Abstract
DNA clamp, a highly conserved ring-shaped protein, binds dsDNA within its central pore. Also, DNA clamp interacts with various nuclear proteins on its front, thereby stimulating their enzymatic activities and biological functions. It has been assumed that the DNA clamp is a functionally single-faced ring from bacteria to humans. Here, we report the crystal structure of the heterotrimeric RAD9-RAD1-HUS1 (9-1-1) checkpoint clamp bound to a peptide of RHINO, a recently identified cancer-related protein that interacts with 9-1-1 and promotes activation of the DNA damage checkpoint. This is the first structure of 9-1-1 bound to its partner. The structure reveals that RHINO is unexpectedly bound to the edge and around the back of the 9-1-1 ring through specific interactions with the RAD1 subunit of 9-1-1. Our finding indicates that 9-1-1 is a functionally double-faced DNA clamp.
Keywords: DNA damage response, structural biology, proliferating cell nuclear antigen (PCNA), checkpoint control, protein structure, protein complex, 9-1-1, DNA clamp, crystal structure, DNA damage checkpoint, protein-protein interaction
Introduction
DNA clamp proteins are widely conserved from bacteria to humans and play a crucial role in DNA metabolism, such as DNA replication and repair (1). Each DNA clamp is a ring-shaped protein that encircles dsDNA within its central pore, enabling it to slide along the dsDNA. In addition, the DNA clamp tethers various proteins involved in DNA metabolism and stimulates their enzymatic activities or biological functions. Bacteria has a single DNA clamp (β-clamp) comprising a homodimeric ring (2), whereas eukaryotes have two kinds of DNA clamp. The first type is proliferating cell nuclear antigen (PCNA),2 a homotrimeric protein (3). Most proteins that associate with PCNA have a PIP-box motif (4, 5); this motif is present in hundreds of proteins, and its binding to PCNA is well-elucidated (6, 7). The PIP-box is bound to the hydrophobic pocket in PCNA, underneath the so-called “interdomain connecting” loop (IDC-loop) that bridges two domains of a single PCNA subunit. The other type of DNA clamp is the heterotrimeric RAD9-RAD1-HUS1 complex (9-1-1). Each subunit of 9-1-1 comprises of a PCNA-like domain, whereas RAD9 additionally has an unstructured C-terminal tail. 9-1-1 functions as a DNA damage–specific clamp that activates the ATR-CHK1 checkpoint pathway (8, 9). Similar to PCNA, the core structure of 9-1-1 lacking the C-terminal tail of RAD9 is a ring-shaped trimer (10–12). Binding of proteins to 9-1-1 is thought to be analogous to binding to PCNA (10, 12); as yet, however, no structure of 9-1-1 bound to its partners has been determined, and thus the underlying mechanism remains unclear.
Owing to their ring-shaped structures, DNA clamp proteins inherently have two faces, a front and a back. To date, all of the partners of β-clamp and PCNA have been found to bind to the same face of the DNA clamp, the so-called C-side; the C termini of PCNA subunits protrude from this side (6). In this study, the C-side is defined as the “front.” Regardless of species, therefore, it has been assumed that clamp-binding proteins bind to the front of the DNA clamp, and thus the DNA clamp is a functionally single-faced ring from bacteria to humans. Indeed, previous reports have suggested that 9-1-1 also utilizes its front to interact with its partners (10, 12, 13). Here, we report the structure of 9-1-1 bound to a peptide derived from RHINO (RAD9, HUS1, RAD1-interacting nuclear orphan protein 1, RHNO1), a recently identified cancer-related protein that interacts with 9-1-1 and promotes activation of CHK1 in response to DNA damage (14–16). This is the first structure of 9-1-1 bound to its partner. Our results provide not only a structural basis for the mechanism of interaction between RHINO and 9-1-1, but also a new finding about molecular interactions on DNA clamp.
