Abstract
The complement systems play an important role in innate and adaptive immunity. In this study, the complement C3 gene, designated CagC3, was cloned and sequenced from Gibel carp (Carassius auratus gibelio). The expression pattern of CagC3 in different tissues of healthy Gibel carp and after challenge with Cyprinid herpesvirus 2 (CyHV-2) were evaluated using quantitative real-time PCR. The full-length CagC3 cDNA was 5131 bp with an ORF of 4950 bp, encoding a predicted protein of 1649 amino acids. The deduced amino acid sequence showed that CagC3 has conserved domains and residues known to be critical for C3 function. Phylogenetic analysis demonstrated that CagC3 clustered with homologs from common carp and grass carp (Ctenopharyngodon idella). CagC3 is expressed in all examined tissues of healthy Gibel carp, with the highest expression in liver. In vivo, after CyHV-2 challenge, CagC3 transcription was significantly upregulated in liver, spleen and kidney with the peaks at 24 hr, 2 d, and 2 d, respectively. In vitro, CagC3 expression in the Gibel carp brain cell line showed the same pattern as that in vivo after stimulation with CyHV-2 or poly(I:C). However, CagC3 expression was downregulated at 24 hr after induction with lipopolysaccharide (LPS), and then reached the peak at 2 d. These results suggest that CagC3 is involved in the innate immune response of Gibel carp to viral infection.
Keywords: cloning, complement component C3, Cyprinid herpesvirus 2 (CyHV-2), Gibel carp (Carassius auratus gibelio), immune response
The complement system participates in host protection, acting as the first line of defense against microbial intruders, promoting inflammatory reactions, and coordinating the adaptive immune response [3, 29]. It contains more than 35 soluble plasma proteins that play key roles in innate and adaptive immunity after activation [7, 35]. The third complement component, C3, is a key protein that plays a major role in host defense against infections [26]. Cleavage of inactive C3 generates a large fragment (C3b) and a smaller fragment (C3a), a crucial step in the complement activation cascade, which can be initiated by one or more of the three distinct pathways, called the alternative, classical and lectin complement pathways [24]. After C3 activation, the three distinct pathways converge into a final common pathway, known as the cytolytic pathway, in which the membrane attack complex forms, mediating lysis of target cells and playing a role in anti-infection immunity. Therefore, C3 is the crossing point of the three pathways for complement activation and acts as one of the bridges linking innate and acquired immunity [4, 20, 23].
In fish, a series of studies have been performed on the gene structure, type, function and expression of C3. A single isotype of C3 has recently been isolated from the spotted wolffish (Anarhichas minor Olafsen) [1], Atlantic cod (Gadus morhua) [14], halibut (Hippoglossus hippoglossus) [14], miiuy croaker (Miichthys miiuy) [34], orange-spotted grouper (Epinephelus coioides) [28], rohu (Labeo rohita) [27] and large yellow croaker (Larimichthys crocea) [40], which, like mammalian C3, were encoded by a single gene. In contrast, multiple forms of C3 have been described in rainbow trout (Salmo gairdneri) [37], gilthead sea bream (Sparus aurata) [36] and common carp (Cyprinus carpio) [22]. Differences in the binding specificity of C3 to various targets and in the hemolytic activity have been reported for trout [37], common carp [36] and sea bream [18]. In cod and halibut [14], C3 is expressed in a wide range of organs, with expression detected in different stages of larval development. However, these results contrast with studies on C3 expression in the spotted wolfish [1], in which C3 could only be detected in the liver. In addition, there are considerable numbers of publications that describe the expression response of C3 after stimulation of teleosts with bacteria, LPS and poly(I:C), but few studies have focused on the expression of C3 after viral infection.
Gibel carp (Carassius auratus gibelio) is a subspecies of crucian carp (Carassius auratus), and is widely cultured in almost the whole of China [42]. However, outbreaks of disease of this species have become of great concern; for example, a severe disease caused by Cyprinid herpesvirus 2 (CyHV-2) has emerged and caused huge economic losses [41]. CyHV-2, also known as herpes-viral hematopoietic necrosis virus of goldfish, is the second member of the genus Cyprinivirus isolated from carp [8]. In this study, the full cDNA encoding the Gibel carp complement component C3 was cloned and its domain architecture, phylogenetic relationships and mRNA expression in vitro and in vivo analyzed. These results provide a foundation for further study of the immunological responses of Gibel carp to CyHV-2 infection and for the control of this disease.
MATERIALS AND METHODS
Animals, cell line and virus
Gibel carp (about 150 g/individual) were obtained from the experimental farm of the Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences. The fish were kept in tanks with circulated and aerated water at 25°C (2 × 1.5 × 1.5 m) for 15 days and fed twice a day with a commercial diet before experiments. Gibel carp brain (GiCB) cell line was established and conserved by our laboratory; cells were grown at 25°C in M199 Medium (M199; Sigma, St. Louis, MO, USA), supplemented with 10% fetal bovine serum (FBS) [17]. CyHV-2 was isolated by our laboratory [41] and propagated in GiCB cells.
