Summary
Most metazoan cells entering mitosis undergo characteristic rounding, which is important for accurate spindle positioning and chromosome separation. Rounding is driven by contractile tension generated by myosin motors in the sub-membranous actin cortex. Recent studies highlight that alongside myosin activity, cortical actin organization is a key regulator of cortex tension. Yet, how mitotic actin organization is controlled remains poorly understood. To address this, we characterized the F-actin interactome in spread interphase and round mitotic cells. Using super-resolution microscopy, we then screened for regulators of cortex architecture and identified the intermediate filament vimentin and the actin-vimentin linker plectin as unexpected candidates. We found that vimentin is recruited to the mitotic cortex in a plectin-dependent manner. We then showed that cortical vimentin controls actin network organization and mechanics in mitosis and is required for successful cell division in confinement. Together, our study highlights crucial interactions between cytoskeletal networks during cell division.
Keywords: actin organization, cortex, cell shape, mitosis, mitotic rounding, vimentin, proteomics, plectin, CDK1, phosphorylation
Highlights
-
•
Comparison of the F-actin interactome in spread interphase and round mitotic cells
-
•
Proteomics identifies vimentin and plectin as key regulators of the mitotic cortex
-
•
Vimentin intermediate filaments localize under the actin cortex in mitosis
-
•
Sub-cortical vimentin regulates actin cortex organization and mechanics in mitosis
Mitotic rounding, which is essential for accurate spindle positioning, is controlled by the sub-membranous cortical actin network. Using proteomics, Serres et al. identify the intermediate filament vimentin as a key regulator of the mitotic cortex. Vimentin forms a sub-cortical layer in mitosis and controls actin cortex organization and mechanics.
Introduction
A precise control of cell shape is central to a wealth of physiological processes, including tissue morphogenesis, cell migration, and cell division. One of the first events of cell division in most animal cells, both in culture and in vivo, is cellular rounding at mitosis entry. Mitotic rounding generates the intracellular space required for accurate spindle assembly and metaphase plate formation (reviewed in Lancaster and Baum, 2014). This is of particular importance for cells dividing in confined environments. In packed epithelia, mitotic rounding defects lead to abnormal spindle orientation and division asymmetries (Chanet et al., 2017, Luxenburg et al., 2011). In cells cultured in 3-dimensional (3D) confining devices, physically preventing rounding leads to spindle assembly defects and delayed mitotic progression (Lancaster et al., 2013). It is thus essential to understand how mitotic rounding is controlled.
Multiple studies converge on identifying the reorganization of cellular actin into a uniform, contractile actomyosin cortex at the cell surface as a key driver of mitotic rounding (reviewed in Champion et al., 2017, Clark and Paluch, 2011, Lancaster and Baum, 2014). Cortical contractility, which increases through mitosis and peaks in metaphase, creates tension at the cell surface and promotes rounding (Clark and Paluch, 2011). At an upstream level, rounding appears to be controlled by the mitotic master regulator CDK1 and its substrate the RhoA-GEF Ect2 (Gavet and Pines, 2010, Maddox and Burridge, 2003, Matthews et al., 2012). However, how exactly these pathways effect the changes in actin organization and cell surface mechanics driving cell shape change remains unclear.
In Drosophila cells, an increase in membrane-to-cortex attachment and cortex stiffness via the ezrin-radixin-moesin (ERM) family protein moesin is essential for rounding (Carreno et al., 2008, Kunda et al., 2008). However, in mammalian cells, although ezrin depletion slightly decreases mitotic tension (Toyoda et al., 2017), ERMs do not appear to be required for rounding (Machicoane et al., 2014). Instead, for many years, cortex tension in mammalian cells had been thought to be primarily controlled by the levels and activity of cortical myosin (Mayer et al., 2010, Ramanathan et al., 2015, Tinevez et al., 2009). However, recent studies, including a screen for regulators of cortex tension (Toyoda et al., 2017), have shown that proteins controlling actin filament length and actin cross-linkers affect cortical tension (Chugh et al., 2017, Ding et al., 2017, Logue et al., 2015, Toyoda et al., 2017). Taken together, it is increasingly clear that the organization of cortical actin is a key regulator of cortex tension (reviewed in Koenderink and Paluch, 2018). Yet, a systematic investigation of how cortex organization in mitosis is controlled has been missing. As a result, our understanding of the regulation of mitotic actin architecture remains fragmented and limited to the role of a handful of selected proteins.
Here, we took an unbiased approach to identify proteins controlling actin cortex organization in mitosis. First, we established a phalloidin affinity matrix and mass spectrometry-based method to determine the proteins that bind filamentous actin in interphase and mitosis. This identified F-actin binding proteins (F-ABPs) specifically enriched in metaphase, when cortical tension is highest. We then conducted a quantitative small interfering RNA (siRNA) screen to test the effect of candidate regulators on mitotic cortex architecture, using thickness, measured by super-resolution microscopy, as a readout. Our screen revealed that the intermediate filament protein vimentin is a key regulator of mitotic cortex thickness. We then found that in mitosis, vimentin relocalizes to the actin cortex in a plectin-dependent manner, while mitotic phosphorylation appears to control vimentin network organization. The sub-cortical vimentin layer appears to mechanically resist the contractility, and possibly the expansion toward the cytoplasm, of the actomyosin cortex. We then show that the sub-cortical vimentin network controls cortical tension and is required for effective rounding and successful division in 3D confinement. Taken together, our study shows that the organization and mechanics of the actin cortex cannot be studied in isolation but must be investigated in the context of the entire cell.
Results
F-Actin Interactome in Interphase and Mitosis
To identify ABPs potentially regulating mitotic cortex organization, we compared which proteins bind actin filaments in mitotic and interphase cells. We developed a pull-down assay to isolate F-ABPs from cellular extracts (Figure 1A) and compared HeLa cells synchronized in mitosis (arrested in metaphase), when cells are rounded with a uniform actin cortex, and in interphase (arrested in G2 phase), when cells adopt spread shapes with prominent lamellipodia and stress fibers (Figures 1A and 1B). We verified that the synchronized cells displayed the expected actin and microtubule distributions (Figures 1B and S1A). We also validated synchronization efficiency by checking the levels of both cyclinB1, a subunit of the cyclin B-CDK1 complex expressed at high levels from G2 to anaphase onset, and phospho-Histone H3, a marker of mitosis (Figure S1B).
Figure 1.
Determining the Interphase and Mitotic F-Actin Interactomes
(A) Schematic representation of the synchronization procedure and the experimental pipeline. Cells were first arrested in S phase by thymidine treatment. For synchronization in interphase, thymidine-pre-treated cells were then released and treated with the CDK1 inhibitor RO3306, yielding a population of cells in G2 phase. For synchronization in metaphase, thymidine-pre-treated cells were submitted to a short treatment with nocodazole, followed by a mitotic shake off and metaphase arrest with the proteasome inhibitor MG132. The drugs used for synchronization were washed out prior to extract preparation.
(B) Representative images of HeLa cells after synchronization following the procedure described in (A). Cells were stained with DAPI (red) to visualize DNA and phalloidin (cyan) to visualize F-actin. Scale bars, 10 μm.
(C) Immunoblot of actin, cyclin B, and phospho-Histone H3 (P-HH3) levels in the different synchronized cell extracts. S, supernatant; P, Pellet.
(D) Colloidal Coomassie staining of an SDS-PAGE gel of synchronized cell extracts and of the control and phalloidin-bead elutions for mass spectrometry (corresponding to one of the mass spectrometry experiments, MS2). The x marks an unrelated lane. The asterisk marks the position of the actin band on the Coomassie gel.
(E) Gene Ontology Panther protein classification of the two mass spectrometry data sets (contaminants and proteins found more enriched in the control bead-bound fractions were removed).
(F) Number of proteins enriched in the phalloidin-bead-bound fractions in the two mass spectrometry analyses (MS1 and MS2) for different phases of the cell cycle (G2, metaphase).
(G) Total number of proteins enriched in the phalloidin-bound fractions and overlap between the two mass spectrometry experiments.
See also Figure S1.
To isolate F-ABPs, we used a phalloidin affinity matrix for mapping F-actin interactomes (Samwer et al., 2013). First, we recovered soluble fractions containing maximal amounts of actin. Cells were lysed in the presence of low doses of the actin depolymerizing drug Latrunculin A and the supernatant was collected. After optimization of lysis conditions, our protocol yielded more than 70% of the cellular actin in the supernatant (Figure S1C). We then applied this lysis protocol to each synchronized cell population and recovered cellular extracts with a comparable amount of actin in each phase (Figure 1C). In order to isolate F-ABPs, we incubated the solubilized extracts with ATP and MgCl2, to induce actin re-polymerization, together with custom-made superparamagnetic beads conjugated with phalloidin, a toxin that binds and stabilizes F-actin (Figure 1A). We verified that the post-translational modifications appeared maintained and that the incubation did not affect the cell-cycle state of the extracts (Figures S1D and S2E). We also confirmed by scanning electron microscopy that our protocol induced actin polymerization in the extracts (Figure S1F). Control and phalloidin-bead-bound fractions were then eluted and subjected to SDS-PAGE and Coomassie staining to assess protein contents (Figures 1D and S1G). As expected, we observed a strong enrichment of actin in the phalloidin-bead elution compared to controls (Figures 1D and S1G). The gels were then trypsin digested and the protein composition of the elutions was analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). We performed two independent experiments (referred to as MS1 and MS2), which identified 37,221 and 30,502 spectra, respectively (corresponding to 396 and 317 proteins) (Table S1). This list was reduced to 290 and 211 proteins, respectively, after removing contaminants and proteins more represented in the control bead elutions (Table S1). As expected, β-actin was the most abundant protein identified (with a mean of 1,967 and 1,902 spectra in MS1 and MS2, respectively). A Gene Ontology analysis revealed that “cytoskeletal proteins” and the “actin family” represented the most enriched molecular classes (Figure 1E). Importantly, the protein class distributions were similar between replicate experiments, suggesting that our assay yielded highly reproducible data (Figure 1E). The number of F-actin binders identified was comparable between the two phases of the cell cycle in both experiments (Figure 1F), with ∼51% of the proteins identified being present in both phases in at least one of the mass spectrometry experiments (Table S1). Overall, 168 F-actin binders were reproducibly identified between replicate experiments (Figure 1G).
Changes in F-Actin Interactome between Interphase and Mitosis
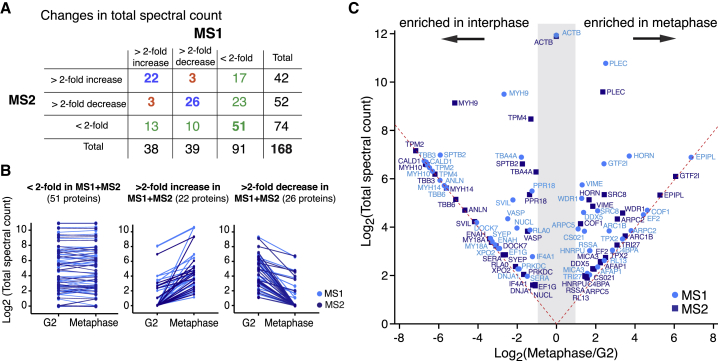
We then explored how the F-actin interactome (thereafter FAI) changes between interphase and mitosis (Table S1). We analyzed changes in protein levels in the F-actin-associated fractions (thereafter FAI abundance), as estimated by total spectral counts, between interphase and metaphase (Figures 2A and 2B). Importantly, we verified that changes in FAI abundance did not simply reflect differences in protein levels in the solubilized extracts (Figure S2). For 67% of the factors identified, FAI abundance changed by less than 2-fold between the two cell cycle phases in one or both experiments (Figure 2A, green), suggesting that actin network composition is relatively similar in interphase and mitosis. Only six proteins (3.5% of total) displayed inconsistent changes, with FAI abundance increasing between interphase and metaphase in one experiment and decreasing in the other; these proteins were excluded from further analysis (Figure 2A, orange). The remaining 48 proteins reproducibly displayed at least 2-fold changes in FAI abundance between interphase and mitosis (Figures 2A (blue) and 2C; Tables 1 and S1).
Figure 2.
Changes in F-Actin Interactome between Interphase and Metaphase
(A) Numbers of proteins displaying increased (>2-fold increase), decreased (>2-fold decrease), or unchanged (<2-fold change) amounts, as estimated by the total spectral count, in the interphase and metaphase FAIs in the two mass spectrometry experiments. Blue: proteins displaying consistent changes of at least 2-fold in the two experiments. Green: proteins displaying less than 2-fold change in one or both experiments. Orange: proteins displaying inconsistent changes between the two experiments.
(B) Graphic representation of the changes in total spectral count, as a readout of protein abundance, between interphase and metaphase for proteins found to bind F-actin in the two mass spectrometry experiments (MS1, light blue and MS2, dark blue). Each line corresponds to an individual protein. Proteins that changed by more than 2-fold in one experiment but not the other are not represented.
(C) FAI abundance changes between interphase and metaphase. The graph displays the total spectral count as a function of the ratio of spectral counts between metaphase and interphase for the 48 proteins that display at least a 2-fold change in abundance in both mass spectrometry experiments (MS1, light blue and MS2, dark blue). Total spectral counts for each protein were normalized to the spectral count of actin-β (ACTB) and multiplied by the mean spectral count of ACTB (used as a reference) for each mass spectrometry experiment. When a protein was only present in interphase or metaphase, its spectral count was set to 1 instead of 0, so that the log of the ratio could be computed; the corresponding proteins fall on the red dashed line in the graph; all spectral counts are reported in Table 1. The gray shaded region highlights the area with less than a 2-fold change in spectral count between interphase and metaphase.
Table 1.
List of Proteins Displaying at Least 2-Fold Changes in F-Actin Interactome Abundance between Interphase and Metaphase
| Protein name | Normalized total Spectral Count |
Mean PAI |
|||
|---|---|---|---|---|---|
| MS1 |
MS2 |
MS1-MS2 | |||
| G2 | Metaphase | G2 | Metaphase | ||
| Enriched in interphase | G2 PAI | ||||
| TPM4 | 87 | 0 | 253 | 103 | 515 |
| TPM2 | 79 | 0 | 143 | 0 | 335 |
| MYH9 | 627 | 99 | 549 | 15 | 259 |
| TBB3 | 106 | 0 | 72 | 0 | 178 |
| CALD1 | 103 | 1 | 96 | 1 | 163 |
| TBA4A | 92 | 27 | 52 | 26 | 145 |
| TBB6 | 48 | 0 | 35 | 0 | 83 |
| PPR18 | 32 | 14 | 29 | 11 | 45 |
| MYH10 | 94 | 0 | 95 | 0 | 41 |
| SPTB2 | 125 | 2 | 75 | 23 | 36 |
| VASP | 17 | 3 | 10 | 4 | 35 |
| ANLN | 61 | 0 | 25 | 0 | 35 |
| RLA0 | 11 | 4 | 6 | 1 | 25 |
| MYH14 | 52 | 0 | 48 | 0 | 22 |
| ENAH | 10 | 0 | 9 | 1 | 14 |
| SVIL | 29 | 6 | 18 | 0 | 12 |
| EF1G | 8 | 1 | 2 | 0 | 10 |
| NUCL | 12 | 3 | 2 | 0 | 9 |
| IF4A1 | 5 | 2 | 3 | 0 | 9 |
| SERA | 3 | 1 | 6 | 0 | 8 |
| DNJA1 | 4 | 0 | 2 | 1 | 7 |
| XPO2 | 8 | 0 | 4 | 0 | 6 |
| DOCK7 | 17 | 1 | 8 | 0 | 5 |
| SYEP | 11 | 1 | 6 | 0 | 5 |
| MY18A | 9 | 0 | 10 | 0 | 4 |
| PRKDC | 4 | 0 | 3 | 0 | 1 |
| Enriched in metaphase | M PAI | ||||
| PLEC | 249 | 1,405 | 0 | 638 | 197 |
| COF1 | 0 | 25 | 5 | 12 | 99 |
| GTF2I | 15 | 83 | 0 | 68 | 70 |
| VIME | 15 | 39 | 8 | 27 | 60 |
| ARPC2 | 1 | 14 | 2 | 18 | 47 |
| SRC8 | 5 | 21 | 6 | 34 | 45 |
| ARPC5 | 5 | 10 | 0 | 3 | 39 |
| WDR1 | 11 | 26 | 2 | 22 | 36 |
| ARC1B | 2 | 13 | 0 | 11 | 29 |
| HORN | 9 | 115 | 6 | 23 | 24 |
| RL13 | 1 | 5 | 1 | 3 | 17 |
| DDX5 | 7 | 18 | 1 | 4 | 16 |
| TRI27 | 0 | 3 | 0 | 9 | 14 |
| EPIPL | 0 | 118 | 0 | 39 | 14 |
| EF2 | 0 | 22 | 0 | 5 | 14 |
| RSSA | 2 | 6 | 0 | 3 | 14 |
| CS021 | 4 | 10 | 0 | 4 | 9 |
| TPX2 | 0 | 10 | 0 | 6 | 9 |
| C4BPA | 0 | 7 | 0 | 4 | 8 |
| HNRPU | 2 | 6 | 0 | 3 | 5 |
| AFAP1 | 1 | 4 | 0 | 4 | 5 |
| MICA3 | 1 | 3 | 0 | 5 | 4 |
Normalized total spectral count (normalization as in Figure 2C) for proteins enriched in the interphase (top part) and metaphase (bottom part) FAIs for the two mass spectrometry repeats (MS1 and MS2). The last column displays the mean protein abundance index (PAI), which is calculated as the ratio of the spectral count of the protein of interest, averaged between the two mass spectrometry repeats, and of the molecular weight of the protein. The PAI corrects for differences in the number of observable peptides due to protein size and is thus an adjusted readout of protein abundance.
Consistent with previous observations (Kordowska et al., 2006, Yamashiro et al., 1990, Yamashiro et al., 1991), Caldesmon 1 (CALD1), a myosin II activity regulator, was associated with F-actin in interphase but not in metaphase (Figure 2C and Table 1). As expected, we also found that Ena/VASP, a key stress fiber regulator, was more associated with F-actin in extracts from interphase cells, where stress fibers dominate actin organization (Ciobanasu et al., 2012, Krause et al., 2002). Our analysis further revealed that myosin heavy chains IIA (MYH9), IIB (MYH10), IIC (MYH14), and myosin XVIII (MY18A), as well as different tropomyosins (TPM2 and TPM4), displayed a higher FAI abundance in interphase than in metaphase. Finally, we observed that several subunits of the Arp2/3 complex (ARC1B, ARPC2, and ARPC5), as well as the intermediate filament protein vimentin and the actin-vimentin binding factor plectin, were enriched in the FAI of metaphase cells. Overall, our FAI mass spectrometry analysis uncovered several potential regulators of actin architecture during the cell cycle.
Regulation of Cortical Actin Organization
We then investigated if proteins displaying increased FAI abundance in mitosis (Table 1) affected mitotic actin cortex architecture. For this, we depleted each identified mitotic hit (Figure 2C and Table 1, excluding two ribosomal proteins) and investigated the effect on cellular actin organization. Target proteins were depleted using siRNA pools and mRNA transcript depletion efficiency was confirmed using qPCR (Figure S3A). We observed that none of the factors tested individually led to an overall disruption of the cortex, suggesting that the formation of a cortical actin network in mitosis is robust to perturbations (Figure S3B). We then measured cortical thickness, a key readout of cortical architecture (Chugh et al., 2017) in the different depletion conditions. As cortical thickness is generally below the resolution limit of conventional light microscopy (Clark et al., 2013), we used direct stochastic optical reconstruction microscopy (dSTORM) (Figures 3A and 3B). As dSTORM resolution is ∼40 nm when imaging at the cell center, significantly lower than cortical thickness, we measured the full width at half maximum (FWHM) of actin profiles across the cortex as a readout of thickness (Figures 3B, 3C, and S3C). Depletion of the actin severing protein cofilin (CFL1) led to an increase in thickness (Figures 3B, 3C, and S3C), consistent with previous observations (Chugh et al., 2017, Clark et al., 2013). We further identified several additional regulators of cortex thickness: the transcription factor GTF2I, the oxydoreductase MICAL3, and the spindle assembly factor TPX2 (Figures 3B and S3C). Surprisingly, we also found that depletion of the intermediate filament protein vimentin, and of the F-actin intermediate filament linker plectin, led to increased cortical thickness, suggesting that intermediate filaments play a role in controlling actin architecture in mitosis (Figures 3B, 3C, and S3C). To our knowledge, how intermediate filaments affect mitotic actin organization has not been investigated. Therefore, we focused on vimentin for the rest of the study.
Figure 3.
Regulation of Cortical Actin Organization
(A) Cortex thickness measurements by super-resolution microscopy. Top: STORM image of a mitotic cell stained with phalloidin-Alexa647. Bottom: the cell was automatically segmented and the mean full width at half maximum (FWHM) of intensity profiles across the actin cortex was measured as a readout of actin cortex thickness. Scale bar, 10 μm.
(B) Actin FWHM after depletion of protein candidates. Relative FWHM values for siRNA-depleted cells (target, black data points) were obtained by dividing the FWHM of individual depleted cells by the median of the corresponding non-targeting control siRNA (NTC, gray datapoints). Individual data points as well as median and interquartile range are plotted. N ≥ 3 independent experiments. Gene names are used, instead of protein names as in Tables 1, S1 and Figure 2. Depletion levels were checked by qPCR (Figure S3A). Unpaired Welch’s t test; p values: ∗∗∗p < 0.001; ∗∗p < 0.01.
(C) Examples of STORM images of the actin cortex for treatments leading to increased FWHM. Scale bars: top panel, 10 μm; bottom panel, 2 μm.
Vimentin Localizes to the Cell Cortex in Mitosis in a Plectin-Dependent Manner and Controls Actin Cortex Organization and Mechanics
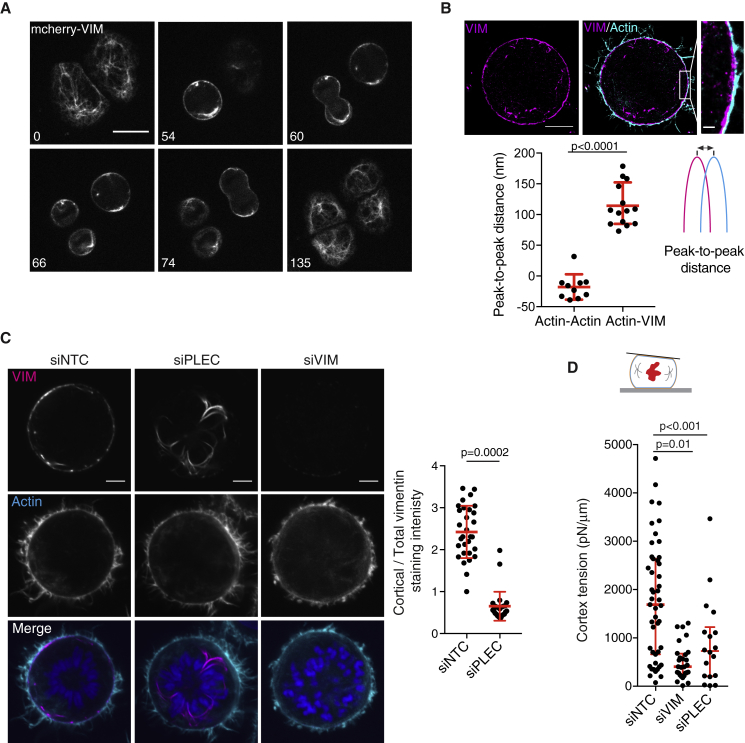
We first characterized vimentin localization and dynamics during mitosis. Previous studies suggested that vimentin networks disassemble during mitosis in some cell types but not in others (Charras et al., 2009, Chou et al., 2003, Duarte et al., 2019). We thus monitored vimentin during the cell cycle in HeLa cells (Video S1). We observed that vimentin distribution changed from a prominent cytoplasmic network in interphase to cytoplasmic aggregates and a clear cortical layer in metaphase (Video S1; Figure 4A). In some cells, the cortical vimentin layer appeared somewhat patchy (Figures 4A and S4A, left panel). 3D visualization revealed that mitotic vimentin formed a network of thick cables at the cell periphery, with a faint vimentin meshwork between the cables (Figure S4A). Finally, we explored the relative localization of vimentin and actin using 2-color dSTORM (Figures 4B and S4B). As also reported in a recent study in several other cell types (Duarte et al., 2019), the vimentin layer was located immediately underneath, and might thus interact with, the actin cortex (Figures 4B and S4B).
Figure 4.
Vimentin Forms a Sub-cortical Layer in Mitosis and Controls Actin Cortex Organization and Mechanics
(A) Representative time lapse of HeLa cells expressing mCherry-vimentin through the cell cycle (images from Video S1). Time in minutes. Scale bar, 20 μm.
(B) Top: representative two-color dSTORM image of vimentin (magenta) and F-actin (cyan) in a mitotic cell. Scale bars, 5 and 1 μm. Bottom: distance between the peaks of the actin and vimentin fluorescence profiles across the cortex (actin-VIM; see STAR Methods for details). To rule out any systematic shift, we also measured peak-to-peak distance for actin profiles where actin was labeled with two differentially labeled phalloidin probes (actin-actin; Figure S4B). Graph: mean ± standard deviation (actin-actin, n = 10 and actin-VIM, n = 14; at least 3 independent experiments). Mann-Whitney test.
(C) Left: representative images of endogenous vimentin localization in mitotic HeLa cells transfected with non-targeting control siRNA (siNTC), siRNA against plectin (siPLEC), or vimentin (siVIM). Scale bars, 5 μm. Right: quantification of vimentin enrichment at the cell cortex. Graph: mean ± standard deviation; 3 independent experiments. siNTC, n = 30 and siPLEC, n = 29. Mann-Whitney test.
(D) Cortex tension in mitotic (prometaphase) cells measured by atomic force microscopy (top schematic, see STAR Methods for details on tension measurement) for cells treated with non-targeting control siRNA (siNTC, n = 47), siRNA against vimentin (siVIM, n = 28), or siRNA against plectin (siPLEC, n = 18). Graph: median and interquartile range; at least 3 independent experiments. Kruskal-Wallis test.
See also Figure S4.
Frames were acquired every 2 min. Scale bar, 20 μm.
We then asked whether plectin, an F-actin-vimentin linker (Wiche et al., 2015) and a key hit in our screen (Figure 3), contributed to recruiting vimentin to the F-actin cortex in mitosis. We observed that plectin formed cytoplasmic networks in interphase cells and reorganized into a cortical layer in mitosis (Figure S4C). We then explored how depleting plectin and vimentin affected their respective localizations (Figures 4C, S4D, and S4E). Interestingly, in plectin-depleted mitotic cells, vimentin did not localize to the cortex but instead accumulated in cytoplasmic bundles (Figure 4C). In contrast, vimentin depletion did not affect mitotic plectin localization (Figure S4E). Thus, plectin appears to be instrumental in recruiting vimentin to the mitotic cortex.
Finally, we explored how the plectin-dependent formation of a vimentin sub-cortical layer affects functional properties of the mitotic actin cortex. The key function of the cortex in mitosis is the generation of high tension driving mitotic rounding. We have shown recently that cortical thickness decreases in mitosis, and this cortex thinning contributes to increasing cortical tension (Chugh et al., 2017). Therefore, we measured cortical tension in vimentin- and plectin-depleted cells using atomic force microscopy. Both treatments resulted in a strong decrease in mitotic cortex tension (Figure 4D), with vimentin depletion bringing cortex tension to levels comparable to the low tensions measured in rounded interphase cells (Figure S4F) (Chugh et al., 2017). As cortical tension also strongly depends on myosin II activity, we verified that vimentin depletion did not significantly affect the levels of total or phosphorylated myosin regulatory light chain (Figures S4G and S4H). Together, these results suggest that in mitotic cells, plectin recruits a vimentin network to the cortex, which results in thinning of the cortical actin layer and contributes to increasing cortical tension.
Phosphorylation Controls the Organization of the Sub-cortical Vimentin Network in Mitosis
We then further explored how the organization of the mitotic sub-cortical vimentin network is regulated. The formation of a sub-cortical vimentin layer in mitosis is not simply a consequence of mitotic rounding, as artificially rounding interphase cells did not lead to recruitment of vimentin to the actin cortex (Figure S5A). This suggests that vimentin reorganization in mitosis is the result of a mitosis-specific signal. The mitotic kinases CDK1 and PLK1 were previously shown to phosphorylate vimentin on Ser56 and Ser83, respectively (Tsujimura et al., 1994, Yamaguchi et al., 2005, Yasui et al., 2001). We thus asked how mitotic kinase activity affects vimentin organization and found that both CDK1 inhibition using RO-3306 and PLK1 inhibition using BI2536 disrupted vimentin organization in mitosis, converting the network of cortical vimentin cables into a more diffuse cortical meshwork (Figure S5B; Video S2; Video S3). Furthermore, staining with phospho-specific antibodies showed that vimentin phosphorylated on Ser56 and Ser83 localized to the cortex in mitosis, at levels similar to but not higher than total vimentin (Figure S5C). Together, these experiments suggest that phosphorylation is unlikely to target vimentin to the cortex but that it could regulate the organization of the sub-cortical vimentin network.
The video displays projected z-stacks for each time point. Frames were acquired every 1 min. Cells were synchronized in pro-metaphase using STLC and treated with the CDK1 inhibitor RO3306 immediately before the movie. Scale bar, 10 μm.
To test this, we generated cell lines stably overexpressing GFP-tagged wild-type vimentin (WT-VIM) or vimentin with single point mutations that either mimic phosphorylation by CDK1 and PLK1 (VIM-56E and VIM-83E, respectively) or abolish CDK1 phosphorylation (VIM-56A) (Figure S5D). All three mutants formed cytoplasmic networks in interphase, suggesting that the mutant vimentins normally integrate into filaments, but VIM-56E-GFP and VIM-83E-GFP also often displayed cytoplasmic aggregates, indicating that the phosphomimetic mutations affect vimentin network organization in interphase (Figure S5E). In mitosis, expression of VIM-WT, -56A, or -83E did not significantly alter cortical vimentin localization (Figure 5A). In contrast, expression of VIM-56E reduced the cortical accumulation of vimentin in mitosis (Figure 5A). Analysis of 3D stacks indicated that phosphomimetic VIM-56E induced the collapse of vimentin cables into aggregates (Figure 5B; Video S4); the aggregates were localized close to the cortex, explaining the patchy appearance of the vimentin staining in single plane images (Figure 5A). Finally, the localization of the mutant vimentins themselves mostly recapitulated the distribution of total vimentin, with the exception of VIM-83E, which displayed a slight decrease in cortical accumulation compared to total vimentin (Figures 5A and S5F). Together, these results indicate that vimentin phosphorylation on Ser56, and to a milder degree on Ser83, controls the organization of the vimentin network in mitosis leading to the formation of a bundle-like meshwork, with excessive phosphorylation resulting in aggregation of vimentin into localized spots.
Figure 5.
Sub-cortical Vimentin Directly Controls Cortical Actin Organization in Mitosis
(A) Left: representative images of total vimentin (immunostaining) in mitotic Hela cells stably expressing GFP-vimentin mutants. Right: corresponding quantification of vimentin enrichment at the cell cortex. Scale bar, 5 μm. Right: graph, mean ± standard deviation; 3 independent experiments. WT, n = 31; 56A, n = 31; 56E, n = 33; and 83E, n = 29. Kruskal-Wallis test; ns, non-significant.
(B) Representative maximum z projections of confocal stacks across mitotic HeLa cells stably expressing vimentin mutants and stained for total vimentin. Scale bar, 10 μm. See Video S3 for 3D visualizations.
(C) Left: examples of STORM images of the actin cortex in cells expressing the different vimentin mutants. Scale bars, 0.5 and 5 μm. Right: values of full width at half maximum (FWHM) of the cortical actin signal in cells expressing the different vimentin mutants. NT, n = 16; WT, n = 26; 56E, n = 20; 56A, n = 19; 83E, and n = 21. 3 independent experiments. Graph: mean ± standard deviation. ANOVA Tukey’s multiple comparison.
(D) Cortex tension in mitotic (prometaphase) cells expressing VIM-WT, VIM-56E, or VIM-83E. Graph: median with interquartile range; 3 independent experiments. WT, n = 60; 56E, n = 29; and 83E, n = 26. Kruskal-Wallis test.
(E) Left: representative time lapses of curvature maps along the cell contour (see STAR Methods for details) during ablation experiments in HeLa cells stably expressing GFP-VIM-WT or GFP-VIM-56E (displayed in insets; yellow circles, ablation region). Right: quantification of the effects of laser ablation in HeLa cells expressing GFP-VIM-WT or GFP-VIM-56E. N = 4 independent experiments for VIM-WT (n = 52 cells) and N = 3 for VIM-56E (n = 57 cells). In some cells (15 cells out of 52 for VIM-WT and 26 cells out of 57 for VIM-56E), ablation elicited bleb formation at the site of ablation (Video S7, right panel); these cells were excluded from the quantifications, as bleb formation indicated ablation disrupted the actin cortex (Tinevez et al., 2009). Scale bars, 5 μm.
See also Figure S5.
The Sub-cortical Vimentin Layer Directly Controls Cortical Actin Organization
We then asked whether expression of the phosphomimetic vimentin mutants affected the actin cortex. Interestingly, we found that actin cortex thickness was increased in cells expressing VIM-56E and VIM-83E (Figure 5C). Furthremore, cortical tension was decreased in cells expressing VIM-56E, though not VIM-83E (Figure 5D), possibly because of the weaker effect of this mutant on actin organization compared to the VIM-56E mutant (Figures 5A and 5B). These results suggest that the sub-cortical vimentin layer directly controls the arrangement of the actin cortex in mitosis.
Because vimentin forms stiff elastic networks, we speculated the sub-cortical vimentin layer might mechanically counteract the cortex. To test this hypothesis, we used laser ablation to locally disrupt the vimentin layer in mitotic cells. We first verified that with our settings, laser ablation did not disrupt the plasma membrane (Video S5). Given the proximity of the vimentin and actin layers, the ablation of the vimentin layer occasionally led to the disruption of the actin cortex, resulting in bleb formation (Video S6, lower panel), consistent with previous observations (Tinevez et al., 2009). When no bleb was observed (indicating that the actin cortex was not disrupted, Video S6), we found that in cells expressing VIM-WT, local ablation focused on sub-cortical vimentin led to local flattening of the cell surface, suggesting a local contraction of the cortex (18 cells out of 35, Figure 5E; Video S7). In contrast, cortex flattening was only seldom observed upon similar laser ablation in cells expressing VIM-56E, where the levels of vimentin at the cortex are reduced (9 cells out of 31, Figure 5E; Video S7). Finally, inhibiting actomyosin contractility with the ROCK inhibitor Y27632 (Tinevez et al., 2009) strongly decreased occurrences of cortex flattening upon sub-cortical ablation (Figure S5G). These experiments suggest that disrupting the sub-cortical vimentin layer locally releases mechanical resistance to cortical forces, leading to a contraction of the overlaying actomyosin cortex and cell surface flattening. We thus propose that the sub-cortical vimentin network could contribute to mitotic cortex thinning by physically resisting the growth of the cortical actin meshwork toward the cytoplasm.
Ablation was performed in HeLa cells stably expressing GFP-vimentin-WT and in presence of Cell Mask to monitor the plasma membrane during ablation (left panel) or in presence of fluorescent dextran in the medium (right panel). The yellow circle represents the site of ablation.
Ablation was performed in HeLa cells stably expressing GFP-vimentin-WT (left panel) and transfected with mCherry-Lifeact to monitor the actin cortex during ablation (right panel). The yellow circle represents the site of ablation.
Ablation was performed in HeLa cells stably expressing GFP-vimentin-WT or -56E. The yellow circle represents the site of ablation. Frames were acquired every 3.26 s and the ablation was performed at 25 s (left panel) and at 9s (middle panel and right panels). Scale bars, 5 μm.
Vimentin Promotes Mitotic Cell Rounding
Finally, we asked if the changes in cortex organization and tension upon vimentin depletion had functional consequences on mitotic rounding. We observed that vimentin-depleted cells were less round in mitosis than control cells (Figure 6A) and that the rounding-up process was slower compared to control cells (Figure 6B; Video S8). This shows that vimentin plays a role in mitotic rounding, possibly by promoting cortex thinning and the concomitant cortex tension increase.
Figure 6.
Vimentin Is Important for Mitotic Cell Rounding
(A) Representative images of control and vimentin-depleted cells in metaphase and quantification of cell roundness (see STAR Methods); R on the images is the roundness coefficient of the cell depicted. Scale bars, 10 μm. Graph: mean ± standard deviation (siNTC, n = 21 and siVIM, n = 23; 3 independent experiments). Non-parametric Mann-Whitney test.
(B) Representative time lapses of mitotic rounding in control and vimentin-depleted HeLa cells stably expressing H2B-mCherry (red) and Lifeact-GFP (cyan). Scale bars, 20 μm. Right: quantification of rounding timing. Graph: mean ± standard deviation (NT, n = 16; siNTC, n = 22; and siVIM, n = 63. 2 independent experiments). NT = non-transfected. ANOVA Tukey’s multiple comparison; ns, non-significant.
(C) Representative images of HeLa cells stably expressing H2B-mCherry to mark the chromosomes, and aplasma membrane marker (membrane targeting domain of LYN tagged with GFP), dividing unconfined or in confinement under a 5 kPa polyacrylamide gel. Scale bars, 10 μm.
(D) Quantification of chromosome segregation defects in control and vimentin-depleted cells during division without confinement (no gel) and in confinement under 5 kPa gels. Percentages of cells with lagging chromosomes, chromosomal bridges or multipolar spindles were quantified in time lapse movies of cells stably expressing H2B-mCherry (N ≥ 3 independent experiments; no gel, n = 64 and n = 65 and 5 kPa gel, n = 112 and n = 101 for siNTC and siVIM, respectively).
(E) Representative images of lagging chromosomes, chromosomal bridges, and multipolar spindles in HeLa cells stably expressing H2B-mCherry (inverted contrast) to mark the chromosomes, dividing in confinement under a 5-kPa polyacrylamide gel. Scale bar, 10 μm.
See also Figure S5H.
Frames were acquired every 2 min. DNA (red); F-actin (cyan); z-projections are diaplayed. Scale bar, 20 μm.
Finally, we investigated whether the rounding defects observed upon vimentin depletion interfered with cell division. We did not observe any apparent defects for cells dividing in a dish (Figures 6C, 6D, and S5H; “no gel” conditions). However, mitotic rounding is particularly crucial for cells dividing in confinement, where external pressure interferes with spindle assembly and chromosome segregation if rounding is defective (Lancaster et al., 2013). We thus confined the cells using 5 kPa polyacrylamide gels mimicking the mechanical environment of a tissue (Figure 6C). This range of stiffness allowed for normal rounding and limited mitotic defects in control cells (Figures 6D and 6E), consistent with previous observations (Lancaster et al., 2013). Strikingly, confined vimentin-depleted cells displayed increased occurrences of chromosome missegregation and multipolar spindles (Figures 6C–6E). Taken together, our experiments indicate that by affecting actin cortex organization and mechanics, vimentin is involved in controlling mitotic cell rounding, essential for division accuracy in 3D confinement.
Discussion
To systematically investigate the regulation of cortical actin organization in mitosis, we established a method to identify proteins that bind filamentous actin in interphase and mitotic cells. Of note, in our protocol, F-actin is first depolymerized for extract preparation and then re-polymerized for recovery of the F-actin binding fraction on phalloidin-coated beads (Figure 1). As a result, the organization of actin filaments on the beads is unlikely to recapitulate actin network organization in the original, spread or rounded cells. Thus, while our assay recovers proteins present and able to bind F-actin in the specific cell-cycle phases investigated, differences in F-actin binding due to the actual organization of cellular networks might be missed. Importantly, our assay cannot disentangle differences in actin binding from differences in protein abundance in the soluble extracts, though comparing differences in protein levels and FAI abundance for a subset of proteins did not reveal any clear correlation (Figure 2). Nonetheless, our FAI analysis identified a list of F-ABP candidates for the regulation of mitotic cortex organization (Table 1).
We then performed a quantitative super-resolution screen to test the effects of the candidate regulators on cortex architecture (Figure 3). Strikingly, we found that the depletion of the intermediate filament protein vimentin and the actin-vimentin linker plectin increase mitotic cortex thickness (Figure 3B). This was surprising, as some studies had suggested that the vimentin network is disassembled in mitosis (Chou et al., 1989, Inagaki et al., 1987, Takai et al., 1996). In the cells used in our study, we observed a clear mitotic vimentin layer directly underneath the actin cortex (Figures 4A and 4B), in agreement with early observations suggesting a cortical vimentin layer in mitosis (Celis et al., 1983), and a recent study showing that vimentin and actin interact in mitosis (Duarte et al., 2019). We further show that the formation of a sub-cortical vimentin layer in mitosis depends on the vimentin-actin linker plectin (Figures 4C and S4E). Mitosis-specific phosphorylation of plectin has been shown to reduce its affinity for vimentin, with a large proportion of cellular plectin becoming soluble (Foisner et al., 1996). Our immunostainings may indicate a large cytoplasmic pool (Figure S4E), which would imply that plectin-driven recruitment to the cortex in mitosis relies on only a portion of cellular plectin.
Our screen identified a number of regulators for which knock down led to actin cortex thickening. In a previous study, we had shown that the actin cortex becomes thinner in mitosis and that this thinning is important for increasing cortical tension (Chugh et al., 2017). Thus, our assay identifies potential regulators of mitotic cortex mechanics. Consistently, the increase in cortical thickness upon vimentin and plectin depletion correlated with a decrease in cortical tension (Figure 4D). Interestingly, a previous study of vimentin contribution to cell mechanics had shown that vimentin does not affect cortical stiffness in spread interphase cells (Guo et al., 2013); this suggests that vimentin cortical localization and function in cortex mechanics could be specific to mitosis. We further found that impairing the formation of the sub-cortical vimentin layer using phosphomimetic vimentin mutants also led to increased cortical thickness and decreased cortical tension (Figures 5C and 5D). Vimentin may control mitotic actin biochemically (Hubert et al., 2011). However, our ablation experiments suggest that the effect of the sub-cortical vimentin layer on the cortical actin network could be at least partly mechanical, with the vimentin network physically resisting the growth of the actin cortex toward the cytoplasm. It will be interesting to investigate how exactly vimentin and actin interact, biochemically and mechanically, at the cortex.
Our study identified vimentin as a key regulator of mitotic cortex tension. High cortical tension in mitosis is thought to be instrumental to support a rounded cell shape, generating space for accurate spindle positioning and chromosome segregation (Ramkumar and Baum, 2016). This is particularly important in confinement, when cells must push against surrounding structures in order to round up (Lancaster et al., 2013). Consistent with this, we observed that vimentin-depleted cells displayed rounding defects (Figures 6A and 6B) and chromosome segregation defects when placed in 3D confinement (Figures 6C–6E). Interestingly, vimentin overexpression is a hallmark of various cancers (Heerboth et al., 2015, Satelli and Li, 2011). Vimentin function in cancer has so far been associated with epithelial-to-mesenchymal transitions and cell migration (Huber et al., 2015, Leduc and Etienne-Manneville, 2017, Mendez et al., 2010). Our data suggest that high vimentin levels might also facilitate cell division in strongly confined and stiff environments, such as those experienced by proliferating cancer cells (Levental et al., 2009).
Taken together, our unbiased approach reveals a key function for vimentin in controlling cortex architecture and mechanics in mitosis. It is also possible that vimentin contributes to mitotic rounding by regulating focal adhesions (Burgstaller et al., 2010, Mendez et al., 2010) or by affecting other mechanical properties of the actin cortex, such as viscosity or turnover. It will be exciting to investigate how the mechanical and biochemical integration of actin networks with intermediate filaments, and possibly other intracellular structures, controls actin organization during mitosis and cell morphogenetic events in general.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies and Dyes | ||
| mouse anti-cyclin B | Santa Cruz Biotechnology | Santa Cruz Biotechnology sc-245; WB 1/1000; RRID:AB_627338 |
| mouse anti-β-actin | Santa Cruz Biotechnology | Santa Cruz Biotechnology sc-47778; WB 1/1000; RRID:AB_2714189 |
| mouse anti-GAPDH | Abcam | Abcam ab8245; WB 1/4000; RRID:AB_2107448 |
| rabbit phospho-Histone H3 S10 | Cell Signaling Tech | Cell Signaling Tech 9701; WB 1/1000; RRID:AB_331535 |
| mouse anti-MRLC pS19 | Cell Signaling Tech | Cell Signaling Tech 3675; WB 1/1000; RRID:AB_2250969 |
| rabbit anti-MRLC | Cell Signaling Tech | Cell Signaling Tech 3672; WB 1/1000; RRID:AB_10692513 |
| rabbit anti-vimentin | Cell Signaling Tech | Cell Signaling Tech 3932; WB 1/1000 – IF 1/100; RRID:AB_2288553 |
| mouse anti-α-tubulin | ThermoFisher Scientific | ThermoFisher Scientific DM1A; WB 1/1000; RRID:AB_1954824 |
| rabbit-anti phospho (Ser)-CDK1 substrate | Cell Signaling Tech | Cell Signaling Tech 9477; WB 1/1000; RRID:AB_2714143 |
| rat anti-GFP | Chromoteck | Chromotek; WB 1/1000; RRID:AB_10773374 |
| rabbit anti-VASP | Cell Signaling Tech | Cell Signaling Tech 3132S; WB 1/1000; RRID:AB_2213393 |
| rabbit anti-MENA | Santa Cruz Biotechnology | Santa Cruz Biotechnology sc-135988; WB 1/1000; RRID:AB_2098633 |
| mouse anti-MYH9 | Abnova | Abnova; WB 1/1000 |
| mouse anti-caldesmon | Abcam | Abcam ab183146; WB 1/1000 |
| mouse anti-fascin | Santa Cruz Biotechnology | Santa Cruz Biotechnology sc-21743; WB 1/1000; RRID:AB_627580 |
| rabbit anti-filamin | Cell Signaling Tech | Cell Signaling Tech 4762S; WB 1/1000; RRID:AB_2106408 |
| rabbit anti-cofilin | Cell Signaling Tech | Cell Signaling Tech 3318S; WB 1/1000; RRID:AB_2080595 |
| Rabbit anti Phospho-Vimentin (Ser83) | Cell Signaling Tech | Cell Signaling Tech #12569; RRID:AB_2797957 |
| Rabbit anti Phospho-Vimentin (Ser56) | Thermo Fisher Scientific | Thermo Fisher Scientific 711212; RRID:AB_2633001 |
| Rabbit anti Vimentin conjugated with Alexa Fluor 647 | Cell Signaling Tech | Cell Signaling Tech #5741; RRID:AB_10695459 |
| Goat anti-WDR1 | Santa Cruz Biotechnology | Santa Cruz Biotechnology sc-160907; WB 1/500; RRID:AB_2215093 |
| rabbit anti-plectin | Thermo Fisher Scientific | Thermo Fisher Scientific #PA556292 IF 1/100; RRID:AB_2645646 |
| mouse anti-α-tubulin | Sigma | Sigma T5168; IF 1/2000; RRID:AB_477579 |
| Phalloidin conjugated with Alexa Fluor 568 | Life Technology | Life Technology A12380 |
| Phalloidin conjugated with Alexa Fluor 647 | Life Technology | Life Technology A22287 |
| DAPI | Thermo Fisher Scientific | Molecular Probes D1306 |
| Alexa Fluor 680 | Thermo Fisher Scientific | Life Technology A21058 |
| Alexa Fluor 790 | Thermo Fisher Scientific | Life Technology A11369 |
| Cell Mask membrane stain | Thermo Fisher Scientific | Invitrogen C10046 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RNAi max lipofectamine | Thermo Fisher Scientific | Thermo Fisher Scientific 13778075 |
| Fugene HD | Promega | Promega E2311 |
| Thymidine | Sigma | Sigma T1895 |
| RO3306 | Merck Millipore | Calbiochem 217699 |
| BI 2536 | Santa Cruz Biotechnology | Santa Cruz Biotechnology sc-364431 |
| Y27632 | Tocris | Tocris 1254 |
| Dextran | Thermo Fisher Scientific | Thermo Fisher Scientific D22914 |
| Non conjugated Phalloidin | Sigma | Sigma – P2141 |
| Experimental Models: Cell Lines | ||
| HeLa Kyoto cells | Research Institute of Molecular Pathology (Vienna, Austria) | N/A |
| Hela Kyoto cells - H2B-mCherry | Su et al., 2011 | N/A |
| Hela Kyoto cells - H2B-mCherry - GFP-Lifeact | This paper | N/A |
| Hela Kyoto cells - GFP-vimentin WT and mutants cell lines | This paper | N/A |
| Oligonucleotides | ||
| SiRNA used in this study | This paper | Table S2 |
| Primers used in this study | This paper | Table S2 |
| point mutations in the human vimentin cDNA 5′- gc agc ctc tac gcc tcg GCC ccg ggc ggc gtg tat gcc a -3′ | This paper | N/A |
| point mutations in the human vimentin cDNA 5′- t ggc ata cac gcc gcc cgg GGC cga ggc gta gag gct gc - 3′ | This paper | N/A |
| point mutations in the human vimentin cDNA 5′- gc agc ctc tac gcc tcg GAA ccg ggc ggc gtg tat gcc a -3′ | This paper | N/A |
| point mutations in the human vimentin cDNA 5′- t ggc ata cac gcc gcc cgg TTC cga ggc gta gag gct gc - 3′ | This paper | N/A |
| point mutations in the human vimentin cDNA 5′- g gtg cgg ctc ctg cag gac GAG gtg gac ttc tcg ctg gcc -3′ | This paper | N/A |
| point mutations in the human vimentin cDNA 5′- ggc cag cga gaa gtc cac CTC gtc ctg cag gag ccg cac c - 3′ | This paper | N/A |
| Recombinant DNA | ||
| Lifeact-GFP plasmid | Matthieu Piel (Institut Curie, France) | N/A |
| pIRESpuro3 Flag–AcGFP- vimentin mutant plasmids | This paper | N/A |
| Software and Algorithms | ||
| Fiji | N/A | https://imagej.nih.gov/ij/ |
| Matlab | Mathworks | MathWorks |
| Prism | GraphPad | GraphPad |
| Icy | N/A | http://icy.bioimageanalysis.org/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ewa Paluch (ekp25@cam.ac.uk).
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Experimental Model and Subject Details
Cell Lines and Culture
All cell lines were grown at 37°C with 5% CO2 in DMEM GlutaMAX, 4.5 g/l glucose (Gibco, Invitrogen/ Life Technologies) supplemented with 10% fetal bovine serum (Sigma), 1% penicillin-streptomycin (Gibco, Invitrogen/ Life Technologies) and regularly tested for mycoplasma. HeLa Kyoto cells were obtained from the Research Institute of Molecular Pathology (Vienna, Austria). For the stable Lifeact cell line, a Lifeact-GFP plasmid kindly provided by Matthieu Piel (Institut Curie) was transfected into HeLa Kyoto cells stably expressing H2B-mCherry (kindly provided by Kuan-Chung Su (Su et al., 2011)) using FuGENE 6 (Roche) according to the manufacturer’s instructions. HeLa Kyoto cells stably expressing H2B-mCherry and GFP-Lifeact were grown in DMEM supplemented with 500 μg/ml G418 (Invitrogen), and 0.35 μg/ml puromycine to maintain selection. For the stable GFP-vimentin cell lines, GFP-vimentin mutant plasmids were transfected into HeLa Kyoto cells using FuGENE HD (Roche) according to the manufacturer’s instructions. Hela Kyoto cells stably expressing GFP-Vimentin were grown in DMEM supplemented with 0.35 μg/ml puromycine to maintain selection. Cell lines have not been authenticated.
Method Details
siRNA Transfection
For siRNA transfection, RNAi max lipofectamine (Invitrogen) reverse transfection was performed according to the manufacturer’s instructions. Final concentrations of 30 nM siRNA were used for all the experiments. SiRNAs used in this study are listed in Table S2.
Preparation of GFP-Vimentin Mutants
All vimentin mutants (S56A, S56E, S83E) were generated using the QuikChange methodology (Stratagene, La Jolla, CA). The following oligonucleotides were used to create the different point mutations in the human vimentin cDNA (capital letters indicate the codon mutated). 56A: Forward: 5’- gc agc ctc tac gcc tcg GCC ccg ggc ggc gtg tat gcc a -3’; Reverse: 5’- t ggc ata cac gcc gcc cgg GGC cga ggc gta gag gct gc - 3’. 56E: Forward: 5’- gc agc ctc tac gcc tcg GAA ccg ggc ggc gtg tat gcc a -3’; Reverse: 5’- t ggc ata cac gcc gcc cgg TTC cga ggc gta gag gct gc - 3’. 83E: Forward: 5’- g gtg cgg ctc ctg cag gac GAG gtg gac ttc tcg ctg gcc -3’; Reverse: 5’- ggc cag cga gaa gtc cac CTC gtc ctg cag gag ccg cac c - 3’. The mutated cDNAs were fully sequenced to confirm the sole presence of intended mutations and inserted into pIRESpuro3 Flag–AcGFP-tagged plasmid.
Cell Synchronization for Mass Spectrometry
To prepare cell extracts from specific phases of the cell cycle, HeLa cells were first synchronized in early S phase with thymidine (Sigma) at 2 mM for 22 hours. For synchronization in G2 phase, cells were released for 6hrs and treated with CDK1 inhibitor RO3306 (Calbiochem) at 10 μM for 3 hours. To synchronize cells in metaphase, the cells were released from a single thymidine block for 6hrs, then treated with nocodazole 50 μg/ml for 4 h, and then with 10 μM of MG132 for 2 h as previously described (Petronczki et al., 2007). In all cases, cells were washed twice with DMEM between each treatment and wash with PBS before processed for experiment.
Extracts Preparation
Around 20 millions of cells were first mechanically lysed for 30min by vortexing and pipetting-up-and-down in 300 μL of buffer containing 10 mM HEPES pH 7.5, 83 mM KCl, 17 mM NaCl and 0.1% Triton X-100 supplemented with proteases and phosphatases inhibitors: Microcystin, 10 μg/ml of Leupeptin, Pepstatin and 10 mM of β-glycerophosphate. To solubilize F-actin network, 200 nM of LatrunculinA was added to the lysis. After centrifugation at 28 000 rpm with TLA-120.1 rotor (Beckman Coulter) for 20 min, pellets were resuspended in 300 μL of same lysis buffer without Triton X-100 and mechanically lysed again for 15 min. After a second centrifugation, pellets were sonicated, both supernatants were pooled and frozen in liquid nitrogen. Latrunculin A was present throughout extract preparation, limiting the possibility of actin loss through repolymerization. Lysis conditions were optimized using cells enriched in mitosis (overnight nocodazole treatment). Protein concentrations were determined using Bradford protein assay reagent (Biorad). For western blotting, supernatant and pellets from the preparation were then resuspended in 5X loading buffer (312.5 mM Tris-HCl pH 6.8, 50% glycerol, 5% SDS, 5% β-mercapto ethanol, 0.25% Bromophenol blue), and boiled for 5 min. Pellets amounts loaded for immunoblotting were adjusted to be the same amount as the supernatant.
Phalloidin Pull-Down Assay
Extracts were cleared by centrifugation at 30,000 g for 5 min and 700 μg of each extract were used per condition. We verified that no significant amount of actin was lost from the extract at this step. Pull-down assay was performed using a phalloidin affinity matrix (phalloidin coupled through a flexible polyethylene glycol linker to superparamagnetic beads) (Samwer et al., 2013). Control and Phalloidin matrix were pre-washed 2 times with 10 mM HEPES pH 7.5, 83 mM KCl, 17 mM NaCl and 0.05% Triton-100. Extracts and beads were incubated for 1 hours at room temperature with 2 mM MgCl2 and 1 mM ATP to induce actin polymerization (Samwer et al., 2013). Of note, the concentration of phalloidin on the beads was sufficient to effectively induce actin polymerization even though a small amount of Latrunculin was present in the extract (see section above). After the incubation, beads were washed 2 or 3 times for 2 min with wash buffer (10 mM HEPES pH 7.5, 83 mM KCl, 17 mM NaCl and 0.05% Triton-100) and eluted using 25 μl of loading buffer (125 mM Tris-HCl pH 6.8, 3% SDS, 50 mM DTT, 1 M sucrose, 0.1% bromophenol blue). Samples were boiled for 3 min, separated by SDS-PAGE (NuPage Invitrogen) and stained by colloidal Coomassie (Neuhoff et al., 1988, Samwer et al., 2013).
Mass Spectrometry and Data Analysis
Coomassie-stained gel bands were excised and subjected to in-gel trypsin digestion, as described previously in (Galan et al., 2014). The resulting peptides were extracted and subjected to capillary LC-MS/MS using a high-resolution Orbitrap Fusion Tribrid (Thermo Scientific, San Jose, CA). Raw files were searched with MaxQuant (Cox and Mann, 2008) software version 1.3.0.3 and searched against the Uniprot Human database (http://www.uniprot.org/) release 2016_02 (17-Feb-2016) including the reversed database with 74508 entries. General false discovery rate (FDR) for peptides was at 1.0% with decoy removal. Searches were performed at a 25-ppm precursor ion tolerance and 1.0 Da fragment ion tolerance, assuming full tryptic digestion with up to three missed cleavages. Correct peptide identifications were distinguished from incorrect using the target-decoy approach, coupled with linear discriminant analysis as described previously. Once peptides were filtered to an initial FDR of 1%, peptides were assembled into proteins and further filtered to a final protein-level FDR of 1%. All proteins of the same family were grouped and isoforms of the same protein were considered as one. Finally, contaminants (e.g. keratins) and proteins more represented in control beads eluates were removed (see Table S1). To estimate the abundance of proteins within eluates, we rely on total spectral counts and a modified Protein Abundance Index (PAI) (Biro et al., 2013, Bovellan et al., 2014, Rappsilber et al., 2002). PAI was calculated based on total spectral counts as follows: PAI = total spectral count/MW, where MW corresponds to the protein molecular weight, which was used to adjust for differences between proteins in the number of observable peptides. Mass spectrometry data were also analyzed using Gene ontology enrichment analysis and panther protein classification system (Mi et al., 2017)
Immunoblotting
Protein samples were subjected to SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). After blocking with PBS containing 0.1% Tween-20 and 5% milk, membranes were incubated overnight with primary antibodies, washed in PBS containing 0.1% Tween-20 and then incubated 1 h with HRP-conjugated secondary antibodies (GE Healthcare; 1/5000). After washes with PBS containing 0.1% Tween-20, immunodectection was performed with Amersham ECL Western Blotting Detection Reagent and Amersham Hyperfilm (GE Healthcare). Membrane stripping was performed using stripping buffer containing glycine, 0.01% SDS and 0.01% Tween pH 2.2. For fluorescent immunoblotting, nitrocellulose membranes (Thermo Scientific) and secondary antibodies conjugated to IRDye 800 (LI-COR) or Alexa Fluor 680 and Alexa Fluor 790 (Life Technology) were used. Immunodetection was performed using an Odyssey infrared imaging system (LI-COR). The following primary antibodies were used: mouse anti-cyclin B (Santa Cruz Biotechnology sc-245; 1/1000); mouse anti-β-actin (Santa Cruz Biotechnology sc-47778; 1/1000); mouse anti-GAPDH (Abcam ab8245; 1/4000); phosphoHistone H3 S10 (Cell Signaling Tech 9701; 1/1000); mouse anti-MRLC pS19 (Cell Signaling Tech 3675; 1/1000); rabbit anti-MRLC (Cell Signaling Tech 3672; 1/1000); rabbit anti-vimentin (Cell Signaling Tech 3932; 1/1000); mouse anti-plectin (Thermo Fisher Scientific PA556292 1/1000); mouse anti-α-tubulin (ThermoFisher Scientific DM1A; 1/1000); rabbit-anti phospho (Ser) CDK1 substrate (Cell Signaling Tech 9477; 1/1000); rat anti-GFP (Chromotek; 1/1000); rabbit anti-VASP (Cell Signaling Tech 3132S; 1/1000); rabbit anti-MENA (Santa Cruz Biotechnology sc-135988; 1/1000); mouse anti-MYH9 (Abnova; 1/1000) ; mouse anti-caldesmon (Abcam ab183146; 1/1000); mouse anti-fascin (Santa Cruz Biotechnology sc-21743; 1/1000), rabbit anti-filamin (Cell Signaling Tech 4762S; 1/1000), rabbit anti-cofilin (Cell Signaling Tech 3318S; 1/1000) and WDR1 (Santa Cruz Biotechnology sc-135988; 1/500).
Immunofluorescence
Cells were seeded in labteck chambers (Thermo Scientific) or on coverslips (No 1.5, 170 nm). For CDK1 or PLK1 inhibition experiments, STLC treated cells were treated with DMSO, RO3306 (Calbiochem) at 10 μM for 20 min or BI2536 (Santa Cruz Biotechnology) at 100 nM for 2 h. Cells were fixed in 4% PFA in PBS containing 0.01 Triton X-100 (PBST) for 15 min at room temperature. After washes in PBST, cells were permeabilized in PBS 0.2% Triton X-100 for 10 min and incubated with primary antibodies for 1h at room temperature. After washes in PBST, cells were then incubated with secondary antibodies and/or phalloidin conjugated with Alexa fluorescent dyes. The following primary antibodies were used: rabbit anti-vimentin (Cell signaling Tech 3932; 1/100); mouse anti-plectin (Thermo Fisher Scientific #PA556292 1/100); Rabbit anti Phospho-vimentin (Ser83) (Cell Signaling Tech 2569; 1/100); Rabbit anti Phospho-vimentin (Ser56) (Thermo Fisher Scientific 711212; 1/100); mouse anti-α-tubulin (Sigma T6024; 1/2000); rabbit anti-vimentin conjugated with Alexa Fluor 647 (Cell Signaling Tech 5741; 1/100). Fluorescent dyes used were Phalloidin conjugated with Alexa Fluor 488 (Life Technology A12379); Phalloidin conjugated with Alexa Fluor 568 (Life Technology A12380); Phalloidin conjugated with Alexa Fluor 647 (Life Technology A22287) and DAPI (Molecular Probes) to stain DNA. Secondary antibodies conjugated to Alexa Fluor 488 or 568 (Life Technology; 1/500) were used for immunofluorescence microscopy detection. Coverslips were mounted using Prolong Gold Antifade (Invitrogen). For cortex localization analysis, a single plane in the middle of the cell was used for the quantification of mean cortex intensity. Cell cortex intensity was calculated using Fiji software as the mean intensity of 0.4 μm2 circles placed all along the cell cortex, normalized by the mean intensity of the entire cell.
For one-color STORM experiments (measurements of cortical thickness screen), cells were synchronized using thymidine (Sigma) at 2 mM for 22 h. Then, cells were rinsed twice and fresh media was added for the release for 9 h. For the one-color STORM experiments on GFP-vimentin mutants, cells were pre-treated with STLC for 6h to enrich for mitotic cells. Cells were then permeabilized and fixed at the same time in 4% PFA, 0.2% Triton X-100 in Cytoskeleton buffer pH to 7.4 (10 mM MES, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA) supplemented with sucrose (Cytoskeleton buffer with sucrose; CBS) for 6 min and then 14 min in 4% PFA in CBS. After 3 washes in CBS, cells were incubated 20 min with phalloidin conjugated with Alexa-647 (Life Technology A22287). Coverslips were mounted using Vectashield (Vectorlabs).
For two-color STORM experiments, cells were seeded onto glass-bottom petri dishes (Ibidi) and fixed, permeabilized and washed as described above for one-color STORM experiments. Cells were then incubated with vimentin antibody conjugated with Alexa 647 (Cell signaling Tech 9856; 1/1000) and phalloidin conjugated with Alexa 568 (Life Technology A12380) for 30 min. For imaging, samples were kept in STORM buffer (Tris base 50 mM, NaCl 100 mM, MEA 1M, supplemented with Glucose, Catalase and Oxidase).
STORM Imaging, Rendering, and Analysis
One-color STORM imaging: For the thickness screen, raw images were acquired on an N-STORM microscope (Nikon) using a x100 objective (1.49 numerical aperture CFI Apo TIRF). DAPI staining was used to select cells in metaphase. For actin cortex thickness measurements in cells expressing GFP-vimentin and GFP-vimentin mutant constructs, raw images were acquired on a Zeiss Elyra microscope in STORM mode using a x100 objective (1.46 numerical aperture alpha Plan-Apochromat). Prometaphase cells were selected based on DNA appearance in bright field microscopy. Coordinates of single molecules in the raw STORM images were detected using the Nikon microscope software (thickness screen) or the Zen software (GFP-vimentin mutants). Drift, chromatic aberration correction and super-resolution rendering were performed using a custom-written script in MATLAB (MathWorks).
Two-color STORM: raw images were acquired on a Zeiss Elyra microscope in STORM mode using a x100 (1.46 numerical aperture alpha Plan-Apochromat) objective. To correct for chromatic aberration and drift during the acquisition, multi-color fluorescent beads (TetraSpeck 0.1 μm diamater, Invitrogen T7279) were added to the samples. Coordinates of single molecules in raw STORM images were detected using the open source software ThunderSTORM (Ovesný et al., 2014). Drift, chromatic aberration correction and super-resolution rendering were done using a custom-written script in MATLAB (MathWorks).
STORM Image Analysis
The cell cortex in each rendered STORM image was detected semi-automatically. First, portions of cell periphery where the cortex is well defined, i.e., without too many microvilli or aggregation of fiducial beads, were manually selected using a custom-written GUI in MATLAB. The cortex was then automatically detected by finding the maximum intensity peak of transverse profiles across the cell contour at every pixel along the regions selected. The segmented cortex was straightened up using a spline and individual intensity linescans across the cortex were obtained.
In one-color STORM experiments, the full width at half maximum (FWHM) of cortex intensity profiles was used to assess cortical thickness. For two-color STORM experiments, the intensity peaks for each protein pair (phalloidin in two colors, or phalloidin and vimentin) were localized and peak-to-peak distance was measured using a custom-written MATLAB script.
Cell Confinement
Confinement experiments were done as previously described (Lancaster et al., 2013) with small alterations. Briefly, polyacrylamide gels (5 kPa rigidity) were polymerized onto 19 mm glass coverslips and incubated in imaging media at 37°C. Gel-covered coverslips were then gently positioned on top of the siRNA treated cells seeded on glass bottom petri dishes (Ibidi), and pressure to confine the cells was exerted using a PDMS column. Time lapse images were taken every 5 min with 4 z-stack of 3 μm step with a Zeiss Observer Z1 microscope widefield timelapse microscope equipped with a 12bit Hamamatsu Orca ER camera and a 60X objective.
Live Cell Imaging and Analysis
All cells were imaged in in phenol-red free and CO2-independent medium (Invitrogen) at 37°C. Hela Kyoto cells stably expressing H2B-mCherry and Lifeact-GFP were imaged using a Nikon TE2000 microscope equipped with a Plan Fluor x60/1.4 DIC H objective (Nikon), a PerkinElmer ERS Spinning disk system, a Digital CCD C4742-80-12AG camera (Hamamatsu), and controlled by Volocity 6.0.1 software. For vimentin dynamics during the cell cycle, CDK1 inhibitor time lapses, and cell rounding experiments, stacks of 7 z-planes 3 μm apart every 2 min. Cell roundness (4∗area/pi∗sqr(major axis)) was calculated using Fiji software. Hela Kyoto cells stably expressing the different GFP-vimentin mutants were imaged with an Olympus FluoView FV1200 Confocal Laser Scanning Microscope using a 60X objective (UPLSAPO60XS) and the FV1200 software. Cortex localization analysis of GFP-vimentin mutants were done the same way as explained in the Immunofluorescence section.