Results and discussion
The RHINO peptide is bound to the edge and around the back of 9-1-1
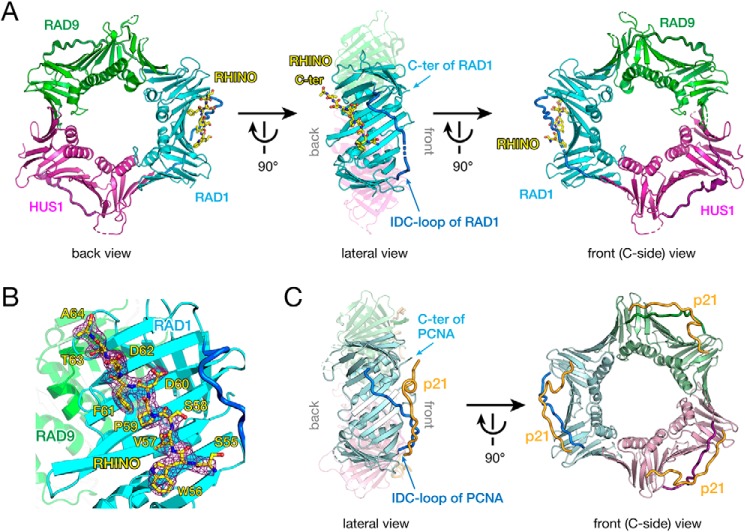
RHINO is known to form a stoichiometric complex with 9-1-1 through interactions with RAD9 lacking the C-terminal tail and RAD1 (16). Residues 55SWVSPDF61 of RHINO are crucial for this interaction with 9-1-1 and activation of the checkpoint (15). Thus, to elucidate details of the interaction between 9-1-1 and its binding proteins, we determined the X-ray crystal structure of the 9-1-1 core ring bound to a RHINO peptide (45SKPIDHSTITSWVSPDFDTA64) at 2.4 Å resolution (Fig. 1A and Table 1). Electron density map shows that residues 55–64 are well-ordered, whereas the N-terminal region (residues 45–54) is disordered (Fig. 1B). In the structure, the RHINO peptide adopts an extended conformation and is bound to the RAD1 subunit. Unexpectedly, the peptide is bound to the edge and around the back of the 9-1-1 ring (Fig. 1A, left). This interaction is distinct from that of known clamp-peptide complexes, such as PCNA in complex with a p21 peptide, where the p21 peptide including the PIP-box is bound to a binding pocket underneath the IDC-loop on the front of PCNA (17) (Fig. 1C).
Figure 1.
A, overall structure of 9-1-1 bound to the RHINO peptide. RAD9, RAD1, and HUS1 subunits are shown as green, cyan, and magenta cartoons, respectively. The IDC-loop of each subunit is shown as a darker-colored thick tube. The RHINO peptide is shown as a yellow stick. Left, back view of 9-1-1 bound to the RHINO peptide. Center, lateral view of 9-1-1 bound to the RHINO peptide. For clarification, RAD9 and HUS1 are shown as semitransparent models. The C terminus and IDC-loop of RAD1 are indicated by arrows. Right, front view (C-side) of 9-1-1 bound to the RHINO peptide. B, the electron density map of the RHINO peptide bound to RAD1. The σ-A weighted 2Fo − Fc map of the peptide contoured at 1σ is shown as a purple cage. RHINO, RAD9, and RAD1 are colored as shown in A. Residues of the RHINO peptide are labeled. C, overall structure of PCNA bound to the p21 peptide (PDB entry 1AXC) as a representative structure of the PCNA-partner complex. Subunits of the homotrimer are shown as light green, pale blue, and pink cartoons. The IDC-loop of each subunit is shown as darker-colored thick tube. The p21 peptide is shown as an orange tube. Left, lateral view of PCNA bound to the p21 peptide. For clarification, far subunits (light green and pink) are shown as transparent models. The C terminus and IDC-loop of PCNA are indicated by arrows. Right, front view (C-side) of PCNA bound to the p21 peptide. The orientation PCNA in each view corresponds to that of 9-1-1 in A.
Table 1.
Data collection and refinement statistics
One crystal was used for a structure. Values in parentheses are for the highest-resolution shell.