Primer design, cloning and sequencing of CagC3
A pair of degenerate primers (CagC3F and CagC3R) was designed to obtain the internal region of CagC3 according to the conserved sequence of complement C3 homologs from other fish [common carp C3-H1 (GenBank accession number BAA36619.1), common carp C3-H2 (BAA36620.1), common carp C3-S (BAA36621.1), and grass carp C3 (AAQ74974.1)]. To obtain the full-length cDNA of CagC3, rapid-amplification of cDNA ends (RACE)-PCR was performed using the Clontech SMART cDNA synthesis kit (following the manufacturer’s instructions) to identify the 5′- and 3′- regions of CagC3. Four specific primers were based on the amplified partial Gibel carp C3 sequence described above. Using nested PCR, primers 3F1 and 3F2 were used for 3′-RACE and 5R1 and 5R2 for 5′-RACE. To confirm the resulting full-length cDNA sequence of CagC3, specific primers (C3F/C3R) were designed. All the primers used in this study are listed in Table 1. Total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Residual DNA was removed by DNase I (Invitrogen) treatment. First strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen) and stored at −20°C. cDNA was used as template in 50-µl PCR reactions; the PCR cycling conditions were: 30 cycles of 94°C for 10 sec, 60°C for 30 sec, and 72°C for 45 sec. The PCR products were purified using the Wizard® SV Gel and PCR Cleanup System (Promega, Madison, WI, USA), ligated into vector pMD19-T (TaKaRa, Dalian, Liaoning, China) and then transformed into Escherichia coli DH5α competent cells. Five randomly selected clones were sequenced by Beijing Tianyi Huiyuan Bioscience & Technology Inc. (Beijing, China).
Table 1. Primer sequences used in this study.
| Name | Primer sequence (5′–3′) | Applications |
|---|---|---|
| CagC3F | ACTKGGCATCTGTGTGGCAGAACC | Homologous cloning |
| CagC3R | GGCCAGTATTCAAYCCAGKTCTKC | Homologous cloning |
| 3F1 | ACCATCATGGTGTTCCAGGCTGTG | First 3′ RACE |
| 5R1 | CGGTCCTTCACCTGCGTGCGATAC | First 5′ RACE |
| 3F2 | CAGGTGCTGAAAGAAGGAACTGAT | Nest 3′ RACE |
| 5R2 | CGCAACAGCGTAAGGATTGGTCAG | Nest 5′ RACE |
| C3F | TGCGGCAGTGATTAGACTCGGTACG | full-length cDNA |
| C3R | TGTACCATTTTTATTCAGACAGTAT | full-length cDNA |
| CagC3-qF1 | AGTGAAATGGTGGAAGCAGAAAG | qRT-PCR |
| CagC3-qR1 | TACGTATACCGAGACATCGAAGG | qRT-PCR |
| CagActin-F | CTCCCCTCAATCCCAAAGCCAA | qRT-PCR |
| CagActin-R | ACACCATCACCAGAATCCATCA | qRT-PCR |
| Universal Primer Mix (UPM) | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | RACE |
| Nest Universal Primer (NUP) | AAGCAGTGGTATCAACGCAGAGT | Nest RACE |
F indicating forward primer and R reverse primer. RACE, rapid-amplification of cDNA ends; qRT-PCR, quantitative real-time-PCR.
Sequence analysis of CagC3
The cDNA of CagC3 was translated into its potential open reading frame (ORF) using the ORF finder algorithm (http://www.ncbi.nlm.nih.gov/gorf/). Identities between CagC3 and other known C3 amino acid sequences were determined by BLAST (NCBI). Domain analyses were carried out with Simple Modular Architecture Research Tools (SMART; http://smart.gembl-heidelberg.de/) [31], Pfam 20.0 (http://pfam.wustl.edu/) [5] and Scanprosite (http://prosite.expasy.org/scanprosite/) [9] programs. Multiple sequence alignments were performed with the CLUSTALW 2.1 program [11]. The predicted mass and isoelectric point (pI) of the putative protein were calculated using the ProtParam tool at the ExPASy website (http://www.expasy.org/tool/protparam/). A phylogenetic tree was constructed with the neighbor-joining (NJ) method in MEGA 5.0 software [39] and the reliability of the analysis was assessed by 1,000 bootstrap replicates.
CagC3 expression in vivo after CyHV-2 infection
To assess the tissue distribution of CagC3 expression in healthy gibel carp, nine different tissues including liver, spleen, kidney, brain, muscle, heart, gill, intestine and skin were collected from every sample fish. To examine CagC3 expression in vivo in different tissues after virus infection, carp in the virus-infected group were injected intraperitoneally with 0.5 ml CyHV-2 (106 TCID50/ml); carp in the control group were injected intraperitoneally with an equal amount of Dulbecco’s phosphate buffered saline (DPBS) (Sigma). Virus-injected carp (n=5 per time point; 40 total) and PBS-injected carp (n=5 per time point; 40 total) were sacrificed at 0, 6, 12 and 24 hr, and 2, 3, 6 and 9 d after challenge.
CagC3 expression in vitro after induction with virus, poly(I:C) or LPS
In vitro, GiCB cells, as a sensitive cell line to CyHV-2 [17], were maintained at 28°C in 6-well culture plates at 1 × 106 cells/well in M199 supplemented with 10% FBS. For viral infection, GiCB cells were infected with CyHV-2 at a multiplicity of infection of 1, 10 µg/ml LPS (final concentration) or 10 µg/ml poly(I:C) (final concentration), respectively. Controls were treated with DPBS. All the above infections were performed in parallel. The cells were collected 6, 12, 24, 48 and 72 hr post-infection. All samples were stored at −80°C until use.
Quantitative real-time PCR expression analysis
In quantitative real-time PCR (qRT-PCR), synthesized cDNA was used as the template; along with two pairs of gene specific primers (CagC3-qF1/CagC3-qR1 or CagActinF/CagActinR) designed using Primer Premier 5.0 software. The qRT-PCR amplifications were carried out in total reaction volumes of 20 µl, containing 10 µl SYBR real-time PCR premixture (BioTeKe, Beijing, China), 1 µl diluted cDNA sample, 1 µl of each primer (0.5 µM), and 7 µl ddH2O, in a Real-Time PCR Detection System (Rotor-Gene 6000, Qiagen, Dusseldorf, Germany). The PCR program was: one cycle of 95°C for 5 min, then 40 cycles of 94°C for 15 sec, 60°C for 15 sec and 72°C for 20 sec. For each cDNA sample, reactions were performed in triplicate and measured by the crossing-point (Ct) values. Expression of β-actin was used as the internal control for analysis of C3 gene expression. The relative CagC3 expression ratio was determined by the 2−ΔΔCT method [16]. The data are expressed as mean ± SD. Expression differences between control and treatment groups were assessed for statistical significance (P<0.05) using Student’s t-test and the SPSS statistical software package.
