ABSTRACT
Alveologenesis is an essential developmental process that increases the surface area of the lung through the formation of septal ridges. In the mouse, septation occurs postnatally and is thought to require the alveolar myofibroblast (AMF). Though abundant during alveologenesis, markers for AMFs are minimally detected in the adult. After septation, the alveolar walls thin to allow efficient gas exchange. Both loss of AMFs or retention and differentiation into another cell type during septal thinning have been proposed. Using a novel Fgf18:CreERT2 allele to lineage trace AMFs, we demonstrate that most AMFs are developmentally cleared during alveologenesis. Lung mesenchyme also contains other poorly described cell types, including alveolar lipofibroblasts (ALF). We show that Gli1:CreERT2 marks both AMFs as well as ALFs, and lineage tracing shows that ALFs are retained in adult alveoli while AMFs are lost. We further show that multiple immune cell populations contain lineage-labeled particles, suggesting a phagocytic role in the clearance of AMFs. The demonstration that the AMF lineage is depleted during septal thinning through a phagocytic process provides a mechanism for the clearance of a transient developmental cell population.
KEY WORDS: FGF signaling, Lung development, Myofibroblast, Lipofibroblast, Lineage tracing, Alveolar macrophage
Summary: During postnatal lung development, two major mesenchymal cell types have different developmental fates: most alveolar myofibroblasts are cleared from the lung by phagocytic cells, while alveolar lipofibroblasts are retained into adulthood.
INTRODUCTION
Lung development results in an organ with the capacity to support efficient gas exchange in a mature animal. This is facilitated by the alveoli, the functional unit of air-gas exchange, located at the distal tips of the respiratory tree. In mice, branching morphogenesis during the embryonic and fetal stages of development generates a distal lung that, at birth, is composed of large undivided saccules (primary septa) that have only ∼10% of the respiratory epithelium of an adult animal (Schittny, 2017). Formation of alveoli (alveologenesis), the final stage of lung development, is divided into two phases in mice. The first phase is septation, which begins at postnatal day 4 (P4) and is defined by the formation of septal ridges (secondary septa), which are extracellular matrix rich invaginations of the saccular wall that subdivide and increase alveolar surface area (Mund et al., 2008). Septal ridge formation concludes by ∼P21 in the mouse. In the second phase of alveolar development, thinning of the newly formed septal walls and resolution of the double capillary network to a single capillary sheet, forms a distinctively thin alveolar wall in the mature lung (Schittny et al., 1998). The second phase of alveolar development begins contemporaneously with the ending of septal formation at ∼P14 and continues until ∼P36. Thinning of the alveolar walls in the second phase is thought to involve apoptosis of lung fibroblasts, although given the cellular heterogeneity of the mesenchymal compartment, the identity of the depleted cells is unclear (Bruce et al., 1999; McGowan and McCoy, 2011; Schittny et al., 1998).
Although the epithelial lineages of the lung are well defined, the fibroblast lineages are not because of a lack of definitive genetic markers, and heterogeneity and plasticity compared with the epithelial lineage (Ahlfeld and Perl, 2017). During alveologenesis, two of the best described mesenchymal lineages are the alveolar myofibroblast (AMF) and the alveolar lipofibroblast (ALF). These two cell types display overlapping and unique markers. AMFs are interstitial contractile cells that are thought to play a role in the construction of septal ridges through the deposition of an elastin-rich extracellular matrix (Boström et al., 1996; Noguchi et al., 1989). AMFs are located at the septal ridges, embedded within elastin fibers, express Pdgfra and the smooth muscle marker α-smooth muscle actin (αSMA, encoded by Acta2), and are required for alveologenesis (Branchfield et al., 2016; Li et al., 2018; McGowan et al., 2008). ALFs are interstitial lipid laden cells that secrete triglycerides, store retinoids and provide trophic support for alveolar type 2 cells (Barkauskas et al., 2013; McGowan et al., 1995; Torday et al., 1995). ALFs are located towards the septal base, associate less with elastin fibers, are marked by the presence of lipid droplets, and also express Pdgfra (McGowan et al., 2008; O'Hare and Sheridan, 1970).
AMFs are derived from lung mesenchymal progenitors during the embryonic stage that express Pdgfra and Gli1 (Li et al., 2015, 2018). Their number peaks during alveologenesis. Interestingly, AMFs are absent in the adult alveolar region based on lack of αSMA+ cells, suggesting either a phenotypic conversion through downregulation of smooth muscle markers or that the cells themselves are actively removed from the lung through cell death (Kapanci et al., 1995; Yamada et al., 2005). Multiple lineage-tracing experiments have been performed, but there is no consensus on the fate of the AMF. Marking Gli1-expressing AMFs determined that few cells remain until adulthood, suggesting cell death, while marking Pdgfra-expressing AMFs determined that the cells survive into adulthood, suggesting that they turn off their smooth muscle markers (Li et al., 2015, 2018). Differences in results could be attributed to different timing of labeling and specificity of lineage-tracing alleles. The mechanism of loss of αSMA+ AMFs after alveologenesis has yet to be fully resolved.
ALFs, like AMFs, increase in number during alveologenesis, peaking at the end of the first phase of alveolar development (Tordet et al., 1981). ALFs are present in the adult lung; however, they contain smaller lipid droplets than the postnatal ALFs (Kaplan et al., 1985). Much work has used real-time expression of PdgfraeGFP to label a population of ALFs, with noted reduction of PdgfraeGFP+ cells at the conclusion of lung development (Endale et al., 2017; Gouveia et al., 2017; McGowan and McCoy, 2011). However, recent short-term lineage studies using Tcf21mCrem or Plin2CreERT2 suggest that the ALF is a relatively stable population of cells, although both of these lineage alleles are expressed in multiple cell types (Ntokou et al., 2017; Park et al., 2019). Therefore, it is still not known whether the reduction in PdgfraeGFP-labeled ALFs after alveologenesis is due to downregulation of ALF markers or to the clearance of cells from the lung. To date, we know of no lineage-tracing experiment that follows the fate of the ALFs into adulthood.
Disruptions to the lung during postnatal development result in an insufficiency of secondary septa, characteristic of the pediatric disorder bronchopulmonary dysplasia (BPD), a major cause of preterm infant morbidity and chronic lung disease (Martinez, 2016; Voynow, 2017). The lung mesenchyme has long been recognized as important in directing alveologenesis (Boström et al., 1996; Kugler et al., 2017; Li et al., 2019, 2017; Nicola et al., 2009; Tsujino et al., 2017) and many mesenchymal proteins are dysregulated in models of BPD (Ahlfeld and Conway, 2012). Further alterations to AMF and ALF numbers are seen in genetic and hyperoxia-induced mouse models of BPD (Branchfield et al., 2016; Choi et al., 2013; Li et al., 2018; Popova et al., 2014; Rehan et al., 2006). Understanding the dynamics of these cell types will be important to assess pathogenic roles in human lung disease.
Fibroblast growth factor (FGF) signaling is critically important for proper lung development at all stages (Bellusci et al., 1997; Colvin et al., 2001; Tichelaar et al., 2000; Usui et al., 2004; Weinstein et al., 1998). The significance of FGF signaling during alveologenesis was highlighted by the dramatic alveolar defect seen in organism-wide or mesenchyme-specific knockouts of Fgf receptors 3 and 4 (Fgfr3 and Fgfr4) (Li et al., 2017; Srisuma et al., 2010; Weinstein et al., 1998). Given the functional importance of FGF signaling, the strategy of genetically tagging cells that express ligand molecules has allowed the identification of developmentally important cell populations in the lung and other organs (El Agha et al., 2014; Huh et al., 2015, 2013; Watson et al., 2017; Yang et al., 2018). Fgf18 is upregulated in the postnatal rodent lung and third trimester human lung during alveologenesis, suggesting that it would be expressed in a cell population with a functional role in alveologenesis (Boucherat et al., 2007; Chailley-Heu et al., 2005; Franco-Montoya et al., 2011).
In this study, we show that AMFs and ALFs have distinct developmental fates at the culmination of alveologenesis. We identify an Fgf18CreERT2-expressing AMF population that represents ∼14% of total cells in the alveolar region during alveologenesis. Through lineage-tracing analysis, we show that the Fgf18CreERT2 AMF lineage decreases by ∼88% by P21. We confirm previous reports that Gli1LacZ (and Gli1CreERT2) marks AMFs but also identify a previously unrecognized contribution to the ALF lineage. Lineage tracing with Gli1CreERT2 shows a lesser decrease in labeled cells compared with the Fgf18CreERT2 lineage trace, with retained cells maintaining the ALF marker ADRP (encoded by Plin2). We also report data identifying multiple immune cell populations that contains lineage-labeled particles, suggesting a role for phagocytosis in the clearance of AMFs.
RESULTS
Fgf18CreERT2 labels alveolar myofibroblasts and alveolar type 1 cells in the postnatal mouse lung
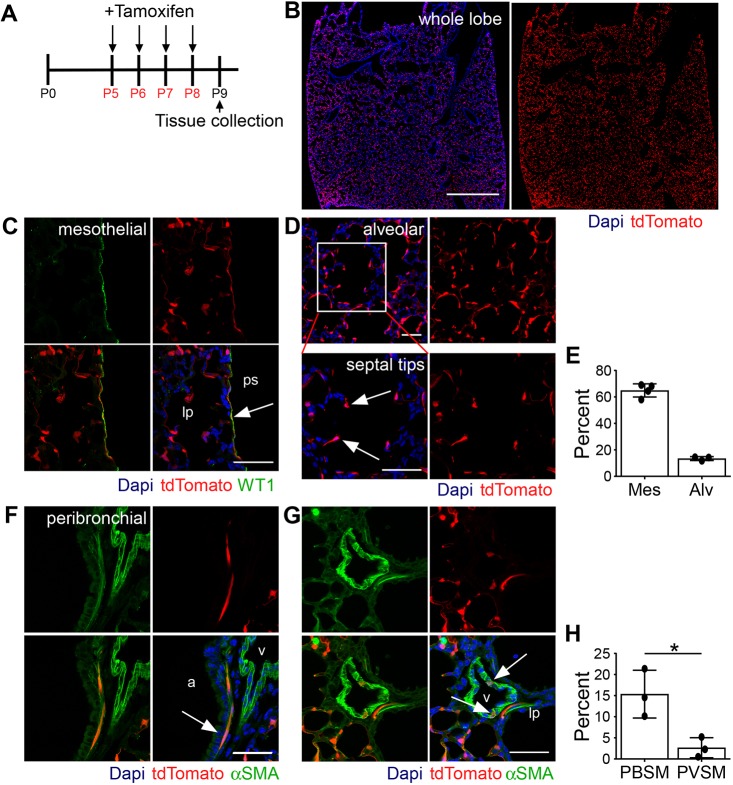
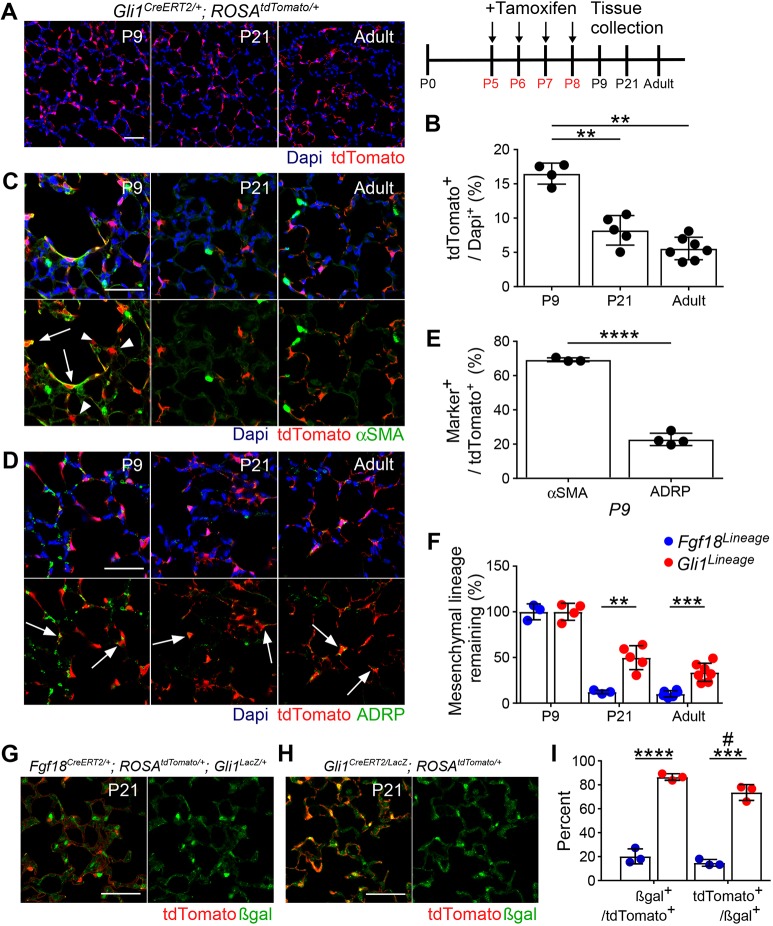
Fgf18 is weakly expressed during fetal rodent lung development but is dramatically and transiently upregulated during the first stage of alveolar development (Chailley-Heu et al., 2005; Franco-Montoya et al., 2011). To identify cells expressing Fgf18 during postnatal lung development, Fgf18CreERT2; ROSAtdTomato (Fgf18Lineage) mice were given daily tamoxifen (Tam) doses from postnatal day 5 (P5) to P8 (Fig. 1A). This dosing regimen maximized the labeling efficiency of Fgf18CreERT2 at the developmental time when the Fgf18 allele is most highly expressed (P3-P10 in mouse and rat lung) (Chailley-Heu et al., 2005; Franco-Montoya et al., 2011). At P9, 24 h after the final Tam dose, Fgf18Lineage-expressing cells were found throughout the lung (Fig. 1B). Importantly, in uninduced controls (no Tam), tdTomato+ cells could not be detected during alveologenesis (P9), at the end of the first stage of alveolar development (P21) or in the adult (Fig. S1). Fgf18Lineage includes mesothelial, alveolar, peribronchial and perivascular cells (Fig. 1C,D,F,G).
