ABSTRACT
Striking similarities between wound healing, epimorphic regeneration and the progression of solid tumors have been uncovered by recent studies. In this Review, we discuss systemic effects of tumorigenesis that are now being appreciated in epimorphic regeneration, including genetic, cellular and metabolic heterogeneity, changes in circulating factors, and the complex roles of immune cells and immune modulation at systemic and local levels. We suggest that certain mechanisms enabling regeneration may be co-opted by cancer to promote growth at primary and metastatic sites. Finally, we advocate that working with a unified approach could complement research in both fields.
KEY WORDS: Cancer, Epimorphic regeneration, Immune system, Solid tumor, Stroma, Wound healing
Summary: Although regeneration and cancer retain certain core differences as research models, emerging studies could allow us to bridge the biological mechanisms found in both and tackle important questions using a unified approach.
Introduction
Over three decades ago, Harold Dvorak synthesized observations from solid tumors and wound healing studies in his seminal paper ‘Tumors: wounds that do not heal’ (Dvorak, 1986). He more recently updated this work (Dvorak, 2015), continuing to elaborate on the hypothesis that aspects of tumor development may be co-opted from wound healing. In particular, the wound healing response, as characterized by Dvorak, consists of hemostasis at the site of injury (i.e. the formation of a blood clot), inflammation involving the infiltration and activation of local immune cells (such as macrophages and neutrophils), angiogenesis, fibroblast invasion, and synthesis of new extracellular matrix (ECM) (Fig. 1A). This is perhaps best exemplified by wound healing of the skin, which ostensibly follows these distinct stages (Singer and Clark, 1999) and requires the close crosstalk of epithelial and innate immune cells throughout the stages of wound healing (reviewed by Brazil et al., 2019). Dvorak thus argues that the biological processes underlying wound healing are co-opted by solid tumors to produce the tumor stroma, which consists of the surrounding immune cells, ECM, vasculature and connective tissue that help facilitate tumor growth.
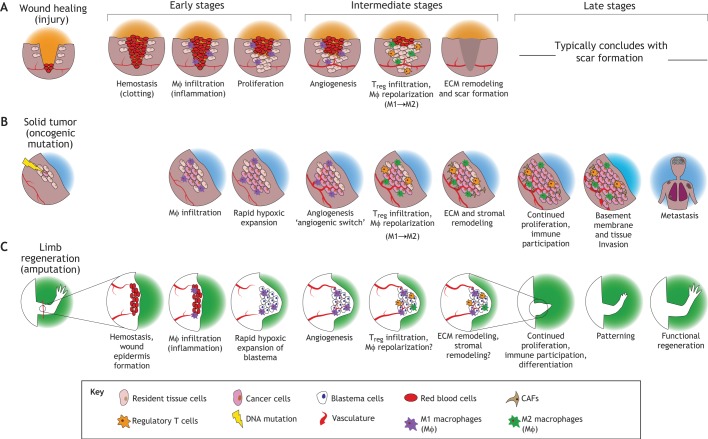
Fig. 1.
Wound healing, solid tumor progression and epimorphic regeneration (e.g. limb regeneration) share several broad characteristics. (A) Wound healing typically begins with hemostasis and clot formation, followed by immune-mediated inflammation (e.g. macrophage ‘MΦ’ infiltration), cell proliferation and angiogenesis (Dvorak, 2015, 1986). Later stages of wound healing include immune suppression of inflammation by regulatory T cells (Tregs) and extracellular matrix (ECM) remodeling. This process typically resolves with scar formation. (B) In cancer, following an oncogenic mutation, the tumor-initiating cell undergoes rapid hypoxic expansion. In certain cancers, this can be paired with immune cell infiltration, neo-vascularization, and ECM and stromal remodeling once the tumor progresses. Later stage tumors can continue to proliferate, invade surrounding tissues and enter the circulation to metastasize. (C) In limb regeneration, amputation is followed by migration of progenitor cells and rapid expansion in a hypoxic environment. Immune cell infiltration and angiogenesis, both of which are required for successful regeneration, also occur in later stages of regeneration. Epimorphic regeneration typically concludes with the differentiation of blastemal progenitors into functional cell types, re-patterning (e.g. of the regenerating limb) and functional regeneration of the body part. CAF, cancer-associated macrophage; RBC, red blood cells.
However, cancers (more specifically solid tumors) rely on rapid, continuous and uncontrolled proliferation stemming from genetic changes. These characteristics make tumorigenesis clearly distinct from the finite process of wound healing. Although a single model of tumor progression fails to acknowledge the breadth of clinical presentation, it is generally appreciated that there are several hallmarks of cancer development and progression (Hanahan and Weinberg, 2011). In brief, an initial genetic alteration (e.g. a mutation in a growth factor receptor) confers high proliferative capacity to the tumor-initiating cell, causing rapid cell division that may lead to the accumulation of additional mutations and confer other properties of apoptotic and immune evasion (Fig. 1B). Recruited immune cells, such as macrophages and local fibroblasts, may be induced to support cancer growth through macrophage polarization (discussed below) and growth-factor related activation, respectively, thus forming the supportive tumor ‘stroma’ (Chen and Song, 2019; Qian and Pollard, 2010). As the resulting solid tumor grows in size, hypoxia and nutrient deprivation may limit tumor growth; this can be overcome during the ‘angiogenic switch’, in which a shifted balance towards pro-angiogenic factors induces neovascularization of the tumor and facilitates the development of larger tumors (Bergers and Benjamin, 2003). Solid tumors may also gain the ability to invade their surround tissue, enter the bloodstream and subsequently colonize distant tissues to form metastases (Steeg, 2016). Thus, whereas wound healing in humans ultimately terminates in the formation of connective scar tissue, solid tumors typically engage in prolonged growth and proliferation, relying on the induced surrounding stroma for support. For the purposes of this Review we focus exclusively on the development and progression of solid tumors, and when we mention ‘cancer’ or ‘tumor’, we refer to solid tumors.
Instead of comparing tumor growth with wound healing alone, we argue that comparisons between tumor growth and more complex, prolonged forms of regeneration may be more informative. Indeed, a compelling comparison can be made between tumor formation and epimorphic regeneration: the process of complex tissue regeneration that relies on the formation of a mass of rapidly proliferating, undifferentiated cells called the blastema. Adult humans do not naturally display profound examples of epimorphic regeneration, although they can regenerate digit tips, as can mice (Farrell et al., 1977). In contrast, various model organisms can perfectly regenerate parts of organs or appendages (e.g. the heart in zebrafish) (Jopling et al., 2010; Poss et al., 2002) or entire bodies from tiny pieces (such as planaria) (Reddien, 2018; Wagner et al., 2011; Zeng et al., 2018). One remarkable example of epimorphic regeneration is limb regeneration in salamanders, which begins with similar processes to wound healing, including hemostasis, inflammation and wound closure (Fig. 1C). Wound healing during limb regeneration results in the formation of a specialized wound epidermis over the site of injury that is required for regeneration and is distinct from typical epidermis (Campbell and Crews, 2008; Mescher, 1976; Thornton, 1957). It remains unclear the extent to which the mechanisms behind generic wound healing and the wound healing that takes place before epimorphic regeneration are the same. Notably, whether immune system participation contributes to the formation of this specialized wound epidermis is unknown (reviewed in Godwin and Rosenthal, 2014). However, signals from the wound epidermis are thought to help orchestrate the accumulation of undifferentiated cells into the hallmark blastema because experimentally blocking wound epidermis formation inhibits regeneration (Goss, 1956; Mescher, 1976; Thornton, 1957). The accumulated cells within the blastema undergo sustained proliferation to produce all the cells contained within the regenerated limb (Whited and Tabin, 2009). Blastema formation is then followed by patterning, during which cells within the blastema differentiate, establish a positional identity and organize into functional structures (McCusker et al., 2014; Purushothaman et al., 2019). Overall, epimorphic regeneration is distinct from typical wound healing because it relies on the formation and outgrowth of the blastema. For simplicity, we will henceforth use the term ‘regeneration’ to refer to epimorphic regeneration.
Although not all tumors are identical, recent research highlights shared aspects between certain tumors and blastema formation that motivate a closer comparison. These include: the engagement of wound healing programs as discussed above (Dvorak, 2015; Whited and Tabin, 2009), accumulation of rapidly proliferating cells (Pitot, 1993; Whited and Tabin, 2009), rapid hypoxic proliferation and expansion (Eales et al., 2016; Simkin et al., 2017a), infiltration and modulation by immune cells (Hui et al., 2017; Leigh et al., 2018; Qian and Pollard, 2010; Zhang et al., 2012), and neovascularization to support larger scale growth (Baeriswyl and Christofori, 2009; Bayliss et al., 2006; Kopp et al., 2006; Ritenour and Dickie, 2017). Additionally, single-cell sequencing (scRNA-seq) data are highlighting the cellular heterogeneity within single tumors (Bernard et al., 2019; Puram et al., 2017; Tirosh et al., 2016; Wagner et al., 2019) and the regenerating blastema (Gerber et al., 2018; Leigh et al., 2018) alike. Moreover, many cancers exhibit intra-tumoral genetic heterogeneity from the expansion of specific subclones, which can ultimately contribute to treatment resistance if new genetic mutations are acquired (Burrell et al., 2013; Kim et al., 2018). Metabolic heterogeneity within a tumor is also thought to be a contributor to varying treatment sensitivity and treatment resistance (Robertson-Tessi et al., 2015; Vander Heiden, 2011), although this has yet to be studied in detail in the context of the blastema.
Of particular note are the local and systemic effects driven by immune inflammatory responses and circulating factors in both regeneration and tumorigenesis (Hui et al., 2017; Krall et al., 2018; McAllister et al., 2008; Rodgers et al., 2017). Functional analyses of recruited macrophages and regulatory T cells (Tregs) are revealing commonalities within the local immune response (Hui et al., 2017; Johnson et al., 2018; Nguyen-Chi et al., 2017; Ojalvo et al., 2009; Petersen et al., 2006; Wenemoser and Reddien, 2010), and the resulting systemic inflammatory responses may regulate distant cell cycle progression during regeneration and metastatic progression. Thus, critical analysis of both the systemic and local participation of immune cells could reveal shared mechanisms and avenues for further research.
In this Review, we argue that new technologies will enable an increasingly deeper and more nuanced understanding of the connections between cancer, wound healing and regeneration. We highlight scRNA-seq studies that have uncovered parallels in cellular and molecular phenotypes between solid tumors and epimorphic regeneration. In addition, we explore parallels in genetic, cellular and metabolic heterogeneity, as well as growth factor dependencies between solid tumors and the regenerating blastema. Finally, we focus on the common roles of immune cells in promoting cell proliferation and establishing a supportive microenvironment in both contexts. Overall, we aim to highlight the similarities between solid tumors and the regenerating blastema to suggest that shared biological mechanisms could lead to improved therapeutic interventions for cancer and regenerative medicine alike.
Single-cell RNA sequencing highlights cellular heterogeneity in solid tumors and blastemas
Recent advances in single-cell RNA sequencing (scRNA-seq) technologies have facilitated the dissection of cell types and states within tissues and whole organisms. A recent study employed scRNA-seq to study the stem cells of planaria, a type of flatworm capable of full body regeneration through the formation of a blastema (Box 1) (Zeng et al., 2018). Below, we discuss the cellular heterogeneity within solid tumors and blastemas formed during axolotl limb regeneration, as revealed by scRNA-seq.