RESULTS
Sequence of CagC3 and homology alignment
The full-length cDNA of CagC3 is 5,131 bp (GenBank accession number: KF110786) and contains an ORF of 4,950 bp, encoding a predicted protein of 1649 amino acids (aa) with an estimated molecular mass of 184.81 kDa. The pI of CagC3 is predicted to be 6.28. A pair of specific primers C3F/C3R (Table 1) was used to confirm the full-length cDNA of CagC3 in the PCR reaction. As shown in Fig. 1, a specific fragment, approximately 5,100 bp, consistent with the expected size was obtained in the PCR reaction with reverse transcriptase and was subsequently confirmed by sequencing analysis, while no band was found in the reaction without reverse transcriptase (Fig. 1).
Fig. 1.
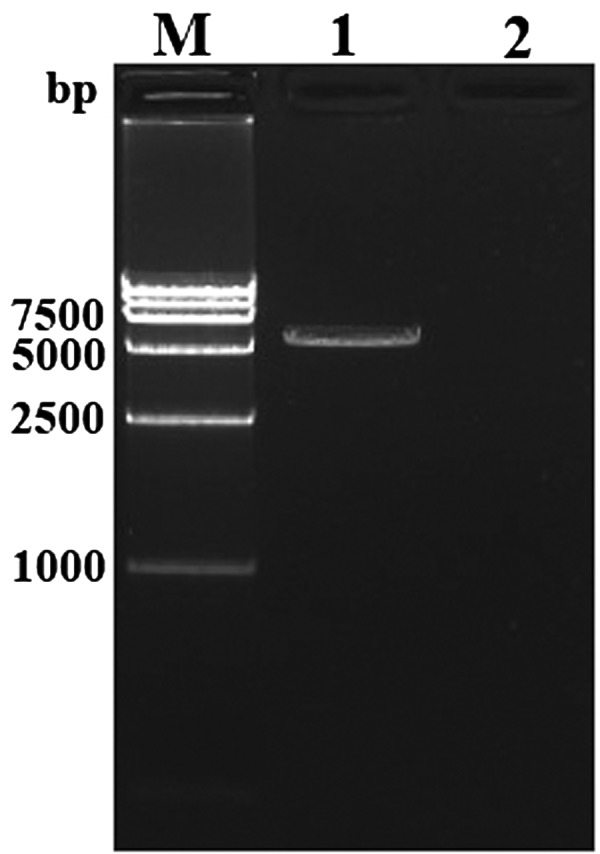
Amplification results of the full-length cDNA of CagC3 by RT-PCR assay. Lane M: DL1.5 kb DNA ladder marker; Lane 1: Amplification of CagC3 in the PCR reaction with reverse transcriptase; Lane 2: Amplification of CagC3 in the PCR reaction without reverse transcriptase.
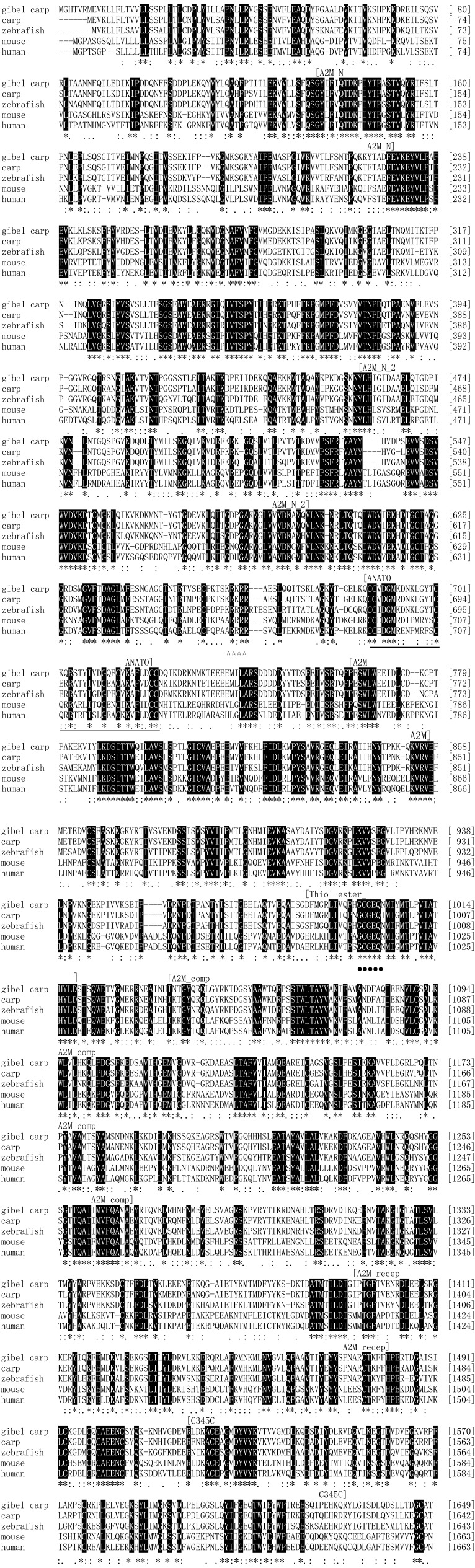
Based on multiple alignment of the predicted amino acid sequence of CagC3 with homologs from other species, the CagC3 sequence includes a putative cleavage site and eight well-conserved domains (Fig. 2). The putative cleavage site is ‘RXXR’ which occurs from aa 662 to 665. The first (N-terminal) domain is alpha-2-macroglobulin (A2M_N), from aa 136 to 230. Moving through the primary structure toward the C-terminus, the second domain is alpha-2-macroglobulin_2 (A2M_N_2, aa 459–600). The next domain, occurring after the cleavage site, is anaphylatoxin (ANATO, aa 687–722). Next is another alpha-2-macroglobulin domain (A2M, aa 765–858), followed from aa 988 to 1018 by an alpha-macroglobulin thioester bond-forming region. Sixth, the complement component region of the alpha-2-macroglobulin family is found (A2M_comp, aa 1040–1272). The next domain is a receptor-binding domain of alpha-2-macroglobulin proteins (A2M_recep, aa 1385–1480). Finally, there is a basic domain called the Netrin C-terminal domain (C345C, aa 1519–1631).