Fig. 1.
Expression pattern of Fgf18CreERT2 in the postnatal lung. (A) Fgf18Lineage mice were injected with Tam daily from P5 to P8 to induce recombination of ROSAtdTomato. Lungs were analyzed at P9. (B) Whole-lobe section showing tdTomato epifluorescence. (C) Colocalization of tdTomato (red) with WT1 (green). Arrow indicates mesothelium. (D) tdTomato+ cells in the alveolar region. Arrows indicate septal tips. (E) Mes, quantification of the percentage of WT1+ mesothelial cells that are tdTomato+; Alv, quantification of the percentage of DAPI+ cells in the alveolar region that are tdTomato+. (F,G) Colocalization of tdTomato (red) with smooth muscle marker αSMA (green) in the (F) peribronchial and (G) perivascular regions. Arrows indicate colocalization. (H) Quantification of the percentage of αSMA+ smooth muscle cells that are tdTomato+ in the peribronchial (PBSM) and perivascular (PVSM) regions. Student's t-test, *P<0.05. lp, lung parenchyma; ps, pleural space; a, airway; v, blood vessel; Mes, mesothelial; Alv, alveolar; PBSM, peribronchial smooth muscle; PVSM, perivascular smooth muscle. DAPI (blue). Scale bars: 1 mm in B; 50 µm in C,D,F,G. Data are mean±s.d. n=4 for C; n=3 for B,D-G.
The majority of Fgf18Lineage cells were in the alveolar region (Fig. 1B). These cells were associated with the growing alveolar septal ridges (Fig. 1D, arrows) and appeared uniform throughout peripheral and central alveolar regions. The majority (65%±5.0%) of WT1+ mesothelial cells, the single-cell layer lining the periphery of the lung, co-labeled with tdTomato (Fig. 1C,E) (Batra and Antony, 2015). This labeling pattern was similar on both medial and lateral surfaces of the lung (Fig. 1B). A small percentage (15±5.7%) of peribronchial cells were Fgf18Lineage+; however, individual airways were highly variable (with 0-78% of cells being Fgf18Lineage+, data from Fig. 1F,H). Perivascular cells, identified by αSMA+ immunostaining, contained very few Fgf18Lineage+ cells (Fig. 1G,H).
To determine the identity of the alveolar cells that expressed Fgf18, Fgf18Lineage+ cells were colocalized with markers of all major alveolar cell types at P9 (Fig. 2). The majority of Fgf18Lineage+ cells co-expressed the AMF marker αSMA (Fig. 2A,H). To support this finding, the PdgfraeGFP allele (Hamilton et al., 2003; McGowan et al., 2008), which marks AMFs with eGFP expression, was bred into Fgf18Lineage mice. 72.0±4.5% of all Fgf18Lineage+ cells in the alveolar region were triple positive with αSMA and PdgfraeGFP (Fig. 2B,H), while only 28±7.8% of PdgfraeGFP+ cells were positive for tdTomato (n=5; data not shown).
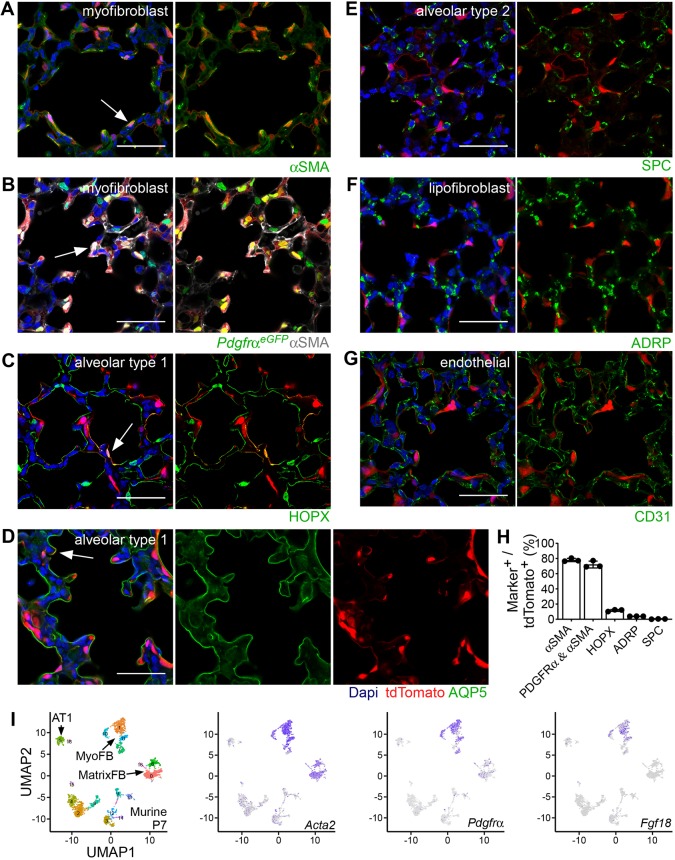
Fig. 2.
Fgf18CreERT2 in the alveolar region labels alveolar myofibroblasts and alveolar type 1 cells. (A-G) Colocalization of tdTomato (red) with markers of the major cell lineage of the distal lung in Fgf18Lineage mice injected with Tam daily from P5 to P8 and collected at P9. (A) Alveolar myofibroblasts (αSMA, green). (B) Alveolar myofibroblasts in Fgf18CreERT2/+; ROSAtdTomato/+; PdgfraGFP/+ mice (PdgfraeGFP, green; αSMA, white). (C) Alveolar type 1 cells (HOPX, green, nuclear and membrane). (D) Alveolar type 1 cells (AQP5, green, membrane). (E) Alveolar type 2 cells (SPC, green). (F) Lipofibroblasts (ADRP, green). (G) Endothelial cells (CD31, green). (H) Quantification of the percentage of tdTomato+ cells that are lineage marker+. (I) Visualization of scRNA-seq data by UMAP plots from postnatal murine lung cells isolated at P7. MyoFB, MatrixFB and AT1 clusters are indicated by arrows (left). Panels for cells expressing individual genes are on the right: Acta2 (purple), Pdgfra (purple) and Fgf18 (purple). DAPI is in blue in A-G. Scale bars: 50 µm. MyoFB, myofibroblast; MatrixFB, matrix fibroblast; AT1, alveolar type 1. Arrows indicate colocalization of signals. Data are mean±s.d. n=3 for A,C-G; n=5 for B.
Some Fgf18Lineage+ cells had long thin processes that suggested expression in alveolar type 1 (AT1) cells. Consistent with this, some Fgf18Lineage+ cells colocalized with the membrane-bound AT1 marker AQP5 and the nuclear and membrane marker HOPX (Jain et al., 2015) (Fig. 2C,D). To quantify Fgf18 expression in AT1 cells, HOPX was used to show that 12±1.3% of Fgf18Lineage+ cells were AT1 cells, while 23±2.3% of AT1 cells were Fgf18Lineage+ (Fig. 2C,H). Fgf18Lineage+ cells showed minimal (3.9%±0.064%) colocalization with the alveolar lipofibroblast marker ADRP (PLIN2) (Fig. 2F) (McGowan et al., 2008; Varisco et al., 2012). Other major cell types in the lung, including alveolar type 2 and endothelial cells, also showed minimal colocalization with tdTomato based on expression of their respective markers, SPC (Sftpc) and CD31 (Pecam1) (Fig. 2E,G).
To further validate the fidelity of the Fgf18CreERT2 allele, we independently analyzed single cell RNA-sequencing (scRNA-seq) data generated by the LungGens consortium from P3 and P7 mouse lung using uniform manifold approximation and projection (UMAP) (Becht et al., 2019; Du et al., 2017, 2015). P7 mouse lung scRNA-seq confirmed that Fgf18 is expressed primarily in AMFs with high levels of Acta2 (which encodes αSMA) (MyoFB, Fig. 2I). Pdgfra displayed broader mesenchymal expression, localizing in AMFs as well as in matrix fibroblasts with low levels of Acta2, a population that likely contains the ALFs (Guo et al., 2019). Earlier in development at P3, Fgf18 is expressed in Acta2 high AMFs (MyoFB) as well as in AT1 cells (Fig. S2A). P1 human lung scRNA-seq generated and analyzed by the LungGens consortium showed Fgf18 almost exclusively in AMFs (MyoFB, Fig. S2B). These results indicate that Fgf18CreERT2-expressing cells include AMFs and AT1 cells in mouse and AMFs in human lung.
Analysis of P7 scRNA-seq showed Fgf18 in a subset of aSMA+ myofibroblasts (Fig. 2I, Acta2). In immunostained histological sections, Fgf18 marks 62.0±10.3% of all αSMA+ cells in the alveolar region, suggesting a subpopulation (data derived from Fig. 2A). However, UMAP plots of the identified myofibroblast subtypes at P7 were not able to separate Fgf18+ cells into a single subcluster (Fig. S2C). These results suggest that Fgf18-expressing AMFs are not a distinct subgroup of AMFs.
Fgf18CreERT2-labeled AMF lineage decreases temporally during development
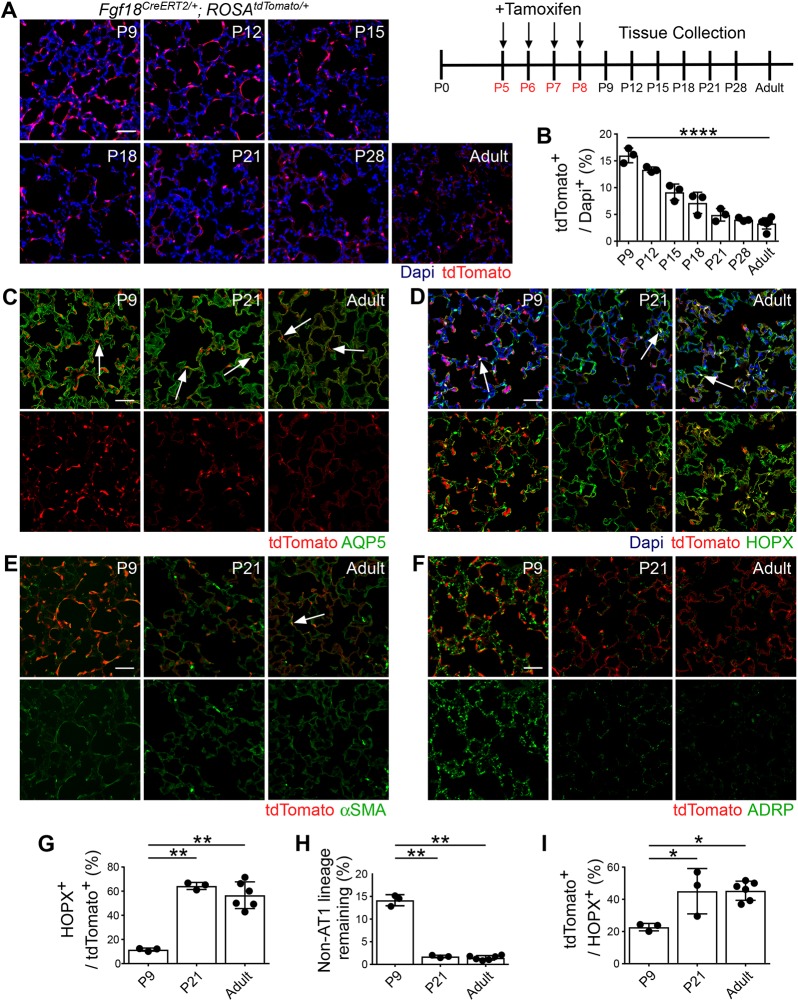
The fate of AMFs was investigated by long-term lineage tracing of Fgf18CreERT2 cells labeled from P5 to P8 (Fig. 3A). Quantification of total alveolar Fgf18Lineage+ cells showed a decreasing trend throughout the first phase of alveolar development that stabilized by P21 (percentage of all cells: P9, 16±1.4%; P21, 4.9±1.2%; adult, 3.3%±1.0%) (Fig. 3B). Most remaining cells were morphologically like AT1 cells, with long thin processes and expression of AQP5 (Fig. 3C). Quantification of the remaining cells in the adult showed that most were AT1 cells using a rabbit HOPX polyclonal antibody (57±10%, Fig. 3D,G; arrows) or a directly conjugated HOPX mouse monoclonal antibody (77±2.3%, Fig. S3). Analysis of the HOPX− (non-AT1 lineage) Fgf18Lineage+ cells revealed a large reduction (88±2.0%) in the mesenchymal-labeled lineage by the end of the first phase (P21) of alveolar development (Fig. 3H). Restricting analysis to only HOPX− Fgf18Lineage+ cells revealed that the few remaining mesenchymal lineage cells (∼4 cells per 20× field) were mostly positive for the fibroblast marker Desmin, with minimal positive cells for defined mesenchymal lineage markers αSMA, ADRP or NG2 (Fig. 3E,F and Fig. S3). A few αSMA+ cells near the more proximal alveolar ducts were tdTomato+ (Fig. 3E, arrow). Fgf18Lineage+ cells in the mesothelial, peribronchial and perivascular compartments displayed similar patterns in the adult compared with the postnatal lung (Fig. S4). Overall, these data suggest that loss of Fgf18Lineage+ cells are restricted to the AMFs. This demonstrates that the transient nature of AMFs is largely due to cell loss from the alveolus as opposed to downregulation of an AMF gene expression program.
