Abstract
The rising prevalence of type 1 diabetes (T1D) over the past decades has been linked to lifestyle changes, but the underlying mechanisms are largely unknown. Recent findings point to gut-associated mechanisms in the control of T1D pathogenesis. In nonobese diabetic (NOD) mice, a model of T1D, diabetes development accelerates after deletion of the Toll-like receptor 4 (TLR4). We hypothesized that altered intestinal functions contribute to metabolic alterations, which favor accelerated diabetes development in TLR4-deficient (TLR4−/−) NOD mice. In 70–90-day-old normoglycemic (prediabetic) female NOD TLR4+/+ and NOD TLR4−/− mice, gut morphology and microbiome composition were analyzed. Parameters of lipid metabolism, glucose homeostasis, and mitochondrial respiratory activity were measured in vivo and ex vivo. Compared with NOD TLR4+/+ mice, NOD TLR4−/− animals showed lower muscle mass of the small intestine, higher abundance of Bacteroidetes, and lower Firmicutes in the large intestine, along with lower levels of circulating short-chain fatty acids (SCFA). These changes are associated with higher body weight, hyperlipidemia, and severe insulin and glucose intolerance, all occurring before the onset of diabetes. These mice also exhibited insulin resistance–related abnormalities of energy metabolism, such as lower total respiratory exchange rates and higher hepatic oxidative capacity. Distinct alterations of gut morphology and microbiota composition associated with reduction of circulating SCFA may contribute to metabolic disorders promoting the progression of insulin-deficient diabetes/T1D development.
Keywords: animal model, autoimmune disease, glucose metabolism, insulin resistance, islet, microbiome, mitochondrial metabolism, type 1 diabetes, Toll-like receptor 4 (TLR4)
Introduction
Type 1 diabetes (T1D)3 is characterized by absolute insulin deficiency resulting from progressive autoimmune destruction of pancreatic beta cells (1, 2). Epidemiological data indicate that not only the prevalence of obesity-related type 2 diabetes (T2D) but also of T1D is continuously increasing (3). In this context, accelerated childhood body weight gain (4) and insulin resistance (5) along with impaired glucose tolerance (6) associate with a higher risk for or earlier onset of T1D (7). Moreover, recent studies suggest that tissue-specific abnormalities of energy metabolism could underlie the insulin resistance not only in T2D but also in rodent models and humans with T1D (8, 9).
Toll-like receptor 4 (TLR4), a dominant member of the family of pattern recognition receptors (10, 11), is an attractive candidate to link diabetes-promoting immunologic and metabolic processes. TLR4 was initially identified as a receptor for lipopolysaccharides (LPS), which are integral components of the cell wall of Gram-negative bacteria (12). Based on this feature, TLR4 contributes to the activation of host defense against microorganisms (12). Meanwhile, however, increasing evidence suggests that TLR4 is also involved in the induction of autoimmunity, including development of T1D (13, 14). The presence of TLR4 on insulin target tissues, such as liver, adipose tissue, and skeletal muscle (15), supports its relevance for diabetes-associated metabolic disorders such as obesity and insulin resistance (16). In fact, elevated TLR4 expression correlates with increased systemic concentrations of free fatty acids (FFA) and glucose (17) and with the degree of insulin resistance in obesity (18, 19). However, the mechanisms underlying TLR4-dependent modulation of the progression of insulin-deficient/T1D are not yet clear.
Recent findings suggest an impact of the intestinal microbiome on the onset and progression of T1D. Patients with T1D have a less diverse and stable gut microbiome than glucose-tolerant humans (20, 21). Moreover, gut microbiome dysbiosis in early childhood associates with an increased risk of progression to T1D (22, 23). Intestinal bacteria are able to modulate the host's metabolism by various mechanisms, specifically by the release of short-chain fatty acids (SCFA), a set of highly-abundant fermentation products derived from the bacterial degradation of complex macrofibrous dietary components (24, 25). SCFA are potent modulators of glucose and energy metabolism (26) and thereby might affect the development of T1D (27).
These findings are in line with the concept of control of the pathogenesis of insulin-deficient/T1D by TLR4 expression status via modification of morphological and functional properties of the gut. Here, we tested the hypothesis that TLR4-dependent alterations of intestinal function affect metabolic homeostasis thereby contributing to accelerated progression of insulin-deficient diabetes. To this end, we used the TLR4-deficient (TLR4−/−) nonobese diabetic (NOD) mouse strain, which shows enhanced insulitis and therefore can serve as a model of accelerated human T1D development (14), that allows us to examine disease-relevant metabolic processes on a genetic background predisposing to insulin-deficient diabetes (28).
Results
Accelerated development of body weight and diabetes in NOD TLR4−/− mice
Food intake, body weight development, and body composition were monitored during the prediabetic period in 70–90-day-old female normoglycemic NOD TLR4+/+ and NOD TLR4−/− mice. Despite comparable food intake (Fig. 1A), NOD TLR4−/− mice showed an accelerated development of body weight (Fig. 1B) associated with slightly greater fat mass than NOD TLR+/+ animals (Fig. 1C). Female NOD TLR4−/− mice manifested diabetes more than 7 weeks earlier (age at disease onset: 152 ± 28 days) than their NOD TLR4+/+ littermates (age at disease onset: 208 ± 40 days) (Fig. 1D). At disease onset, hyperglycemia was more pronounced in NOD TLR4−/− than in NOD TLR4+/+ mice (Fig. 1E).
Figure 1.
Accelerated body weight development and higher fat mass in the prediabetic state as well as younger age and higher blood glucose levels at diabetes onset in NOD TLR4−/− mice. Food intake (A), body weight (B), and body composition (C) were determined in normoglycemic 70–90-day-old female TLR4-expressing (TLR4+/+) and TLR4-deficient (TLR4−/−) NOD mice. The age at diabetes onset (D) was defined as the first of 2 consecutive days with blood glucose levels above 250 mg/dl. Initial blood glucose concentrations (E) were documented at diabetes manifestation. Data are presented as means ± S.E. (n = 9–13 per group). *, p < 0.05; ***, p < 0.001 by Student's t test.
Altered gut morphology and gut microbiota in NOD TLR4−/− mice
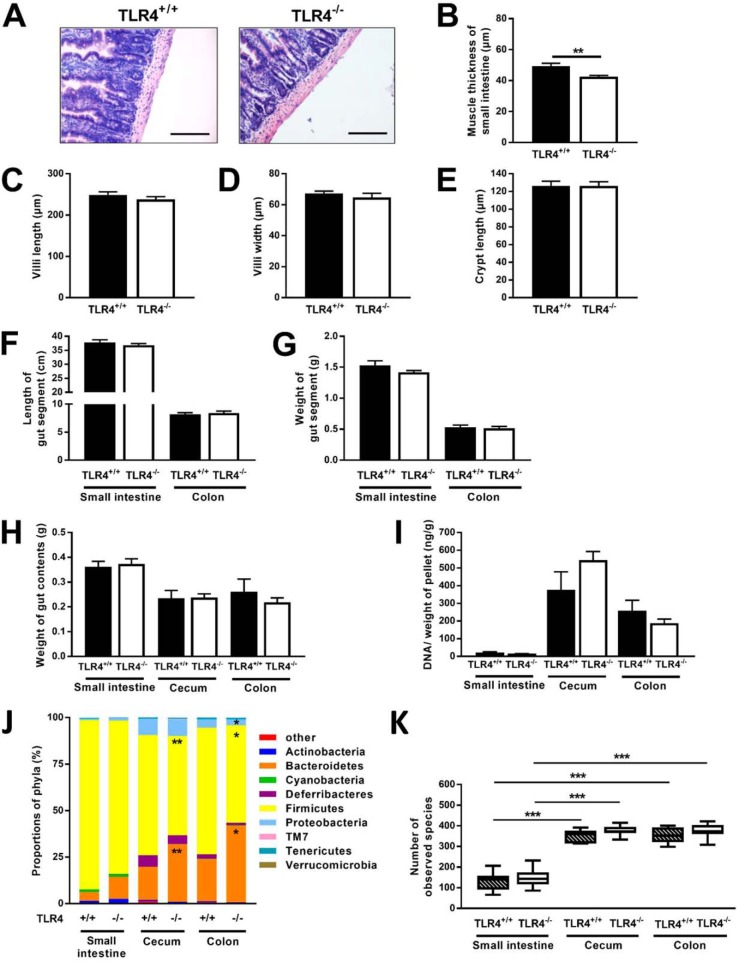
Detailed analyses of gut morphology showed a proportional reduction of both longitudinal and circular muscle layers of the small intestine of NOD TLR4−/− mice (Fig. 2A) resulting in lower total muscle thickness of the gut segment (Fig. 2B). The TLR4 expression status neither affected the dimensions of small intestinal villi and crypts (Fig. 2, C–E) nor the length and weight of the gut segments (Fig. 2, F and G) or weight of gut contents (Fig. 2H).