Laser Ablation Experiments
Ablation experiments were performed on an Olympus FluoView FV1200 Confocal Laser Scanning Microscope using a 60X objective (UPLSAPO60XS) as previously described (Tinevez et al., 2009). A 405 nm laser (PicoQuant) delivering picosecond pulses with nominal power 3 mW was used with the maximal speed setting corresponding to ∼3 s between time points. Pulses were delivered for ∼2 s in a spiraling movement within a circular region with a diameter of ∼400 nm focused on the sub-cortical vimentin layer. To confirm that our ablation settings did not result in puncture of the cell membrane, CellMaskTM Deep Red Plasma membrane stain (Thermo Fisher Scientific) or 10 KDa non-cell permeable dextran conjugated to Alexa 647 was added to the media prior to the experiment. For the ROCK inhibitor experiment, cells were pre-treated with 10 μM Y27632 for 15 min. For vimentin ablation, the laser was focused on sub-cortical the vimentin layer; areas of bright and low intensities of vimentin were chosen randomly and no difference in cell behavior was observed between the different areas.
To compute cell curvature (Figure 5E), the cell was segmented by adding a Gaussian filter to the image and then automatically thresholding with Otsu's method. Contour points of the cell from the binary image were extracted (∼1000 points/cell) and for each point a circle through that point was fitted. To reduce local noise that highly affects curvature measurements, a circle to each point was fitted using the 60th point before and after the point. Extracted local curvature (1/radius of the fitted circles) was then plotted along the cell contour.
AFM Measurements
For cortical tension measurements, cells were transfected with siRNA 48 hours prior to the experiment. Cells were then treated with STLC 4 h before the first measurement to enrich for mitotic cells. Interphase cells were pre-treated with thymidine for 22h and detached using trypsin before the measurement. Cells were placed in glass bottom dishes (Mattek) in phenol-red free and CO2-independent medium (Invitrogen) and incubated with 0.01% CellMaskTM Deep Red Plasma membrane Stain (Thermo Fisher Scientific). Mitotic and interphase cells were then selected based on DNA appearance in bright field microscopy. Atomic force microscopy tension measurements were performed using Tipless silicon cantilevers (ARROW-TL1Au-50) with a nominal spring constant of 0.03 N/m, mounted on a JPK CellHesion module (JPK Instruments) on an IX81 inverted confocal microscope (Olympus). Sensitivity was calibrated by acquiring a force curve on a glass coverslip, and the spring constant was calibrated by the thermal noise fluctuation method. Constant height mode was selected and a Z-length parameter of 35 μm was used with a setpoint force set at 25 nN. The measurement was carried out by first lowering the tipless cantilever on the glass in an empty area next to the cell to detect the position of the substrate. The cantilever was then positioned above the cell and a compression was applied for 250 seconds. During the constant height compression, the force acting on the cantilever was recorded. After initial force relaxation, the resulting force value was used to extract surface tension.
The calculation of cortex tension is based on (Fischer-Friedrich et al., 2014) and described in (Chugh et al., 2017). Briefly, neglecting the angle of the cantilever with respect to the dish (∼8°) and assuming negligible adhesion between cell, dish and cantilever, the force balance at the contact point reads:
where rmid is the radius of the maximum cross-sectional area of the selected cell, rc is the radius of the contact area of the cell with the cantilever and F is the force exerted by the cell on the cantilever. To avoid errors due to direct measurement of rc, the contact radius was calculated using the following formula (Stewart et al., 2012):
where Ac = π rc2 is the contact area between the cell and the cantilever, Amid is the cell maximum cross-sectional area and hcell is the cell height.
Scanning Electron Microscopy (SEM)
Synchronized extracts were supplemented with 20 μM soluble phalloidin (Sigma), 2 mM MgCl2 and 1 mM ATP to promote F-actin polymerization. The mixture (10 μl) was sandwiched between coverslips spaced with parafilm strips as previously described (Plastino et al., 2004). Samples were incubated 1 h at room temperature and then fixed with glutaraldehyde 1%. Preparation for scanning electron microscopy was performed as previously described in (Bovellan et al., 2014, Chugh et al., 2017). Briefly, after the first fixation samples were fixed a second time with 0.1% Tannic Acid for 5 min, washed 5 times with water and incubated with 0.1 % uranyl-acetate for 10 min. After 5 washes with water, samples were then dehydrated with serial ethanol dilutions, dried in a critical point dryer, coated with a thin layer of approximately 7 nm thick of Gold using a Quorum sputter coater and then viewed and images recorded using a FEI Quanta 200 FEG ESEM, operated at 5 kV in the high vacuum mode.
RNA Extraction and RT-qPCR
Total RNA from Hela Kyoto cells was extracted using RNeasy Mini Kit (Qiagen) 48h after siRNA transfection. Total RNA was reverse-transcribed using a cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, United States of America) following the manufacturer’s instructions. Gene expression levels of endogenous controls GAPDH and beta-actin (ACTB) were determined using pre-validated TaqMan Gene Expression Assays (Applied Biosystems). Expression levels of queried and control genes were determined using assays designed with the Universal Probe Library from Roche. The PCR reactions were carried out on an ABI Real Time 7900HT cycler and analyzed with SDS 2.2 software. qPCR was carried out with TaqMan Gene Expression Master Mix (Applied Biosystems), and all samples were tested in duplicate. mRNA levels of queried genes were normalized to the averaged levels of GAPDH and ACTB, and relative abundance was calculated by normalizing values from cells treated with control and targeted siRNA pools by dividing by the control value. All the genes tested and the corresponding primers are listed in Table S2.
Quantification and Statistical Analysis
3D visualization videos were generated using Icy software. dSTORM sata were analyased using Matlab (MathWorks). All the other image analysis was carried out in Fiji. Graphs and statistical tests were produced in GraphPad Prism. Normally distributed data sets were analyzed using the unpaired t test, or ANOVA. Non-normal data sets were analyzed using Mann-Whitney or Kruskal-Wallis test. Details of the statistical tests used, exact value of n, definition error bars on graphs and number of experiments performed are all detailed in the legends of the figures.
Data and Code Availability
Data and custom-written codes developed for data analysis are available upon request from the lead contact.
The software used for STORM rendering and analysis is described in (Truong Quang et al., submitted).
Acknowledgments
We thank Dennis Bray, Kevin Chalut, and members of the Paluch lab, in particular Dani Bodor, for feedback on the work and manuscript. We thank Wolfram Pönisch for help with analyzing cell curvatures; Mark Turmaine for help with scanning electron microscopy; Andrew Vaughan, Ki Hng, and Catherine Scherer for support with light and STORM microscopy; Davide Cassani for technical help with AFM; and Jürgen Schünemann (MPI for Biophysical Chemistry) for HPLC-purifying phalloidin derivatives. We thank Éric Bonneil and Pierre Thibault for expert technical help and advice on mass spectrometry analysis and peptide quantification. We acknowledge support from the Medical Research Council UK (MRC programme award MC_UU_00012/5, M.P.S., B.A.T.Q., U.P., and E.K.P.), the Human Frontier Science Program (Young Investigator grant RGY 66/2013 to G.C., P.P.R., and E.K.P.), the European Research Council (starting grant 311637-MorphoCorDiv to E.K.P.), Cancer Research UK (M.P. and M.P.S.), a BBSRC ALERT16 equipment grant (BB/R000042 to G.C.) for purchase of AFM instrumentation, and an operating grant from the Canadian Institute of Health Research to P.P.R. (MOP-142374). P.P.R. is supported by a FRQS Senior investigator career award.
Authors Contributions
M.P.S., M.P., P.P.R., and E.K.P. designed the research; M.P.S. carried out most of the experiments and analyzed the data. M.S. and D.G. developed and provided the phalloidin matrix; B.A.T.Q. developed the analysis tools for STORM and analyzed the 2-color STORM experiment; G.L. carried out the qPCR, helped with mass spectrometry, and performed the mutagenesis; U.P. carried out SEM experiments and helped with other experiments and data analysis; M.P.S. and E.K.P. wrote the paper; D.G., G.C., and P.P.R. provided technical support and conceptual advice. Funding Acquisition, G.C., M.P., P.P.R., and E.K.P. All authors discussed the results and manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: January 9, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2019.12.011.
Contributor Information
Philippe P. Roux, Email: philippe.roux@umontreal.ca.
Ewa K. Paluch, Email: ekp25@cam.ac.uk.
Supplemental Information
References
- Biro M., Romeo Y., Kroschwald S., Bovellan M., Boden A., Tcherkezian J., Roux P.P., Charras G., Paluch E.K. Cell cortex composition and homeostasis resolved by integrating proteomics and quantitative imaging. Cytoskeleton (Hoboken) 2013;70:741–754. doi: 10.1002/cm.21142. [DOI] [PubMed] [Google Scholar]
- Bovellan M., Romeo Y., Biro M., Boden A., Chugh P., Yonis A., Vaghela M., Fritzsche M., Moulding D., Thorogate R. Cellular control of cortical actin nucleation. Curr. Biol. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller G., Gregor M., Winter L., Wiche G. Keeping the vimentin network under control: cell-matrix adhesion-associated plectin 1f affects cell shape and polarity of fibroblasts. Mol. Biol. Cell. 2010;21:3362–3375. doi: 10.1091/mbc.E10-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno S., Kouranti I., Glusman E.S., Fuller M.T., Echard A., Payre F. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J. Cell Biol. 2008;180:739–746. doi: 10.1083/jcb.200709161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis J.E., Larsen P.M., Fey S.J., Celis A. Phosphorylation of keratin and vimentin polypeptides in normal and transformed mitotic human epithelial amnion cells: behavior of keratin and vimentin filaments during mitosis. J. Cell Biol. 1983;97:1429–1434. doi: 10.1083/jcb.97.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion L., Linder M.I., Kutay U. Cellular reorganization during mitotic entry. Trends Cell Biol. 2017;27:26–41. doi: 10.1016/j.tcb.2016.07.004. [DOI] [PubMed] [Google Scholar]
- Chanet S., Sharan R., Khan Z., Martin A.C. Myosin 2-induced mitotic rounding enables columnar epithelial cells to interpret cortical spindle positioning cues. Curr. Biol. 2017;27:3350–3358.e3. doi: 10.1016/j.cub.2017.09.039. [DOI] [PubMed] [Google Scholar]
- Charras G.T., Mitchison T.J., Mahadevan L. Animal cell hydraulics. J. Cell Sci. 2009;122:3233–3241. doi: 10.1242/jcs.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.H., Khuon S., Herrmann H., Goldman R.D. Nestin promotes the phosphorylation-dependent disassembly of vimentin intermediate filaments during mitosis. Mol. Biol. Cell. 2003;14:1468–1478. doi: 10.1091/mbc.E02-08-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.H., Rosevear E., Goldman R.D. Phosphorylation and disassembly of intermediate filaments in mitotic cells. Proc. Natl. Acad. Sci. USA. 1989;86:1885–1889. doi: 10.1073/pnas.86.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P., Clark A.G., Smith M.B., Cassani D.A.D., Dierkes K., Ragab A., Roux P.P., Charras G., Salbreux G., Paluch E.K. Actin cortex architecture regulates cell surface tension. Nat. Cell Biol. 2017;19:689–697. doi: 10.1038/ncb3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciobanasu C., Faivre B., Le Clainche C. Actin dynamics associated with focal adhesions. Int. J. Cell Biol. 2012;2012:941292. doi: 10.1155/2012/941292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.G., Dierkes K., Paluch E.K. Monitoring actin cortex thickness in live cells. Biophys. J. 2013;105:570–580. doi: 10.1016/j.bpj.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.G., Paluch E. Mechanics and regulation of cell shape during the cell cycle. Results Probl. Cell Differ. 2011;53:31–73. doi: 10.1007/978-3-642-19065-0_3. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Ding W.Y., Ong H.T., Hara Y., Wongsantichon J., Toyama Y., Robinson R.C., Nédélec F., Zaidel-Bar R. Plastin increases cortical connectivity to facilitate robust polarization and timely cytokinesis. J. Cell Biol. 2017;216:1371–1386. doi: 10.1083/jcb.201603070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte S., Viedma-Poyatos Á., Navarro-Carrasco E., Martínez A.E., Pajares M.A., Pérez-Sala D. Vimentin filaments interact with the actin cortex in mitosis allowing normal cell division. Nat. Commun. 2019;10:4200. doi: 10.1038/s41467-019-12029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Friedrich E., Hyman A.A., Jülicher F., Müller D.J., Helenius J. Quantification of surface tension and internal pressure generated by single mitotic cells. Sci. Rep. 2014;4:6213. doi: 10.1038/srep06213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R., Malecz N., Dressel N., Stadler C., Wiche G. M-phase-specific phosphorylation and structural rearrangement of the cytoplasmic cross-linking protein plectin involve p34cdc2 kinase. Mol. Biol. Cell. 1996;7:273–288. doi: 10.1091/mbc.7.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.A., Geraghty K.M., Lavoie G., Kanshin E., Tcherkezian J., Calabrese V., Jeschke G.R., Turk B.E., Ballif B.A., Blenis J. Phosphoproteomic analysis identifies the tumor suppressor PDCD4 as a RSK substrate negatively regulated by 14–3-3. Proc. Natl. Acad. Sci. USA. 2014;111:E2918–E2927. doi: 10.1073/pnas.1405601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O., Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Ehrlicher A.J., Mahammad S., Fabich H., Jensen M.H., Moore J.R., Fredberg J.J., Goldman R.D., Weitz D.A. The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophys. J. 2013;105:1562–1568. doi: 10.1016/j.bpj.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerboth S., Housman G., Leary M., Longacre M., Byler S., Lapinska K., Willbanks A., Sarkar S. EMT and tumor metastasis. Clin. Transl. Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber F., Boire A., López M.P., Koenderink G.H. Cytoskeletal crosstalk: when three different personalities team up. Curr. Opin. Cell Biol. 2015;32:39–47. doi: 10.1016/j.ceb.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Hubert T., Vandekerckhove J., Gettemans J. Unconventional actin conformations localize on intermediate filaments in mitosis. Biochem. Biophys. Res. Commun. 2011;406:101–106. doi: 10.1016/j.bbrc.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Inagaki M., Nishi Y., Nishizawa K., Matsuyama M., Sato C. Site-specific phosphorylation induces disassembly of vimentin filaments in vitro. Nature. 1987;328:649–652. doi: 10.1038/328649a0. [DOI] [PubMed] [Google Scholar]
- Koenderink G.H., Paluch E.K. Architecture shapes contractility in actomyosin networks. Curr. Opin. Cell Biol. 2018;50:79–85. doi: 10.1016/j.ceb.2018.01.015. [DOI] [PubMed] [Google Scholar]
- Kordowska J., Hetrick T., Adam L.P., Wang C.L. Phosphorylated l-caldesmon is involved in disassembly of actin stress fibers and postmitotic spreading. Exp. Cell Res. 2006;312:95–110. doi: 10.1016/j.yexcr.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Krause M., Bear J.E., Loureiro J.J., Gertler F.B. The Ena/VASP enigma. J. Cell Sci. 2002;115:4721–4726. doi: 10.1242/jcs.00218. [DOI] [PubMed] [Google Scholar]
- Kunda P., Pelling A.E., Liu T., Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 2008;18:91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- Lancaster O.M., Baum B. Shaping Up to divide: coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin. Cell Dev. Biol. 2014;34:109–115. doi: 10.1016/j.semcdb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Lancaster O.M., Le Berre M., Dimitracopoulos A., Bonazzi D., Zlotek-Zlotkiewicz E., Picone R., Duke T., Piel M., Baum B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev. Cell. 2013;25:270–283. doi: 10.1016/j.devcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Leduc C., Etienne-Manneville S. Intermediate filaments join the action. Cell Cycle. 2017;16:1389–1390. doi: 10.1080/15384101.2017.1345230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue J.S., Cartagena-Rivera A.X., Baird M.A., Davidson M.W., Chadwick R.S., Waterman C.M. Erk regulation of actin capping and bundling by Eps8 promotes cortex tension and leader bleb-based migration. eLife. 2015;4:e08314. doi: 10.7554/eLife.08314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C., Pasolli H.A., Williams S.E., Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat. Cell Biol. 2011;13:203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machicoane M., de Frutos C.A., Fink J., Rocancourt M., Lombardi Y., Garel S., Piel M., Echard A. SLK-dependent activation of ERMs controls LGN-NuMA localization and spindle orientation. J. Cell Biol. 2014;205:791–799. doi: 10.1083/jcb.201401049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox A.S., Burridge K. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J. Cell Biol. 2003;160:255–265. doi: 10.1083/jcb.200207130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews H.K., Delabre U., Rohn J.L., Guck J., Kunda P., Baum B. Changes in Ect2 localization couple actomyosin-dependent cell shape changes to mitotic progression. Dev. Cell. 2012;23:371–383. doi: 10.1016/j.devcel.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M., Depken M., Bois J.S., Jülicher F., Grill S.W. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature. 2010;467:617–621. doi: 10.1038/nature09376. [DOI] [PubMed] [Google Scholar]
- Mendez M.G., Kojima S., Goldman R.D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- Ovesný M., Křížek P., Borkovec J., Svindrych Z., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Glotzer M., Kraut N., Peters J.M. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 2007;12:713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Plastino J., Olivier S., Sykes C. Actin filaments align into hollow comets for rapid VASP-mediated propulsion. Curr. Biol. 2004;14:1766–1771. doi: 10.1016/j.cub.2004.09.054. [DOI] [PubMed] [Google Scholar]
- Ramanathan S.P., Helenius J., Stewart M.P., Cattin C.J., Hyman A.A., Muller D.J. Cdk1-dependent mitotic enrichment of cortical myosin II promotes cell rounding against confinement. Nat. Cell Biol. 2015;17:148–159. doi: 10.1038/ncb3098. [DOI] [PubMed] [Google Scholar]
- Ramkumar N., Baum B. Coupling changes in cell shape to chromosome segregation. Nat. Rev. Mol. Cell Biol. 2016;17:511–521. doi: 10.1038/nrm.2016.75. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A.I., Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer M., Dehne H.J., Spira F., Kollmar M., Gerlich D.W., Urlaub H., Görlich D. The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. EMBO J. 2013;32:1886–1902. doi: 10.1038/emboj.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A., Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M.P., Toyoda Y., Hyman A.A., Müller D.J. Tracking mechanics and volume of globular cells with atomic force microscopy using a constant-height clamp. Nat. Protoc. 2012;7:143–154. doi: 10.1038/nprot.2011.434. [DOI] [PubMed] [Google Scholar]
- Su K.C., Takaki T., Petronczki M. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell. 2011;21:1104–1115. doi: 10.1016/j.devcel.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Takai Y., Ogawara M., Tomono Y., Moritoh C., Imajoh-Ohmi S., Tsutsumi O., Taketani Y., Inagaki M. Mitosis-specific phosphorylation of vimentin by protein kinase C coupled with reorganization of intracellular membranes. J. Cell Biol. 1996;133:141–149. doi: 10.1083/jcb.133.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.Y., Schulze U., Salbreux G., Roensch J., Joanny J.F., Paluch E. Role of cortical tension in bleb growth. Proc. Natl. Acad. Sci. USA. 2009;106:18581–18586. doi: 10.1073/pnas.0903353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda Y., Cattin C.J., Stewart M.P., Poser I., Theis M., Kurzchalia T.V., Buchholz F., Hyman A.A., Müller D.J. Genome-scale single-cell mechanical phenotyping reveals disease-related genes involved in mitotic rounding. Nat. Commun. 2017;8:1266. doi: 10.1038/s41467-017-01147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura K., Ogawara M., Takeuchi Y., Imajoh-Ohmi S., Ha M.H., Inagaki M. Visualization and function of vimentin phosphorylation by cdc2 kinase during mitosis. J. Biol. Chem. 1994;269:31097–31106. [PubMed] [Google Scholar]
- Wiche G., Osmanagic-Myers S., Castañón M.J. Networking and anchoring through plectin: a key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 2015;32:21–29. doi: 10.1016/j.ceb.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Goto H., Yokoyama T., Silljé H., Hanisch A., Uldschmid A., Takai Y., Oguri T., Nigg E.A., Inagaki M. Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J. Cell Biol. 2005;171:431–436. doi: 10.1083/jcb.200504091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S., Yamakita Y., Ishikawa R., Matsumura F. Mitosis-specific phosphorylation causes 83K non-muscle caldesmon to dissociate from microfilaments. Nature. 1990;344:675–678. doi: 10.1038/344675a0. [DOI] [PubMed] [Google Scholar]
- Yamashiro S., Yamakita Y., Hosoya H., Matsumura F. Phosphorylation of non-muscle caldesmon by p34cdc2 kinase during mitosis. Nature. 1991;349:169–172. doi: 10.1038/349169a0. [DOI] [PubMed] [Google Scholar]
- Yasui Y., Goto H., Matsui S., Manser E., Lim L., Nagata Ki K., Inagaki M. Protein kinases required for segregation of vimentin filaments in mitotic process. Oncogene. 2001;20:2868–2876. doi: 10.1038/sj.onc.1204407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frames were acquired every 2 min. Scale bar, 20 μm.
The video displays projected z-stacks for each time point. Frames were acquired every 1 min. Cells were synchronized in pro-metaphase using STLC and treated with the CDK1 inhibitor RO3306 immediately before the movie. Scale bar, 10 μm.
Ablation was performed in HeLa cells stably expressing GFP-vimentin-WT and in presence of Cell Mask to monitor the plasma membrane during ablation (left panel) or in presence of fluorescent dextran in the medium (right panel). The yellow circle represents the site of ablation.
Ablation was performed in HeLa cells stably expressing GFP-vimentin-WT (left panel) and transfected with mCherry-Lifeact to monitor the actin cortex during ablation (right panel). The yellow circle represents the site of ablation.
Ablation was performed in HeLa cells stably expressing GFP-vimentin-WT or -56E. The yellow circle represents the site of ablation. Frames were acquired every 3.26 s and the ablation was performed at 25 s (left panel) and at 9s (middle panel and right panels). Scale bars, 5 μm.
Frames were acquired every 2 min. DNA (red); F-actin (cyan); z-projections are diaplayed. Scale bar, 20 μm.
Data Availability Statement
Data and custom-written codes developed for data analysis are available upon request from the lead contact.
The software used for STORM rendering and analysis is described in (Truong Quang et al., submitted).