.0%) | 10 (100%) | |
| Other tumors | 5 | 0 (0.0%) | 5 (100%) | |
| Non-tumor | 50 | 0 (0.0%) | 50 (100%) | |
| Inflammation | 30 | 0 (0.0%) | 30 (100%) | |
| Other diseases | 10 | 0 (0.0%) | 10 (100%) | |
| Healthy | 10 | 0 (0.0%) | 10 (100%) | |
Fig. 2.
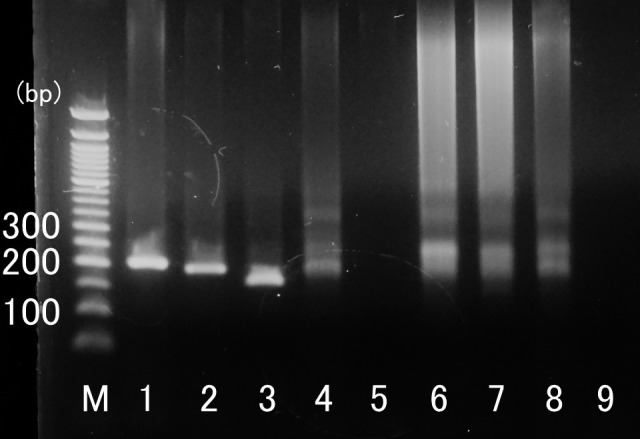
Example of representative PCR results obtained with each group of samples. Lanes 1–3: B-cell lymphoma; Lane 4: T-cell lymphoma; Lane 5: mesothelioma; Lane 6: mastitis; Lane 7: ventricular septal defect; Lane 8: healthy; Lane 9: negative control (DW); M: molecular weight marker.
The 68% sensitivity of PCR analysis as a diagnostic tool for bovine B-cell lymphoma was lower than that for PCR analysis of B-cell clonality in other species (83–100%, 96%, and 84% in humans, dogs, and cats, respectively) [2, 4, 7, 9]. The lower sensitivity of clonality analysis using PCR for IgH gene rearrangement in the present study may be due to primer mismatches. For instance, in cats, clonality of B-cell neoplasms were detected by singleplex and multiplex PCR using 11 primers; in dogs, B-cell lymphocytic malignancies were detected by PCR using 3 primers [2, 4]. Because limited information on bovine IgH sequences was available in the present study, further investigation is needed for cattle IgH sequences. Adding new primers may improve PCR sensitivity for IgH gene clonality in cattle.
With the exception of those with bovine B-cell lymphoma, all cattle with any other diseases exhibited polyclonal population of B-cells by PCR, with a PCR specificity for IgH gene clonality of 100% in the present study. However, diversity of the IgH repertoire might be reduced in reactive lymph nodes by predominance of several antigen-selected subclones [9]. Further investigation is required to elucidate how the various infectious diseases may influence a clonality analysis using PCR for IgH gene rearrangement in cattle. Moreover, because the present study based diagnoses in Non-tumor groups on clinical and macroscopic examination without histopathological examination, additional studies on IgH gene clonality in reactive and normal lymph nodes with histological examination are needed.
The present study found no correlation between histological subclassification of B-cell lymphoma and results of clonality analysis using PCR for IgH gene rearrangement. However, the smaller sample size may have led to an incorrect evaluation of the relationship between histological subclassification of B-cell lymphoma and result of PCR for IgH gene rearrangement. Further investigation is required to clarify the relationship between histological subclassification of B-cell lymphoma and the results of the clonality analysis using PCR for IgH gene rearrangement.
Acknowledgments
We thank the veterinarians of the Iwate and Tokachi Agriculture Mutual Aid Association and Obihiro Meat Hygiene Inspection Center for sampling, and Mr. Tetsuharu Oshima and Ms. Shizuka Kayou of the Laboratory of Veterinary Internal Medicine of Obihiro University of Agriculture and Veterinary Medicine for technical assistance.