Figure 2.
Altered gut morphology and gut microbiome composition in NOD TLR4−/− mice. In the normoglycemic 70–90–day-old female TLR4-expressing (TLR4+/+) and TLR4-deficient (TLR4−/−) NOD mice, gut morphology was analyzed from H&E-stained thin sections of the small intestine (A, bar = 100 μm), and the total thickness of gut muscle layer (B), villi length (C), villi width (D), and crypt length (E) were determined morphometrically. The length (F) and weight (G) of small intestine and colon, the weight of the contents of the small intestine, cecum, and colon (H), as well as the ratio of bacterial DNA amount per g of gut contents (I) were determined. Data are presented as means ± S.E. (n = 5–16 per group). **, p < 0.01 by Student's t test. Composition of bacterial phyla in small intestine, cecum, and colon (J) was determined by NGS, and the numbers of bacterial species (α diversity) in the distinct gut segments (K) were calculated. Data are shown as means ± S.E. Numbers of bacterial species are presented as boxplots (lowest value, lower quartile, median, upper quartile, and highest value) (n = 8–15 per group). *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test or one-way ANOVA.
Effects of TLR4 deletion on gut microbiome composition were assessed using bacterial DNA isolated from small intestine, cecum, and colon. The gut segments contained comparable microbial biomass in NOD TLR4+/+ and NOD TLR4−/− mice as estimated from the ratio of isolated bacterial DNA/pellet of gut contents (Fig. 2I). Employing next generation sequencing (NGS) revealed an increased abundance of Bacteroidetes and a decrease of Firmicutes in the cecum and colon of NOD TLR4−/− mice compared with the corresponding gut segments of NOD TLR4+/+ animals (Fig. 2J and Table S1). In line with the high quantity of bacteria in cecum and colon, these gut segments exhibited the highest α-diversity (Fig. 2K).
Assessing intestinal inflammation from frequency and distribution of CD3+ lymphocytes in the gut wall revealed no differences between NOD TLR4+/+ and NOD TLR4−/− mice (Fig. S1, A and B), which resembled the distribution pattern in corresponding tissue samples of age- and sex-matched normal healthy C57BL10 mice (data not shown). NOD TLR4−/− mice had a higher activity of matrix metalloproteinase-2 (MMP-2), a marker of extracellular matrix integrity (Fig. S1C), but unchanged levels of other mediators of systemic inflammation (Fig. S1, D and E). Mucispirillum content correlated with plasma glucagon like peptide (GLP)-2 only in NOD TLR4−/− mice, again indicating impaired gut integrity of NOD TLR4−/− mice (Fig. S2).
Higher LPS levels and altered lipid and SCFA profiles in NOD TLR4−/− mice
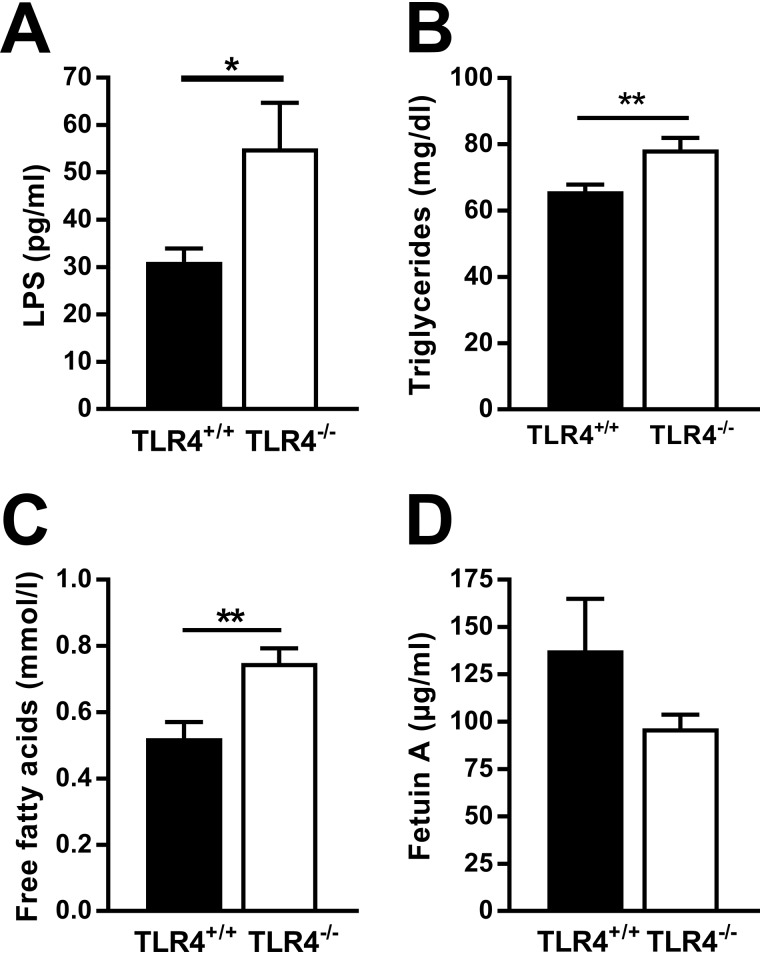
As gut wall integrity and intestinal microbiome composition strongly affect the availability of bacterial products in the host's circulation, we determined the levels of plasma components reflecting absorption of gut-derived bacterial products and lipids. When compared with NOD TLR4+/+ mice, NOD TLR4−/− mice showed higher plasma concentrations of LPS (Fig. 3A), a cell wall component of Gram-negative bacteria that can be incorporated into chylomicrons. NOD TLR4−/− mice also had higher levels of triglycerides (TG) and FFA than NOD TLR4+/+ mice in plasma (Fig. 3, B and C). We next measured circulating levels of fetuin A in serum, an abundant liver-derived glycoprotein, acting as an adaptor to enable the interaction of FFA with TLR4 (29). However, both NOD TLR4+/+ and NOD TLR4−/− mice showed high but comparable fetuin A concentrations (Fig. 3D).
Figure 3.
Effect of the TLR4 expression status on peripheral levels of LPS, lipids, and fetuin A. LPS concentrations (A) were determined in the plasma of nonfasted 70–90-day-old female TLR4-expressing (TLR4+/+) and TLR4-deficient (TLR4−/−) NOD mice. Plasma concentrations of triglycerides (B) and free fatty acids (C) and serum fetuin A levels (D) were determined in 6-h–fasted NOD mice. Data are presented as means ± S.E. (n = 3–7 per group). *, p < 0.05; **, p < 0.01 by Student's t test.
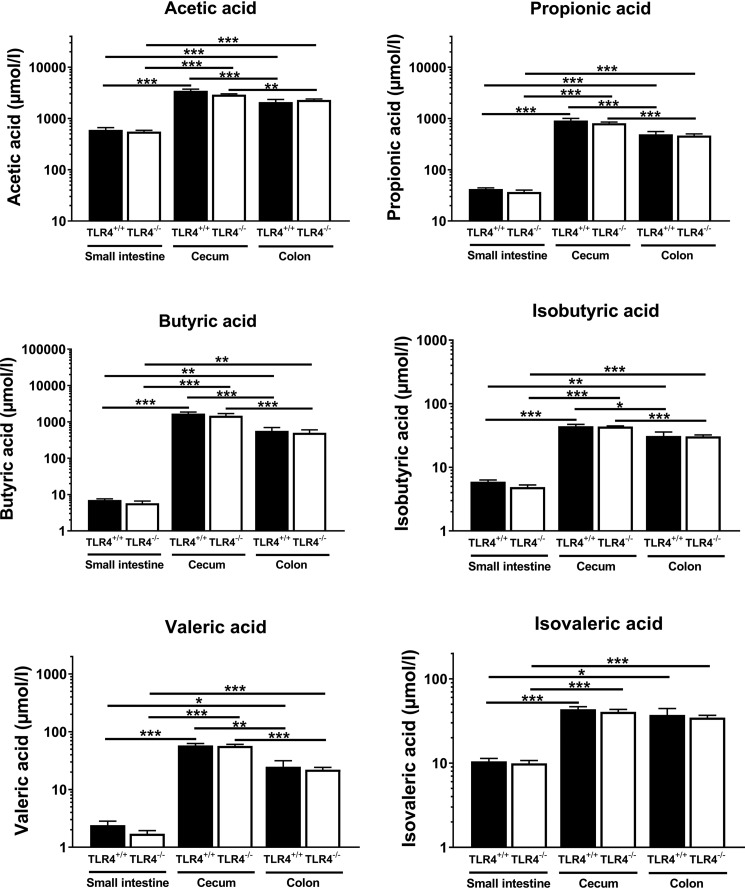
As intestinal bacteria-derived SCFA are potent modulators of host energy metabolism, we determined their concentrations in the lumen of small intestine, cecum, and colon using GC-MS. Highest SCFA concentrations were found in the two distal gut segments with acetic acid, propionic acid, and butyric acid as the most abundant species (Fig. 4). No differences were observed in SCFA concentrations in gut segments from TLR4+/+ and NOD TLR4−/− mice.
