Abstract
Endocrine disrupting compounds (EDCs) have the potential to cause adverse effects on wild-life and human health. Two important EDCs are the synthetic estrogen 17α-ethynylestradiol (EE2) and bisphenol-A (BPA) both of which are xenoestrogens (XEs) as they bind the estrogen receptor and dis-rupt estrogen physiology in mammals and other vertebrates. In the recent years the influence of XEs on oncogenes, specifically in relation to breast and prostate cancer has been the subject of considerable study.
Methodology:
In this study, healthy primary human prostate epithelial cells (PrECs) were exposed to environmentally relevant concentrations of BPA (5nM and 25nM BPA) and interrogated using a whole genome microarray.
Results:
Exposure to 5 and 25nM BPA resulted in 7,182 and 7,650 differentially expressed (DE) genes, respectively in treated PrECs. Exposure to EE2 had the greatest effect on the PrEC transcriptome (8,891 DE genes).
Conclusion:
We dissected and investigated the nature of the non-estrogenic gene signature associated with BPA with a focus on transcripts relevant to epigenetic modifications. The expression of transcripts encoding nuclear hormone receptors as well as histone and DNA methylation, modifying enzymes were significantly perturbed by exposure to BPA.
Keywords: Endocrine disruptor (ED), xenoestrogen (XE), bisphenol-A (BPA), microarray, prostate epithelial cells, meta-analysis, epigenomic biomarkers
1. INTRODUCTION
Endocrine disrupting compounds (EDCs) are chemicals found in many consumer products that mimic natural hormones [1]. EDCs have been shown to disrupt normal endocrine function by binding nuclear hormone receptors including estrogen receptors and activating estrogen related signaling pathways, thus they represent an emerging problem for ecosystem and human health. A specific subset of EDCs, the xenoestrogens (XEs), can mimic 17β-estradiol (E2), the female estrogen [2]. One such XE is bisphenol-A (BPA), a chemical building block used to strengthen polycarbonate plastic products [3]. It is a global contaminant found in the environment at concentrations up to 92 nM in surface waters [4]. The primary source of BPA exposure is dietary. Canned foods, beer and sodas are frequently coated with BPA-containing epoxy films and BPA monomers can leach into the contents of the can. BPA has also been found in fast-food hamburgers [5].
Even though the prostate gland is androgen-dependent and develops during puberty under androgen control, estrogens also play a part in normal and malignant prostate development [6]. During the 3rd trimester of pregnancy, maternal estrogens allow the growth of the prostate gland in men [7], and it has previously been shown that embryos exposed to the exogenous estrogen diethylstilbestrol (DES) in that critical window present prostate hyperplasia as newborns [8].
In humans, estrogen receptor alpha (ERα) is encoded by the gene ESR1 (EStrogen Receptor 1) and estrogen receptor beta (ERβ) by the gene ESR2 (EStrogen Receptor 2). Prostate cells express ERα and ERβ, which are localized characteristically in stroma and epithelium, respectively [9]. Pasquali et al. [10] have provided evidence for the co-expression of both ESR1 and ESR2 transcripts in normal epithelial cells in primary cultures. In both human and rodent prostate, ERβ is more highly expressed than ERα, while ERβ is found in both epithelial and (to a lesser extent) stromal cells, ERα is generally localized to the stroma [9].
Estrogen used to be the foundation of prostate cancer therapy prior to the understanding that it was also associated with a high risk of serious cardiovascular complications, which ultimately limited its clinical use [11]. The indirect therapeutic effect of estrogen was primarily mediated by suppression of the hypothalamic-pituitary-gonadal axis and by direct effects on the Leydig cells in testis, which together lead to decreased serum levels of testosterone and castration like effects in prostate [12]. These studies also reported that estrogen had direct “toxic” effects on prostate.
In rodents, perinatal or neonatal exposure to 17β-estradiol (estradiol) or other estrogens can lead to “imprinting” of prostate and disrupt normal morphogenesis and inhibit prostate growth via ERα, resulting in aberrant prostate function and morphology in the adult prostate, as well as increased proliferation, inflammation and dysplasia, and cause epithelial hyperplasia including drastic alterations in the expression pattern of steroid receptors [13-15]. Prolonged treatment of adult rodents with estrogens also leads to epithelial metaplasia, prostatic intraepithelial neoplasia (PIN)-like lesions and even adenocarcinoma of the prostate highlighting the role of estrogen in prostate cancer development [9]. Chronic exposure to chlordecone, an estrogenic organochloride pesticide commonly used in the farming of bananas, is also associated with increased risk of prostate cancer [16].
BPA binds and activates the estrogen receptors (ERα and ERβ) [17, 18]. Additionally, BPA can stimulate other nuclear hormone receptors including estrogen-related receptor gamma (ESRRG) [19], the androgen receptor (NR3C4) [20], thyroid hormone receptors (THRs) [21], G-protein-coupled receptors (GPER) [22], glucocorticoid and mineralocorticoid receptors (NR3C1 and NR3C2) [23, 24], and the pregnane X receptor (NR1/2) [25]. BPA has also been shown to interfere with endocannabinoid receptors CNR1 and CNR2 [26, 27]. This promiscuous affinity for multiple nuclear hormone and G-protein coupled receptors results in BPA impacting multiple physiological processes in both humans and wildlife.
In the United States, BPA has been detected in 93% of 2,517 urine samples collected from both adults and children, indicating pervasive exposure to this contaminant [3, 28, 29]. Data from multiple sources indicate that its ubiquitous presence and thereby continuous exposure in humans cause adverse health effects including premature puberty [30], reduced fertility [31-33], obesity [34-37], metabolic diseases [38] and cancer [39-42]. The initial study to assess BPA’s estrogenic effects demonstrated that it increased the growth rate of MCF-7 cells [43]. Low (nM) concentrations of BPA exhibit estrogenic activity [44, 45]. Furthermore, a BPA metabolite, 4-methyl-2,4-bis(4-hydroxyphenyl)pent-1-ene (MBP) exhibits transcriptional activity at low nM concentrations and suggests that metabolism of BPA to MBP may explain some of the adverse effects observed [46]. Furthermore, a comparison of 3D structural models of human ERα and ERβ with both MBP and BPA revealed that MBP, but not BPA, had key interactions with amino acid residues important in E2 binding in human ERα and ERβ [47].
Since the late 1980s, prostate cancer has been the second leading cause of death due to cancer in men [48]. BPA has since been linked to increased risks for prostate cancer (PCa) by affecting cellular proliferation and prostate cancer cell migration [49]. Additionally, BPA has been shown to promote PCa by disrupting centrosome amplification and microtubule dynamics that contribute to neoplastic transformation of prostate cells [50, 51]. Moreover, a correlation exists between men affected by PCa and higher levels of BPA in urine than those without the disease, suggesting that urinary BPA levels may have prognostic value for PCa [51].
Interestingly, early-life exposure to BPA during critical developmental windows has also been shown to cause epigenetic alterations in the prostate, and, in so doing, promote prostate disease in men as they get older [52, 53]. BPA alters the epigenome by interfering with the expression of histone modifying enzymes for example, the histone deacetylase SIRT1, and the histone methyltransferase SET8, which cause gene expression modifications in PCa cells [54]. Cheong et al. [55] suggested that DNA methylation changes may be an epigenetic signature associated with increased cancer susceptibility in the adult gland due to early-life exposure.
The objective of this study was to perform a systems level analysis of the effects of environmentally relevant doses of BPA on the transcriptome of healthy primary human prostate epithelial cells (PrECs). As a positive estrogenic control for these experiments, we selected 17α-ethinylestradiol (EE2), an artificial analogue of E2, commonly used in birth control pills and a well-characterized XE [56, 57]. To our knowledge, this is the first in vitro study that (1) examines the effects of low dose BPA exposure on the healthy human prostate cell transcriptome using a system level approach, and (2) characterizes non-estrogenic effects of BPA by distinguishing how BPA exposure differs from EE2, (3) defines the difference between a low (5 nM) and higher dose (25 nM) of BPA on primary prostate epithelial cells (PrECs), and (4) specifically examines the effects of BPA on the expression of genes encoding histone and DNA methylation modifying enzymes.
2. MATERIALS AND METHODS
2.1. Primary Human Cells
Primary prostate epithelial cells (PrECs) derived from a healthy 23-year old male with no history of prostate disease were obtained from CloneticsTM (San Diego, CA). The PrEC used in this study were cryopreserved in the second passage as proliferating cultures and were not an immortalized cell line. Telomerase reverse transcriptase (TERT) expression negative immortalizes various normal cells, including prostate epithelial and stromal-derived cells in culture. The cells used in this study were TERT expression negative. PrECs were cultured in Clonetics™ PrEGM™ Prostate Epithelial Cell Growth Medium, with a confluence of 70-90% in 0.2 mL/well 96 well plates- and approximately 0.1 x 106 cells for each well - and supplemented with 10% fetal bovine serum, 2mM glutamine, and 1% penicillin/streptomycin. Within the first passage, PrECs were exposed to either 5 nM BPA, 25 nM BPA, or 0.1 nM EE2 for 24 hours. A control group received no treatment.
2.2. RNA Extraction and Microarray Analysis
RNA was extracted using TRIzol reagent (Invitrogen) and the extracted RNA was further purified using the RNeasy Mini kit (Qiagen, Valencia, CA). RNA was assessed via absorbance readings (OD) at 260nm using an ND-1000 (Nanodrop, Wilmington, DE). RNA integrity was examined with the 6000 Nano LabChip assay from Agilent, (Santa Clara, CA). Only RNA samples with an RNA integrity number (RIN) score of >7.0 indicating intact RNA were used for microarray experiments (RIN is a measure of the degradation of RNA sample) [58, 59]. The samples were then sent to the UCSD BIOGEM core for Illumina Beadarray processing. HumanHT-12 v4 Expression BeadChip Kits were used for the array experiments (Illumina, San Diego). RNA labeling and microarray hybridization have been described in detail elsewhere [60]. Microarray experiments were performed with three biological replicates. The arrays were scanned on the Illumina BeadArray Reader, a confocal-type imaging system with 532 (cye3) nm laser illuminations. Image analysis and data extraction was carried out as in accordance with Illumina specifications. Preliminary data analysis and QC was carried out using GenomeStudio version 1.8.0 (Illumina Inc.). All array data have been deposited in the EBI ArrayExpress Database accession number E-MTAB-7576.
2.3. Gene and Systems Level Analysis
Microarray data were analyzed (Fig. 1) using the Pipeline for Integrated Microarray Expression and Normalization Toolkit (PIMENTo) [61, 62]. The tool follows best practices for array data analyses and streamlines the processes necessary for gene expression analysis. It is built with the R programming language and leverages several open-source packages available through CRAN and Bioconductor, routinely used for microarray analyses. PIMENTo enables researchers to perform complex tasks with a minimal number of operations. It allows the user to review necessary data inputs, provides (1) data pre-processing and quality control, (2) normalization to remove the technical variability across arrays data [63, 64], (3) data visualization and (4) differential expression (DE) analysis. Three biological replicate cultures exposed to BPA or EE2 were used in the analyses. Expression level data from the Illumina Bead Studio software were normalized using a loess algorithm [42]. Probes, whose expression level that exceeded a threshold value in at least one sample, are called detected. The threshold value was found by inspection from the distribution plots of (log) expression levels. As one of the biological replicates was performed over 9 months after the other two we used the Bioconductor package 'limma' to remove technical batch effects. Limma allows significant differentially expressed genes to be determined based on the p-values, adjusted p-values or false discovery rate (FDR) estimates. The methodology follows the premise that transcripts are sorted according to their q-value, which is the smallest false discovery rate (FDR) at which the transcript is called significant. FDR is the expected fraction of false positive tests among significant tests. We set FDR at 0.1. We then search for groups of genes that collectively enrich GO terms or pathways. It should also be noted that the systems level analysis we performed (GO and Pathway) was itself subjected to FDR testing thereby adding rigor to the data analysis.
Fig. (1).
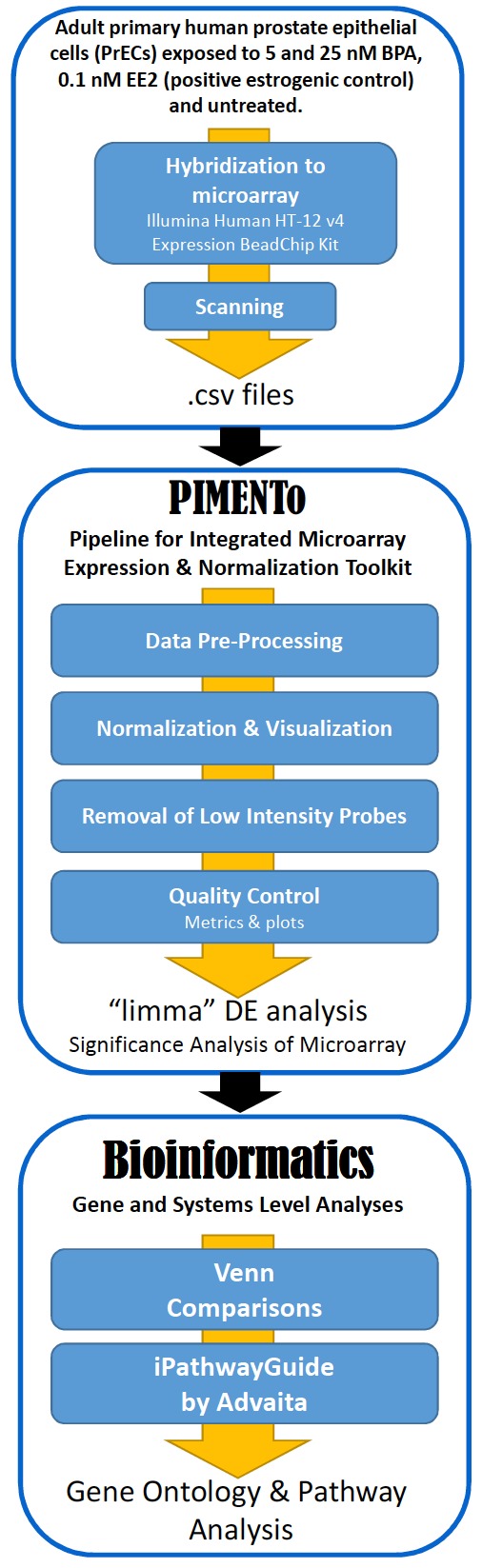
Bioinformatics workflow schematic. .
This was conducted using iPathwayGuide (Advaita Bioinformatics) [65]. Venn comparisons were performed using VENNY 2.1 online tool [66].
3. RESULTS
3.1. The Transcriptome of PrECs Reveals an Altered Signature in Response to BPA (Low and High) and EE2 Exposures
Using Venn diagrams, the list of significant differentially expressed (DE) microarray probes (q ≤ 0.1) after exposures to 5 and 25 nM BPA as well as 0.1 nM EE2 (positive estrogenic control) relative to control PrECs were compared (Fig. 2A). Exposure to EE2 had the strongest effect on the PrEC transcriptome and resulted in 11,241 DE probes corresponding to 8,891 transcripts. Exposure to 5 and 25 nM BPA perturbed the expression patterns of 8,876 and 9,525 probes corresponding to 7,182 and 7,650 unique transcripts respectively, indicating that BPA also impacts the PrECs transcriptome. The number of array probes with an absolute log2 fold change differences of 1 (linear fold-change of 2) in PrECs exposed to BPA and EE2 compared to control cells were 821, 755 and 715 for EE2, 5nM and 25 nM BPA respectively (Fig. S1). All three exposures have 7,011 DE probes in common (Fig. 2B, intersection) while the 5 nM BPA exposure has 548 unique DE probes (orange), the 25 nM BPA has 593 unique DE probes (green) and EE has 2,389 unique DE probes (purple). Together, the low and high doses of BPA affected 1,839 DE probes not shared with the EE2 signature.
Fig. (2).
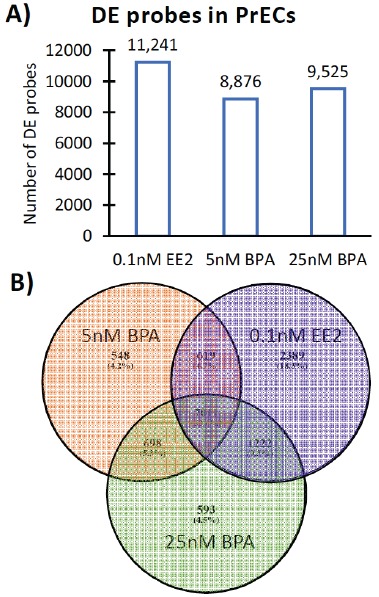
Effect of BPA and EE2 exposure on the PrECs transcriptome as determined by Illumina Bead Arrays. (A) Total number of DE microarray probes (q ≤ 0.1) in PrECs after exposure to EE2 (positive estrogenic control) and BPA (5 nM BPA and 25 nM BPA). (B) Overlap of DE microarray probes across the different exposures of BPA (5 nM: orange; 25 nM: green) and EE2 (0.1 nM EE2: purple) in PrECs. .
3.2. Meta-analysis (Advaita iPathwayGuide)
Meta-analysis carried out using iPathwayGuide using unique significant DE genes for all three exposures revealed that 24 pathways were commonly enriched (Fig. 3). Amongst the 24 commonly enriched pathways all pathways were related to metabolism (Table 1, orange), DNA replication and cell cycle (light green), cancer (light yellow), inflammation and immune response (light blue), protein biogenesis (purple), biological adhesion (pink). The TGF-β signaling pathway (dark green), which is involved in many cellular processes including cell growth, cell differentiation, apoptosis, cellular homeostasis and other cellular functions, was also enriched in all three exposures.
Fig. (3).
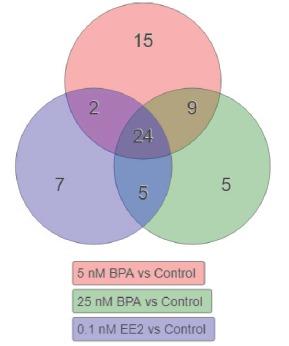
Meta-analysis - Biological pathways enriched in PrECs exposed to BPA (5 and 25 nM) and EE2 (positive estrogenic control, 0.1 nM). Shared and unique pathways are presented as a Venn Diagram. .
Table 1. Pathways that are commonly enriched in PrECs exposed to 5 nM BPA, 25 nM BPA and 0.1 nM EE2. Orange: pathways related to metabolism pathways, green: pathways related to DNA replication and cell cycle, yellow: pathways related to cancer, light blue: pathways related to inflammation and immune response, purple: pathways related to protein biogenesis, dark green: signaling pathways, pink: pathways related to biological adhesion. (The color version of the table is available in the electronic copy of the article).
| Pathways Common to All 3 Exposures |
5 nM BPA
(p-value) |
25 nM BPA
(p-value) |
0.1 nM EE2
(p-value) |
|---|---|---|---|
| Alanine, aspartate and glutamate metabolism | 5.11E-03 | 1.07E-02 | 9.81E-03 |
| Biosynthesis of amino acids | 1.36E-04 | 8.12E-04 | 1.70E-03 |
| Carbon metabolism | 2.98E-05 | 1.82E-04 | 1.04E-03 |
| Cell cycle | 9.63E-04 | 1.82E-03 | 1.37E-02 |
| Central carbon metabolism in cancer | 6.05E-04 | 1.01E-02 | 6.84E-03 |
| Chemical carcinogenesis | 2.68E-03 | 7.53E-03 | 3.01E-02 |
| Cytokine-cytokine receptor interaction | 5.00E-03 | 2.97E-03 | 1.22E-02 |
| DNA replication | 3.88E-06 | 1.56E-08 | 1.46E-07 |
| ECM-receptor interaction | 5.28E-06 | 6.99E-06 | 4.75E-04 |
| HTLV-I infection | 2.67E-03 | 1.69E-03 | 2.86E-03 |
| Leukocyte transendothelial migration | 4.34E-02 | 1.81E-02 | 3.71E-02 |
| Metabolic pathways | 5.32E-03 | 1.39E-03 | 4.28E-04 |
| Mismatch repair | 3.63E-02 | 4.50E-04 | 7.97E-05 |
| Natural killer cell mediated cytotoxicity | 1.60E-02 | 3.43E-02 | 4.87E-02 |
| Protein digestion and absorption | 4.46E-03 | 3.00E-03 | 3.44E-02 |
| Proteoglycans in cancer | 1.12E-02 | 1.06E-02 | 6.08E-03 |
| Purine metabolism | 1.20E-02 | 2.28E-04 | 6.75E-04 |
| Pyrimidine metabolism | 6.38E-03 | 1.06E-03 | 3.82E-03 |
| Regulation of actin cytoskeleton | 3.32E-02 | 2.24E-02 | 2.58E-02 |
| Ribosome biogenesis in eukaryotes | 6.62E-04 | 1.01E-03 | 1.28E-02 |
| RNA transport | 2.28E-02 | 3.28E-02 | 2.45E-02 |
| Small cell lung cancer | 4.79E-02 | 1.74E-02 | 1.70E-02 |
| TGF-beta signaling pathway | 4.30E-02 | 3.21E-02 | 4.19E-02 |
Meta-analysis also revealed 276 Biological Process (BP) terms that were significantly enriched as part of the Gene Ontology (GO) analysis. Table 2 displays the top 30 BP terms that relate to inflammation and immune response (light blue), metabolic pathways (orange), embryonic development (dark yellow), angiogenesis and blood circulation (turquoise), DNA replication and cell cycle (light green), biological adhesion (pink), cancer (light yellow), and protein biogenesis (purple). This suggests that BPA has a strong estrogenic effect similar to EE2 on several biological pathways and processes.
Table 2. Top 30 GO biological process terms that are commonly enriched in PrECs exposed to 5 nM BPA, 25 nM BPA and 0.1 nM EE2. Orange: BP terms related to metabolic pathways, green: BP terms related to DNA replication and cell cycle, light yellow: BP terms related to cancer, dark yellow: BP terms related to embryonic development, light blue: BP terms related to inflammation and immune response, purple: BP terms related to protein biogenesis, turquoise: BP terms related to angiogenesis and blood circulation, pink: pathways related to biological adhesion. (The color version of the table is available in the electronic copy of the article).
|
Biological Process Terms Common
to All 3 Exposures |
5 nM BPA
(p-value) |
25 nM BPA
(p-value) |
0.1 nM EE2
(p-value) |
|---|---|---|---|
| Acute inflammatory response | 3.81E-02 | 1.29E-02 | 2.62E-02 |
| Acylglycerol metabolic process | 3.20E-02 | 2.12E-02 | 7.64E-03 |
| ADP metabolic process | 5.60E-04 | 5.75E-03 | 3.30E-02 |
| Alditol metabolic process | 1.48E-03 | 1.67E-02 | 2.13E-03 |
| Alpha-amino acid metabolic process | 1.83E-02 | 6.40E-03 | 4.39E-02 |
| Ameboidal-type cell migration | 2.64E-03 | 1.49E-02 | 2.58E-02 |
| Aminoglycan catabolic process | 3.25E-03 | 3.59E-03 | 1.58E-02 |
| Anatomical structure development | 8.20E-07 | 1.90E-04 | 1.00E-04 |
| Anatomical structure formation involved in morphogenesis | 2.20E-04 | 2.69E-03 | 4.29E-03 |
| Anatomical structure homeostasis | 8.87E-03 | 2.34E-02 | 1.78E-02 |
| Anatomical structure morphogenesis | 1.40E-04 | 9.77E-03 | 1.01E-02 |
| Angiogenesis | 2.50E-05 | 9.50E-05 | 3.10E-05 |
| Animal organ development | 6.00E-07 | 3.90E-06 | 1.00E-04 |
| Animal organ morphogenesis | 5.76E-03 | 3.09E-03 | 4.65E-03 |
| Animal organ regeneration | 1.57E-03 | 9.07E-03 | 1.85E-02 |
| ATP generation from ADP | 3.70E-04 | 8.19E-03 | 3.75E-02 |
| Attachment of spindle microtubules to kinetochore | 2.08E-02 | 1.60E-03 | 2.64E-02 |
| Biological adhesion | 6.60E-04 | 2.60E-04 | 4.23E-02 |
| Blood circulation | 2.46E-03 | 2.50E-05 | 2.63E-03 |
| Blood vessel development | 1.40E-04 | 1.20E-04 | 1.10E-04 |
| Blood vessel morphogenesis | 9.30E-05 | 1.30E-04 | 5.00E-04 |
| C-terminal protein amino acid modification | 2.50E-03 | 4.89E-02 | 2.02E-02 |
| C-terminal protein lipidation | 1.34E-03 | 3.43E-02 | 2.22E-02 |
| Calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 2.61E-02 | 3.80E-02 | 1.24E-02 |
| Carbohydrate biosynthetic process | 2.13E-02 | 4.60E-04 | 1.39E-03 |
| Carbohydrate metabolic process | 3.50E-05 | 2.40E-05 | 9.25E-03 |
| Carboxylic acid biosynthetic process | 9.28E-03 | 3.70E-05 | 9.00E-04 |
| Carboxylic acid metabolic process | 4.40E-05 | 1.60E-05 | 6.80E-04 |
| Cardiovascular system development | 3.10E-04 | 9.30E-05 | 1.60E-04 |
| Cell adhesion | 4.80E-04 | 2.80E-04 | 3.80E-02 |
Additionally, the meta-analysis revealed that each of the three exposures affected PrECs differently with each yielding an exclusive gene expression signature. Exposure to 5 nM BPA perturbed 15 biological pathways (Fig. 3, orange), including apelin signaling (Table 3A, dark green) a key regulator of angiogenesis/cell proliferation/energy metabolism regulation, cell metabolism (orange), fat digestion/absorption and lysosome (white), focal adhesion (pink), neurotransmission and taste transduction (grey) and inflammation/immune response (light blue). Exposure to 25 nM BPA enriched 5 unique pathways (Fig. 3, green) related to protein synthesis (Table 3B, purple) and metabolism (orange) as well as antifolate resistance and the Fanconi anemia pathway (white). Exposure to EE2, the positive estrogenic control, affected 7 unique pathways (Fig. 3, purple), including circadian rhythm (Table 3C, white), metabolism (orange) and Wnt/HIF-1/PPAR signaling (dark green).
Table 3. Pathways that are uniquely enriched in PrECs exposed to 5 nM BPA, 25 nM BPA and 0.1 nM EE2. Orange: pathways related to metabolic and hormone biosynthesis, purple: pathways related to protein biogenesis, pink: pathways related to biological adhesion, grey: pathways related to neurotransmission and taste transduction, light blue: pathways related to inflammation and immune response, dark green: cell signaling pathways. (The color version of the table is available in the electronic copy of the article).
| A) Pathways Unique to 5 nM BPA | p-value |
|---|---|
| Apelin signaling pathway | 4.52E-02 |
| Drug metabolism - cytochrome P450 | 3.71E-02 |
| Fat digestion and absorption | 2.34E-02 |
| Focal adhesion | 9.05E-03 |
| Fructose and mannose metabolism | 4.72E-03 |
| Gap junction | 3.78E-02 |
| Glutamatergic synapse | 4.73E-02 |
| Glycolysis / Gluconeogenesis | 1.10E-03 |
| Lysosome | 2.46E-02 |
| Metabolism of xenobiotics by cytochrome P450 | 1.62E-02 |
| Phenylalanine metabolism | 4.25E-02 |
| Pyruvate metabolism | 8.55E-03 |
| Rheumatoid arthritis | 4.93E-02 |
| Serotonergic synapse | 2.73E-02 |
| Taste transduction | 2.69E-02 |
| B) Pathways unique to 25 nM BPA | p-value |
| Antifolate resistance | 1.90E-02 |
| Fanconi anemia pathway | 2.60E-02 |
| Ribosome | 9.87E-03 |
| Thiamine metabolism | 3.52E-02 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 3.52E-02 |
| C) Pathway name unique to 0.1 nM EE2 | p-value |
| Circadian rhythm | 4.26E-02 |
| Glycosaminoglycan degradation | 3.68E-02 |
| Glyoxylate and dicarboxylate metabolism | 9.81E-03 |
| HIF-1 signaling pathway | 3.11E-02 |
| PPAR signaling pathway | 4.89E-02 |
| Steroid hormone biosynthesis | 2.44E-02 |
| Wnt signaling pathway | 1.91E-02 |
Together these results suggest that each exposure has a unique impact on specific biological pathways. For instance, 5 nM BPA has a strong impact on neurological pathways such as gap junction, glutamatergic synapse and serotonergic synapse, a signature that is not observed in 25 nM BPA and 0.1 nM EE2 exposures. These findings highlight the non-estrogenic signature of BPA exposure.
3.3. Expression Levels of BPA-activated Receptors in PrECs: Treated Versus Basal Expression
As BPA has been previously shown to bind and activate several receptors other than ERα and ERβ (listed in the introduction), we examined their expression after exposure to BPA and EE2 and compared all exposures to the control group in the meta-analysis. We determined that ESR1 was upregulated only by the 25nM BPA exposure (Fig. 4), and ESR2 remained unaffected (not present in meta-analysis). Both ESRRA and THRA were upregulated after exposure to BPA and EE2, with 25nM BPA causing a robust increase in expression similar to the positive control EE2 (Fig. 4). At the other end of the spectrum, BPA and EE2 exposures profoundly decreased GPER, NR3C1 and APLN expression levels and even the low-level BPA exposure caused similar decreases compared to EE2 (Fig. 4).
Fig. (4).
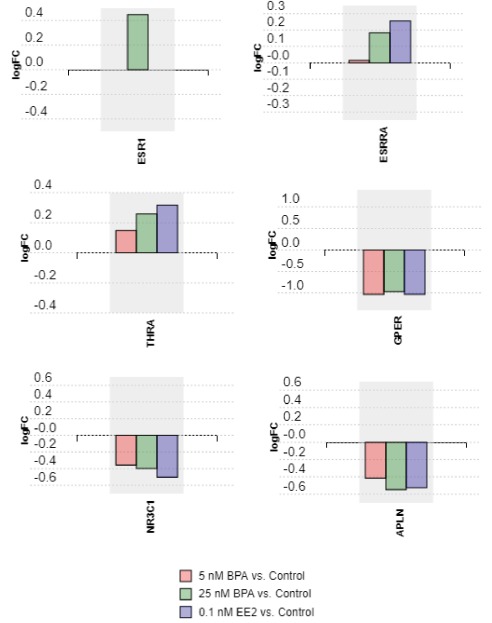
Analysis of nuclear hormone receptor transcripts, G protein-coupled estrogen receptor 1 and the endogenous ligand for the G-protein coupled apelin receptor in EE2 and BPA exposed cells relative to controls. .
3.4. Effects of BPA Exposure on Histone and DNA Methylation Modifying Enzymes
To assess the effects of BPA on epigenetic factors, we compiled a list of all histone and DNA methylation modifying enzymes present in our DE analysis and generated a bar plot (Fig. 5 and Supplementary Table S1 (286.3KB, pdf) ). Two DNA methyltransferases (DNMT1 and DNMT3B), as well as 2 histone deacetylases (HDAC4 and SIRT5), 5 histone methyltransferases (EZH2, SETD2, SETD6, SETMAR, SUV39H1) and UHRF1 were present in our DE gene list and upregulated in all 3 exposures. In contrast, 2 histone deacetylases (HDAC9 and SIRT2) and 3 histone methyltransferases (MLL3, SETD5, SMYD3) were downregulated in all 3 exposures. Certain genes were only significantly upregulated (EHMT2, SETD8) and downregulated (HDAC3, HDAC5, HDAC8, SIRT1) by the positive estrogenic control EE2, while other genes were upregulated (SETD1A) or downregulated (MLL5) by both EE2 and 25nM BPA exposures. HDAC7A was only upregulated in 25nM BPA exposure.
Fig. (5).
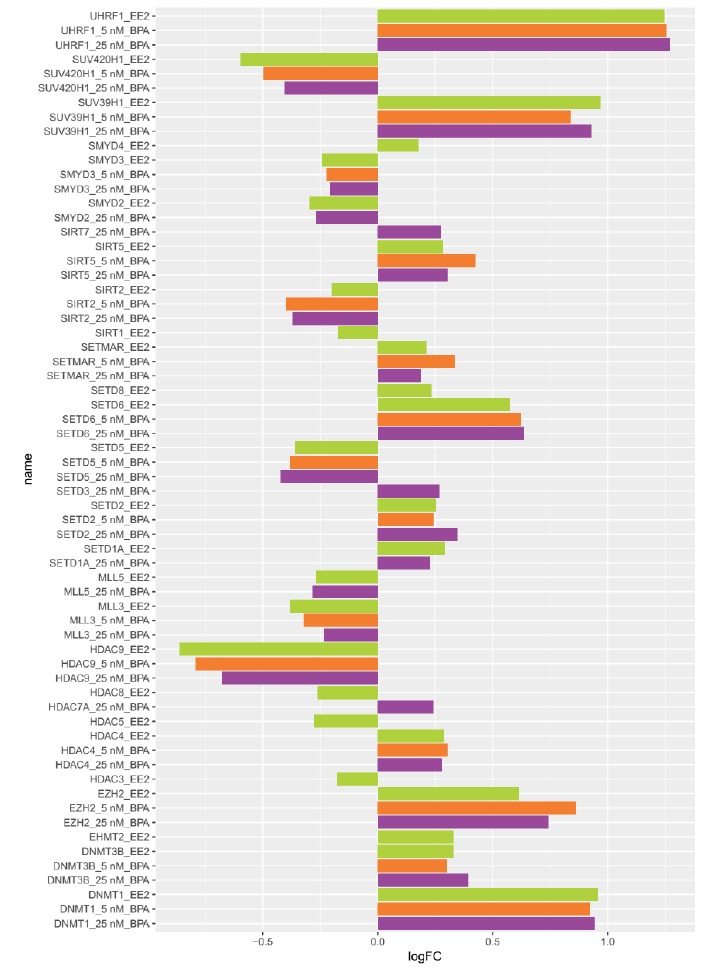
Expression levels of transcripts encoding histone and DNA methylation modifying enzymes after exposure to 5 nM BPA (orange), 25 nM BPA (purple) and 0.1 nM EE2 (green). Expression is shown as log Fold Change (logFC) relative to the control for genes with a q-value ≤ 0.1. Genes not DE in the comparisons are not shown in the plot. .
4. DISCUSSION
4.1. BPA Induces Estrogen-like Response in PrECs
We demonstrated that the synthetic estrogen EE2, used here as estrogenic control, and BPA both have strong impacts on transcripts encoding proteins belonging to fundamental signaling pathways and biological processes in PrECs comparable to the effects of estrogen reported above, confirming that they can both be categorized as xenoestrogens. BPA (5 and 25 nM) and EE2 (0.1nM) shared approximately 54% of the DE genes and 24 biological pathways in our in vitro study. There is evidence from rodent models and human prostate cell lines that BPA can influence carcinogenesis by modulating prostate cancer cell proliferation, and for some tumors, stimulating progression [67, 68]. Our data showed that all three exposures enriched cell cycle, DNA replication and mismatch repair biological pathways, suggesting that BPA and EE2 induce at a molecular level the potential for cancer development and progression in prostate cells. This data is consistent with previous studies showing that BPA induces prostate cancer (PCa) cells migration via modulation of the ion channel protein expression involved in calcium entry and in cancer cell migration [49].
Additionally, all three exposures perturbed several biological pathways related to angiogenesis and blood circulation. In fact, new growth in the vascular network is important since the proliferation, as well as metastatic spread, of cancer cells depends on an adequate supply of oxygen and nutrients and the removal of waste products [69]. BPA has been shown previously to promote angiogenesis later in life following prenatal exposure [70], suggesting that early life exposure to BPA can imprint the epigenome and increase cancer risk later in life. Prins et al. [71] reported that perinatal exposure to BPA or to estradiol increases the susceptibility to develop prostate cancer either spontaneously or after a second estrogenic exposure during adulthood.
Our data showed a strong genomic signature of metabolic pathways with all exposures. Prenatal exposure to BPA exhibited important effects in adult males including food intake, an increase in body weight and liver weight, abdominal adipocyte mass, number and volume, and in serum leptin and insulin, and a decrease in serum adiponectin and in glucose tolerance [72], highlighting that BPA is both an endocrine and metabolic disruptor [7].
Inflammation and immune response pathways were also enriched by all three exposures. This is consistent with several studies demonstrating that pro- and anti-inflammatory response, as well as the production of natural killer cells, vary according to estrogen levels [73]. More specifically, our system level analysis revealed that all three exposures commonly affected cytokine-cytokine receptor interaction, leukocyte transendothelial migration and natural killer cell mediated cytotoxicity biological pathways as well as BP GO terms related to acute inflammatory response. BPA has been shown to induce an inflammation-like response in human adipocytes by inducing the release of inflammatory factors, such as IL-6 and IFN-γ, and activate pro-inflammatory pathways including JNK, JAK/STAT and NFκB pathways [74].
4.2. Non-estrogenic Effects of BPA in PrECs
The main goal of this study was to tease out the non-estrogenic effects of BPA in PrECs by examining the unique genomic signature BPA has in contrast to the positive estrogenic control EE2. Additionally, we also aimed to compare the 5 and 25 nM exposures. Our in vitro study revealed that BPA exposure (both at 5 and 25 nM) results in a BPA specific signature and that approximately 14% of the DE genes are strictly affected by BPA (and not by EE2), suggesting that BPA also induces a non-estrogenic response in PrECs. PrECs treated with 5 and 25 nM BPA had approximately the same number of DE genes (7,182 and 7,650 DE genes respectively) but 5 nM BPA perturbed about 3 times as many biological pathways and 4 times as many predicted miRNAs than 25 nM BPA did, suggesting that at low doses BPA has a greater biological impact on the PrEC transcriptome. The higher dose of BPA affected pathways related to protein biogenesis and metabolism-hormone biosynthesis whereas the lower dose had more far reaching effects including perturbation of the apelin signaling pathway, focal adhesion, glycolysis/gluconeogenesis biological pathways.
The apelin signaling pathway was the most significantly enriched pathway in the gene list that is unique to the 5 nM BPA exposure, suggesting that BPA interferes with apelin expression or binds to the G-protein-coupled apelin receptor (APJ). Apelin and APJ are implicated in different key physiological processes such as angiogenesis, cell proliferation and energy metabolism regulation [75, 76]. Wan et al. [77] have recently described a miR-224/apelin axis that regulates cell invasion and migration in prostate cancer. Additionally, a role in the regulation of blood pressure, cardiac function, and fluid homeostasis via the renin-angiotensin-aldosterone
system (RAS) has been suggested [78]. Given that a locally based RAS exists in the prostate [79], further examination of the effect of BPA on the apelin signaling pathway and how apelin interferes with the RAS in the prostate is required to gain a better understanding of the impact BPA can have on this organ. No direct evidence exists yet of direct interaction between BPA and APJ but a recent study demonstrated that 1 nM BPA exposure increased apelin expression and secretion in ovarian carcinoma cell lines via activation of the peroxisome proliferator-activated receptor γ (PPARγ) [80]. Of
note is the fact it has been shown previously that PPARγ is expressed on prostate-derived cells [81] and that BPA can bind to PPARγ [82].
Focal adhesion was another biological pathway significantly enriched in PrECs exposed to 5 nM BPA. This is consistent with another study in which BPA caused alterations in the focal adhesion pathway following fetal exposure in the peri-ductal stroma of mice [83]. This pathway is important for cell motility, cell proliferation, cell differentiation, regulation of gene expression and cell survival as these biological processes are regulated by cell-matrix adhesion [84, 85]. A recent study has shown that BPA promotes migration, invasion, and an increase in the number of focal contacts in breast cancer cells and induces activation of FAK, Src, and ERK2 migration-promoting kinases [86].
Exposure to 5 nM BPA also enriched the gap junction pathway. Gap junctions contain intercellular channels that allow direct communication between the cytosolic compartments of adjacent cells and the direct transfer of small molecules including ions, amino acids, nucleotides, second messengers and other metabolites essential for many physiological events, including embryonic development, electrical coupling, metabolic transport, apoptosis, and tissue homeostasis [87, 88]. It has previously been shown that compared to normal cells, gap‐junctional communication in malignant cells is either reduced (or undetectable) via a cAMP‐dependent signal transduction pathway [89]. Normal PrECs express mRNA transcripts for connexin-32 (GJB1), connexin-40 (GJA5) and connexin-43 (GJA1) whereas in malignant cells, the expression of GJA1 and GJB1 are decreased [90], signifying that as cancer cells become autonomous, a decrease in intercellular communicative activity occurs. In our datasets, expression of both GJB1 and GJA5 were not detected and GJA1 was significantly down-regulated in PrECs exposed to BPA and EE2 relative to control (Supplementary Fig S2 (286.3KB, pdf) ). This suggests that exposures to BPA and EE2 shifted the profile of normal PrECs towards a malignant signature associated with perturbation of gap junction communication.
To further examine this signature, we have to consider other BPA-activated receptors expressed in PrECs. BPA has been shown to bind to the androgen receptor (AR) present in prostate cells, causing an anti-androgenic effect [91-93]. Given that androgens and AR play a pivotal role in the expression of the male phenotype, this BPA-induced anti-androgen effect could have far-reaching impacts on male reproduction and physiology.
The nuclear estrogen-related receptor-gamma (ERRγ) receptor also has a good affinity for BPA and is expressed in PrECs [94-96]. ERRγ is an inducible transcription factor and
a downstream mediator of endocrine and metabolic signals such as glucagon, insulin, and endocannabinoids [97]. Nuclear receptors mediate gene transcription through interactions with coactivators and corepressors, a group of proteins that modulates chromatin structure and the recruitment of the basal transcription machinery [98, 99]. ERRγ is an in vivo receptor of BPA. In zebrafish during development ERRγ mediates BPA-induced malformations in otoliths, structures found in the saccule and utricle of the inner ear [19]. BPA strongly binds to ERRγ in a dose-dependent manner with an IC50 value of 13.1 nM [95]. We did not observe any perturbations in ERRγ expression levels upon EE2 or BPA treatments suggesting that BPA exposure does not influence the expression of ERRγ. Few studies have examined the function of ERR in the prostate but recent studies have shown that it is a crucial mediator of multiple endocrine and metabolic signals and it plays important roles in pathological conditions such as insulin resistance, alcoholic liver injury, and cardiac hypertrophy, and controls energy metabolism in the heart, skeletal muscle, and pancreatic β cells [97]. ERRγ has been shown to regulate mitochondrial-oxidative metabolism and promote angiogenesis, mitochondrial biogenesis, and oxidative remodeling [100, 101].
4.3. Effects of BPA on Histone and DNA Methylation Modifying Enzymes
The epigenetic effects of BPA were first described by Dolinoy et al. (2007) who showed that BPA exposure during pregnancy in Agouti viable (Avy) yellow mice causes hypomethylation of DNA on the epigenome of the offspring and a change in coat color [102]. Doherty et al. (2011) demonstrated that BPA promotes translation of the histone methyltransferase EZH2 (Enhancer of Zeste Homolog 2), which increases the histone modification H3K27me3, a mark of transcriptional repression [39, 103, 104]. Doshi et al. (2011) reported that neonatal BPA exposure in male rats led to hypermethylation of the promoter region of ESR1 and ESR2 and downregulation of both ESRs expression in adult testes, and increased DNMT3a and 3b expression by 2 fold, highlighting that BPA induces aberrant DNA methylation [105]. Santangeli et al. (2016) also described enrichment of H3K4me3 and H3K27me3 histone modifications in genes of interest and increased DNMT1 and DNMT3 expression levels in BPA exposed zebrafish [106]. Our data is consistent with these findings since we observed a significant increase in EZH2, DNMT1 and DNMT3B after exposure to BPA and EE2.
The gene encoding the transcription factor ubiquitin-like with PHD and ring finger domains 1 (UHRF1) controls the expression of topoisomerase IIα (TOP2A) and is involved in a wide range of physiological and pathological phenomena, including cancer development and metastasis, by controlling gene expression through regulating epigenetic mechanisms, including DNA methylation, histone deacetylation, histone methylation, and histone ubiquitination [107, 108]. In our datasets, UHRF1 is upregulated in all three exposures, a signature that is observed in many types of cancer, and contributes to cancer cell activation through hyper-methylation of tumor-suppressor genes such as BRCA1, CDKN2A, p73, and RASSF1, making UHRF1 a progression marker in cancer [109].
CONCLUSION
Low levels of BPA and EE2, comparable to those routinely detected in the environment, critically affect global transcriptomic responses in prostate epithelial cells. Comparing and contrasting BPA to EE2, we were able to tease out the estrogenic from non-estrogenic transcriptomic responses that BPA induces. Similar to EE2, (1) BPA impacts expression of genes belonging to fundamental pathways that regulates cell proliferation, cell cycle, DNA replication, mismatch repair and angiogenesis, all of which are relevant for cancer development and progression in prostate cells, (2) BPA impacts endocrine and metabolic transcripts in prostate cells, and (3) BPA perturbs expression of transcripts belonging to inflammation and immune response pathways.
Interestingly, BPA also has a non-estrogenic signature in PrECs and we showed in this study that BPA has profound effects on transcripts related to biological pathways involved in regulation of energy metabolism, protein biogenesis and metabolism-hormone biosynthesis, focal adhesion and gap junctions. Additionally, BPA perturbed the expression of the receptors AR and ERRγ, suggesting that its effects are driven via other receptors beside estrogen receptors.
We also show here that BPA can also deregulate expression of key epigenetic factors including EZH2, DNMT1, DNMT3B and UHRF1, highlighting that BPA-induced transcriptional perturbations have the potential to have an influence at the level of the epigenome.
In conclusion, we note that this study focused on an in vitro model using primary prostate cells and differences may exist with the in vivo BPA exposure response. The results presented here provide a compelling rationale for the need for future studies on BPA exposure in human tissues, using newer technologies such as RNAseq and mass spectrometry.
ACKNOWLEDGEMENTS
We thank the staff and faculty at the MUSC Center for Genomic Medicine and the UCSD Biomedical Genomics Facility (BIOGEM). We are grateful to Dr. Michael Baker, Dr. William B. Glen and Dr. E. Starr Hazard for many useful discussions.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All the experimental procedures were approved by the Research Protocol Oversight Committee at The University California, San Diego.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available at http://grande.sobs.qub.ac.uk/data, microarray data has been deposited in the EBI ArrayExpress Database, accession number E-MTAB-7576.
FUNDING
This work was supported by the funding from SC EPSCoR (Gary Hardiman, 2017) and start-up funding from the College of Medicine at the Medical University of South Carolina and Queen’s University Belfast.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Diamanti-Kandarakis E., Bourguignon J.P., Giudice L.C., Hauser R., Prins G.S., Soto A.M., Zoeller R.T., Gore A.C. Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocr. Rev. 2009;30(4):293–342. doi: 10.1210/er.2009-0002. [http://dx.doi.org/10.1210/er.2009-0002]. [PMID: 19502515]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterni I., Granchi C., Minutolo F. Risks and benefits related to alimentary exposure to xenoestrogens. Crit. Rev. Food Sci. Nutr. 2016;57(16):3384–3404. doi: 10.1080/10408398.2015.1126547. [PMID: 26744831]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang J.H., Kondo F., Katayama Y. Human exposure to bisphenol A. Toxicology. 2006;226(2-3):79–89. doi: 10.1016/j.tox.2006.06.009. [http://dx.doi.org/10.1016/j.tox.2006.06.009]. [PMID: 16860916]. [DOI] [PubMed] [Google Scholar]
- 4.Belfroid A., van Velzen M., van der Horst B., Vethaak D. Occurrence of bisphenol A in surface water and uptake in fish: Evaluation of field measurements. Chemosphere. 2002;49(1):97–103. doi: 10.1016/s0045-6535(02)00157-1. [http://dx.doi.org/10.1016/S0045-6535(02)00157-1]. [PMID: 12243336]. [DOI] [PubMed] [Google Scholar]
- 5.Rubin B.S., Bisphenol A. An endocrine disruptor with widespread exposure and multiple effects. J. Steroid Biochem. Mol. Biol. 2011;127(1-2):27–34. doi: 10.1016/j.jsbmb.2011.05.002. [http://dx.doi.org/10.1016/j.jsbmb.2011.05.002]. [PMID: 21605673]. [DOI] [PubMed] [Google Scholar]
- 6.Prins G.S., Korach K.S. The role of estrogens and estrogen receptors in normal prostate growth and disease. Steroids. 2008;73(3):233–244. doi: 10.1016/j.steroids.2007.10.013. [http://dx.doi.org/10.1016/j.steroids.2007.10.013]. [PMID: 18093629]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenichel P., Chevalier N., Brucker-Davis F., Bisphenol A. An endocrine and metabolic disruptor. Ann. Endocrinol. (Paris) 2013;74(3):211–220. doi: 10.1016/j.ando.2013.04.002. [http://dx.doi.org/10.1016/j.ando.2013.04.002]. [PMID: 23796010]. [DOI] [PubMed] [Google Scholar]
- 8.Sugimura Y., Cunha G.R., Yonemura C.U., Kawamura J. Temporal and spatial factors in diethylstilbestrol-induced squamous metaplasia of the developing human prostate. Hum. Pathol. 1988;19(2):133–139. doi: 10.1016/s0046-8177(88)80340-x. [http://dx.doi.org/10.1016/S0046-8177(88)80340-X]. [PMID: 3343029]. [DOI] [PubMed] [Google Scholar]
- 9.Härkönen P.L., Mäkelä S.I. Role of estrogens in development of prostate cancer. J. Steroid Biochem. Mol. Biol. 2004;92(4):297–305. doi: 10.1016/j.jsbmb.2004.10.016. [http://dx.doi.org/10.1016/j.jsbmb.2004.10.016]. [PMID: 15663993]. [DOI] [PubMed] [Google Scholar]
- 10.Pasquali D., Rossi V., Esposito D., Abbondanza C., Puca G.A., Bellastella A., Sinisi A.A. Loss of estrogen receptor β expression in malignant human prostate cells in primary cultures and in prostate cancer tissues. J. Clin. Endocrinol. Metab. 2001;86(5):2051–2055. doi: 10.1210/jcem.86.5.7441. [http://dx.doi.org/10.1210/jc.86.5.2051]. [PMID: 11344205]. [DOI] [PubMed] [Google Scholar]
- 11.Cox R.L., Crawford E.D. Estrogens in the treatment of prostate cancer. J. Urol. 1995;154(6):1991–1998. [http://dx.doi.org/10.1016/S0022-5347(01)66670-9]. [PMID: 7500443]. [PubMed] [Google Scholar]
- 12.Huggins C., Scott W.W., Hodges C.V. Studies on Prostatic Cancer. III. The effects of fever, of desoxycorticosterone and of estrogen on clinical patients with metastatic carcinoma of the prostate. J. Urol. 1941;46(5):997–1006. [http://dx.doi.org/10.1016/S0022-5347(17)71004-X]. [Google Scholar]
- 13.Huang L., Pu Y., Alam S., Birch L., Prins G.S. Estrogenic regulation of signaling pathways and homeobox genes during rat prostate development. J. Androl. 2004;25(3):330–337. doi: 10.1002/j.1939-4640.2004.tb02796.x. [http://dx.doi.org/10.1002/j.1939-4640.2004.tb02796.x]. [PMID: 15064308]. [DOI] [PubMed] [Google Scholar]
- 14.Prins G.S., Birch L., Couse J.F., Choi I., Katzenellenbogen B., Korach K.S. Estrogen imprinting of the developing prostate gland is mediated through stromal estrogen receptor α: Studies with alphaERKO and betaERKO mice. Cancer Res. 2001;61(16):6089–6097. [PMID: 11507058]. [PubMed] [Google Scholar]
- 15.Risbridger G., Wang H., Young P., Kurita T., Wang Y.Z., Lubahn D., Gustafsson J.A., Cunha G. Evidence that epithelial and mesenchymal estrogen receptor-α mediates effects of estrogen on prostatic epithelium. Dev. Biol. 2001;229(2):432–442. doi: 10.1006/dbio.2000.9994. [http://dx.doi.org/10.1006/dbio.2000.9994]. [PMID: 11150243]. [DOI] [PubMed] [Google Scholar]
- 16.Multigner L., Ndong J.R., Giusti A., Romana M., Delacroix-Maillard H., Cordier S., Jégou B., Thome J.P., Blanchet P. Chlordecone exposure and risk of prostate cancer. J. Clin. Oncol. 2010;28(21):3457–3462. doi: 10.1200/JCO.2009.27.2153. [http://dx.doi.org/10.1200/JCO.2009.27.2153]. [PMID: 20566993]. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Burns K.A., Arao Y., Luh C.J., Korach K.S. Differential estrogenic actions of endocrine-disrupting chemicals bisphenol A, bisphenol AF, and zearalenone through estrogen receptor α and β in vitro. Environ. Health Perspect. 2012;120(7):1029–1035. doi: 10.1289/ehp.1104689. [http://dx.doi.org/10.1289/ehp.1104689]. [PMID: 22494775]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews J.B., Twomey K., Zacharewski T.R. In vitro and in vivo interactions of bisphenol A and its metabolite, bisphenol A glucuronide, with estrogen receptors α and β. Chem. Res. Toxicol. 2001;14(2):149–157. doi: 10.1021/tx0001833. [http://dx.doi.org/10.1021/tx0001833]. [PMID: 11258963]. [DOI] [PubMed] [Google Scholar]
- 19.Tohmé M., Prud’homme S.M., Boulahtouf A., Samarut E., Brunet F., Bernard L., Bourguet W., Gibert Y., Balaguer P., Laudet V. Estrogen-related receptor γ is an in vivo receptor of bisphenol A. FASEB J. 2014;28(7):3124–3133. doi: 10.1096/fj.13-240465. [http://dx.doi.org/10.1096/fj.13-240465]. [PMID: 24744145]. [DOI] [PubMed] [Google Scholar]
- 20.Sohoni P., Sumpter J.P. Several environmental oestrogens are also anti-androgens. J. Endocrinol. 1998;158(3):327–339. doi: 10.1677/joe.0.1580327. [http://dx.doi.org/10.1677/joe.0.1580327]. [PMID: 9846162]. [DOI] [PubMed] [Google Scholar]
- 21.Zoeller R.T. Environmental chemicals as thyroid hormone analogues: New studies indicate that thyroid hormone receptors are targets of industrial chemicals? Mol. Cell. Endocrinol. 2005;242(1-2):10–15. doi: 10.1016/j.mce.2005.07.006. [http://dx.doi.org/10.1016/j.mce.2005.07.006]. [PMID: 16150534]. [DOI] [PubMed] [Google Scholar]
- 22.Bouskine A., Nebout M., Brücker-Davis F., Benahmed M., Fenichel P. Low doses of bisphenol A promote human seminoma cell proliferation by activating PKA and PKG via a membrane G-protein-coupled estrogen receptor. Environ. Health Perspect. 2009;117(7):1053–1058. doi: 10.1289/ehp.0800367. [http://dx.doi.org/10.1289/ehp.0800367]. [PMID: 19654912]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sargis R.M., Johnson D.N., Choudhury R.A., Brady M.J. Environmental endocrine disruptors promote adipogenesis in the 3T3-L1 cell line through glucocorticoid receptor activation. Obesity (Silver Spring) 2010;18(7):1283–1288. doi: 10.1038/oby.2009.419. [http://dx.doi.org/10.1038/oby.2009.419]. [PMID: 19927138]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birceanu O., Mai T., Vijayan M.M. Maternal transfer of bisphenol A impacts the ontogeny of cortisol stress response in rainbow trout. Aquat. Toxicol. 2015;168:11–18. doi: 10.1016/j.aquatox.2015.09.002. [http://dx.doi.org/10.1016/j.aquatox.2015.09.002]. [PMID: 26398930]. [DOI] [PubMed] [Google Scholar]
- 25.Sui Y., Park S.H., Helsley R.N., Sunkara M., Gonzalez F.J., Morris A.J., Zhou C. Bisphenol A increases atherosclerosis in pregnane X receptor-humanized ApoE deficient mice. J. Am. Heart Assoc. 2014;3(2):e000492. doi: 10.1161/JAHA.113.000492. [http://dx.doi.org/10.1161/JAHA.113.000492]. [PMID: 24755147]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forner-Piquer I., Mylonas C.C., Calduch-Giner J., Maradonna F., Gioacchini G., Allarà M., Piscitelli F., Di Marzo V., Pérez-Sánchez J., Carnevali O. Endocrine disruptors in the diet of male Sparus aurata: Modulation of the endocannabinoid system at the hepatic and central level by Di-isononyl phthalate and Bisphenol A. Environ. Int. 2018;119:54–65. doi: 10.1016/j.envint.2018.06.011. [http://dx.doi.org/10.1016/j.envint.2018.06.011]. [PMID: 29933238]. [DOI] [PubMed] [Google Scholar]
- 27.Martella A., Silvestri C., Maradonna F., Gioacchini G., Allarà M., Radaelli G., Overby D.R., Di Marzo V., Carnevali O. Bisphenol A induces fatty liver by an endocannabinoid-mediated positive feedback loop. Endocrinology. 2016;157(5):1751–1763. doi: 10.1210/en.2015-1384. [http://dx.doi.org/10.1210/en.2015-1384]. [PMID: 27014939]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schug T.T., Janesick A., Blumberg B., Heindel J.J. Endocrine disrupting chemicals and disease susceptibility. J. Steroid Biochem. Mol. Biol. 2011;127(3-5):204–215. doi: 10.1016/j.jsbmb.2011.08.007. [http://dx.doi.org/10.1016/j.jsbmb.2011.08.007]. [PMID: 21899826]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calafat A.M., Ye X., Wong L.Y., Reidy J.A., Needham L.L. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ. Health Perspect. 2008;116(1):39–44. doi: 10.1289/ehp.10753. [http://dx.doi.org/10.1289/ehp.10753]. [PMID: 18197297]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howdeshell K.L., Hotchkiss A.K., Thayer K.A., Vandenbergh J.G., vom Saal F.S. Exposure to bisphenol A advances puberty. Nature. 1999;401(6755):763–764. doi: 10.1038/44517. [http://dx.doi.org/10.1038/44517]. [PMID: 10548101]. [DOI] [PubMed] [Google Scholar]
- 31.Cabaton N.J., Wadia P.R., Rubin B.S., Zalko D., Schaeberle C.M., Askenase M.H., Gadbois J.L., Tharp A.P., Whitt G.S., Sonnenschein C., Soto A.M. Perinatal exposure to environmentally relevant levels of bisphenol A decreases fertility and fecundity in CD-1 mice. Environ. Health Perspect. 2011;119(4):547–552. doi: 10.1289/ehp.1002559. [http://dx.doi.org/10.1289/ehp.1002559]. [PMID: 21126938]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salian S., Doshi T., Vanage G. Neonatal exposure of male rats to Bisphenol A impairs fertility and expression of sertoli cell junctional proteins in the testis. Toxicology. 2009;265(1-2):56–67. doi: 10.1016/j.tox.2009.09.012. [http://dx.doi.org/10.1016/j.tox.2009.09.012]. [PMID: 19782717]. [DOI] [PubMed] [Google Scholar]
- 33.Vitku J., Heracek J., Sosvorova L., Hampl R., Chlupacova T., Hill M., Sobotka V., Bicikova M., Starka L. Associations of bisphenol A and polychlorinated biphenyls with spermatogenesis and steroidogenesis in two biological fluids from men attending an infertility clinic. Environ. Int. 2016;89-90:166–173. doi: 10.1016/j.envint.2016.01.021. [http://dx.doi.org/10.1016/j.envint.2016.01.021]. [PMID: 26863184]. [DOI] [PubMed] [Google Scholar]
- 34.Manikkam M., Tracey R., Guerrero-Bosagna C., Skinner M.K. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 2013;8(1):e55387. doi: 10.1371/journal.pone.0055387. [http://dx.doi.org/10.1371/journal.pone.0055387]. [PMID: 23359474]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed S., Atlas E. Bisphenol S- and bisphenol A-induced adipogenesis of murine preadipocytes occurs through direct peroxisome proliferator-activated receptor gamma activation. Int. J. Obes. 2016;40(10):1566–1573. doi: 10.1038/ijo.2016.95. [http://dx.doi.org/10.1038/ijo.2016.95]. [PMID: 27273607]. [DOI] [PubMed] [Google Scholar]
- 36.Grün F., Blumberg B. Perturbed nuclear receptor signaling by environmental obesogens as emerging factors in the obesity crisis. Rev. Endocr. Metab. Disord. 2007;8(2):161–171. doi: 10.1007/s11154-007-9049-x. [http://dx.doi.org/10.1007/s11154-007-9049-x]. [PMID: 17657605]. [DOI] [PubMed] [Google Scholar]
- 37.Ohlstein J.F., Strong A.L., McLachlan J.A., Gimble J.M., Burow M.E., Bunnell B.A. Bisphenol A enhances adipogenic differentiation of human adipose stromal/stem cells. J. Mol. Endocrinol. 2014;53(3):345–353. doi: 10.1530/JME-14-0052. [http://dx.doi.org/10.1530/JME-14-0052]. [PMID: 25143472]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.vom Saal F.S., Myers J.P. Bisphenol A and risk of metabolic disorders. JAMA. 2008;300(11):1353–1355. doi: 10.1001/jama.300.11.1353. [http://dx.doi.org/10.1001/jama.300.11.1353]. [PMID: 18799451]. [DOI] [PubMed] [Google Scholar]
- 39.Doherty L.F., Bromer J.G., Zhou Y., Aldad T.S., Taylor H.S. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BPA) increases EZH2 expression in the mammary gland: An epigenetic mechanism linking endocrine disruptors to breast cancer. Horm. Cancer. 2010;1(3):146–155. doi: 10.1007/s12672-010-0015-9. [http://dx.doi.org/10.1007/s12672-010-0015-9]. [PMID: 21761357]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prins G.S., Birch L., Tang W.Y., Ho S.M. Developmental estrogen exposures predispose to prostate carcinogenesis with aging. Reprod. Toxicol. 2007;23(3):374–382. doi: 10.1016/j.reprotox.2006.10.001. [http://dx.doi.org/10.1016/j.reprotox.2006.10.001]. [PMID: 17123779]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuomo D., Porreca I., Cobellis G., Tarallo R., Nassa G., Falco G., Nardone A., Rizzo F., Mallardo M., Ambrosino C. Carcinogenic risk and Bisphenol A exposure: A focus on molecular aspects in endoderm derived glands. Mol. Cell. Endocrinol. 2017;457:20–34. doi: 10.1016/j.mce.2017.01.027. [http://dx.doi.org/10.1016/j.mce.2017.01.027]. [PMID: 28111205]. [DOI] [PubMed] [Google Scholar]
- 42.Gomez A.L., Delconte M.B., Altamirano G.A., Vigezzi L., Bosquiazzo V.L., Barbisan L.F., Ramos J.G., Luque E.H., Muñoz-de-Toro M., Kass L. Perinatal exposure to bisphenol A or diethylstilbestrol increases the susceptibility to develop mammary gland lesions after estrogen replacement therapy in middle-aged rats. Horm. Cancer. 2017;8(2):78–89. doi: 10.1007/s12672-016-0282-1. [http://dx.doi.org/10.1007/s12672-016-0282-1]. [PMID: 28078498]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan A.V., Stathis P., Permuth S.F., Tokes L., Feldman D. Bisphenol-A: An estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology. 1993;132(6):2279–2286. doi: 10.1210/endo.132.6.8504731. [http://dx.doi.org/10.1210/endo.132.6.8504731]. [PMID: 8504731]. [DOI] [PubMed] [Google Scholar]
- 44.Welshons W.V., Nagel S.C., vom Saal F.S. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology. 2006;147(Suppl. 6):S56–S69. doi: 10.1210/en.2005-1159. [http://dx.doi.org/10.1210/en.2005-1159]. [PMID: 16690810]. [DOI] [PubMed] [Google Scholar]
- 45.vom Saal F.S., Welshons W.V. Large effects from small exposures. II. The importance of positive controls in low-dose research on bisphenol A. Environ. Res. 2006;100(1):50–76. doi: 10.1016/j.envres.2005.09.001. [http://dx.doi.org/10.1016/j.envres.2005.09.001]. [PMID: 16256977]. [DOI] [PubMed] [Google Scholar]
- 46.Okuda K., Takiguchi M., Yoshihara S. In vivo estrogenic potential of 4-methyl-2,4-bis(4-hydroxyphenyl)pent-1-ene, an active metabolite of bisphenol A, in uterus of ovariectomized rat. Toxicol. Lett. 2010;197(1):7–11. doi: 10.1016/j.toxlet.2010.04.017. [http://dx.doi.org/10.1016/j.toxlet.2010.04.017]. [PMID: 20435109]. [DOI] [PubMed] [Google Scholar]
- 47.Baker M.E., Chandsawangbhuwana C. 3D models of MBP, a biologically active metabolite of bisphenol A, in human estrogen receptor α and estrogen receptor β. PLoS One. 2012;7(10):e46078. doi: 10.1371/journal.pone.0046078. [http://dx.doi.org/10.1371/journal.pone.0046078]. [PMID: 23056236]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jemal A., Tiwari R.C., Murray T., Ghafoor A., Samuels A., Ward E., Feuer E.J., Thun M.J. American Cancer Society. Cancer statistics, 2004. CA Cancer J. Clin. 2004;54(1):8–29. doi: 10.3322/canjclin.54.1.8. [http://dx.doi.org/10.3322/canjclin.54.1.8]. [PMID: 14974761]. [DOI] [PubMed] [Google Scholar]
- 49.Derouiche S., Warnier M., Mariot P., Gosset P., Mauroy B., Bonnal J.L., Slomianny C., Delcourt P., Prevarskaya N., Roudbaraki M. Bisphenol A stimulates human prostate cancer cell migration via remodelling of calcium signalling. Springerplus. 2013;2(1):54. doi: 10.1186/2193-1801-2-54. [http://dx.doi.org/10.1186/2193-1801-2-54]. [PMID: 23450760]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho S.M., Rao R., To S., Schoch E., Tarapore P. Bisphenol A and its analogues disrupt centrosome cycle and microtubule dynamics in prostate cancer. Endocr. Relat. Cancer. 2017;24(2):83–96. doi: 10.1530/ERC-16-0175. [http://dx.doi.org/10.1530/ERC-16-0175]. [PMID: 27998958]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarapore P., Ying J., Ouyang B., Burke B., Bracken B., Ho S.M. Exposure to bisphenol A correlates with early-onset prostate cancer and promotes centrosome amplification and anchorage-independent growth in vitro. PLoS One. 2014;9(3):e90332. doi: 10.1371/journal.pone.0090332. [http://dx.doi.org/10.1371/journal.pone.0090332]. [PMID: 24594937]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho S-M., Tang W.Y., Belmonte de Frausto J., Prins G.S. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66(11):5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [http://dx.doi.org/10.1158/0008-5472.CAN-06-0516]. [PMID: 16740699]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prins G.S., Tang W.Y., Belmonte J., Ho S.M. Developmental exposure to bisphenol A increases prostate cancer susceptibility in adult rats: Epigenetic mode of action is implicated. Fertil. Steril. 2008;89(Suppl. 2):e41. doi: 10.1016/j.fertnstert.2007.12.023. [http://dx.doi.org/10.1016/j.fertnstert.2007.12.023]. [PMID: 18308059]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burton K., Shaw L., Morey L.M. Differential effect of estradiol and bisphenol A on Set8 and Sirt1 expression in prostate cancer. Toxicol. Rep. 2015;2:817–823. doi: 10.1016/j.toxrep.2015.01.016. [http://dx.doi.org/10.1016/j.toxrep.2015.01.016]. [PMID: 28962417]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheong A., Zhang X., Cheung Y.Y., Tang W.Y., Chen J., Ye S.H., Medvedovic M., Leung Y.K., Prins G.S., Ho S.M. DNA methylome changes by estradiol benzoate and bisphenol A links early-life environmental exposures to prostate cancer risk. Epigenetics. 2016;11(9):674–689. doi: 10.1080/15592294.2016.1208891. [http://dx.doi.org/10.1080/15592294.2016.1208891]. [PMID: 27415467]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhl H. Pharmacology of estrogens and progestogens: Influence of different routes of administration. Climacteric. 2005;8(Suppl. 1):3–63. doi: 10.1080/13697130500148875. [http://dx.doi.org/10.1080/13697130500148875]. [PMID: 16112947]. [DOI] [PubMed] [Google Scholar]
- 57.Schug T.T., Johnson A.F., Birnbaum L.S., Colborn T., Guillette L.J., Jr, Crews D.P., Collins T., Soto A.M., Vom Saal F.S., McLachlan J.A., Sonnenschein C., Heindel J.J. Minireview: Endocrine disruptors: Past lessons and future directions. Mol. Endocrinol. 2016;30(8):833–847. doi: 10.1210/me.2016-1096. [http://dx.doi.org/10.1210/me.2016-1096]. [PMID: 27477640]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006;7(1):3. doi: 10.1186/1471-2199-7-3. [http://dx.doi.org/10.1186/1471-2199-7-3]. [PMID: 16448564]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bratslavsky G., Sanford T., Srinivasan R., Aprelikova O., Liu J., Quezado M., Merino M., Linehan W.M. Differential genetic expression in large versus small clear cell renal cell carcinoma: Results from microarray analysis. J. Cancer. 2011;2:271–279. doi: 10.7150/jca.2.271. [http://dx.doi.org/10.7150/jca.2.271]. [PMID: 21611108]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozak I., Sasik R., Freeman W.R., Sprague L.J., Gomez M.L., Cheng L., El-Emam S., Mojana F., Bartsch D.U., Bosten J., Ayyagari R., Hardiman G. A degenerative retinal process in HIV-associated non-infectious retinopathy. PLoS One. 2013;8(9):e74712. doi: 10.1371/journal.pone.0074712. [http://dx.doi.org/10.1371/journal.pone.0074712]. [PMID: 24069333]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. https://github.com/MUSC-CGM/PIMENTo
- 62.Nash T., Huff M., Glen W.B., Jr, Hardiman G. Pipeline for Integrated Microarray Expression Normalization Tool Kit (PIMENTo) for tumor microarray profiling experiments. Methods Mol. Biol. 2019;1908:153–168. doi: 10.1007/978-1-4939-9004-7_11. [http://dx.doi.org/10.1007/978-1-4939-9004-7_11]. [PMID: 30649727]. [DOI] [PubMed] [Google Scholar]
- 63.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [http://dx.doi.org/10.1093/nar/gkv007]. [PMID: 25605792]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smyth G. Limma: Linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [http://dx.doi.org/10.1007/0-387-29362-0_23] [Google Scholar]
- 65.Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C., Georgescu C., Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–1545. doi: 10.1101/gr.6202607. [http://dx.doi.org/10.1101/gr.6202607]. [PMID: 17785539]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oliveros J.C. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 Available from: http://bioinfogp.cnb.csic.es/tools/venny.
- 67.Keri R.A., Ho S.M., Hunt P.A., Knudsen K.E., Soto A.M., Prins G.S. An evaluation of evidence for the carcinogenic activity of bisphenol A. Reprod. Toxicol. 2007;24(2):240–252. doi: 10.1016/j.reprotox.2007.06.008. [http://dx.doi.org/10.1016/j.reprotox.2007.06.008]. [PMID: 17706921]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prins G.S. Endocrine disruptors and prostate cancer risk. Endocr. Relat. Cancer. 2008;15(3):649–656. doi: 10.1677/ERC-08-0043. [http://dx.doi.org/10.1677/ERC-08-0043]. [PMID: 18524946]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishida N., Yano H., Nishida T., Kamura T., Kojiro M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006;2(3):213–219. doi: 10.2147/vhrm.2006.2.3.213. [http://dx.doi.org/10.2147/vhrm.2006.2.3.213]. [PMID: 17326328]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Durando M., Kass L., Perdomo V., Bosquiazzo V.L., Luque E.H., Muñoz-de-Toro M. Prenatal exposure to bisphenol A promotes angiogenesis and alters steroid-mediated responses in the mammary glands of cycling rats. J. Steroid Biochem. Mol. Biol. 2011;127(1-2):35–43. doi: 10.1016/j.jsbmb.2011.04.001. [http://dx.doi.org/10.1016/j.jsbmb.2011.04.001]. [PMID: 21513798]. [DOI] [PubMed] [Google Scholar]
- 71.Prins G.S., Tang W.Y., Belmonte J., Ho S.M. Perinatal exposure to oestradiol and bisphenol A alters the prostate epigenome and increases susceptibility to carcinogenesis. Basic Clin. Pharmacol. Toxicol. 2008;102(2):134–138. doi: 10.1111/j.1742-7843.2007.00166.x. [http://dx.doi.org/10.1111/j.1742-7843.2007.00166.x]. [PMID: 18226066]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Angle B.M., Do R.P., Ponzi D., Stahlhut R.W., Drury B.E., Nagel S.C., Welshons W.V., Besch-Williford C.L., Palanza P., Parmigiani S., vom Saal F.S., Taylor J.A. Metabolic disruption in male mice due to fetal exposure to low but not high doses of bisphenol A (BPA): Evidence for effects on body weight, food intake, adipocytes, leptin, adiponectin, insulin and glucose regulation. Reprod. Toxicol. 2013;42:256–268. doi: 10.1016/j.reprotox.2013.07.017. [http://dx.doi.org/10.1016/j.reprotox.2013.07.017]. [PMID: 23892310]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Straub R.H. The complex role of estrogens in inflammation. Endocr. Rev. 2007;28(5):521–574. doi: 10.1210/er.2007-0001. [http://dx.doi.org/10.1210/er.2007-0001]. [PMID: 17640948]. [DOI] [PubMed] [Google Scholar]
- 74.Valentino R., D’Esposito V., Passaretti F., Liotti A., Cabaro S., Longo M., Perruolo G., Oriente F., Beguinot F., Formisano P. Bisphenol-A impairs insulin action and up-regulates inflammatory pathways in human subcutaneous adipocytes and 3T3-L1 cells. PLoS One. 2013;8(12):e82099. doi: 10.1371/journal.pone.0082099. [http://dx.doi.org/10.1371/journal.pone.0082099]. [PMID: 24349194]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eyries M., Siegfried G., Ciumas M., Montagne K., Agrapart M., Lebrin F., Soubrier F. Hypoxia-induced apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ. Res. 2008;103(4):432–440. doi: 10.1161/CIRCRESAHA.108.179333. [http://dx.doi.org/10.1161/CIRCRESAHA.108.179333]. [PMID: 18617693]. [DOI] [PubMed] [Google Scholar]
- 76.Bertrand C., Valet P., Castan-Laurell I. Apelin and energy metabolism. Front. Physiol. 2015;6:115. doi: 10.3389/fphys.2015.00115. [http://dx.doi.org/10.3389/fphys.2015.00115]. [PMID: 25914650]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan Y., Zeng Z.C., Xi M., Wan S., Hua W., Liu Y.L., Zhou Y.L., Luo H.W., Jiang F.N., Zhong W.D. Dysregulated microRNA-224/apelin axis associated with aggressive progression and poor prognosis in patients with prostate cancer. Hum. Pathol. 2015;46(2):295–303. doi: 10.1016/j.humpath.2014.10.027. [http://dx.doi.org/10.1016/j.humpath.2014.10.027]. [PMID: 25532941]. [DOI] [PubMed] [Google Scholar]
- 78.Kleinz M.J., Davenport A.P. Emerging roles of apelin in biology and medicine. Pharmacol. Ther. 2005;107(2):198–211. doi: 10.1016/j.pharmthera.2005.04.001. [http://dx.doi.org/10.1016/j.pharmthera.2005.04.001]. [PMID: 15907343]. [DOI] [PubMed] [Google Scholar]
- 79.Chow L., Rezmann L., Catt K.J., Louis W.J., Frauman A.G., Nahmias C., Louis S.N. Role of the renin-angiotensin system in prostate cancer. Mol. Cell. Endocrinol. 2009;302(2):219–229. doi: 10.1016/j.mce.2008.08.032. [http://dx.doi.org/10.1016/j.mce.2008.08.032]. [PMID: 18824067]. [DOI] [PubMed] [Google Scholar]
- 80.Hoffmann M., Fiedor E., Ptak A. Bisphenol A and its derivatives tetrabromobisphenol A and tetrachlorobisphenol A induce apelin expression and secretion in ovarian cancer cells through a peroxisome proliferator-activated receptor gamma-dependent mechanism. Toxicol. Lett. 2017;269:15–22. doi: 10.1016/j.toxlet.2017.01.006. [http://dx.doi.org/10.1016/j.toxlet.2017.01.006]. [PMID: 28111160]. [DOI] [PubMed] [Google Scholar]
- 81.Nwankwo J.O., Robbins M.E. Peroxisome proliferator-activated receptor- γ expression in human malignant and normal brain, breast and prostate-derived cells. Prostaglandins Leukot. Essent. Fatty Acids. 2001;64(4-5):241–245. doi: 10.1054/plef.2001.0266. [http://dx.doi.org/10.1054/plef.2001.0266]. [PMID: 11418018]. [DOI] [PubMed] [Google Scholar]
- 82.Li L., Wang Q., Zhang Y., Niu Y., Yao X., Liu H. The molecular mechanism of bisphenol A (BPA) as an endocrine disruptor by interacting with nuclear receptors: Insights from molecular dynamics (MD) simulations. PLoS One. 2015;10(3):e0120330. doi: 10.1371/journal.pone.0120330. [http://dx.doi.org/10.1371/journal.pone.0120330]. [PMID: 25799048]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wadia P.R., Cabaton N.J., Borrero M.D., Rubin B.S., Sonnenschein C., Shioda T., Soto A.M. Low-dose BPA exposure alters the mesenchymal and epithelial transcriptomes of the mouse fetal mammary gland. PLoS One. 2013;8(5):e63902. doi: 10.1371/journal.pone.0063902. [http://dx.doi.org/10.1371/journal.pone.0063902]. [PMID: 23704952]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gilmore A.P., Romer L.H. Inhibition of focal adhesion kinase (FAK) signaling in focal adhesions decreases cell motility and proliferation. Mol. Biol. Cell. 1996;7(8):1209–1224. doi: 10.1091/mbc.7.8.1209. [http://dx.doi.org/10.1091/mbc.7.8.1209]. [PMID: 8856665]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waters A.M. Inhibition of Focal Adhesion Kinase (FAK) Leads to Decreased Cell Survival in Rhabdomyosarcoma Cells in Vitro and in Vivo. 2015 AAP National Conference and Exhibition. 2015. [Google Scholar]
- 86.Castillo Sanchez R., Gomez R., Perez Salazar E. Bisphenol a induces migration through a GPER-, FAK-, Src-, and ERK2-dependent pathway in MDA-MB-231 breast cancer cells. Chem. Res. Toxicol. 2016;29(3):285–295. doi: 10.1021/acs.chemrestox.5b00457. [http://dx.doi.org/10.1021/acs.chemrestox.5b00457]. [PMID: 26914403]. [DOI] [PubMed] [Google Scholar]
- 87.Tong X., Han X., Yu B., Yu M., Jiang G., Ji J., Dong S. Role of gap junction intercellular communication in testicular leydig cell apoptosis induced by oxaliplatin via the mitochondrial pathway. Oncol. Rep. 2015;33(1):207–214. doi: 10.3892/or.2014.3571. [http://dx.doi.org/10.3892/or.2014.3571]. [PMID: 25355463]. [DOI] [PubMed] [Google Scholar]
- 88.Mayan M.D., Gago-Fuentes R., Carpintero-Fernandez P., Fernandez-Puente P., Filgueira-Fernandez P., Goyanes N., Valiunas V., Brink P.R., Goldberg G.S., Blanco F.J. Articular chondrocyte network mediated by gap junctions: Role in metabolic cartilage homeostasis. Ann. Rheum. Dis. 2015;74(1):275–284. doi: 10.1136/annrheumdis-2013-204244. [http://dx.doi.org/10.1136/annrheumdis-2013-204244]. [PMID: 24225059]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mehta P.P. Gap‐junctional communication in normal and neoplastic prostate epithelial cells and its regulation by cAMP. 1. Vol. 15. Molecular Carcinogenesis: Published in cooperation with the University of Texas MD Anderson Cancer Center; 1996. pp. 18–32. [http://dx.doi.org/10.1002/(SICI)1098-2744(199601)15:13.0.CO;2-O]. [DOI] [PubMed] [Google Scholar]
- 90.Habermann H., Ray V., Habermann W., Prins G.S. Alterations in gap junction protein expression in human benign prostatic hyperplasia and prostate cancer. J. Urol. 2002;167(2 Pt 1):655–660. doi: 10.1016/S0022-5347(01)69118-3. [http://dx.doi.org/10.1016/S0022-5347(01)69118-3]. [PMID: 11792947]. [DOI] [PubMed] [Google Scholar]
- 91.Sun H., Xu L.C., Chen J.F., Song L., Wang X.R. Effect of bisphenol A, tetrachlorobisphenol A and pentachlorophenol on the transcriptional activities of androgen receptor-mediated reporter gene. Food Chem. Toxicol. 2006;44(11):1916–1921. doi: 10.1016/j.fct.2006.06.013. [http://dx.doi.org/10.1016/j.fct.2006.06.013]. [PMID: 16893599]. [DOI] [PubMed] [Google Scholar]
- 92.Tan M.H., Li J., Xu H.E., Melcher K., Yong E.L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015;36(1):3–23. doi: 10.1038/aps.2014.18. [http://dx.doi.org/10.1038/aps.2014.18]. [PMID: 24909511]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wetherill Y.B., Petre C.E., Monk K.R., Puga A., Knudsen K.E. The xenoestrogen bisphenol A induces inappropriate androgen receptor activation and mitogenesis in prostatic adenocarcinoma cells. Mol. Cancer Ther. 2002;1(7):515–524. [PMID: 12479269]. [PubMed] [Google Scholar]
- 94.Okada H., Tokunaga T., Liu X., Takayanagi S., Matsushima A., Shimohigashi Y. Direct evidence revealing structural elements essential for the high binding ability of bisphenol A to human estrogen-related receptor-γ. Environ. Health Perspect. 2008;116(1):32–38. doi: 10.1289/ehp.10587. [http://dx.doi.org/10.1289/ehp.10587]. [PMID: 18197296]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takayanagi S., Tokunaga T., Liu X., Okada H., Matsushima A., Shimohigashi Y. Endocrine disruptor bisphenol A strongly binds to human estrogen-related receptor γ (ERRgamma) with high constitutive activity. Toxicol. Lett. 2006;167(2):95–105. doi: 10.1016/j.toxlet.2006.08.012. [http://dx.doi.org/10.1016/j.toxlet.2006.08.012]. [PMID: 17049190]. [DOI] [PubMed] [Google Scholar]
- 96.Yu S., Wang X., Ng C.F., Chen S., Chan F.L. ERRgamma suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res. 2007;67(10):4904–4914. doi: 10.1158/0008-5472.CAN-06-3855. [http://dx.doi.org/10.1158/0008-5472.CAN-06-3855]. [PMID: 17510420]. [DOI] [PubMed] [Google Scholar]
- 97.Misra J., Kim D-K., Choi H-S. ERRγ: A junior orphan with a senior role in metabolism. Trends Endocrinol. Metab. 2017;28(4):261–272. doi: 10.1016/j.tem.2016.12.005. [http://dx.doi.org/10.1016/j.tem.2016.12.005]. [PMID: 28209382]. [DOI] [PubMed] [Google Scholar]
- 98.McKenna N.J., Lanz R.B., O’Malley B.W. Nuclear receptor coregulators: Cellular and molecular biology. Endocr. Rev. 1999;20(3):321–344. doi: 10.1210/edrv.20.3.0366. [PMID: 10368774]. [DOI] [PubMed] [Google Scholar]
- 99.Giguère V. To ERR in the estrogen pathway. Trends Endocrinol. Metab. 2002;13(5):220–225. doi: 10.1016/s1043-2760(02)00592-1. [http://dx.doi.org/10.1016/S1043-2760(02)00592-1]. [PMID: 12185669]. [DOI] [PubMed] [Google Scholar]
- 100.Pei L., Mu Y., Leblanc M., Alaynick W., Barish G.D., Pankratz M., Tseng T.W., Kaufman S., Liddle C., Yu R.T., Downes M., Pfaff S.L., Auwerx J., Gage F.H., Evans R.M. Dependence of hippocampal function on ERRγ-regulated mitochondrial metabolism. Cell Metab. 2015;21(4):628–636. doi: 10.1016/j.cmet.2015.03.004. [http://dx.doi.org/10.1016/j.cmet.2015.03.004]. [PMID: 25863252]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fan W., He N., Lin C.S., Wei Z., Hah N., Waizenegger W., He M.X., Liddle C., Yu R.T., Atkins A.R., Downes M., Evans R.M. ERRγ promotes angiogenesis, mitochondrial biogenesis, and oxidative remodeling in PGC1α/β-deficient muscle. Cell Rep. 2018;22(10):2521–2529. doi: 10.1016/j.celrep.2018.02.047. [http://dx.doi.org/10.1016/j.celrep.2018.02.047]. [PMID: 29514081]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dolinoy D.C., Huang D., Jirtle R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA. 2007;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [http://dx.doi.org/10.1073/pnas.0703739104]. [PMID: 17670942]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [http://dx.doi.org/10.1038/nature04431]. [PMID: 16357870]. [DOI] [PubMed] [Google Scholar]
- 104.Zhu Z., Edwards R.J., Boobis A.R. Increased expression of histone proteins during estrogen-mediated cell proliferation. Environ. Health Perspect. 2009;117(6):928–934. doi: 10.1289/ehp.0800109. [http://dx.doi.org/10.1289/ehp.0800109]. [PMID: 19590685]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doshi T., Mehta S.S., Dighe V., Balasinor N., Vanage G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology. 2011;289(2-3):74–82. doi: 10.1016/j.tox.2011.07.011. [http://dx.doi.org/10.1016/j.tox.2011.07.011]. [PMID: 21827818]. [DOI] [PubMed] [Google Scholar]
- 106.Santangeli S., Maradonna F., Gioacchini G., Cobellis G., Piccinetti C.C., Dalla Valle L., Carnevali O. BPA-induced deregulation of epigenetic patterns: Effects on female zebrafish reproduction. Sci. Rep. 2016;6:21982. doi: 10.1038/srep21982. [http://dx.doi.org/10.1038/srep21982]. [PMID: 26911650]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hopfner R., Mousli M., Jeltsch J.M., Voulgaris A., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60(1):121–128. [PMID: 10646863]. [PubMed] [Google Scholar]
- 108.Unoki M., Brunet J., Mousli M. Drug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem. Pharmacol. 2009;78(10):1279–1288. doi: 10.1016/j.bcp.2009.05.035. [http://dx.doi.org/10.1016/j.bcp.2009.05.035]. [PMID: 19501055]. [DOI] [PubMed] [Google Scholar]
- 109.Bronner C., Krifa M., Mousli M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem. Pharmacol. 2013;86(12):1643–1649. doi: 10.1016/j.bcp.2013.10.002. [http://dx.doi.org/10.1016/j.bcp.2013.10.002]. [PMID: 24134914]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
The data supporting the findings of the article is available at http://grande.sobs.qub.ac.uk/data, microarray data has been deposited in the EBI ArrayExpress Database, accession number E-MTAB-7576.