REFERENCES
- 1.Angelos J. A., Thurmond M. C.2015. Bovine lymphoma. pp. 1070–1073. In: Large Animal Internal Medicine, 5th ed. (Smith, B. P. ed.), Elsevier, St. Louis. [Google Scholar]
- 2.Burnett R. C., Vernau W., Modiano J. F., Olver C. S., Moore P. F., Avery A. C.2003. Diagnosis of canine lymphoid neoplasia using clonal rearrangements of antigen receptor genes. Vet. Pathol. 40: 32–41. doi: 10.1354/vp.40-1-32 [DOI] [PubMed] [Google Scholar]
- 3.Maezawa M., Watanabe K., Horiuchi N., Matsumoto K., Kobayashi Y., Inokuma H.2018. A clinical case of enzootic bovine leucosis in a 13-month-old Holstein heifer. Jpn. J. Vet. Res. 66: 209–150. [Google Scholar]
- 4.Mochizuki H., Nakamura K., Sato H., Goto-Koshino Y., Sato M., Takahashi M., Fujino Y., Ohno K., Uchida K., Nakayama H., Tsujimoto H.2011. Multiplex PCR and Genescan analysis to detect immunoglobulin heavy chain gene rearrangement in feline B-cell neoplasms. Vet. Immunol. Immunopathol. 143: 38–45. doi: 10.1016/j.vetimm.2011.05.030 [DOI] [PubMed] [Google Scholar]
- 5.Nishimori A., Konnai S., Okagawa T., Maekawa N., Goto S., Ikebuchi R., Nakahara A., Chiba Y., Ikeda M., Murata S., Ohashi K.2017. Identification of an atypical enzootic bovine leukosis in Japan by using a novel classification of bovine leukemia based on immunophenotypic analysis. Clin. Vaccine Immunol. 24: e00067–e17. doi: 10.1128/CVI.00067-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schatz D. G., Oettinger M. A., Schlissel M. S.1992. V(D)J recombination: molecular biology and regulation. Annu. Rev. Immunol. 10: 359–383. doi: 10.1146/annurev.iy.10.040192.002043 [DOI] [PubMed] [Google Scholar]
- 7.Segal G. H., Wittwer C. T., Fishleder A. J., Stoler M. H., Tubbs R. R., Kjeldsberg C. R.1992. Identification of monoclonal B-cell populations by rapid cycle polymerase chain reaction. A practical screening method for the detection of immunoglobulin gene rearrangements. Am. J. Pathol. 141: 1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson B. S., Goodrick E. R.2018. Miscellaneous infectious diseases. pp. 737–783. In: Diseases of Dairy Cattle, 3rd ed. (Peek, S. F. and Divers, T. J. eds.), Elsevier, St. Louis. [Google Scholar]
- 9.van Dongen J. J. M., Langerak A. W., Brüggemann M., Evans P. A. S., Hummel M., Lavender F. L., Delabesse E., Davi F., Schuuring E., García-Sanz R., van Krieken J. H. J. M., Droese J., González D., Bastard C., White H. E., Spaargaren M., González M., Parreira A., Smith J. L., Morgan G. J., Kneba M., Macintyre E. A.2003. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17: 2257–2317. doi: 10.1038/sj.leu.2403202 [DOI] [PubMed] [Google Scholar]
- 10.Washburn K. E., Streeter R. N., Lehenbauer T. W., Snider T. A., Rezabek G. B., Ritchey J. W., Meinkoth J. H., Allison R. W., Rizzi T. E., Boileau M. J.2007. Comparison of core needle biopsy and fine-needle aspiration of enlarged peripheral lymph nodes for antemortem diagnosis of enzootic bovine lymphosarcoma in cattle. J. Am. Vet. Med. Assoc. 230: 228–232. doi: 10.2460/javma.230.2.228 [DOI] [PubMed] [Google Scholar]


