Abstract
Introduction: The Mechanism of laser therapy and also its safety are 2 important features of the application of different types of lasers in medicine. This study aims to investigate the critically affected genes after the treatment of squamous cell carcinoma patients.
Methods: The gene expression profiles of 4 squamous cell carcinoma patients that were treated via chemoradiotherapy (CRT) plus the laser and 3 similar patients without laser exposure from Gene Expression Omnibus (GEO) were downloaded and were screened to find critical genes via network analysis. The STRING database, Cytoscape software, and the Clue GO plug-in of Cytoscape software were used.
Results: The genes HSX70 and NCC27 were determined as neighbors and HSPA1B, CLIC1, RAB13, PPIF, and LCE3D as hub genes. The over-expression of LCE3D was interpreted as the side effect of laser therapy. Apoptosis and the cell cycle were the dominant biological processes regulated by the HSP molecules in the laser-treated patients.
Conclusion: The laser affected the main biological processes and simultaneously issued side effects
Keywords: Laser therapy, Squamous cell carcinoma HSPA1B, LCE3D
Introduction
The application of high-dose lasers and light devices in oncology, dermatology, and other fields of medicine is a basic therapeutic procedure. Different types of activities like surgery and skin rejuvenation have been established by using laser therapy. Photobiomodulation therapy is the term that is tied to the clinical application of low-dose biophotonic which can lead to lessening pain and inflammation and also wound healing.1 There are a large number of documents about the clinical application of low-level laser therapy and photobiomodulation therapy to various tissues and organs, indicating the growing feature of low-level laser therapy in medicine.2-4 As laser therapy application in medicine is rising, its safety and effectiveness have also been investigated by researchers to approve the related devices and methods.5,6 In this regard, the understanding of the molecular mechanism of laser therapy is an attractive subject. There are many documents about the molecular base of laser effect in medicine and biology.7-9 An important feature of the molecular mechanism is gene expression regulation in the laser-irradiated samples. In such samples, laser irradiation affects gene expression and therefore, some genes up-regulate and other ones down-regulate.10,11 In the case of laser irradiation, a large number of genes may get dysregulated. A proper method for screening many genes to find the critical one is PPI network analysis.12,13 In network analysis, all related elements such as genes are included in the scale-free connected unit. In this mode of connection, each one of the elements or nodes is linked to the different kinds and numbers of the other nodes and plays a special role in the construction of the network. The topological analysis of the network provides important information: the identification of hub nodes and also bottlenecks.14-16 These critical nodes are considered as key elements, which are involved in the studied phenomenon.17 In the present study, the critical DEGs of squamous cell carcinoma patients after laser therapy are investigated to understand the main affected biochemical pathways.
Methods
The patients who were ≥18 years old and diagnosed with the squamous cell carcinoma were studied and grouped into 4 patients in the laser group (LG) and 3 patients in the placebo group (PG). Two repeats of each sample were included in the analysis. The patients were treated via chemoradiotherapy (CRT) plus a laser and a placebo. A diode laser characterized by Lsae III® indium phosphide, gallium, and aluminum with emission in λ660 nm was applied for photobiomodulation therapy. The collected buccal smears of the patients on day 1 (before CRT) and day 10 of CRT were used for RNA extraction and cDNA microarray analysis. More details of the procedure are available in the published document of Antunes et al.18 Data were downloaded from GEO under the title of GSE94833/GPL6244. Statistical analysis was done via GEO2R and the samples and controls were matched. The top 250 significant DEGs were considered for more analysis. The valid DEGs (P value <0.05 and fold change >1.5) were imported in the “Universal Interaction Database Client” data source and searched via the “Search by ID (gene/protein/compound ID)” search mode by Cytoscape software v3.7.1. The PPI network was constructed by Cytoscape software; the nodes were connected in an undirected model of connections.
The network was analyzed by the Network Analyzer application of Cytoscape software to determine the central nodes. The top nodes based on the degree value were determined as hubs. The action map including activation, inhibition, binding, expression, catalysis, and ptmod was illustrated for hub nodes via the CluePedia plug-in of Cytoscape.
Results
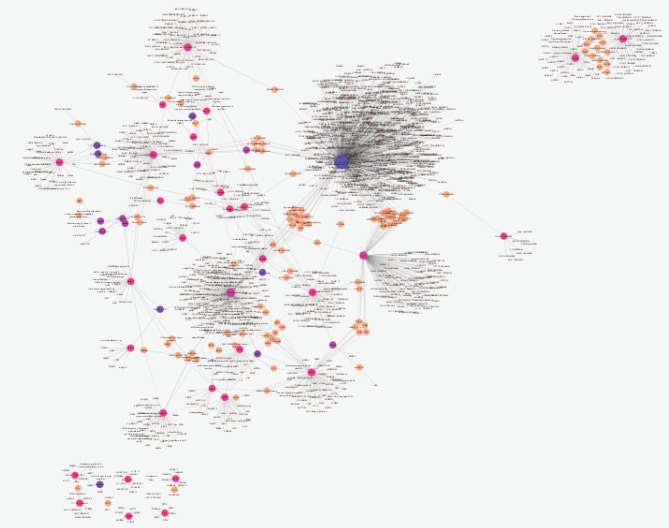
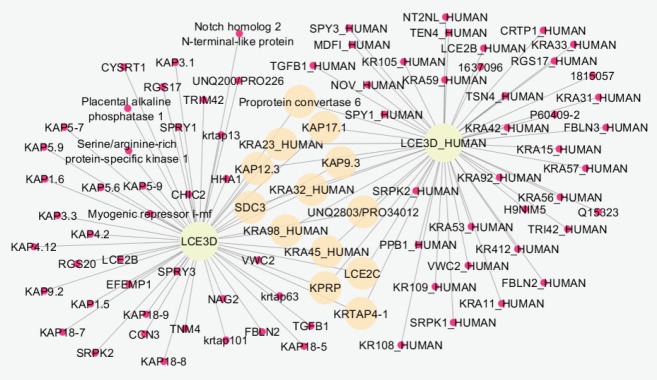
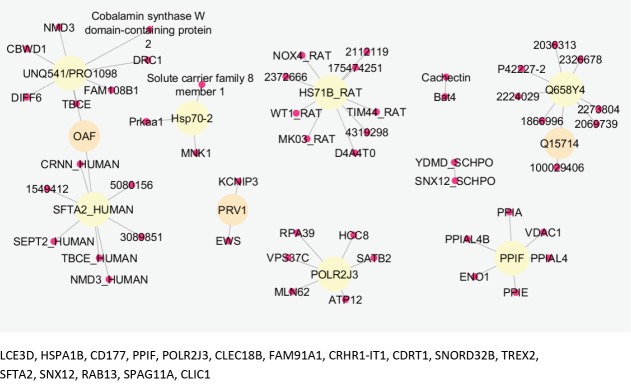
Comparing data requires statistical matching. As it is shown in Figure 1, the distribution of gene expression profiles is median-centered; therefore, the samples are comparable. Among 250 requested differentially expressed genes (DEGs), 111 characterized ones were downloaded and 16 recognized individuals were significant with fold change >1.5 (see Figure 2). The fold change of FAM91A1 and CRHR1-IT1 was almost 1.5. The results from the searched data in the “Universal Interaction Database Client” data source are tabulated in Table 1. As it is shown in this table, 8 databases which contained different types of data were activated. The most found data corresponded to the iRefIndex database. This database provides protein interaction records from several databases, including BIND, BioGrid, DIP, HPRD, IntAct, MINT, MPact, MPPI, and OPHID.19 Since the number of records may be repeated in the databases, the network including 1447 nodes and 1535 edges organized in 11 connected components was created (see Figure 3). Due to a large number of nodes, Figure 3 is not informative; therefore, 2 parts of this figure including the connected components except the main connected component are shown in Figures 4 and 5 with a suitable resolution. LCE3D and PPIF, the two queried DEGs, are highlighted in Figures 4 and 5 respectively.
Figure 1.
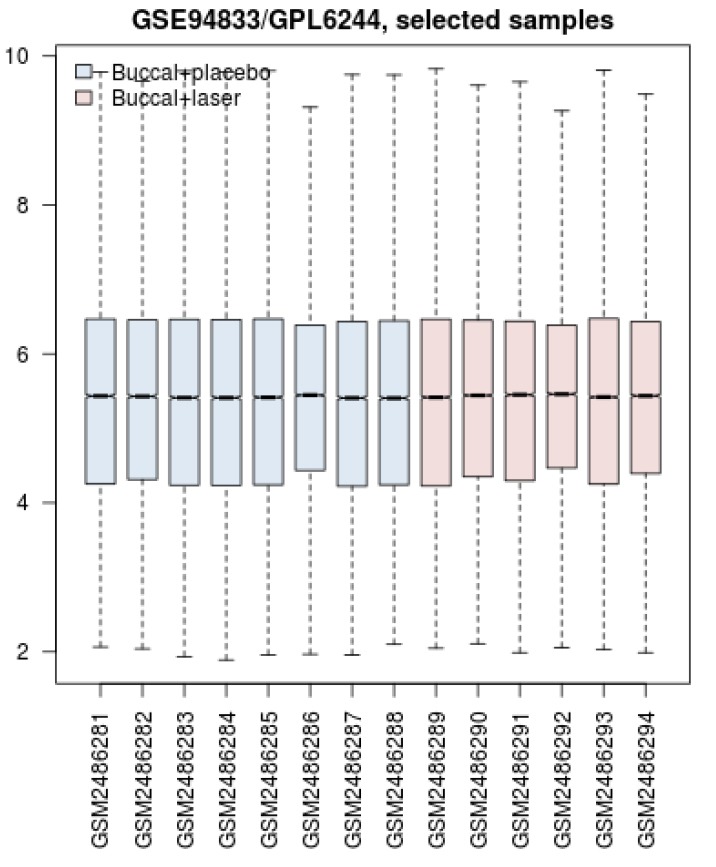
Box Plot Comparison Between the Samples. Blue and red colors refer to the gene expression profiles of the control group (placebo applied) and the laser-treated group respectively. The distribution of gene expression amounts for the 4 quarters of genes is illustrated.
Figure 2.
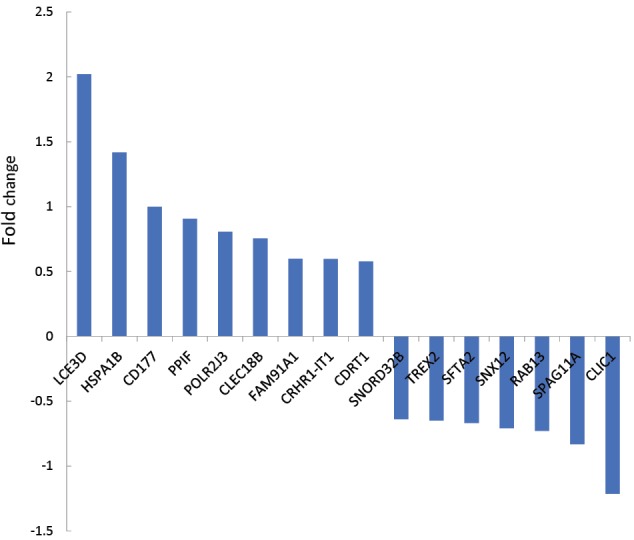
Fold Change of 16 Recognized Significant DEGs Relative to the Laser-Treated Buccal Samples in Comparison With Controls. Negative fold changes refer to down-regulation.
Table 1. Different Types of Activated Databases in Response to the Searched Valid 16 DEGs via the STRING Database .
| Database Name | Records Found |
| iRefIndex | 2499 |
| IMEX | 683 |
| IntAct | 599 |
| MINT | 146 |
| UniProt | 74 |
| Reactome-FIs | 30 |
| bhf-ucl | 4 |
| HPIDb | 3 |
Figure 3.
PPI Network Including 1447 Nodes and 1535 Edges Organized in 11 Connected Components of the Buccal Laser-Treated Samples Relative to the Controls.
Figure 4.
A Connected Component From the Right-up Part of Figure 3.
Figure 5.
The connected component which is presented in the down part of Figure 3.
The list of queried DEGs with a degree value above 3 including HSPA1B, CLIC1, RAB13, PPIF, LCE3D, FAM91A1, TREX2, SNX12, SFTA2, POLR2J3, and CD175 is shown in Table 2. As it is presented in Table 2, the degree values of DEGs were distributed in four categories: 5-9, 20-21, 53-79, and 577. We selected the last two classes as hub-DEGs and the other elements of the network (except the queried ones) that were characterized by a degree value above 53 with a human source were considered as hub nodes. In this regard, HSX70 and NCC27 with degree values of 174 and 61 respectively were identified as hubs. Activation, inhibition, binding, expression, catalysis, and ptmod actions for hubs revealed that HSX70 and NCC27 were not recognized by CluePedia and only there were binding, catalysis, and ptmod actions between HSPA1B and PPIF (see Figure 6). Since HSPA1B was the top hub, it reacted to the action map, and also its expression change was significant; its connections with the other neighbors were searched in the STRING database (see Figure 7).
Table 2. The List of Queried DEGs With a Degree Value >3 .
| Name | Degree |
| HSPA1B | 577 |
| CLIC1 | 97 |
| RAB13 | 63 |
| PPIF | 54 |
| LCE3D | 53 |
| FAM91A1 | 21 |
| TREX2 | 20 |
| SNX12 | 9 |
| SFTA2 | 8 |
| POLR2J3 | 6 |
| CD175 | 5 |
Figure 6.
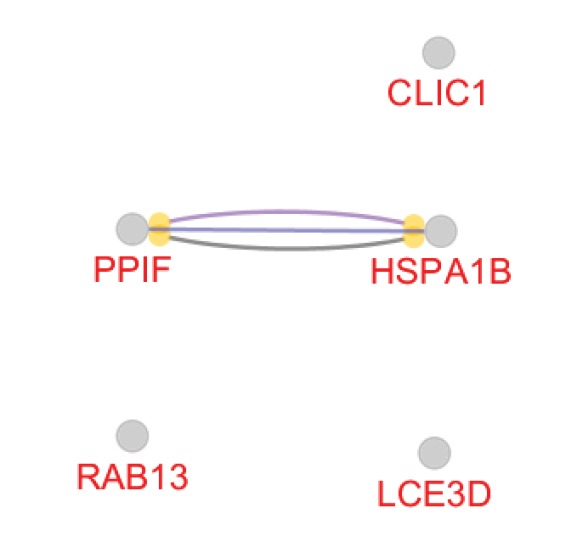
The Action Map of Hub Nodes. There are binding, catalysis, and PTMD connections between PPIF and HSPA1B.
Figure 7.
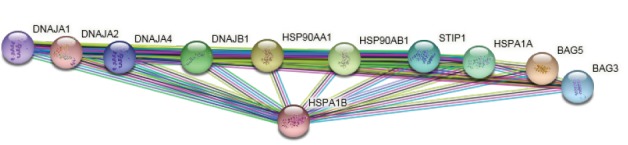
HSPA1B and its first 10 neighbors (https://string-db.org/cgi/network.pl?taskId=6wJNOXvF3W4E).
Discussion
Based on the box plot analysis, the distribution of gene expression profiles of the samples is statistically comparable. In Figure 1, not only bars are median-centered but also distribution profiles follow a similar pattern. Therefore, it is expected that there are a number of DEGs that discriminate the profiles. These significant valid DEGs which are shown in Figure 2 include 16 individuals. LCE3D, HSPA1B, and CLIC1 are the DEGs that are characterized by most expression changes relative to the other DEGs. However, the amount of expression change of the studied genes is an important criterion in proteomics and their interaction properties are the critical feature that should be considered. In the network analysis, the topographic aspects of the constructed network provide informative data to better understand the role of the network elements in the promotion of the studied condition such as disease. The construction of a network including 1447 nodes and 1535 edges indicates a wide range of interactions around the queried DEGs. As it is illustrated in Figures 3-5 and Table 2, the queried DEGs are ranked based on the number of linkages with the other nodes. HSPA1B as the top hub DEG in Table 2 is connected to 577 nodes (about 40% of all nodes of the network). Considering the significant rank of expression change of HSPA1B and the top degree value, it can be implied that this chaperon is introduced as a critical gene in response to laser treatment.
HSPA1B is an intronless gene that encodes a 70 kDa heat shock protein. This protein and several other HSP molecules are known as the heat shock protein family. Like the other HSPs, this heat shock protein is involved in protein stabilization and folding in the cells. The role of HSPA1B in the development of the hepatocellular carcinoma, the urinary tract infection, chronic heart failure, noncognitive symptoms in late-onset Alzheimer’s disease, and several other diseases are investigated and discussed in detail.20-25 Based on a report by Schröder et al, the heat shock protein 70 genotypes HSPA1B and HSPA1L are two important regulators of cytokine concentrations and interfere with consequences after major injury.26
As it is shown in Figure 7, different types of genes are related to HSPA1B. Based on the extracted data from the STRING database, DnaJ homolog subfamily A member 1 plays a role in protein transport into mitochondria via its role as a co-chaperone. This protein as a co-chaperone for HSPA1B in response to cellular stress protects cells against apoptosis. DnaJ homolog subfamily A member 2 as a co-chaperone of Hsc70 stimulates ATP hydrolysis and the folding of unfolded proteins which are mediated by HSPA1A/B. Heat shock protein HSP 90-alpha/beta is involved in cell cycle control and signal transduction. The BAG family molecular chaperone regulator 3, the other co-chaperone for HSP70, has anti-apoptotic activity. It can be concluded HSPA1B is involved in the critical protecting activity in response to stress.
The second top hub that is identified by a degree value of 174 is HSX70, a heat shock protein which is involved in the response by cells to stress, including hyperthermia, hypoxia, and injury.27 The role of this heat shock protein in neurodegenerative diseases is also reported.28 This protein is not the queried one and is highlighted among the neighbor individuals.
The late cornified envelope-3 D gene (LCE3D) that has undergone most expression changes among the queried genes is another hub which is characterized by a degree value of 53. The correlation between psoriasis which is one of the most common inflammatory skin diseases worldwide and LCE3D has been investigated and reported by researchers.29,30 As Titova et al report, intense terahertz (THz) pulse radiation suppresses epidermal differentiation complex (EDC) genes such as the SPRR family and S100 genes that encode calcium-binding proteins and multiple cornified envelope genes. In this report, it is expressed that S100A11, S100A12, SPRR1B, SPRR2B, SPRR2C, SPRR3, involucrin (IVL), and LCE3D are associated with cutaneous squamous cell carcinomas and are up-regulated in the patients.31 It can be concluded that the up-regulation of LCE3D in the present study is related to the side effect of laser therapy. On the other hand, treatment with laser radiation not only is ineffective in inhibiting cancer but also increases one of the factors that are related to cancer. This can be due to the fact that the laser is not capable of molecular targeting.
Chloride ion channel protein (CLIC1) the most down-regulated gene is the other important DEGthat is appeared as hub with a degree value of 97. The over-expression of CLIC1 in a gastric carcinoma is reported by Chen et al.32 Therefore, the intense down-regulation of CLIC1 in this study can be related to the treatment procedure.
Conclusion
The findings revealed that network analysis is a useful method to provide detailed information about a convoluted complex system such as laser therapy. In this study, there was a logical correlation between the expression change and the centrality parameter of the studied DEGs. Laser therapy suppressed some dysregulated genes in the patient but simultaneously activated the nasty ones such as the LCE3D gene. It can be concluded that there are benefits in the application of the laser beside some side effects that should be considered to protect the body against possible damage.
Ethical Considerations
Not applicable.
Conflict of Interests
The authors declare no conflict of interest.
Acknowledgment
The Proteomics Research Center of Shahid Beheshti University of Medical Sciences supports this research.
Please cite this article as follows: Mansouri V, Rezaei-Tavirani M, Zadeh-Esmaeel MM, Rezaei-Tavirani S, Razzaghi M, Okhovatian F, et al. Analysis of laser therapy effects on squamous cell carcinoma patients: a system biology study. J Lasers Med Sci. 2019;10(suppl 1):S1-S6. doi:10.15171/jlms.2019.S1.
References
- 1.Arany PR. Craniofacial wound healing with photobiomodulation therapy: new insights and current challenges. J Den Res. 2016;95(9):977–84. doi: 10.1177/0022034516648939. [DOI] [PubMed] [Google Scholar]
- 2.Leal Junior ECP, Lopes-Martins RAB, Dalan F, Ferrari M, Sbabo FM, Generosi RA. et al. Effect of 655-nm low-level laser therapy on exercise-induced skeletal muscle fatigue in humans. Photomed Laser Surg. 2008;26(5):419–24. doi: 10.1089/pho.2007.2160. [DOI] [PubMed] [Google Scholar]
- 3.Ekizer A, Uysal T, Güray E, Akkuş D. Effect of LED-mediated-photobiomodulation therapy on orthodontic tooth movement and root resorption in rats. Lasers Med Sci. 2015;30(2):779–85. doi: 10.1007/s10103-013-1405-3. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins D, Abrahamse H. Effect of multiple exposures of low-level laser therapy on the cellular responses of wounded human skin fibroblasts. Photomed Laser Surg. 2006;24(6):705–14. doi: 10.1089/pho.2006.24.705. [DOI] [PubMed] [Google Scholar]
- 5.Kulekcioglu S, Sivrioglu K, Ozcan O, Parlak M. Effectiveness of low‐level laser therapy in temporomandibular disorder. Scand J Rheumatol. 2003;32(2):114–8. doi: 10.1080/03009740310000139. [DOI] [PubMed] [Google Scholar]
- 6.Zivin JA, Albers GW, Bornstein N, Chippendale T, Dahlof B, Devlin T. et al. Effectiveness and safety of transcranial laser therapy for acute ischemic stroke. Stroke. 2009;40(4):1359–64. doi: 10.1161/STROKEAHA.109.547547. [DOI] [PubMed] [Google Scholar]
- 7.Karu TI. Molecular mechanism of the therapeutic effect of low-intensity laser radiation. Lasers Life Sci. 1988;2(1):53–74. [Google Scholar]
- 8.Gao X, Xing D. Molecular mechanisms of cell proliferation induced by low power laser irradiation. J Biomed Sci. 2009;16(1):4. doi: 10.1186/1423-0127-16-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KC. The photobiological basis of low level laser radiation therapy. Laser Ther. 1991;3(1):19–24. doi: 10.5978/islsm.91-OR-03. [DOI] [Google Scholar]
- 10.Nomura K, Yamaguchi M, Abiko Y. Inhibition of interleukin-1β production and gene expression in human gingival fibroblasts by low-energy laser irradiation. Lasers Med Sci. 2001;16(3):218–23. doi: 10.1007/PL00011358. [DOI] [PubMed] [Google Scholar]
- 11.Gavish L, Perez L, Gertz SD. Low‐level laser irradiation modulates matrix metalloproteinase activity and gene expression in porcine aortic smooth muscle cells. Lasers Surg Med. 2006;38(8):779–86. doi: 10.1002/lsm.20383. [DOI] [PubMed] [Google Scholar]
- 12.Safaei A, Rezaei Tavirani M, Zamanian Azodi M, Lashay A, Mohammadi SF, Ghasemi Broumand M. et al. Diabetic retinopathy and laser therapy in rats: A protein-protein interaction network analysis. J Lasers Med Sci. 2017;8(1):S20–S21. doi: 10.15171/jlms.2017.s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rezaei-Tavirani M, Rezaei Tavirani M, Zamanian Azodi M, Razzaghi M, Moravvej Farshi H. Evaluation of skin response after erbium: yttrium–aluminum–garnet laser irradiation: a network analysis approach. J Lasers Med Sci. 2019;10(3):194–99. doi: 10.22037/jlms.v10i3.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santolini M, Barabási AL. Predicting perturbation patterns from the topology of biological networks. Proc Natl Acad Sci USA. 2018;115(27):E6375–E83. doi: 10.1073/pnas.1720589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albert R, Barabási AL. Statistical mechanics of complex networks. Rev Mod Phys. 2002;74(1):47–97. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- 16.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014;7(1):17–31. doi: 10.22037/ghfbb.v7i1.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khayer N, Zamanian-Azodi M, Mansouri V, Ghassemi-Broumand M, Rezaei-Tavirani M, Heidari MH. et al. Oral squamous cell cancer protein-protein interaction network interpretation in comparison to esophageal adenocarcinoma. Gastroenterol Hepatol Bed Bench. 2017;10(2):118–24. doi: 10.22037/ghfbb.v0i0.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antunes HS, Wajnberg G, Pinho MB, Jorge NAN, de Moraes JLM, Stefanoff CG. et al. cDNA microarray analysis of human keratinocytes cells of patients submitted to chemoradiotherapy and oral photobiomodulation therapy: pilot study. Lasers Med Sci. 2018;33(1):11–18. doi: 10.1007/s10103-017-2313-8. [DOI] [PubMed] [Google Scholar]
- 19.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9(1):405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito Y, Ando A, Ando H, Ando J, Saijoh Y, Inoko H. et al. Genomic structure of the spermatid-specific hsp70 homolog gene located in the class III region of the major histocompatibility complex of mouse and man. J Biochem. 1998;124(2):347–53. doi: 10.1093/oxfordjournals.jbchem.a022118. [DOI] [PubMed] [Google Scholar]
- 21.Jeng JE, Tsai JF, Chuang LY, Ho MS, Lin ZY, Hsieh MY. et al. Heat shock protein A1B 1267 polymorphism is highly associated with risk and prognosis of hepatocellular carcinoma: a case-control study. Medicine (Baltimore) 2008;87(2):87–98. doi: 10.1097/MD.0b013e31816be95c. [DOI] [PubMed] [Google Scholar]
- 22.Milner CM, Campbell RD. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32(4):242–51. doi: 10.1007/bf00187095. [DOI] [PubMed] [Google Scholar]
- 23.Karoly E, Fekete A, Banki NF, Szebeni B, Vannay A, Szabo AJ. et al. Heat shock protein 72 (HSPA1B) gene polymorphism and Toll-like receptor (TLR) 4 mutation are associated with increased risk of urinary tract infection in children. Pediatr Res. 2007;61(3):371–4. doi: 10.1203/pdr.0b013e318030d1f4. [DOI] [PubMed] [Google Scholar]
- 24.Gombos T, Förhécz Z, Pozsonyi Z, Jánoskuti L, Prohászka Z. Interaction of serum 70-kDa heat shock protein levels and HspA1B (+ 1267) gene polymorphism with disease severity in patients with chronic heart failure. Cell Stress Chaperones. 2008;13(2):199–206. doi: 10.1007/s12192-007-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarimón J, Bertranpetit J, Boada M, Tàrraga L, Comas D. HSP70-2 (HSPA1B) is associated with noncognitive symptoms in late-onset Alzheimer’s disease. J Geriatr Psychiatry Neurol. 2003;16(3):146–50. doi: 10.1177/0891988703256051. [DOI] [PubMed] [Google Scholar]
- 26.Schröder O, Schulte K-M, Ostermann P, Röher HD, Ekkernkamp A, Laun RA. Heat shock protein 70 genotypes HSPA1B and HSPA1L influence cytokine concentrations and interfere with outcome after major injury. Crit Care Med. 2003;31(1):73–9. doi: 10.1097/00003246-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 27.Harrison PJ, Procter AW, Exworthy T, Roberts GW, Najlerahim A, Barton AJ. et al. Heat shock protein (hsx70) mRNA expression in human brain: effects of neurodegenerative disease and agonal state. Neuropathol Appl Neurobiol. 1993;19(1):10–21. doi: 10.1111/j.1365-2990.1993.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 28.Oka N, Akiguchi I, Nagao M, Nishio T, Kawasaki T, Kimura J. Expression of endothelial leukocyte adhesion molecule‐1 (ELAM‐1) in chronic inflammatory demyelinating polyneuropathy. Neurology. 1994;44(5):946–50. doi: 10.1212/wnl.44.5.946. [DOI] [PubMed] [Google Scholar]
- 29.Xianfa T, Min L, Chengyao Z. Correlation of rs4085613 polymorphism within the late cornified envelope 3D gene with some clinical features of psoriasis vulgaris in Chinese Han population. Acta Universitatis Medicinalis Anhui. 2009;17(3):337–340. [Google Scholar]
- 30.Shen C, Gao J, Yin X, Sheng Y, Sun L, Cui Y. et al. Association of the late cornified envelope-3 genes with psoriasis and psoriatic arthritis: a systematic review. J Genet Genomics. 2015;42(2):49–56. doi: 10.1016/j.jgg.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Titova LV, Ayesheshim AK, Golubov A, Rodriguez-Juarez R, Woycicki R, Hegmann FA. et al. Intense THz pulses down-regulate genes associated with skin cancer and psoriasis: a new therapeutic avenue? Sci Rep. 2013;3:2363. doi: 10.1038/srep02363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CD, Wang CS, Huang YH, Chien KY, Liang Y, Chen WJ. et al. Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics. 2007;7(1):155–67. doi: 10.1002/pmic.200600663. [DOI] [PubMed] [Google Scholar]