Figure 4.
Effect of the TLR4 expression status on intraluminal concentrations of SCFA in gut segments of NOD mice. Concentrations of SCFA were determined in the lumen of the small intestine, cecum, and colon of nonfasted 70–90-day-old female TLR4-expressing (TLR4+/+) and TLR4-deficient (TLR4−/−) NOD mice. Data are presented as means ± S.E. (n = 8–10 per group). *, p < 0.05; **, p < 0.01; ***, p < 0.001 by ANOVA.
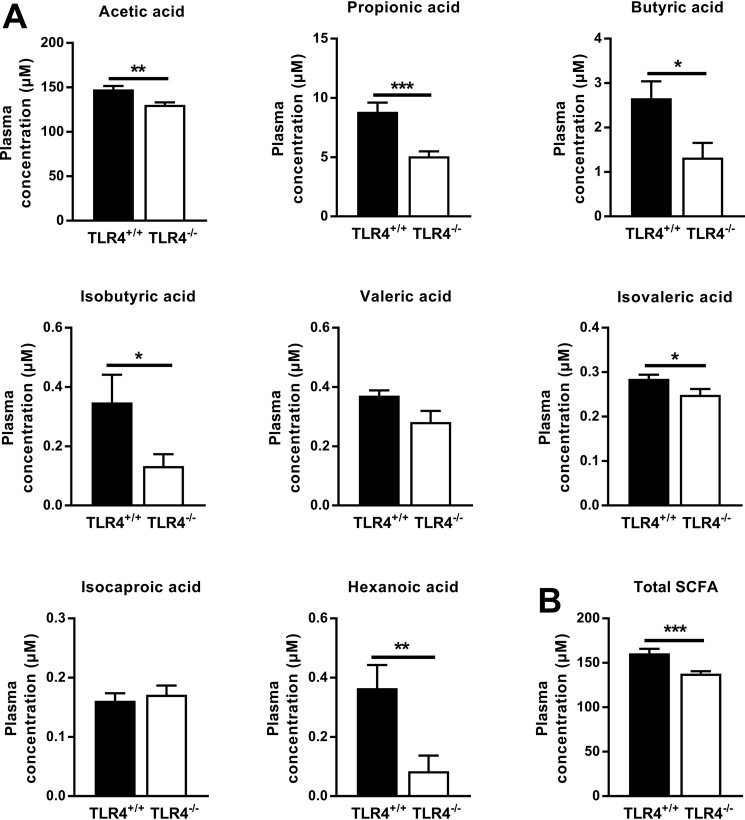
In the plasma of both NOD TLR4+/+ and NOD TLR4−/− mice, acetic acid, propionic acid, and butyric acid were also identified as the most abundant SCFA. However, NOD TLR4−/− mice had 12, 43, and 68% lower concentrations of acetic acid, propionic acid, and butyric acid, respectively (Fig. 5A). Only low concentrations of isobutyric acid, valeric and isovaleric acid, isocaproic acid, and hexanoic acid were detectable. The total concentration of peripheral SCFA was markedly lower in NOD TLR4−/− than in NOD TLR4+/+ mice (p < 0.001) (Fig. 5B).
Figure 5.
Effect of the TLR4 expression status on peripheral levels of SCFA. Concentrations of SCFA were determined in plasma of nonfasted 70–90-day-old female TLR4-expressing (TLR4+/+) and TLR4-deficient (TLR4−/−) NOD mice. Data of individual (A) and total (B) SCFA are presented as means ± S.E. (n = 10 per group). *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test.
Impaired energy homeostasis and glucose metabolism in NOD TLR4−/− mice
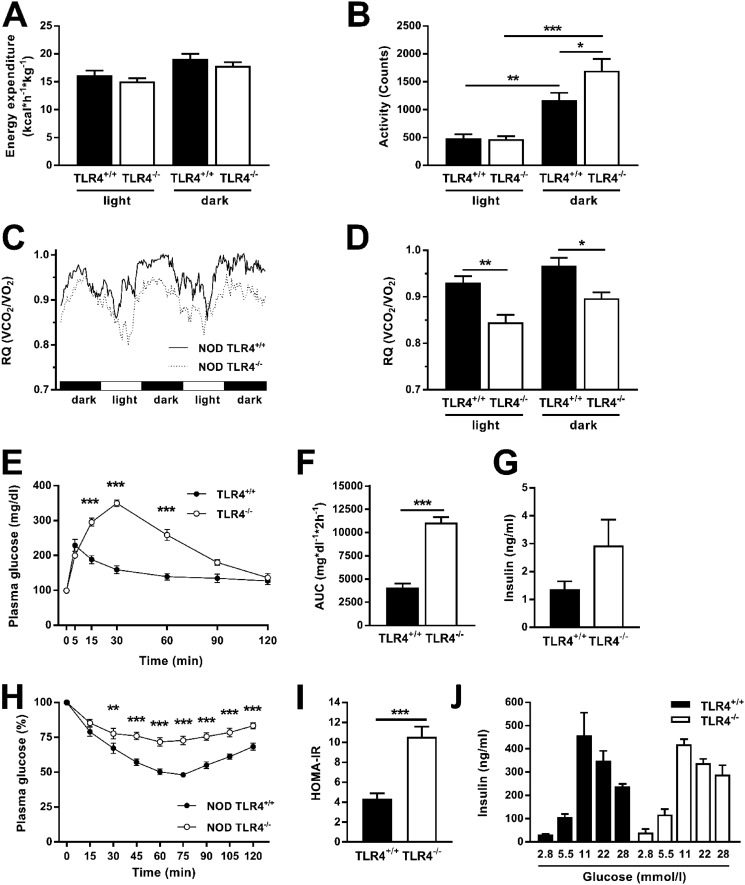
Metabolic phenotyping during three dark and two light phases revealed lower energy expenditure during light phases, occurring independently of the TLR4 expression status (Fig. 6A). Physical activity followed a similar pattern, showing comparable low levels during the light phase and 3–4-fold increases in the dark phase (Fig. 6B). Interestingly, NOD TLR4−/− mice had greater maximum physical activity than NOD TLR4+/+ mice during the dark phase. All mice also exhibited the typical pattern of circadian rhythmicity of the respiratory exchange rate (RQ) with maximum values in the dark phases (Fig. 6, C and D). NOD TLR4−/− mice had consistently lower RQ values than NOD TLR4+/+ mice (Fig. 6D), suggesting increased utilization of dietary fat as energy source.
Figure 6.
Effect of the TLR4 expression status on energy balance and glucose homeostasis of NOD mice. Energy expenditure (A), physical activity (B), and RQ (C and D) were monitored in normoglycemic 70–90-day-old female TLR4-expressing (TLR4+/+) and TLR4-deficient (TLR4−/−) NOD mice over three dark and two light phases. Glucose tolerance was determined by ipGTT (E and F), and the serum insulin concentrations of the animals were quantified by ELISA (G). Insulin tolerance was determined by ipITT (H). HOMA-IR was calculated from fasting blood glucose and insulin levels (I). Glucose-stimulated insulin release was determined from isolated cultivated islets of NOD TLR4+/+ and NOD TLR4−/− mice (J). Data are presented as means or means ± S.E. (n = 3–10 per group). *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test or one-way ANOVA.
