Abstract
Given their extensive role in cell signalling, GPCRs are significant drug targets; despite this, many of these receptors have limited or no available prophylaxis. Novel drug design and discovery significantly rely on structure determination, of which GPCRs are typically elusive. Progress has been made thus far to produce sufficient quantity and quality of protein for downstream analysis. As such, this review highlights the systems available for recombinant GPCR expression, with consideration of their advantages and disadvantages, as well as examples of receptors successfully expressed in these systems. Additionally, an overview is given on the use of detergents and the styrene maleic acid (SMA) co-polymer for membrane solubilisation, as well as purification techniques.
Keywords: Expression, Purification, GPCR, SMALP, Review
Highlights
-
•
1713% increase in unique membrane protein structures since 2000.
-
•
Variety of expression methods available to GPCR researchers, now including the eyes of Drosophila melanogaster.
-
•
Development of novel solubilisation strategies including the styrene maleic acid co-polymer.
1. Introduction
As the largest family of membrane proteins in the human genome, G protein-coupled receptors (GPCRs) are widely studied due to their involvement in normotypical and pathological cell signalling profiles [1]. Characteristically, as shown in Fig. 1, these seven-transmembrane receptors undergo a conformational change upon activation by a ligand, allowing propagation of signalling cascades within the cell [2].
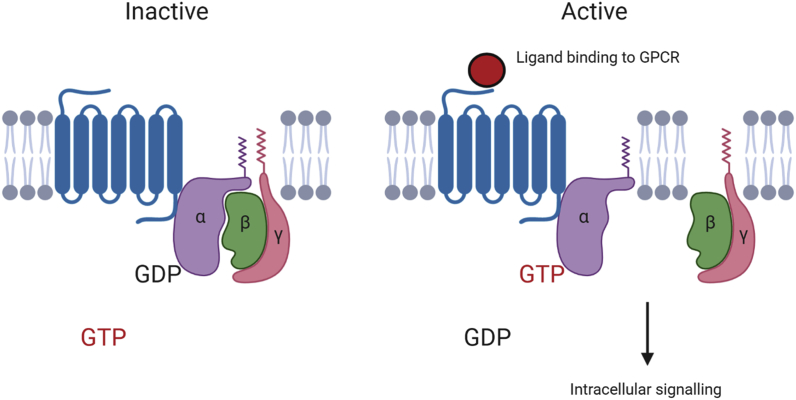
Fig. 1.
Ligand induced activation of a G protein-coupled receptor (GPCR). GPCRs (blue) are transmembrane receptors which activate intracellular signalling pathways, through coupling to G proteins. These heterotrimeric proteins consist of three subunits denoted α, β and γ, and are classically activated by a ligand induced conformational change in the GPCR [2]. This movement is proposed to involve a rotational transmembrane helix reorientation, exposing an intracellular binding cleft [3]. GDP is exchanged for GTP on Gα, while the βγ complex splits away and is able to signal independently of the Gα subunit. Humans encode 18, 5 and 12 different α, β and γ subunits, respectively. These combine into a variety of stimulatory (Gs) or inhibitory (Gi/q) effects on pathways including those dependent on adenylyl cyclase and phospholipase C. Created with Biorender.com.
Understanding the relationship between a GPCR's structure and function will aid further development of ortho- and allosteric molecules against these receptors to affect their pharmacology. While approximately half of all drugs target GPCRs, this is only reflected in a 5% coverage of these receptors, providing significant scope for further structure-based novel drug discovery [4].
While the structure of some GPCRs have been successfully determined, many challenges remain in this field. These include the concepts of homo-dimerization, heteromeric protein-protein interactions and the structural complexity of important motifs [5] – of note, the folding and flexibility of the ligand-binding domain of family B GPCRs [6]. While computational biology has greatly enhanced the versatility of studying GPCR structures [7], classical techniques are also often employed including x-ray crystallography [8], cryo-electron microscopy [9] and nuclear magnetic resonance spectroscopy [10]. A significant drawback to these methods lies in the initial requirement for a high yield and purity of mature, folded target protein.
These challenges have propelled the development and implementation of recombinant membrane protein expression, solubilisation and purification systems over the last two decades - contributing, in no small part, to the increase in resolved structures of membrane proteins in the same time-frame [11]. This review will provide a current summary of the methodology, benefits and drawbacks of the expression systems available to GPCR researchers (Table 1), as well as an overview of applicable solubilisation and purification techniques.
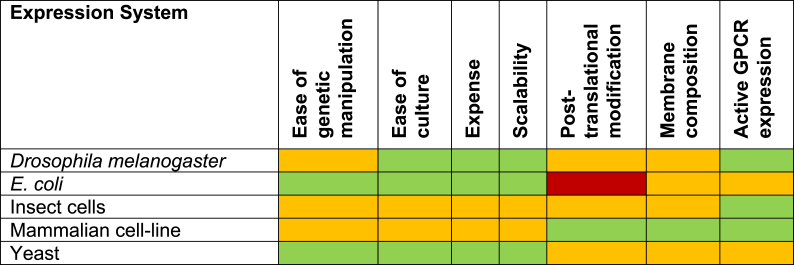
Table 1.
A comparison of expression systems for GPCRs. A qualitative assessment of considerations linked to recombinant protein expression systems. Green = positive, amber = moderate, red = negative. While E. coli and yeast were historically favourable due to ease of genetic manipulation, culture and scalability, recent developments in insect and mammalian lines have increased their use. Standard expression vectors can now be grown in litre volumes in lines including Sf9, expi293 and expiCHO at comparable cost to produce milligram quantities of receptor. The use of Drosophila is an emerging yet promising method requiring further attention.
2. Expression systems
2.1. E. coli
Native expression of GPCRs is well-known to be restricted to eukaryotic organisms [12] yet E. coli has proved an attractive host for expression and purification of a subset of receptors [13]. E. coli has become a laboratory workhorse for a number of reasons. Firstly, decades of work have led to well-characterised, rapidly growing cells which are easy to culture and strains have been optimised for protein expression, including membrane protein-specific strains [14]. E. coli can be easily grown across a range of scales allowing fermentation to produce large quantities of protein, although it should be noted that there is not always a linear relationship between culture volume and product yield [14]. The genetic tractability of E. coli allows a variety of expression plasmids to be used to tune protein expression levels. This can be particularly important with membrane proteins when saturation of the translocon can be a rate-limiting step [13,15]. Indeed, high level expression can lead to formation of inclusion body and refolding of GPCRs from such environments has met with limited success [16]. Some of these problems can be overcome with judicial strain selection and expression at lower temperatures [17]. However, due to its prokaryotic nature, E. coli does not possess a number of features that can be essential for GPCR function. There is a lack of post-translational modification, including glycosylation, which can be essential for ligand binding [18]. Despite this, there are several examples of active receptor expression [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34]] including the neurotensin and cannabinoid CB2 receptors. The use of E. coli in this sense can also be supported by the ability of deglycosylated receptors [35] to bind ligand, and protein engineering for stability [36]. Additionally, the lipid membrane environment may not include essential components such as cholesterol [37] and contains a very different lipidome to eukaryotic cells – there is clear evidence for lipid-dependent GPCR activity [38].
Despite these clear limitations [39], there have been a number of reports of GPCRs being successfully expressed in E. coli [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34]]. Unmodified GPCRs tend to have low stability and may aggregate in such systems [33]. A key strategy for successful expression and correct folding of GPCRs in E. coli is the use of fusion partners [40]. These serve both to direct the correct insertion of the receptor into the membrane whilst also increasing its overall solubility, thereby aiding both expression and purification [26]. Additionally, strategies such as selective mutagenesis to introduce stabilising mutations and the use of insertions or truncations has proven successful in some cases [17,33]. Indeed, the genetic tractability of E. coli can be used to select for variants with increased stability and expression even for relatively intractable receptors [[41], [42], [43]].
There are a number of advantages of the use of E. coli for downstream applications. It is relatively easy to conduct isotopic labelling experiments such that the subsequent protein can be used for NMR studies [10]. It should, however, be noted that the relatively low expression levels of GPCRs in E. coli is further impacted by such labelling strategies [10]. However, through optimised expression it has been reported that GPCR expression of up to 50 mg/L can be achieved [40]. The genetic amenability and tools available for E. coli open possibilities to select GPCR variants with enhanced expression and stability, generate those “locked” in a particular conformation, and also to, potentially, engineer those with completely novel functions [44]. Despite its prokaryotic nature, E. coli has clear potential for at least a subset of GPCRs.
2.2. Yeast
The fission yeast S. pombe and baker's yeast S. cerevisiae are important tools to express and investigate the signalling and stability of GPCRs [[45], [46], [47], [48], [49], [50], [51]], however, the methylotrophic yeast, Pichia pastoris (reclassified as Komagataella phaffii), is favored for the overexpression of GPCRs for structural studies [52]. High yields of functional receptors have been expressed [39], including the adenosine 2a receptor [[53], [54], [55]], 5HT5A receptor, beta2-adrenergic receptor [56] and muscarinic acetylcholine receptor M2 subtype (CHRM2) [57]. In addition, high-resolution crystal structures of the histamine H1 receptor [58] and the adenosine 2a receptor in complex with an antibody Fab fragment [59] have been obtained using the P. pastoris expression system as well as other membrane proteins.
This has been feasible due to the ease of manipulation and stable integration of expression vectors into P. pastoris coupled with its ability to grow to high cell densities on glycerol and to utilize methanol as the sole carbon source [60]. This system allows high levels of protein expression to be induced under the tightly controlled AOX1 promoter [61,62]. Other promoters are also available, including the constitutive glyceraldehyde-3-phosphate dehydrogenase (GAP) promoter as well as emerging novel methanol-inducible, non-methanol inducible and constitutive promoters [63,64]. Several expression vectors and strains are commercially available to optimize protein expression. Commonly used vectors are the pPIC9K, pPICZ or pPICZalpha, the latter of which contains an α-MF signal sequence derived from S. cerevisiae to enhance protein secretion. Expression vectors generally contain geneticin, kanamycin or zeocin resistance genes, and auxotrophic markers have also been used for GPCR expression [52,60]. Strains frequently used include the wild type X-33 strain, protease deficient strains (SMD1163, SMD1168 and SMD1165) and auxotrophic strains GS115 and KM71 [11,52,60].
P. pastoris is able to perform post translational modifications such as disulphide bond formation, and N and O-linked glycosylation. N-linked glycosylation occurs at the Asn-X-Ser/Thr motif on extracellular domains of GPCRs [65], and in some cases is required for cell surface expression [66], ligand binding and cell signalling [67]. While early steps of P. pastoris N-linked glycosylation are similar to the process in mammalian cells [68], it may potentially hypermannosylate the protein which can lead to misfolding, although this is less extensive than in S. cerevisiae [69]. It may also glycosylate residues of protein where this would not naturally occur [69,70]. Some GPCRs expressed in P. pastoris have therefore been engineered with these sites removed to facilitate crystallisation [58,59]. A study by Yurugi-Kobayashi et al., 2009 [71] which analysed glycosylation-deficient GPCRs demonstrated that while some receptors were expressed with lower functional levels, others were expressed at levels suitable for structural studies when this approach was combined with culture optimisation. P. pastoris strains and vectors with humanized N-glycosylation have been developed [68,72], but have not as of yet been applied to GPCR expression. Other modifications to GPCRs expressed by P. pastoris include codon optimisation, N and C-terminal truncations [54,59] and T4-lysozyme fusion to intracellular loop 3 (ICL3) [58].
In contrast to mammalian cells, yeast membranes contain ergosterol rather than cholesterol. Membrane cholesterol is thought to be required for the correct function of some GPCRs, and may cause direct conformational changes or indirectly alter membrane properties. Crystal structures suggest that some GPCRs contain specific cholesterol binding sites [[73], [74], [75]]. A humanized P. pastoris strain has been engineered which synthesizes cholesterol [76], and could be of benefit for GPCR expression. Cholesteryl hemi-succinate, a cholesterol derivative, can also be added to maintain stability [35, 55, 58].
In summary, yeast possess several advantages over other expression systems, including the ability to perform eukaryotic post-translational modifications while being capable of rapid growth to high cell densities on a large scale in relatively cheap media [77]; these aspects make yeast an appealing host, and their use for GPCR expression has significantly improved knowledge of GPCR structure and function.
2.3. Insect cell-line (Sf9, Sf21, Hi5)
In GPCR structural studies, insect cells are the most commonly used expression system to achieve milligram quantities of protein [35,78]. Expression is achieved via infection with a recombinant form of Autographica californica; a multiple nuclear polyhedrosis virus. This baculovirus infects the cells and drives the production of the protein of interest, usually via the polyhedrin promoter [79]. The majority of GPCR studies use the Spodoptera frugiperda (Sf9) or Sf21 cell-line, in preference to Trichoplusia ni (High Five) cells [78]. However protein expression levels can vary between different cell-lines so screening of cell lines is necessary when using insect cells for structural studies. The advantages of insect cell expression include growth in serum-free shaker cultures, which decreases costs and enables relatively easy scale-up, as well as high yield and the ability to perform most post-translational modifications [79].
A range of different systems have been developed to generate the recombinant baculovirus [79]. Once generated the virus requires titration in order to achieve an appropriate multiplicity of infection (MOI); an excess of virus will kill the cells before they can be harvested [79]. This step is often the most difficult as how to directly quantify virus is unclear and often inaccurate. Viral plaque assays are commonly used but take a minimum of two weeks and, often, their accuracy is questionable [79]. Various alternative approaches have been devised, including flow cytometry and qPCR, but cost, time and variability can still be problematic [80]. Many researchers find that using the virus directly to express protein in Sf9 cells and quantifying the expression level is quicker and more accurate. However, once the virus has been generated and titrated it can be stored for a number of years at 4 °C and much longer at −80 °C. More virus can also be generated by infecting the cells with a high MOI and collecting the cell culture media five days post-infection [81]. Initially, it can take up to a month to produce enough baculovirus to drive large scale expression however, once the virus has been generated, protein can be expressed within a week.
One potential disadvantage of insect cell expression arises from differences in lipid composition compared to mammalian cells. Insect cell membranes are low in cholesterol, have very high phosphatidylinositol content and no phosphatidylserine [35], and as noted below, protein function is highly dependent on lipid environment [82]. There have also been reports that a proportion of the protein produced can be misfolded [83], or that the lytic pathway of viral infection can cause protein degradation [81]. However, overall it remains one of the key approaches for GPCR overexpression.
2.4. Mammalian cell-line (HEK293, COS)
Membrane proteins are often expressed in insect, bacterial or yeast expression systems due to their high protein yield and expression, which is advantageous for structural studies [84]. High resolution human membrane protein structures have been solved from recombinant proteins derived from these sources, but the protein conformation and modifications may differ from a human protein expressed in human cells. To address these issues, mammalian cell-lines capable of expressing a desired protein have been trialled [85]. The selection of a specific cell-line is determined by whether the expression system represents the near-native environment in which the desired protein is endogenously expressed [84]. GPCRs are heavily post-translationally modified, therefore expressing human GPCRs in mammalian cells is often ideal to characterise their function and pharmacology [35]. For structural biology however, post-translational modifications can be detrimental during crystal formation. Glycosylation sites affect formation of ordered crystals due to the flexibility and heterogeneity of glycan residues [35]. This issue can be solved by mutating the N-glycosylation sites, provided that the conformation of the receptor is still stable [35]. Alternatively, the use of the GnTI− line which lacks N-acetylglucosaminyl transferase I activity would possibly enable further control of complex glycans [86].
The native environment in which GPCRs are expressed has an impact on the conformation and pharmacology of these receptors. The phospholipid composition of the native lipid bilayer has an allosteric effect on GPCRs. In the case of the human beta 2 adrenergic receptor in different liposomes, synthetic phosphatidylglycerol stabilised the active conformation of the receptor [82]. Due to variations in lipid composition between cell-lines of different expression systems, choosing a human cell-line that has a similar lipid composition to that of the endogenous GPCR is important for receptor pharmacology. As cholesterol can allosterically modulate GPCRs [87], its replacement with ergosterol in yeast expression systems can be detrimental. In the case of the human μ-opioid GPCR, ergosterol constrain the receptor in an inactive state, whereas cholesterol stabilises the active state [88]. Post-translational modification is important for GPCR function, where mammalian cells have the correct enzymes for phosphorylation and palmitoylation of human GPCRs [35].
Yields of recombinant protein in non-mammalian expression systems are often higher than mammalian expression systems, therefore optimising the GPCR gene construct for expression is an important first step [35]. Note, however, that higher levels of expression does not necessarily correlate with functional expression and a robust assay for ligand binding and/or signalling can be essential in any optimisation process [83]. This may take the form of traditional radioligand or fluorescent binding assay or could employ NanoBRET e.g. using β1AR tagged with NanoLuc at the N terminus [89]. In any case, GPCR gene constructs are often codon optimised for mammalian cell expression [90]. Kozak sequences (GCCACCATGG) and signal peptide sequences can be fused to the 5‘ end of the GPCR construct to enhance protein expression and cell surface delivery [90,91]. Subsequently, the optimised construct can be ligated into a plasmid vector, which can be transfected into mammalian cells transiently, or be used to create stable cell lines [35]. While transient transfections with popular reagents give detectable expression at the 48 h mark (on average), this can be extended. The BacMam technology uses a modified baculovirus to give expression within 4–6 h of transduction, lasting up to 5–14 days [92]. Finally, not only are stable lines more reproducible in terms of expression levels, inducible lines have been shown to improve the correct folding of GPCRs when compared to insect cells [83].
Overall, immortalised mammalian cell-lines are useful to study human GPCRs in their wild-type or mutated form. The immortalised human embryonic kidney 293 (HEK293) can transiently express recombinant proteins and is amongst the most popular human cell-line to use [93,94]. HEK293 cells exist as adherent cells or suspension cells; the latter are grown at a higher density, which is useful for protein production [93]. Table 2 summarises the currently known 3D structures of GPCRs derived from expression in mammalian cell-lines, as of 30/8/19.
Table 2.
3D structures of recombinant GPCRs derived from mammalian cell-line expression. Database query generated with MemProtMD at https://blanco.biomol.uci.edu/mpstruc/Accessed 5/9/19.
| GPCR | Organism | Cell-Line | Resolution, Å | PDB Entry |
|---|---|---|---|---|
| Angiotensin type II receptor | H. sapiens | Expi293F | 2.90 | 6DO1 |
| CB1 cannabinoid receptor | H. sapiens | HEK293F | 2.80 | 5TGZ |
| Cytomegalovirus US28 | H. sapiens | HEK | 2.89 | 4XT1 |
| Leukotriene B4 receptor | C. porcellus | HEK293 | 3.70 | 5X33 |
| Rhodopsin | B. taurus | Cos | 3.40 | 2J4Y |
| HEK293S-GnTI- | 3.30 | 4A4M | ||
| HEK293S | 2.36 | 6FK6 | ||
| HEK293 | 4.38 | 6QNO | ||
| H. sapiens | HEK293S | 3.30 | 4ZWJ | |
| H. adansoni | HEK293 | 2.14 | 6I9K | |
| Smoothened receptor | H. sapiens | HEK293S-GnTI- | 3.20 | 5L7D |
| HEK293S | 3.84 | 6OT0 | ||
| M. musculus | HEK293 | 2.80 | 6O3C |
2.5. Drosophila melanogaster
Each of the conventional expression systems detailed above are not without their limitations. One of the major drawbacks associated with all of these systems is the build-up of immature proteins in the intracellular membranes caused by the cell's failure to properly fold and transport the mature GPCR to the cell surface. This issue can lead to inadequate yields for structural studies thus limiting our understanding of GPCR structure and function [95,96] and obtaining adequate yields of the mature GPCR often requires optimisation of the expression conditions, increasing cost.
The fruit-fly, Drosophila melanogaster, has recently been utilised as an attractive alternative expression system to overcome some of these problems. The system takes advantage of the unique properties and architecture of the fly eye which consists of photoreceptor cells (PRCs) containing membrane stacks called rhabdomeres [97] therefore providing a large surface area for expression and folding of large amounts of membrane-associated proteins [98].
Heterologous expression of proteins within the PRCs is achieved using the well-established GAL4-UAS system [99]. This system allows the tissue-specific expression of transgenes by exploiting the use of the yeast GAL4 protein, a transcription factor that specifically binds to an Upstream Activating Sequence (UAS) to drive expression of its target genes. To express a specific protein in a particular tissue type within the fly, two strains are mated together: the driver-strain which expresses GAL4 from a tissue-specific promoter and the UAS strain which contains the transgene of interest cloned downstream of the GAL4 UAS (Fig. 2). In the resulting offspring, the transgene will be expressed in those specific cells that contain GAL4 [99]. By using a driver-strain that specifically expresses GAL4 within the fly eye, heterologous GPCR expression can be restricted to the PRCs. Generating transgenic flies is relatively easy and comparable in cost to other conventional expression systems. Also, Drosophila culture media for rearing experimental animals is relatively inexpensive and the need to work in sterile conditions is eliminated when working with flies [98].
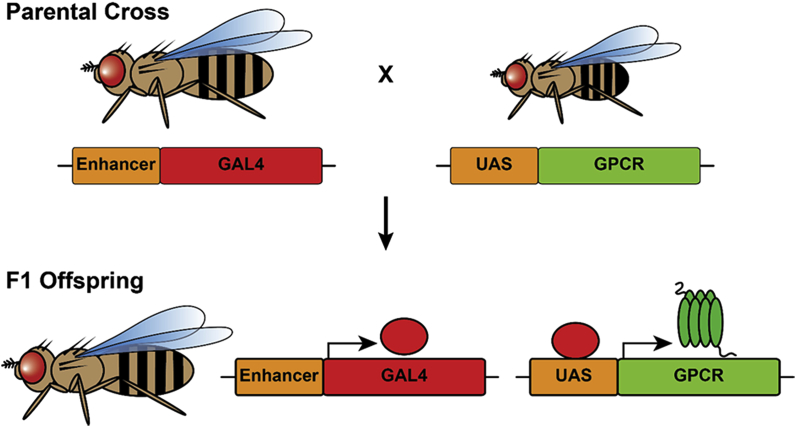
Fig. 2.
The Drosophila GAL4-UAS system for targeted gene expression. The GAL4-UAS system can be used for targeted expression of GPCRs within the Drosophila photoreceptor cells (PRCs). To obtain flies expressing a gene of interest in a tissue specific pattern, two Drosophila strains are mated together in the parental cross. The driver strain expresses the yeast GAL4 protein from a tissue-specific enhancer/promoter. The UAS strain contains the gene of interest cloned downstream of the GAL4 Upstream Activating Sequence (UAS). The resulting F1 offspring will express GAL4 protein in a tissue-restricted pattern which will bind to the UAS sequences upstream of the gene of interest to drive its expression in those specific cells. By using a driver strain that expresses GAL4 specifically within the fly eye, heterologous GPCR expression can be restricted to the PRCs.
Drosophila PRCs have been successfully used to express a number of GPCRs [100]; the Drosophila metabotropic glutamate receptor, DmGluRA being first reported. Overexpression of DmGluRA in PRCs resulted in higher yields of mature receptor than obtained using other conventional methods, including insect cell culture. Moreover, toxicity effects of DmGluRA overexpression were not observed in the host cells, overcoming a major limitation of other expression systems. Expressing mammalian mGluRs in the fly eye produced similar yields as expressing DmGluRA, suggesting that this system can be used to express foreign GPCRs from other species including human, rat and Chlamydomonas [98]. Furthermore, expression of the other two classes of GPCRs have now also been successfully reported using this system [100]. Importantly, it should also be noted that scale-up of expression, as required for downstream processes such as crystallisation, can be readily achieved and can sometimes be an issue for other cell culture-based expression systems [77].
Although the use of Drosophila for GPCR expression overcomes several of the major drawbacks associated with more conventional expression systems, it is not without its own limitations. Firstly, this method requires access to fly genetics expertise and facilities for Drosophila culture that if unavailable will require the need for collaboration with specialised laboratories that can provide these services [98]. Additionally, although this system is capable of post-translationally modifying proteins there are differences in some of the modifications that occur in the fly that may be important for GPCR function. An example of this is N-glycosylation, which tends to be less complex in insects and lacking in extended antennae compared to mammals [101,102]. Furthermore, regarding purification of membrane-associated proteins expressed in the fly eye, there is currently a lack of reports describing the use of detergent-free purification methods such as SMALPs using this system [98,100]. This will be an important future development due to the problems of membrane protein stabilisation associated with using detergents [103,104]. Yet despite these apparent shortfalls, Drosophila could prove to be a cheaper and more efficient alternative for functional GPCR expression and purification.
3. Solubilisation and purification
3.1. Detergents
An important barrier to studying GPCRs is the need to solubilise and purify these membrane proteins away from their native bilayer [105]. Ideally, this process should simultaneously retain target proteins in their folded, functional conformations for further in vitro study. Surfactant detergents are able to solubilise and extract membrane proteins due to their amphiphilic nature, improving the aqueous solubility of the protein [106]. A plethora of detergents are commercially available with different physicochemical properties; often, a screen is best performed to identify optimal detergents, likely on a protein-by-protein basis [107].
Briefly, detergents fall into three classes based on their polar head group – ionic, zwitterionic and non-ionic. Ionic detergents such as SDS are regarded as harsh, zwitterions are milder (LDAO) while non-ionic detergents are considered mild. The described harshness is derived from the efficacy of disrupting intra- and inter-molecular interactions. While some factors can be scrutinised, such as the critical micelle concentration (CMC) and hydrophilic-lipophilic balance (HLB), some detergents clearly perform well [108]. Overall, the non-ionic alkyl maltopyranoside detergents DM and DDM have been most successful in contributing to the resolution of membrane protein structures (approximately 45%) [109], and can be regarded as an evidence-based starting point [11,55,58,59,110].
Detergent monomers will, above the CMC, associate with a biological membrane and undergo a transbilayer mechanism to flip from the outer to inner leaflet [111]. This leads to the formation of lipid-detergent micelles (Fig. 3), the efficacy of which depends on the HLB and size/polarity of the detergent molecule. At this point, the bilayer falls apart resulting in the solubilisation of the membrane. A GPCR's protrusion from the bilayer can aid in the incorporation of detergent monomers due to disruptions in the ordered lipid arrangement. However, this can be opposed by the notion of detergent-resistant membranes [112] – especially in regards to GPCR populations in cholesterol-rich lipid rafts [113].
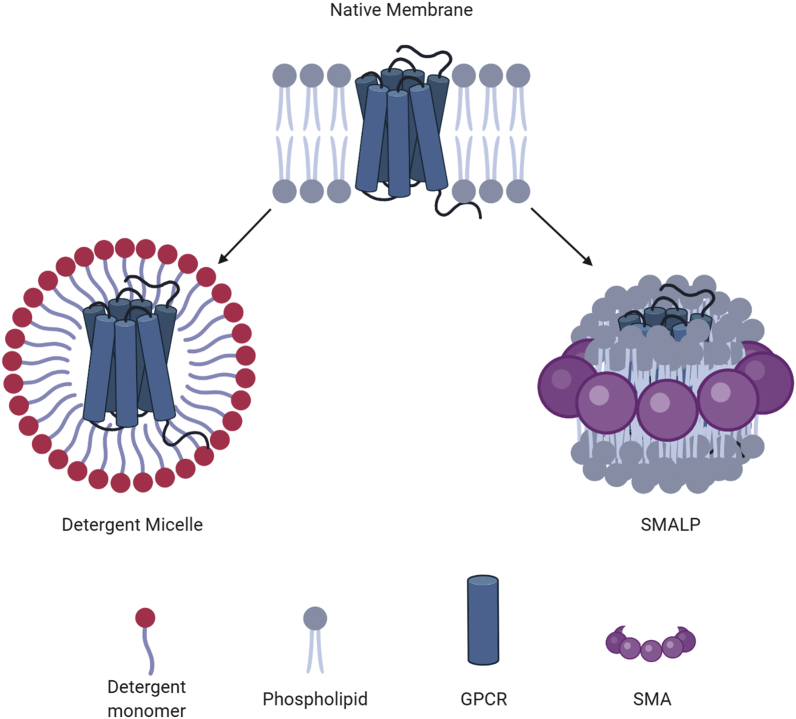
Fig. 3.
Solubilised GPCR in a detergent micelle compared to a SMALP. These diagrams show the interactions of the phospholipid bilayer (grey) or detergent monomers (red) with a GPCR (blue). Importantly, the difference between a detergent micelle and SMALP is shown, with regards to the retention of the GPCR's annular lipids when surrounded by the SMA co-polymer (purple). Created with Biorender.com.
Following solubilisation, it is often necessary to reduce or remove excess detergent to enable purification and further structural or functional analysis. Several methods are sufficient to achieve this including dialysis, size exclusion and affinity chromatography [111].
Despite their utility thus far, detergents are not without their limitations. Understanding of GPCR structure/function has led to the acknowledgement of the native lipid environment. Not only do the lipids surrounding a receptor provide lateral pressure, directly bound lipids can also be essential to influence active/inactive conformations [55]. As such, detergent micelles do not exert the same lateral pressure, and in some cases, the directly bound lipids required for stability/function are removed. It is therefore preferential to adopt techniques which retain these important components to represent a more biologically realistic reflection of GPCRs and their surroundings; and to prevent destabilisation or inactivation during solubilisation [114].
While several new detergents are being designed/developed, other advances reviewed elsewhere include amphipols [115], nano discs [116] and co-polymers including DIBMA [117] and SMA [118]. Largely, detergents must be chosen empirically, which can be an expensive and protein-demanding approach.
3.2. SMALPs
GPCRs are one of the classes of protein that have most frequently defied the attempts by biochemists to purify and characterise them. Poor thermal stability is often blamed for this, and considerable effort and resources have been expended to generate thermostabilized versions of GPCRs, in particular for structural studies [119]. An alternative outlook is that GPCRs are destabilised by detergents. Replacing these with better membrane mimetics could prevent destabilisation of the proteins. Amongst the alternatives that have been proposed to meet this need are styrene maleic acid lipid particles (SMALPs). Styrene maleic acid (SMA) is an amphipathic co-polymer that, when added to lipids, spontaneously assembles into nanoparticles of ∼10 nm diameter [120,121]. These nanoparticles consist of SMA polymer surrounding a patch of lipid bilayer (Fig. 3). When SMA is added to biological membranes a similar self-assembly process forms polymer-bound lipid particles containing membrane proteins [[122], [123], [124], [125], [126]].
One of the first reported successes using the SMALP method was the purification and functional characterisation of the adenosine-2A receptor (A2AR) which was overexpressed in both human epithelial kidney (HEK) cells and Pichia pastoris (Table 3) [55]. This demonstrated that the protein could be rapidly and effectively purified in a detergent-free manner.
Table 3.
Summary of published methods for the solubilisation of GPCRs by styrene maleic acid and related polymers.
| Protein name | GPCR class | Downstream analysis | Source membrane | Polymer used for solubilisation | Reference |
|---|---|---|---|---|---|
| A2aR | A | Radioligand binding assays, thermal stability | Human endothelial kidney (HEK 293T) cells; Pichia pastoris | 2:1 SMA (SMA2000P) | [55] |
| CTR | B | Radioligand binding | Cos cells | [131] | |
| GHS-1aR | A | Ligand binding via FRET; arrestin recruitment; GTP binding assays | Asolectin proteoliposomes | 2:1 SMA (SMA2000P) | [129] |
| HWbR | A | Crystallisation | Escherichia coli | 3:1 SMA (SMA3000P) | [130] |
| MT1R | A | Ligand binding; G protein activation; arrestin recruitment | P. pastoris | 2:1 SMA (SMA2000P) | [129] |
| V1aR | A | Radioligand binding | HEK 293T | 2:1 SMI | [127,131] |
The ligand-binding properties of the A2AR-SMALPs were used to assay its stability under a variety of conditions. Notably, A2AR-SMALPs withstood more than 5 freeze-thaw cycles without reduction in their ligand binding ability. Likewise, A2AR-SMALPs and A2AR in the membrane both retained 75% of their specific ligand binding capacity after up to 15 days of incubation at 4 °C. In detergent, this binding declined to 0% by day 3. Similarly at 37 °C the stability of A2AR in SMALPs far outstripped that of the detergent-solubilised sample. This remarkable stability under a range of conditions makes A2AR-SMALPs a much more flexible and useful reagent than the detergent-solubilised equivalent.
A2AR was also used in a study of an alternative styrene co-polymer: styrene maleimide (SMI). This has some similar properties and architecture to SMA, but is acid-compatible and can be used in buffers with pH < 7.8 [127]. By contrast SMA is soluble only above pH 5.8, and is more usually used in buffers of pH 8. A2AR-SMILPs had ligand-binding properties equivalent to the protein in the cell membrane, indicating that SMI also has potential as a reagent for the detergent-free purification of GPCRs. In the same study, the vasopressin receptor (V1aR) also retained its specific ligand-binding properties in SMILPs.
The ability to bind ligands is not the only indicator of GPCR function. Perhaps of more importance is the ability of a purified GPCR to recruit/signal to G-proteins and initiate intracellular signalling cascades [128]. One study using SMA has demonstrated that the melatonin receptor (MT1R) and the ghrelin receptor (GHS-R1a) in SMALPs are capable of G-protein activation, arrestin recruitment and ligand binding [129].
To date, there is only one high resolution structure of a GPCR in SMALPs, Haloquadratum walsbyi bacteriorhodopsin (HWbR). This structure was solved at 2.0 Å resolution using the in meso crystallisation (lipidic cubic phase) methodology [130]. Hence, it is likely that the protein-SMALPs integrated into the bilayers of the cubic phase prior to crystallogenesis. Lipids are visible in the structure, but these are identifiable as monooleins, the lipids used to assemble the cubic phase. In a parallel experiment, bR purified using detergent had a remarkably similar structure to the structure derived from SMA-solubilised bR. Therefore in this case there is an argument that using SMA did not provide additional structural information compared to using detergent. By contrast, a recent structure was solved by cryo-electron microscopy at 3.4 Å resolution of a bacterial respiratory supercomplex purified using SMA [131]. This structure did show specific native lipids bound to the protein, which may be of relevance in understanding the subtleties of its structure and function. This hints that cryo-EM may be a viable approach for solving structures of membrane proteins retaining their native lipids.
Following solubilisation, several purification methods may be employed which have been reviewed elsewhere [132]. Summarised in Table 4, these include gel filtration, ion exchange and affinity chromatography, of which the latter is most popular. While affinity to antibodies or ligands such as lectin can be utilised for membrane proteins, GPCRs expressed in the systems discussed have utilised a range of purification tags. These include poly-histidine [55,58], FLAG [56,133], HA [134], Strep-Tactin [135], Rho [136] and EF1 [137] tags among others. Following detection and affinity chromatography purification, it is possible to obtain the quantities of functional material required for structural studies with these tags [58,59].
Table 4.
Pros and cons of purification techniques available for GPCRs.
| Purification Technique | Pros | Cons |
|---|---|---|
| Affinity chromatography [139,140] | Can be used if protein molecular weight, charge or hydrophobicity is unknown. High affinity binding can result in high sample purity. |
May require a tag or terminal fusion. Washing may remove weakly bound molecules. SMALPs are sensitive to divalent cations. |
| Gel filtration [141,142] | Efficient separation of large and small molecules. Minimal elution volume. No sample loss. |
Only separated on size. May require further techniques. Limited resolution due to short chromatogram timescale. |
| Ion exchange [139] | Only one charge-based interaction. Predictable elution pattern. |
Inconsistency between columns. Limited to ionizable groups. |
More recent advancements include the use of mini-G proteins to study GPCRs in their active conformations [138]. These are engineered GTPase domains of the Gα subunits of G proteins and stabilise the active conformation of the receptor. Not only have they been shown to form stable complexes purified by SEC, N-terminal fusion with GFP allows for successful detection of coupling by FSEC [138]. Such reagents provide huge potential for state-selective purifications.
4. Conclusion
GPCRs remain a challenging component of the membrane protein structural biology field. While the sources of difficulty are gradually being lessened as understanding and technology advance, the dearth of structural information is limiting novel drug design and discovery [4]. Computational biology has greatly enhanced the ability to predict and manipulate GPCR structure, and how this affects their functions. However, in silico experiments remain only a component of the holistic study of membrane proteins; expression and purification are largely required before downstream biochemical and biophysical analysis [8,9,143,144].
As such, and discussed in this review, the expression systems available to GPCR researchers each come with their own benefits and drawbacks. While ease of culture and genetic amenability are undoubtedly attractive qualities, they clearly do not entirely make up for biologically important characteristics such as post-translational modifications. There will always seemingly remain a compromise with the expression system of choice, if only the expense. Regardless of these drawbacks, each traditional system will be preferred for application to certain techniques. For example, post-translational modification may be less desired with regards to crystallisation, but more so for trafficking and functionality.
An interesting alternative to consider is cell-free expression [145]. As cell lysate is used, problems such as toxicity and sequestering of protein to inclusion bodies is avoided. Additionally, this technique allows for modification of GPCRs with unnatural amino acids [146] and is a useful method for NMR labelling [10]. Finally, expression in the eyes of Drosophila offers a promising solution for a scalable production of functional recombinant membrane proteins (Table 5). Currently, as of September 2019, only five PDB entries were derived from expression in Drosophila – none of which were GPCRs. Future work to broaden the diversity and characterisation of varied GPCRs would invaluably reinforce the use of this emerging technique. Overall, the field is currently in a much stronger position than a few decades ago, and will undoubtedly continue to build upon the methods reviewed here.
Table 5.
Expression levels of recombinant GPCRs in the photoreceptor cells of Drosophila melanogaster. Data obtained from Panneels et al., 2011 [98]. MP = membrane protein.
| GPCR | Organism | Expression level, pmol/mg total MP |
|---|---|---|
| CCR5 Chemokine receptor | H. sapiens | 555 |
| DmGluRA Metabotropic glutamate receptor | D. melanogaster | 226 |
| mGluR5 Metabotropic glutamate receptor | R. norvegicus | 192 |
| Rh1 Rhodopsin | D. melanogaster | 502 |
| V2R Vasopressin receptor | H. sapiens | >1000 |
Acknowledgements
We are grateful for support from BBSRC (BB/N007417/1, BB/P025927/1, BB/P022685/1, BB/M016668/1, BB/L015846/1, BB/M007529/1, BB/R02152X/1, BB/S004696/1 and BB/T001488/1), H2020 ERA CoBioTech (under grant agreement No. 722361) and H2020 MSCA COFUND (under grant agreement No. 847419).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pep.2019.105524.
Contributor Information
Daniel N. Wiseman, Email: d.wiseman1@aston.ac.uk.
Abigail Otchere, Email: otcherea@aston.ac.uk.
Jaimin H. Patel, Email: patelj47@aston.ac.uk.
Romez Uddin, Email: uddinr3@aston.ac.uk.
Naomi L. Pollock, Email: n.pollock@bham.ac.uk.
Sarah J. Routledge, Email: s.routledge@aston.ac.uk.
Alice J. Rothnie, Email: a.rothnie@aston.ac.uk.
Cathy Slack, Email: c.slack@aston.ac.uk.
David R. Poyner, Email: d.r.poyner@aston.ac.uk.
Roslyn M. Bill, Email: r.m.bill@aston.ac.uk.
Alan D. Goddard, Email: a.goddard@aston.ac.uk.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kobilka B. G protein coupled receptor structure and activation. Biochim. Biophys. Acta. 2007;1768(4):794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bockaert J., Pin J.P. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18(7):1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trzaskowski B., Latek D., Yuan S., Ghoshdastider U., Debinski A., Filipek S. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr. Med. Chem. 2012;19(8):1090–1109. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos R., Ursu O., Gaulton A., Bento P.A., Donadi R.S., Bologa C.G., Harlsson A., Al-Lazikani B., Hersey A., Oprea T.I., Overington J.P. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ladds G., Goddard A.D., Hill C., Thornton S., Davey J. Differential effects of RGS proteins on Gαq and Gα11 activity. Cell. Signal. 2007;19(1):103–113. doi: 10.1016/j.cellsig.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Liang Y.L., Khoshouei M., Deganutti G., Glukhova A., Koole C., Peat T.S., Radjainia M., Plitzko J.M., Baumeister W., Miller L.J., Hay D.L., Christopoulos A., Reynolds C.A., Wooten D., Sexton P.M. Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature. 2018;561:492–497. doi: 10.1038/s41586-018-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coudrat T., Simms J., Christopoulos A., Wootten D., Sexton P.M. Improving virtual screening of G protein-coupled receptors via ligand-directed modelling. PLoS Comput. Biol. 2017;13(11) doi: 10.1371/journal.pcbi.1005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birch J., Axford D., Foadi J., Meyer A., Eckhardt A., Thielmann Y., Moraes I. The fine art of integral membrane protein crystallisation. Methods. 2018;147:150–162. doi: 10.1016/j.ymeth.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Thongin N., Kargas V., Clews J., Ford R.C. Cryo-electron microscopy of membrane proteins. Methods. 2018;147:176–186. doi: 10.1016/j.ymeth.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Tapaneeyakorn S., Goddard A.D., Oates J., Willis C.L., Watts A. Solution-and solid-state NMR studies of GPCRs and their ligands. Biochim. Biophys. Acta Biomembr. 2011;1808(6):1462–1475. doi: 10.1016/j.bbamem.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Dilworth M.V., Piel M.S., Bettany K.E., Ma P., Luo J., Sharples D., Poyner D.R., Gorss S.R., Moncoq K., Henderson P.J.F., Miroux B. Microbial expression systems for membrane proteins. Methods. 2018;147:3–39. doi: 10.1016/j.ymeth.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 12.de Mendoza A., Sebe-Pedros A., Ruiz-Trillo I. The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biology and Evolution. 2014;6(3):606–619. doi: 10.1093/gbe/evu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Kuipers G., Niemiec L., Baumgarten T., Slotboom D.J., de Gier J.W., Hjelm H. High-level production of membrane proteins in E. coli BL21 (DE3) by omitting the inducer IPTG. Microb. Cell Factories. 2015;14(1):p142. doi: 10.1186/s12934-015-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260(3):289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 15.Wagner S., Baars L., Ytterberg A.J., Klussmeier A., Wagner C.S., Nord O., Nygren P.A., van Wijk K.J., de Gier J.W. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteom. 2007;6(9):1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Baneres J.L., Martin A., Hullot P., Girard J.P., Rossi J.C., Parello J. Structure-based analysis of GPCR function: conformational adaptation of both agonist and receptor upon leukotriene B4 binding to recombinant BLT1. J. Mol. Biol. 2003;329(4):801–814. doi: 10.1016/s0022-2836(03)00438-8. [DOI] [PubMed] [Google Scholar]
- 17.Link A.J., Skretas G., Strauch E.M., Chari N.S., Georgiou G. Efficient production of membrane-integrated and detergent-soluble G protein-coupled receptors in Escherichia coli. Protein Sci. 2008;17(10):1857–1863. doi: 10.1110/ps.035980.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheatley M., Hawtin S.R. Glycosylation of G-protein-coupled receptors for hormones central to normal reproductive functioning: its occurrence and role. Hum. Reprod. Update. 1999;5(4):356–364. doi: 10.1093/humupd/5.4.356. [DOI] [PubMed] [Google Scholar]
- 19.Bertin B., Freissmuth M., Breyer R.M., Schutz W., Strosberg A.D., Marullo S. Functional expression of the human serotonin 5-HT1A receptor in Escherichia coli. Ligand binding properties and interaction with recombinant G protein alpha-subunits. J. Biol. Chem. 1992;267(12):8200–8206. [PubMed] [Google Scholar]
- 20.Freissmuth M., Selzer E., Marullo S., Schutz W., Strosberg A.D. Expression of two human beta-adrenergic receptors in Escherichia coli: functional interaction with two forms of the stimulatory G protein. PNAS USA. 1991;88(19):8548–8552. doi: 10.1073/pnas.88.19.8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa H., Haga T. Expression of functional M2 muscarinic acetylcholine receptor in Escherichia coli. J. Biochem. 2000;127(1):151–161. doi: 10.1093/oxfordjournals.jbchem.a022577. [DOI] [PubMed] [Google Scholar]
- 22.Grisshammer R., Duckworth R., Henderson R. Expression of a rat neurotensin receptor in Escherichia coli. Biochem. J. 1993;295(Pt 2):571–576. doi: 10.1042/bj2950571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill R.A., Sillence M.N. Improved membrane isolation in the purification of beta 2-adrenoceptors from transgenic Escherichia coli. Protein Expr. Purif. 1997;10(1):162–167. doi: 10.1006/prep.1997.0732. [DOI] [PubMed] [Google Scholar]
- 24.Kiefer H., Krieger J., Olszewski J.D., von Heijne G., Prestwich G.D., Breer H. Expression of an olfactory receptor in Escherichia coli: purification, reconstitution, and ligand binding. Biochemistry. 1996;35(50):16077–16084. doi: 10.1021/bi9612069. [DOI] [PubMed] [Google Scholar]
- 25.Yeliseev A.A., Wong K.K., Soubias O., Gawrisch K. Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 2005;14(10):2638–2653. doi: 10.1110/ps.051550305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Attrill H., Harding P.J., Smith E., Ross S., Watts A. Improved yield of a ligand-binding GPCR expressed in E. coli for structural studies. Protein Expr. Purif. 2009;64(1):32–38. doi: 10.1016/j.pep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Baneres J.L., Mesnier D., Martin A., Joubert L., Dumuis A., Bockaert J. Molecular characterization of a purified 5-HT4 receptor: a structural basis for drug efficacy. J. Biol. Chem. 2005;280(21):20253–20260. doi: 10.1074/jbc.M412009200. [DOI] [PubMed] [Google Scholar]
- 28.Busuttil B.E., Turney K.L., Frauman A.G. The expression of soluble, full-length, recombinant human TSH receptor in a prokaryotic system. Protein Expr. Purif. 2001;23(3):369–373. doi: 10.1006/prep.2001.1519. [DOI] [PubMed] [Google Scholar]
- 29.Inagaki S., Ghirlando R., White J.F., Gvozdenovic-Jeremic J., Northup J.K., Grisshammer R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012;417(1–2):95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krepkiy D., Wong K., Gawrisch K., Yeliseev A. Bacterial expression of functional, biotinylated peripheral cannabinoid receptor CB2. Protein Expr. Purif. 2006;49(1):60–70. doi: 10.1016/j.pep.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Tian C., Breyer R.M., Kim H.J., Karra M.D., Friedman D.B., Karpay A., Sanders C.R. Solution NMR spectroscopy of the human vasopressin V2 receptor, a G protein-coupled receptor. J. Am. Chem. Soc. 2005;127(22):8010–8011. doi: 10.1021/ja051161b. [DOI] [PubMed] [Google Scholar]
- 32.Weiss H.M., Grisshammer R. Purification and characterization of the human adenosine A (2a) receptor functionally expressed in Escherichia coli. Eur. J. Biochem. 2002;269(1):82–92. doi: 10.1046/j.0014-2956.2002.02618.x. [DOI] [PubMed] [Google Scholar]
- 33.Mallipeddi S., Zvonok N., Makriyannis A. Expression, purification and characterization of the human cannabinoid 1 receptor. Sci. Rep. 2018;8(1):2935. doi: 10.1038/s41598-018-19749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalke K., Huyghe C., Lichiere J., Graviere M.E., Siponen M., Sciara G., Lepaul I., Wagner R., Magg C., Rudolph R., Cambillau C., Desmyter A. Mammalian G protein-coupled receptor expression in Escherichia coli: II. Refolding and biophysical characterization of mouse cannabinoid receptor 1 and human parathyroid hormone receptor 1. Anal. Biochem. 2010;401(1):74–80. doi: 10.1016/j.ab.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Milić D., Veprintsev D.B. Large-scale production and protein engineering of G protein-coupled receptors for structural studies. Front. Pharmacol. 2015;31(6):66. doi: 10.3389/fphar.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertheleme N., Chae P.S., Singh S., Mossakowska D., Hann M.M., Smith K.J., Hubbard J.A., Dowell S.J., Byrne B. Unlocking the secrets of the gatekeeper: methods for stabilizing and crystallizing GPCRs. Biochim. Biophys. Acta Biomembr. 2013;1828(11):2583–2591. doi: 10.1016/j.bbamem.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Goddard A.D., Watts A. Regulation of G protein-coupled receptors by palmitoylation and cholesterol. BMC Biol. 2012;10:p27. doi: 10.1186/1741-7007-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goddard A.D., Dijkman P.M., Adamson R.J., Watts A. Lipid-dependent GPCR dimerization. Methods Cell Biol. 2013;117:341–357. doi: 10.1016/B978-0-12-408143-7.00018-9. [DOI] [PubMed] [Google Scholar]
- 39.Lundstrom K., Wagner R., Reinhart C., Desmyter A., Cherouati N., Magnin T., Zeder-Lutz G., Courtot M., Prual C., Andre N., Hassaine G., Michel H., Cambillau C., Pattus F. Structural genomics on membrane proteins: comparison of more than 100 GPCRs in 3 expression systems. J. Struct. Funct. Genom. 2006;7(2):77–91. doi: 10.1007/s10969-006-9011-2. [DOI] [PubMed] [Google Scholar]
- 40.Petrovskaya L.E., Shulga A.A., Bocharova O.V., Ermolyuk Y.S., Kryukova E.A., Chupin V.V., Blommers M.J., Arseniev A.S., Kirpichnikov M.P. Expression of G-protein coupled receptors in Escherichia coli for structural studies. Biochemistry (Mosc.) 2010;75(7):881–891. doi: 10.1134/s0006297910070102. [DOI] [PubMed] [Google Scholar]
- 41.Dodevski I., Pluckthun A. Evolution of three human GPCRs for higher expression and stability. J. Mol. Biol. 2011;408(4):599–615. doi: 10.1016/j.jmb.2011.02.051. [DOI] [PubMed] [Google Scholar]
- 42.Klenk C., Ehrenmann J., Schutz M., Pluckthun A. A generic selection system for improved expression and thermostability of G protein-coupled receptors by directed evolution. Sci. Rep. 2016;6:p21294. doi: 10.1038/srep21294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar C.A., Dodevski I., Kenig M., Dudli S., Mohr A., Hermans E., Pluckthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. PNAS USA. 2008;105(39):14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skretas G., Georgiou G. Engineering G protein-coupled receptor expression in bacteria. PNAS USA. 2008;105(39):14747–14748. doi: 10.1073/pnas.0807741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ficca A.G., Testa L., Tocchini Valentini G.P. The human beta 2-adrenergic receptor expressed in Schizosaccharomyces pombe retains its pharmacological properties. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1995;377(2):140–144. doi: 10.1016/0014-5793(95)01330-x. [DOI] [PubMed] [Google Scholar]
- 46.Sander P., Grunewald S., Reilnder H., Michel H. Expression of the human D2s dopamine receptor in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe: a comparative study. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1994;344:41–46. doi: 10.1016/0014-5793(94)00335-1. [DOI] [PubMed] [Google Scholar]
- 47.Iguchi Y., Ishii J., Nakayama H., Ishikura A., Izawa K., Tanaka T., Ogino C., Kondo A. Control of signalling properties of human somatostatin receptor subtype-5 by additional signal sequences on its amino-terminus in yeast. J. Biochem. 2010;147(6):875–p884. doi: 10.1093/jb/mvq023. [DOI] [PubMed] [Google Scholar]
- 48.Hashi H., Nakamura Y., Ishii J., Kondo A. Modifying expression modes of human neurotensin receptor type 1 alters sensing capabilities for agonists in yeast signaling biosensor. Biotechnol. J. 2018;13(4) doi: 10.1002/biot.201700522. [DOI] [PubMed] [Google Scholar]
- 49.Togawa S., Ishii J., Ishikura A., Tanaka T., Ogino C., Kondo A. Importance of asparagine residues at positions 13 and 26 on the amino-terminal domain of human somatostatin receptor subtype-5 in signalling. J. Biochem. 2010;147(6):867–p873. doi: 10.1093/jb/mvq022. [DOI] [PubMed] [Google Scholar]
- 50.Weston C., Poyner D.R., Patel V., Dowell S., Ladds G. Investigating G protein signalling bias at the glucagon-like peptide-1 receptor in yeast. Br. J. Pharmacol. 2014;171(15):3651–p3665. doi: 10.1111/bph.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ladds G., Davis K., Hillhouse E.W., Davey J. Modified yeast cells to investigate the coupling of G protein-coupled receptors to specific G proteins. Mol. Microbiol. 2003;47(3):781–792. doi: 10.1046/j.1365-2958.2003.03336.x. [DOI] [PubMed] [Google Scholar]
- 52.Byrne B. Pichia pastoris as an expression host for membrane protein structural biology. Curr. Opin. Struct. Biol. 2015;32:9–17. doi: 10.1016/j.sbi.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Fraser N.J. Expression and functional purification of a glycosylation deficient version of the human adenosine 2a receptor for structural studies. Protein Expr. Purif. 2006;49(1):129–p137. doi: 10.1016/j.pep.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Singh S., Gras A., Fiez-Vandal C., Ruprecht J., Rana R., Martinez M., Strange P.G., Wagner R., Byrne B. Large-scale functional expression of WT and truncated human adenosine A2A receptor in Pichia pastoris bioreactor cultures. Microb. Cell Factories. 2008;7:p28. doi: 10.1186/1475-2859-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamshad M., Charlton J., Lin Y.P., Routledge S.J., Bawa Z., Knowles T.J., Overduin M., Dekker N., Dafforn T.R., Bill R.M., Poyner D.R., Wheatley M. G-protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent. Biosci. Rep. 2015;35(2) doi: 10.1042/BSR20140171. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss H.M., Haase W., Michel H., Reiländer H. Comparative biochemical and pharmacological characterization of the mouse 5HT5A 5-hydroxytryptamine receptor and the human beta2-adrenergic receptor produced in the methylotrophic yeast Pichia pastoris. Biochem. J. 1998;330(Pt3):1137–1147. doi: 10.1042/bj3301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asada H., Uemura T., Yurugi-Kobayashi T., Shiroishi M., Shimamura T., Tsujimoto H., Ito K., Sugawara T., Nakane T., Nomura N., Murata T., Haga T., Iwata S., Kobayashi T. Evaluation of the Pichia pastoris expression system for the production of GPCRs for structural analysis. Microb. Cell Factories. 2011;22(10):24. doi: 10.1186/1475-2859-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimamura T., Shiroishi M., Weyand S., Tsujimoto H., Winter G., Katritch V., Abagyan R., Cherezov V., Liu W., Han G.W., Kobayashi T., Stevens R.C., Iwata S. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475(7354):65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hino T., Arakawa T., Iwanari H., Yurugi-Kobayashi T., Ikeda-Suno C., Nakada-Nakura Y., Kusano-Arai O., Weyand S., Shimamura T., Nomura N., Cameron A.D., Kobayashi T., Hamakubo T., Iwata S., Murata T. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482(7384):237–p240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahmad M., Hirz M., Pichler H., Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014;98(12):5301–p5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cregg J.M., Barringer K.J., Hessler A.Y., Madden K.R. Pichia pastoris as a host system for transformations. Mol. Cell. Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bollok M., Resina D., Valero F., Ferrer P. Recent patents on the Pichia pastoris expression system: expanding the toolbox for recombinant protein production. Recent Pat. Biotechnol. 2009;3:192–201. doi: 10.2174/187220809789389126. [DOI] [PubMed] [Google Scholar]
- 63.Prielhofer R., Maurer M., Klein J., Wenger J., Kiziak C., Gasser B., Mattanovich D. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb. Cell Factories. 2013;12:5. doi: 10.1186/1475-2859-12-5. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu N., Zhu J., Zhu Q., Xing Y., Cai M., Jiang T., Zhou M., Zhang Y. Identification and characterization of novel promoters for recombinant protein production in yeast Pichia pastoris. Yeast. 2018;35(5):379–385. doi: 10.1002/yea.3301. [DOI] [PubMed] [Google Scholar]
- 65.Dong C., Filipeanu C.M., Duvernay M.T., Wu G. Regulation of G protein-coupled receptor export trafficking. Biochim. Biophys. Acta. 2007;1768(4):853–p870. doi: 10.1016/j.bbamem.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Q., Miller L.J., Dong M. Role of N-linked glycosylation in biosynthesis, trafficking, and function of the human glucagon-like peptide 1 receptor. Am. J. Physiol. Endocrinol. Metabol. 2010;299(1):E62–E68. doi: 10.1152/ajpendo.00067.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nehring R.B., Richter D., Meyerhof W. Glycosylation affects agonist binding and signal transduction of the rat somatostatin receptor subtype 3. J. Phys. 2000;94(3–4):185–192. doi: 10.1016/s0928-4257(00)00203-5. [DOI] [PubMed] [Google Scholar]
- 68.Choi B.K., Bobrowicz P., Davidson R.C., Hamilton S.R., Kung D.H., Li H., Miele R.G., Nett J.H., Wildt S., Gerngross T.U. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. PNAS USA. 2003;100(9):5022–p5027. doi: 10.1073/pnas.0931263100. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vieira Gomes A.M., Souza Carmo T., Silva Carvalho L., Mendonça Bahia F., Parachin N.S. Comparison of yeasts as hosts for recombinant protein production. Microorganisms. 2018;6(2):E38. doi: 10.3390/microorganisms6020038. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laron Z. Insulin-like growth factor 1 (IGF-1): a growth hormone. Mol. Pathol. 2001;54:311–316. doi: 10.1136/mp.54.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yurugi-Kobayashi T., Asada H., Shiroishi M., Shimamura T., Funamoto S., Katsuta N., Ito K., Sugawara T., Tokuda N., Tsujimoto H., Murata T., Nomura N., Haga K., Haga T., Iwata S., Kobayashi T. Comparison of functional non-glycosylated GPCRs expression in Pichia pastoris. Biochem. Biophys. Res. Commun. 2009;380(2):271–276. doi: 10.1016/j.bbrc.2009.01.053. [DOI] [PubMed] [Google Scholar]
- 72.Jacobs P.P., Geysens S., Vervecken W., Contreras R., Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat. Protoc. 2009;4:58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- 73.Pucadyil T.J., Chattopadhyay A. Cholesterol modulates ligand binding and G-protein coupling to serotonin (1A) receptors from bovine hippocampus. Biochim. Biophys. Acta. 2004;1663(1–2):188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 74.Hanson M.A., Cherezov V., Griffith M.T., Roth C.B., Jaakola V.-P., Chien E.Y.T., Velasquez J., Kuhn P., Stevens R.C. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paila Y.D., Tiwari S., Chattopadhyay A. Are specific nonannular cholesterol binding sites present in G-protein coupled receptors? Biochim. Biophys. Acta. 2009;1788(2):295–302. doi: 10.1016/j.bbamem.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 76.Hirz M., Richter G., Leitner E., Wriessnegger T., Pichler H. A novel cholesterol-producing Pichia pastoris strain is an ideal host for functional expression of human Na,K-ATPase α3β1 isoform. Appl. Microbiol. Biotechnol. 2013;97(21):9465–p9478. doi: 10.1007/s00253-013-5156-7. [DOI] [PubMed] [Google Scholar]
- 77.Macauley-Patrick S., Fazenda M.L., McNeil B., Harvey L.M. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22(4):249–p270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 78.Saarenpaa T., Jaakola V.P., Goldman A. Baculovirus-mediated expression of GPCRs in insect cells. Methods Enzymol. 2015;556:185–218. doi: 10.1016/bs.mie.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 79.McKenzie E.A., Abbott W.M. Expression of recombinant proteins in insect and mammalian cells. Methods. 2018;147:40–49. doi: 10.1016/j.ymeth.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 80.Roldao A., Oliveira R., Carrondo M.J., Alves P.M. Error assessment in recombinant baculovirus titration: evaluation of different methods. J. Virol Methods. 2009;159(1):69–80. doi: 10.1016/j.jviromet.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 81.Aloia A.L., Glatz R.V., McMurchie E.J., Leifert W.R. GPCR expression using baculovirus-infected Sf9 cells. Methods Mol. Biol. 2009;552:115–129. doi: 10.1007/978-1-60327-317-6_8. [DOI] [PubMed] [Google Scholar]
- 82.Dawaliby R., Trubbia C., Delporte C., Masureel M., van Antwerpen P., Kobilka B.K., Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12(1):35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas J.A., Tate C.G. Quality control in eukaryotic membrane protein overproduction. J. Mol. Biol. 2014;426(24):4139–4154. doi: 10.1016/j.jmb.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andrell J., Tate C.G. Overexpression of membrane proteins in mammalian cells for structural studies. Mol. Membr. Biol. 2013;30(1):52–63. doi: 10.3109/09687688.2012.703703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang L., Teng G.M.K., Chan E.Y.M., Au S.W.N., Wise H., Lee S.S.T., Cheung W.-T. Impact of cell type and epitope tagging on heterologous expression of G-protein coupled receptor: a systematic study on angiotensin type II receptor. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu B., Chakraborty R., Eilers M., Dakshinamurti S., O'Neil J.D., Smith S., Bhullar R.P., Chelikani P. High-level expression, purification and characterization of a constitutively active thromboxane A2 receptor polymorphic variant. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0076481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guixa-Gonzalez R., Albasanz J.L., Rodriguez-Espigares I., Pastor M., Sanz F., Marti-Solano M., Manna M., Martinez-Seara H., Hildebrand P.W., Martin M., Selent J. Membrane cholesterol access into a G-protein-coupled receptor. Nat. Commun. 2017;8:p14505. doi: 10.1038/ncomms14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lagane B., Gaibelet G., Meilhoc E., Masson J.M., Cezanne L., Lopez A. Role of sterols in modulating the human mu-opioid receptor function in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275(43):33197–33200. doi: 10.1074/jbc.C000576200. [DOI] [PubMed] [Google Scholar]
- 89.Soave M., Stoddart L., Brown A., Woolard J., Hill S.J. Use of a new proximity assay (NanoBRET) to investigate the ligand-binding characteristics of three fluorescent ligands to the human β1-adrenoceptor expressed in HEK-293 cells. Pharmacol. Res. Perspect. 2016;4(5) doi: 10.1002/prp2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chakraborty R., Xu B., Bhullar R.P., Chelikani P. Expression of G protein-coupled receptors in Mammalian cells. Methods Enzymol. 2015;556:267–281. doi: 10.1016/bs.mie.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 91.Zucchelli S., Patrucco L., Persichetti F., Gustincich S., Cotella D. Engineering translation in mammalian cell factories to increase protein yield: the unexpected use of long non-coding SINEUP RNAs. Comput. Struct. Biotechnol. J. 2016;14:404–410. doi: 10.1016/j.csbj.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu Y.-C. Baculovirus vectors for gene delivery: a review. Curr. Gene Ther. 2008;8(1):54–65. doi: 10.2174/156652308783688509. [DOI] [PubMed] [Google Scholar]
- 93.Thomas P., Smart T.G. HEK293 cell line: a vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods. 2005;51(3):187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 94.Subedi G.P., Johnson R.W., Moniz H.A., Moremen K.W., Barb A. High yield expression of recombinant human proteins with the transient transfection of HEK293 cells in suspension. J. Vis. Exp. 2015;(106) doi: 10.3791/53568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Griffith D.A., Delipala C., Leadsham J., Jarvis S.M., Oesterhelt D. A novel yeast expression system for the overproduction of quality-controlled membrane proteins. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2003;553(1–2):45–50. doi: 10.1016/s0014-5793(03)00952-9. [DOI] [PubMed] [Google Scholar]
- 96.Panneels V., Eroglu C., Cronet P., Sinning I. Pharmacological characterization and immunoaffinity purification of metabotropic glutamate receptor from Drosophila overexpressed in Sf9 cells. Protein Expr. Purif. 2003;30(2):275–282. doi: 10.1016/s1046-5928(03)00100-1. [DOI] [PubMed] [Google Scholar]
- 97.Kumar J.P., Ready D.F. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121(12):4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- 98.Panneels V., Kock I., Krijnse-Locker J., Rezgaoui M., Sinning I. Drosophila photoreceptor cells exploited for the production of eukaryotic membrane proteins: receptors, transporters and channels. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0018478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phelps C.B., Brand A.H. Ectopic gene expression in Drosophila using GAL4 system. Methods. 1998;14(4):367–379. doi: 10.1006/meth.1998.0592. [DOI] [PubMed] [Google Scholar]
- 100.Eroglu C., Cronet P., Panneels V., Beaufils P., Sinning I. Functional reconstitution of purified metabotropic glutamate receptor expressed in the fly eye. EMBO Rep. 2002;3(5):491–496. doi: 10.1093/embo-reports/kvf088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bernaudat F., Frelet-Barrand A., Pochon N., Dementin S., Hivin P., Boutigny S., Rioux J.B., Salvi D., Seigneurin-Berny D., Richaud P., Joyard J., Pignol D., Sabaty M., Desnos T., Pebay-Peyroula E., Darrouzet E., Vernet T., Rolland N. Heterologous expression of membrane proteins: choosing the appropriate host. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0029191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schiller B., Hykollari A., Yan S., Paschinger K., Wilson I.B. Complicated N-linked glycans in simple organisms. J. Biol. Chem. 2012;393(8):661–673. doi: 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee S.C., Knowles T.J., Postis V.L., Jamshad M., Parslow R.A., Lin Y.P., Goldman A., Sridhar P., Overduin M., Muench S.P., Dafforn T.R. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11(7):1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 104.Prive G.G. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41(4):388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 105.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta Biomembr. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 106.le Maire M., Champeil P., Moller J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 107.Arachea B.T., Sun Z., Potente N., Malik R., Isailovic D., Viola R.E. Detergent selection for enhanced extraction of membrane proteins. Protein Expr. Purif. 2012;86:12–20. doi: 10.1016/j.pep.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 108.Anatrace Detergents and their uses in membrane protein science. https://www.anatrace.com/Technical-Documentation/Catalogs/Anatrace-Detergents-Booklet-FINAL Pdf available at:
- 109.Lyons J.A., Shahsavar A., Paulsen P.A., Pederson B.P., Nissen P. Expression strategies for structural studies of eukaryotic membrane proteins. Curr. Opin. Struct. Biol. 2016;38:137–144. doi: 10.1016/j.sbi.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 110.Parker J.L., Newstead S. Current trends in α-helical membrane protein crystallization: an update. Protein Sci. 2012;21:1358–1365. doi: 10.1002/pro.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anandan A., Vrielink A. Detergents in membrane protein purification and crystallisation. In: Moraes I., editor. vol. 922. 2016. pp. 13–28. (The Next Generation in Membrane Protein Structure Determination. Advances in Experimental Medicine and Biology). [DOI] [PubMed] [Google Scholar]
- 112.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. Resistance of cell membranes to different detergents. PNAS USA. 2003;100(10):5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sezgin E., Levental I., Mayor S., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Popot J.L. Amphipols, nanodiscs and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 115.Popot J.L., Berry E.A., Charvolin D., Creuzenet C., Ebel C., Engelman D.M., Flöetenmeyer M., Giusti F., Gohon Y., Hervé P., Hong Q., Lakey J.H., Leonard K., Shuman H.A., Timmins P., Warschawski D.E., Zito F., Zoonens M., Pucci B., Tribet C. Amphipols: polymeric surfactants for membrane biology research. Cell. Mol. Life Sci. 2003;60(8):1559–1574. doi: 10.1007/s00018-003-3169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McLean M.A., Gregory M.C., Sligar S.G. Nanodiscs: a controlled bilayer surface for the study of membrane proteins. Annu. Rev. Biophys. 2018;47:107–124. doi: 10.1146/annurev-biophys-070816-033620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oluwole A.O., Danielczak B., Meister A., Babalola J.O., Vargas C., Keller S. Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer. Angew Chem. Int. Ed. Engl. 2017;56(7):1919–1924. doi: 10.1002/anie.201610778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stroud Z., Hall S.C.L., Dafforn T.R. Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights. Methods. 2018;147:106–117. doi: 10.1016/j.ymeth.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 119.Muk S., Ghosh S., Achuthan S., Chen X., Yao X., Sandhu M., Griffor M.C., Fennell K.F., Che Y., Shanmugasundaram V., Qiu X., Tate C.G., Vaidehi N. 2019. Machine learning for prioritization of thermostabilizing mutations for G-Protein Coupled Receptors. BioRXiv. 715375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Knowles T.J., Finka R., Smith C., Lin Y.-P., Dafforn T., Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131(22):7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 121.Lee S.C., Knowles T.J., Postis V.L.G., Jamshad M., Parslow R.A., Lin Y.-P., Goldman A., Sridhar P., Overduin M., Muench S.P., Dafforn T.R. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 122.Scheidelaar S., Koorengevel M.C., Pardo J.D., Meeldijk J.D., Breukink E., Killian J.A. Molecular model for the solubilisation of membranes into nanodisks by styrene maleic acid copolymers. Biophys. J. 2015;108(2):279–290. doi: 10.1016/j.bpj.2014.11.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pollock N.L., Lee S.C., Patel J.H., Gulamhussein A.A., Rothnie A.J. Structure and function of membrane proteins encapsulated in a polymer-bound lipid bilayer. Biochim. Biophys. Acta Biomembr. 2017;1860(4):809–817. doi: 10.1016/j.bbamem.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 124.Simon K.S., Pollock N.L., Lee S.C. Membrane protein nanoparticles: the shape of things to come. Biochem. Soc. Trans. 2018;46(6):1495–1504. doi: 10.1042/BST20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee S.C., Pollock N.L. Membrane proteins: is the future disc shaped? Biochem. Soc. Trans. 2016;44(4):1011–1018. doi: 10.1042/BST20160015. [DOI] [PubMed] [Google Scholar]
- 126.Wheatley M., Charlton J., Jamshad M., Routledge S.J., Bailey S., La-Borde P.J., Azam M.T., Logan R.T., Bill R.M., Dafforn T.R., Poyner D.R. GPCR–styrene maleic acid lipid particles (GPCR–SMALPs): their nature and potential. Biochem. Soc. Trans. 2015;44(2):619–623. doi: 10.1042/BST20150284. [DOI] [PubMed] [Google Scholar]
- 127.Hall S.C.L., Tognoloni C., Charlton J., Bragginton E.C., Rothnie A.J., Sridhar P., Wheatley M., Knowles T.J., Arnold T., Edler K.J., Dafforn T.R. An acid-compatible co-polymer for the solubilisation of membranes and proteins into lipid bilayer-containing nanoparticles. Nanoscale. 2018;10:10609–10619. doi: 10.1039/c8nr01322e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Damian M., Pons V., Renault P., M'Kadmi C., Delort B., Hartmann L., Kaya A.I., Louet M., Gagne D., Slah K.B.H., Denoyelle S., Ferry G., Boutin J.A., Wagner R., Fehrentz J.-A., Martinez J., Marie J., Floquet N., Gales C., Mary S., Hamm H.E., Baneres J.-L. GHSR-D2R heteromerization modulates dopamine signalling through and effect on G protein conformation. Proc. Natl. Acad. Sci. 2018;115(17):4501–4506. doi: 10.1073/pnas.1712725115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Logez C., Damian M., Legros C., Dupre C., Guery M., Mary S., Wagner R., M'Kadmi C., Nosjean O., Fould B., Marie J., Fehrentz J.-A., Martinez J., Ferry G., Boutin J.A., Baneres J.-L. Detergent-free isolation of functional G-Protein Coupled Receptors into nanomatric lipid particles. Biochemistry. 2015;55:38–48. doi: 10.1021/acs.biochem.5b01040. [DOI] [PubMed] [Google Scholar]
- 130.Broecker J., Eger B.T., Ernst O.P. Crystallogenesis of membrane proteins mediated by polymer-bounded lipid nanodiscs. Structure. 2017;25(2):99384–99392. doi: 10.1016/j.str.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 131.Charlton J. University of Birmingham; Birmingham: 2016. Solubilisation and Characterisation of G Protein-Coupled Receptors Using Styrene Maleic Acid Polymer.https://etheses.bham.ac.uk//id/eprint/6525/1/Charlton16PhD.pdf PhD thesis. Viewed 13/8/19. [Google Scholar]
- 132.Lin S.H., Guidotti G. Purification of membrane proteins. Methods Enzymol. 2009;463:619–629. doi: 10.1016/S0076-6879(09)63035-4. [DOI] [PubMed] [Google Scholar]
- 133.André N., Cherouati N., Prual C., Steffan T., Zeder-Lutz G., Magnin T., Pattus F., Michel H., Wagner R., Reinhart C. Enhancing functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci. 2006;15(5):1115–1126. doi: 10.1110/ps.062098206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McLatchie L.M., Fraser N.J., Main M.J., Wise A., Brown J., Thompson N., Solari R., Lee M.G., Foord S.M. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393(6683):p333. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 135.Yeliseev A., Zoubak L., Schmidt T.G.M. Application of Strep-Tactin XT for affinity purification of Twin-Strep-tagged CB2, a G protein-coupled cannabinoid receptor. Protein Expr. Purif. 2017;131:109–118. doi: 10.1016/j.pep.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Locatelli-Hoops S., Yeliseev A., Gawrisch K., Gorshkova I. Surface plasmon resonance applied to G protein-coupled receptors. Biomed. Spectrosc. Imaging. 2013;2(3):155–181. doi: 10.3233/BSI-130045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mhurchu N.N., Zoubrak L., McGauran G., Linse S., Yeliseev A., O'Connell D.J. Simplifying G protein-coupled receptor isolation with a calcium-dependent fragment complementation affinity system. Biochemistry. 2018;57(30):4383–4390. doi: 10.1021/acs.biochem.8b00469. [DOI] [PubMed] [Google Scholar]
- 138.Nehme R., Carpenter B., Singhal A., Strege A., Edwards P.C., White C.F., Du H., Grisshammer R., Tate C.G. Mini-G proteins: novel tools for studying GPCRs in their active conformation. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ninfa A., Ballou D., Benore M. second ed. Wiley; 2009. Fundamental Laboratory Approaches for Biochemistry and Biotechnology. [DOI] [PubMed] [Google Scholar]
- 140.Kumar P. second ed. Pathfinder Publication; New Delhi: 2018. Fundamentals and Techniques of Biophysics and Molecular Biology. [Google Scholar]
- 141.Garret R.H., Grisham C.M. fifth ed. Brooks/Cole, Cengage Learning; Belmont, CA: 2013. Biochemistry. [Google Scholar]
- 142.Skoog D.A., Holler F.J., Crouch S.R. sixth ed. Thomson Brooks/Cole; Belmont, CA: 2006. ‘Liquid Chromatography’ in Principles of Instrumental Analysis. [Google Scholar]
- 143.Castell O.K., Dijkman P.M., Wiseman D.N., Goddard A.D. Single molecule fluorescence for membrane proteins. Methods. 2018;147:221–228. doi: 10.1016/j.ymeth.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 144.Uddin R., Simms J., Poyner D.R. Functional characterisation of G protein-coupled receptors. Methods. 2018;147:213–220. doi: 10.1016/j.ymeth.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 145.Sobhanifar S., Reckel S., Junge F., Schwarz D., Kai L., Karbyshev M., Lohr F., Bernhard F., Dotsch V. J. Biomol. NMR. 2010;46(1):33–43. doi: 10.1007/s10858-009-9364-5. [DOI] [PubMed] [Google Scholar]
- 146.Muranaka N., Miura M., Taira H., Hohsaka T. Incorporation of unnatural non-α-amino acids into the N terminus of proteins in a cell-free translation system. Chembiochem. 2007;8(14):1650–1653. doi: 10.1002/cbic.200700249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.