| Parameters | 9-1-1–RHINO |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 52.9, 136.1, 154.0 |
| Resolution (Å) | 19.80–2.40 (2.48–2.40) |
| Rmerge | 0.062 (0.733) |
| Rmeas | 0.067 (0.796) |
| I/σI | 21.1 (2.3) |
| CC½ | 99.9 (77.9) |
| Completeness (%) | 99.6 (99.5) |
| No. of reflections | 44,421 (6,970) |
| Redundancy | 6.7 (6.7) |
| Refinement | |
| Resolution (Å) | 19.8–2.40 |
| No. of reflections | 44,415 |
| Rwork | 0.2184 |
| Rfree | 0.2581 |
| No. of atoms | |
| 9-1-1 | 6,120 |
| RHINO peptide | 79 |
| Water | 68 |
| B-factors | |
| 9-1-1 | 63.5 |
| RHINO peptide | 74.0 |
| Water | 53.9 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (degrees) | 0.70 |
| PDB entry | 6J8Y |
The RHINO peptide specifically interacts with the RAD1 subunit
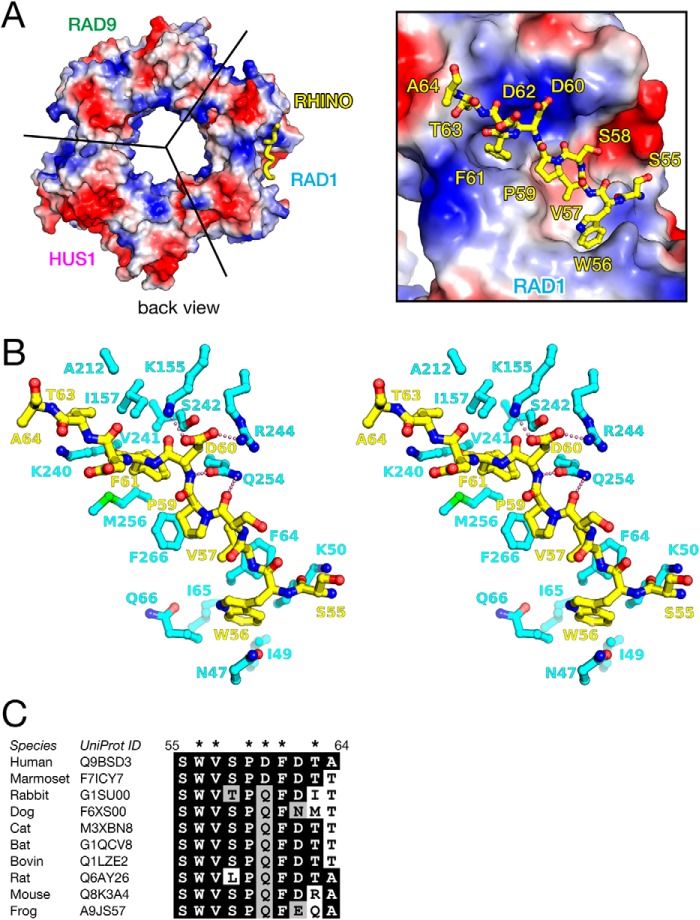
The RHINO peptide is accommodated in the pocket on the edge and around the back of the RAD1 subunit of 9-1-1, and the chemical properties of the bound peptide are complementary to the charge distribution of the binding pocket (Fig. 2A). The hydrophobic side chains of Trp56, Val57, Pro59, and Phe61 of RHINO interact significantly with the pocket of RAD1 through van der Waals (vdW) contacts, as follows (Fig. 2B). Trp56 of RHINO contacts with the side chain of Phe64 of RAD1 by a face-to-edge interaction. Val57, Pro59, and Phe61 of RHINO contact with the hydrophobic side chains of Phe64, Phe266, and Met256 of RAD1, respectively, by CH-π interactions. The methyl group of Thr63 of RHINO is also accommodated into the hydrophobic pocket. In addition to these vdW interactions, the acidic side chain of Asp60 of RHINO forms ion pairs with the basic side chains of Lys155 and Arg244 of RAD1 (Fig. 2B), which contribute to the basic patch on the surface (Fig. 2A). Furthermore, hydrogen bonds between the peptide bond of Pro59-Asp60 of RHINO and the side chain of Gln254 of RAD1 are also observed. The bidentate hydrogen bond by Gln254 retains the conformation of the peptide bond, thereby probably stabilizing the conformation for binding. Furthermore, the residues of RHINO involved in interaction with RAD1 are highly conserved (Fig. 2C).
Figure 2.
A, binding of the RHINO peptide on the surface of RAD1. Left, the overall structure of 9-1-1 is shown as surface model with electrostatic potential. The RHINO peptide bound to RAD1 is shown as a yellow thick tube. The orientation of 9-1-1 is identical to that of the left panel of Fig. 1A. Right, close-up view of binding of the RHINO peptide on the surface of RAD1. The RHINO peptide is shown as a yellow stick model. B, stereo view of detailed interactions between the RHINO peptide and RAD1. The RHINO peptide and RAD1 are shown in yellow and cyan stick models, respectively. Electrostatic interactions between RHINO and RAD1 are shown by pink dots. C, sequence alignment of the RAD1-binding region of RHINO. Identical and homologous residues are highlighted by black and gray backgrounds, respectively. Side chains involved in remarkable interactions with RAD1 are indicated by asterisks.