Intraperitoneal glucose tolerance tests (ipGTT) revealed severe glucose intolerance with elevation of both glucose peak (Fig. 6E) and overall glucose appearance, as measured from the area under the glucose concentration curve, in NOD TLR4−/− mice (Fig. 6F). Of note, peak glucose levels occurred 25 min later, pointing to delayed glucose absorption in the TLR4-deficient mice. Serum insulin concentration, measured at the end of the ipGTT, was not appropriately increased in NOD TLR4−/−, indicating impaired glucose-dependent insulin secretion (Fig. 6G). To assess insulin sensitivity in more detail, the animals underwent an intraperitoneal insulin tolerance test (ipITT), which showed marked whole-body insulin resistance in NOD TLR4−/− mice (Fig. 6H). In addition, in line with hepatic insulin resistance these mice had higher HOMA-IR values (Fig. 6I). To examine glucose-responsive insulin secretion, we isolated islets of Langerhans from pancreata of the mice. These studies revealed comparable dose-dependent glucose-stimulated insulin release in NOD TLR4+/+ and NOD TLR4−/− mice (Fig. 6J).
Higher hepatic oxidative capacity but unchanged muscle oxidative capacity in NOD TLR4−/− mice
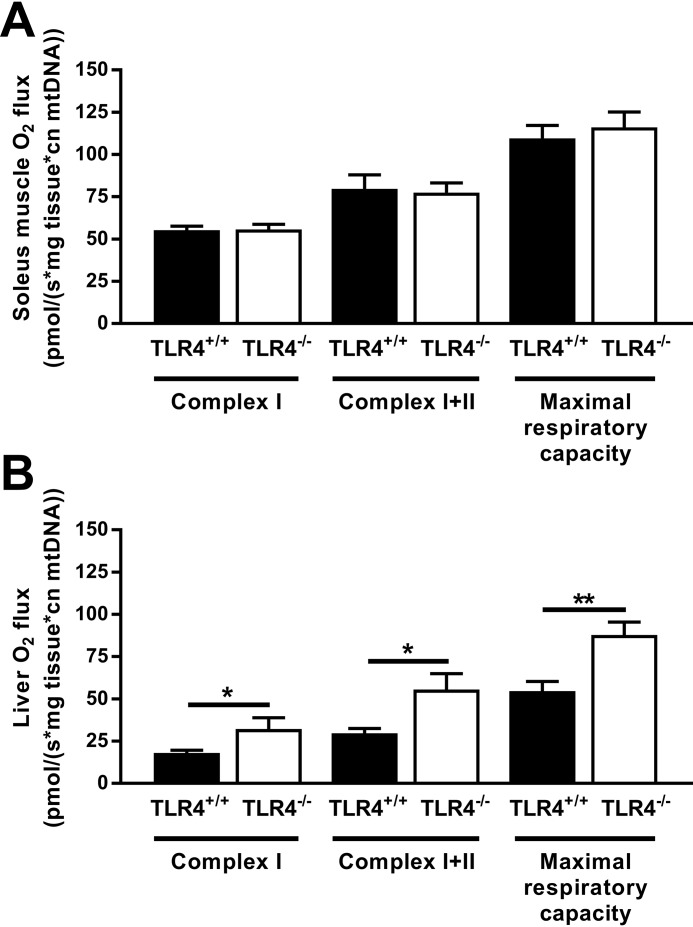
Reduced RQ and glucose tolerance along with higher plasma lipids in NOD TLR4−/− mice indicate abnormal energy metabolism. For the direct measurement of substrate-dependent oxidation, we employed high-resolution respirometry on samples from soleus muscle and liver to quantify the mitochondrial oxygen fluxes via complex I and complexes I and II and to assess maximum respiratory capacity (Fig. 7). In soleus muscle, TLR4 expression status did not affect oxygen fluxes (Fig. 7A). In contrast, livers from NOD TLR4−/− mice showed 60–86% higher rates of oxygen flux in complex I and complexes I and II as well as maximal respiratory capacity compared with those obtained from NOD TLR4+/+ mice (Fig. 7B). This indicates hepatic mitochondrial adaptation to greater glucose and lipid availability as reported previously in insulin-resistant rodent models and humans (8, 30).
Figure 7.
Effect of the TLR4 expression status on mitochondrial function in NOD mice. Mitochondrial respiration capacity through complex I and complexes I and II as and the maximal respiratory capacities of the electron transport system were determined in the soleus muscle (A) and the liver (B) of NOD TLR4+/+ and NOD TLR4−/− mice. Respiratory data were individually corrected for mitochondrial content. Data are presented as means ± S.E. (n = 3–7 per group). *, p < 0.05; **, p < 0.01 by Student's t test.
Discussion
These data show that profound alterations of gut morphology and microbiota and tissue-specific abnormalities of energy metabolism associate with altered availability of SCFA and precede accelerated diabetes progression in a model of T1D. This study also suggests a complex relationship between TLR4-dependent intestinal abnormalities and altered inter-organ communication by an imbalance between SCFA and FFA resulting in disease-promoting conditions in the prediabetic state.
The earlier onset of diabetes and higher blood glucose levels at disease onset in NOD TLR4−/− mice confirmed the diabetes-accelerating effect of TLR4 deficiency in this NOD mouse model (14). This study provides evidence that TLR4 deficiency promotes the development of diabetes under conditions of early alterations of gut microbiota composition and gut morphology before disease onset.
The TLR4 expression status determines important structural and functional properties of the intestinal system. By controlling the proliferation of smooth muscle cells, this receptor is involved in the regulation of muscle tissue expansion (31). As intestinal smooth muscle cells express functional TLR4 (32), TLR4 deficiency may foster decreasing the thickness of the intestinal muscle layer as observed in the NOD TLR4−/− mice in this study. Reduced intestinal muscle thickness is known to impair gut motility (33). Moreover, the contractile function of intestinal smooth muscle cells also depends on the appropriate expression of TLR4 (34). Thus, TLR4 deficiency likely promotes prolongation of the intestinal transit time of gut contents (35) as reported recently in metabolically healthy C57BL6 mice carrying a homozygous TLR4 defect (36).
Consequently, reduced gut motility and prolonged intestinal transit time cause alterations of the digestive process and the accumulation of absorbable food components (37), thereby modulating the survival conditions for distinct bacterial communities. A recent study demonstrated such an association between delayed intestinal transit and alterations of gut microbiome composition (38). In this study, NOD TLR4−/− mice showed an imbalance of the two most abundant bacterial phyla when compared with NOD TLR4+/+ mice. Bacteroidetes were increased while Firmicutes were decreased in the distal gut segments of normoglycemic TLR4-deficient mice at an age of 70–90 days, i.e. before the age of diabetes-onset. This finding corresponds to observations in patients with T1D and in children at increased risk for T1D showing a predominance of distinct Bacteroidetes species in their gut microbiome prior to the clinical onset of auto-immunity (21). Of note, some previous studies suggested an involvement of gut microbiota in the development of insulin-deficient diabetes (3, 39, 40).
In this study, NOD TLR4−/− mice further exhibited signs of impaired barrier function of the intestinal wall, indicated by increased activity of MMP2, a dominant member of the metalloproteinase family involved in tissue remodeling (41). Of note, expression of metalloproteinases is also up-regulated in the gut of patients with T1D pointing to disturbed tissue integrity (42). Furthermore, NOD TLR4−/− mice had higher plasma concentrations of bacterial LPS, indicative of an enhanced translocation of the toxin from the gut lumen to the circulation. The increased abundance of Bacteroidetes, which are mainly composed of Gram-negative species, might further contribute to the elevated peripheral LPS levels in these animals. Bacterial products, including LPS, can reduce self-tolerance thereby promoting the development of organ-specific autoimmunity (43). In line, augmented translocation of bacterial products from the gut lumen into the circulation may particularly contribute to the accelerated progression of beta-cell damage and insulin-deficient diabetes observed in NOD TLR4−/− mice (14, 39). Indeed, the 70–90-day-old prediabetic NOD TLR4−/− mice exhibited impaired glucose-induced insulin secretion during the ipGTT.