Box 1. Tetraspanins in planarian neoblasts and cancer.
Planaria rely exclusively on a population of stem cells called neoblasts, characterized by high piwi expression, to regenerate their tissues. While single-cell transplantation studies have identified neoblasts as adult pluripotent cells (Wagner et al., 2011), more recent studies have uncovered transcriptional heterogeneity within the neoblast population that suggest potential lineage differentiation (Scimone et al., 2014). scRNA-seq has revealed that a transmembrane protein, tetraspanin 1 (TSPAN-1), is enriched in pluripotent neoblasts, as demonstrated by restoration of viability and regenerative potential in a lethally irradiated planarian following the transplantation of a single piwi-1+/TSPAN-1+ neoblast (Zeng et al., 2018). Interestingly, expression of TSPAN-1 is nearly undetectable in homeostasis but becomes upregulated in response to injury, and TSPAN-1 knockdown by RNAi significantly inhibits neoblast mobilization to the wound site and regeneration of tissue, demonstrating the importance of this tetraspanin to pluripotent stem cell self-renewal and planarian epimorphic regeneration.
Tetraspanins also contribute to tumor initiation, promotion and progression (Hemler, 2014). For example, tetraspanin CD151 is elevated in human squamous cell carcinomas, and expression of Cd151 increased rates of tumor initiation and progression in a mouse model of chemical-induced skin carcinogenesis (Li et al., 2013). A recent meta-analysis showed overexpression of CD151 in particular to be a negative prognostic factor for overall survival (Zeng et al., 2017). Ablation of tetraspanin 12 (TSPAN12) in a mouse xenograft model of mammary carcinoma significantly reduces tumor growth and metastasis (Knoblich et al., 2014). Tetraspanin 1 has been shown to be upregulated in cervical, pancreatic and prostate cancer cells (Hölters et al., 2013; Hou et al., 2015; Munkley et al., 2017). Overexpression of tetraspanin 1 protein increases prostate cancer cell migration, while knockdown by siRNA decreased viability in vitro (Munkley et al., 2017).
The importance of tetraspanins in stem cell function during planarian regeneration, and in cancer initiation, migration and progression, could suggest shared mechanisms of migratory regulation and proliferation. Tetraspanins can modulate downstream signaling of certain growth factor receptors (Charrin et al., 2014), suggesting a possible mechanism of action. Further research into the function of TSPAN-1 and other tetraspanins is warranted to understand whether analogous downstream signaling could be regulating self-renewal and proliferation in planarian neoblasts, and whether similar mechanisms are shared between planarian regeneration and various mammalian cancer types.
Intra-tumoral cellular heterogeneity
Single-cell analysis of cancers is revealing a high degree of intra-tumoral heterogeneity within solid tumors. Indeed, scRNA-seq data of tumors from various cancers, including head and neck squamous cell carcinoma, melanoma, breast cancer and pancreatic ductal adenocarcinoma, all reveal high epithelial heterogeneity, possibly representing different stages of differentiation (Bernard et al., 2019; Puram et al., 2017; Tirosh et al., 2016; Wagner et al., 2017). Interestingly, these studies have highlighted the presence of specialized subpopulations that can exhibit transcriptional programs associated with drug resistance and become enriched after treatment (Tirosh et al., 2016). In addition, some subpopulations exhibit aspects of an epithelial-to-mesenchymal transition (EMT) program (Puram et al., 2017; Wagner et al., 2017), which is associated with resistance to chemotherapy and increased metastasis (Fischer et al., 2015).
Moreover, these studies have revealed the presence of supportive stromal cell populations, such as cancer-associated fibroblasts (CAFs), and immune cells, such as macrophages and T cells (Puram et al., 2017; Tirosh et al., 2016). Indeed, CAFs are thought to be critical regulators of cancer progression through secretion of growth factors and cell-cell interactions (Bremnes et al., 2011; Erez et al., 2010). Specifically, CAFs secrete growth factors, including hepatocyte growth factor (HGF), transforming growth factor β (TGFβ), epidermal growth factor (EGF) and insulin-like growth factor 1 (IGF1), that ultimately promote malignant cell proliferation, chemotherapy resistance and immunosuppression (Bremnes et al., 2011; Chen and Song, 2019; Komohara and Takeya, 2017) (Fig. 2A,B); interestingly, these molecules also promote wound healing (summarized in Table 1). CAFs also promote inflammation, angiogenesis and macrophage recruitment in a NF-kB signaling-dependent manner (Erez et al., 2010). scRNA-seq studies paired with histological analysis have shown that cell-cell interactions between peripheral malignant tumor cells and CAFs induce a pre-EMT transcriptional program (Puram et al., 2017). Therefore, in the context of the tumor microenvironment, CAFs can provide positional and proliferation cues for cells within the tumor. Tumor-associated macrophages (TAMs) also secrete various factors that support tumor progression, including angiogenic factors such as vascular endothelial growth factor A (VEGFA), as well as immunosuppressive factors such as interleukin 10 (IL10), TGFβ and arginase, thus highlighting the importance of stromal interactions and local signaling events in driving tumor growth (Fig. 2A,B).
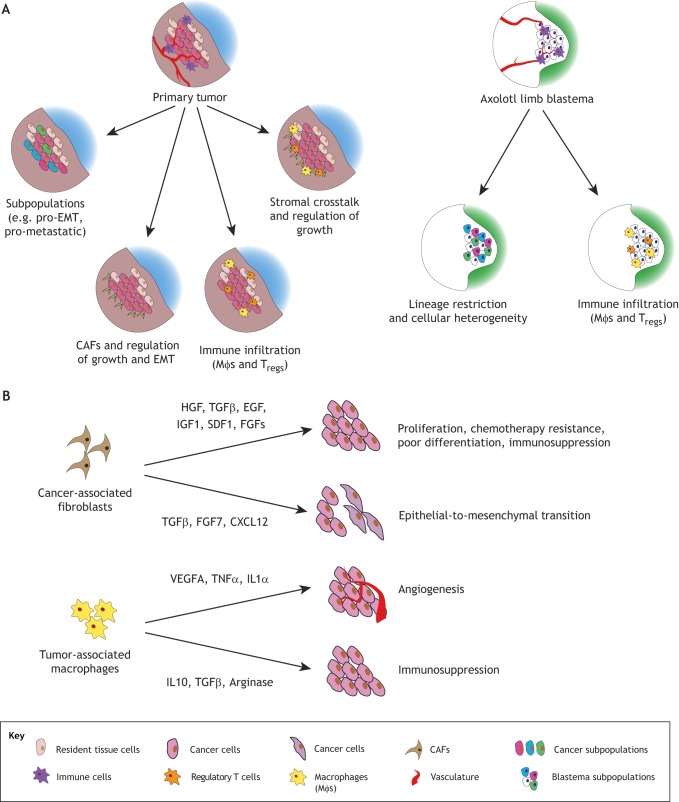
Fig. 2.
Single-cell sequencing could reveal parallel processes governing tumor and blastemal outgrowth. (A) Single-cell RNA-sequencing (scRNA-seq) of tumor samples has highlighted heterogeneity of the tumor itself, with some subpopulations exhibiting greater metastatic potential or promoting epithelial- to-mesenchymal transition (EMT) transcriptional programs. Additionally, stromal cells, including cancer-associated fibroblasts (CAFs), help to create a supportive microenvironment for tumor proliferation through cell-cell interaction and secretion of growth factors and cytokines. scRNA-seq of the axolotl limb blastema has revealed cellular heterogeneity in terms of lineage restriction, as well as persistent infiltration of immune cells such as macrophages and T cells. (B) CAF-derived secretion of factors such as hepatocyte growth factor (HGF), transforming growth factor β (TGFβ), epidermal growth factor (EGF), insulin-like growth factor 1 (IGF1) and stromal-derived factor 1 (SDF1) are associated with increasing tumor proliferation, chemotherapy resistance, poor differentiation, immunosuppression and EMT. Additionally, tumor-associated macrophages (TAMs) have been found to secrete factors such as vascular endothelial growth factor (VEGF) to induce angiogenesis, as well as promote immunosuppression and immune evasion of the tumor through the secretion of interleukin 10 (IL10), TGFβ and arginase. Comparing stromal signaling and growth-coordinating signals between the cancer and regeneration may be informative for future research.
Table 1.
Known systemic factors in cancer have existing roles in regeneration
scRNA sequencing of the axolotl limb blastema
The axolotl is a salamander that has been studied extensively for its ability to regenerate. scRNA-seq performed on the blastema during axolotl limb regeneration has identified similar aspects of differentiation-state heterogeneity within cell lineages seen in solid tumors, but without EMT signatures (Gerber et al., 2018; Leigh et al., 2018). Blastema formation during axolotl limb regeneration is thought to involve some dedifferentiation of certain cell types local to the site of injury, followed by differentiation into the cell types required to replace the limb (Kragl et al., 2009). This model is generally supported by single-cell data, which has specifically shown that a connective tissue lineage enters a blastema-specific state, before achieving multipotency and then undergoing controlled differentiation (Gerber et al., 2018). Pseudotime analysis of blastema cells at various time points during regeneration suggests that other cell types likely remain lineage restricted (Leigh et al., 2018). However, the data also indicate that even among similar or possibly identical cell differentiation trajectories, heterogeneity in the timing of transitions is likely (Leigh et al., 2018). Additionally, scRNA-seq has identified putative macrophage and T-cell populations that persist throughout the various stages of regeneration (Leigh et al., 2018), corroborating various functional studies that suggest important roles for recruited macrophages and myeloid cells in successful regeneration (Godwin et al., 2013; Tsai et al., 2019). Thus, as in tumors, scRNA-seq of regenerating blastemas during axolotl limb regeneration has revealed a high degree of cellular heterogeneity, likely linked to the differentiation state of many cell types, as well as to the infiltration of immune cells (Fig. 2A).
Whereas solid tumors are dependent on the formation of a supportive microenvironment and stroma for growth, it is unclear whether a similar stroma or microenvironment is established during blastemal outgrowth. Although it is known that many of the positional and proliferative signals (such as FGFs, BMPs and Wnts) during limb regeneration originate from the wound epidermis (McCusker et al., 2015), intra-blastema signaling pathways also likely exist, and defining these is an important goal. For example, the identification of a fibroblast-like cell type during blastema formation (Leigh et al., 2018) could suggest that populations of cells within the blastema are important for cell-cell interactions and may engage in local signaling to promote proliferation – not unlike the role of CAFs in promoting tumor outgrowth (Bremnes et al., 2011; Bruzzese et al., 2014). Selective ablation and manipulation of specific genes within specific blastema cells will enable these types of relationships to be uncovered, but this type of work is currently technologically challenging.
Meanwhile, insights from intra-tumoral signaling, and specifically the role of CAFs, may be informative (Fig. 2B). Although one potential source of instructive signaling during regeneration could be the transient fibroblast-like population mentioned above (Leigh et al., 2018), further studies will elucidate whether intra-blastemal cells engage in pro-proliferative signaling in the same manner as CAFs. Additionally, while macrophages are important for various forms of epimorphic regeneration (Godwin et al., 2017, 2013; Nguyen-Chi et al., 2017), the exact mechanisms of macrophage participation during regeneration are not known. One recent study identified myeloid participation and IL8 signaling as important for axolotl limb regeneration (Tsai et al., 2019), suggesting that distinct immune compartments play important roles in regeneration; however, further mechanistic investigation is required. Insights from TAM and immune cell participation in angiogenesis and immunosuppression could serve as a starting point for future study into immune involvement in regeneration; e.g. by assessing for secretion of pro-angiogenic factors such as VEGF or immunosuppressive factors such as IL10 and TGFβ.