Fig. 2.
Multiple sequence alignment of CagC3 in comparison with C3 sequences of other vertebrates. Carp (Cyprinus carpio C3-H1, accession no. BAA36619.1), zebrafish (Danio rerio C3a, NP_571317.1), mouse (Mus musculus C3, NP_033908.2) and human (Homo sapiens C3, AAA85332.1) C3 protein sequences were compared using ClustalW (v. 2.1) and MEGA 5 software. Amino acid numbering is indicated on the right. Identical residues are indicated by black shading and with an asterisk (*). Conserved and semi-conserved residues are indicated by semicolons (:) and periods (.), respectively. Names and boundaries of protein domains are shown above the sequences. Characteristic domains and motifs are highlighted below the sequences: the ANATO domain has six canonical cysteine residues at conserved positions (solid black line); the thioester domain (●); the RKRR cleavage site of the alpha and beta chains (☆).
Deduced amino acid sequence comparisons indicate that CagC3 shows 81%, 77%, 52% and 51% identities with the C3 proteins of grass carp, zebrafish, grouper (Epinephelus coioides) and medaka (Oryzias latipes), respectively. The highest amino acid identity is 91% with common carp C3-H1. However, the identities with the proteins from tammar wallaby (Macropus eugenii) and human (Homo sapiens) are only 44%.
Phylogenetic analysis of CagC3
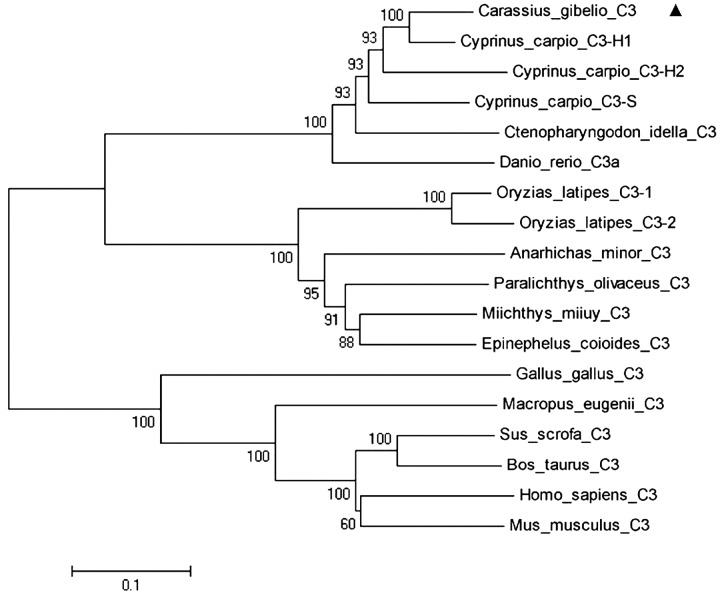
The relationships among vertebrate C3 sequences were revealed in a phylogenetic tree (Fig. 3). The unrooted phylogenetic tree divided into two groups: fish and mammals. The fish group has two branches, and the CagC3 sequence belongs to the branch containing cyprinid fish. According to the phylogenetic tree, CagC3 is most closely related to that from common carp, and has high similarity with homologs from zebrafish and grass carp. The tree was consistent with traditional taxonomy and phylogenetic transition.
Fig. 3.
Phylogenetic tree of C3 from Gibel carp and other species constructed with the neighbor-joining (NJ) method in MEGA 5. The deduced amino acid sequences used and their GenBank accession numbers are: Carassius auratus gibelio C3 (KF110786), marked with ▲; Cyprinus carpio C3-H1 (BAA36619.1); Cyprinus carpio C3-H2 (BAA36620.1); Cyprinus carpio C3-S (BAA36621.1); Ctenopharyngodon idella C3 (AAQ74974.1); Danio rerio C3.1 (NP_571317.1); Oryzias latipes C3-1 (NP_001098552.1); Oryzias latipes C3-2 (NP_001098553.1); Anarhichas minor C3 (CAC29154.1); Paralichthys olivaceus C3 (BAA88901.1); Miichthys miiuy C3 (AFC89899.1); Epinephelus coioides C3 (BAA88901.1); Gallus gallus C3 (NP_990736.1); Macropus eugenii C3 (C3 AAW69835.1); Sus scrofa C3 (NP_999174.1); Bos taurus C3 (CAJ31249.1); Homo sapiens C3 (AAA85332.1); and Mus musculus C3 (NP_033908.2). The reliability of the analysis was assessed by 1,000 bootstrap replicates; bootstrap support percentages are shown at nodes.
Expression of CagC3 in vivo after infection with CyHV-2
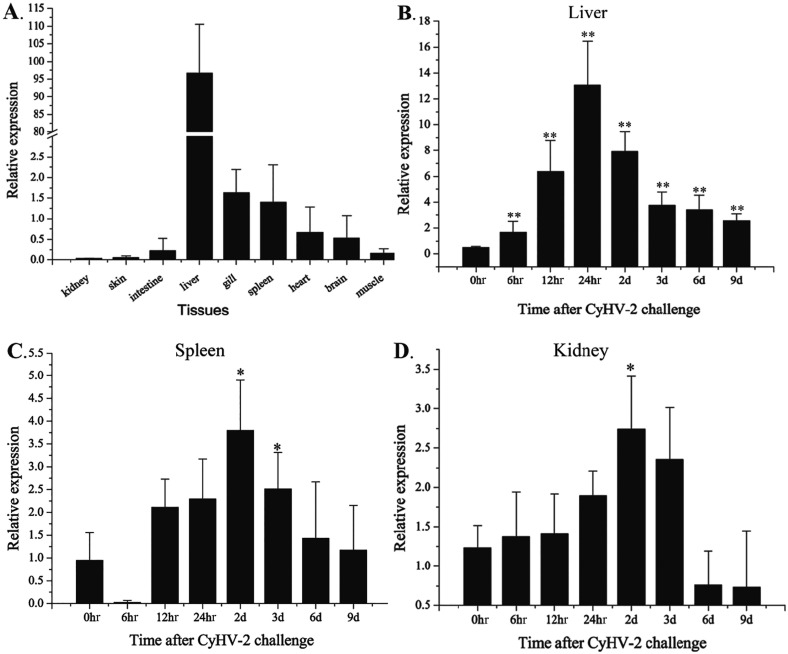
The relative expression levels of CagC3 in various tissues were determined by qRT-PCR. A fragment consisting of 123 bp was amplified from each of the tissues. Retrieval and sequencing results verified the detection of the CagC3 fragment. The expression of CagC3 was observed in all nine normal (healthy) tissues examined (Fig. 4A). However, the expression profiles of CagC3 in different tissues showed significant differences. CagC3 mainly expressed in the liver, was moderately expressed in the gill, spleen, heart and brain, and was expressed at a low level in muscle, intestine, skin and kidney.
Fig. 4.
Expression of CagC3 in Gibel carp determined by qRT-PCR. A, Expression in nine tissues of healthy fish (kidney, intestine, liver, gill, spleen, brain, heart, muscle and skin). Expression after challenge with CyHV-2 in (B) liver, (C) spleen, and (D) kidney. All data were normalized to β-actin. The data are expressed as means ± SD (n=5) and were analyzed by calculating 2−ΔΔCT. Significant differences (*P<0.05; **P<0.01) in CagC3 expression in the challenge groups are relative to the expression in that tissue at 0 hr.
In vivo, after challenge with CyHV-2, the highest expression of CagC3 was observed in liver, and high expression levels were also observed in spleen and kidney (Fig. 4). In the liver, the expression level increased significantly from 0 to 24 hr post-infection, reached the peak at 24 hr, then decreased sharply from 24 hr to 3 d and maintained a stable level from 3 to 9 d (Fig. 4B). In the spleen (Fig. 4C), expression of CagC3 increased sharply from 6 to 12 hr, reached the peak at 2 d and then decreased from 2 to 9 d. In the kidney (Fig. 4D), the expression of CagC3 increased from 0 to 24 hr, reached the peak level at 2 d, and remained relatively high at 3 d. Subsequently, the expression of CagC3 decreased sharply and the level was low from 6 to 9 d.
Expression of CagC3 in vitro after induction with virus, poly(I:C) or LPS
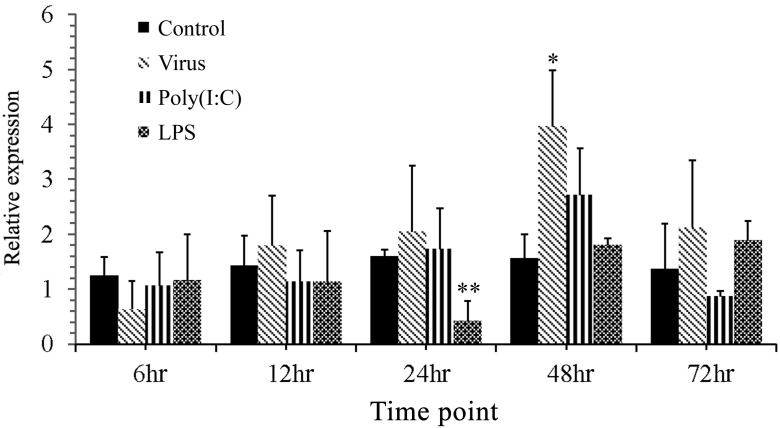
In vitro, the mRNA expression of CagC3 in GiCB cells infected with CyHV-2 increased significantly at 48 hr and declined at 72 hr. Similarly, mRNA expression levels of CagC3 in GiCB cells induced by poly(I:C) were obviously elevated at 24 hr compared with the control group, reached the peak at 48 hr, and recovered to the normal level at 72 hr. Interestingly, LPS induced different CagC3 mRNA expression trends: the mRNA expression fell from 6 to 24 hr, reached the minimum at 24 hr, then rose sharply from 24 to 72 hr (Fig. 5).
Fig. 5.
Expression of CagC3 mRNA in Gibel carp brain (GiCB) cells induced by CyHV-2, LPS or poly(I:C) challenge, determined by qRT-PCR at various time points. All data were normalized to β-actin. The data are expressed as means ± SD (n=5) and were analyzed by calculating 2−ΔΔCT. Significant differences (*P<0.05; **P<0.01) in CagC3 expression are relative to the expression in control group.
DISCUSSION
As the central component of the complement system, C3 may play an important role in innate and acquired immunity. C3 has been investigated in many teleost species [2]. In contrast to mammals, almost all teleost fish studied thus far have been found to possess multiple forms of C3: two isoforms have been found in Oryzias latipes [12], Dicentrarchus labrax [18] and Trematomus bernacchii [19], five in Sparus aurata [36], eight in Oncorhynchus mykiss [37], and four in Cyprinus carpio [22] and Danio rerio [6]. In the present study, the complement component C3 gene from Gibel carp was cloned and characterized and the expression profiles were analyzed for the first time. However, it should be noted that this study examined only one isoform of C3, and whether there are other isoforms of C3 in Gibel carp is worthy of further investigation.
Analysis of the deduced amino acid sequence showed the presence of eight well-conserved domains in CagC3, as observed in other species. The structures and functions of the eight domains were similar to the complement component C3 of gilthead sea bream [18], which showed the typical domain architecture of alpha-2-macroglobulin family proteins. It is worth noting that the five-residue thioester domain (GCGEQ), which is thought to be critical in the formation, stability and function of this region [15], is highly conserved in CagC3. Moreover, previous study revealed that Pro1007 and Pro1020 in human C3 are necessary for stable thioester formation [10]. Our analysis indicated that Pro1007 is conserved in CagC3, whereas Pro1020 is replaced by Leu (Fig. 2). Similar results were also found in trout C3-1, C3- 3 and C3-4 isoforms [43]. In addition, in human C3, residues His1126 and Glu1128 have been shown to be critical in determining the binding specificity of C3 to its target surface [15, 21]. These two residues are also conserved in Gibel carp, as His1115 and Glu1117, respectively (Fig. 2). In trout, the His residue is conserved in the C3-1 and C3-3 isoforms, whereas in C3-4 this residue is substituted by Thr. It has been suggested that differences in these residues account, at least in part, for the observed variations in the binding efficiencies of the various trout C3 isoforms [43].
Analysis of the CagC3 ANATO and C345C domains revealed that these domains both contain six canonical cysteine residues which are known to be significant for their functions [38]. The potential α/β processing signal RKRR is present in CagC3 (Fig. 2). Previous studies have suggested that ‘RXXR’ is the cleavage site where C3 proteins are processed into the two-subunit chain structure [25]. The C3-convertase cleavage site and the residues that align to the two cofactor I cleavage sites (Arg–Ser1304, Arg–Ser1321 in the human C3 numbering) are also conserved in CagC3 (Arg–Ser1292 and Arg–Ser1309). Similar residue conservation was also found in Labeo rohita C3 (LRC3) [27]. These sequence data suggest that CagC3 functions in a similar way to the C3 molecules of other animals. Analysis of the deduced amino acid sequence of CagC3, multiple sequence alignment and phylogenetic analysis showed that the structure of CagC3 is similar to counterparts in other vertebrates, with 91% identity with common carp C3-H1.
In vivo, C3 can constitutively express in tissues including liver, spleen, kidney, brain, muscle, heart, gill, intestine and skin. Generally, a great deal of C3 is present in liver, which has been suggested to be the main site of C3 production in mammals and fish [1, 2]. These results were consistent with the previous reports on other teleost fish, such as common carp [22], Gilthead seabream [18], miiuy croaker [34], orange-spotted grouper [28], rohu [27] and large yellow croaker [40]. Spleen and kidney are very important lymphoid organs in teleost fish, and serve similar roles to bone marrow and lymph nodes in mammals [33]. After infection of Gibel carp with CyHV-2, the expression of CagC3 in liver, spleen and kidney varied significantly, consistent with previous studies in miiuy croaker [34], rainbow trout [32] and common carp [30]. Interestingly, the expression in liver reached its peak at 24 hr post-challenge, which was earlier than in spleen and kidney. This may imply that response to virus stimulation occurs firstly in liver; the same result has been found in Atlantic halibut [13] and miiuy croaker [34]. Although the increase was lower than in liver, the expression levels of C3 in the kidney and spleen were significantly higher than the levels in the control group and reached their peak 2 d after infection of Gibel carp with CyHV-2. These results indicate that organs other than the liver also play an important role in the complement immune system.
In vitro, GiCB cells were challenged with CyHV-2, poly(I:C) and LPS, showing different CagC3 mRNA expression patterns. The RNA expression of CagC3 from GiCB stimulated with LPS fell to the minimum in the first 24 hr, then increased sharply; on the contrary, the RNA expression of CagC3 post CyHV-2 or poly(I:C) challenge increased for the first 2 d. These differences in the gene induction patterns indicate that the complement system was activated by distinct pathways. CyHV-2 is a DNA virus and poly(I:C) is a synthetic analog of double-stranded RNA, so we suggest that the Gibel carp complement system could be activated similarly by DNA virus infection and RNA, at least in vitro.
In all, the full-length cDNA of the complement component C3 of Gibel carp was cloned and sequenced for the first time. The deduced amino acid sequence of CagC3 showed typical structural properties similar to other teleosts. Although only one isoform of complement component C3 is identified in the present study, presence of more isoforms cannot be ruled out. CagC3 genes were expressed in various tissues with the highest level in liver. Significant upregulation of CagC3 expression was observed in liver, spleen, kidney and the GiCB cell line after infection with CyHV-2, suggesting that CagC3 might play an important role in the immune response to viral infections in Gibel carp. The results reported herein provide a basis for further studies of CagC3 in the innate response and disease defense, which may potentially be useful for the control of viral disease caused by CyHV-2.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31802346), the Key Project of Scientific & Technological Innovation of Hubei Province (2018ABA101), and the Earmarked Fund for China Agriculture Research System (CARS-45-16).
REFERENCES
- 1.Abelseth T. K., Stensvåg K., Espelid S., Nygaard R., Ellingsen T., Bøgwald J., Dalmo R. A.2003. The spotted wolffish (Anarhichas minor Olafsen) complement component C3: isolation, characterisation and tissue distribution. Fish Shellfish Immunol. 15: 13–27. doi: 10.1016/S1050-4648(02)00135-3 [DOI] [PubMed] [Google Scholar]
- 2.Boshra H., Li J., Sunyer J. O.2006. Recent advances on the complement system of teleost fish. Fish Shellfish Immunol. 20: 239–262. doi: 10.1016/j.fsi.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 3.Carroll M. C., Isenman D. E.2012. Regulation of humoral immunity by complement. Immunity 37: 199–207. doi: 10.1016/j.immuni.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll M. C.2004. The complement system in regulation of adaptive immunity. Nat. Immunol. 5: 981–986. doi: 10.1038/ni1113 [DOI] [PubMed] [Google Scholar]
- 5.Finn R. D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Eddy S. R., Sonnhammer E. L. L., Bateman A.2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34: D247–D251. doi: 10.1093/nar/gkj149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forn-Cuní G., Reis E. S., Dios S., Posada D., Lambris J. D., Figueras A., Novoa B.2014. The evolution and appearance of C3 duplications in fish originate an exclusive teleost c3 gene form with anti-inflammatory activity. PLoS One 9: e99673. doi: 10.1371/journal.pone.0099673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasque P.2004. Complement: a unique innate immune sensor for danger signals. Mol. Immunol. 41: 1089–1098. doi: 10.1016/j.molimm.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 8.Hanson L., Dishon A., Kotler M.2011. Herpesviruses that infect fish. Viruses 3: 2160–2191. doi: 10.3390/v3112160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulo N., Bairoch A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P. S., Pagni M., Sigrist C. J. A.2006. The PROSITE database. Nucleic Acids Res. 34: D227–D230. doi: 10.1093/nar/gkj063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaac L., Isenman D. E.1992. Structural requirements for thioester bond formation in human complement component C3. Reassessment of the role of thioester bond integrity on the conformation of C3. J. Biol. Chem. 267: 10062–10069. [PubMed] [Google Scholar]
- 11.Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J.1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23: 403–405. doi: 10.1016/S0968-0004(98)01285-7 [DOI] [PubMed] [Google Scholar]
- 12.Kuroda N., Naruse K., Shima A., Nonaka M., Sasaki M.2000. Molecular cloning and linkage analysis of complement C3 and C4 genes of the Japanese medaka fish. Immunogenetics 51: 117–128. doi: 10.1007/s002510050020 [DOI] [PubMed] [Google Scholar]
- 13.Lange S., Bambir S. H., Dodds A. W., Bowden T., Bricknell I., Espelid S., Magnadóttir B.2006. Complement component C3 transcription in Atlantic halibut (Hippoglossus hippoglossus L.) larvae. Fish Shellfish Immunol. 20: 285–294. doi: 10.1016/j.fsi.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 14.Lange S., Dodds A. W., Magnadóttir B.2004. Isolation and characterization of complement component C3 from Atlantic cod (Gadus morhua L.) and Atlantic halibut (Hippoglossus hippoglossus L.). Fish Shellfish Immunol. 16: 227–239. doi: 10.1016/S1050-4648(03)00081-0 [DOI] [PubMed] [Google Scholar]
- 15.Law S. K., Dodds A. W.1997. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 6: 263–274. doi: 10.1002/pro.5560060201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak K. J., Schmittgen T. D.2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ CT Method. Methods 25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 17.Ma J., Jiang N., LaPatra S. E., Jin L., Xu J., Fan Y., Zhou Y., Zeng L.2015. Establishment of a novel and highly permissive cell line for the efficient replication of cyprinid herpesvirus 2 (CyHV-2). Vet. Microbiol. 177: 315–325. doi: 10.1016/j.vetmic.2015.04.006 [DOI] [PubMed] [Google Scholar]
- 18.Mauri I., Roher N., MacKenzie S., Romero A., Manchado M., Balasch J. C., Béjar J., Alvarez M. C., Tort L.2011. Molecular cloning and characterization of European seabass (Dicentrarchus labrax) and Gilthead seabream (Sparus aurata) complement component C3. Fish Shellfish Immunol. 30: 1310–1322. doi: 10.1016/j.fsi.2011.03.013 [DOI] [PubMed] [Google Scholar]
- 19.Melillo D., Varriale S., Giacomelli S., Natale L., Bargelloni L., Oreste U., Pinto M. R., Coscia M. R.2015. Evolution of the complement system C3 gene in Antarctic teleosts. Mol. Immunol. 66: 299–309. doi: 10.1016/j.molimm.2015.03.247 [DOI] [PubMed] [Google Scholar]
- 20.Morgan B. P., Marchbank K. J., Longhi M. P., Harris C. L., Gallimore A. M.2005. Complement: central to innate immunity and bridging to adaptive responses. Immunol. Lett. 97: 171–179. doi: 10.1016/j.imlet.2004.11.010 [DOI] [PubMed] [Google Scholar]
- 21.Nagar B., Jones R. G., Diefenbach R. J., Isenman D. E., Rini J. M.1998. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science 280: 1277–1281. doi: 10.1126/science.280.5367.1277 [DOI] [PubMed] [Google Scholar]
- 22.Nakao M., Mutsuro J., Obo R., Fujiki K., Nonaka M., Yano T.2000. Molecular cloning and protein analysis of divergent forms of the complement component C3 from a bony fish, the common carp (Cyprinus carpio): presence of variants lacking the catalytic histidine. Eur. J. Immunol. 30: 858–866. doi: [DOI] [PubMed] [Google Scholar]
- 23.Nielsen C. H., Leslie R. G. Q.2002. Complement’s participation in acquired immunity. J. Leukoc. Biol. 72: 249–261. [PubMed] [Google Scholar]
- 24.Nonaka M., Smith S. L.2000. Complement system of bony and cartilaginous fish. Fish Shellfish Immunol. 10: 215–228. doi: 10.1006/fsim.1999.0252 [DOI] [PubMed] [Google Scholar]
- 25.Nonaka M.2011. The complement C3 protein family in invertebrates. ISJ-Invert. Surviv. J. 8: 21–32. [Google Scholar]
- 26.Prado-Álvarez M., Rotllant J., Gestal C., Novoa B., Figueras A.2009. Characterization of a C3 and a factor B-like in the carpet-shell clam, Ruditapes decussatus. Fish Shellfish Immunol. 26: 305–315. doi: 10.1016/j.fsi.2008.11.015 [DOI] [PubMed] [Google Scholar]
- 27.Pushpa K., Gireesh-Babu P., Rajendran K. V., Purushothaman C. S., Dasgupta S., Makesh M.2014. Molecular cloning, sequencing and tissue-level expression of complement C3 of Labeo rohita (Hamilton, 1822). Fish Shellfish Immunol. 40: 319–330. doi: 10.1016/j.fsi.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 28.Qi Z. H., Liu Y. F., Wang W. N., Wu X., Xin Y., Lu Y. F., Wang A. L.2011. Molecular characterization and functional analysis of a complement C3 molecule in the orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 31: 1284–1290. doi: 10.1016/j.fsi.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 29.Ricklin D., Hajishengallis G., Yang K., Lambris J. D.2010. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11: 785–797. doi: 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saeij J. P., de Vries B. J., Wiegertjes G. F.2003. The immune response of carp to Trypanoplasma borreli: kinetics of immune gene expression and polyclonal lymphocyte activation. Dev. Comp. Immunol. 27: 859–874. doi: 10.1016/S0145-305X(03)00083-1 [DOI] [PubMed] [Google Scholar]
- 31.Schultz J., Milpetz F., Bork P., Ponting C. P.1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864. doi: 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigh J., Lindenstrøm T., Buchmann K.2004. The parasitic ciliate Ichthyophthirius multifiliis induces expression of immune relevant genes in rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 27: 409–417. doi: 10.1111/j.1365-2761.2004.00558.x [DOI] [PubMed] [Google Scholar]
- 33.Solem S. T., Stenvik J.2006. Antibody repertoire development in teleosts—a review with emphasis on salmonids and Gadus morhua L. Dev. Comp. Immunol. 30: 57–76. doi: 10.1016/j.dci.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 34.Sun Y., Wang R., Xu T.2013. Conserved structural complement component C3 in miiuy croaker Miichthys miiuy and their involvement in pathogenic bacteria induced immunity. Fish Shellfish Immunol. 35: 184–187. doi: 10.1016/j.fsi.2013.04.028 [DOI] [PubMed] [Google Scholar]
- 35.Sunyer J. O., Lambris J. D.1999. Complement. In: Encyclopedia of Life Sciences, Macmillan References Limited, London. [Google Scholar]
- 36.Sunyer J. O., Tort L., Lambris J. D.1997. Structural C3 diversity in fish: characterization of five forms of C3 in the diploid fish Sparus aurata. J. Immunol. 158: 2813–2821. [PubMed] [Google Scholar]
- 37.Sunyer J. O., Zarkadis I. K., Sahu A., Lambris J. D.1996. Multiple forms of complement C3 in trout that differ in binding to complement activators. Proc. Natl. Acad. Sci. USA 93: 8546–8551. doi: 10.1073/pnas.93.16.8546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki M. M., Satoh N., Nonaka M.2002. C6-like and C3-like molecules from the cephalochordate, amphioxus, suggest a cytolytic complement system in invertebrates. J. Mol. Evol. 54: 671–679. doi: 10.1007/s00239-001-0068-z [DOI] [PubMed] [Google Scholar]
- 39.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H., Qi P., Guo B., Li J., He J., Wu C., Gul Y.2015. Molecular characterization and expression analysis of a complement component C3 in large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 42: 272–279. doi: 10.1016/j.fsi.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 41.Xu J., Zeng L., Zhang H., Zhou Y., Ma J., Fan Y.2013. Cyprinid herpesvirus 2 infection emerged in cultured gibel carp, Carassius auratus gibelio in China. Vet. Microbiol. 166: 138–144. doi: 10.1016/j.vetmic.2013.05.025 [DOI] [PubMed] [Google Scholar]
- 42.Xue M., Cui Y. B.2001. Effect of several feeding stimulants on diet preference by juvenile gibel carp (Carassius auratus gibelio), fed diets with or without partial replacement of fish meal by meat and bone meal. Aquaculture 198: 281–292. doi: 10.1016/S0044-8486(00)00602-5 [DOI] [Google Scholar]
- 43.Zarkadis I. K., Sarrias M. R., Sfyroera G., Sunyer J. O., Lambris J. D.2001. Cloning and structure of three rainbow trout C3 molecules: a plausible explanation for their functional diversity. Dev. Comp. Immunol. 25: 11–24. doi: 10.1016/S0145-305X(00)00039-2 [DOI] [PubMed] [Google Scholar]