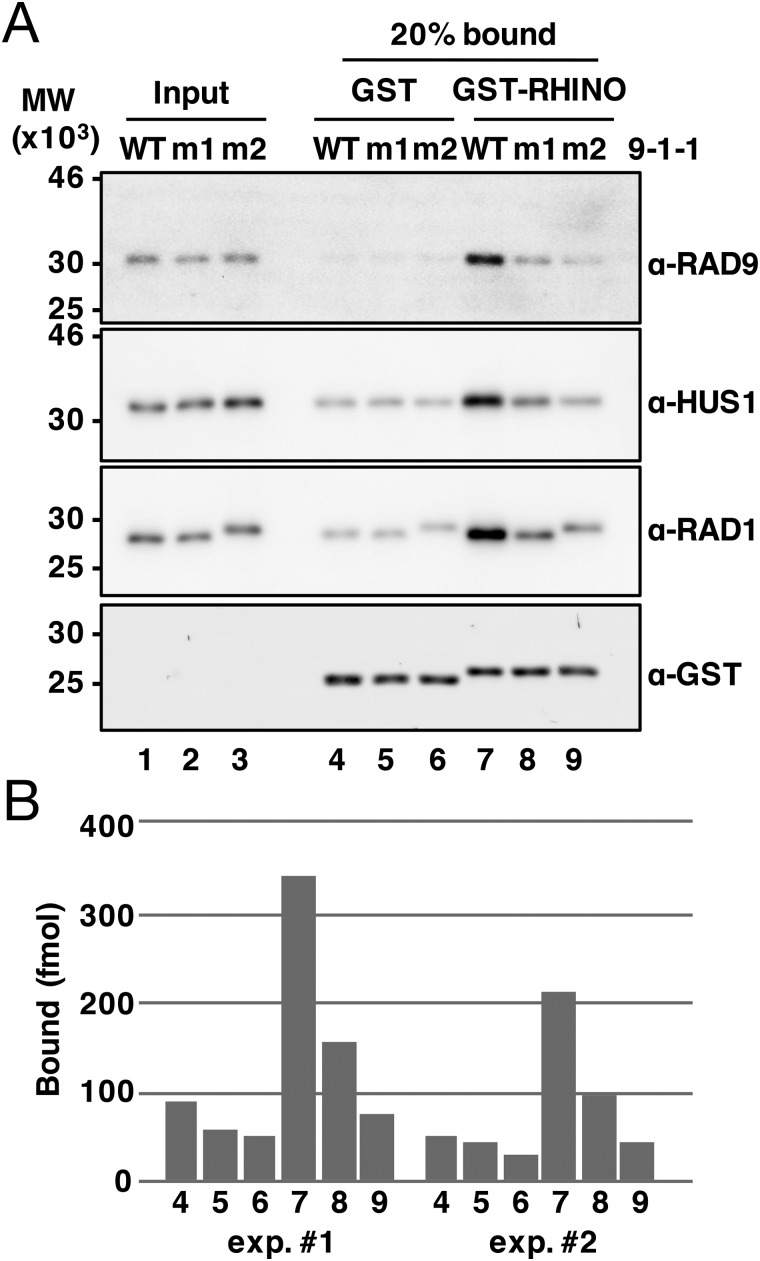
For functional validation of the present structure, we performed interaction analysis using a GST pulldown assay with triple mutations (m1 (K155A/R244A/Q254A) or m2 (F64A/M256A/F266A)) in the RAD1 subunit (Fig. 3). First, we confirmed binding of RHINO(45–64) to 9-1-1 (Fig. 3, lane 7). The triple mutations in Lys155, Arg244, and Gln254, which are engaged in electrostatic interactions with the RHINO peptide (Fig. 2B), reduced the binding (Fig. 3, lane 8). Furthermore, the triple mutations in Phe64, Met256, and Phe266, which are involved in vdW contacts with the RHINO peptide (Fig. 2B), largely reduced the binding (Fig. 3, lane 9). These results are in good agreement with the crystal structure, and thus we conclude that the RHINO peptide is specifically bound to the pocket of RAD1. In addition, the pulldown assay might indicate that vdW contacts are more crucial for the binding.