Understanding the sources of differentiation signals in the blastema may yield insight into how differentiation is controlled in malignant tumor cells. While understanding control over tumor differentiation has had limited therapeutic success, given the difficulties associated with differentiation therapy (reviewed by de Thé, 2018), a molecular understanding of tumor cell dedifferentiation could include potential risk factors for cancer progression. Insights from stromal crosstalk in cancer could potentially inform future research in regeneration.
Genetic heterogeneity
Genetic heterogeneity in cancer can arise from intercellular genetic variation caused by random mutations, followed by selective expansion of genetic subclones that have a phenotypic advantage (Burrell et al., 2013). For example, microdissections and DNA testing of various regions within advanced colorectal adenocarcinoma demonstrated that many tumors exhibit topographical genetic heterogeneity (Baisse et al., 2001), and this trend has been demonstrated for many different types of cancer (McGranahan and Swanton, 2017). Drug treatment is thought to preferentially clear sensitive clones, while allowing resistant subclones to dominate the tumor. This leads to an initial treatment response, but eventual progression or recurrence (McGranahan and Swanton, 2017). In particular, LINE-1 retrotransposon elements can contribute to tumorigenesis in various ways, including insertion-mediated inhibition or activation of gene expression, 3′ transduction (co-transcription) and mediation of pseudogene formation (reviewed by Xiao-Jie et al., 2016). Analysis of transposon insertion from whole-genome sequencing data in five different cancer types revealed LINE-1 insertions in genes that are commonly mutated in cancer in epithelial tumors, highlighting a role for LINE-1 elements in tumorigenesis (Lee et al., 2012) and possibly in facilitating drug resistance. Thus, genetic heterogeneity and transposable elements can contribute to tumorigenesis and drug resistance.
Genetic heterogeneity between cells has not been studied extensively during regeneration, but could yield important insights into how highly regenerative animals may use it as a pro-regenerative mechanism or might counteract it during the highly proliferative state to protect genomic integrity. In particular, one study showed that LINE-1 transcription is induced in regenerating axolotl limb blastemas and leads to increased LINE-1 transposition (Zhu et al., 2012a). This study indicates that genomic instability may be induced during regeneration, although it also suggests that repeated injury might exacerbate this instability and, if so, the effects on regeneration are currently unknown. Interestingly, piwi (a crucial mediator of transposon silencing) homologs are upregulated during limb regeneration, and knockdown leads to increased cell death, decreased cell proliferation and significantly delayed regeneration (Zhu et al., 2012a,b). These studies could suggest that LINE-1 activation during regeneration induces co-activation of transposon silencing machinery involving piwi to maintain genomic integrity, and that unhindered LINE-1 activation may lead to cell death. The concurrent activation of LINE-1 elements with upregulation of piwi could suggest an evolutionary mechanism to maintain genomic integrity that may be conserved in cancer. Indeed, piwi and its corresponding piwi-interacting RNAs are abnormally expressed in various cancers (Liu et al., 2019; Weng et al., 2019) but their combined function in cancer remains poorly understood. One possibility is that high piwi expression may represent an attempt to defend against genomic instability in high-grade tumors, but that failure to maintain this genomic stability leads to high expression with rapid tumor progression. Interactions between LINE-1 and piwi in the regenerative context could provide a basis for studying the interaction between rapid cell proliferation and retrotransposon elements, and they may yield insight into the function of piwi in cancer.
Metabolic cues and heterogeneity
Yet another characteristic of solid tumors that may be translatable to the blastema is metabolic heterogeneity. A canonical hallmark of cancer is upregulated aerobic glycolysis and fermentation of glucose into lactate (commonly known as the Warburg effect) (Vander Heiden et al., 2009). More recent analysis suggests that metabolic and spatial heterogeneity within tumors may be tied to genetic mutations, epigenetic changes and stochastic cell state transitions (Strickaert et al., 2017). For example, one study tracked the repopulation dynamics of single lineages within 10 human colorectal cancers through serial xenografts in mice, with and without chemotherapy treatment (Kreso et al., 2013). Despite genetic stability throughout transplantation, the proliferation and chemotherapy resistance of cells originating from the same sub-clone is highly variable. This work indicates that both intracellular epigenetic and metabolic alterations in response to changes in the microenvironment may affect response to therapy (reviewed in Caiado et al., 2016). In addition, metabolic ‘coupling’ between ovarian cancer cells and adipocytes (Nieman et al., 2011), and between cancer-associated fibroblasts (CAFs) and breast cancer cells (Rattigan et al., 2012), highlights the role of surrounding stromal cells in determining the metabolic status of tumor cells. Varying levels of hypoxia from inadequate perfusion also contribute to metabolic heterogeneity (Eales et al., 2016; Terry et al., 2017). For example, transcriptional signatures related to hypoxia-inducible factor 1α (HIF1α) have been detected in scRNA-seq data of primary glioblastoma (Patel et al., 2014). Thus, metabolic heterogeneity can arise within solid tumors as a result of metabolic cues and nutrient concentrations (i.e. oxygen) within the stromal microenvironment, and conceivably all of these could be influenced by non-cell-autonomous effects between tumor cells and stromal cells.
While metabolic heterogeneity has not been studied extensively in the field of epimorphic regeneration, hypoxia is appreciated as an early characteristic of blastema formation. Specifically, the early blastema during newt and axolotl limb regeneration is avascular (Mescher, 1996; Peadon and Singer, 1966), while neovascularization in axolotl occurs later in blastema formation and relies on VEGF signaling (Ritenour and Dickie, 2017). Using qPCR at different time points of the regenerating gecko tail, it was shown that HIF1α mRNA is upregulated early, highlighting the initial hypoxic state during blastema formation (Novianti et al., 2019). Additionally, slice cultures of the regenerating mouse digit-tip blastema revealed that dynamic changes in oxygen may contribute to bone regeneration – namely, that initial hypoxic events in the blastema followed by increasing oxygen concentrations from vascularization may promote bone mineralization (Sammarco et al., 2014). Thus, while hypoxia has been appreciated on a gross level in epimorphic regeneration, it may be worthwhile investigating whether intra-blastemal heterogeneity may be arising from an oxygen gradient. This could be investigated by assessing differential activation of HIF1α-mediated transcriptional programs within cells of the blastema. Additionally, metabolic heterogeneity within the blastema could be a regulator of cell proliferation as it is in cancer. Initial investigations could assess whether glycolytic metabolism is upregulated or altered in the blastema (e.g. assess for the Warbug effect), and whether cells within the blastema exhibit distinct metabolic signatures from one another.
Secretion of growth factors by both the primary tumor and blastema
Approaching both cancer and regeneration as systemic processes could suggest that primary tumors and blastemas secrete factors that promote local cell proliferation and growth while also having systemic effects. Indeed, recent studies are beginning to reveal that cancers secrete various growth factors and cytokines that recruit cells to the site of the primary tumor to support proliferation, but also help to prime distant niches for metastatic colonization (discussed below and reviewed by McAllister and Weinberg, 2014). Similarly, recent research has revealed HGF to be a systemic growth factor that activates distant stem cells during muscle regeneration in mice (Rodgers et al., 2017). Systemic cell cycle activation is also seen in axolotl (Johnson et al., 2018) and planaria (Wenemoser and Reddien, 2010), and is induced by wound healing following injury; however, the underlying growth factor or proliferative signals in these two models have not been elucidated. Coincident findings that circulating factors enact systemic effects during both cancer and regeneration could suggest shared mechanisms, requiring further investigation into the source, identity and functions of secreted factors. A summary of secreted factors and their functions in cancer and regenerative models is provided in Table 1. However, the data highlights that further research defining changes in circulating factors during regeneration is needed. Below, we discuss two promising candidates for cross-disciplinary study: VEGFA and HGF.
VEGF
VEGF is a well-characterized growth factor principally associated with angiogenesis during development and wound healing (reviewed in Apte et al., 2019), but is also a key regulator of angiogenesis and neovascularization in cancer (Carmeliet, 2005). Dvorak's early hypothesis and subsequent work in the field helped to highlight the role of VEGF in promoting vascular permeability and early extravasation of fibrinogen, which ultimately serves as the basis for stroma formation in cancer (Dvorak, 2015, 1986). Tumor-derived VEGF acts as part of the ‘angiogenic switch’ (Baeriswyl and Christofori, 2009), which induces neovascularization and recruits bone marrow-derived cells (BMCs), such as endothelial progenitor cells, hemangiocytes and macrophages, to promote tumor growth (Gao and Mittal, 2009; Kopp et al., 2006). The surprising participation of BMCs in tumor growth was demonstrated by experiments involving hematopoietic stem cell transplantation, in which a small percentage (0.5-12%) of tumor endothelial cells were found to originate from donor cells (Gao and Mittal, 2009; Nolan et al., 2007). Similar to its role in cancer, signaling of VEGF through binding to the VEGF receptor (VEGFR) in regeneration facilitates regenerative angiogenesis and is required for zebrafish heart and tail fin regeneration, while inhibition of VEGFR signaling delays axolotl tail regeneration (Bayliss et al., 2006; Marín-Juez et al., 2016; Ritenour and Dickie, 2017). Indeed, although the early blastema is relatively hypoxic (Seifert and Muneoka, 2018), vascularization is required for continued growth and complete regeneration of lost tissue (Bayliss et al., 2006). The requirement of VEGF-mediated angiogenesis for prolonged growth of the blastema and complete regeneration thus seems strikingly similar to the ‘angiogenic switch’ described in cancer progression.
Whereas the angiogenesis during cancer progression can involve recruitment of BMCs (Gao and Mittal, 2009), it is currently unknown whether revascularization of the blastema during regeneration relies solely upon local endothelial cells and vasculature or whether bone marrow recruitment also occurs. Previous findings regarding axolotl digit regeneration have suggested that cells from connective tissue within 500 µm of the amputation plane are recruited to form the blastema (Currie et al., 2016), leading to the hypothesis that only local progenitors are recruited for regeneration (Butler, 1935; Butler and O'Brien, 1942). Given the similar functions of VEGF in promoting neovascularization of a tumor and the blastema, it may be possible that distant or circulating endothelial progenitors may also be recruited to the regenerating blastema through VEGF signaling. Importantly, hematopoiesis in the axolotl occurs in the liver and spleen, rather than in bone marrow (Lopez et al., 2014); thus, recruited cells may originate from these organs. Further studies evaluating whether neovascularization of the blastema recruits distant cells could challenge the hypothesis that only cells local to injury are recruited to form the blastema and would draw closer parallels between blastema formation and tumor progression. Additionally, recruited cells could serve additional roles beyond serving as progenitors, including secreting factors to promote progenitor cell activity within the blastema. The possibility of distantly recruited cells contributing to blastema formation should therefore be investigated.
HGF
HGF is a growth factor that signals through the MET receptor tyrosine kinase and is involved in liver development and the regeneration of various tissues (Nakamura et al., 2011). However, recent research indicates that the HGF-MET signaling pathway is exploited to promote cancer cell invasion and metastasis (Gherardi et al., 2012). HGF is known to be secreted by CAFs, where it promotes cancer growth and drug resistance by promoting survival and proliferation (Owusu et al., 2017). Indeed, HGF confers resistance to BRAF inhibitors in previously sensitive mutant melanoma cells (Wilson et al., 2012). High serum levels of HGF are correlated with poorer prognosis and cancer metastasis (Matsumoto et al., 2017), suggesting that circulating HGF could have a systemic effect in promoting metastatic colonization and growth.
In the context of wound healing, HGF activator (HGFA) is a systemic protease that is activated in response to injury and cleaves HGF into its active form. Injuring the muscle in one leg of a mouse primes stem cells to enter the cell cycle in the opposite/contralateral leg, and this priming is linked to systemic HGFA activation and increased HGF cleaving activity (Rodgers et al., 2017). Administration of activated HGFA leads to rapid tissue repair following subsequent injury, suggesting that HGFA primes stem cells to respond quickly to injury (Rodgers et al., 2017). In both axolotl (Johnson et al., 2018) and planaria (Wenemoser and Reddien, 2010), systemic cell cycle entry has been observed following injury, suggesting that this is a conserved wound response across species. Although the roles of HGF in wound healing and systemic stem cell priming have been characterized, it is still unclear whether HGF may play a role during epimorphic regeneration. The role of HGF in inducing systemic cell cycle entry could represent a mechanistic explanation for the correlation between high HGF serum levels and poor prognosis for individuals with cancer. One possibility is that circulating HGF facilitates systemic cell cycle entry and proliferation as part of a conserved systemic wound-healing response. If so, HGF may mediate metastasis through a co-opted injury response. Further research into whether injury-induced activation of HGF can promote metastatic outgrowth is warranted, and a direct link between tumor secretion of HGF and metastatic colonization should be assessed.
Immune system participation in epimorphic regeneration and solid tumors
Macrophage infiltration and repolarization
Local immune cell infiltration appears to play a role in modulating proliferation and outgrowth in cancer and epimorphic regeneration alike. For example, high levels of macrophage infiltration is associated with poor patient prognosis in cancer (Zhang et al., 2012) and TAMs are thought to perform a diverse array of functions, including promoting tumor cell invasion, migration and angiogenesis, and suppressing anti-tumor cell immunity (Qian and Pollard, 2010; Schouppe et al., 2012). Macrophages are conventionally divided into M1 (pro-inflammatory) and M2 (anti-inflammatory and immune-modulatory) categories (Mantovani and Sica, 2010), although gene profiling analysis suggests that TAM diversity extends beyond this binary categorization with specialized macrophages for particular functions (Ojalvo et al., 2009). It is postulated that initially pro-inflammatory M1 macrophages are recruited to the site of the cancer, but are then repolarized by the tumor microenvironment through exposure to factors such as interleukin (IL) 4, IL10 and IL1β, and TGFβ to adopt a M2-like pro-tumorigenic identity (Pollard, 2004) (Fig. 3A). Repolarized macrophages are a crucial component of the ‘angiogenic switch’, helping secrete pro-angiogenic factors such as VEGF or producing ECM-degrading proteases that release angiogenic factors previously sequestered in the ECM (De Palma et al., 2005; Riabov et al., 2014).
Fig. 3.
Macrophage depolarization occurs both during blastemal and tumor outgrowth. (A) During zebrafish tail regeneration, macrophages expressing M1-associated transcripts are recruited during and throughout regeneration. Some of these macrophages then shift to expressing M2-associated transcripts during the tissue remodeling stage. In cancer, pro-inflammatory M1 macrophages are initially recruited to the tumor, but are ‘educated’ by the microenvironment to adopt a pro-tumorigenic M2 phenotype. These similarities could suggest that the mechanisms employed during wound healing and tissue remodeling may also be employed by cancer to control and re-educate during tumor outgrowth, highlighting a potential similarity between cancer and regeneration. However, the source of macrophage re-polarizing signals during regeneration is unknown. (B) Regulators of macrophage repolarization include Toll-like receptor (TLR) activation, interferon γ (IFNγ), transforming growth factor β (TGFβ) and various interleukins. Assessing expression or activation of these signaling pathways could be a first step in understanding molecular controls of macrophage repolarization during regeneration. CAFs, cancer-associated fibroblasts.
Macrophages are required for epimorphic regeneration in various models, including the axolotl limb and heart (Godwin et al., 2017, 2013), zebrafish tail fin regeneration (Fig. 3A) (Petrie et al., 2014), ear injury in African spiny mice (Simkin et al., 2017a), as well as digit tip regeneration in mice (Simkin et al., 2017b). Single-cell analyses and tracking using fluorescent labelling have revealed that macrophages persist throughout wound healing and blastema formation in both axolotl limb regeneration and zebrafish fin regeneration (Leigh et al., 2018; Nguyen-Chi et al., 2017), suggesting that macrophages may play prolonged roles in facilitating regeneration. Indeed, macrophage depletion at early and late stages in axolotl limb regeneration showed that early depletion prevents regeneration, whereas late depletion merely delays regeneration (Godwin et al., 2013).
Interestingly, macrophage subtypes resembling M1 and M2 classifications have been identified in zebrafish, based on their expression of tumor necrosis factor α (TNF-α) and TGFβ, respectively, as well as other characteristic cytokines and receptors (Mantovani et al., 2002; Nguyen-Chi et al., 2015). In response to tail fin injury, some of the macrophages that migrate to the site of injury adopt an initial M1 phenotype, but gradually shift to a M2 phenotype during the tissue remodeling stage (Nguyen-Chi et al., 2015). Early ablation of macrophages irrespective of subtype prevents blastema formation in this zebrafish model, while late stage ablation leads to regenerative defects but no change in proliferation or cell death (Nguyen-Chi et al., 2017). Together, these results support a stage-dependent view of macrophage contribution to regeneration, not unlike what has been observed in cancer progression. Another study comparing successful neonatal cardiac regeneration to non-regenerative adult hearts in mice has found that distinct lineages of macrophages may be responsible for pro-reparative versus pro-inflammatory programs (Lavine et al., 2014). These studies highlight the role of macrophages in promoting regeneration and indicate that distinct macrophage subtypes may each play a unique role during the regenerative process.
Parallels in recruitment and conversion of macrophages from M1 to M2 subtype could suggest similar inflammatory and immunomodulatory cues in both tumoral and blastemal microenvironments. In cancer, a combination of microenvironmental cues, including hypoxia, cytokine exposure and even cell-to-cell contact with CAFs, is believed to control the shift from anti-tumor to pro-angiogenic and proliferative programs in macrophages (Komohara and Takeya, 2017; Pollard, 2004) (Fig. 3B). A deeper understanding of the molecular and micro-environmental cues within the blastema is needed to identify the signals responsible for shifting macrophage identity. Crosstalk between blastema cells and infiltrating macrophages could potentially control the shift from a pro-inflammatory program to an immunosuppressive, pro-angiogenic program; however, this requires additional research. Thus, the origin of immunomodulatory signals in the blastema remains to be investigated but could be guided by known macrophage dynamics and re-polarizing signals during cancer progression.
Systemic inflammatory responses modulate metastatic growth
Recent studies have shown that both wound healing associated with surgeries and tumors themselves can induce systemic inflammatory responses that modulate the growth and survival of distant metastases (Castaño et al., 2018; Krall et al., 2018). In particular, one study demonstrated that primary breast cancer tumors induce a systemic IL1β-mediated inflammatory response that suppresses the outgrowth of distantly seeded, metastasis-inducing cells (Castaño et al., 2018). Knockdown of the IL1 receptor (IL1-R) in distantly seeded tumor cells makes them insensitive to this systemic inflammatory response and thus causes them to proliferate in spite of the presence of a primary tumor. Local inhibition of inflammation at the primary tumor site by embedding primary tumor cells in Matrigel with an IL1R1 antagonist leads to differentiation and growth of distant metastases, suggesting that attenuation of inflammation at the site of the tumor prevents the suppressive systemic inflammatory response. These findings suggest that a local inflammatory response at the site of the tumor may trigger an IL1β-systemic immune activation that prevents metastatic colonization and growth (Fig. 4A).
Fig. 4.

Systemic modulation of cell cycle entry occurs in both regeneration and cancer. (A) In cancer, a systemic pro-inflammatory interleukin 1β (IL1β) response has been shown to suppress the growth of distant metastases, whereas a separate systemic wound response following surgery has been associated with metastatic progression. This surgery-induced metastatic progression is characterized by mobilization of neutrophils and monocytes, as well as increased levels of circulating interleukin 6 (IL6), granulocyte colony-stimulating factor (G-CSF) and cytokine CCL2. (B) In various regenerative organisms, including axolotl, planaria and zebrafish, systemic responses have been observed following injury, either manifesting as systemic cell cycle entry following injury or mobilization of pro-regenerative immune cells such as Tregs. These observed similarities could suggest that a common, systemic wound-healing response is employed by cancer to promote the growth of metastases, and that this systemic response normally helps to facilitate regeneration in other contexts.
However, these findings must be considered in the context of another study that found systemic inflammation following injury to promote the growth of distant metastases (Krall et al., 2018). In this study, the authors use a mammary carcinoma model and a melanoma cell line to demonstrate that surgical wounding triggers a systemic inflammatory response characterized by increased circulating levels of IL6, granulocyte colony-stimulating factor (G-CSF) and the cytokine CCL2, ultimately leading to the progression of distant metastases. Additionally, inflammatory monocytes are mobilized into circulation in response to surgical injury, which may increase their recruitment to distant metastases and subsequent differentiation into TAMs (Krall et al., 2018). Increased numbers of TAMs may subsequently promote tumor outgrowth through immunosuppression by increasing expression of programmed cell death-ligand 1 (PD-L1), an immune evasion regulator that is the target of novel immunotherapies (Le et al., 2015; Rizvi et al., 2015). Other recent studies have highlighted how TAMs may release key metabolites, such as cholesterol or pyrmidines, to promote cancer cell growth and division (Goossens et al., 2019; Halbrook et al., 2019). Neutrophils, which are an immune cell type commonly associated with pro-inflammatory responses, have also been shown to play a role in promoting metastasis. One study found that neutrophil depletion during the expected period of metastatic colonization leads to significantly fewer tumor metastases. Furthermore, neutrophil-derived leukotrienes can aid metastatic colonization by stimulating proliferation of cancer cells with high metastatic potential (Wculek and Malanchi, 2015). Additionally, systemic inflammation caused by auto-immune, collagen-induced arthritis, which results in higher levels of circulating IL17 and IL6, increases the chance of metastasis in a mouse model of mammary cancer (Roy et al., 2011). Thus, wound healing and the associated inflammatory response appears to trigger a systemic response through macrophage and neutrophil mobilization that promotes metastatic growth, distinct from the anti-proliferative immune response initiated by a primary tumor (Fig. 4A).
In considering the contrary effects of the tumor itself versus wound healing on metastatic growth, one hypothesis is that tumors themselves evoke an anti-metastatic inflammatory wound-healing response, but that a distinct wound-healing response triggered by surgical intervention – or the tumor itself – can promote metastasis (Fig. 4A). Considering the examples of systemic cell cycle activation during wound healing in regenerative models, this wound healing response could be conserved across evolution. While primary tumors may induce a systemic IL1β inflammatory response that prevents their own metastasis (Castaño et al., 2018), a distinct wound healing response (characterized by IL6, G-CSF and CCL2) could be triggered by surgical resection or the tumor itself during cancer progression to promote the growth of distant metastases (Krall et al., 2018). Indeed, Dvorak puts forward several lines of evidence suggesting that cancers engage a local wound-healing response to develop the surrounding stroma (Dvorak, 2015). It stands to reason that some aspect of a systemic wound-healing response might be engaged to promote metastatic growth. Understanding whether a similar systemic inflammatory response is triggered during regeneration could facilitate research into a possible parallel between tumor progression and regeneration, and possibly reveal how this response is typically resolved at the end of wound healing. The molecular basis for resolution could then serve as a therapeutic avenue for controlling cancer metastasis.
While tumor and blastemal microenvironments are likely to be different, it is possible that similar interactions or local signaling between the blastema and the immune system could modulate systemic inflammation. Below, we discuss the evidence for a systemic, pro-proliferative inflammatory response activated during the regeneration process, which might be analogous to those observed in tumor conditions.
Systemic immune responses during regeneration
Although systemic immune responses to regeneration have been less well studied, immune cell mobilization and recruitment are crucial for regeneration and likely modulate cell proliferation and scarring. For example, macrophage ablation prevents blastema formation during limb regeneration in axolotl (Godwin et al., 2013) and fin regeneration in zebrafish (Nguyen-Chi et al., 2017), supporting the idea that wound healing can at least trigger an immune cell recruitment and a local pro-proliferative inflammatory response. Additionally, ablation of macrophages results in scarring rather than regeneration after axolotl limb amputation (Godwin et al., 2013), suggesting that macrophages may function to prevent scarring during regeneration. Regeneration of various organs in zebrafish requires mobilization and secretion of organ-specific factors by regulatory T cells (Tregs). Interestingly, infiltration of tumors with Tregs has been associated with cancer progression, poor patient prognosis and a higher incidence of metastasis (Bates et al., 2006; Miracco et al., 2007; Petersen et al., 2006; Sato et al., 2005), likely due to associated effects of immunosuppression. However, Tregs also promote stem cell differentiation and tissue regeneration through secretion of growth factors and expression of signaling ligands (Ali et al., 2017; Castiglioni et al., 2015; Hui et al., 2017), suggesting possible additional roles in promoting cancer progression. One possibility is that Tregs could be co-opted in cancer to express proliferative signals to promote tumor growth and expansion. Intra-tumoral Tregs should therefore be interrogated for expression of these pro-regenerative factors [e.g. Ntf3, Nrg1 and Igf1 (Hui et al., 2017), and Jag1 (Ali et al., 2017)] as potential simulators of malignant cell proliferation. In parallel, the participation of Tregs or immunosuppressive and pro-proliferative T cells should be investigated in other examples of epimorphic regeneration, such as salamander limb regeneration. Insights into immune participation in regeneration could inform future cancer therapeutics potentially targeting Tregs.
Beyond immune cell mobilization and recruitment, several studies have demonstrated systemic cell cycle entry following injury that may be mediated by immune-mediated signals. In particular, injury in both axolotl and planaria triggers systemic cell cycle activation (Johnson et al., 2018; Wenemoser and Reddien, 2010) (Fig. 4B). Although the exact mechanism of activation is unknown, it is possible that this effect is immune-cell mediated; indeed, planaria are believed to have a rudimentary innate immune system (reviewed by Peiris et al., 2014). It is possible that this systemic activation is due to a circulating factor such as HGF, as discussed above, or to another as yet unidentified circulating factor. However, it is also possible that immune cells transmit a pro-proliferative signal systemically; for example, through the IL-β-mediated inflammatory response (Castaño et al., 2018) or by increasing levels of stimulatory cytokines that increase pro-proliferative immune recruitment in distant tissues, such as IL6, G-CSF or CCL2 – as observed in cancer (Krall et al., 2018). If a role of these cytokines is found in the systemic wound healing response in axolotl or planaria, it could suggest that typical wound healing may differ from the wound healing that precedes epimorphic regeneration, and that cancers may exploit the latter. Conversely, growth factors (such as HGF) should be investigated in the models of metastases used in the studies above because, although systemic inflammation could be one aspect of the wound healing response that modulates metastatic progression, it remains possible that other circulating growth factors could play a role.
One model is that during cancer progression the initial, metastasis-suppressing, IL1β-mediated immune response suppresses the growth of distant cells in favor of the formation of a tumor-supportive stroma (Fig. 4B). This metastasis-suppressing immune response can be negated or overcome, either spontaneously during tumor development or by surgical resection, in favor of a distinct wound healing response that stimulates metastatic tumor cell proliferation and growth, characterized by IL6, G-CSF or CCL2. This latter wound healing response could be similar to the phenomenon of systemic cell cycle activation observed during regeneration in highly regenerative organisms, and thus the two may share common signaling pathways and similarly engage immune cells. Therefore, immune participation in systemic cell cycle entry should be investigated in regenerative models, and circulating growth factors should be assessed in models of wound-induced metastatic progression. Given similarities in control over distant cell cycle entry and proliferation, these two biological processes warrant investigation in parallel.
It should be noted that the observation of distant cell cycle entry following injury in mice (Rodgers et al., 2017, 2014) may contradict this model, because mice are not considered highly regenerative organisms. However, these studies could suggest that it is the type of injury that triggers one wound healing response versus the other. The model used in these studies relied on chemical injury of mice muscle using barium chloride injection, which could have led to complex tissue injury involving more than just muscle fibers. In the study of systemic cell cycle activation in axolotl, only complex tissue injury, such as amputation, elicits robust cell cycle entry in an uninjured, contralateral limb (Johnson et al., 2018). These studies could suggest that the extent of tissue damage, and not just innate capability for regeneration, could determine the kind of wound healing response that is triggered. Further research into whether or not distinct wound healing responses exist across models of regeneration and if they are harnessed by solid tumors would serve to concretize models of regeneration in the study of biological processes underlying cancer.
Conclusions and future perspectives
This survey of similarities between cancer progression and epimorphic regeneration reveals opportunities to advance our understanding of one using insights from the other. On the one hand, knowledge from cancer could allow us to challenge critical assumptions about regeneration, such as the possibility that distantly recruited endothelial progenitors contribute to the regenerating blastema, the relatively unexplored concept of blastema cell crosstalk and signaling, or the possible roles of blastemal cell genetic and metabolic heterogeneity in driving cell proliferation. On the other hand, regeneration may be an informative model for cancer research, highlighting the roles of various growth factors in stimulating local and systemic cell proliferation, as well as the role of the immune system in promoting local cell proliferation during regeneration. Importantly, parallel analysis of the two processes may highlight complexities of wound healing responses associated with either the regenerative capability of an organism or the extent of tissue damage. Translating our molecular understanding of wound responses in solid tumor progression and metastases to regenerative models will facilitate a deeper understanding into the mechanisms of wound healing and regeneration, and the evolutionary bases for such responses. Regenerative models may also provide another system to study macrophage repolarization and Tregs modulation of cell proliferation. Research that is conscious of both disciplines will help uncover therapeutic avenues for preventing metastasis or unveil changes in circulating growth factors and cytokines that can be used in regenerative medicine. While regeneration and cancer retain certain core differences as research models, emerging studies may allow us to bridge the biological mechanisms found in both and tackle important questions using a unified approach.
Acknowledgements
We are very grateful for the critical comments provided by three anonymous reviewers and for the editorial work of Alex Eve in preparing this Review.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The authors research is funded by the National Institutes of Health (1DP2HD087953-01) and the Richard and Susan Smith Family Foundation (Odyssey Award). Deposited in PMC for release after 12 months.
References
- Ali N., Zirak B., Rodriguez R. S., Pauli M. L., Truong H.-A., Lai K., Ahn R., Corbin K., Lowe M. M., Scharschmidt T. C. et al. (2017). Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell 169, 1119-1129. 10.1016/j.cell.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte R. S., Chen D. S. and Ferrara N. (2019). VEGF in signaling and disease: beyond discovery and development. Cell 176, 1248-1264. 10.1016/j.cell.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeriswyl V. and Christofori G. (2009). The angiogenic switch in carcinogenesis. Semin. Cancer Biol. 19, 329-337. 10.1016/j.semcancer.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Baisse B., Bouzourene H., Saraga E. P., Bosman F. T. and Benhattar J. (2001). Intratumor genetic heterogeneity in advanced human colorectal adenocarcinoma. Int. J. Cancer 93, 346-352. 10.1002/ijc.1343 [DOI] [PubMed] [Google Scholar]
- Bates G. J., Fox S. B., Han C., Leek R. D., Garcia J. F., Harris A. L. and Banham A. H. (2006). Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. 24, 5373-5380. 10.1200/JCO.2006.05.9584 [DOI] [PubMed] [Google Scholar]
- Bayliss P. E., Bellavance K. L., Whitehead G. G., Abrams J. M., Aegerter S., Robbins H. S., Cowan D. B., Keating M. T., O'Reilly T., Wood J. M. et al. (2006). Chemical modulation of receptor signaling inhibits regenerative angiogenesis in adult zebrafish. Nat. Chem. Biol. 2, 265-273. 10.1038/nchembio778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G. and Benjamin L. E. (2003). Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer 3, 401-410. 10.1038/nrc1093 [DOI] [PubMed] [Google Scholar]