Intestinal bacteria release SCFA, which can enter the circulation thereby representing a possibly important link between gut microbiome and host metabolism (24, 26). Interestingly, NOD TLR4−/− mice exhibited profound changes in the composition of gut bacteria, dominated by a decrease in the abundance of Firmicutes and an increase of Bacteroidetes. However, this shift in the intestinal microbial community did not result in altered SCFA composition in the gut lumen of these mice. Nevertheless, NOD TLR4−/− mice showed lower plasma SCFA levels with a most pronounced decline in butyrate. As butyrate stabilizes tight junctions in gastrointestinal epithelia, lower butyrate concentration may further contribute to destabilizing the gut barrier in NOD TLR4−/− mice (44). Reduced SCFA levels in blood but not in the gut of NOD TLR4−/− mice may result from altered peripheral consumption of SCFA. These findings further suggest that SCFA in the circulation rather than in the gut lumen are decisive for the progression of insulin-deficient diabetes. Of note, a recent study demonstrates the relevance of peripheral but not fecal SCFA for regulation of insulin sensitivity and lipolysis in humans (45). Moreover, the observation of reduced peripheral SCFA concentrations in NOD TLR4−/− mice with accelerated diabetes development are in line with results of a study in children on the protective effect of SCFA in early-onset T1D (27).
The key metabolic effects of SCFA result from binding to specific FFA receptors, which are involved in the control of intracellular lipid turnover (46). Activation of FFA receptors stimulates fatty acid oxidation and inhibits lipolysis resulting in reduced plasma FFA concentrations and decreased body weight (47). In the NOD TLR4−/− mice, lower plasma SCFA were associated with increases in plasma FFA, TG, body weight, and fat mass despite unaltered food uptake in the prediabetic period. Augmented availability of (long-chain) FFA, mainly mediated by the diacylglycerol/protein kinase C pathway (26, 48), is most likely responsible for the marked whole-body insulin resistance in NOD TLR4−/− mice. This view is supported by the finding that NOD TLR4−/− mice exhibit low-insulin sensitivity along with lower butyrate, but higher FFA levels than the insulin-sensitive NOD TLR4+/+ mice.
The elevation of circulating FFA may result from excessive production and/or impaired mitochondrial oxidation (49). Interestingly, NOD TLR4−/− mice featured increased oxidative capacity only in liver and not in skeletal muscle. Altered substrate availability might have contributed to the increased hepatic oxygen flux, supporting our recent findings of augmented hepatic mitochondrial respiration in prediabetic NOD mice (8) and in severely obese humans (30). Of note, this will promote hepatic oxidative stress and challenge hepatocellular anti-oxidant defense mechanisms and further worsen energy metabolism and insulin resistance (30). Recent studies confirm abnormalities of hepatic mitochondrial function and energy homeostasis also in patients with overt T1D (49). In addition, the liver-derived protein, fetuin A, could have served as a mediator between hyperlipidemia and metabolic balance, due to its described function as adaptor mediating FFA binding to TLR4 interfering with insulin signaling (50). However, fetuin A levels were independent of the TLR4 expression status and tissue-specific mitochondrial function in these mice. Likewise, hepatic mitochondrial adaptation occurs independently of fetuin A in obese humans (30). Thus, these data exclude a relevant role of fetuin A-FFA–TLR4 interaction in the observed metabolic changes.
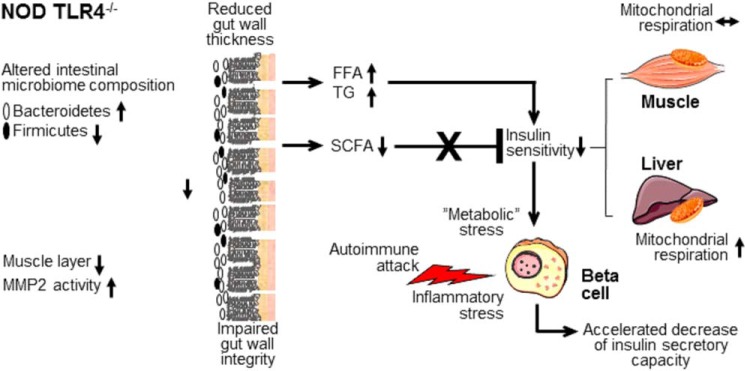
The profound metabolic disturbances in normoglycemic NOD TLR4−/− mice support the view that the metabolic disturbances mediated by TLR4 deficiency affect the pathogenesis of the disease already at an early, prediabetic stage. According to the model outlined in Fig. 8, NOD TLR4−/− mice exhibit an altered composition of gut bacteria characterized by an increased abundance of Bacteroidetes and a decrease of Firmicutes. The animals further show impaired gut morphology due to muscular atrophy and increased MMP2 activity. Increased peripheral levels of (long-chain) FFA and TG further point to an altered absorption of nutrients and an impaired gut barrier in NOD TLR4−/− mice. Altered substrate availability as a consequence of hyperlipidemia results in impaired insulin sensitivity associated with increased respiratory activity of hepatic mitochondria. Decreased peripheral levels of SCFA, particularly butyrate, which are linked to lower body weight and improved glycemic control, are insufficient to counteract the impairment of insulin sensitivity and to normalize glucose homeostasis. Under conditions of impaired insulin sensitivity, an increased demand for insulin might impose further “metabolic” stress on beta cells that are already exposed to enhanced inflammatory stress mediated by the rapidly-progressing insulitis as observed in prediabetic NOD TLR4−/− mice (14). This accelerates beta-cell failure and thereby progression to insulin-deficient diabetes in NOD TLR4−/− mice.
Figure 8.
Model of the gut-associated mechanisms underlying the accelerated progression of insulin-deficient diabetes in NOD TLR4−/− mice. Insulin-sensitive metabolic processes modulate the development of T1D, specifically the accelerated progression of diabetes in the NOD TLR4−/− mouse resulting from altered microbiota, which lead to impaired metabolic homeostasis, thereby promoting insulin-deficient diabetes.
In conclusion, NOD TLR4−/− mice allowed us to identify the associations between distinct alterations of gut morphology and microbiome composition with metabolic disorders promoting the progression of insulin-deficient diabetes/T1D. Accelerated diabetes development likely results from altered hepatic respiratory activity and impaired insulin sensitivity. Circulating SCFA may serve as important mediators in these processes. Taken together, these findings identify TLR4-dependent metabolic pathways as promising targets for intervention strategies aimed at the preservation of beta-cell function in individuals at risk of T1D.
Experimental procedures
Animals
NOD mice were from the breeding colony at the German Diabetes Center. TLR4-expressing C57BL/10ScSn (C57BL/10 TLR4+/+) and C57BL/10ScCr mice lacking TLR4 expression (C57BL/10 TLR4−/−), due to a spontaneous deletion of the TLR4-encoding region (51), were from the Max-Planck-Institute for Immunobiology and Epigenetics, Freiburg, Germany. The TLR4 defect was backcrossed onto the NOD background for more than 12 generations, and heterozygous littermates were intercrossed to generate NOD TLR4+/+ and TLR4−/− animals (14). Unless otherwise indicated, the animals had free access to water and standard diet (Ssniff M-Z Extrudat, 4.5% fat; SSNIFF Spezialdiäten GmbH, Soest, Germany). All experiments were performed with co-housed female mice at the age of 70–90 days derived from breeding pairs carrying a heterozygous TLR4 defect. Female mice were monitored for the development of diabetes until 220 days of age. Animals were considered diabetic with blood glucose levels (measured on an EPOS Analyzer 5060, Eppendorf, Hamburg, Germany) exceeding 250 mg/dl on 2 consecutive days. All experiments were conducted in accordance with the Principles of Laboratory Animal Care (GV-SOLAS, Society for Laboratory Animal Science) and were approved by the ethics committee on animal welfare of the State of North Rhine Westphalia.
Indirect calorimetry and body composition
Mice were individually placed in an 8-chamber indirect calorimetry system (PhenoMaster, TSE Systems, Bad Homburg, Germany). After 24 h of adaptation, parameters of indirect calorimetry (RQ), physical activity (interruption of IR light beam), and the intake of water and food were simultaneously analyzed for each mouse over 48 h (8). Body composition of mice was assessed by an Echo MRI body composition analyzer system (EchoMRITM-100 System, Echo Medical Systems, Houston, TX) allowing the quantification of fat and lean body mass (52).
Glucose and insulin tolerance tests
The ipGTT was performed in 70–90-day-old animals. It was commenced after 6 h of fasting by injecting 2 g/kg body weight glucose and measuring the glucose levels in blood obtained from the tail vein before (baseline) and 5, 15, 30, 60, 90, and 120 min after injection. The ipITT was performed in 70–90-day-old animals after 6 h of fasting by intraperitoneally injecting 0.75 units/kg body weight human fast-acting insulin (Insuman Rapid, Sanofi-Aventis, Frankfurt am Main, Germany). Glucose was measured in blood collected from the tail tip immediately before (baseline) and 15, 30, 45, 60, 75, 90, 105, and 120 min after the injection of insulin (53). Homeostatic model assessment–insulin resistance was calculated from fasting blood glucose and insulin concentrations according to the formula: HOMA-IR = ((fasting blood glucose (mg/dl) × (fasting insulin concentration (units/ml))/405 (54).