Figure 3.
A, GST pulldown assay. 50 pmol of GST-RHINO(45–64) (lanes 7–9) or GST alone (lanes 4–6) was immobilized on GSH-Sepharose 4B beads and incubated with 100 pmol of purified WT 9ΔC-1-1 (WT) or its derivatives, m1 (K155A/R244A/Q254A in RAD1) and m2 (F64A/M256A/F266A in RAD1). Input 9-1-1 (20 fmol, lanes 1–3) and bound proteins (lanes 1–9) were analyzed by immunoblotting. B, the intensity of the HUS1 band in each lane was quantified, and the amount of bound HUS1 was plotted. The pulldown assay was performed twice, and results of quantification were shown (exp.#1 and exp.#2).
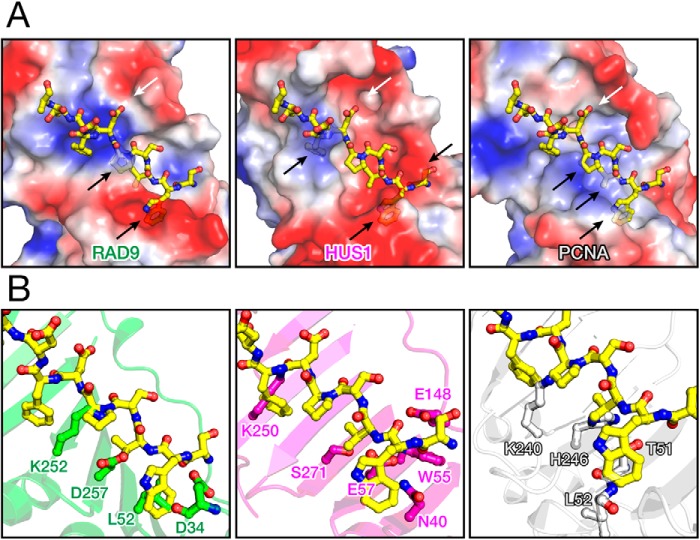
In contrast to PCNA, 9-1-1 is a heterotrimer, and thus the molecular surface of each subunit possesses different physicochemical properties, whereas RAD9, HUS1, and PCNA also possess large concaves that seem to be available for a binding site on their backs and edges. Thus, by structural superimposition of RAD9, HUS1, and PCNA with RAD1 bound to the RHINO peptide, we built hypothetical structures of the RHINO peptide bound to the backs and edges of RAD9, HUS1, and PCNA (Fig. 4). These models suggest that the RHINO peptide is unlikely to be bound to these surfaces because of steric and electrostatic conflicts. In fact, it has been reported that HUS1 and PCNA show no binding to RHINO (16). Furthermore, the mutation of 55SWVSPDF61 to 55AAAAAA61 in RHINO abolishes its interaction with 9-1-1 (15), and RHINO forms a stoichiometric complex with 9-1-1 (16). These results strongly suggest that the interactions between RHINO and RAD1 observed in this study are dominant in formation of the 9-1-1–RHINO complex, although RHINO might possess a minor binding region to interact with RAD9. Our present finding is consistent with a functional study of RHINO mutants (15), thereby indicating that the interactions of RHINO with the back and edge of RAD1 observed here is essential for activation of the ATR-CHK1 checkpoint pathway by RHINO.
Figure 4.
A, hypothetical models of RHINO peptide bound to RAD9 (left), HUS1 (center), and PCNA (right). Electrostatic potentials are mapped on the molecular surfaces of RAD9, HUS1, and PCNA. Each molecular surface is shown as a semitransparent model to show steric conflicts. The RHINO peptide is also shown as the same model as in Fig. 2A. Steric and electrostatic conflicts are shown by black and white arrows, respectively, in each panel. B, steric clashes in the hypothetical models. Typical residues of RAD9 (green), HUS1 (magenta), and PCNA (gray) implying steric clashes with RHINO (yellow) are shown by stick models.
9-1-1 is a functionally double-faced DNA clamp
DNA clamps are loaded on dsDNA by specific clamp loaders, which are pentameric protein complexes classified into the AAA+ ATPase family (18). Eukaryotes have four clamp loaders: RFC, CTF18-RFC, ELG1-RFC, and RAD17-RFC (19). RFC, the primary clamp loader for PCNA, consists of one large and four small subunits: RFC1 and RFC2–5, respectively. The others consist of the RFC1 paralog as a large subunit and CTF18, ELG1, and RAD17, associated with universal small subunits, RFC2–5. The crystal structure of yeast homolog of the RFC-PCNA complex has revealed that RFC also binds to the front of PCNA for ring opening and loading of PCNA on dsDNA (20). In contrast to PCNA, 9-1-1 is loaded on dsDNA by RAD17-RFC (21). Based on sequence similarity and crystal structures, plausible models of interaction between RFC17-RFC and 9-1-1 have been proposed (10, 12, 13). These models suggest that RAD17-RFC also interacts with the front of 9-1-1. A preliminary result also implies that FEN1 nuclease might be bound to the front of RAD1 (12). Here, our structure reveals that the RHINO specifically interacts with the edge and back of 9-1-1 for activation of the checkpoint. Therefore, 9-1-1 could be a functionally double-faced DNA clamp and thereby might enable multiple protein-protein interactions in signaling of the DNA damage checkpoint. Although RHINO is unlikely to interact with the edge and back of PCNA (Fig. 4), we cannot exclude the possibility that PCNA utilizes its edge or back for protein-protein interactions. Further studies are needed to discuss that possibility.
Conclusion
In this study, we have determined the crystal structure of 9-1-1 in complex with the RHINO peptide. Most surprisingly, the structure reveals that 9-1-1 utilizes its edge and back to interact with RHINO. Our unexpected finding first sheds light on the other side of 9-1-1, thereby indicating that 9-1-1 is a functionally double-faced DNA clamp and fundamentally changing our view of DNA clamps. The structure also reveals detailed interactions between 9-1-1 and RHINO and thus could provide a new clue for developing chemical compounds or artificial peptides to down-regulate the ATR-CHK1 checkpoint pathway for cancer therapy.
Experimental procedures
Protein production
The gene encoding human truncated RAD9 (residues 1–270), RAD9ΔC, was inserted into the pCOLA vector. The whole genes encoding HUS1 (residues 1–280) and RAD1 (residues 1–282) were inserted into the pETDuet vector, which incorporated a His tag at the N terminus of HUS1. Escherichia coli BL21(DE3) harboring the two vectors was cultured in lysogeny broth medium at 37 °C to an optical density of about 0.8 at 660 nm, 0.2 mm isopropyl 1-thio-β-d-galactopyranoside was added, and the cells were then cultured at 25 °C overnight. The cells were harvested by centrifugation, washed with buffer I (20 mm HEPES-NaOH, pH 7.4, and 500 mm NaCl), and resuspended in buffer II (50 mm HEPES-NaOH, pH 6.8, and 250 mm NaCl). After sonication, the cell lysate was clarified by centrifugation for 1 h at 4 °C (48,300 × g). Subsequent purification was carried out at 4 °C. The supernatant was applied to a HiTrap Heparin column (GE Healthcare), and bound protein was eluted with a linear gradient of 250 mm to 1 m NaCl. The eluent was diluted with buffer III (50 mm Tris-HCl, pH 8.5, and 200 mm NaCl) and applied to an anion-exchange column, HiTrap Q column (GE Healthcare). The bound protein was eluted with a linear gradient of 200 mm to 1 m NaCl. The eluted protein was passed through a size-exclusion column, HiLoad 16/600 Superdex200 (GE Healthcare), equilibrated with buffer IV (20 mm HEPES-NaOH, pH 7.4, and 100 mm NaCl), and then concentrated to 15–20 mg/ml using a Vivaspin concentrator (Sartorius). The purity of the RAD9ΔC-RAD1-HUS1 complex (9ΔC-1-1) was confirmed by SDS-PAGE with Coomassie Brilliant Blue staining. In the present study, RAD9ΔC and 9ΔC-1-1 are simply termed RAD9 and 9-1-1, respectively, unless otherwise noted. For interaction analysis, site-directed mutations in RAD1 (m1 (K155A/R244A/Q254A) or m2 (F64A/M256A/F266A)) were introduced by a PCR-based mutagenesis. Recombinant 9ΔC-1-1 with RAD1 mutations was produced and purified by a procedure similar to that of the WT. All purified proteins were frozen in liquid N2 and stored at −80 °C until use.
Crystallization and structure determination
The RHINO peptide (45SKPIDHSTITSWVSPDFDTA64) was commercially synthesized (TORAY Research Center, Inc.) and dissolved in buffer IV. A 10-fold molar excess of peptide was incubated with 0.13 mm 9-1-1 and 8.8 mm DTT. Crystallization was performed by the vapor-diffusion method. Rod crystals were obtained with a reservoir solution of 0.1 m Bistris propane, pH 7.5, 0.1 m sodium citrate, and 16% PEG 3350 at 20 °C. Crystals were transferred to a buffer containing the reservoir solution and 25% sucrose. X-ray diffraction data were collected at Photon Factory beamline BL-17A (Tsukuba, Japan) with an Eiger X16M single-photon-counting detector (DECTRIS). Diffraction data were processed with the program XDS (22). The crystal structure of 9-1-1 bound to the RHINO peptide was determined by molecular replacement method using the program PHASER (23). Binding of the RHINO peptide to the RAD1 subunit was confirmed by difference Fourier map, and the structure of the peptide was built by the program Coot (24). The structure was manually improved and refined by the program PHENIX (25). The final electron density of the RHINO peptide is shown in Fig. 1B. Data collection and refinement statistics are given in Table 1. Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (entry 6J8Y).
GST pulldown assay
GST or GST-fused RHINO (residues 45–64) termed GST-RHINO(45–64) was expressed by E. coli BL21(DE3) by conventional induction with isopropyl 1-thio-β-d-galactopyranoside. The cell lysates expressing GST or GST-RHINO(45–64) were incubated with GSH-Sepharose 4B (GE Healthcare) beads for 2 h at 4 °C. The beads were washed four times with buffer H (25 mm HEPES-NaOH (pH 7.8), 150 mm NaCl, 1 mm EDTA, 10% glycerol, 0.01% Nonidet P-40, 0.1 mm phenylmethylsulfonyl fluoride, and 2 μg/ml leupeptin). The beads immobilized with ∼50 pmol of GST or GST-RHINO(45–64) were incubated with 100 pmol of 9ΔC-1-1 (WT) or its mutants (m1 and m2) for 2 h at 4 °C in 25 μl of buffer H, followed by five washes with the same buffer. The bound proteins were eluted by boiling in SDS sample buffer (50 mm Tris-HCl (pH 6.8), 0.1 m DTT, 2% SDS, 0.05% bromphenol blue, and 10% glycerol), separated by 12.5% SDS-PAGE, and analyzed by immunoblotting using the indicated antibodies. The band intensities were quantified by ImageJ software (National Institutes of Health).
Antibodies
The rabbit anti-HUS1 and anti-RAD1 antibodies were gifts from Dr. Katsunori Sugimoto (State University of New Jersey). The rabbit anti-RAD9, mouse anti-GST, horseradish peroxidase–conjugated anti-rabbit IgG, and Alexa 647–conjugated anti-mouse IgG antibodies were purchased from LifeSpan Biosciences, Santa Cruz Biotechnology, Bio-Rad, and Jackson ImmunoResearch Laboratories, respectively.
Figure preparation
Figures of protein structures were prepared with the program PyMOL (Schrödinger, LLC). All figures were modified with the program Illustrator (Adobe Systems).
Author contributions
K. H., N. I., R. T., E. O., H. S., and A. H. investigation; K. H., E. O., and H. H. validation; H. H. conceptualization; K. H. and E. O. data curation; K. H. and E. O. formal analysis; K. H. and H. H. supervision; K. H. and H. H. funding acquisition; E. O. and H. H. visualization; E. O. and H. H. writing-original draft; H. H. project administration; K. H., E. O., Y. I., A. H., and H. H. writing-review and editing.
Acknowledgments
We thank the beamline staff of Photon Factory for kind support in data collection. Human cDNAs of RAD9, RAD1, and HUS1 were gifts from Dr. Toshiki Tsurimoto (Kyushu University).
This work was supported by Japan Society for the Promotion of Science (JSPS)/MEXT KAKENHI Grants 25291017, 16H04755, and 17H06014 (to H. H.) and Grants 15K18491 and 17K07314 (to K. H.) and by the Collaborated Research Program as the Visiting Fellow of the Institute for Protein Research, Osaka University, VFCR-14–02 (to H. H.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 6J8Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PCNA
- proliferating cell nuclear antigen
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- IDC-loop
- interdomain connecting loop
- vdW
- van der Waals
- PDB
- Protein Data Bank.
References
- 1. Yao N. Y., and O'Donnell M. E. (2016) Evolution of replication machines. Crit. Rev. Biochem. Mol. Biol. 51, 135–149 10.3109/10409238.2015.1125845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kong X. P., Onrust R., O'Donnell M., and Kuriyan J. (1992) Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69, 425–437 10.1016/0092-8674(92)90445-I [DOI] [PubMed] [Google Scholar]
- 3. Krishna T. S., Kong X. P., Gary S., Burgers P. M., and Kuriyan J. (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 10.1016/0092-8674(94)90014-0 [DOI] [PubMed] [Google Scholar]
- 4. Warbrick E. (1998) PCNA binding through a conserved motif. Bioessays 20, 195–199 [DOI] [PubMed] [Google Scholar]
- 5. Warbrick E. (2000) The puzzle of PCNA's many partners. Bioessays 22, 997–1006 [DOI] [PubMed] [Google Scholar]
- 6. Moldovan G. L., Pfander B., and Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 7. Mailand N., Gibbs-Seymour I., and Bekker-Jensen S. (2013) Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14, 269–282 10.1038/nrm3562 [DOI] [PubMed] [Google Scholar]
- 8. Parrilla-Castellar E. R., Arlander S. J., and Karnitz L. (2004) Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Rep. 3, 1009–1014 10.1016/j.dnarep.2004.03.032 [DOI] [PubMed] [Google Scholar]
- 9. Eichinger C. S., and Jentsch S. (2011) 9-1-1: PCNA's specialized cousin. Trends Biochem. Sci. 36, 563–568 10.1016/j.tibs.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Doré A. S., Kilkenny M. L., Rzechorzek N. J., and Pearl L. H. (2009) Crystal structure of the Rad9-Rad1-Hus1 DNA damage checkpoint complex—implications for clamp loading and regulation. Mol. Cell 34, 735–745 10.1016/j.molcel.2009.04.027 [DOI] [PubMed] [Google Scholar]
- 11. Sohn S. Y., and Cho Y. (2009) Crystal structure of the human rad9-hus1-rad1 clamp. J. Mol. Biol. 390, 490–502 10.1016/j.jmb.2009.05.028 [DOI] [PubMed] [Google Scholar]
- 12. Xu M., Bai L., Gong Y., Xie W., Hang H., and Jiang T. (2009) Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J. Biol. Chem. 284, 20457–20461 10.1074/jbc.C109.022384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Venclovas C., Colvin M. E., and Thelen M. P. (2002) Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 11, 2403–2416 10.1110/ps.0214302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J. W., Fukukawa C., Ueda K., Nishidate T., Katagiri T., and Nakamura Y. (2010) Involvement of C12orf32 overexpression in breast carcinogenesis. Int. J. Oncol. 37, 861–867 10.3892/ijo_00000737 [DOI] [PubMed] [Google Scholar]
- 15. Cotta-Ramusino C., McDonald E. R 3rd, Hurov K., Sowa M. E., Harper J. W., and Elledge S. J. (2011) A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 332, 1313–1317 10.1126/science.1203430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindsey-Boltz L. A., Kemp M. G., Capp C., and Sancar A. (2015) RHINO forms a stoichiometric complex with the 9-1-1 checkpoint clamp and mediates ATR-Chk1 signaling. Cell Cycle 14, 99–108 10.4161/15384101.2014.967076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., and Kuriyan J. (1996) Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87, 297–306 10.1016/S0092-8674(00)81347-1 [DOI] [PubMed] [Google Scholar]
- 18. Kelch B. A. (2016) The lord of the rings: structure and mechanism of the sliding clamp loader. Biopolymers 105, 532–546 10.1002/bip.22827 [DOI] [PubMed] [Google Scholar]
- 19. Ohashi E., and Tsurimoto T. (2017) Functions of multiple clamp and clamp-loader complexes in eukaryotic DNA replication. Adv. Exp. Med. Biol. 1042, 135–162 10.1007/978-981-10-6955-0_7 [DOI] [PubMed] [Google Scholar]
- 20. Bowman G. D., O'Donnell M., and Kuriyan J. (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 10.1038/nature02585 [DOI] [PubMed] [Google Scholar]
- 21. Bermudez V. P., Lindsey-Boltz L. A., Cesare A. J., Maniwa Y., Griffith J. D., Hurwitz J., and Sancar A. (2003) Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. U.S.A. 100, 1633–1638 10.1073/pnas.0437927100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., and Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]