Fig. 3.
Most Fgf18CreERT2-labeled alveolar myofibroblasts are cleared and alveolar type 1 cells remain at the end of alveologenesis. (A) Fgf18Lineage mice were injected with Tam daily from P5 to P8 and collected throughout postnatal lung development and in the adult (≥8 weeks). (B) Quantification of the percentage of DAPI+ cells in the alveolar region that are tdTomato+. n=3 for P9, P12, P15, P18, P21 and P28; n=6 for adult. One-way ANOVA, ****P=3.4×10−10. (C-F) Colocalization of tdTomato (red) in lineage-traced mice with: (C) alveolar type 1 membrane marker AQP5 (green); (D) alveolar type 1 nuclear and membrane marker HOPX (green); (E) alveolar myofibroblast marker αSMA (green); and (F) lipofibroblast marker ADRP (green). (G) Quantification of the percentage of tdTomato+ cells that are HOPX+ in lineage-traced mice. P9 data are repeated from Fig. 2H. One-way ANOVA, P=4.5×10−5. **α<0.01, Tukey's HSD. n=3 for P9 and P21; n=6 for adult. (H) Quantification of the percentage of remaining mesenchymal lineage tdTomato+ cells in lineage-traced mice. Remaining mesenchymal lineage is calculated from the percentage of tdTomato+/DAPI+ cells (Fig. 3B) with alveolar type 1-labeled cells removed (Fig. 3G). One-way ANOVA, P=2.3×10−9. **α<0.01, Tukey's HSD. n=3 for P9 and P21; n=6 for adult. (I) Quantification of the percentage of HOPX+ cells that are tdTomato+ in lineage-traced mice. One-way ANOVA, P=0.008. *α<0.05, Tukey's HSD. n=3 for P9 and P21; n=6 for adult. DAPI (blue). Scale bars: 50 µm. Arrows indicate colocalization of signals. Data are mean±s.d.
Coinciding with the decrease in AMFs was an increase in the proportion of AT1 cells that were in the Fgf18Lineage (Fig. 3I). This suggests either proliferation of Fgf18Lineage AT1 cells, activity of Cre-mediated recombination after P9 or transdifferentiation of Fgf18Lineage cells into the AT1 lineage. A minimal number of AT1 cells incorporated EdU from P9 to P21, ruling out proliferation (Fig. S5A). However, Fgf18Lineage mice induced at later time points also labeled AT1 cells, which supports the possibility that residual Tam led to Cre-mediated recombination in AT1 cells after P9 (Fig. S5B-D).
Fgf18 expression is dynamic during the first few days of postnatal lung development, increasing dramatically at P3 in rodents (Chailley-Heu et al., 2005; Franco-Montoya et al., 2011). To address the lineage of cells marked by Fgf18CreERT2 before this upregulation, we induced Fgf18Lineage mice with a single injection of Tam at P1 and collected mice at P2 (to define the starting population), at P7 (during alveologenesis) and at P21 (the time of a previously identified decline in AMFs) (Fig. S6). A single injection labeled fewer total starting cells than the P5-8 regime (P2, 4.0±2.4%, Fig. S6A,B versus P9, 16±1.4%, Fig. 3B) and a greater proportion of the tdTomato+ cells were HOPX+ AT1 cells (P2, 41±7.4%, Fig. S6C versus P9, 12±1.3%, Fig. 2C,H). Of labeled cells at P7, 40±5.1% (n=5, from Fig. S6D) were αSMA+ AMFs; however, many fewer total AMFs were labeled between comparable timepoints with the single injection (P7, 2.7±1.8%, derived from Fig. S6B,D versus the P5-8 regime P9, 12±0.75%, derived from Figs 2H and 3B). The total Fgf18Lineage+ cells did not increase by P7, and there was no reduction by P21 (Fig. S6A,B). However, we did notice an increase in the ratio of Fgf18Lineage+ cells that were AT1 cells by P21 (Fig. S6C,E). Interestingly, at P7 and P21 there was also an increased ratio of total AT1 cells labeled compared with P2 (Fig. S6F). Accounting for the increased number of AT1-labeled cells did not reveal a significant reduction in the tdTomato+ remaining non-AT1 lineage cells (Fig. S6G). Both P1 induction and P5-8 induction had a similar amount of total non-AT1 lineage cells remaining at P21 (Fig. S6G versus Fig. 3H). Remaining non-AT1 lineage cells were often found in what appeared to be alveolar ducts.
These results demonstrate that earlier induction of Fgf18Lineage mice preferentially marks AT1 cells and some AMFs. The AT1 population appears stable or increasing with age and we do not see a significant loss of the non-AT1 cell population, likely owing to labeling many fewer total cells, high variability between mice and labeling a population that was negative for HOPX and αSMA (∼20% of labeled cells).
Fgf18CreERT2 marks a subset of Gli1+ mesenchymal cells
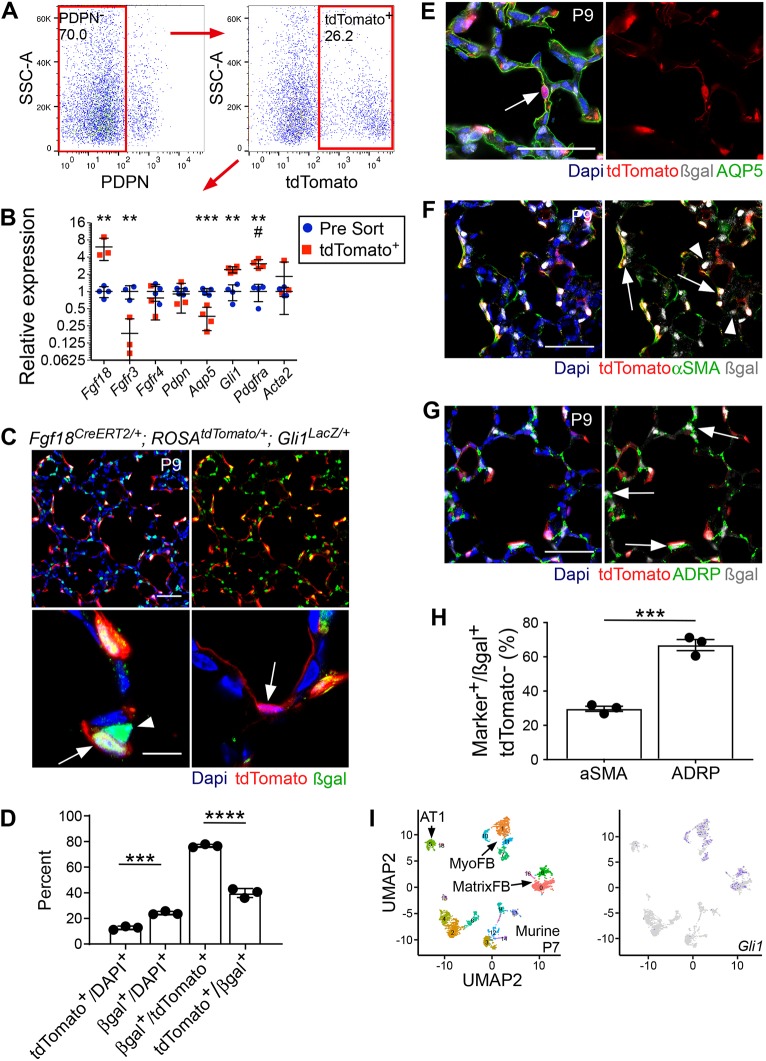
To determine whether transdifferentiation of AMFs into the AT1 lineage was occurring, a complementary lineage trace of AMFs that excludes initial targeting of AT1 cells was desired. Gli1 reporters mark αSMA+, Pdgfra+ AMFs with no noted expression in AT1 cells (Kugler et al., 2017; Li et al., 2015). Tandem flow cytometry-qRT-PCR of Fgf18Lineage+ lung mesenchymal cells at P9 confirmed enrichment of target cells (Fgf18), depletion of AT1 cells (Aqp5) and enrichment of the AMF markers (Pdgfra and Gli1) (Fig. 4A,B). To confirm that Gli1 marks the same AMF population as Fgf18CreERT2, Gli1LacZ was bred into Fgf18Lineage mice and mice induced from P5 to P8 were collected at P9 (Fig. 4C). Gli1LacZ is a reporter allele of Gli1 that marks its transcriptional expression (Bai et al., 2002). The majority (77±1.2%) of Fgf18Lineage+ cells colocalized with Gli1-driven β-galactosidase (βgal) expression (Fig. 4D). The remaining Fgf18Lineage+, Gli1LacZ− cells appeared to be AT1 cells based on morphology (Fig. 4C; bottom right panel; arrow) and colocalized with AQP5 (Fig. 4E). Confirming that Gli1LacZ is not expressed in AT1 cells, only one cell out of 77 AQP5+ tdTomato+ cells was βgal+ in sections from three mice (data not shown).
Fig. 4.
Gli1 marks Fgf18Lineage+ alveolar myofibroblasts and Fgf18Lineage− alveolar lipofibroblasts. (A) Fgf18Lineage mice were injected with Tam daily from P5 to P8 and collected at P9 for tandem fluorescence-activated cell sorting-qRT-PCR. Samples were depleted from endothelial (CD31+) and hematopoietic (CD45+) cells before sorting. Sorted cells were gated against PDPN+ to remove AT1 cells (left) and for tdTomato+ (right) to enrich for lineage-marked AMFs. (B) Relative gene expression (pre-sort control cells versus collected tdTomato+ cells). Student's t-test, **P<0.01; ***P<0.001. Mann–Whitney, #u=0.029. n=4 for all samples except for pre-sort control (Fgfr3) and tdTomato+ (Fgf18, Fgfr3, Fgfr4, Pdpn and Acta2) where n=3. (C) Fgf18CreERT2/+; ROSAtdTomato/+; Gli1LacZ/+ mice were injected with Tam daily from P5 to P8 and collected at P9. Arrow (bottom, left) indicates colocalization of signals. Arrowhead (bottom, left) indicates a βgal+, tdTomato− cell. Arrow (bottom, right) indicates a tdTomato+ cell with AT1 morphology that is βgal−. (D) Quantification of the percentage of tdTomato+/DAPI+ versus βgal+/DAPI+ cells and βgal+/tdTomato+ versus tdTomato+/βgal+ cells in the alveolar region. Student's t-test, ***P<0.001; ****P<0.0001. n=3. (E-G) Colocalization of tdTomato (red) and βgal (white) with: (E) AT1 marker AQP5 (green), (F) alveolar myofibroblast marker αSMA (green) and (G) lipofibroblast marker ADRP (green). Arrows indicate colocalization of βgal with lineage marker; arrowhead indicates absence of colocalization with αSMA. (H) Quantification of the percentage of βgal+; tdTomato− cells that colocalize with αSMA and ADRP. Student's t-test, ***P<0.001. n=3. (I) Visualization of scRNA-seq data by UMAP plots from postnatal murine lung cells isolated at P7. MyoFB and MatrixFB clusters are indicated (left) and Gli1-expressing cells are indicated in purple (right). DAPI is in blue in C,E-G. Scale bars: 50 µm in C (top), E-G; 10 µm in C (bottom). MyoFB, myofibroblast; MatrixFB, matrix fibroblast; AT1, alveolar type 1. Data are mean±s.d.
Gli1LacZ targeted an overall greater percent of the total lung cell population than did Fgf18Lineage (24±1.3% versus 13±1.4%; Fig. 4D) at P9. Fgf18Lineage−, Gli1LacZ+ cells were located towards the base of the secondary septa, characteristic of ALFs (Fig. 4C, bottom left panel, arrowhead) (McGowan et al., 2008). Staining with αSMA and ADRP revealed that the majority of Fgf18Lineage− Gli1LacZ+ cells were ADRP+ ALFs (67±5.6%) and the remaining cells were αSMA+ AMFs (30±2.6%) (Fig. 4F-H). P7 mouse lung scRNA-seq analyzed by UMAP confirmed Gli1 expression in AMFs as well as in matrix fibroblasts, a population that likely contains the ALFs (Fig. 4I) (Du et al., 2017, 2015; Guo et al., 2019). EdU incorporation demonstrated that Fgf18Lineage+, Gli1LacZ+ AMFs were less proliferative than Fgf18Lineage−, Gli1LacZ+ ALFs (2.1±0.70% versus 3.6±0.27% EdU+), suggesting a functional difference in cellular fate (Fig. S7). These data demonstrate that Gli1LacZ marks the same AMF population as the Fgf18Lineage; however, it also marks a separate, yet unrecognized, population of ALFs in the postnatal lung.