Glucose-stimulated insulin secretion from isolated islets
Pancreatic islets were isolated from normoglycemic female mice by collagenase digestion (Serva, Heidelberg, Germany), as described previously (54). The islets were exposed to increasing glucose concentrations (2.8–28 mm) in Hanks' balanced salt solution for 24 h (37 °C, 5% (v/v) CO2). The amount of insulin released from the cultivated islets was measured by ELISA (Mouse Insulin ELISA kit, Mercodia, Uppsala, Sweden).
Morphological analysis and immunofluorescence staining of intestinal tissue
Paraformaldehyde-fixed frozen thin sections of the small intestine were stained with hematoxylin and eosin. Images were acquired using a Leica DMRBE inverted microscope equipped with an Olympus DP73 digital color camera. CellSens imaging software version 1.7 (Olympus Life Science, Hamburg, Germany) was used for morphometric analyses of muscle thickness, villi length and width, as well as crypt length. Paraformaldehyde-fixed thin sections of small intestinal tissue were incubated with a polyclonal rabbit anti-CD3 antibody (DAKO, Hamburg, Germany) followed by incubation with a secondary Alexa Fluor 488–conjugated donkey anti-rabbit antibody (Life Technologies, Inc., Darmstadt, Germany) and Hoechst 33342 dye (Sigma, Steinheim, Germany) for nuclei staining.
Assessment of MMP-2 activity
MMP-2 activity was assessed by gelatin zymography in homogenates of intestinal tissue, as described previously (55). Collagenase served as a positive control for proteinase activity. Band intensities were quantified using the software Fiji (National Institute of Health, Bethesda, MD). Resulting values were used as relative arbitrary units for MMP activity.
High-resolution respirometry
Ex vivo mitochondrial function was measured by high-resolution respirometry (Oxygraph-2k, Oroboros Instruments, Innsbruck, Austria) as described previously (8). Briefly, fresh liver and soleus muscle samples were permeabilized by saponin treatment, and defined respiratory states were obtained by the following multiple substrate–inhibitor titration protocol: 2 mm malate, 10 mm pyruvate, 10 mm glutamate, and 2.5 mm ADP for state 3 respiration of complex I; 10 mm succinate for combined state 3 respiration of complex I and II; 10 μm cytochrome c for mitochondrial membrane integrity check; carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (stepwise increments of 0.25 μm up to the final concentration of 1.25 μm) for maximal respiratory capacity, namely state u, and 2.5 μm antimycin A. Addition of cytochrome c did not increase oxygen consumption indicating integrity of the outer mitochondrial membrane after saponin permeabilization. The results of the analyses were expressed as pmol/(s × mg) after correction for mitochondrial copy numbers as described (8).
Extraction of genomic DNA and profiling of the 16S rRNA gene by NGS
Genomic DNA was isolated from gut contents by applying the DNA stool mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. DNA concentrations and the OD260/280 ratio for quality control were determined using the NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). A minimum OD260/280 ratio of 1.9 was regarded as sufficient for further analyses.
The bacterial DNA samples were profiled by sequencing of the V4 region of the 16S rRNA gene on an Illumina MiSeq (Illumina RTA version 1.17.28; MCS version 2.5) using 515 forward and 806 reverse primers designed for dual indexing and the V2 kit (2 × 250 bp paired-end reads). For details of DNA isolation, next generation sequencing, and evaluation, see supporting information (Extraction of genomic DNA and profiling of the 16S rRNA gene by Next Generation Sequencing (NGS)).
Measurement of SCFA
SCFA were extracted from plasma samples and from samples of the contents of ileum, cecum, and colon and analyzed as described previously (56–58). The acids were quantified using GC-MS. Adequate internal standards of the SCFA were included in all steps of the preparation and analysis procedures. SCFA concentrations were quantified from calibration curves of SCFA that underwent the same preparation and analysis procedures.
Laboratory analyses
Plasma TG (Roche/Hitachi, Roche Diagnostics, Mannheim, Germany) and FFA (Wako Chemicals GmbH, Neuss, Germany) were assayed photometrically. Serum insulin (Mouse Insulin ELISA kit, Mercodia) and fetuin A (Mouse Fetuin A/AHSG DuoSet, R&D Systems, Abingdon, UK) were quantified by ELISA. Serum cytokines were measured with a multiplex screening assay (Biotechne, Wiesbaden, Germany) using the BioPlex analyzer (Bio-Rad, Munich, Germany) according to the manufacturer's protocol. Plasma LPS was quantified using the Limulus amebocyte lysate assay (Lonza, Basel, Switzerland) according to the manufacturer's protocol.
Statistical analyses
Data are expressed as mean values ± S.E., and statistical analysis was performed using nonlinear regression, ANOVA with Bonferroni post hoc analysis, or Student's t test. Differences were considered statistically significant at p < 0.05. All statistical analyses were performed using the Prism software package version 4 (GraphPad Software, San Diego, CA).
Author contributions
M.-C. S., A. L. R., C.W., J. H., K. K., J. A.-C., A. S., M. B., F. B., and V. B. data curation; M.-C. S., A. L. R., C. W., J. H., K. K., M. B., F. B., and V. B. formal analysis; M.-C. S., F. B., and V. B. funding acquisition; M.-C. S., A. L. R., C. W., J. H., T. J., K. K., J. A.-C., A. S., M. B., F. B., and V. B. investigation; M.-C. S., A. L. R., C. W., J. H., T. J., K. K., A. S., M. B., F. B., and V. B. methodology; M.-C. S., F. B., and V. B. writing-original draft; A. L. R., C. W., J. H., T. J., K. K., J. A.-C., A. S., F. B., and V. B. visualization; T. J., F. B., V. B., and M. R. conceptualization; F. B. and V. B. resources; F. B. software; F. B., V. B., and M. R. supervision; V. B. and M. R. project administration; V. B. and M. R. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Valentina Tremaroli and Rozita Akrami for excellent bioinformatics support and Gilles Séquaris, Jörg Kotzka, Birgit Knebel, Jürgen Weiss, Peter Nowotny, Waltraud Fingberg, Kay Jeruschke, and Bärbel Gruhl for excellent technical assistance. We further thank Xuan-Mai Petterson and K. Sreekumaran Nair (Mayo Clinic, Rochester, MN) for support with the plasma SCFA analyses.
This work was supported by the Ministry for Culture and Science of the State of North Rhine-Westphalia (MKW and NRW); the German Federal Ministry of Health (BMG); the Federal Ministry for Research (BMBF) to the German Center for Diabetes Research (DZD e.V.); European Funds for Regional Development Grant EFRE-0400191; EUREKA Eurostars-2 Grant E! 113230 DIA-PEP; German Science Foundation (DFG) Grant CRC/SFB 1116/2 B12; the Schmutzler Stiftung, Germany; the German Diabetes Association (DDG); the German Society for Mucosal Immunology and Microbiome (DGMIM); and the Wallenberg Laboratory, Department of Molecular and Clinical Medicine, University of Gothenburg, Sweden. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
- T1D
- type 1 diabetes
- T2D
- type 2 diabetes
- ANOVA
- analysis of variance
- SCFA
- short-chain fatty acid
- NOD
- nonobese diabetic
- TLR4
- Toll-like receptor 4
- LPS
- lipopolysaccharide
- FFA
- free fatty acid
- NGS
- next generation sequencing
- TG
- triglyceride
- RQ
- respiratory exchange rate
- ipGTT
- intraperitoneal glucose tolerance test
- ipITT
- intraperitoneal insulin tolerance test
- HOMA-IR
- homeostatic model assessment-insulin resistance
- MMP-2
- matrix metalloproteinase-2.
