ABSTRACT
Embryonic interneuron development underlies cortical function and its disruption contributes to neurological disease. Yet the mechanisms by which viable interneurons are produced from progenitors remain poorly understood. Here, we demonstrate dosage-dependent requirements of the exon junction complex component Magoh for interneuron genesis in mouse. Conditional Magoh ablation from interneuron progenitors, but not post-mitotic neurons, depletes cortical interneuron number through adulthood, with increased severity in homozygotes. Using live imaging, we discover that Magoh deficiency delays progenitor mitotic progression in a dosage-sensitive fashion, with 40% of homozygous progenitors failing to divide. This shows that Magoh is required in progenitors for both generation and survival of newborn progeny. Transcriptome analysis implicates p53 signaling; moreover, p53 ablation in Magoh haploinsufficient progenitors rescues apoptosis, completely recovering interneuron number. In striking contrast, in Magoh homozygotes, p53 loss fails to rescue interneuron number and mitotic delay, further implicating mitotic defects in interneuron loss. Our results demonstrate that interneuron development is intimately dependent upon progenitor mitosis duration and uncover a crucial post-transcriptional regulator of interneuron fate relevant for neurodevelopmental pathologies.
This article has an associated ‘The people behind the papers’ interview.
KEY WORDS: Interneuron, RNA binding, Mitosis, Apoptosis, Progenitor, p53, Mouse
Highlighted Article: Genetics, live imaging and genomics demonstrate that the RNA-binding protein Magoh, which controls progenitor mitosis, interneuron generation and survival, is required in a dosage-dependent manner for interneuron development.
INTRODUCTION
The cerebral cortex is integral to higher cognition, including analytical thought and decision making. Disruptions in neurotransmission are hallmarks of multiple neurological disorders, including autism, epilepsy and schizophrenia. These disorders are thought to result from an imbalance in the number of excitatory glutamatergic neurons and inhibitory GABAergic interneurons (Takesian and Hensch, 2013; Marín, 2012). Importantly, cortical interneuron number and function depend upon proper generation and survival of GABAergic inhibitory interneurons during embryonic development.
In mammals, cortical interneurons are generated in the ganglionic eminence, primarily in the medial (MGE) and caudal (CGE) ganglionic eminences (Fig. 1A) (Bandler et al., 2017; Hu et al., 2017b; Sultan and Shi, 2018; Anderson et al., 1997; Sultan et al., 2016). In mice, cortical interneuron neurogenesis is accomplished between embryonic days (E) 10 and 17. Within the ventricular zone (VZ) and sub-ventricular zone (SVZ), cortical interneurons are produced by radial glia and intermediate progenitors (IPs), respectively (Petros et al., 2015; Turrero Garcia and Harwell, 2017). The MGE gives rise to the majority of cortical interneurons, including somatostatin (SST) and parvalbumin (PV) subclasses. SST interneurons are born earlier with peak generation around E12.5 whereas PV interneuron generation peaks around E14 (Bandler et al., 2017; Inan et al., 2012). Single cell sequencing reveals that heterogeneous progenitors contribute to this interneuron diversity (Mayer et al., 2018; Mi et al., 2018; Chen et al., 2017). Further, manipulation of progenitor attributes, such as polarity and excitability, can bias production of interneuron subtypes (Petros et al., 2015; Wamsley and Fishell, 2017). Yet there is still a limited understanding of interneuron neurogenesis, including the role of the cell cycle in fate specification (Glickstein et al., 2007; Ross, 2011). In this context, clonal live imaging of individual progenitors and progeny is invaluable, but these approaches have not been previously implemented for investigation of interneurons.
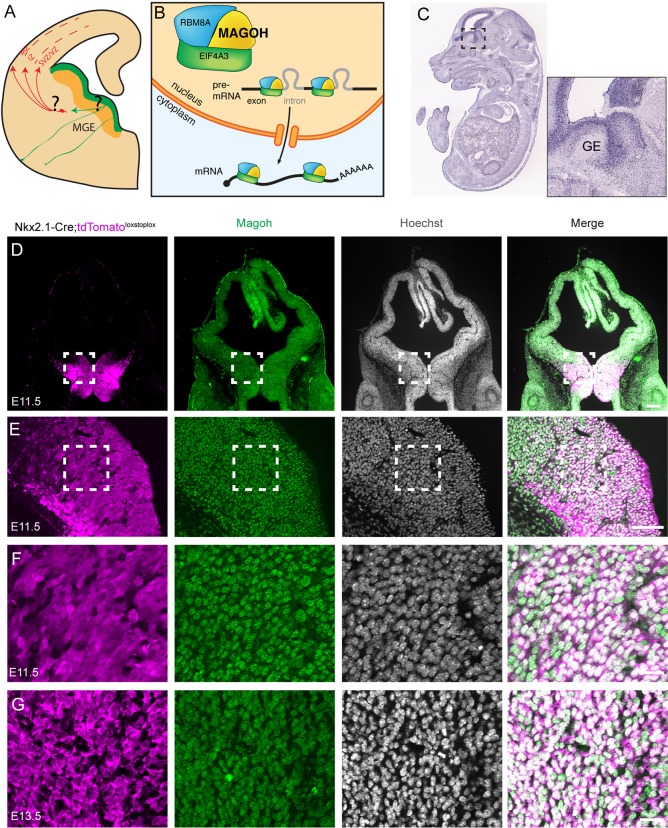
Fig. 1.
Magoh is expressed within germinal zones of the mouse embryonic ganglionic eminence. (A) Schematic of embryonic interneuron development depicting the medial ganglionic eminence (MGE), where progenitors (green) produce interneurons (red), which then migrate into the cortex. This study investigates requirements for Magoh in cellular processes demarcated by questions marks. (B) Schematic of the exon junction complex (EJC) bound to pre-mRNA and mRNA in the nucleus and cytoplasm, respectively. (C) Magoh in situ hybridization of an E14.5 embryo with boxed region (right) depicting robust expression in progenitor zones of the ganglionic eminences (GE). (D-F) Coronal sections of E11.5 Nkx2.1-Cre;RosaloxSTOPloxtdTomato brains co-immunostained for Magoh (green), RFP (tdTomato, magenta) and Hoechst DNA stain (white). The boxed areas in D and E are magnified in E and F, respectively. (G) High-magnification images of E13.5 MGE co-stained as in F. Scale bars: 200 µm (D); 50 µm (E); 25 µm (F,G).
Following their genesis, newborn interneurons migrate tangentially through the marginal, intermediate and subventricular zones of the dorsal telencephalon. Once in the dorsal telencephalon, interneurons migrate radially along cortical radial glial scaffolds to populate layers composed of excitatory neurons. They do so clonally such that lineage-specific populations ultimately reside in different laminar locations (Ciceri et al., 2013; Brown et al., 2011; Sultan et al., 2016; Harwell et al., 2015; Mayer et al., 2015).
Molecular and genetic studies have implicated specific transcription factors, signaling pathways and morphogenic gradients in subtype interneuron generation, survival and migration (Mayer et al., 2018; Mi et al., 2018; Hu et al., 2017b; Tischfield et al., 2017; Guo and Anton, 2014). These intrinsic and extrinsic factors function within radial glia, IPs and/or post-mitotic interneurons to modulate development. Yet, with the exception of a few examples (Wamsley et al., 2018), roles for post-transcriptional regulation in interneuron development remain largely unexplored.
The exon junction complex (EJC) is an outstanding candidate for influencing interneuron development post-transcriptionally (Fig. 1B). The EJC, composed of core components Magoh, Rbm8a and Eif4a3, binds to mRNA to regulate nonsense-mediated decay (NMD) and mRNA localization, translation and splicing (Kataoka et al., 2000; van Eeden et al., 2001; Shibuya et al., 2004; Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). Importantly, human mutations including copy number variations (CNVs) in EJC components are associated with intellectual disability, autism and microcephaly (McMahon et al., 2016). These neurological disorders can arise from aberrant development of interneurons and excitatory neurons; although to date most investigations of microcephaly have only focused on glutamatergic neurogenesis (Gilmore and Walsh, 2012). Notably, the EJC, and in particular Magoh, is crucial for the production of excitatory neurons, but its role in inhibitory neurogenesis is entirely unknown (McMahon et al., 2014, 2016; Mao et al., 2015, 2016; Pilaz et al., 2016). This is a significant gap in our knowledge as we posit that dysregulation of interneuron generation may contribute to EJC-dependent disease etiology. Altogether, this begs the question of whether Magoh functions in interneuron development.
Here, we use mouse models to demonstrate essential dosage-sensitive requirements for Magoh in interneuron generation. We discover that reduced expression of Magoh in interneuron progenitors, but not post-mitotic interneurons, depletes interneurons within the neocortex. Using transcriptomics, genetics, and live imaging, we pinpoint disrupted mitosis and p53 (Trp53) signaling mechanisms. Our findings implicate Magoh as a crucial regulator of interneuron development and further provide clues for how EJC dysfunction may contribute to neurological disorders in humans.
RESULTS
Magoh is highly expressed in MGE progenitors and controls interneuron number
To first assess roles for Magoh in GABAergic interneuron development, we examined Magoh expression in the developing brain. By in situ hybridization, Magoh transcript was highly enriched in both ganglionic eminences at E14.5 (Fig. 1C) (Visel et al., 2004). To assess Magoh protein expression, Nkx2.1-Cre; Rosa26loxStoplox-tdTomato mice were used to label radial glia of the MGE, beginning at E9.5 (Xu et al., 2008). Using immunofluorescence, Magoh expression was evident throughout the developing ganglionic eminence at E11.5 and E13.5, spanning the early and middle developmental stages of neurogenesis (Fig. 1D,E). A higher-magnification view reflects Magoh expression in most Tomato+ cells at both stages (Fig. 1F,G).This expression pattern was independently validated with previously published mouse single cell expression data at E14.5 and postnatal day (P) 0 (Fig. S1A) (Loo et al., 2019). Magoh transcript was also highly expressed within interneuron progenitors of the human fetal cortex (Nowakowski et al., 2017) (Fig. S1B). This indicates that Magoh expression in interneuron progenitors is conserved between mice and humans.
To test the requirement of Magoh in MGE progenitors, we next conditionally depleted Magoh in radial glia using Nkx2.1-Cre. Again, we utilized tdTomato to label Nkx2.1-Cre expressing progenitors and their interneuron progeny. In order to monitor interneuron number, we quantified tdTomato+ cells that had migrated into the dorsal telencephalon by E14.5 and E18.5 (Fig. 2A,D). At E14.5, compared with control (Nkx2.1-Cre;ROSAloxStoplox-tdTomato), Magoh conditional heterozygote (cHet; Nkx2.1-Cre; ROSAloxStoplox-tdTomato; Magohlox/+) brains displayed a 30% reduction of cortical interneurons (Fig. 2A-C). By E18.5, cHets displayed a more severe and significant 50% reduction of interneurons within the dorsal neocortex compared with control (P<0.001) (Fig. 2D-F). To determine if Magoh controls generation of GABAergic interneurons in a dosage-dependent fashion, we quantified interneuron number in Magoh conditional knockout (cKO; Nkx2.1-Cre;ROSAloxStoplox-tdTomato; Magohlox/lox) mice. Conditional homozygosity for Magoh led to an even more striking loss of cortical interneurons, with 85% and 80% depletion at E14.5 and E18.5, respectively (Fig. 2A-F). These data demonstrate that Magoh is essential for cortical interneuron number in a dosage-dependent fashion.
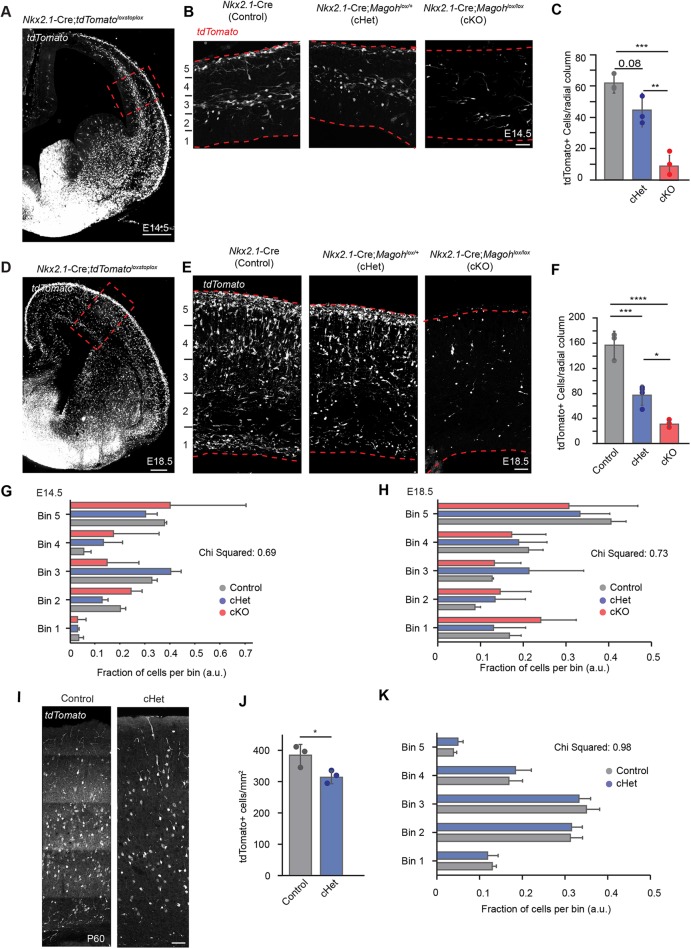
Fig. 2.
Magoh depletion from MGE progenitors results in a dosage-dependent loss of cortical interneurons in embryos and adults. (A,D) Coronal sections of Nkx2.1-Cre;RosaloxSTOPloxtdTomato E14.5 (A) and E18.5 (D) cortices depicting Cre-derived tdTomato+ interneurons. (B,E) High-magnification images of the boxed regions in A and D of E14.5 (B) and E18.5 (E) Nkx2.1-Cre;RosaloxSTOPloxtdTomato (control, left), Nkx2.1-Cre;Magohlox/+; RosaloxSTOPloxtdTomato (cHet, middle), and Nkx2.1-Cre;Magohlox/lox;RosaloxSTOPloxtdTomato (cKO, right). (C,F) Quantification of total tdTomato+ cells per 250 µm wide radial column for the indicated genotypes at E14.5 (C) and E18.5 (F). (G,H) Quantification of total tdTomato+ cells in five equally spaced bins, with Bin 5 at the pia and Bin 1 at the ventricle at E14.5 (G) and E18.5 (H). (I) Coronal sections of P60 control (left) and cHet (right) cortices depicting Cre-derived tdTomato+ interneurons (white). (J) Quantification of tdTomato+ cell density in 350 µm wide radial column of control and conditional heterozygous P60 cortex. (K) Quantification of tdTomato+ cell distribution in five equally spaced bins as in G,H. For all graphs, individual dots are biological replicates and error bars represent s.d.; a.u., arbitrary units. For B,E,I, cortices are oriented with ventricle at the bottom. A-H: n=3-4 brains each, ≥2 sections/brain; I-K: n=3 brains each, n=6 sections/brain. C,F: ANOVA with Tukey post-hoc, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; G: χ2=0.692; H: χ2=0.918; J: two-tailed unpaired Student's t-test, *P=0.0385; K: χ2=0.98. Scale bars: 300 µm (A,D); 50 µm (B,E,I).
Interneurons pursue distinct tangential migratory paths into the cortex before moving radially along radial glia. Further, interneurons born early in development populate deeper layers of the cortex whereas later-born interneurons populate more superficial regions (Miller, 1985; Fairen et al., 1986; Peduzzi, 1988). We therefore asked if Magoh controls interneuron distribution in the cortex as a proxy for investigating its role in specifying early- versus late-born interneuron populations. For this, we quantified the relative proportion of remaining tdTomato+ interneurons within five equivalent bins spanning the cortical VZ to the pia. Neither heterozygotes nor homozygotes showed a significant difference in interneuron distribution compared with controls at E14.5 or at E18.5 (χ2=0.69 and 0.73, respectively) (Fig. 2G,H). These data indicate that although Magoh depletion reduces overall interneuron number, the remaining interneurons are still distributed normally within a radial column of the cortex. This suggests that development of both early- and late-born interneurons are equally affected by Magoh depletion.
We next asked if interneuron loss persists into adulthood. For this, cortical interneuron number was quantified in Nkx2.1-Cre; ROSAloxStoplox-tdTomato and Nkx2.1-Cre; ROSAloxStoplox-tdTomato; Magohlox/+ adult brains. Nkx2.1-Cre;Magohlox/lox mutants failed to survive into adulthood, presumably because of Nkx2.1-Cre expression outside the brain, thus precluding analysis of this genotype. tdTomato+ cells were quantified in the neocortex of 2-month-old (P60) adult mice. The number of Nkx2.1-derived GABAergic interneurons in Magoh haploinsufficient adult brains was significantly lower than that in control littermates by 18% (P<0.05) (Fig. 2I,J). The fold-reduction observed postnatally was markedly less than at embryonic stages, suggesting there may be compensatory mechanisms at play postnatally. As in the embryonic neocortex, binning analysis showed no bias in interneurons populating a specific cortical layer (Fig. 2K). However, quantification of SST and PV interneurons showed a slight, albeit not quite significant, shift in the ratio of these populations, with trends towards more PV and fewer SST interneurons (Fig. S2A-D). This suggests that Magoh could function in subtype fate specification and/or differential control of SST- and PV-producing progenitors (Hu et al., 2017b). Taken together, these findings demonstrate that Magoh is essential for establishing proper cortical interneuron number into adulthood, and functions in a dosage-dependent fashion.
Magoh controls interneuron progenitor mitosis and progeny survival
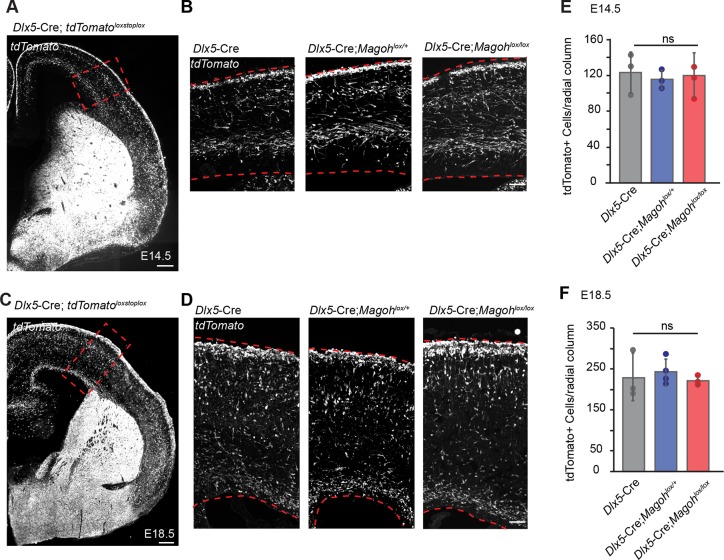
We reasoned that reduced cortical interneuron number could result from Magoh expression in progenitors and/or its requirements for migration. To discriminate between these possibilities, we depleted Magoh from migrating post-mitotic interneurons by crossing Magohlox/lox mice with Dlx5/6-Cre, which is expressed in newly born post-mitotic neurons prior to their migration into the cortex (Monory et al., 2006; Wang et al., 2010). Quantification of tdTomato+ neurons within the neocortex revealed similar numbers of interneurons in control, Dlx5/6-Cre;Magohlox/+ and Dlx5/6-Cre;Magohlox/lox E14.5 and E18.5 brains (Fig. 3). As Dlx5/6-Cre is expressed in both MGE- and CGE-derived interneurons, Magoh could differentially control survival and migration of both populations. We assessed apoptosis in the ventral telencephalon using cleaved caspase 3 (CC3), an apoptosis marker. However, we observed no cell death in either ganglionic eminence for the cHet or cKO. Moreover, we did not observe any striking difference in the pattern of migratory tdTomato+ interneurons into the cortex (Fig. S3). Together with the Nkx2.1-Cre depletion experiments (Fig. 2), these data argue that, although interneuron progenitors are exquisitely sensitive to Magoh levels, Magoh is largely dispensable in post-mitotic interneurons for their tangential migration and survival.
Fig. 3.
Magoh is dispensable in post-mitotic cortical interneurons for their migration and number. (A,C) Coronal sections of Dlx5/6-Cre;RosaloxSTOPloxtdTomato E14.5 (A) and E18.5 (C) cortices showing Cre-derived interneurons. (B,D) High-magnification images of the boxed regions in A and C of E14.5 (B) and E18.5 (D) Dlx5/6-Cre; RosaloxSTOPloxtdTomato (left), Dlx5/6-Cre; Magohlox/+;RosaloxSTOPloxtdTomato (middle) and Dlx5/6-Cre; Magohlox/lox;RosaloxSTOPloxtdTomato (right). Brains are oriented with ventricle at the bottom. (E,F) Quantification of total tdTomato+ cells per 250 µm-wide radial column at E14.5 (E) and E18.5 (F). n=3 for all conditions; ≥2 sections/embryo. ANOVA with Tukey post-hoc: P=0.891 (E), P=0.793 (F); ns, not significant. Individual dots represent biological replicates. Error bars represent s.d. Scale bars: 200 µm (A); 300 µm (C); 50 µm (B,D).
The genetic experiments using Nkx2.1-Cre and Dlx5/6-Cre drivers argue that Magoh controls interneuron number by modulating progenitor behavior. But how might Magoh regulate progenitors? We postulated that there could be defects in cell proliferation or cell survival. Notably, siRNA-mediated ablation of Magoh in immortalized cells causes a drastic mitotic arrest (Silver et al., 2010; Ishigaki et al., 2013). Likewise, in the cortex, Magoh haploinsufficiency prolongs mitosis of excitatory progenitors (McMahon et al., 2014; Pilaz et al., 2016; Silver et al., 2010). This mitotic defect is associated with increased neuronal apoptosis and depletion of the progenitor pool, which ultimately results in fewer neurons. Given these observations, we hypothesized that in the MGE, Magoh similarly controls interneuron number by influencing progenitor mitosis and subsequent interneuron survival.
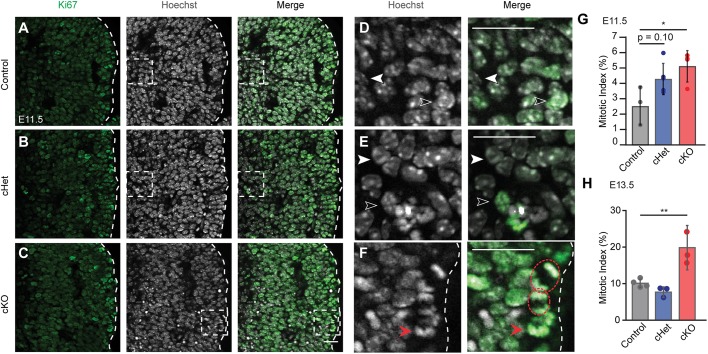
To test this possibility, we quantified the mitotic status of MGE progenitors in vivo. For this, we measured the fraction of mitotic cells within the population of Ki67+ cycling progenitors. This mitotic index was calculated for control (Nkx2.1-Cre), cHet (Nkx2.1-Cre;Magohlox/+) and cKO (Nkx2.1-Cre;Magohlox/lox) embryos (Fig. 4A-C). E11.5 was assessed as this marks a highly proliferative stage of development. We assessed mitosis by quantifying Ki67+ progenitors exhibiting a mitotic morphology, evidenced by Hoechst staining (Fig. 4D-F). Compared with control, E11.5 Magoh haploinsufficient progenitors showed a 1.7-fold increase in mitotic index, although this was not significant (P=0.10) (Fig. 4G). In comparison, and consistent with dosage sensitivity, cKO progenitors showed a significant twofold increase in mitotic index relative to E11.5 control (P=0.03) (Fig. 4G). At E13.5, there was no significant difference in mitotic index for cHets; however, cKOs maintained a significant twofold increase compared with controls (Fig. 4H). These data demonstrate that Magoh controls progenitor mitotic progression in a dosage-dependent fashion in vivo. Thus, similar to immortalized cells and cortical progenitors, MGE progenitor mitosis is exquisitely sensitive to Magoh levels.
Fig. 4.
Genetic depletion of Magoh causes dosage-dependent mitotic delay of MGE progenitors. (A-C) Coronal sections of E11.5 Nkx2.1-Cre;RosaloxSTOPloxtdTomato (control) (A), Nkx2.1-Cre;Magohlox/+;RosaloxSTOPloxtdTomato (cHet) (B) and Nkx2.1-Cre;Magohlox/lox;RosaloxSTOPloxtdTomato (cKO) (C) MGE stained for Ki67 (green/left) and Hoechst DNA stain (white/middle). Dashed line demarcates ventricular zone with ventricle at right. (D-F) High-magnification images of the boxed areas in A-C. White filled arrowheads, Ki67− cells; unfilled arrowheads, Ki67+ cells; red filled arrowheads, prometaphase cells; red dashed circles, anaphase cells. (G,H) Quantification of mitotic index (mitotic cells/total Ki67+ cells) at E11.5 (G) and E13.5 (H). E11.5: n=3 control, n=4 cHet, n=3 cKO; E13.5: n=3 control and cHet, n=3 cKO; ≥2 sections/embryo. ANOVA with Tukey post-hoc: *P<0.05, **P<0.01. Individual dots represent biological replicates and error bars represent s.d. Scale bars: 50 µm (A-C); 25 µm (D-F).
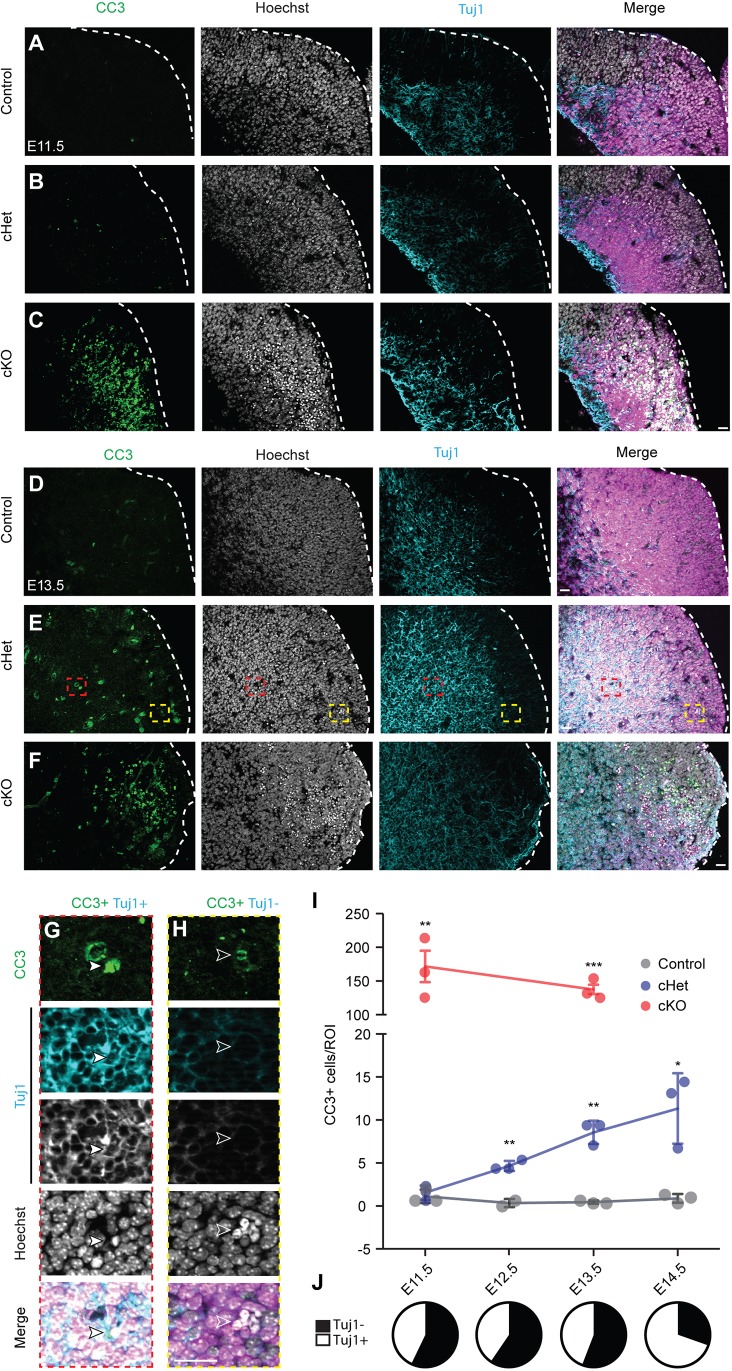
In many systems, including the Magoh-deficient cortex, mitotic defects are associated with cell death (McMahon et al., 2014; Pilaz et al., 2016; Silver et al., 2010). Most relevant, apoptosis is implicated in interneuron development, as exemplified by Couptf1/2 (Nr2f1/2) and Lhx6 mutants (Denaxa et al., 2018; Hu et al., 2017a). We thus postulated that excessive cell death could contribute to interneuron loss in Magoh mutants. To assess this possibility, we performed immunostaining for the apoptosis marker CC3 in Nkx2.1-Cre control, cHet and cKO embryos between E10.5 and E14.5 (Fig. 5A-F). There was negligible cell death observed in the MGE of control embryos between these stages (Fig. 5I, Fig. S4). In cHets, apoptosis significantly increased as development proceeded (Fig. 5I, Fig. S4). Apoptosis was evident in a small fraction of cells at E11.5 but initiated primarily at E12.5 and continued to increase through E14.5. In cKOs, apoptosis initiated even earlier, at E10.5, and was more profound than that observed in cHets (e.g. 15-fold higher at E13.5) (Fig. 5I, Fig. S4A). We then asked whether apoptosis was evident in both Tuj1 (Tubb3)+ neurons and Tuj1− cells (presumed progenitors). Quantification of E11.5, E12.5 and E13.5 cHets showed that one-third of apoptotic cells were Tuj1+ neurons (Fig. 5G,H,J). Notably, by E14.5, the majority of apoptotic cells were neurons (Fig. 5J). These results indicate that Magoh deficiency causes apoptosis in a dosage-dependent fashion. Taken together with the observation that interneuron number is unaffected in Dlx5/6-Cre;Magoh cHet and cKO embryos (Fig. 2), these experiments indicate that Magoh acts within progenitors to control survival of interneuron progeny and presumed progenitor progeny.
Fig. 5.
Genetic depletion of Magoh causes dosage-dependent apoptosis of progenitors and newborn interneurons. (A-F) Coronal sections of E11.5 (A-C) and E13.5 (D-F) Nkx2.1-Cre;RosaloxSTOPloxtdTomato (control) (A,D), Nkx2.1-Cre;Magohlox/+;RosaloxSTOPloxtdTomato (cHet) (B,E) and Nkx2.1-Cre;Magohlox/lox;RosaloxSTOPloxtdTomato (cKO) (C,F) MGE stained for CC3 (green/left), Hoechst DNA stain (white/middle), and Tuj1 (cyan/right), and tdTomato (magenta). Dashed line demarcates the ventricular zone with ventricle to the right. (G,H) High-magnification images of red and yellow boxed areas in E. Filled white arrowheads, CC3+ Tuj1+ co-labeled cells; white unfilled arrowheads, CC3+ Tuj1- single-labeled cells. (I) Quantification of total CC3+ cells within defined ROI of E11.5-E14.5 control (gray), cHet (blue) and cKO (red) (n=3 for all conditions, 3 sections/embryo). An ROI of 200 µm×200 µm or 200 µm×300 µm was quantified for E11.5 and ≥E12.5 embryos, respectively, beginning at the ventricle of the eminence. *P<0.05, **P<0.01, ***P<0.001. (J) Pie charts representing the proportion of Tuj1+ (white) or Tuj1− (black) CC3+ cells in the cHet at E11.5-E14.5. Unpaired two-tailed Student's t-test: cHet: E12.5 P=0.004, E13.5 P=0.008, E14.5 P=0.046; cKO: E11.5 P=0.006, E13.5 P=0.0008. Individual dots represent biological replicates and error bars represent s.d. Scale bars: 25 µm.
Live imaging reveals autonomous requirements for Magoh in MGE progenitors for interneuron production and survival in a dosage-dependent fashion
The in vivo data indicate that Magoh is required for progenitor mitosis and viability, and for interneuron number and survival. We sought to pinpoint the mechanisms by which Magoh controls progenitors and to further challenge the notion that interneuron apoptosis results from the requirement for Magoh in progenitors. Thus, we developed a live-imaging assay to monitor both MGE progenitor divisions and the fate of their direct progeny (Fig. 6A). We based this assay upon an experimental paradigm previously established for the cortex that enables high-temporal resolution of progenitor and cell fate dynamics (Pilaz et al., 2016). In brief, using this assay we sought to quantify mitosis duration and couple this to cell and viability fates of immediate progeny.
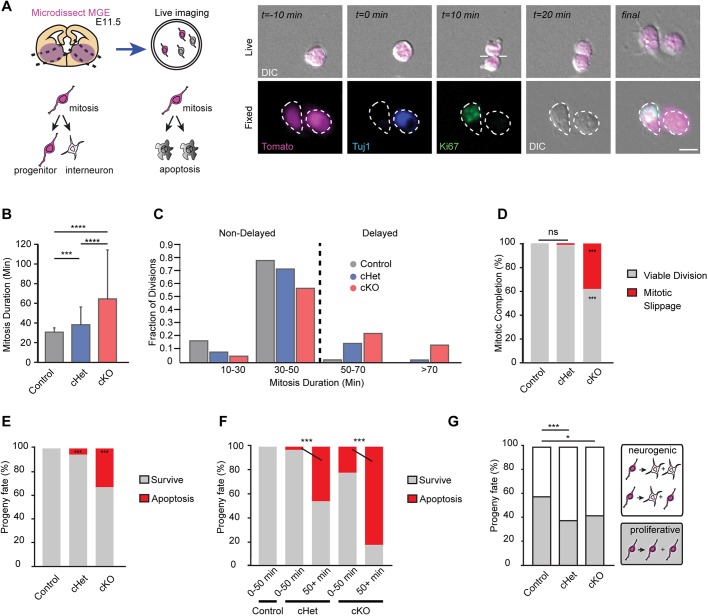
Fig. 6.
Live-imaging analysis of Magoh-depleted interneuron progenitors reveals autonomous requirements for mitotic progression, survival and cell fate. (A) Schematic of 24 h clonal live-imaging analysis with fixed analysis of progeny cell fate (example on the right). Dashed lines in images represent cell borders. (B-G) Analyses of Nkx2.1-Cre; RosaloxSTOPloxtdTomato (control), Nkx2.1-Cre; Magohlox/+; RosaloxSTOPloxtdTomato (cHet) and Nkx2.1-Cre; Magohlox/lox; RosaloxSTOPloxtdTomato (cKO). (B) Quantification of average mitosis duration (minutes) of tdTomato+ cells. (C) Histogram of mitosis duration (minutes). A threshold of delayed mitosis at 50 min (dashed line) is based upon control progenitors completing mitosis within this period. (D) Quantification of the proportion of viable (gray) and non-viable (red) divisions. Non-viable divisions characterized as mitotic slippage were due to unsuccessful cytokinesis and subsequent cell death. (E,F) Quantification of the proportion of progenitor progeny surviving (gray) or undergoing apoptosis (red) amongst all progeny (E) or relative to mitosis duration (F). (G) Quantification of the proportion of progeny fate undergoing neurogenic (white) or proliferative (gray) divisions. Neuronal fate was determined post-imaging by Tuj1+ staining and progenitor fate was determined by Ki67+Tuj1− staining. ANOVA with Tukey post-hoc: *P<0.05, **P<0.01, ***P<0.001 (B). χ2 analysis with post-hoc Bonferroni-adjusted P-values represented by asterisks: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (B,D-G). ns, not significant. Error bars represent s.d. Experiments represent two live-imaging sessions, two litters; control n=3 embryos (182 cells), cHet n=3 embryos (219 cells), cKO n=2 embryos (57 cells). Scale bar: 10 µm.
Dissociated progenitors were generated from E11.5 Nkx2.1-Cre (control), Nkx2.1-Cre;Magohlox/+ (cHet) and Nkx2.1-Cre;Magohlox/lox (cKO) embryos. Live imaging was carried out as described in Materials and Methods. Live imaging over a 24-h period revealed that cHet progenitors exhibited a slight but significantly longer mitosis relative to control (average 31±7 min versus 38±17 min, P=0.002) (Fig. 6B,C). This finding is consistent with the trend towards a higher mitotic index observed at E11.5 in cHets in vivo (Fig. 4E). Notably, in cKO progenitors the average mitosis duration was significantly longer compared with control and cHet (average 64±48 min, P<0.0001 each) (Fig. 6B). These data demonstrate that the live-imaging clonal assay recapitulates dosage-dependent requirements of Magoh for MGE progenitor mitosis. It also highlights the sensitivity of live imaging for assaying progenitor mitosis duration.
Using this live-imaging assay, we then monitored the fate of Magoh-deficient progenitors, and specifically their ability to complete mitosis. Although Magoh haploinsufficient progenitors displayed delayed mitotic progression, 99% of these ultimately completed cell division (Fig. 6D). This is in sharp contrast to cKO, in which viable divisions were evident in only 62% of progenitors. Instead, 38% of cKO progenitors underwent mitotic slippage, in which cells exit a prolonged mitosis and either senesce or undergo cell death (Blagosklonny, 2007; Castedo et al., 2004). Notably, this link between mitotic delay and subsequent cell death phenocopies the mitotic catastrophe caused by Magoh siRNA depletion in immortalized cells (Silver et al., 2010; Ishigaki et al., 2013; Pilaz et al., 2016). Altogether, these data indicate that whereas virtually all cHets complete divisions, this is not the case for cKOs, in which a significant fraction of progenitors fail to divide. This provides a plausible mechanism for interneuron depletion in the absence of Magoh.
Using this live-imaging assay, we then asked whether those Magoh cHet and cKO progenitors that complete divisions produce viable progeny. To monitor progeny, we used differential interference contrast (DIC) microscopy for live imaging, as previously described (Pilaz et al., 2016). Relative to control, cHet progenitors showed a slight (5%) but significant increase in production of non-viable progeny (Fig. 6E). This parallels slight apoptosis in vivo at the same stage, E11.5 (Fig. 5G). Consistent with the exquisite sensitivity of progenitors to Magoh dosage, approximately one-third of all progeny derived from cKO progenitors were non-viable (Fig. 6E). Thus, both cKO progenitors and their direct progeny are subject to cell death.
We then asked whether survival of newborn progeny is linked to abnormally long mitosis. Because virtually all control tdTomato+ progenitors completed mitosis within 50 min (n=360/362), we used a threshold for mitotic delay of 50 min or longer for these analyses (Fig. 6C). Strikingly, the likelihood of producing non-viable progeny increased significantly with mitoses greater than 50 min (Fig. 6F). This correlation parallels findings in the cortex, where mitotically delayed precursors also preferentially produce apoptotic progeny (Pilaz et al., 2016). Taken together, these live-imaging experiments reinforce the dosage-dependent requirement of Magoh for progenitor mitotic progression and also demonstrate that mitotic length is directly correlated with production of viable interneurons. Therefore, these findings provide important mechanistic insights into in vivo apoptosis phenotypes. In cHets, apoptosis of newborn interneurons and progenitors is linked to aberrant progenitor divisions. Yet apoptosis was significantly more severe in cKOs; this striking phenotype is attributed to death of progenitors via catastrophe and, independently, death of progeny.
A large fraction of cHet and cKO progenitors produce viable progeny, which begs the question of what is the fate of these mutant daughter cells? Thus, we asked whether Magoh-deficient progenitors undergo appropriate neurogenic and proliferative divisions to generate interneurons or two progenitors, respectively. For this, we performed 24-h live imaging of progenitors followed by immunostaining for Ki67 (progenitors) and Tuj1 (neurons) (Fig. 6A). We then assigned fates of progeny to progenitor divisions. Approximately 40% of control MGE progenitor divisions were neurogenic, resulting in production of at least one interneuron, and 60% were proliferative, resulting in production of two new progenitors. In contrast, both Magoh cHet and cKO mutant progenitors underwent more neurogenic and fewer proliferative divisions (60 and 40%, respectively) (Fig. 6G). Thus, reduced Magoh dosage in MGE progenitors disrupts their divisions, resulting in direct production of interneurons at the expense of new progenitors. This reduction of interneuron progenitors is predicted to ultimately deplete the available precursor pool and thereby drastically affect interneuron generation at subsequent stages of development.
Altogether these live-imaging experiments demonstrate that Magoh-deficient progenitors exhibit delayed mitotic progression, reduced proliferative divisions and increased apoptotic divisions. Collectively, these phenotypes contribute to decreased interneuron production and the observed loss of cortical interneurons in Magoh cHet and cKO brains. In addition, a substantial fraction of cKO progenitors fail to complete cell division, which exacerbates interneuron loss in this genotype. More broadly, these findings highlight mitotic duration as a conserved mechanism for impacting interneuron fate and viability.
Transcriptome analysis of Magoh-deficient interneuron progenitors highlights upregulated p53 signaling
Having exposed cell biological mechanisms at play in MGE progenitors, we sought to further elucidate how Magoh depletion affects progenitor mitosis and subsequent apoptosis at a molecular level. For this, we quantified transcriptome alterations associated with Magoh deficiency. In order to avoid potential confounding effects of apoptosis evident in cKO embryos and to pinpoint the earliest onset of progenitor defects, we focused on E11.5 Nkx2.1-Cre; Magohlox/+ embryos. We microdissected the ventral telencephalon from Nkx2.1-Cre and Nkx2.1-Cre; Magohlox/+ tdTomato+ embryos and subsequently used fluorescence-activated cell sorting (FACs) to purify tdTomato+ cells. Samples were subjected to transcriptome profiling (n=2 Magoh mutants, n=4 control) (Fig. 7A). As expected, Magoh mutants showed a reduction in Magoh mRNA expression (average 34%) by RNA-seq and qPCR (Fig. S5A).
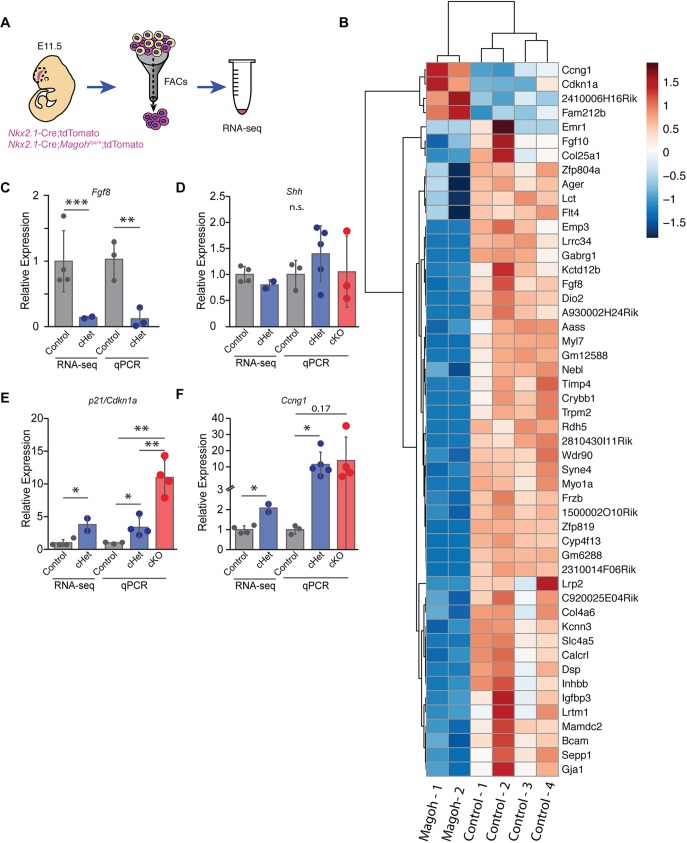
Fig. 7.
Transcriptome analysis of Magoh-depleted interneuron progenitors. (A) Diagram of transcriptome analysis including microdissection of E11.5 MGE from Nkx2.1-Cre; RosaloxSTOPloxtdTomato (control) and Nkx2.1-Cre; Magohlox/+; RosaloxSTOPloxtdTomato (cHet) embryos followed by FACS of tdTomato+ Nkx2.1-Cre-expressing cells, and RNA sequencing. (B) Heatmap representation of differentially expressed transcripts between control and cHets (n=4 and n=2 respectively, FDR q<0.05), with key on the right. (C-F) Validation of RNA sequencing results using independent biological replicates for cHet and cKO (n=3 control, n=5 cHet, n=3 cKO). Replicate expression normalized to endogenous control transcript levels and β-actin. Unpaired two-tailed Student's t-test, FDR-adjusted q-value (RNA-seq): Fgf8 P=0.009, q=2.16e-06 (C), Cdkn1a cHet: P=0.038, q=0.016; cKO P=0.17 (E), Ccng1 cHet: P=0.0.04, q=0.014; cKO: P=0.004; cHet versus cKO: P=0.004 (F). *P<0.05, **P<0.01, ***P<0.001. n.s., not significant. Individual dots represent biological replicates and error bars represent s.d.
The transcriptome profiling revealed that of 18,065 detected coding and non-coding transcripts only 0.25% were differentially expressed (FDR, q<0.05). This relatively limited impact on the transcriptome parallels that seen in the Magoh haploinsufficient cortex as well as the orthologous mago homozygous Drosophila mutant (Mao et al., 2016; Roignant and Treisman, 2010). Magoh is also implicated in the regulation of RNA splicing (Wang et al., 2014; Malone et al., 2014), and controls exon skipping by suppressing cryptic exonic splicing sites, including via recursive splicing (RS) (Boehm et al., 2018; Blazquez et al., 2018). Transcriptome analysis of Magoh mutant cortices indicates that RS may be relevant for cortical development (Blazquez et al., 2018). We therefore probed whether Magoh controls RS in interneurons, by examining Hnrpdl and Dnmbp, two previously noted Magoh RS targets that are expressed in the brain (Fig. S5B-D) (Yoshizawa et al., 2003; Visel et al., 2004). Strikingly, both transcripts showed dosage-dependent evidence of RS. This finding further supports the role of Magoh in post-transcriptional regulation of interneuron development.
We next focused on those transcripts for which expression levels were Magoh dependent. Amongst these, four were upregulated and 46 were downregulated, compared with control (Fig. 7B). We noted significant reductions in Fgf8, a canonical regulator of interneuron development (Storm et al., 2006). This effect was validated using RT-qPCR for independent samples (n=3 biological replicates each) (Fig. 7C). Fgf8 controls interneuron development via Shh; however, we did not observe any alteration in Shh levels, either by transcriptome analysis or RT-qPCR (Fig. 7D).
Notably, of the four transcripts upregulated in Magoh haploinsufficient progenitors, two are well-established p53-signaling targets, Ccng1 and Cdkn1a (p21), both of which promote p53-dependent cell-cycle arrest in response to cellular stress (Hafner et al., 2019). Validation of these p53 targets using independent samples (n=3 biological replicates each) confirmed fourfold and twofold increased expression of Cdkn1a and Ccng1, respectively (Fig. 7E,F).
Given the dosage-dependent requirements of Magoh for interneuron development, we next investigated whether transcript levels of these signaling pathways were more dramatically affected in the cKO (Fig. 7D-F). For this, we performed RT-qPCR on FACs-isolated E11.5 tdTomato+ Magoh cKO interneurons. Remarkably, comparing Magoh cHet with cKO, there was an even greater increase in levels of both p21 and Ccng1 transcripts. Notably, cKO did not affect Shh levels, further reinforcing that this transcript (and possibly this pathway) is not controlled by Magoh. Taken together, this transcriptome analysis indicates that upregulation of p53 signaling may contribute to cortical interneuron loss, in a dosage-dependent fashion.
Magoh mediates survival of newborn interneurons and progenitors via p53 signaling
Notably, p53 targets were amongst the transcripts prominently increased following Magoh haploinsufficiency in the MGE. This prompted us to investigate the role of p53 activation in vivo as an effector for interneuron loss. To assess p53 activation, we measured stable p53 accumulation using immunostaining as a proxy, as previously (Pilaz et al., 2016). We observed robust p53 activation in Magoh mutant MGE in a dosage-sensitive fashion, with significant 25-fold increases in cHets and 100-fold increases in the cKO at both E11.5 and E13.5 (Fig. 8A-G). This is consistent with a dosage-dependent increase in transcript levels of p53 targets. We then examined co-expression of p53+ cells with cytoplasmic Tuj1 to assess which cells were activated (Fig. 8D-G). p53 activation was most prevalent in Tuj1− cells (presumed progenitors), suggesting it is an early defect associated with Magoh deficiency (Fig. 8F,G). Consistent with this notion, the timing of p53 activation at E11.5 in cHets precedes the onset of apoptosis, which is most prominent at E12.5 (Fig. 5G, Fig. S4B,C).
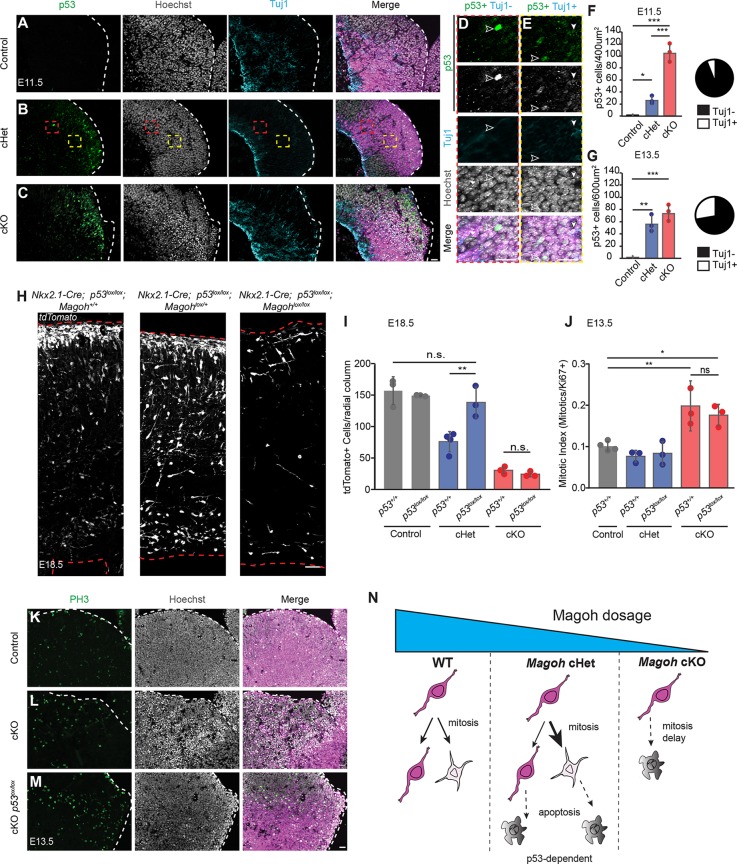
Fig. 8.
Magoh depletion within interneuron progenitors causes dosage-dependent p53 activation, and conditional p53 loss rescues cHet but not cKO cortical interneuron loss. (A-C) Coronal sections from E11.5 Nkx2.1-Cre; RosaloxSTOPloxtdTomato (control) (A), Nkx2.1-Cre; Magohlox/+; RosaloxSTOPloxtdTomato (cHet) (B) and Nkx2.1-Cre; Magohlox/lox; RosaloxSTOPloxtdTomato (cKO) (C) MGE with tdTomato representing Cre-recombinase-expressing cells (magenta) co-immunostained for p53 (green/left), Hoechst DNA staining (white/middle) and Tuj1 (cyan/right). (D,E) High-magnification images of the red (D) and yellow (E) boxed areas in B. Filled white arrowheads, p53+Tuj1+ co-labeled cells; white unfilled arrowheads, p53+Tuj1− single-labeled cells. (F,G) Quantification of E11.5 (F) and E13.5 (G) total p53+ cells in a ROI of 200 µm×200 µm or 200 µm×300 µm for E11.5 and E13.5, respectively. Pie charts represent the proportion of Tuj1+ (white) or Tuj1− (black) p53+ cells at E11.5 (n=22 cells) (D) and E13.5 (n=an average of 55.6 cells) (E) in cHet. (H) Coronal section of E18.5 neocortex from control (left), cHet (middle) and cKO (right) on a p53lox/lox background, stained for RFP representing Cre-derived interneurons. (I) Quantification of total tdTomato+ cells within a 250 µm wide radial cortical column from control, cHet and cKO embryos on p53+/+ or p53lox/lox backgrounds (n=3, control;p53+/+; n=3, control;p53lox/lox; n=4 cHet;p53+/+; n=3, cHet;p53lox/lox; n=3, cKO;p53+/+; n=3 cKO;p53lox/lox; ≥2 sections/embryo). (J) Quantification of mitotic index within a 200 µm×300 µm region of E13.5 MGE from control, cHet and cKO embryos on p53+/+ or p53lox/lox backgrounds. (n=3, cKO; p53+/+; n=3 others; 3 sections/embryo). (K-M) Coronal sections of E13.5 control (K), cKO (L) and cKO;p53lox/lox (M) MGE stained for PH3 and Hoechst DNA staining. (N) Model depicting the dosage-dependent role of Magoh in cortical interneuron progenitors. cHets exhibit p53-dependent cortical interneuron loss via apoptosis of progenitor progeny. cKOs exhibit a more severe loss of cortical interneurons due to p53-independent mitotic catastrophe. WT, wild type. ANOVA with Tukey post-hoc: *P<0.05, **P<0.01, ***P<0.001. n.s., not significant. Individual dots represent biological replicates and error bars represent s.d. Dashed lines in A-C,H,K-M demarcate the ventricular zone with ventricle at the right. Scale bars: 25 µm (A-C); 50 µm (F); 25 µm (K-M).
To test whether the loss of cortical interneurons in Magoh cHets is p53 dependent, we crossed conditional Magoh mutants onto a Nkx2.1-Cre; RosaloxSTOPloxtdTomato; p53lox/lox background (Marino et al., 2000). Interneurons were quantified using lineage tracing of tdTomato+ progeny at E18.5. Importantly, p53lox/lox alone did not affect cortical interneuron number, suggesting that p53 levels are largely dispensable for normal embryonic cortical interneuron number (Fig. 8H,I). Strikingly, conditional p53 deletion in a Magoh conditional haploinsufficient background completely rescued cortical interneuron number (P<0.01) (Fig. 8H,I). Reinforcing this rescue, ANOVA analysis demonstrated no significant difference between control and Magohlox/+;p53 compound mutants. To understand the mechanism of rescue, we measured apoptosis in E13.5 compound mutants. As expected, apoptosis was completely rescued in the compound mutants (Fig. S6A-C). This genetic experiment demonstrates that p53-mediated apoptosis is the major mechanism for interneuron loss in Magoh heterozygotes.
Relative to heterozygotes, Magoh homozygotes exhibited more profound phenotypes, including more substantial apoptosis and marked mitotic defects resulting in failed progenitor divisions. We thus postulated that p53 activation could also explain the more substantial interneuron loss in cKO mice. Similar to compound cHets, apoptosis was completely rescued in Magohlox/lox;p53 compound knockout mutants (Fig. S6A,C). However, in contrast to the heterozygotes, surprisingly, p53 loss was insufficient to rescue cortical interneuron number in Magoh; p53 compound homozygotes (Fig. 8H,I). This suggests that p53 upregulation and increased apoptosis alone are insufficient to deplete interneuron number in the complete absence of Magoh.
Given that p53 was abundantly upregulated in Magoh cKO, we sought to understand why p53 loss failed to rescue interneuron loss. Compared with cHets, cKO showed a substantial mitotic delay. Thus, we hypothesized that p53 may fail to rescue the cKO because these mitotic defects are p53 independent. To test this possibility, we quantified the mitotic index of E13.5 p53 alone and Magoh;p53 compound mutants (Fig. 8J). p53 loss alone did not affect baseline mitotic index and resembled control. Likewise, Magoh cHet;p53 compound mutants showed no significant mitotic delay, as was observed for E13.5 Magoh cHet alone (Fig. 4F). We predicted that if mitotic defects are p53 independent Magoh cKO; p53 compound mutants should exhibit equivalent mitotic indices to Magoh cKO alone. Indeed, similar to Magoh cKO, the Magoh cKO; p53 compound mutants exhibited comparably high mitotic indices (Fig. 8J). Thus, p53 loss failed to rescue the mitotic defect of Magoh-deficient MGE progenitors. Notably, the mitotic cells in Magoh cKO; p53 compound mutant brains were ectopically distributed within the MGE, as evidenced by cell morphology (Hoechst staining) and phospho-histone H3 staining (Fig. 8K-M, Fig. S6C). This aberrant distribution resembled that of apoptotic cells observed in Magoh cKO MGE but not LGE (compare with Fig. 5). This observation suggests that in the absence of p53, cells that were destined for an apoptotic fate instead remain stuck in mitosis. The sustained mitotic defect in these mice thus provides a plausible explanation for why p53 loss fails to rescue interneuron number. Altogether, this result strongly implicates progenitor mitotic defects as a major contributor to interneuron loss in Magoh homozygotes.
DISCUSSION
Proper development of GABAergic interneurons is essential for cortical function, yet the key mechanisms by which progenitors produce interneurons are poorly understood. Here, we demonstrate a novel layer of post-transcriptional regulation via the EJC component Magoh in the control of cell fate specification of interneurons. Using both in vitro and in vivo analyses, we discover that cortical interneuron progenitors are exquisitely sensitive to Magoh dosage for proper mitotic progression, as well as for generation and survival of newborn interneurons (Fig. 8N). We further demonstrate that p53 signaling is integral to Magoh-mediated regulation of apoptosis but not mitosis progression. Strikingly, this genetic analysis exposes differential mechanisms at play in heterozygous and homozygous mutant animals. Our study provides important insights into the mechanisms underlying neurological disorders driven by mutations of EJC components. More broadly, these findings highlight fundamental roles for progenitor mitosis progression and p53 signaling in the development of GABAergic cortical interneurons.
Our study highlights the importance of precise regulation of progenitor mitosis duration for interneuron development. By coupling fixed analysis with a new assay for live imaging of interneuron fate, we discover that prolonged mitotic duration is linked to progeny apoptosis and death of dividing progenitors. Coupled with our previous findings in cortical progenitors (Pilaz et al., 2016), this discovery implicates aberrant mitosis duration in cell fate specification in other brain regions. Importantly, in contrast to cortical progenitors, far less is known about the contribution of cell cycle control to interneuron production. Studies of the ganglionic eminences have implicated G1 duration in cell fate; however, the role of mitosis duration in interneuron fate has thus far been unappreciated (Brown et al., 2011; Ross, 2011). Given this, our findings raise a number of fascinating questions. Does mitosis perturbation impact all MGE progenitors equivalently? Does mitosis duration causally alter interneuron fate? Further, does mitotic progression contribute to increased apoptosis and altered cell fate in other genetic models of interneuron development and disease? This latter point is of interest as microcephaly is heavily linked to mitosis dysregulation (Gilmore and Walsh, 2012), yet most studies of microcephaly mechanisms have primarily investigated the impact on excitatory neurons.
Given the profound phenotypes associated with reduced Magoh dosage, we conclude that Magoh is a major regulator of interneuron development. This adds Magoh to a growing list of RNA-binding factors, including Rbfox1, that mediate post-transcriptional control during interneuron development (Wamsley et al., 2018). We did not observe widespread steady-state transcriptome changes in E11.5 heterozyogotes. Going forward, to understand better how Magoh cKO differentially controls interneuron development, unbiased transcriptome analysis may be valuable. However, it is also likely that Magoh controls interneuron development via diverse RNA regulatory mechanisms, including splicing; Indeed, our study suggests that comprehensive analysis of splicing, including recursive splicing events, may give valuable insights into how Magoh controls interneuron genesis.
From transcriptome and immunofluorescence analysis we pinpoint p53 signaling upregulation amongst the earliest defects in Magoh-deficient progenitors, driving pathogenic levels of apoptosis. Thus, we employed genetic experiments to test whether p53 dysregulation drives interneuron loss. The picture that emerged was surprising and allowed us to discriminate contributions of prolonged mitosis and apoptosis for Magoh-mediated interneuron development. Magoh conditional heterozygotes displayed a 50% reduction in cortical interneurons, and this was entirely explained by p53-dependent apoptosis. In contrast, in Magoh conditional knockouts, there was a more severe 80% loss of interneurons, which was not rescued by p53 loss. Instead, in the complete absence of both Magoh and p53, mitotic cells that were destined to undergo cell death were ectopic. Based upon the live-imaging assays, we posit that these aberrant Magoh-deficient mitotic progenitors fail to produce sufficient numbers of interneurons, and thus interneurons remain depleted. Together, this exposes key apoptotic and mitotic mechanisms by which Magoh controls interneuron development in a dosage-dependent fashion.
Our study demonstrates conserved requirements for Magoh in progenitors, pointing to parallel functions in the developing MGE and cortex. Indeed, for both excitatory and inhibitory progenitors, Magoh promotes mitosis progression, viability of newborn neurons, and neurogenic and proliferative divisions (Mao et al., 2016; McMahon et al., 2014; Pilaz et al., 2016; Silver et al., 2010). Further, for development of both neuronal populations, Magoh mediates neuronal survival by acting within the precursor, and not the post-mitotic neuron (Pilaz et al., 2016). This suggests fascinating cellular requirements of Magoh for neuronal cell survival. In the developing cortex, Magoh functions in the EJC to control excitatory neuron production (Mao et al., 2016). Given Rbm8a and Eif4a3 are also enriched in the MGE (Visel et al., 2004), it will be exciting to investigate whether these proteins work in concert with Magoh in a complex to control interneuron generation.
Although Magoh has conserved roles in excitatory and interneuron radial glia it remains unclear to what extent Magoh is required post-mitotically. We demonstrate that post-mitotic depletion of Magoh from migrating interneurons is dispensable for generation of the appropriate final number. As Dlx5/6-Cre marks both the MGE and CGE, Magoh could differentially impact these populations. However, neither cell survival nor migratory patterns from either eminence were disrupted in this context. It is plausible that Magoh post-mitotic depletion causes postnatal defects, including altered interneuron circuit formation and function. Indeed, we observed a slight but not quite significant difference in interneuron subtype number. Understanding the nature of this difference will be interesting, including whether it is due to a fate shift, and/or selective requirements of Magoh for survival and proliferation of progenitors that produce SST+ interneurons.
Mutations in EJC components are linked to neurodevelopmental pathologies, yet the mechanisms underlying these disorders are poorly understood (McMahon et al., 2016). Copy number variations in EJC components are linked to intellectual disability (Nguyen et al., 2013). Moreover, RBM8A loss of function is associated with thrombocytopenia with absent radius (TAR) syndrome, and EIF4A3 hypomorphic mutations cause Richieri-Costa–Pereira (RCP) syndrome(Brunetti-Pierri et al., 2008; Mefford et al., 2008; Albers et al., 2012; Favaro et al., 2014; Bertola et al., 2017). Both disorders present with neurological impairment, including microcephaly, and language and learning deficits. Previous studies demonstrate that defective excitatory neuron generation may contribute to these pathologies (Mao et al., 2015; McMahon et al., 2016; Zou et al., 2015). The current study now provides a more complete picture, implicating EJC-mediated control of both glutamatergic and GABAergic development in disease etiology. Altogether, future investigation into how Magoh and its binding partners control interneuron development promises to give new insights into post-transcriptional regulation of cortical interneuron development and disease.
MATERIALS AND METHODS
Ethics statement
All experiments were performed in agreement with the guidelines from the Division of Laboratory Animal Resources from Duke University School of Medicine and the Institutional Animal Care and Use Committee of Duke University.
Mouse husbandry
Plug dates were defined as E0.5 on the morning the plug was identified. Magohlox/lox animals were previously described (McMahon et al., 2014). The following lines were obtained from The Jackson Laboratory: Nkx2.1-Cre (The Jackson Laboratory, 008661), ROSAloxSTOPloxTdTomato (The Jackson Laboratory, 007905), Dlx5a-Cre (The Jackson Laboratory, 008199), Trp53flox (The Jackson Laboratory, 008462). Genotyping for these alleles was performed using The Jackson Laboratory protocols.
Immunofluorescence
Embryonic brains were fixed overnight in 4% paraformaldehyde (PFA) at 4°C, submerged in 30% sucrose overnight, and embedded in NEG-50 (Fisher Scientific, 22-110-617). Frozen sections (20 μm) were generated on a cryotome and stored at −80°C . Sections were permeabilized with 0.25% Triton X-100 for 10 min at room temperature followed by blocking with 10% normal goat serum (NGS) in PBS for 1 h at room temperature. Sections were incubated with primary antibodies overnight at 4°C , and secondary antibodies at room temperature for 30 min. Cultured cells were fixed for 10 min in 4%PFA, permeabilized with 0.25% Triton X-100 for 10 min at room temperature, blocked using 10% NGS for 30 min, incubated with primary antibodies for 90 min at room temperature and secondary antibodies for 25 min at room temperature. Adult animals were transcardially perfused with 1× PBS followed by 4% PFA. Adult brains were incubated in 4% PFA overnight at 4°C, cryoprotected in 30% sucrose, then frozen in NEG-50. Cryotome sections (40 μm) were permeabilized and blocked in blocking solution (10% NGS, 0.3% Triton X-100, 1× PBS) for >1 h. Sections were incubated in blocking solution with primary antibodies overnight at 4°C, washed in 1× PBS and incubated in blocking solution with secondary antibodies at room temperature for 30-60 min. As needed for some experiments, anti-RFP was used (across all conditions). Images were captured using a Zeiss Axio Observer Z.1 equipped with an Apotome for optical sectioning. Tiled images (Fig. 1D, 2A,D,I and 3A,C) were acquired and stitched using Zen software. The following primary antibodies were used: mouse anti-TUJ1 (Biolegend, 801202, 1:1000), rabbit anti-CC3 (Cell Signaling Technology, 9661, 1:250), rat anti-somatostatin (Millipore, MAB354, 1:100), rabbit anti-parvalbumin (Abcam, ab11247, 1:400), rabbit anti-RFP (Rockland, 600-401-379S, 1:500), rabbit anti-Magoh (Proteintech, 12347-1-AP, 1:75), anti-p53 (Leica, CM5, 1:500), rabbit anti-Ki67 (Cell Signaling Technology, 12202, 1:250), rat anti-tdTomato (Kerafast, EST203, 1:250), rabbit anti-PH3 (Millipore, 06-570, 1:500). Secondary antibodies were Alexa Fluor 488-, 594- and 647-conjugated (Thermo Fisher Scientific, A32731, A32723, A32740, A32742 and A32728, 1:500).
Expression analysis
E11.5 tdTomato+ telencephalon was FACs sorted using a B-C Astrios cell sorter directly into RLT buffer (Qiagen RNAeasy kit; Qiagen 74034). RNA was extracted using the RNAeasy kit (Qiagen, 74034). cDNA was synthesized using iScript kit (Bio-Rad, 1788090) following the manufacturer’s protocol. qPCR was performed in triplicate using Taqman probes (Life Technologies): Magoh (Mm00487546_m1) and GAPDH (Mm99999915_G1). Sybr Green iTaq (Bio-Rad, 172-5121) was performed using the following primer sets: β-actin (5′ forward- AGATCAAGATCATTGCTCCT and 3′ reverse- CCTGCTTGCTGATCCACATC), Cdkn1a (5′ forward- TGACAGATTTCTATCACTCCAAGC and 3′ reverse- ACTTTAAGTTTGGAGACTGGGAGA), Ccng1 (5′ forward- TTGGCTTTGACACGGAGACA and 3′ reverse- AAGCAGCTCAGTCCAACACA), Fgf8 (5′ forward- CCCGACATCATATTTAAGGATGAGGA and 3′ reverse- ACTCCAGGCCACTGGTTCA), Shh (5′ forward- TTGGCTTTGACACGGAGACA and 3′ reverse- AAGCAGCTCAGTCCAACACA), Hnrnpdl (5′ forward- GCAACAACAGAAAGGAGGCA and 3′ reverse- CCCAGCGTCCTCCTTTAGTA) and Dnmbp (5′ forward- TGGAGAAGAGAGCCAAGGTG and 3′ reverse- AGGTGCTTCTGGGTCTTCTC).
RNA sequencing and bioinformatic analysis
E11.5 tdTomato+ telencephalon was FACS sorted using a B-C Astrios cell sorter directly into RLT buffer (Qiagen RNAeasy kit; Qiagen 74034) and frozen at −80°C. RNA was extracted using the RNAeasy kit (Qiagen, 74034). Libraries were prepared using NuGEN mRNA Seq with AnyDeplete Globin (AnyDeplete step was skipped during the library preparation protocol). Libraries were sequenced using Illumina HiSeq4000 paired-end 150 bp reads. RNA-seq data were processed using the TrimGalore toolkit which employs Cutadapt to trim low-quality bases and Illumina sequencing adapters from the 3′ end of the reads (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore). Only reads that were 20 nt or longer after trimming were kept for further analysis. Reads were mapped to the GRCm38v73 version of the mouse genome and transcriptome using the STAR RNA-seq alignment tool (Dobin et al., 2012; Kersey et al., 2012). Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool (https://htseq.readthedocs.io/en/release_0.11.1/). Only genes that had at least ten reads in any given library were used in subsequent analysis. Normalization and differential expression was carried out using the DESeq2 Bioconductor package with the R statistical programming environment (Love et al., 2014; Huber et al., 2015) (www.r-project.org). The false discovery rate (FDR) was calculated to control for multiple hypothesis testing. Gene set enrichment analysis was performed to identify pathways associated with altered gene expression for each of the comparisons performed. Sequencing data have been deposited in Gene Expression Omnibus under accession number GSE142422.
Live-imaging experiments
E11.5 ganglionic eminences were isolated from either Nkx2.1-Cre; RosaloxSTOPloxtdTomato (control), Nkx2.1-Cre; Magohlox/+; RosaloxSTOPloxtdTomato (cHet) or Nkx2.1-Cre; Magohlox/lox; RosaloxSTOPloxtdTomato (cKO) embryos. Tissue was prepared in a single cell suspension by trypsinization (10-15 min with 0.25% Trypsin-EDTA) (Thermo Fisher Scientific, 25200056), and mechanical dissociation by pipetting through a fire-polished pasteur pipette. Isolated cells were plated in neural progenitor media (DMEM with glucose, sodium pyruvate, B-27, N2, N-acetyl cystein and bFGF). Cells were plated in either a 6- or 12-well glass-bottom dish (MatTek) and given 1-2 h to settle at 37°C in 5% CO2 prior to imaging. Images were captured every 10 min for 18 h using a 20× magnification on a Zeiss Axio Observer Z.1 equipped with a XL multi S1 incubation chamber, CO2 module S, temperature module S, and humidity control. Mitosis duration, catastrophe and viability were identified by morphology (rounding of cells and visual identification of condensation of chromatin). Fate determination was performed post-imaging by fixation with 4% PFA and immunostaining for Ki67 and Tuj1. Analyses were similar to those previously described (Pilaz et al., 2016).
Fixed quantification
Image analysis was conducted using ImageJ or Zen software (Zeiss Microscopy). Quantification within the medial ganglionic eminence utilized a 200 µm×200 µm region of interest (ROI) and a 200 µm×300 µm ROI for E11.5 and ≥E12.5 embryos, respectively, beginning at the ventricle of the eminence. Quantification within the cortex utilized a 250 µm- and 350 µm-wide radial column spanning pia to ventricle, for embryonic and post-natal analyses, respectively (Figs 2, 3 and 8). For binning analysis, a radial column was divided into five equally sized bins spanning from the pial to the ventricular surface (Fig. 2). Mitotic cells were classified by presence of bright Ki67 and Hoechst 33342 staining for characteristic chromatin pattern (Fig. 4). Apoptotic cells were classified by bright CC3 staining and Hoechst to detect stereotypical apoptotic morphology, as well by the presence or absence of Tuj1 staining (Fig. 5). Because Tuj1 is cytoplasmic, TuJ1+ cells were defined when the entire cell cytoplasm was positive for this antibody. p53+ cells were classified by both bright and moderate signal co-expressed with Tuj1 (delineated as above) (Fig. 8). Between two and four sections were analyzed per embryo depending on the experiment, with at least three biological replicates per condition. All analyses were performed blind to genotype, and in some cases repeated by multiple individuals. In order to maintain consistency, for a given marker, images were quantified by one individual blinded to genotype within the same session. Images were captured with identical exposures.
Supplementary Material
Acknowledgements
We thank members of the Silver lab for helpful discussions and careful reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.J.M., D.L.S.; Methodology: C.J.S., J.J.M., D.L.S.; Validation: C.J.S., J.J.M., L.D.S., D.L.S.; Formal analysis: C.J.S., J.J.M., L.D.S., D.L.S.; Investigation: C.J.S., J.J.M., L.D.S., D.L.S.; Resources: C.J.S., J.J.M., D.L.S.; Data curation: C.J.S., J.J.M., D.L.S.; Writing - original draft: C.J.S., J.J.M., D.L.S.; Writing - review & editing: C.J.S., J.J.M., D.L.S.; Visualization: C.J.S., J.J.M., D.L.S.; Supervision: D.L.S.; Project administration: D.L.S.; Funding acquisition: D.L.S.
Funding
This work was funded by the National Institutes of Health (NS083897, NS098176, NS110388 and MH119813 to D.L.S.). Deposited in PMC for release after 12 months.
Data availability
RNA-sequencing data have been deposited in Gene Expression Omnibus under accession number GSE142422.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.182295.supplemental
References
- Albers C. A., Paul D. S., Schulze H., Freson K., Stephens J. C., Smethurst P. A., Jolley J. D., Cvejic A., Kostadima M., Bertone P. et al. (2012). Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 44, 435-439. 10.1038/ng.1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. A., Eisenstat D. D., Shi L. and Rubenstein J. L. (1997). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474-476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D., Udell C. M., Lavoie H., Baril C., Lefrançois M., Chagnon P., Gendron P., Caron-Lizotte O., Bonneil E., Thibault P. et al. (2010). The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143, 251-262. 10.1016/j.cell.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Bandler R. C., Mayer C. and Fishell G (2017). Cortical interneuron specification: the juncture of genes, time and geometry. Curr. Opin. Neurobiol. 42, 17-24. 10.1016/j.conb.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertola D. R., Hsia G., Alvizi L., Gardham A., Wakeling E., Yamamoto G. L., Honjo R. S., Oliveira L. A. N., Di Francesco R. C., Perez B. A. et al. (2017). Richieri-Costa-Pereira syndrome: expanding its phenotypic and genotypic spectrum. Clin. Genet. 1-28. [DOI] [PubMed] [Google Scholar]
- Blagosklonny M. V. (2007). Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle (Georgetown, Tex) 6, 70-74. 10.4161/cc.6.1.3682 [DOI] [PubMed] [Google Scholar]
- Blazquez L., Emmett W., Faraway R., Pineda J. M. B., Bajew S., Gohr A., Haberman N., Sibley C. R., Bradley R. K., Irimia M. et al. (2018). Exon junction complex shapes the transcriptome by repressing recursive splicing. Mol. Cell 72, 496-509.e9. 10.1016/j.molcel.2018.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm V., Britto-Borges T., Steckelberg A. L., Singh K. K., Gerbracht J. V., Gueney E., Blazquez L., Altmuller J., Dieterich C. and Gehring N. H. (2018). Exon junction complexes suppress spurious splice sites to safeguard transcriptome integrity. Mol. Cell 72, 482-495.e7. 10.1016/j.molcel.2018.08.030 [DOI] [PubMed] [Google Scholar]
- Brown K. N., Chen S., Han Z., Lu C.-H., Tan X., Zhang X.-J., Ding L., Lopez-Cruz A., Saur D., Anderson S. A. et al. (2011). Clonal production and organization of inhibitory interneurons in the neocortex. Science (New York, NY) 334, 480-486. 10.1126/science.1208884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N., Berg J. S., Scaglia F., Belmont J., Bacino C. A., Sahoo T., Lalani S. R., Graham B., Lee B., Shinawi M. et al. (2008). Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 40, 1466-1471. 10.1038/ng.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M., Perfettini J. L., Roumier T., Valent A., Raslova H., Yakushijin K., Horne D., Feunteun J., Lenoir G., Medema R. et al. (2004). Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene 23, 4362-4370. 10.1038/sj.onc.1207572 [DOI] [PubMed] [Google Scholar]
- Chen Y. J., Friedman B. A., Ha C., Durinck S., Liu J., Rubenstein J. L., Seshagiri S. and Modrusan Z (2017). Single-cell RNA sequencing identifies distinct mouse medial ganglionic eminence cell types. Sci. Rep. 7, 45656 10.1038/srep45656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciceri G., Dehorter N., Sols I., Huang Z. J., Maravall M. and Marin O (2013). Lineage-specific laminar organization of cortical GABAergic interneurons. Nat. Neurosci. 16, 1199-1210. 10.1038/nn.3485 [DOI] [PubMed] [Google Scholar]
- Denaxa M., Neves G., Rabinowitz A., Kemlo S., Liodis P., Burrone J. and Pachnis V (2018). Modulation of apoptosis controls inhibitory interneuron number in the cortex. Cell Rep. 22, 1710-1721. 10.1016/j.celrep.2018.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2012). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairen A., Cobas A. and Fonseca M (1986). Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J. Comp. Neurol. 251, 67-83. 10.1002/cne.902510105 [DOI] [PubMed] [Google Scholar]
- Favaro F. P., Alvizi L., Zechi-Ceide R. M., Bertola D., Felix T. M., De Souza J., Raskin S., Twigg S. R. F., Weiner A. M. J., Armas P. et al. (2014). A noncoding expansion in EIF4A3 causes richieri-costa-pereira syndrome, a craniofacial disorder associated with limb defects. Am. J. Hum. Genet. 94, 120-128. 10.1016/j.ajhg.2013.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore E. C. and Walsh C. A. (2012). Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdiscip. Rev. Dev. Biol. 2, 461-478. 10.1002/wdev.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickstein S. B., Moore H., Slowinska B., Racchumi J., Suh M., Chuhma N. and Ross M. E. (2007). Selective cortical interneuron and GABA deficits in cyclin D2-null mice. Development 134, 4083-4093. 10.1242/dev.008524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. and Anton E. S. (2014). Decision making during interneuron migration in the developing cerebral cortex. Trends Cell Biol. 24, 342-351. 10.1016/j.tcb.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner A., Bulyk M. L., Jambhekar A. and Lahav G (2019). The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20, 199-210. 10.1038/s41580-019-0110-x [DOI] [PubMed] [Google Scholar]
- Harwell C. C., Fuentealba L. C., Gonzalez-Cerrillo A., Parker P. R. L., Gertz C. C., Mazzola E., Garcia M. T., Alvarez-Buylla A., Cepko C. L. and Kriegstein A. R. (2015). Wide dispersion and diversity of clonally related inhibitory interneurons. Neuron 87, 999-1007. 10.1016/j.neuron.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. S., Vogt D., Lindtner S., Sandberg M., Silberberg S. N. and Rubenstein J. L. R. (2017a). Coup-TF1 and Coup-TF2 control subtype and laminar identity of MGE-derived neocortical interneurons. Development 144, 2837-2851. 10.1242/dev.150664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. S., Vogt D., Sandberg M. and Rubenstein J. L. (2017b). Cortical interneuron development: a tale of time and space. Development 144, 3867-3878. 10.1242/dev.132852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., Carvalho B. S., Bravo H. C., Davis S., Gatto L., Girke T. et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115-121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan M., Welagen J. and Anderson S. A. (2012). Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb. Cortex 22, 820-827. 10.1093/cercor/bhr148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Nakamura Y., Tatsuno T., Hashimoto M., Shimasaki T., Iwabuchi K. and Tomosugi N (2013). Depletion of RNA-binding protein RBM8A (Y14) causes cell cycle deficiency and apoptosis in human cells. Exp. Biol. Med. 238, 889-897. 10.1177/1535370213494646 [DOI] [PubMed] [Google Scholar]
- Kataoka N., Yong J., Kim V. N., Velazquez F., Perkinson R. A., Wang F. and Dreyfuss G (2000). Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6, 673-682. 10.1016/S1097-2765(00)00065-4 [DOI] [PubMed] [Google Scholar]
- Kersey P. J., Staines D. M., Lawson D., Kulesha E., Derwent P., Humphrey J. C., Hughes D. S., Keenan S., Kerhornou A., Koscielny G. et al. (2012). Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 40, D91-D97. 10.1093/nar/gkr895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L., Simon J. M., Xing L., Mccoy E. S., Niehaus J. K., Guo J., Anton E. S. and Zylka M. J. (2019). Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134 10.1038/s41467-018-08079-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Mestdagh C., Akhtar J., Kreim N., Deinhard P., Sachidanandam R., Treisman J. and Roignant J.-Y. (2014). The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes Dev. 28, 1786-1799. 10.1101/gad.245829.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Pilaz L.-J., Mcmahon J. J., Golzio C., Wu D., Shi L., Katsanis N. and Silver D. L. (2015). Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J. Neurosci. 35, 7003-7018. 10.1523/JNEUROSCI.0018-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., McMahon J. J., Tsai Y.-H., Wang Z. and Silver D. L. (2016). Haploinsufficiency for core exon junction complex components disrupts embryonic neurogenesis and causes p53-mediated microcephaly. PLoS Genet. 12, e1006282-e27. 10.1371/journal.pgen.1006282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O. (2012). Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 13, 107-120. 10.1038/nrn3155 [DOI] [PubMed] [Google Scholar]
- Marino S., Vooijs M., Van Der Gulden H., Jonkers J. and Berns A (2000). Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994-1004. [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Jaglin X. H., Cobbs L. V., Bandler R. C., Streicher C., Cepko C. L., Hippenmeyer S. and Fishell G (2015). Clonally related forebrain interneurons disperse broadly across both functional areas and structural boundaries. Neuron 87, 989-998. 10.1016/j.neuron.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Hafemeister C., Bandler R. C., Machold R., Batista Brito R., Jaglin X., Allaway K., Butler A., Fishell G. and Satija R (2018). Developmental diversification of cortical inhibitory interneurons. Nature 555, 457-462. 10.1038/nature25999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmahon J. J., Shi L. and Silver D. L. (2014). Generation of a Magoh conditional allele in mice. Genesis (New York, NY : 2000) 52, 752-758. 10.1002/dvg.22788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon J. J., Miller E. E. and Silver D. L. (2016). The exon junction complex in neural development and neurodevelopmental disease. Int. J. Dev. Neurosci. 55, 117-123. 10.1016/j.ijdevneu.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H. C., Sharp A. J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V. K., Crolla J. A., Baralle D. et al. (2008). Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl. J. Med. 359, 1685-1699. 10.1056/NEJMoa0805384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi D., Li Z., Lim L., Li M., Moissidis M., Yang Y., Gao T., Hu T. X., Pratt T., Price D. J. et al. (2018). Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81-85. 10.1126/science.aar6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. W. (1985). Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain Res. 355, 187-192. 10.1016/0165-3806(85)90040-9 [DOI] [PubMed] [Google Scholar]
- Monory K., Massa F., Egertová M., Eder M., Blaudzun H., Westenbroek R., Kelsch W., Jacob W., Marsch R., Ekker M. et al. (2006). The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron 51, 455-466. 10.1016/j.neuron.2006.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. S., Kim H.-G., Rosenfeld J. A., Shen Y., Gusella J. F., Lacassie Y., Layman L. C., Shaffer L. G. and Gécz J (2013). Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum. Mol. Genet. 22, 1816-1825. 10.1093/hmg/ddt035 [DOI] [PubMed] [Google Scholar]
- Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S. J., Velmeshev D. et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science (New York, NY) 358, 1318-1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduzzi J. D. (1988). Genesis of GABA-immunoreactive neurons in the ferret visual cortex. J. Neurosci. 8, 920-931. 10.1523/JNEUROSCI.08-03-00920.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros T. J., Bultje R. S., Ross M. E., Fishell G. and Anderson S. A. (2015). Apical versus basal neurogenesis directs cortical interneuron subclass fate. Cell Rep. 13, 1090-1095. 10.1016/j.celrep.2015.09.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L.-J., Mcmahon J. J., Miller E. E., Lennox A. L., Suzuki A., Salmon E. and Silver D. L (2016). Prolonged mitosis of neural progenitors alters cell fate in the developing Brain. Neuron 89, 83-99. 10.1016/j.neuron.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J.-Y. and Treisman J. E. (2010). Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143, 238-250. 10.1016/j.cell.2010.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M. E. (2011). Cell cycle regulation and interneuron production. Dev. Neurobiol. 71, 2-9. 10.1002/dneu.20823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T., Tange T. Ø., Sonenberg N. and Moore M. J (2004). eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11, 346-351. 10.1038/nsmb750 [DOI] [PubMed] [Google Scholar]
- Silver D. L., Watkins-Chow D. E., Schreck K. C., Pierfelice T. J., Larson D. M., Burnetti A. J., Liaw H.-J., Myung K., Walsh C. A., Gaiano N. et al. (2010). The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci. 13, 551-558. 10.1038/nn.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm E. E., Garel S., Borello U., Hebert J. M., Martinez S., Mcconnell S. K., Martin G. R. and Rubenstein J. L. (2006). Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133, 1831-1844. 10.1242/dev.02324 [DOI] [PubMed] [Google Scholar]
- Sultan K. T. and Shi S.-H. (2018). Generation of diverse cortical inhibitory interneurons. Wiley Interdiscip. Rev. Dev. Biol. 7 10.1002/wdev.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan K. T., Han Z., Zhang X. J., Xianyu A., Li Z., Huang K. and Shi S. H. (2016). Clonally related GABAergic interneurons do not randomly disperse but frequently form local clusters in the forebrain. Neuron 92, 31-44. 10.1016/j.neuron.2016.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takesian A. E. and Hensch T. K. (2013). Balancing plasticity/stability across brain development. Prog. Brain Res. 207, 3-34. 10.1016/B978-0-444-63327-9.00001-1 [DOI] [PubMed] [Google Scholar]
- Tischfield D. J., Kim J. and Anderson S. A. (2017). Atypical PKC and notch inhibition differentially modulate cortical interneuron subclass fate from embryonic stem cells. Stem Cell Reports 8, 1135-1143. 10.1016/j.stemcr.2017.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrero Garcia M. and Harwell C. C. (2017). Radial glia in the ventral telencephalon. FEBS Lett. 591, 3942-3959. 10.1002/1873-3468.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eeden F. J., Palacios I. M., Petronczki M., Weston M. J. and St Johnston D. (2001). Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J. Cell Biol. 154, 511-523. 10.1083/jcb.200105056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Thaller C. and Eichele G (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552-D556. 10.1093/nar/gkh029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamsley B. and Fishell G (2017). Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat. Rev. Neurosci. 18, 299-309. 10.1038/nrn.2017.30 [DOI] [PubMed] [Google Scholar]
- Wamsley B., Jaglin X. H., Favuzzi E., Quattrocolo G., Nigro M. J., Yusuf N., Khodadadi-Jamayran A., Rudy B. and Fishell G. (2018). Rbfox1 mediates cell-type-specific splicing in cortical interneurons. Neuron 100, 846-859.e7. 10.1016/j.neuron.2018.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dye C. A., Sohal V., Long J. E., Estrada R. C., Roztocil T., Lufkin T., Deisseroth K., Baraban S. C. and Rubenstein J. L. (2010). Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J. Neurosci. 30, 5334-5345. 10.1523/JNEUROSCI.5963-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Murigneux V. and Le Hir H. E. (2014). Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol. 15, 551 10.1186/s13059-014-0551-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Tam M. and Anderson S. A (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16-29. 10.1002/cne.21529 [DOI] [PubMed] [Google Scholar]
- Yoshizawa M., Sone M., Matsuo N., Nagase T., Ohara O., Nabeshima Y. and Hoshino M (2003). Dynamic and coordinated expression profile of dbl-family guanine nucleotide exchange factors in the developing mouse brain. Gene Expr. Patterns 3, 375-381. 10.1016/S1567-133X(03)00002-4 [DOI] [PubMed] [Google Scholar]
- Zou D., Mcsweeney C., Sebastian A., Reynolds D. J., Dong F., Zhou Y., Deng D., Wang Y., Liu L., Zhu J. et al. (2015). A critical role of RBM8a in proliferation and differentiation of embryonic neural progenitors. Neural Dev. 10, 18 10.1186/s13064-015-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.