Gli1CreERT2-expressing mesenchymal cell lineage is partially retained throughout development
Using Gli1CreERT2 and ROSAtdTomato, a complementary lineage-tracing experiment was performed (Gli1Lineage) (Fig. 5A). Gli1Lineage+ cells were never observed to colocalize with AT1 markers, confirming that transdifferentiation was not occurring (Fig. S8A). However, total alveolar Gli1Lineage+ cells decreased by P21, showing that some of the Gli1Lineage+ cells were lost (Fig. 5A,B). Staining with αSMA and ADRP confirmed that Gli1Lineage marks AMFs and ALFs at P9 (Fig. 5C,D,E). In adults, few remaining cells expressed αSMA (Fig. 5C); however, some retained expression of ADRP (Fig. 5D, arrows). Remaining Gli1Lineage+ cells were immediately adjacent to the CD31+ capillary endothelium (Fig. S8B) but were negative for the pericyte marker NG2 (Fig. S8C). A greater proportion of the mesenchymal Gli1Lineage+ cells were retained at P21 and into adulthood compared with mesenchymal Fgf18Lineage+ cells (Fig. 5F).
Fig. 5.
Most Gli1CreERT2-labeled alveolar myofibroblasts are cleared at the end of alveologenesis, whereas alveolar lipofibroblasts remain. (A) Gli1CreERT2/+; ROSAtdTomato/+ mice were injected with Tam daily from P5 to P8 and lungs were analyzed at P9, P21 and in the adult (≥8 weeks). (B) Quantification of the percentage of DAPI+ cells in the alveolar region that are tdTomato+. One-way ANOVA, P=9.8×10−7. **α<0.01, Tukey's HSD. n=4 (P9), n=5 (P21) and n=7 (adult). (C,D) Colocalization of tdTomato (red) in lineage-traced mice with: (C) alveolar myofibroblast marker αSMA (green) and (D) alveolar lipofibroblast marker ADRP (green). (E) Quantification of the percentage of tdTomato+ cells that colocalize with αSMA and ADRP at P9. Student's t-test, ****P<0.0001. n=3 (αSMA), n=4 (ADRP). Arrows indicate localization of signals. Arrowheads indicate tdTomato+ αSMA− cells. (F) Quantification of the percentage of remaining mesenchymal lineage tdTomato+ cells in Fgf18Lineage and Gli1Lineage mice. Fgf18 data from Fig. 3H and Gli1 data from Fig. 5D are normalized to P9 for comparison. The entire Gli lineage was considered mesenchymal as there was no contribution to other lineages. Student's t-test, **P<0.01; ***P<0.001. Fgf18 lineage: n=3 for P9 and P21, n=6 for adult. Gli1 lineage: n=4 (P9), n=5 (P21), n=7 (adult). (G,H) Fgf18CreERT2/+; ROSAtdTomato/+; Gli1LacZ/+ mice (G) and Gli1CreERT2/LacZ; ROSAtdTomato/+ mice (H) were injected with Tam daily from P5 to P8 and lungs were collected at P21. Colocalization of tdTomato (red) with βgal (green). (I) Quantification of the percentage of βgal+/tdTomato+ cells and tdTomato+/βgal+ cells in the alveolar region in Fgf18Lineage versus Gli1Lineage mice. Student's t-test, ***P<0.001; ****P<0.0001. Mann–Whitney, #u=0.10. n=3 DAPI (blue). Scale bars: 50 µm. Data are mean±s.d.
Gli1LacZ-expressing cells are decreased but present in the lung at the end of the first stage of alveolar development (Liu et al., 2013b). To further follow the fate of Gli1-expressing AMFs and ALFs, the Gli1LacZ allele was bred into both lineage-tracing models (Fgf18Lineage and Gli1Lineage) and mice were induced with Tam from P5 to P8 and lungs were analyzed at P21 (Fig. 5G,H). A smaller proportion of the Fgf18Lineage+ cells retained Gli1LacZ expression than Gli1Lineage+ cells (20±6.3% versus 86±2.8%; Fig. 5I), supporting a model in which Fgf18-labeled AMFs are lost. Furthermore, the majority of Gli1LacZ+ cells were also Gli1Lineage+ (74±6.6%), demonstrating that most of the identified Gli1LacZ-expressing cells at the end of the first stage of alveologenesis arise from initially labeled ALFs. It should be noted that combination of the Gli1CreERT2 and Gli1LacZ alleles results in a homozygous knockout of Gli1 (Gli1CreERT2/LacZ), a transcription factor of the Hedgehog (HH) signaling pathway. Though Gli1−/− mice are viable and show no overt lung phenotype (Park et al., 2000), HH signaling is a positive regulator of αSMA expression and of proliferation in AMFs (Kugler et al., 2017). The combination of these two alleles in a lineage trace (Fig. 5G) could result in labeled AMFs that do not behave as wild type, complicating interpretation. Furthermore, Gli1 itself is a transcriptional target of GLI1, suggesting that homozygous removal of Gli1 could affect the levels of the Gli1LacZ reporter expression (Vokes et al., 2007). Given the differing starting mesenchymal populations in the two lineage-tracing experiments (Fgf18Lineage, AMFs; Gli1Lineage, AMFs and ALFs), these data, taken together, support a model in which most AMFs are cleared from the alveolus and most of the ALFs are retained.
Multiple immune cell populations phagocytose particles from labeled AMFs
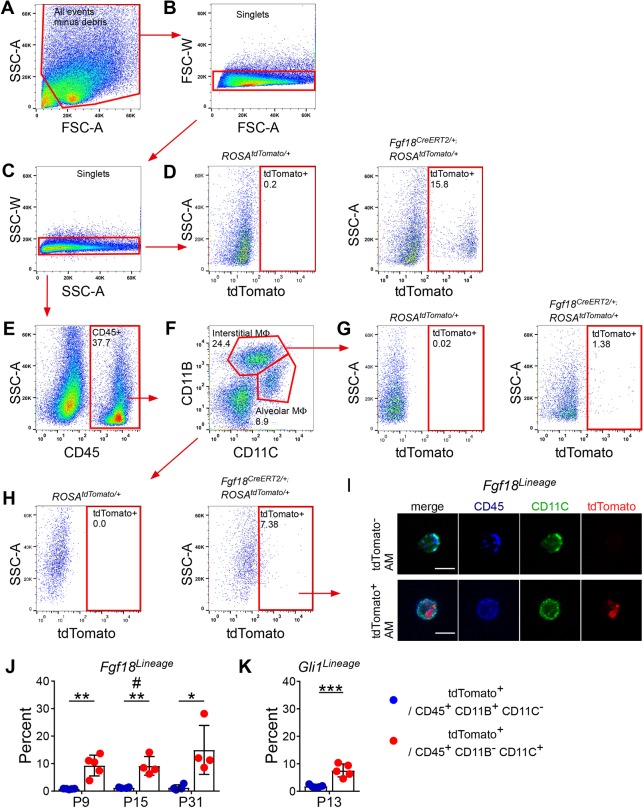
To determine whether AMFs undergo apoptosis, Fgf18Lineage-labeled lungs were assayed by TUNEL labeling. TUNEL labeling detected few (∼1%) apoptotic cells in P9 through adult time points, and very few of these TUNEL-positive cells were in the Fgf18Lineage in early postnatal timepoints and none were detected in the Fgf18Lineage at P21 and adult time points (Fig. S9A,B). Early stages of apoptosis can be detected by monitoring the translocation of phosphatidylserine to the outer cell membrane through labeling with Annexin V, a protein that specifically binds phosphatidylserine (Zhang et al., 1997). Annexin V labeling detected a larger percentage of early apoptotic cells compared with late apoptotic cells in the P9 Fgf18Lineage lungs (10±3.0%; Fig. S9C,D). As a positive control, incubation for 2-6 h with camptothecin, a topoisomerase inhibitor, was able to shift the early apoptotic population to a late apoptotic/necrotic population (data not shown). We next considered whether clearance of AMFs could be mediated through phagocytosis by macrophages (Li et al., 2003; Nagata, 2018; Schittny et al., 1998). We reasoned that phagocytosis of dying AMFs could be assayed through detection of tdTomato+ particles within macrophages in the lung. The intracellular pan-macrophage marker CD68 was used to identify lung macrophages. Immunostaining with CD68 identified Fgf18Lineage+ AMFs located adjacent to macrophages at P9 and P18, but not in adult lung (Fig. S10). Additionally, the membrane-bound pan-macrophage marker F4/80 identified macrophages that fully surround tdTomato+ Dapi+ puncta, indicative of phagocytosis of a Fgf18Lineage+ cell (Movie 1). To quantify phagocytosis of Fgf18Lineage-labeled cells, alveolar (AMΦ) and interstitial (IMΦ) macrophages were isolated by flow cytometry, and further sorted for cells that contain tdTomato+ particles at P9, P15 and P31 (Fig. 6A-H). Both IMΦ (CD45+, CD11B+ and CD11C−) and AMΦ (CD45+, CD11B− and CD11C+) were isolated that were also positive for the tdTomato fluorophore (Fig. 6G,H,J). Interestingly, the number of tdTomato-containing macrophages was relatively constant over time, with ∼1-2% of IMΦ and ∼7-10% of AMΦ being tdTomato positive (Fig. 6J). Confocal microscopy of sorted AMΦ revealed tdTomato+ particles within the positively scored cells and absent in the negatively scored cells (Fig. 6I). We also sorted Gli1Lineage lungs and confirmed the presence of tdTomato+ particles in IMΦs and AMΦs (Fig. 6K; Fig. S11).
Fig. 6.
Alveolar macrophages phagocytose particles from Fgf18CreERT2-labeled cells. (A-H) Fgf18CreERT2 ROSAtdTomato/+ mice were injected with Tam daily from P5 to P8 and collected on P9, P15 or P31 for fluorescence-activated cell sorting. Sorted cells were gated against (A) debris and (B,C) doublets. (D) Gating strategy to identify tdTomato+ cells, set on control ROSAtdTomato/+, mice showing enrichment in Fgf18CreERT2/+; ROSAtdTomato/+ mice. (E-H) Sequential gating strategy to identify populations of tdTomato+ interstitial macrophages (CD45+ CD11B+ CD11C−) and alveolar macrophages (CD45+ CD11B− CD11C+). (G,H; left) Fluorescence minus one (FMO) control stained with all antibodies but without the tdTomato fluorophore were used as a negative control. All representative plots were generated from P9 sorted cells except FMOs, which were generated from P31 sorted cells. (I) Image of pooled flow-sorted cells from P9 tdTomato− and tdTomato+ alveolar macrophages. (J,K) Quantification of the percentage of interstitial macrophages (CD45+ CD11B+ CD11C−) and alveolar macrophages (CD45+ CD11B− CD11C+) that were gated as tdTomato+ in (J) Fgf18CreERT2/+; ROSAtdTomato/+ mice and (K) Gli1CreERT2/+; ROSAtdTomato/+ mice induced from P5 to P8 and collected on P13. Student's t-test, *P<0.05; **P<0.01; ***P<0.001. Mann–Whitney, #u=0.029. n=5 (P9, P13), n=4 (P15, P31). Scale bars: 10 µm. Data are mean±s.d. MΦ, macrophage.
Our initial AMΦ antibody panel enriched for AMΦs but also contained dendritic cells. To confirm that true AMΦs were positive for tdTomato particles, Siglec-F, a marker that, together with CD11C, provides the most accurate identification of true alveolar macrophages, was used (Fig. S12) (Misharin et al., 2013). Both AMΦs (CD45+, CD11B−, CD11C+ and Siglec-F+) and dendritic cells (CD45+, CD11B−, CD11C+ and Siglec-F−) were isolated that were positive for the tdTomato fluorophore, suggesting that multiple phagocytic populations are able in ingest proteins from Fgf18Lineage+ cells.
DISCUSSION
Mesenchymal thinning is an essential component of postnatal lung development (Schittny, 2017). Generation of exquisitely thin alveolar walls is necessary to maximize gas diffusion between the atmosphere and blood. Though this phenomenon has long been appreciated, the kinetics of mesenchymal cell disappearance are just now being explored (Li et al., 2015, 2018; McGowan and McCoy, 2011). To address the differences in developmental trajectories of two major mesenchymal cell types in the lung, we employed combinatorial lineage tracing to ascertain that most AMFs are cleared from the lung by the conclusion of alveologenesis, while ALFs persist into adulthood.
Fgf18CreERT2 marks multiple cell lineages in the postnatal lung
It has been difficult to find a genetic marker that specifically targets AMFs due to the promiscuity of available lineage-tracing alleles and the lack of AMF-specific antibodies (Ahlfeld and Perl, 2017). Using an Fgf18CreERT2 allele (Hagan et al., 2019), we identified the cellular lineages expressing Fgf18 in situ during postnatal lung development. Fgf18CreERT2 abundantly labels cells in the lineage diverse alveolar region and contributes to most mesothelial and a few peribronchial smooth muscle cells. In lung mesenchyme, Fgf18CreERT2 is expressed almost exclusively in AMFs. In the alveolar region, Fgf18CreERT2 also labels ∼23% of AT1 cells and a small percentage of other cell types. These results were consistent with analysis of LungGENS scRNA-seq data of murine lung, which showed Fgf18 expression in AMFs and AT1 cells with minimal contribution to other mesenchymal cell types at P3 and more restricted expression to AMFs at P7 (Du et al., 2017, 2015). Fgf18CreERT2 labeling of AT1 cells between P5 and P8 suggests that it is marking AT1 cells just as they are turning off Fgf18 expression or that Fgf18 is lowly expressed in these cells and not detectable by scRNA-seq.