References
- 1. Atkinson M. A., Eisenbarth G. S., and Michels A. W. (2014) Type 1 diabetes. Lancet 383, 69–82 10.1016/S0140-6736(13)60591-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Belle T. L., Coppieters K. T., and von Herrath M. G. (2011) Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol. Rev. 91, 79–118 10.1152/physrev.00003.2010 [DOI] [PubMed] [Google Scholar]
- 3. Knip M., and Siljander H. (2016) The role of the intestinal microbiota in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 12, 154–167 10.1038/nrendo.2015.218 [DOI] [PubMed] [Google Scholar]
- 4. Verbeeten K. C., Elks C. E., Daneman D., and Ong K. K. (2011) Association between childhood obesity and subsequent type 1 diabetes: a systematic review and meta-analysis. Diabet. Med. 28, 10–18 10.1111/j.1464-5491.2010.03160.x [DOI] [PubMed] [Google Scholar]
- 5. Xu P., Cuthbertson D., Greenbaum C., Palmer J. P., Krischer J. P., and Diabetes Prevention Trial-Type 1 Study Group. (2007) Role of insulin resistance in predicting progression to type 1 diabetes. Diabetes Care 30, 2314–2320 10.2337/dc06-2389 [DOI] [PubMed] [Google Scholar]
- 6. Chase H. P., Cuthbertson D. D., Dolan L. M., Kaufman F., Krischer J. P., Schatz D. A., White N. H., Wilson D. M., and Wolfsdorf J. (2001) First-phase insulin release during the intravenous glucose tolerance test as a risk factor for type 1 diabetes. J. Pediatr. 138, 244–249 10.1067/mpd.2001.111274 [DOI] [PubMed] [Google Scholar]
- 7. Fourlanos S., Narendran P., Byrnes G. B., Colman P. G., and Harrison L. C. (2004) Insulin resistance is a risk factor for progression to type 1 diabetes. Diabetologia 47, 1661–1667 10.1007/s00125-004-1507-3 [DOI] [PubMed] [Google Scholar]
- 8. Jelenik T., Séquaris G., Kaul K., Ouwens D. M., Phielix E., Kotzka J., Knebel B., Weiss J., Reinbeck A. L., Janke L., Nowotny P., Partke H. J., Zhang D., Shulman G. I., Szendroedi J., and Roden M. (2014) Tissue-specific differences in the development of insulin resistance in a mouse model for type 1 diabetes. Diabetes 63, 3856–3867 10.2337/db13-1794 [DOI] [PubMed] [Google Scholar]
- 9. Kacerovsky M., Brehm A., Chmelik M., Schmid A. I., Szendroedi J., Kacerovsky-Bielesz G., Nowotny P., Lettner A., Wolzt M., Jones J. G., and Roden M. (2011) Impaired insulin stimulation of muscular ATP production in patients with type 1 diabetes. J. Intern. Med. 269, 189–199 10.1111/j.1365-2796.2010.02298.x [DOI] [PubMed] [Google Scholar]
- 10. Pasare C., and Medzhitov R. (2004) Toll-like receptors: linking innate and adaptive immunity. Microbes Infect. 6, 1382–1387 10.1016/j.micinf.2004.08.018 [DOI] [PubMed] [Google Scholar]
- 11. Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., and Flier J. S. (2006) TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Invest. 116, 3015–3025 10.1172/JCI28898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beutler B. (2002) TLR4 as the mammalian endotoxin sensor. Curr. Top. Microbiol. Immunol. 270, 109–120 10.1007/978-3-642-59430-4_7 [DOI] [PubMed] [Google Scholar]
- 13. Pasare C., and Medzhitov R. (2003) Toll-like receptors: balancing host resistance with immune tolerance. Curr. Opin. Immunol. 15, 677–682 10.1016/j.coi.2003.09.002 [DOI] [PubMed] [Google Scholar]
- 14. Gülden E., Ihira M., Ohashi A., Reinbeck A. L., Freudenberg M. A., Kolb H., and Burkart V. (2013) Toll-like receptor 4 deficiency accelerates the development of insulin-deficient diabetes in nonobese diabetic mice. PLoS ONE 8, e75385 10.1371/journal.pone.0075385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin C., Henao-Mejia J., and Flavell R. A. (2013) Innate immune receptors: key regulators of metabolic disease progression. Cell Metab. 17, 873–882 10.1016/j.cmet.2013.05.011 [DOI] [PubMed] [Google Scholar]
- 16. Dasu M. R., Devaraj S., Park S., and Jialal I. (2010) Increased toll-like receptor (TLR) activation and TLR ligands in recently diagnosed type 2 diabetic subjects. Diabetes Care 33, 861–868 10.2337/dc09-1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dasu M. R., and Jialal I. (2011) Free fatty acids in the presence of high glucose amplify monocyte inflammation via Toll-like receptors. Am. J. Physiol. Endocrinol. Metab. 300, E145–E154 10.1152/ajpendo.00490.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reyna S. M., Ghosh S., Tantiwong P., Meka C. S., Eagan P., Jenkinson C. P., Cersosimo E., Defronzo R. A., Coletta D. K., Sriwijitkamol A., and Musi N. (2008) Elevated toll-like receptor 4 expression and signaling in muscle from insulin-resistant subjects. Diabetes 57, 2595–2602 10.2337/db08-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jialal I., Huet B. A., Kaur H., Chien A., and Devaraj S. (2012) Increased toll-like receptor activity in patients with metabolic syndrome. Diabetes Care 35, 900–904 10.2337/dc11-2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Endesfelder D., zu Castell W., Ardissone A., Davis-Richardson A. G., Achenbach P., Hagen M., Pflueger M., Gano K. A., Fagen J. R., Drew J. C., Brown C. T., Kolaczkowski B., Atkinson M., Schatz D., Bonifacio E., et al. (2014) Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes 63, 2006–2014 10.2337/db13-1676 [DOI] [PubMed] [Google Scholar]
- 21. Brown C. T., Davis-Richardson A. G., Giongo A., Gano K. A., Crabb D. B., Mukherjee N., Casella G., Drew J. C., Ilonen J., Knip M., Hyoty H., Veijola R., Simell T., Simell O., Neu J., et al. (2011) Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 6, e25792 10.1371/journal.pone.0025792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kostic A. D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A. M., Peet A., Tillmann V., Pöhö P., Mattila I., Lähdesmäki H., Franzosa E. A., Vaarala O., de Goffau M., Harmsen H., et al. (2015) The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harbison J. E., Roth-Schulze A. J., Giles L. C., Tran C. D., Ngui K. M., Penno M. A., Thomson R. L., Wentworth J. M., Colman P. G., Craig M. E., Morahan G., Papenfuss A. T., Barry S. C., Harrison L. C., and Couper J. J. (2019) Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: a prospective cohort study. Pediatr. Diabetes 20, 574–583 10.1111/pedi.12865 [DOI] [PubMed] [Google Scholar]
- 24. Canfora E. E., Jocken J. W., and Blaak E. E. (2015) Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 11, 577–591 10.1038/nrendo.2015.128 [DOI] [PubMed] [Google Scholar]
- 25. Koh A., De Vadder F., Kovatcheva-Datchary P., and Bäckhed F. (2016) From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 26. Gancheva S., Jelenik T., Álvarez-Hernandez E., and Roden M. (2018) Interorgan metabolic crosstalk in human insulin resistance. Physiol. Rev. 98, 1371–1415 10.1152/physrev.00015.2017 [DOI] [PubMed] [Google Scholar]
- 27. Vatanen T., Franzosa E. A., Schwager R., Tripathi S., Arthur T. D., Vehik K., Lernmark Å., Hagopian W. A., Rewers M. J., She J. X., Toppari J., Ziegler A. G., Akolkar B., Krischer J. P., Stewart C. J., et al. (2018) The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 10.1038/s41586-018-0620-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chaparro R. J., and Dilorenzo T. P. (2010) An update on the use of NOD mice to study autoimmune (type 1) diabetes. Expert. Rev. Clin. Immunol. 6, 939–955 10.1586/eci.10.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pal D., Dasgupta S., Kundu R., Maitra S., Das G., Mukhopadhyay S., Ray S., Majumdar S. S., and Bhattacharya S. (2012) Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 18, 1279–1285 10.1038/nm.2851 [DOI] [PubMed] [Google Scholar]
- 30. Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., Herder C., Carstensen M., Krausch M., Knoefel W. T., Schlensak M., and Roden M. (2015) Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 21, 739–746 10.1016/j.cmet.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 31. Yin Q., Jiang D., Li L., Yang Y., Wu P., Luo Y., Yang R., and Li D. (2017) LPS promotes vascular smooth muscle cells proliferation through the TLR4/Rac1/Akt signalling pathway. Cell. Physiol. Biochem. 44, 2189–2200 10.1159/000486024 [DOI] [PubMed] [Google Scholar]