- Bernard V., Semaan A., Huang J., San Lucas F. A., Mulu F. C., Stephens B. M., Guerrero P. A., Huang Y., Zhao J., Kamyabi N. et al. (2019). Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin. Cancer Res. 25, 2194-2205. 10.1158/1078-0432.CCR-18-1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil J. C., Quiros M., Nusrat A. and Parkos C. A. (2019). Innate immune cell–epithelial crosstalk during wound repair. J. Clin. Invest. 129, 2983-2993. 10.1172/JCI124618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremnes R. M., Dønnem T., Al-Saad S., Al-Shibli K., Andersen S., Sirera R., Camps C., Marinez I. and Busund L.-T. (2011). The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J. Thorac. Oncol. 6, 209-217. 10.1097/JTO.0b013e3181f8a1bd [DOI] [PubMed] [Google Scholar]
- Bruzzese F., Hagglof C., Leone A., Sjoberg E., Roca M. S., Kiflemariam S., Sjoblom T., Hammarsten P., Egevad L., Bergh A. et al. (2014). Local and systemic protumorigenic effects of cancer-associated fibroblast-derived GDF15. Cancer Res. 74, 3408-3417. 10.1158/0008-5472.CAN-13-2259 [DOI] [PubMed] [Google Scholar]
- Burrell R. A., McGranahan N., Bartek J. and Swanton C. (2013). The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338-345. 10.1038/nature12625 [DOI] [PubMed] [Google Scholar]
- Butler E. G. (1935). Studies on limb regeneration in X-rayed amblystoma larvae. Anat. Rec. 62, 295-307. 10.1002/ar.1090620308 [DOI] [Google Scholar]
- Butler E. G. and O'Brien J. P. (1942). Effects of localized x-radiation on regeneration of the urodele limb. Anat. Rec. 84, 407-413. 10.1002/ar.1090840408 [DOI] [Google Scholar]
- Caiado F., Silva-Santos B. and Norell H. (2016). Intra-tumour heterogeneity - going beyond genetics. FEBS J. 283, 2245-2258. 10.1111/febs.13705 [DOI] [PubMed] [Google Scholar]
- Campbell L. J. and Crews C. M. (2008). Molecular and cellular basis of regeneration and tissue repair: wound epidermis formation and function in urodele amphibian limb regeneration. Cell. Mol. Life Sci. 65, 73-79. 10.1007/s00018-007-7433-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. (2005). VEGF as a key mediator of angiogenesis in cancer. Oncology 69, 4-10. 10.1159/000088478 [DOI] [PubMed] [Google Scholar]
- Castaño Z., San Juan B. P., Spiegel A., Pant A., DeCristo M. J., Laszewski T., Ubellacker J. M., Janssen S. R., Dongre A., Reinhardt F. et al. (2018). IL-1β inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat. Cell Biol. 20, 1084-1097. 10.1038/s41556-018-0173-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni A., Corna G., Rigamonti E., Basso V., Vezzoli M., Monno A., Mondino A., Wagers A. J., Manfredi A. A. and Rovere-Querini P. (2015). FOXP3+ T cells recruited to sites of sterile skeletal muscle injury regulate the fate of satellite cells and guide effective tissue regeneration. PLoS One 10, e0128094 10.1371/journal.pone.0128094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F. and Jazwinska A. (2012). The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development 139, 1921-1930. 10.1242/dev.078543 [DOI] [PubMed] [Google Scholar]
- Charrin S., Jouannet S., Boucheix C. and Rubinstein E. (2014). Tetraspanins at a glance. J. Cell Sci. 127, 3641-3648. 10.1242/jcs.154906 [DOI] [PubMed] [Google Scholar]
- Chen X. and Song E. (2019). Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 18, 99-115. 10.1038/s41573-018-0004-1 [DOI] [PubMed] [Google Scholar]
- Cortez-Retamozo V., Etzrodt M., Newton A., Ryan R., Pucci F., Sio S. W., Kuswanto W., Rauch P. J., Chudnovskiy A., Iwamoto Y. et al. (2013). Angiotensin II drives the production of tumor-promoting macrophages. Immunity 38, 296-308. 10.1016/j.immuni.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie J. D., Kawaguchi A., Traspas R. M., Schuez M., Chara O. and Tanaka E. M. (2016). Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev. Cell 39, 411-423. 10.1016/j.devcel.2016.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Goldstone A. B., Wang H., Farry J., D'Amato G., Paulsen M. J., Eskandari A., Hironaka C. E., Phansalkar R., Sharma B. et al. (2019). A unique collateral artery development program promotes neonatal heart regeneration. Cell 176, 1128-1142.e18. 10.1016/j.cell.2018.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma M., Venneri M. A., Galli R., Sergi L. S., Politi L. S., Sampaolesi M. and Naldini L. (2005). Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell 8, 211-226. 10.1016/j.ccr.2005.08.002 [DOI] [PubMed] [Google Scholar]
- de Thé H. (2018). Differentiation therapy revisited. Nat. Rev. Cancer 18, 117-127. 10.1038/nrc.2017.103 [DOI] [PubMed] [Google Scholar]
- Dvorak H. F. (1986). Tumors: wounds that do not heal. N. Engl. J. Med. 315, 1650-1659. 10.1056/NEJM198612253152606 [DOI] [PubMed] [Google Scholar]
- Dvorak H. F. (2015). Tumors: wounds that do not heal–Redux. Cancer Immunol. Res. 3, 1-11. 10.1158/2326-6066.CIR-14-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eales K. L., Hollinshead K. E. R. and Tennant D. A. (2016). Hypoxia and metabolic adaptation of cancer cells. Oncogenesis 5, e190-e190 10.1038/oncsis.2015.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabets M., Gifford A. M., Scheel C., Nilsson B., Reinhardt F., Bray M.-A., Carpenter A. E., Jirström K., Magnusson K., Ebert B. L. et al. (2011). Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J. Clin. Invest. 121, 784-799. 10.1172/JCI43757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez N., Truitt M., Olson P. and Hanahan D. (2010). Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-κB-dependent manner. Cancer Cell 17, 135-147. 10.1016/j.ccr.2009.12.041 [DOI] [PubMed] [Google Scholar]
- Farrell R. G., Disher W. A., Nesland R. S., Palmatier T. H. and Truhler T. D. (1977). Conservative management of fingertip amputations. J. Am. Coll. Emerg. Physicians 6, 243-246. 10.1016/S0361-1124(77)80461-9 [DOI] [PubMed] [Google Scholar]
- Fischer K. R., Durrans A., Lee S., Sheng J., Li F., Wong S. T. C., Choi H., El Rayes T., Ryu S., Troeger J. et al. (2015). Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472-476. 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdames J. A., Zuniga-Traslavina C., Reyes A. E. and Feijóo C. G. (2014). Gcsf-Chr19 promotes neutrophil migration to damaged tissue through blood vessels in zebrafish. J. Immunol. 193, 372-378. 10.4049/jimmunol.1303220 [DOI] [PubMed] [Google Scholar]
- Gao D. and Mittal V. (2009). The role of bone-marrow-derived cells in tumor growth, metastasis initiation and progression. Trends Mol. Med. 15, 333-343. 10.1016/j.molmed.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S. et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi E., Birchmeier W., Birchmeier C. and Woude G. V. (2012). Targeting MET in cancer: rationale and progress. Nat. Rev. Cancer 12, 89-103. 10.1038/nrc3205 [DOI] [PubMed] [Google Scholar]
- Gilbert R. W. D., Vickaryous M. K. and Viloria-Petit A. M. (2013). Characterization of TGFβ signaling during tail regeneration in the leopard Gecko (Eublepharis macularius): Tgfβ Expression During Tissue Regeneration. Dev. Dyn. 242, 886-896. 10.1002/dvdy.23977 [DOI] [PubMed] [Google Scholar]
- Godwin J. W. and Rosenthal N. (2014). Scar-free wound healing and regeneration in amphibians: Immunological influences on regenerative success. Differentiation 87, 66-75. 10.1016/j.diff.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Godwin J. W., Pinto A. R. and Rosenthal N. A. (2013). Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 110, 9415-9420. 10.1073/pnas.1300290110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin J. W., Debuque R., Salimova E. and Rosenthal N. A. (2017). Heart regeneration in the salamander relies on macrophage-mediated control of fibroblast activation and the extracellular landscape. NPJ Regen. Med. 2, 22 10.1038/s41536-017-0027-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens P., Rodriguez-Vita J., Etzerodt A., Masse M., Rastoin O., Gouirand V., Ulas T., Papantonopoulou O., Van Eck M., Auphan-Anezin N. et al. (2019). Membrane cholesterol efflux drives tumor-associated macrophage reprogramming and tumor progression. Cell Metab. 29, 1376-1389.e4. 10.1016/j.cmet.2019.02.016 [DOI] [PubMed] [Google Scholar]
- Goss R. J. (1956). Regenerative inhibition following limb amputation and immediate insertion into the body cavity. Anat. Rec. 126, 15-27. 10.1002/ar.1091260103 [DOI] [PubMed] [Google Scholar]
- Grange C., Tapparo M., Collino F., Vitillo L., Damasco C., Deregibus M. C., Tetta C., Bussolati B. and Camussi G. (2011). Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic Niche. Cancer Res. 71, 5346-5356. 10.1158/0008-5472.CAN-11-0241 [DOI] [PubMed] [Google Scholar]
- Halbrook C. J., Pontious C., Kovalenko I., Lapienyte L., Dreyer S., Lee H.-J., Thurston G., Zhang Y., Lazarus J., Sajjakulnukit P. et al. (2019). Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 29, 1390-1399.e6. 10.1016/j.cmet.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646-674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hara M., Yuasa S., Shimoji K., Onizuka T., Hayashiji N., Ohno Y., Arai T., Hattori F., Kaneda R., Kimura K. et al. (2011). G-CSF influences mouse skeletal muscle development and regeneration by stimulating myoblast proliferation. J. Exp. Med. 208, 715-727. 10.1084/jem.20101059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori K., Heissig B., Tashiro K., Hongo T., Tateno M., Shieh J.-H., Hackett N. R., Quitoriano M. S., Crystal R. G., Rafii S. et al. (2001). Plasma elevation of stromal cell-derived factor-1 induces mobilization of mature and immature hematopoietic progenitor and stem cells. Blood 97, 3354-3360. 10.1182/blood.V97.11.3354 [DOI] [PubMed] [Google Scholar]
- Hayashiji N., Yuasa S., Miyagoe-Suzuki Y., Hara M., Ito N., Hashimoto H., Kusumoto D., Seki T., Tohyama S., Kodaira M. et al. (2015). G-CSF supports long-term muscle regeneration in mouse models of muscular dystrophy. Nat. Commun. 6, 6745 10.1038/ncomms7745 [DOI] [PubMed] [Google Scholar]
- Hemler M. E. (2014). Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49-60. 10.1038/nrc3640 [DOI] [PubMed] [Google Scholar]
- Hiratsuka S., Watanabe A., Aburatani H. and Maru Y. (2006). Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 8, 1369-1375. 10.1038/ncb1507 [DOI] [PubMed] [Google Scholar]
- Hölters S., Anacker J., Jansen L., Beer-Grondke K., Dürst M. and Rubio I. (2013). Tetraspanin 1 promotes invasiveness of cervical cancer cells. Int. J. Oncol. 43, 503-512. 10.3892/ijo.2013.1980 [DOI] [PubMed] [Google Scholar]
- Hou F.-Q., Lei X.-F., Yao J.-L., Wang Y.-J. and Zhang W. (2015). Tetraspanin 1 is involved in survival, proliferation and carcinogenesis of pancreatic cancer. Oncol. Rep. 34, 3068-3076. 10.3892/or.2015.4272 [DOI] [PubMed] [Google Scholar]
- Huang Y., Harrison M. R., Osorio A., Kim J., Baugh A., Duan C., Sucov H. M. and Lien C.-L. (2013). Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS ONE 8, e67266 10.1371/journal.pone.0067266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S. P., Sheng D. Z., Sugimoto K., Gonzalez-Rajal A., Nakagawa S., Hesselson D. and Kikuchi K. (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659-672.e5. 10.1016/j.devcel.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Johnson K., Bateman J., DiTommaso T., Wong A. Y. and Whited J. L. (2018). Systemic cell cycle activation is induced following complex tissue injury in axolotl. Dev. Biol. 433, 461-472. 10.1016/j.ydbio.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A. and Belmonte J. C. I. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609. 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber M., Papalazarou V., Rouni G., Papageorgopoulou E., Papalois A. and Kostourou V. (2015). Angiotensin II inhibitor facilitates epidermal wound regeneration in diabetic mice. Front. Physiol. 6, 170 10.3389/fphys.2015.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.-Y., Halvorsen O. J., Gravdal K., Bhattacharya N., Lee J. M., Liu N. W., Johnston B. T., Johnston A. B., Haukaas S. A., Aamodt K. et al. (2009). Prosaposin inhibits tumor metastasis via paracrine and endocrine stimulation of stromal p53 and Tsp-1. Proc. Natl. Acad. Sci. USA 106, 12115-12120. 10.1073/pnas.0903120106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Takahashi H., Lin W.-W., Descargues P., Grivennikov S., Kim Y., Luo J.-L. and Karin M. (2009). Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 457, 102-106. 10.1038/nature07623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Gao R., Sei E., Brandt R., Hartman J., Hatschek T., Crosetto N., Foukakis T. and Navin N. E. (2018). Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879-893.e13. 10.1016/j.cell.2018.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich K., Wang H.-X., Sharma C., Fletcher A. L., Turley S. J. and Hemler M. E. (2014). Tetraspanin TSPAN12 regulates tumor growth and metastasis and inhibits β-catenin degradation. Cell. Mol. Life Sci. 71, 1305-1314. 10.1007/s00018-013-1444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komohara Y. and Takeya M. (2017). CAFs and TAMs: maestros of the tumour microenvironment: interactions of CAFs and TAMs. J. Pathol. 241, 313-315. 10.1002/path.4824 [DOI] [PubMed] [Google Scholar]
- Kopp H.-G., Ramos C. A. and Rafii S. (2006). Contribution of endothelial progenitors and proangiogenic hematopoietic cells to vascularization of tumor and ischemic tissue. Curr. Opin. Hematol. 13, 175-181. 10.1097/01.moh.0000219664.26528.da [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H. H. and Tanaka E. M. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60-65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Krall J. A., Reinhardt F., Mercury O. A., Pattabiraman D. R., Brooks M. W., Dougan M., Lambert A. W., Bierie B., Ploegh H. L., Dougan S. K. et al. (2018). The systemic response to surgery triggers the outgrowth of distant immune-controlled tumors in mouse models of dormancy. Sci. Transl. Med. 10, eaan3464 10.1126/scitranslmed.aan3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A., O'Brien C. A., van Galen P., Gan O. I., Notta F., Brown A. M. K., Ng K., Ma J., Wienholds E., Dunant C. et al. (2013). Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 339, 543-548. 10.1126/science.1227670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov H. S., Marsh T., Markens B. A., Castano Z., Greene-Colozzi A., Hay S. A., Brown V. E., Richardson A. L., Signoretti S., Battinelli E. M. et al. (2012). Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov. 2, 1150-1165. 10.1158/2159-8290.CD-12-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine K. J., Epelman S., Uchida K., Weber K. J., Nichols C. G., Schilling J. D., Ornitz D. M., Randolph G. J. and Mann D. L. (2014). Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc. Natl. Acad. Sci. USA 111, 16029-16034. 10.1073/pnas.1406508111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T., Uram J. N., Wang H., Bartlett B. R., Kemberling H., Eyring A. D., Skora A. D., Luber B. S., Azad N. S., Laheru D. et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509-2520. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L. J., Lohr J. G., Harris C. C., Ding L., Wilson R. K. et al. (2012). Landscape of somatic retrotransposition in human cancers. Science 337, 967-971. 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh N. D., Dunlap G. S., Johnson K., Mariano R., Oshiro R., Wong A. Y., Bryant D. M., Miller B. M., Ratner A., Chen A. et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 9, 5153 10.1038/s41467-018-07604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M., Gatien S., Finnson K., Desmeules S., Villiard E., Pilote M., Philip A. and Roy S. (2007). Transforming growth factor beta signaling is essential for limb regeneration in axolotls. PLoS One 2, e1227 10.1371/journal.pone.0001227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Yang X. H., Xu F., Sharma C., Wang H.-X., Knoblich K., Rabinovitz I., Granter S. R. and Hemler M. E. (2013). Tetraspanin CD151 plays a key role in skin squamous cell carcinoma. Oncogene 32, 1772-1783. 10.1038/onc.2012.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Tsao S. W., Chan K. W., Ludwig D. L., Novosyadlyy R., Li Y. Y., He Q. Y. and Cheung A. L. M. (2014). Id1-Induced IGF-II and its autocrine/endocrine promotion of esophageal cancer progression and chemoresistance–implications for IGF-II and IGF-IR-targeted therapy. Clin. Cancer Res. 20, 2651-2662. 10.1158/1078-0432.CCR-13-2735 [DOI] [PubMed] [Google Scholar]
- Liu J., Johnson K., Li J., Piamonte V., Steffy B. M., Hsieh M. H., Ng N., Zhang J., Walker J. R., Ding S. et al. (2011). Regenerative phenotype in mice with a point mutation in transforming growth factor beta type I receptor (TGFBR1). Proc. Natl. Acad. Sci. USA 108, 14560-14565. 10.1073/pnas.1111056108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Dou M., Song X., Dong Y., Liu S., Liu H., Tao J., Li W., Yin X. and Xu W. (2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18, 123 10.1186/s12943-019-1052-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D., Lin L., Monaghan J. R., Cogle C. R., Bova F. J., Maden M. and Scott E. W. (2014). Mapping hematopoiesis in a fully regenerative vertebrate: the axolotl. Blood 124, 1232-1241. 10.1182/blood-2013-09-526970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Yonemochi Y., Nakajyo Y., Hidaka H., Ikeda T. and Ando Y. (2017). CXCL12 and osteopontin from bone marrow-derived mesenchymal stromal cells improve muscle regeneration. Sci. Rep. 7, 3305 10.1038/s41598-017-02928-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A. and Sica A. (2010). Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 22, 231-237. 10.1016/j.coi.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Mantovani A., Sozzani S., Locati M., Allavena P. and Sica A. (2002). Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549-555. 10.1016/S1471-4906(02)02302-5 [DOI] [PubMed] [Google Scholar]
- Marín-Juez R., Marass M., Gauvrit S., Rossi A., Lai S.-L., Materna S. C., Black B. L. and Stainier D. Y. R. (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 113, 11237-11242. 10.1073/pnas.1605431113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Burdon P. C. E., Bridger G., Gutierrez-Ramos J.-C., Williams T. J. and Rankin S. M. (2003). Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity 19, 583-593. 10.1016/S1074-7613(03)00263-2 [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Umitsu M., De Silva D. M., Roy A. and Bottaro D. P. (2017). Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 108, 296-307. 10.1111/cas.13156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister S. S. and Weinberg R. A. (2014). The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 16, 717-727. 10.1038/ncb3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister S. S., Gifford A. M., Greiner A. L., Kelleher S. P., Saelzler M. P., Ince T. A., Reinhardt F., Harris L. N., Hylander B. L., Repasky E. A. et al. (2008). Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell 133, 994-1005. 10.1016/j.cell.2008.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C., Lehrberg J. and Gardiner D. (2014). Position-specific induction of ectopic limbs in non-regenerating blastemas on axolotl forelimbs: inducing regeneration in non-regenerative wounds. Regeneration 1, 27-34. 10.1002/reg2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C., Bryant S. V. and Gardiner D. M. (2015). The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods: the Axolotl Limb Blastema. Regeneration 2, 54-71. 10.1002/reg2.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N. and Swanton C. (2017). Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613-628. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- Mescher A. L. (1976). Effects on adult newt limb regeneration of partial and complete skin flaps over the amputation surface. J. Exp. Zool. 195, 117-127. 10.1002/jez.1401950111 [DOI] [PubMed] [Google Scholar]
- Mescher A. L. (1996). The cellular basis of limb regeneration in urodeles. Int. J. Dev. Biol. 50, 785-795. [PubMed] [Google Scholar]
- Miracco C., Mourmouras V., Biagioli M., Rubegni P., Mannucci S., Monciatti I., Cosci E., Tosi P. and Luzi P. (2007). Utility of tumour-infiltrating CD25+FOXP3+ regulatory T cell evaluation in predicting local recurrence in vertical growth phase cutaneous melanoma. Onc. Rep. 18, 1115-1122. 10.3892/or.18.5.1115 [DOI] [PubMed] [Google Scholar]
- Munkley J., McClurg U. L., Livermore K. E., Ehrmann I., Knight B., Mccullagh P., Mcgrath J., Crundwell M., Harries L. W., Leung H. Y. et al. (2017). The cancer-associated cell migration protein TSPAN1 is under control of androgens and its upregulation increases prostate cancer cell migration. Sci. Rep. 7, 5249 10.1038/s41598-017-05489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Sakai K., Nakamura T. and Matsumoto K. (2011). Hepatocyte growth factor twenty years on: much more than a growth factor: hepatocyte growth factor. J. Gastroenterol. Hepatol. 26, 188-202. 10.1111/j.1440-1746.2010.06549.x [DOI] [PubMed] [Google Scholar]
- Nguyen-Chi M., Laplace-Builhe B., Travnickova J., Luz-Crawford P., Tejedor G., Phan Q. T., Duroux-Richard I., Levraud J.-P., Kissa K., Lutfalla G. et al. (2015). Identification of polarized macrophage subsets in zebrafish. eLife 4, e07288 10.7554/eLife.07288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Chi M., Laplace-Builhé B., Travnickova J., Luz-Crawford P., Tejedor G., Lutfalla G., Kissa K., Jorgensen C. and Djouad F. (2017). TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell Death Dis. 8, e2979-e2979 10.1038/cddis.2017.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman K. M., Kenny H. A., Penicka C. V., Ladanyi A., Buell-Gutbrod R., Zillhardt M. R., Romero I. L., Carey M. S., Mills G. B., Hotamisligil G. S. et al. (2011). Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498-1503. 10.1038/nm.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi M. A., Spencer C. A., Leung D. H. and Leung T. H. (2018). Aging suppresses skin-derived circulating SDF1 to promote full-thickness tissue regeneration. Cell Rep. 24, 3383-3392.e5. 10.1016/j.celrep.2018.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan D. J., Ciarrocchi A., Mellick A. S., Jaggi J. S., Bambino K., Gupta S., Heikamp E., McDevitt M. R., Scheinberg D. A., Benezra R. et al. (2007). Bone marrow-derived endothelial progenitor cells are a major determinant of nascent tumor neovascularization. Genes Dev. 21, 1546-1558. 10.1101/gad.436307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novianti T., Juniantito V., Jusuf A. A., Arida E. A., Jusman S. W. A. and Sadikin M. (2019). Expression and role of HIF-1α and HIF-2α in tissue regeneration: a study of hypoxia in house gecko tail regeneration. Organogenesis 15, 69-84. 10.1080/15476278.2019.1644889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojalvo L. S., King W., Cox D. and Pollard J. W. (2009). High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am. J. Pathol. 174, 1048-1064. 10.2353/ajpath.2009.080676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu B., Galemmo R., Janetka J. and Klampfer L. (2017). Hepatocyte growth factor, a key tumor-promoting factor in the tumor microenvironment. Cancers 9, 35 10.3390/cancers9040035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L. et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396-1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peadon A. M. and Singer M. (1966). The blood vessels of the regenerating limb of the adult newt, Triturus. J. Morphol. 118, 79-89. 10.1002/jmor.1051180106 [DOI] [PubMed] [Google Scholar]
- Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C. M. et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883-891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris T. H., Hoyer K. K. and Oviedo N. J. (2014). Innate immune system and tissue regeneration in planarians: An area ripe for exploration. Semin. Immunol. 26, 295-302. 10.1016/j.smim.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R. P., Campa M. J., Sperlazza J., Conlon D., Joshi M.-B., Harpole D. H. and Patz E. F. (2006). Tumor infiltrating Foxp3+ regulatory T-cells are associated with recurrence in pathologic stage I NSCLC patients. Cancer 107, 2866-2872. 10.1002/cncr.22282 [DOI] [PubMed] [Google Scholar]
- Petrie T. A., Strand N. S., Yang C.-T., Rabinowitz J. S. and Moon R. T. (2014). Macrophages modulate adult zebrafish tail fin regeneration. Development 141, 2581-2589. 10.1242/dev.098459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitot H. C. (1993). The molecular biology of carcinogenesis. Cancer 72, 962-970. [DOI] [PubMed] [Google Scholar]
- Pollard J. W. (2004). Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 4, 71-78. 10.1038/nrc1256 [DOI] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G. and Keating M. T. (2002). Heart regeneration in Zebrafish. Science 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Puram S. V., Tirosh I., Parikh A. S., Patel A. P., Yizhak K., Gillespie S., Rodman C., Luo C. L., Mroz E. A., Emerick K. S. et al. (2017). Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611-1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman S., Elewa A. and Seifert A. W. (2019). Fgf-signaling is compartmentalized within the mesenchyme and controls proliferation during salamander limb development. eLife 8, e48507 10.7554/eLife.48507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.-Z. and Pollard J. W. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39-51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattigan Y. I., Patel B. B., Ackerstaff E., Sukenick G., Koutcher J. A., Glod J. W. and Banerjee D. (2012). Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp. Cell Res. 318, 326-335. 10.1016/j.yexcr.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W. (2018). The cellular and molecular basis for planarian regeneration. Cell 175, 327-345. 10.1016/j.cell.2018.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabov V., Gudima A., Wang N., Mickley A., Orekhov A. and Kzhyshkowska J. (2014). Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front. Physiol. 5, 75 10.3389/fphys.2014.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritenour A. M. and Dickie R. (2017). Inhibition of vascular endothelial growth factor receptor decreases regenerative angiogenesis in axolotls. Anat. Rec. 300, 2273-2280. 10.1002/ar.23689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi N. A., Hellmann M. D., Snyder A., Kvistborg P., Makarov V., Havel J. J., Lee W., Yuan J., Wong P., Ho T. S. et al. (2015). Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 348, 124-128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson-Tessi M., Gillies R. J., Gatenby R. A. and Anderson A. R. A. (2015). Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res. 75, 1567-1579. 10.1158/0008-5472.CAN-14-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. T., King K. Y., Brett J. O., Cromie M. J., Charville G. W., Maguire K. K., Brunson C., Mastey N., Liu L., Tsai C.-R. et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature 510, 393-396. 10.1038/nature13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. T., Schroeder M. D., Ma C. and Rando T. A. (2017). HGFA is an injury-regulated systemic factor that induces the transition of stem cells into G alert. Cell Rep. 19, 479-486. 10.1016/j.celrep.2017.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L. D., Ghosh S., Pathangey L. B., Tinder T. L., Gruber H. E. and Mukherjee P. (2011). Collagen induced arthritis increases secondary metastasis in MMTV-PyV MT mouse model of mammary cancer. BMC Cancer 11, 365 10.1186/1471-2407-11-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammarco M. C., Simkin J., Fassler D., Cammack A. J., Wilson A., Van Meter K. and Muneoka K. (2014). Endogenous bone regeneration is dependent upon a dynamic oxygen event: endogenous bone regeneration is dependent upon a dynamic oxygen event. J. Bone Miner. Res. 29, 2336-2345. 10.1002/jbmr.2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato E., Olson S. H., Ahn J., Bundy B., Nishikawa H., Qian F., Jungbluth A. A., Frosina D., Gnjatic S., Ambrosone C. et al. (2005). Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. 102, 18538-18543. 10.1073/pnas.0509182102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sceneay J., Chow M. T., Chen A., Halse H. M., Wong C. S. F., Andrews D. M., Sloan E. K., Parker B. S., Bowtell D. D., Smyth M. J. et al. (2012). Primary tumor hypoxia recruits CD11b+/Ly6Cmed/Ly6G+ immune suppressor cells and compromises NK cell cytotoxicity in the premetastatic niche. Cancer Res. 72, 3906-3911. 10.1158/0008-5472.CAN-11-3873 [DOI] [PubMed] [Google Scholar]
- Schouppe E., De Baetselier P., Van Ginderachter J. A. and Sarukhan A. (2012). Instruction of myeloid cells by the tumor microenvironment: open questions on the dynamics and plasticity of different tumor-associated myeloid cell populations. OncoImmunology 1, 1135-1145. 10.4161/onci.21566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Kravarik K. M., Lapan S. W. and Reddien P. W. (2014). Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 3, 339-352. 10.1016/j.stemcr.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A. W. and Muneoka K. (2018). The blastema and epimorphic regeneration in mammals. Dev. Biol. 433, 190-199. 10.1016/j.ydbio.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaei F., Wu X., Malik A. K., Zhong C., Baldwin M. E., Schanz S., Fuh G., Gerber H.-P. and Ferrara N. (2007). Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat. Biotechnol. 25, 911-920. 10.1038/nbt1323 [DOI] [PubMed] [Google Scholar]
- Simkin J., Gawriluk T. R., Gensel J. C. and Seifert A. W. (2017a). Macrophages are necessary for epimorphic regeneration in African spiny mice. eLife 6, e24623 10.7554/eLife.24623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin J., Sammarco M. C., Marrero L., Dawson L. A., Yan M., Tucker C., Cammack A. and Muneoka K. (2017b). Macrophages are required to coordinate mouse digit tip regeneration. Development 144, 3907-3916. 10.1242/dev.150086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A. J. and Clark R. A. (1999). Cutaneous wound healing. N. Engl. J. Med. 341, 738-746. 10.1056/NEJM199909023411006 [DOI] [PubMed] [Google Scholar]
- Steeg P. S. (2016). Targeting metastasis. Nat. Rev. Cancer 16, 201-218. 10.1038/nrc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickaert A., Saiselet M., Dom G., De Deken X., Dumont J. E., Feron O., Sonveaux P. and Maenhaut C. (2017). Cancer heterogeneity is not compatible with one unique cancer cell metabolic map. Oncogene 36, 2637-2642. 10.1038/onc.2016.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski F. K., Nahrendorf M., Etzrodt M., Wildgruber M., Cortez-Retamozo V., Panizzi P., Figueiredo J.-L., Kohler R. H., Chudnovskiy A., Waterman P. et al. (2009). Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612-616. 10.1126/science.1175202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry S., Buart S. and Chouaib S. (2017). Hypoxic stress-induced tumor and immune plasticity, suppression, and impact on tumor heterogeneity. Front. Immunol. 8, 1625 10.3389/fimmu.2017.01625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C. S. (1957). The effect of apical cap removal on limb regeneration in Amblystoma larvae. J. Exp. Zool. 134, 357-381. 10.1002/jez.1401340209 [DOI] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S. M., Wadsworth M. H., Treacy D., Trombetta J. J., Rotem A., Rodman C., Lian C., Murphy G. et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189-196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. L., Baselga-Garriga C. and Melton D. A. (2019). Blastemal progenitors modulate immune signaling during early limb regeneration. Development 146, dev169128 10.1242/dev.169128 [DOI] [PubMed] [Google Scholar]
- Tsarouchas T. M., Wehner D., Cavone L., Munir T., Keatinge M., Lambertus M., Underhill A., Barrett T., Kassapis E., Ogryzko N. et al. (2018). Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages in zebrafish spinal cord regeneration. Nat. Commun. 9, 4670 10.1038/s41467-018-07036-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou C.-L., Peters W., Si Y., Slaymaker S., Aslanian A. M., Weisberg S. P., Mack M. and Charo I. F. (2007). Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 117, 902-909. 10.1172/JCI29919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G. (2011). Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10, 671-684. 10.1038/nrd3504 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. and Thompson C. B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029-1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. E., Wang I. E. and Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811-816. 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I., Wang H., Weissert P. M., Straube W. L., Shevchenko A., Gentzel M., Brito G., Tazaki A., Oliveira C., Sugiura T. et al. (2017). Serum proteases potentiate BMP-induced cell cycle re-entry of dedifferentiating muscle cells during newt limb regeneration. Dev. Cell 40, 608-617.e6. 10.1016/j.devcel.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Wagner J., Rapsomaniki M. A., Chevrier S., Anzeneder T., Langwieder C., Dykgers A., Rees M., Ramaswamy A., Muenst S., Soysal S. D. et al. (2019). A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330-1345.e18. 10.1016/j.cell.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasgewatte Wijesinghe D. K., Mackie E. J. and Pagel C. N. (2019). Normal inflammation and regeneration of muscle following injury require osteopontin from both muscle and non-muscle cells. Skelet. Muscle 9, 6 10.1186/s13395-019-0190-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wculek S. K. and Malanchi I. (2015). Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 528, 413-417. 10.1038/nature16140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D. and Reddien P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979-991. 10.1016/j.ydbio.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng W., Li H. and Goel A. (2019). Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim. Biophys. Acta Rev. Cancer 1871, 160-169. 10.1016/j.bbcan.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited J. L. and Tabin C. J. (2009). Limb regeneration revisited. J. Biol. 8, 5 10.1186/jbiol105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. R., Fridlyand J., Yan Y., Penuel E., Burton L., Chan E., Peng J., Lin E., Wang Y., Sosman J. et al. (2012). Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505-509. 10.1038/nature11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler I. G., Sims N. A., Pettit A. R., Barbier V., Nowlan B., Helwani F., Poulton I. J., van Rooijen N., Alexander K. A., Raggatt L. J. et al. (2010). Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 116, 4815-4828. 10.1182/blood-2009-11-253534 [DOI] [PubMed] [Google Scholar]
- Wright D. E., Wagers A. J., Gulati A. P., Johnson F. L. and Weissman I. L. (2001). Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933-1936. 10.1126/science.1064081 [DOI] [PubMed] [Google Scholar]
- Xiao-Jie L., Hui-Ying X., Qi X., Jiang X. and Shi-Jie M. (2016). LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet. Med. 18, 431-439. 10.1038/gim.2015.119 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Galvez S., Tiwari S., Rezk B. M., Semprun-Prieto L., Higashi Y., Sukhanov S., Yablonka-Reuveni Z. and Delafontaine P. (2013). Angiotensin II inhibits satellite cell proliferation and prevents skeletal muscle regeneration. J. Biol. Chem. 288, 23823-23832. 10.1074/jbc.M112.449074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P., Wang Y.-H., Si M., Gu J.-H., Li P., Lu P.-H. and Chen M.-B. (2017). Tetraspanin CD151 as an emerging potential poor prognostic factor across solid tumors: a systematic review and meta-analysis. Oncotarget 8, 5592-5602. 10.18632/oncotarget.13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A., Li H., Guo L., Gao X., McKinney S., Wang Y., Yu Z., Park J., Semerad C., Ross E. et al. (2018). Prospectively isolated Tetraspanin+ neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593-1608.e20. 10.1016/j.cell.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.-W., Liu L., Gong C.-Y., Shi H.-S., Zeng Y.-H., Wang X.-Z., Zhao Y.-W. and Wei Y.-Q. (2012). Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS ONE 7, e50946 10.1371/journal.pone.0050946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Kuo D., Nathanson J., Satoh A., Pao G. M., Yeo G. W., Bryant S. V., Voss S. R., Gardiner D. M. and Hunter T. (2012a). Retrotransposon long interspersed nucleotide element-1 (LINE-1) is activated during salamander limb regeneration. Dev. Growth Differ. 54, 673-685. 10.1111/j.1440-169X.2012.01368.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Pao G. M., Satoh A., Cummings G., Monaghan J. R., Harkins T. T., Bryant S. V., Randal Voss S., Gardiner D. M. and Hunter T. (2012b). Activation of germline-specific genes is required for limb regeneration in the Mexican axolotl. Dev. Biol. 370, 42-51. 10.1016/j.ydbio.2012.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]