Analysis of scRNA-seq data of human lung at P1 showed that FGF18 expression in AMFs is conserved in humans. Notably, at this timepoint scRNA-seq could not distinguish AMFs from other smooth muscle lineages. As there is minimal contribution of the Fgf18Lineage to alveolar lipofibroblasts, peribronchial and perivascular smooth muscle, we propose that Fgf18 is a potential lineage marker that could be used to identify AMFs.
The finding that the Fgf18Lineage includes a subset of AT1 cells was unexpected but suggests a potential role for AT1 cells in regulating alveologenesis through production of FGF18. Additionally, there is heterogeneity of the AT1 lineage based on Igfbp2 expression with functional differences in regeneration capacity after injury (Wang et al., 2018). Interestingly, Fgf18Lineage-labeled AT1 cells at P1 increase in proportion throughout the first phase of alveolar development (Fig. S6E). Although this could be solely due to residual Tam, it seems unlikely, as an increase was seen between the P7 and P21 collection timepoints. Future studies could determine whether Fgf18CreERT2 also marks a functionally distinct AT1 subtype during lung development.
Most AMFs but not ALFs are cleared from the lung by the end of the first phase of alveolar development
Using lineage tracing, we show that most Fgf18Lineage+; Gli1+ AMFs are rapidly cleared from the lung by P21. Previously, Gli1CreERT2 was used to lineage trace AMFs with the conclusion that few cells remain into adulthood (Li et al., 2015). Although the conclusion from this study is consistent with our observed clearance of Fgf18Lineage-labeled AMFs, we found that Gli1CreERT2 (and Gli1LacZ) additionally mark ALFs, and that ALFs persist into adulthood. Differences in the labeled starting populations are likely due to subtle differences in experimental design. Our labeling scheme used a greater number of Tam induction days, a more-sensitive ROSA reporter (ROSAtdTomato versus ROSAmTmG) (Liu et al., 2013a), and visualization of endogenous fluorescence, as opposed to an antibody detection intermediate step, all likely contributing to marking a greater proportion of cells (Liu et al., 2013a). Kugler et al. (2017) showed that Gli1LacZ marks a more inclusive mesenchymal population (interstitial fibroblasts and AMFs) during alveolar development, supporting our observed labeling of ALFs. Additionally, Moiseenko et al. (2017) showed that embryonic induction of Gli1CreERT2 marks ALFs, suggesting that the ability of the Gli1CreERT2 allele to label only AMFs is dependent on the timing of induction. Furthermore, analysis of LungGENS scRNA-seq data identified Gli1 expression in AMFs as well as other mesenchymal cell types during alveologenesis, supporting our conclusion that Gli1CreERT2 marks multiple mesenchymal cell types.
A PdgfrartTA allele has also been used to lineage trace AMFs (Li et al., 2018). This study showed that labeled cells persisted into the adult and concluded that AMFs downregulate their smooth muscle markers and persist as an undifferentiated mesenchymal cell. However, based on our analysis, we hypothesize that the PdgfrartTA lineage trace is marking an additional mesenchymal cell outside of the AMF lineage due to the following reasons: (1) this study induced the lineage trace over a long period of time (P0-20); (2) the identity of the marked cells at the end of the labeling period was not defined; (3) other Pdgfra alleles have been shown to mark both AMFs and ALFs to varying degrees (Endale et al., 2017; Ntokou et al., 2015); and (4) LungGENS scRNA-seq data shows that Pdgfra is expressed in both AMFs and matrix fibroblasts (which contain ADRP-expressing cells) at the time when the lineage trace was induced.
Our Gli1Lineage data show that ALFs are labeled initially, retained in the adult, continue to express ADRP and express Gli1LacZ. In the adult, mesenchymal cells are located in the anatomical location of the developmental AMFs, and are associated with the extracellular matrix proteins elastin and collagen (Yamada et al., 2005). Our data suggest that these remaining mesenchymal cells are ALFs. Lineage tracing with the recently developed Tcf21mCrem or Plin2CreERT2 alleles that mark ALFs (Ntokou et al., 2017; Park et al., 2019) could be informative; however, both of these lineage alleles are also expressed in other cell types.
The promiscuity of genetic mesenchymal markers that label both AMFs and ALFs is thought to be due to the plasticity of the precursor cell type. This is best illustrated through injury models. In an adult pneumonectomy model, PdgfraeGFP+ cells label αSMA− cells (likely ALFs), which upregulate αSMA, becoming more ‘myofibroblast like’ during alveolar regeneration (Green et al., 2016; Perl and Gale, 2009). Additionally, adult ALFs give rise to pathogenic αSMA+ myofibroblasts during bleomycin-induced fibrosis and then revert back to αSMA− ADRP+ ALFs (El Agha et al., 2017). Neonatal hypoxia leads to an increase of myofibroblasts and a decrease of ALFs suggesting transdifferentiation in a model of BPD (Li et al., 2018). In normal development, plasticity between AMFs and ALFs is speculated (Branchfield et al., 2016). Our data indicate that during alveologenesis these lineages are distinct, with no ability of Fgf18CreERT2-labeled AMFs to adopt an ALF phenotype.
Multiple immune populations phagocytose lineage-labeled particles
Although we were unable to observe a significant amount of apoptosis in the AMF lineage using the TUNEL assay, it is likely that these cells are still undergoing apoptosis, then are rapidly cleared and thus not efficiently detected. Phosphatidylserine is the most common signal presented on the surface of an apoptotic cell to allow for recognition by a phagocyte (Fadok et al., 1992) and is present earlier in the apoptosis pathway than DNA fragmentation (Zhang et al., 1997). Supporting this, ∼10% of Fgf18Lineage cells at P9 are labeled by Annexin V, a protein that binds phosphatidylserine. Inability to detect apoptotic αSMA+ AMFs in tissue sections is consistent with other reports (Yamada et al., 2005). Furthermore, clearance of apoptotic cells by macrophages is efficient and quick in normal tissue (Surh and Sprent, 1994). Our results suggest that AMFs are undergoing apoptosis and are rapidly cleared from the lung. Flow cytometry data shows that from an early time in postnatal lung development (P9), AMΦs and dendritic cells are positive for the phagocytosed tdTomato lineage marker. The lack of temporal dynamics in the number of tdTomato+ AMΦ when the underlying population of AMFs are changing suggests that the tdTomato particles are long lived within the phagocyte or there is a contribution from the AT1 labeled cells. Although Fgf18Lineage and Gli1Lineage both mark multiple cell types, the observation that tdTomato particles are seen in AMΦ in both experiments strongly suggests that the AMF is a common source of these phagocytosed particles. It is interesting that tdTomato particles are preferentially in AMΦs and dendritic cells compared with IMΦs. This is counterintuitive based on macrophage subpopulations anatomic locations. IMΦs are located near AMFs in the interstitium while AMΦs are in the alveolar lumen separated from AMFs by an epithelial barrier. Dendritic cells are located throughout the respiratory tract (Condon et al., 2011). tdTomato particles in AMΦs suggest that dying/dead AMFs are extruded into the alveolar lumen or that AMΦs are crossing into the interstitium.
Mac2+ macrophages are known to migrate from the interstitium to the alveolar lumen in the first week of postnatal life (Jones et al., 2013; Tan and Krasnow, 2016). Little is known about AMΦ function during alveologenesis. Our data suggest that they have a novel function in the clearance of AMFs. Macrophages in other tissues are involved in developmental tissue remodeling; however, ablation studies in the lung note no developmental phenotype resulting from the removal of AMΦs (Kalymbetova et al., 2018; Lang and Bishop, 1993; Shibata et al., 2001). It is also possible that the role of AMΦs in lung development is redundant or compensated for by other phagocytic cells, including interstitial macrophages and dendritic cells.
Limitations
A limitation of our methods is the reliance on cell counts generated largely from histological sections normalized to total cellular number. Using these methods, we were not able to distinguish an absolute decrease in a cell population of interest from a relative decrease due to proliferation of other cell populations. The decrease in the proportion of AMFs in the lung may be due to proliferation of other cell types, but this is unlikely to be the sole explanation, as terminally differentiated labeled AT1 cells were seen throughout the alveolar region and their population was stable from P21 to the adult. Moreover, during alveologenesis, Fgf18Lineage AMFs were seen lining most alveoli; however, in the adult there were many alveoli that completely lacked non-epithelial Fgf18Lineage cells. A further limitation was the reliance on tamoxifen induction to label cells. Although the half-life of tamoxifen is only ∼10 h it has been shown to mediate recombination for several days or weeks after the final administration (Jahn et al., 2018; Reinert et al., 2012). Our results showing an increase in the total numbers of tdTomato+-labeled AT1 cells without any detectable proliferation, suggest that our tamoxifen regimen is continuing to mediate recombination for at least a few days after the final injection on P8. However, this does not impact the conclusion that most Fgf18Lineage+ AMFs are cleared from the lung (as this would only increase labeling), but it does allow for the possibility that the Gli1Lineage is marking additional Gli1CreERT2-expressing cells after the final tamoxifen injection on P8.
Conclusion
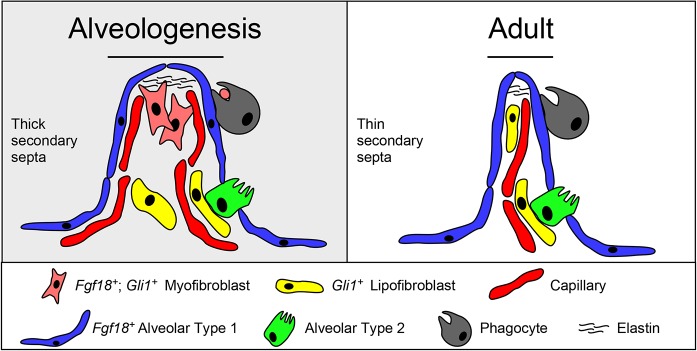
This study demonstrates that the AMF lineage is depleted during alveolar septal wall thinning involving a phagocytic process, providing a mechanism for clearance of a transient developmental cell population (Fig. 7). This phenomenon is potentially relevant to other systems where developmentally important cell types are absent in the adult. Understanding the signals that control this clearance may be important to understanding pathogenic mechanisms in human lung disease, such as bronchopulmonary dysplasia, where abnormally thickened alveolar walls are a pathological outcome (Coalson, 2006; Thibeault et al., 2003). Future studies will be needed to define the molecular mechanisms initiating the depletion of AMFs and how they might contribute to pediatric lung diseases.
Fig. 7.
Model for mesenchymal dynamics during alveologenesis. (Left) During the first phase of alveologenesis, the distal lung is characterized by having a thick, immature secondary septa that contains alveolar myofibroblasts, alveolar lipofibroblasts and a double-wall capillary plexus. The heterogenous mesenchymal cells are thought to be necessary for construction of the elastin-rich extracellular matrix. (Right) At the end of the second phase of alveologenesis, the distal lung is characterized by having a thin, fully mature secondary septa containing alveolar lipofibroblasts, but not alveolar myofibroblasts, accompanied by a resolution of the capillary plexus. Monocyte-derived phagocytes (alveolar macrophages and dendritic cells) are present in the lung throughout alveologenesis and contain lineage-labeled particles from alveolar myofibroblasts at least through postnatal day 31, suggesting a role in removal of dead alveolar myofibroblasts through a phagocytic process.
MATERIALS AND METHODS
Mice
All mice (Mus musculus) were housed in a pathogen-free barrier facility. All studies were performed under a protocol approved by the Institutional Animal Care at Washington University in St Louis (Approval No. 20160013). Mice of both sexes were used. Mice were maintained on a mixed C57BL/6J×129X1/SvJ genetic background. Mouse strains, including Fgf18CreERT2, ROSAtdTomato, PdgfraeGFP, Gli1CreERT2 and Gli1LacZ have been previously described (Ahn and Joyner, 2004; Bai et al., 2002; Hagan et al., 2019; Hamilton et al., 2003; Madisen et al., 2010). Postnatal day 0 (P0) was assigned as the day of birth. Adult mice were 8 weeks of age or older.
Tamoxifen administration
Unless otherwise stated, tamoxifen was administered to neonates by intraperitoneal injection at a dose of 150 µg on four sequential days beginning at postnatal day 5 (P5). Tamoxifen (Sigma; T5648) was prepared at a concentration of 20 µg/µl dissolved in corn oil or sunflower seed oil.
Sample preparation
At designated collection times, mice were sacrificed with an overdose of a cocktail containing ketamine and xylazine, perfused with PBS through the right ventricle, and the lungs were fixed via intratracheal inflation with 10% neutral buffered formalin (VWR; 89370) at a pressure of 20 cm H2O or were inflated with a syringe at earlier timepoints. Lungs were fixed overnight at 4°C with gentle agitation. For samples that contained the Gli1LacZ allele, lungs were fixed for 1.5 h on ice. Samples were washed twice in PBS, cut into lobes, and cryoprotected in 15% sucrose in PBS overnight at 4°C with gentle agitation and then in 30% sucrose overnight at 4°C with gentle agitation. Lungs were patted dry, equilibrated in Tissue-Tek OCT Compound (VWR; 4583) for 20 min, embedded and frozen on dry ice. Generally, the left lobe was used for analysis. Frozen sections were cut at 6 µm with a cryostat, dried at room temperature and stored at −80°C until use.
Immunofluorescence
Slides were warmed for 10 min at room temperature and washed in PBS before being permeabilized in PBS+0.5% Triton X-100 (Sigma, X-100) for 15 min. Sections were blocked using PBS+0.1% Tween 20 (Sigma, P9416) and 5% donkey serum for 20 min. Primary antibodies were diluted in blocking buffer, applied to tissue, covered with parafilm and incubated overnight at 4°C in a humidified chamber. Sections were washed three times in PBS. Secondary antibody was diluted into PBS+0.1% Tween 20, applied to tissue, covered with parafilm and incubated for 1 h at room temperature. Slides were washed three times in PBS, mounted in Vectashield antifade mounting medium with DAPI (ThermoFisher; NC9524612), sealed with nail polish and stored at 4°C until imaged.
Antibodies
The antibodies used were: Alexa Fluor-conjugated secondary antibodies (1:300, ThermoFisher), mouse anti-αSMA conjugated to FITC (1:100, Sigma, F3777) (El Agha et al., 2017), mouse anti-αSMA (1:100, Dako; M0851) (Guzy et al., 2017), chicken anti-β-galactosidase (1:500, Abcam; ab9361) (Huh and Ornitz, 2010), rabbit anti-HOPX (1:200, Santa Cruz; sc-30216) (Barkauskas et al., 2013), mouse anti-HOPX conjugated to Alexa-647 (1:100, Santa Cruz; sc398703) (Volckaert et al., 2019), goat anti-SP-C (1:1000, Santa Cruz, sc-7706) (Chung and Hogan, 2018), rabbit anti-ADFP (1:100, Abcam, ab52356) (El Agha et al., 2017), goat anti-WT1 (1:100, Santa Cruz, sc-15421) (Lee et al., 2017), rat anti-CD31 (1:50, Dianova; DIA-310) (Oladipupo et al., 2018), rabbit anti-NG2 (1:200, MilliporeSigma; ab5320) (Kato et al., 2018), rat anti-CD68 (1:200, Bio-Rad, MCA1957) (Menon and Fisher, 2015), rat anti-F4/80 (1:100, Bio-Rad, MCA497R) (Hasan et al., 2013) and rabbit anti-Desmin (1:100, Abcam, ab8592) (Walton et al., 2016).
Cell proliferation assay
For short-term proliferation analysis EdU (GoldBio; E-980-100) was administered to neonates by intraperitoneal injection at a dose of 100 µg per gram body weight. Tissue was harvested 3 h after injection. For cumulative proliferation analysis, EdU was injected i.p. at a dose of 100 µg per gram body weight daily from P9-P21. EdU was visualized by the Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Thermo Fisher, C10640).
Cell death assay
Frozen sections were prepared as described above. Slides were assayed by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) using the DeadEnd Fluorometric TUNEL system (Promega; G3250).
Flow cytometry
Mice were sacrificed with an overdose of a cocktail containing ketamine and xylazine, perfused with PBS through the right ventricle and the lungs were inflated with a digest media containing 50 units/ml dispase (Beckton Dickinson; 354235) and 0.01 mg/ml DNase I (Roche; 10104159001). Inflated lungs were transferred to a digest media containing 100 mg/ml collagenase/dispase (Roche, 10269638) and 0.01 mg/ml DNase I, and stored on ice until all mice were dissected. Lungs were minced into small pieces using clean razor blades and digested for 45 min at 37°C with gentle agitation. Digestion enzymes were inactivated with FACS buffer (PBS+10% FBS+1 mM EDTA), samples were filtered through a 100 µM cell strainer and then a 40 µM cell strainer. Red blood cells were lysed with ACK lysis buffer (Thermo Fisher, A1049201) and cells were resuspended in FACS buffer. Non-specific binding of Fc receptors was blocked using TruStain FcX (1:50, BioLegend; 101319) (Ge et al., 2010).
Sorting of macrophage expressing tdTomato particles
Cells were stained with anti-CD45 conjugated to BV421 (1:100, BioLegend, 103133) (Podd et al., 2006), anti-CD11B conjugated to APC/Cy7 (1:100, BioLegend, 101225) (Glass et al., 2013) and anti-CD11C conjugated to Alexa 647 (1:100, BioLegend, 117314) (Collins et al., 2016). BV421+; APC/Cy7+; Alexa 647− cells were identified as interstitial macrophages. BV421+; APC/Cy7−; Alexa 647+ cells were identified as alveolar macrophages. Cells were sorted using a SY3200 Synergy cell sorter (Sony) with a 100 µM nozzle directly into FACS buffer on ice. Four cell populations were collected, the interstitial and alveolar macrophages with or without tdTomato fluorescence. Single channel controls were used to set compensation at time of sort and fluorescence minus one control (FMO, lacking tdTomato) was used as negative control (collected at P15 and P31 only). Plotted percentage of tdTomato-positive cells in each population was taken directly from the sorted number of cells. A minimum of 50,000 cells were sorted per sample for each of the alveolar and interstitial macrophage populations. To distinguish between alveolar macrophages and dendritic cells, samples, prepared as described above, were additionally sorted based on binding to Siglec-F conjugated to PerCP-Cy5.5 (1:100, BD Biosciences, 565526) (Hernández-Santos et al., 2018). Statistics on Siglec-F sort were generated using FlowJo software (Beckton Dickinson). Representative plots were generated using FlowJo software (Beckton Dickinson). Sorted cells were pooled from all timepoint matched mice, pelleted, fixed in 4% PFA, resuspended in FACS buffer and stored at 4°C. Slides, if necessary, were generated by centrifugation at 800 g and resuspending in a small volume of Vectashield antifade mounting medium without DAPI (ThermoFisher, H1000NB), sealed with nail polish and stored at 4°C until imaged. Cells were visualized for BV421, Alexa 647 and tdTomato fluorescence.
Annexin V apoptosis assay
Cells were prepared as described above. Cells were stained with Annexin V conjugated to APC (1:20, Biolegend, 640919) (Cao et al., 2016) and Zombie Violet viability dye (Biolegend, 77477) in Annexin V binding buffer (Biolegend, 422201) according to the manufacturer's recommendation. Cells were analyzed on a FACScan flow cytometer (BD Bioscience). Single channel controls were used to set compensation after analysis. A positive control was generated by incubation of cells with 20 µM camptothecin (Sigma, C9911) for 2-6 h at 37°C (n=1). Compensation, statistics and representative plots were generated using FlowJo software (Beckton Dickinson).
Sorting of mesenchymal cells expressing tdTomato, RNA isolation and quantitative real-time PCR
Endothelial and hematopoietic cells were depleted by magnetic bead separation using Dynabeads sheep anti-Rat IgG (Thermo Fisher; 11035) precoated with anti-CD31 (BioLegend, 102404) and anti-CD45 (BioLegend, 103101). Remaining cells were stained with anti-PDPN conjugated to APC/Cy7 (BioLegend, 127417). Pre-sort control cells were taken at this point and stored at 4°C until RNA was extracted. tdTomato+; APC/Cy7− cells were sorted using a Sony SY3200 Synergy cell sorter with a 100 µM nozzle directly into ice-cold FACS buffer.
Cells collected by flow cytometry and pre-sort control cells were spun down at 800 g and RNA was isolated using the Picopure RNA Isolation Kit (Thermo Fisher, KIT0204) with on-column DNA digest. cDNA was synthesized using Bio-Rad iScript Reverse Transcription Supermix (Bio-Rad; 1708840). mRNA expression was determined on a StepOnePlus Real-Time PCR System (Thermo Fisher, 4376600) using TaqMan Fast Advanced Master Mix (Thermo Fisher, 4444557) and Taqman assay probes. Samples were run in technical triplicates with mean Ct value used. If a technical triplicate differed greater than 0.5Ct from the mean Ct that replicate was discarded, and the mean was recalculated from the two remaining technical replicates. Samples were first run with Gapdh to determine relative RNA concentrations and cDNA was diluted to standardize input concentration. mRNA levels of experimental genes were normalized to Gapdh using the standard ΔCt method and data were reported as fold change relative to pre-sort control.
Taqman assay probes
Taqman assay probes used were Gapdh (Thermo Fisher, Mm99999915_g1), Fgf18 (Thermo Fisher, Mm00433286_m1), Fgfr3 (Thermo Fisher, Mm00433294_m1), Fgfr4 (Thermo Fisher, Mm00433314_m1), Aqp5 (Thermo Fisher, Mm00437578_m1), Pdpn (Thermo Fisher, Mm01348912_g1), Pgfra (Thermo Fisher, Mm00440701_m1), Gli1 (Thermo Fisher, Mm00494654_m1) and Acta2 (Thermo Fisher, Mm00725412_m1).
Single cell RNA sequencing data analyses
To assess cell type-specific expression of endogenous Fgf18 and Gli1 in the postnatal mouse and human lung, the online database LungGENS was used. The mouse P3 and P7 Drop-seq dataset was downloaded from LungGENS. Cells with fewer than 500 expressed genes were removed and genes expressed in fewer than 30 cells were excluded from analyses. R package Seurat V3.0 (Satija et al., 2015) was used to cluster and visualize cells. The clustering of major cell types were calculated using the first 10 principal components, and then visualized by the uniform manifold approximation and projection (UMAP) feature reduction method (Becht et al., 2019). Cell clusters were identified based on marker gene expression. Mesenchymal cells express Col1a1, AT1 cells express Aqp5 and HopX, AMFs express high levels of Acta2, and matrix fibroblasts express Tcf21 and Fgf10. Subcluster analysis on P7 data was performed on Acta2 high clusters 1, 10 and 11. Images for human P1 FGF18 expression were generated directly from the Lung Gene Expression Analysis (LGEA) Web Portal (research.cchmc.org/pbge/lunggens last accessed 05/2019).
Imaging
Most fluorescent microscopy was performed using a Zeiss LSM-700 Confocal Microscope and ZEN software (single plane, unless otherwise noted) (Carl Zeiss). Whole-lobe fluorescent images were analyzed using a NanoZoomer 2.0HT slide scanning system with a 40× objective (Hamamatsu). Whole-lobe images were processed in NanoZoomer Digital Pathology (NDP.view) software or ImageJ. In all cases eGFP and tdTomato were detected by native fluorescence. Images are representative of at least three mice unless otherwise stated.
Measurements and cell quantification
Cell quantifications were made using ImageJ (Cell Counter plug-in by Dr Kurt De Vos). Unless otherwise stated, five 20× images were used to count the ratio of labeled cells. Alveolar regions were defined as distal regions of the lung that contained no airways or large vessels, and included cells at the alveolar tips and within the alveolar walls. Quantification of lineage reporter was always dependent on reporter colocalizing with nuclear DAPI staining for scoring as positive. Quantification of variability of Fgf18Lineage in the peribronchial region was calculated by quantifying five 20× images located on large airways in three mice with n=15 (individual airways). Quantification of the non-AT1 lineage remaining (Fig. 3H) was calculated directly from the percentage of tdTomato+/DAPI+ cells (Fig. 3B) at P9, P21 and adult, with the percentage of AT1-labeled cells subtracted from each sample (Fig. 3G). Quantification of the mesenchymal lineage remaining (Fig. 3F) repeats the Fgf18 data from Fig. 3H but is transformed with P9 set as the reference (100%) to allow comparison with the Gli1 data. Gli1 data is repeated from Fig. 5D but transformed with P9 set as the reference (100%) to allow comparison with the Fgf18 data. The entire Gli1 lineage was considered mesenchymal as there was no contribution to other lineages.
Experimental design, statistical analysis and plotting
Sample size was defined based on our previous experiments. Sample size (n) represents the number of mice unless otherwise noted. Significant differences in mean values between two sets of data were calculated by using a two-tailed t-test. If data were not normally distributed by a Shapiro–Wilk test, a Mann–Whitney test was also used. # indicates Mann–Whitney test was performed, u value is reported in the figure legend. Significant differences in mean values between more than two sets of normally distributed data were calculated using a one-way ANOVA with Tukey's HSD for pair-wise post-hoc analysis. Data are represented as mean±s.d. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. All figures were made in Canvas X (Canvas GFX). Plots were generated in Prism 7 (GraphPad Software).
Supplementary Material
Acknowledgements
We thank Drs Brody, Woo, Yin and Yang for critically reading the manuscript; W. Lewis for animal caretaking; D. Schweppe for help with flow cytometry. Whole slide imaging was supported by the Hope Center Alafi Neuroimaging Core at Washington University (NIH Shared Instrumentation Grant 1S10RR027552). We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St Louis, MO, for the use of the Siteman Flow Cytometry core. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant (P30 CA091842).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.S.H., D.M.O.; Methodology: A.S.H., D.M.O.; Formal analysis: A.S.H., B.Z.; Investigation: A.S.H.; Resources: D.M.O.; Writing - original draft: A.S.H., D.M.O.; Writing - review & editing: A.S.H., D.M.O.; Supervision: D.M.O.; Funding acquisition: D.M.O.
Funding
This work was supported by the National Institutes of Health (R01HL111190 to D.M.O.) and the American Heart Association (16PRE26960002 and 18PRE34030091 to A.S.H.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.181032.supplemental
References
- Ahlfeld S. K. and Conway S. J. (2012). Aberrant signaling pathways of the lung mesenchyme and their contributions to the pathogenesis of bronchopulmonary dysplasia. Birth Defects Res. A Clin. Mol. Teratol 94, 3-15. 10.1002/bdra.22869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfeld S. K. and Perl A. K. (2017). A “GLI-tch” in alveolar myofibroblast differentiation. Am. J. Respir. Cell Mol. Biol. 57, 261-262. 10.1165/rcmb.2017-0148ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. and Joyner A. L. (2004). Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505-516. 10.1016/j.cell.2004.07.023 [DOI] [PubMed] [Google Scholar]
- Bai C. B., Auerbach W., Lee J. S., Stephen D. and Joyner A. L. (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753-4761. [DOI] [PubMed] [Google Scholar]
- Barkauskas C. E., Cronce M. J., Rackley C. R., Bowie E. J., Keene D. R., Stripp B. R., Randell S. H., Noble P. W. and Hogan B. L. M. (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025-3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra H. and Antony V. B. (2015). Pleural mesothelial cells in pleural and lung diseases. J. Thorac. Dis. 7, 964-980. 10.3978/j.issn.2072-1439.2015.02.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F. and Newell E. W. (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38-44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bellusci S., Grindley J., Emoto H., Itoh N. and Hogan B. L. (1997). Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124, 4867-4878. [DOI] [PubMed] [Google Scholar]
- Boström H., Willetts K., Pekny M., Levéen P., Lindahl P., Hedstrand H., Pekna M., Hellström M., Gebre-Medhin S., Schalling M. et al. (1996). PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85, 863-873. 10.1016/S0092-8674(00)81270-2 [DOI] [PubMed] [Google Scholar]
- Boucherat O., Benachi A., Barlier-Mur A.-M., Franco-Montoya M.-L., Martinovic J., Thébaud B., Chailley-Heu B. and Bourbon J. R. (2007). Decreased lung fibroblast growth factor 18 and elastin in human congenital diaphragmatic hernia and animal models. Am. J. Respir. Crit. Care. Med. 175, 1066-1077. 10.1164/rccm.200601-050OC [DOI] [PubMed] [Google Scholar]
- Branchfield K., Li R., Lungova V., Verheyden J. M., McCulley D. and Sun X. (2016). A three-dimensional study of alveologenesis in mouse lung. Dev. Biol. 409, 429-441. 10.1016/j.ydbio.2015.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce M. C., Honaker C. E. and Cross R. J. (1999). Lung Fibroblasts Undergo Apoptosis Following Alveolarization. Am. J. Respir. Cell Mol. Biol. 20, 228-236. 10.1165/ajrcmb.20.2.3150 [DOI] [PubMed] [Google Scholar]
- Cao W., Guo J., Wen X., Miao L., Lin F., Xu G., Ma R., Yin S., Hui Z., Chen T. et al. (2016). CXXC finger protein 1 is critical for T-cell intrathymic development through regulating H3K4 trimethylation. Nat. Commun. 7, 11687 10.1038/ncomms11687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailley-Heu B., Boucherat O., Barlier-Mur A.-M. and Bourbon J. R. (2005). FGF-18 is upregulated in the postnatal rat lung and enhances elastogenesis in myofibroblasts. Am. J. Physiol. Lung Cell Mol. Physiol. 288, L43-L51. 10.1152/ajplung.00096.2004 [DOI] [PubMed] [Google Scholar]
- Choi C. W., Kim B. I., Mason S. N., Potts-Kant E. N., Brahmajothi M. V. and Auten R. L. (2013). Intra-amniotic LPS amplifies hyperoxia-induced airway hyperreactivity in neonatal rats. Pediatr. Res. 74, 11-18. 10.1038/pr.2013.58 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chung M.-I. and Hogan B. L. M. (2018). Ager-CreERT2: a new genetic tool for studying lung alveolar development, homeostasis, and repair. Am. J. Respir. Cell Mol. Biol. 59, 706-712. 10.1165/rcmb.2018-0125OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coalson J. J. (2006). Pathology of Bronchopulmonary Dysplasia. Semin. Perinatol. BPD: State of the Art 30, 179-184. 10.1053/j.semperi.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Collins N., Jiang X., Zaid A., Macleod B. L., Li J., Park C. O., Haque A., Bedoui S., Heath W. R., Mueller S. N. et al. (2016). Skin CD4+ memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat. Commun. 7, 11514 10.1038/ncomms11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin J. S., White A. C., Pratt S. J. and Ornitz D. M. (2001). Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 128, 2095-2106. [DOI] [PubMed] [Google Scholar]
- Condon T. V., Sawyer R. T., Fenton M. J. and Riches D. W. H. (2011). Lung dendritic cells at the innate-adaptive immune interface. J. Leukoc. Biol. 90, 883-895. 10.1189/jlb.0311134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Guo M., Whitsett J. A. and Xu Y. (2015). “LungGENS”: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax 70, 1092-1094. 10.1136/thoraxjnl-2015-207035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Kitzmiller J. A., Sridharan A., Perl A. K., Bridges J. P., Misra R. S., Pryhuber G. S., Mariani T. J., Bhattacharya S., Guo M. et al. (2017). Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72, 481-484. 10.1136/thoraxjnl-2016-209598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E., Herold S., Alam D. A., Quantius J., MacKenzie B., Carraro G., Moiseenko A., Chao C.-M., Minoo P., Seeger W. et al. (2014). Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development 141, 296-306. 10.1242/dev.099747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E., Moiseenko A., Kheirollahi V., De Langhe S., Crnkovic S., Kwapiszewska G., Szibor M., Kosanovic D., Schwind F., Schermuly R. T. et al. (2017). Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 20, 261-273.e3. 10.1016/j.stem.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endale M., Ahlfeld S., Bao E., Chen X., Green J., Bess Z., Weirauch M. T., Xu Y. and Perl A. K. (2017). Temporal, spatial, and phenotypical changes of PDGFRα expressing fibroblasts during late lung development. Dev. Biol. 425, 161-175. 10.1016/j.ydbio.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok V. A., Voelker D. R., Campbell P. A., Cohen J. J., Bratton D. L. and Henson P. M. (1992). Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148, 2207-2216. [PubMed] [Google Scholar]
- Franco-Montoya M.-L., Boucherat O., Thibault C., Chailley-Heu B., Incitti R., Delacourt C. and Bourbon J. R. (2011). Profiling target genes of FGF18 in the postnatal mouse lung: possible relevance for alveolar development. Physiol. Genomics 43, 1226-1240. 10.1152/physiolgenomics.00034.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. N., Bahaie N. S., Kang B. N., Hosseinkhani M. R., Ha S. G., Frenzel E. M., Liu F.-T., Rao S. P. and Sriramarao P. (2010). Allergen-induced airway remodeling is impaired in galectin-3–deficient mice. J. Immunol. 185, 1205-1214. 10.4049/jimmunol.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass A. M., Wolf B. J., Schneider K. M., Princiotta M. F. and Taffet S. M. (2013). Connexin43 is dispensable for phagocytosis. J. Immunol. 190, 4830-4835. 10.4049/jimmunol.1202884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia L., Betsholtz C. and Andrae J. (2017). Expression analysis of platelet-derived growth factor receptor alpha and its ligands in the developing mouse lung. Physiol. Rep. 5, e13092 10.14814/phy2.13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Endale M., Auer H. and Perl A.-K. T. (2016). Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor α kinase activity. Am. J. Respir. Cell Mol. Biol. 54, 532-545. 10.1165/rcmb.2015-0095OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Du Y., Gokey J. J., Ray S., Bell S. M., Adam M., Sudha P., Perl A. K., Deshmukh H., Potter S. S. et al. (2019). Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat. Commun. 10, 37 10.1038/s41467-018-07770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy R. D., Li L., Smith C., Dorry S. J., Koo H. Y., Chen L. and Ornitz D. M. (2017). Pulmonary fibrosis requires cell-autonomous mesenchymal fibroblast growth factor (FGF) signaling. J. Biol. Chem. 292, 10364-10378. 10.1074/jbc.M117.791764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan A. S., Boylan M., Smith C., Perez-Santamarina E., Kowalska K., Hung I. H., Lewis R. M., Hajihosseini M. K., Lewandoski M. and Ornitz D. M. (2019). Generation and validation of novel conditional flox and inducible Cre alleles targeting fibroblast growth factor 18 (Fgf18). Dev. Dyn. 248, 882-893. 10.1002/dvdy.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton T. G., Klinghoffer R. A., Corrin P. D. and Soriano P. (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 23, 4013-4025. 10.1128/MCB.23.11.4013-4025.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S. A., Eksteen B., Reid D., Paine H. V., Alansary A., Johannson K., Gwozd C., Goring K.-A. R., Vo T., Proud D. et al. (2013). Role of IL-17A and neutrophils in fibrosis in experimental hypersensitivity pneumonitis. J. Allergy Clin. Immunol. 131, 1663-1673.e5. 10.1016/j.jaci.2013.01.015 [DOI] [PubMed] [Google Scholar]
- Hernández-Santos N., Wiesner D. L., Fites J. S., McDermott A. J., Warner T., Wüthrich M. and Klein B. S. (2018). Lung epithelial cells coordinate innate lymphocytes and immunity against pulmonary fungal infection. Cell Host Microbe 23, 511-522.e5. 10.1016/j.chom.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.-H. and Ornitz D. M. (2010). β-catenin deficiency causes DiGeorge syndrome-like phenotypes through regulation of Tbx1. Development 137, 1137-1147. 10.1242/dev.045534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.-H., Närhi K., Lindfors P. H., Häärä O., Yang L., Ornitz D. M. and Mikkola M. L. (2013). Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes Dev. 27, 450-458. 10.1101/gad.198945.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S.-H., Warchol M. E. and Ornitz D. M. (2015). Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. eLife 4, e05921 10.7554/eLife.05921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn H. M., Kasakow C. V., Helfer A., Michely J., Verkhratsky A., Maurer H. H., Scheller A. and Kirchhoff F. (2018). Refined protocols of tamoxifen injection for inducible DNA recombination in mouse astroglia. Sci. Rep. 8, 5913 10.1038/s41598-018-24085-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Barkauskas C. E., Takeda N., Bowie E. J., Aghajanian H., Wang Q., Padmanabhan A., Manderfield L. J., Gupta M., Li D. et al. (2015). Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 6, 6727 10.1038/ncomms7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. V., Williams T. M., Walker K. A., Dickinson H., Sakkal S., Rumballe B. A., Little M. H., Jenkin G. and Ricardo S. D. (2013). M2 macrophage polarisation is associated with alveolar formation during postnatal lung development. Respir. Res. 14, 41 10.1186/1465-9921-14-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalymbetova T. V., Selvakumar B., Rodríguez-Castillo J. A., Gunjak M., Malainou C., Heindl M. R., Moiseenko A., Chao C.-M., Vadász I., Mayer K. et al. (2018). Resident alveolar macrophages are master regulators of arrested alveolarization in experimental bronchopulmonary dysplasia. J. Pathol. 245, 153-159. 10.1002/path.5076 [DOI] [PubMed] [Google Scholar]
- Kapanci Y., Desmouliere A., Pache J. C., Redard M. and Gabbiani G. (1995). Cytoskeletal protein modulation in pulmonary alveolar myofibroblasts during idiopathic pulmonary fibrosis. Possible role of transforming growth factor beta and tumor necrosis factor alpha. Am. J. Respir. Crit. Care. Med. 152, 2163-2169. 10.1164/ajrccm.152.6.8520791 [DOI] [PubMed] [Google Scholar]
- Kaplan N. B., Grant M. M. and Brody J. S. (1985). The lipid interstitial cell of the pulmonary alveolus. Age and species differences. Am. Rev. Respir. Dis. 132, 1307-1312. [DOI] [PubMed] [Google Scholar]
- Kato K., Diéguez-Hurtado R., Park D. Y., Hong S. P., Kato-Azuma S., Adams S., Stehling M., Trappmann B., Wrana J. L., Koh G. Y. et al. (2018). Pulmonary pericytes regulate lung morphogenesis. Nat. Commun. 9, 2448 10.1038/s41467-018-04913-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler M. C., Loomis C. A., Zhao Z., Cushman J. C., Liu L. and Munger J. S. (2017). Sonic Hedgehog signaling regulates myofibroblast function during alveolar septum formation in murine postnatal lung. Am. J. Respir. Cell Mol. Biol. 57, 280-293. 10.1165/rcmb.2016-0268OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R. A. and Bishop J. M. (1993). Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell 74, 453-462. 10.1016/0092-8674(93)80047-I [DOI] [PubMed] [Google Scholar]
- Lee H. W., Khan S. Q., Khaliqdina S., Altintas M. M., Grahammer F., Zhao J. L., Koh K. H., Tardi N. J., Faridi M. H., Geraghty T. et al. (2017). Absence of miR-146a in podocytes increases risk of diabetic glomerulopathy via up-regulation of ErbB4 and Notch-1. J. Biol. Chem. 292, 732-747. 10.1074/jbc.M116.753822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. O., Sarkisian M. R., Mehal W. Z., Rakic P. and Flavell R. A. (2003). Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302, 1560-1563. 10.1126/science.1087621 [DOI] [PubMed] [Google Scholar]
- Li C., Li M., Li S., Xing Y., Yang C.-Y., Li A., Borok Z., De Langhe S. and Minoo P. (2015). Progenitors of secondary crest myofibroblasts are developmentally committed in early lung mesoderm. Stem Cells 33, 999-1012. 10.1002/stem.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Herriges J. C., Chen L., Mecham R. P. and Sun X. (2017). FGF receptors control alveolar elastogenesis. Development 144, 4563-4572. 10.1242/dev.149443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Bernau K., Sandbo N., Gu J., Preissl S. and Sun X. (2018). Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response. eLife 7, e36865 10.7554/eLife.36865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lee M. K., Gao F., Webster S., Di H., Duan J., Yang C.-Y., Bhopal N., Peinado N., Pryhuber G. et al. (2019). Secondary crest myofibroblast PDGFRα controls the elastogenesis pathway via a secondary tier of signaling networks during alveologenesis. Development 146, dev176354 10.1242/dev.176354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Willet S. G., Bankaitis E. D., Xu Y., Wright C. V. E. and Gu G. (2013a). Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation. Genesis 51, 436-442. 10.1002/dvg.22384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Kugler M. C., Loomis C. A., Samdani R., Zhao Z., Chen G. J., Brandt J. P., Brownell I., Joyner A. L., Rom W. N. et al. (2013b). Hedgehog signaling in neonatal and adult lung. Am. J. Respir. Cell Mol. Biol. 48, 703-710. 10.1165/rcmb.2012-0347OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F. D. (2016). Early-life origins of chronic obstructive pulmonary disease. N. Engl. J. Med. 375, 871-878. 10.1056/NEJMra1603287 [DOI] [PubMed] [Google Scholar]
- McGowan S. E. and McCoy D. M. (2011). Fibroblasts expressing PDGF-receptor-alpha diminish during alveolar septal thinning in mice. Pediatr. Res. 70, 44-49. 10.1203/PDR.0b013e31821cfb5a [DOI] [PubMed] [Google Scholar]
- McGowan S. E., Harvey C. S. and Jackson S. K. (1995). Retinoids, retinoic acid receptors, and cytoplasmic retinoid binding proteins in perinatal rat lung fibroblasts. Am. J. Physiol. Lung Cell. Mol. Physiol. 269, L463-L472. 10.1152/ajplung.1995.269.4.L463 [DOI] [PubMed] [Google Scholar]
- McGowan S. E., Grossmann R. E., Kimani P. W. and Holmes A. J. (2008). Platelet-derived growth factor receptor-alpha-expressing cells localize to the alveolar entry ring and have characteristics of myofibroblasts during pulmonary alveolar septal formation. Anat. Rec. 291, 1649-1661. 10.1002/ar.20764 [DOI] [PubMed] [Google Scholar]
- Menon P. and Fisher E. A. (2015). Immunostaining of macrophages, endothelial cells and smooth muscle cells in the atherosclerotic mouse aorta. Methods Mol. Biol. 1339, 131-148. 10.1007/978-1-4939-2929-0_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin A. V., Morales-Nebreda L., Mutlu G. M., Budinger G. R. S. and Perlman H. (2013). Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 49, 503-510. 10.1165/rcmb.2013-0086MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenko A., Kheirollahi V., Chao C.-M., Ahmadvand N., Quantius J., Wilhelm J., Herold S., Ahlbrecht K., Morty R. E., Rizvanov A. A. et al. (2017). Origin and characterization of alpha smooth muscle actin-positive cells during murine lung development. Stem Cells 35, 1566-1578. 10.1002/stem.2615 [DOI] [PubMed] [Google Scholar]
- Mund S. I., Stampanoni M. and Schittny J. C. (2008). Developmental alveolarization of the mouse lung. Dev. Dyn. 237, 2108-2116. 10.1002/dvdy.21633 [DOI] [PubMed] [Google Scholar]
- Nagata S. (2018). Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 36, 489-517. 10.1146/annurev-immunol-042617-053010 [DOI] [PubMed] [Google Scholar]
- Nicola T., Hagood J. S., James M. L., MacEwen M. W., Williams T. A., Hewitt M. M., Schwiebert L., Bulger A., Oparil S., Chen Y.-F. et al. (2009). Loss of Thy-1 inhibits alveolar development in the newborn mouse lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L738-L750. 10.1152/ajplung.90603.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A., Reddy R., Kursar J. D., Parks W. C. and Mecham R. P. (1989). Smooth muscle isoactin and elastin in fetal bovine lung. Exp. Lung Res. 15, 537-552. 10.3109/01902148909069617 [DOI] [PubMed] [Google Scholar]
- Ntokou A., Klein F., Dontireddy D., Becker S., Bellusci S., Richardson W. D., Szibor M., Braun T., Morty R. E., Seeger W. et al. (2015). Characterization of the platelet-derived growth factor receptor-α-positive cell lineage during murine late lung development. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L942-L958. 10.1152/ajplung.00272.2014 [DOI] [PubMed] [Google Scholar]
- Ntokou A., Szibor M., Rodríguez-Castillo J. A., Quantius J., Herold S., El Agha E., Bellusci S., Salwig I., Braun T., Voswinckel R. et al. (2017). A novel mouse Cre-driver line targeting Perilipin 2-expressing cells in the neonatal lung. Genesis 55, e23080 10.1002/dvg.23080 [DOI] [PubMed] [Google Scholar]
- O'Hare K. H. and Sheridan M. N. (1970). Electron microscopic observations on the morphogenesis of the albino rat lung, with special reference to pulmonary epithelial cells. Am. J. Anat. 127, 181-205. 10.1002/aja.1001270205 [DOI] [PubMed] [Google Scholar]
- Oladipupo S. S., Kabir A. U., Smith C., Choi K. and Ornitz D. M. (2018). Impaired tumor growth and angiogenesis in mice heterozygous for Vegfr2 (Flk1). Sci. Rep. 8, 1-10. 10.1038/s41598-018-33037-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M. and Joyner A. L. (2000). Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593-1605. [DOI] [PubMed] [Google Scholar]
- Park J., Ivey M. J., Deana Y., Riggsbee K., Sörensen E., Schwabl V., Sjöberg C., Hjertberg T., Park G. Y., Swonger J. M. et al. (2019). The Tcf21 lineage constitutes the lung lipofibroblast population. Am. J. Physiol. Lung Cell Mol. Physiol. 316, L872-L885. 10.1152/ajplung.00254.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A.-K. T. and Gale E. (2009). FGF signaling is required for myofibroblast differentiation during alveolar regeneration. Am. J. Physiol. Lung Cell Mol. Physiol. 297, L299-L308. 10.1152/ajplung.00008.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podd B. S., Thoits J., Whitley N., Cheng H.-Y., Kudla K. L., Taniguchi H., Halkias J., Goth K. and Camerini V. (2006). T cells in cryptopatch aggregates share TCR γ variable region junctional sequences with γδ T cells in the small intestinal epithelium of mice. J. Immunol. 176, 6532-6542. 10.4049/jimmunol.176.11.6532 [DOI] [PubMed] [Google Scholar]
- Popova A. P., Bentley J. K., Cui T. X., Richardson M. N., Linn M. J., Lei J., Chen Q., Goldsmith A. M., Pryhuber G. S. and Hershenson M. B. (2014). Reduced platelet-derived growth factor receptor expression is a primary feature of human bronchopulmonary dysplasia. Am. J. Physiol. Lung Cell. Mol. Physiol. 307, L231-L239. 10.1152/ajplung.00342.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan V. K., Wang Y., Patel S., Santos J. and Torday J. S. (2006). Rosiglitazone, a peroxisome proliferator-activated receptor-γ agonist, prevents hyperoxia-induced neonatal rat lung injury in vivo. Pediatr. Pulmonol. 41, 558-569. 10.1002/ppul.20407 [DOI] [PubMed] [Google Scholar]
- Reinert R. B., Kantz J., Misfeldt A. A., Poffenberger G., Gannon M., Brissova M. and Powers A. C. (2012). Tamoxifen-induced Cre-loxP recombination is prolonged in pancreatic islets of adult mice. PLoS ONE 7, e33529 10.1371/journal.pone.0033529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittny J. C. (2017). Development of the lung. Cell Tissue Res. 367, 427-444. 10.1007/s00441-016-2545-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittny J. C., Djonov V., Fine A. and Burri P. H. (1998). Programmed cell death contributes to postnatal lung development. Am. J. Respir. Cell Mol. Biol. 18, 786-793. 10.1165/ajrcmb.18.6.3031 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Zsengeller Z., Otake K., Palaniyar N. and Trapnell B. C. (2001). Alveolar macrophage deficiency in osteopetrotic mice deficient in macrophage colony-stimulating factor is spontaneously corrected with age and associated with matrix metalloproteinase expression and emphysema. Blood 98, 2845-2852. 10.1182/blood.V98.9.2845 [DOI] [PubMed] [Google Scholar]
- Srisuma S., Bhattacharya S., Simon D. M., Solleti S. K., Tyagi S., Starcher B. and Mariani T. J. (2010). Fibroblast growth factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis. Am. J. Respir. Crit. Care Med. 181, 838-850. 10.1164/rccm.200904-0544OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh C. D. and Sprent J. (1994). T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 372, 100 10.1038/372100a0 [DOI] [PubMed] [Google Scholar]
- Tan S. Y. S. and Krasnow M. A. (2016). Developmental origin of lung macrophage diversity. Development 143, 1318-1327. 10.1242/dev.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibeault D. W., Mabry S. M., Ekekezie I. I., Zhang X. and Truog W. E. (2003). Collagen scaffolding during development and its deformation with chronic lung disease. Pediatrics 111, 766-776. 10.1542/peds.111.4.766 [DOI] [PubMed] [Google Scholar]
- Tichelaar J. W., Lu W. and Whitsett J. A. (2000). Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J. Biol. Chem. 275, 11858-11864. 10.1074/jbc.275.16.11858 [DOI] [PubMed] [Google Scholar]
- Torday J., Hua J. and Slavin R. (1995). Metabolism and fate of neutral lipids of fetal lung fibroblast origin. Biochim. Biophys. Acta Lipids Lipid Metab. 1254, 198-206. 10.1016/0005-2760(94)00184-Z [DOI] [PubMed] [Google Scholar]
- Tordet C., Marin L. and Dameron F. (1981). Pulmonary di-and-triacylglycerols during the perinatal development of the rat. Experientia 37, 333-334. 10.1007/BF01959845 [DOI] [PubMed] [Google Scholar]
- Tsujino K., Li J. T., Tsukui T., Ren X., Bakiri L., Wagner E. and Sheppard D. (2017). Fra-2 negatively regulates postnatal alveolar septation by modulating myofibroblast function. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L878-L888. 10.1152/ajplung.00062.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui H., Shibayama M., Ohbayashi N., Konishi M., Takada S. and Itoh N. (2004). Fgf18 is required for embryonic lung alveolar development. Biochem. Biophys. Res. Commun. 322, 887-892. 10.1016/j.bbrc.2004.07.198 [DOI] [PubMed] [Google Scholar]
- Varisco B. M., Ambalavanan N., Whitsett J. A. and Hagood J. S. (2012). Thy-1 signals through PPARγ to promote lipofibroblast differentiation in the developing lung. Am. J. Respir. Cell Mol. Biol. 46, 765-772. 10.1165/rcmb.2011-0316OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W. J. R., Davidson E. H., Wong W. H. and McMahon A. P. (2007). Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977-1989. 10.1242/dev.001966 [DOI] [PubMed] [Google Scholar]
- Volckaert T., Yuan T., Yuan J., Boateng E., Hopkins S., Zhang J.-S., Thannickal V. J., Fässler R. and Langhe S. P. D. (2019). Hippo signaling promotes lung epithelial lineage commitment by curbing Fgf10 and β-catenin signaling. Development 146, dev166454 10.1242/dev.166454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voynow J. A. (2017). “New” bronchopulmonary dysplasia and chronic lung disease. Paediatr. Respir. Rev. 24, 17-18. 10.1016/j.prrv.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Walton K. D., Whidden M., Kolterud Å., Shoffner S. K., Czerwinski M. J., Kushwaha J., Parmar N., Chandhrasekhar D., Freddo A. M., Schnell S. et al. (2016). Villification in the mouse: Bmp signals control intestinal villus patterning. Development 143, 427-436. 10.1242/dev.130112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang Z., Huang H., Li J., Wang Z., Yu Y., Zhang C., Li J., Dai H., Wang F. et al. (2018). Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc. Natl. Acad. Sci. USA 115, 2407-2412. 10.1073/pnas.1719474115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C., Shimogori T. and Puelles L. (2017). Mouse Fgf8-Cre-LacZ lineage analysis defines the territory of the postnatal mammalian isthmus. J. Comp. Neurol. 525, 2782-2799. 10.1002/cne.24242 [DOI] [PubMed] [Google Scholar]
- Weinstein M., Xu X., Ohyama K. and Deng C. X. (1998). FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development 125, 3615-3623. [DOI] [PubMed] [Google Scholar]
- Yamada M., Kurihara H., Kinoshita K. and Sakai T. (2005). Temporal expression of alpha-smooth muscle actin and drebrin in septal interstitial cells during alveolar maturation. J. Histochem. Cytochem. 53, 735-744. 10.1369/jhc.4A6483.2005 [DOI] [PubMed] [Google Scholar]
- Yang L. M., Huh S.-H. and Ornitz D. M. (2018). FGF20-expressing, Wnt-responsive olfactory epithelial progenitors regulate underlying turbinate growth to optimize surface area. Dev. Cell 46, 564-580.e5. 10.1016/j.devcel.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Gurtu V., Kain S. R. and Yan G. (1997). Early detection of apoptosis using a fluorescent conjugate of annexin V. BioTechniques 23, 525-531. 10.2144/97233pf01 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.