- 32. Rumio C., Besusso D., Arnaboldi F., Palazzo M., Selleri S., Gariboldi S., Akira S., Uematsu S., Bignami P., Ceriani V., Ménard S., and Balsari A. (2006) Activation of smooth muscle and myenteric plexus cells of jejunum via Toll-like receptor 4. J. Cell. Physiol. 208, 47–54 10.1002/jcp.20632 [DOI] [PubMed] [Google Scholar]
- 33. Uranga J. A., Garcia-Martinez J. M., Garcia-Jimenez C., Vera G., Martin-Fontelles M. I., and Abalo R. (2017) Alterations in the small intestinal wall and motor function after repeated cisplatin in rat. Neurogastroenterol. Motil. 29, 10.1111/nmo.13047 [DOI] [PubMed] [Google Scholar]
- 34. Forcén R., Latorre E., Pardo J., Alcalde A. I., Murillo M. D., and Grasa L. (2015) Toll-like receptors 2 and 4 modulate the contractile response induced by serotonin in mouse ileum: analysis of the serotonin receptors involved. Neurogastroenterol. Motil. 27, 1258–1266 10.1111/nmo.12619 [DOI] [PubMed] [Google Scholar]
- 35. Anitha M., Vijay-Kumar M., Sitaraman S. V., Gewirtz A. T., and Srinivasan S. (2012) Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143, 1006–1016.e4 10.1053/j.gastro.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caputi V., Marsilio I., Cerantola S., Roozfarakh M., Lante I., Galuppini F., Rugge M., Napoli E., Giulivi C., Orso G., and Giron M. C. (2017) Toll-like receptor 4 modulates small intestine neuromuscular function through nitrergic and purinergic pathways. Front. Pharmacol. 8, 350 10.3389/fphar.2017.00350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasubuchi M., Hasegawa S., Hiramatsu T., Ichimura A., and Kimura I. (2015) Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 7, 2839–2849 10.3390/nu7042839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y., Jin Y., Li J., Zhao L., Li Z., Xu J., Zhao F., Feng J., Chen H., Fang C., Shilpakar R., and Wei Y. (2018) Small bowel transit and altered gut microbiota in patients with liver cirrhosis. Front. Physiol. 9, 470 10.3389/fphys.2018.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A., Gordon J. I., and Chervonsky A. V. (2008) Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greiner T. U., Hyötyläinen T., Knip M., Bäckhed F., and Orěšic M. (2014) The gut microbiota modulates glycaemic control and serum metabolite profiles in non-obese diabetic mice. PLoS ONE 9, e110359 10.1371/journal.pone.0110359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodríguez D., Morrison C. J., and Overall C. M. (2010) Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim. Biophys. Acta 1803, 39–54 10.1016/j.bbamcr.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 42. Bister V., Kolho K. L., Karikoski R., Westerholm-Ormio M., Savilahti E., and Saarialho-Kere U. (2005) Metalloelastase (MMP-12) is upregulated in the gut of pediatric patients with potential celiac disease and in type 1 diabetes. Scand. J. Gastroenterol. 40, 1413–1422 10.1080/00365520510023918 [DOI] [PubMed] [Google Scholar]
- 43. Waldner H., Collins M., and Kuchroo V. K. (2004) Activation of antigen-presenting cells by microbial products breaks self- tolerance and induces autoimmune disease. J. Clin. Invest. 113, 990–997 10.1172/JCI19388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ploger S., Stumpff F., Penner G. B., Schulzke J. D., Gäbel G., Martens H., Shen Z., Günzel D., and Aschenbach J. R. (2012) Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann. N.Y. Acad. Sci. 1258, 52–59 10.1111/j.1749-6632.2012.06553.x [DOI] [PubMed] [Google Scholar]
- 45. Müller M., Hernández M. A. G., Goossens G. H., Reijnders D., Holst J. J., Jocken J. W. E., van Eijk H., Canfora E. E., and Blaak E. E. (2019) Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci. Rep. 9, 12515 10.1038/s41598-019-48775-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le Poul E., Loison C., Struyf S., Springael J. Y., Lannoy V., Decobecq M. E., Brezillon S., Dupriez V., Vassart G., Van Damme J., Parmentier M., and Detheux M. (2003) Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 278, 25481–25489 10.1074/jbc.M301403200 [DOI] [PubMed] [Google Scholar]
- 47. Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T., Terasawa K., Kashihara D., Hirano K., Tani T., Takahashi T., Miyauchi S., Shioi G., Inoue H., and Tsujimoto G. (2013) The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 4, 1829 10.1038/ncomms2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szendroedi J., Yoshimura T., Phielix E., Koliaki C., Marcucci M., Zhang D., Jelenik T., Müller J., Herder C., Nowotny P., Shulman G. I., and Roden M. (2014) Role of diacylglycerol activation of PKCtheta in lipid-induced muscle insulin resistance in humans. Proc. Natl. Acad. Sci. U.S.A. 111, 9597–9602 10.1073/pnas.1409229111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gancheva S., Bierwagen A., Kaul K., Herder C., Nowotny P., Kahl S., Giani G., Klueppelholz B., Knebel B., Begovatz P., Strassburger K., Al-Hasani H., Lundbom J., Szendroedi J., Roden M., and German Diabetes Study (GDS) Group. (2016) Variants in genes controlling oxidative metabolism contribute to lower hepatic ATP independent of liver fat content in type 1 diabetes. Diabetes 65, 1849–1857 10.2337/db16-0162 [DOI] [PubMed] [Google Scholar]
- 50. Heinrichsdorff J., and Olefsky J. M. (2012) Fetuin-A: the missing link in lipid-induced inflammation. Nat. Med. 18, 1182–1183 10.1038/nm.2869 [DOI] [PubMed] [Google Scholar]
- 51. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., and Beutler B. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- 52. Schallschmidt T., Lebek S., Altenhofen D., Damen M., Schulte Y., Knebel B., Herwig R., Rasche A., Stermann T., Kamitz A., Hallahan N., Jähnert M., Vogel H., Schürmann A., Chadt A., and Al-Hasani H. (2018) Two novel candidate genes for insulin secretion identified by comparative genomics of multiple backcross mouse populations. Genetics 210, 1527–1542 10.1534/genetics.118.301578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Burghoff S., Flögel U., Bongardt S., Burkart V., Sell H., Tucci S., Ikels K., Eberhard D., Kern M., Klöting N., Eckel J., and Schrader J. (2013) Deletion of CD73 promotes dyslipidemia and intramyocellular lipid accumulation in muscle of mice. Arch. Physiol. Biochem. 119, 39–51 10.3109/13813455.2012.755547 [DOI] [PubMed] [Google Scholar]
- 54. Burkart V., Wang Z. Q., Radons J., Heller B., Herceg Z., Stingl L., Wagner E. F., and Kolb H. (1999) Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nat. Med. 5, 314–319 10.1038/6535 [DOI] [PubMed] [Google Scholar]
- 55. Toth M., and Fridman R. (2001) Assessment of gelatinases (MMP-2 and MMP-9 by gelatin zymography. Methods Mol. Med. 57, 163–174 10.1385/1-59259-136-1:163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moreau N. M., Goupry S. M., Antignac J. P., Monteau F. J., Le Bizec B. J., Champ M. M., Martin L. J., and Dumon H. J. (2003) Simultaneous measurement of plasma concentrations and 13C-enrichment of short-chain fatty acids, lactic acid and ketone bodies by gas chromatography coupled to mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 784, 395–403 10.1016/S1570-0232(02)00827-9 [DOI] [PubMed] [Google Scholar]
- 57. Weickert M. O., Arafat A. M., Blaut M., Alpert C., Becker N., Leupelt V., Rudovich N., Mohlig M., and Pfeiffer A. F. (2011) Changes in dominant groups of the gut microbiota do not explain cereal-fiber induced improvement of whole-body insulin sensitivity. Nutr. Metab. 8, 90 10.1186/1743-7075-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. García-Villalba R., Giménez-Bastida J. A., García-Conesa M. T., Tomás-Barberán F. A., Carlos Espín J., and Larrosa M. (2012) Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J. Sep. Sci. 35, 1906–1913 10.1002/jssc.201101121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 Auf'm Hennekamp 65, D-40225 Düsseldorf, Germany. Tel.:
49-211-3382-201; Fax:
49-211-3382-691; E-mail:
Auf'm Hennekamp 65, D-40225 Düsseldorf, Germany. Tel.:
49-211-3382-201; Fax:
49-211-3382-691; E-mail: