Abstract
Delay discounting (DD), which is the tendency to discount the value of delayed versus current rewards, is elevated in a constellation of diseases and behavioral conditions. We performed a genome-wide association study of DD using 23,127 research participants of European ancestry. The most significantly associated SNP was rs6528024 (P = 2.40 × 10−8), which is located in an intron of the gene GPM6B. We also showed that 12% of the variance in DD was accounted for by genotype, and that the genetic signature of DD overlapped with attention-deficit/hyperactivity disorder, schizophrenia, major depression, smoking, personality, cognition, and body weight.
Keywords: Behavioral genetics, Genome-wide association studies, RDoC, Impulsivity, Decision making, Serotonin, GPM6B
Delay discounting refers to the extent to which an organism devalues rewards that are delayed and is thus a fundamental aspect of impulse control1,2. In humans, greater delay discounting is associated with a number of psychiatric disorders and health conditions including attention-deficit/hyperactivity disorder (ADHD)3, substance use disorders4 and obesity5. DD is included in the Research Domain Criteria (RDoC) initiative6, which views psychiatric disorders as extremes of normal tendencies, and is intended to foster biological analyses of behavior. While numerous genetic studies have examined psychiatric diseases, much less work has been done on the genetic basis of RDoC traits such as DD.
In collaboration with the direct-to-consumer genetics company 23andMe, Inc., we performed the first genome-wide association study (GWAS) of DD by testing the association between millions of common single nucleotide polymorphisms (SNPs; Supplementary Table 1) and DD. Our sample consisted of 23,217 male and female adult research participants of European ancestry (see Supplementary Table 2 for demographic information). Participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (www.eandireview.com). We measured DD using the well-validated Monetary Choice Questionnaire7, which generates hyperbolic discounting functions (k; Supplementary Tables 3 and 4). We observed strong phenotypic correlations between DD and demographic and substance use variables that were measured in the same cohort (Supplementary Table 5); however, these correlations do not differentiate genetic and environmental influences. Age was not significantly correlated with DD, however females showed greater DD compared to males (r = 0.11, P < 0.0001). BMI was positively correlated with DD (r = 0.11, P < 0.0001). Several measures of cigarette and cannabis use were also positively correlated with DD (r = 0.05–0.09, P < 0.0001); however, surprisingly, heaviest lifetime alcohol use in a 30-day period was negatively correlated with DD (r = −0.07, P < 0.0001) and scores on the Alcohol Use Disorder Identification Test (AUDIT), which is used to screen for alcoholism, were not correlated with DD (r = 0.003, P > 0.5), perhaps due to low rates of alcohol use in this population.
Twin studies of DD have shown that identical twins are more concordant than non-identical twins, yielding narrow-sense heritability estimates from 46% to 62%8. We used the phenotype and genotype data from our cohort of unrelated participants to calculate chip heritability (i.e., the proportion of variance accounted for by SNPs9), which was estimated to be at 12.2% (± 1.7%, P = 5.84 × 10−14).
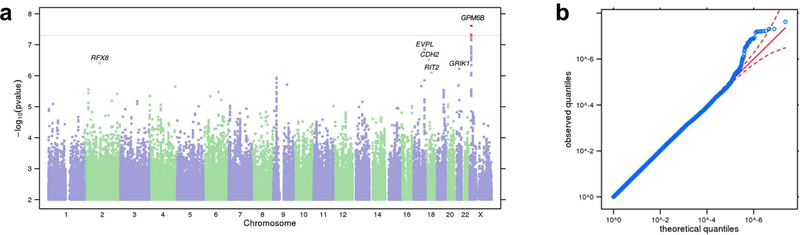
To perform a GWAS, we tested each variant with a linear regression assuming an additive genetic model that included age, sex, the first five genetic principal components, and indicator variables for genotype platforms as covariates (Figure 1; Supplementary Tables 6). The most significant association was at the SNP rs6528024, located on the X-chromosome (P = 2.40 × 10−8; β = −0.10, SE = 0.02; minor allele frequency (MAF) = 0.03; Supplementary Fig. 1). Meta-analysis of rs6528024 using an independent cohort of 928 participants in the Genes for Good study strengthened this association (P = 1.44 × 10−8; β = −0.10, SE = 0.02). rs6528024 is in an intron of the gene GPM6B (Neuronal Membrane Glycoprotein M6B), which has been previously implicated in the internalization of the serotonin transporter10. Gpm6b-knockout mice exhibit deficient prepulse inhibition and an altered response to the 5-HT2A/C agonist DOI11. Serotonergic signaling has also been extensively implicated in DD12–14. Furthermore, GPM6B mRNA levels are downregulated in the brains of depressed suicide victims15. Because rs6528024 is located on the X-chromosome, we re-analyzed it separately in males and females. Although the association with rs6528024 was stronger in males (β = −0.11, SE = 0.02, P = 9.82 × 10−7) than in females (β = −0.08, SE = 0.03, P = 5.70 × 10−3), meta-analyzing males and females supported the original finding (β = −0.10, SE = 0.02, P = 2.81 × 10−8). Several other SNPs showed suggestive associations (Supplementary Table 7), including rs2665993 (P = 1.40 × 10−7, β = −0.04, SE = 0.01; MAF = 0.38; Supplementary Fig. 2). Our results did not support any of the previously published candidate gene studies of DD (reviewed in16; Supplementary Table 8).
Figure 1. Results of GWAS on DD.
(a) Manhattan plot of GWAS results for DD in the 23andMe cohort. The y-axis shows the minimum P-values (−log10) of 11,508,756 SNPs, and the x-axis shows their chromosomal positions. The minimum P-values were obtained by linear regression analysis with adjustment for age, gender, genotyping platform and first five principal components for genotype. The horizontal line denotes genome-wide significance (P < 5 × 10−8). The statistical tests used were two-sided. (b) QQ plot of DD showing the degree of inflation of all test statistics. The results have been adjusted for a genomic control inflation factor λ = 1.022 (sample size = 23,217).
We used S-PrediXcan17 to test the association between predicted gene expression from the genetic data and DD. This approach identified a positive correlation between DD and predicted expression of CDK3 in the hippocampus (FDR 0.05; Supplementary Table 9); CDK3 is near to rs2665993 (Supplementary Fig. 2).
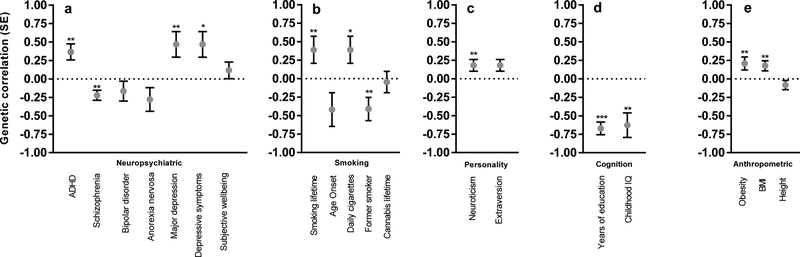
Whereas phenotypic correlations, which involve two traits measured in the same sample, are driven by both genetic and environmental factors, genetic correlations between traits can be obtained by exploiting genetic similarities between two different cohorts. We used LD score regression18 to obtain genetic correlations involving DD (Fig. 2 and Supplementary Table 10). Phenotypic correlations between DD and ADHD were already established19; but we showed, for the first time, that these traits are also positively genetically correlated (rg= 0.37, SE = 0.11, P = 7.76 − 10−4), demonstrating that DD meets the first three criteria necessary to be considered an endophenotype for ADHD20. We identified an unexpected positive genetic correlation with major depressive disorder (MDD) (rg= 0.47, SE = 0.17, P = 6.87 × 10−3) and an equally unexpected negative genetic correlation between DD and schizophrenia (SCZ, rg= −0.22, SE = 0.07, P = 1.16 × 10−3). In contrast, ADHD and SCZ are known to be positively correlated (rg= 0.23, P = 9.0 × 10−3; LDHub). These results highlight an advantage of the RDoC approach – examining individual domains of function may reveal differences between two disorders, even though part of the genetic predisposition to those disorders may be similar. Our interpretation is that the genetic variants that underlie the similarity between ADHD and SCZ have little overlap with the genetic variants that underlie their associations with DD.
Figure 2. Genetic correlations (rg, SE) between DD and several traits.
(a) neuropsychiatric, (b) smoking, (c) personality, (d) cognition, (e) anthropomorphic. * P < 0.05, ** P < 0.01, *** P < 0.0001 (see Supplementary Table 10 for exact P values). The statistical tests used were two-sided; see Supplementary Table 10 for the sample sizes used for each trait.
We also observed a positive genetic correlation between DD and lifetime smoking (rg= 0.32, SE = 0.12, P = 7.98 × 10−3), and a negative genetic correlation with former smoker status (rg= −0.41, SE = 0.16, P = 8.89 × 10−3). Our interpretation of these results is that higher DD facilitates smoking initiation and impedes cessation, showing that DD influences multiple stages of drug abuse vulnerability. We identified a positive genetic correlation between DD and neuroticism (rg= 0.18, SE = 0.08, P = 2.25 × 10−2). DD showed negative correlations with three cognitive measures: college attainment (rg= −0.93, SE = 0.15, P = 3.0 × 10−10), years of education (rg= −0.67, SE = 0.09, P = 7.9 × 10−15) and childhood IQ (rg= −0.63, SE = 0.17, P = 1.63 × 10−4). It is tempting to view college attainment and years of education as examples of working towards delayed rewards; however, the genetic correlation with childhood IQ is inconsistent with this interpretation. Finally, DD was genetically correlated with BMI (rg= 0.18, SE = 0.07, P = 8.93 × 10−3), suggesting that higher DD may promote excessive eating. As expected, height, which is not strongly influenced by an individual’s behavior, was not genetically correlated with DD (rg= −0.08, SE = 0.06, P = 1.77 × 10−1).
We have reported the largest genetic study of DD ever undertaken. The unit of analysis in psychiatric genetic studies has traditionally been disease diagnosis, which cannot be easily mapped to discrete brain circuits. Instead, we have focused on DD, which is a fundamental process that can be studied at molecular, cellular and systems levels. Our results indicate that DD is influenced by numerous genetic variants and would likely benefit from an even larger sample size. Unlike studies of disease traits, which require careful diagnosis and ascertainment, we were able to rapidly obtain a large cohort for which genotype data were available. Consistent with the core goals of RDoC, our approach shows how genetic studies of DD can be used to gain insight into the biology of neuropsychiatric diseases.
ONLINE METHODS
Sample.
All participants were drawn from the customer base of 23andMe, Inc., a direct-to-consumer genetics company. Participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (www.eandireview.com). Over the course of approximately four months in 2015, more than 25,000 23andMe research participants responded to survey questions that were part of a study of the genetics of decision-making designed by Drs. Abraham Palmer and James MacKillop. In this paper we examine only the 23,127 participants who were of European ancestry (> 97% as determined through an analysis of local ancestry21), for whom DD data were available, and who were not excluded for any of the reasons described below. The concepts of randomized data collection and analysis and blinding of experiments do not apply to our study design. Socio-demographic details of this cohort are described in the Supplementary Table 2.
Genotyping, Quality Control and imputation.
DNA extraction and genotyping were performed on saliva samples by CLIA-certified and CAP-accredited clinical laboratories of Laboratory Corporation of America. Quality control, imputation, and genome-wide analysis were performed by 23andMe.
Samples were genotyped on a 23andMe custom genotyping array platform (Illumina HumanHap550+ Bead chip V1 V2, OmniExpress+ Bead chip V3, Custom array V4). Samples had a minimum call rates of 98.5%. A total of 1,050,074 SNPs and InDels (Insertion/Deletion) were genotyped across all platforms (Supplementary Table 1; see22–24 for extended genotyping and sample quality details).
A maximal set of unrelated individuals was chosen for the analysis using a segmental identity-by-descent (IBD) estimation algorithm25 to ensure that only unrelated individuals were included in the sample. Individuals were defined as related if they shared more than 700 cM IBD, including regions where the two individuals shared either one or both genomic segments IBD. This level of relatedness (~20% of the genome) corresponds to approximately the minimal expected sharing between first cousins in an outbred population.
We imputed participant genotype data against the September 2013 release of 1000 Genomes phase 1 version 3 reference haplotypes. We phased and imputed data for each genotyping platform separately. We phased using an internally developed phasing tool, Finch, which implements the Beagle haplotype graph-based phasing algorithm26, modified to separate the haplotype graph construction and phasing steps. In preparation for imputation, we split phased chromosomes into segments of no more than 10,000 genotyped SNPs, with overlaps of 200 SNPs. We excluded SNPs with Hardy-Weinberg equilibrium P < 10−20, call rate < 95%, or with large allele frequency discrepancies compared to European 1000 Genomes reference data. Frequency discrepancies were identified by computing a 2 × 2 table of allele counts for European 1000 Genomes samples and 2000 randomly sampled 23andMe customers with European ancestry, and identifying SNPs with a χ2 P < 10−15. We imputed each phased segment against all-ethnicity 1000 Genomes haplotypes (excluding monomorphic and singleton sites) using Minimac227, using 5 rounds and 200 states for parameter estimation. After quality control, we analyzed 11,508,756 SNPs.
For the X chromosome, we built separate haplotype graphs for the non-pseudoautosomal region and each pseudoautosomal region, and these regions were phased separately. We then imputed males and females together using Minimac2, as with the autosomes, treating males as homozygous pseudo-diploids for the non-pseudoautosomal region.
For tests using imputed data, we use the imputed dosages rather than best-guess genotypes. We imputed HLA allele dosages from SNP genotype data using HIBAG28. We imputed alleles for HLA-A, B, C, DPB1, DQA1, DQB1, and DRB1 loci at four-digit resolution. To test associations between HLA allele dosages and phenotypes, we performed linear regression using the same set of covariates used in the SNP based GWAS. We performed separate association tests for each imputed allele. HLA analysis did not identify any significant signal and hence are not described in the main text.
Delay discounting.
Participants completed the online Monetary Choice Questionnaire (MCQ), a widely used 27-item measure of monetary DD preferences7. For each item, individuals were presented with a choice between a smaller immediate reward and larger delayed reward at one of three magnitudes (small, $25–35; intermediate, $50–60; large, $75–85; see Supplementary Table 3). The overall response pattern was used to infer temporal discounting functions (k) ranging from .00016–.2529, with larger values reflecting greater devaluation of delayed rewards (higher bias for immediate gratification). Values of k were obtained for the three reward magnitudes and were averaged to obtain the final dependent measure. Most research participants (~98%) were highly consistent, suggesting good task effort; individuals with < 80% concordance among the three magnitudes were excluded from further analyses. In addition, we added three items to the 27-items that make up the MCQ that were intended to detect careless responding (items 8, 17 and 24 in Supplementary Table 3). Research participants who answered any of these items inappropriately were excluded. If subjects responded randomly, 87.5% would have answered one of these three items inappropriately, whereas the actual rate of inappropriate responses was 2.10%. A total of 550 research participants were excluded from our sample due to low consistency (< 80%) or inappropriate responses. Distributions of the DD scores are shown in the Supplementary Table 4. Visual inspection of the distribution revealed that k was not normally distributed; we used a log-10 transformation, which is commonly employed to approximate a normal distribution for k. The ascertainment of research participants required being a 23andMe customer and voluntary participation in this research project; we acknowledge that these factors may have circumscribed the distribution of DD.
Additional demographic and phenotypic measures.
Age, sex and other demographic data about the research participants were already available. Additional potentially relevant variables were ascertained as part of the same survey. For example, to evaluate heavy drinking and/or active alcohol abuse or dependence, participants completed The Alcohol Use Disorder Identification Test (AUDIT)30, a ten-item questionnaire that screens for excessive drinking. We also collected additional measures of drug use quantity and frequency (alcohol, cigarettes, cannabis), and estimates of caffeine intake (number of coffee cups per day) by including items from the PhenX toolkit. Results are shown in Supplementary Table 2. Rates of current alcohol and drug use were low in this cohort; this is an advantage because DD has often been studied in populations with higher levels of drug use, raising questions about whether DD is altered by past or present drug use or even the need for money to buy drugs.
Genome-wide association analysis.
We performed association tests by linear regression assuming an additive model using a 23andMe internally developed pipeline. See the Life Sciences Reporting Summary for extended details. Sample size was set at 25,000 because this was larger than comparable well-powered studies of similar phenotypes. Using Quanto (http://biostats.usc.edy/Quanto.html) we estimated that with a sample size of 25,000, allele frequency of 0.05, mean trait value of −2.25 (SD= 0.73) and effect size (beta) of −0.1 to 0.1, we had 89% power to detect significant (5 × 10–8) associations. We included age (inverse-normal transformed), sex, the top five principal components of genotype, and indicator variables for genotype platforms as covariates (Supplementary Table 6). We did not include substance use traits as covariates for two reasons. First, substance-use was very low in this population (Supplementary Table 2). Second, including a heritable trait such as substance use as a cofactor may remove genetic signal31. To derive principal components of genotype, we used a 23andMe internally developed pipeline that uses ~440K genotyped autosomal variants (minor allele frequency, MAF, in European > 0.01 and HLA/inversion removed). For quality control of genotyped GWAS results, we removed SNPs with minor allele frequency of <0.1%, a Hardy-Weinberg P <10−20 in Europeans, or a call rate of < 90%. We also removed SNPs that were only genotyped on the 23andMe V1 platform, due to limited sample size, and SNPs on chrM or chrY because currently many of these are not called reliably. Using trio data, we removed SNPs that failed a test for parent-offspring transmission; specifically, we regressed the child’s allele count against the mean parental allele count and removed SNPs with fitted β < 0.6 and P < 10−20 for a test of β < 1. We also tested genotyped SNPs for genotype date effects, and removed SNPs with P < 10−50 by ANOVA of SNP genotypes against a factor dividing genotyping date into 20 roughly equal-sized buckets. For imputed GWAS results, we removed SNPs with average r2 < 0.5 or minimum r2 < 0.3 in any imputation batch, as well as SNPs that had strong evidence of an imputation batch effect. The batch effect test is an F test from an ANOVA of the SNP dosages against a factor representing imputation batch; we removed results with P < 1 × 10−50. We also removed linear regression results for SNPs with MAF < 0.1% because tests of low frequency variants can be sensitive to violations of the regression assumption of normally distributed residuals. GWAS analysis included genotyped and imputed SNPs. We performed separate male and a female linear regression analyses to identify sex-specific effects at rs6528024, which was the most significantly associated SNP (Supplementary Fig. 1).
Replication and meta-analysis using Genes for Good.
To validate the association with rs6528024, we examined the association of that SNP with the DD in an independent cohort of 928 individuals of European ancestry (mean age = 41.03, SD = 13.94, ~70% female, 89% completing at least some college) from Genes for Good. Participants completed a shorter version of the MCQ (nine items corresponding to the intermediate reward magnitude). k values were calculated as described above. Genotyping, quality control, imputation and GWAS analyses were performed by Genes for Good. Briefly, for quality control of genotyped SNPs, SNPs with minor allele frequency of <0.1%, a Hardy-Weinberg P < 10−4 in Europeans, or a call rate of < 99% were excluded. A set of unrelated individuals were used for the analyses, as determined by KING (http://varianttools.sourceforge.net/Pipeline/KING). Participant genotype data were imputed against the September 2013 release of 1000 Genomes phase 1 version 3 reference haplotypes, phased with ShapeIt232, and imputed with Minimac327. Association tests were performed by linear regression assuming an additive model using EPACTS, with age (ordinal), sex and five principal components as covariates. We meta-analyzed rs6528024 using an inverse variance meta-analysis in METAL33.
Estimation of variance in DD explained by the genotyped SNPs.
To estimate the proportion of phenotypic variance explained (‘chip-heritability’; hg2), we used a genomic restricted maximum likelihood (GREML) method implemented in Genetic Complex Trait Analysis (GCTA)34. In brief, the GREML method estimates the proportion of variation in a phenotype that is due to all SNPs, and exploits the fact that genotypic similarity (i.e., “relatedness”, measured using genotyped SNPs) will be correlated with phenotypic similarity for heritable traits. Distantly related individuals with pair-wise relationships were filtered at two thresholds (KIBS < 0.05 and KIBS < 0.025). We included age (inverse-normalization), self-reported sex (male/female), genotyping platform and first five principal components for genotype as covariates. GREML analyses were run using only directly genotyped SNPs to construct the GRM.
Chip-heritability using LD Score Regression.
We also used a second method to measure chip heritability of DD that is implemented by Linkage Disequilibrium Score Regression Coefficient (LDSC)35. To standardize the input file (GWAS summary statistics), we followed quality controls as implemented by the LDSC python software package. We used pre-calculated LD scores (“eur_w_ld_chr/” files18; MHC region excluded) for each SNP using individuals of European ancestry from the 1000 Genomes Project, suitable for LD score analysis in European populations. We restricted the analysis to well-imputed SNPs: the SNPs were filtered to HapMap3 SNPs36, and were required to have a MAF above 1%. InDels, structural variants, strand-ambiguous SNPs, and SNPs with extremely large effect sizes (χ2 > 80) were removed. In addition, this approach allowed us to distinguish between genomic inflation attributed to polygenic signal and confounding biases such as population stratification or polygenicity (LD Score regression intercept > 1)18,35. As expected under polygenicity, we observed inflation of the median test statistic (Mean χ2 = 1.046), and adjusted for a genomic control inflation factor λ (the ratio of the observed median χ2 to that expected by chance) = 1.022. LD score intercept of 1.013 (SE = 0.01) suggested that deviation from the null was due to a polygenic structure rather than inflation due to population structure biases37.
Phenotypic and genetic correlation analyses.
We examined two distinct types of correlations: phenotypic correlations, where both variables were measured in the same individuals, and genetic correlations, where we used DD data from this cohort in conjunction with summary statistics for GWAS conducted in other cohorts. The interpretation of these is different, since phenotypic correlations can be due to a combination of genetic and non-genetic factors, whereas genetic correlations measure only genetically driven correlations.
We used bivariate correlations to examine the phenotypic correlations among variables of interest (age, gender, race, education, annual household and drug-related phenotypes), and to identify significant covariates for inclusion in subsequent analyses (Supplementary Table 5).
We calculated genetic correlations (rg) between DD and 28 other traits or diseases using LDSC. References for the datasets used are described in the Supplementary Table 10. Files were standardized using the pipeline described above. We did not constrain the intercepts in our analysis, as we could not quantify the exact amount of sample overlap. We used False Discovery Rate (FDR) to correct for multiple testing38.
Gene-based association using transcriptome data with S-PrediXcan.
Expression quantitative trait loci (eQTLs) are genomic loci that contribute to heritable variation in mRNA levels. SNPs or chromosomal regions might influence DD because they influence the expression of a particular gene (often, but not always, a nearby gene). We used S-PrediXcan17 to identify specific eQTL-linked genes associated with DD. This approach uses genetic information to predict gene expression levels in various brain tissues, and tests whether the predicted gene expression correlates with DD. S-PrediXcan17 uses pre-computed tissue weights from the Genotype-Tissue Expression (GTEx) project database (https://www.gtexportal.org/) as the reference transcriptome dataset. As input data, we included our GWAS summary statistics, transcriptome tissue data and covariance matrices of the SNPs within each gene model (based on HapMap SNP set; available to download at the PredictDB Data Repository) from 10 brain tissues: anterior cingulate cortex, caudate basal ganglia, cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens basal ganglia, and putamen basal ganglia. Genes were filtered if they did not contain SNPs from our dataset, or if the gene’s correlation with the predicted expression model (R2) was < 0.01. We applied FDR to correct for multiple testing across all brain tissues (N= 31,722)38. S-PrediXcan does not include the X chromosome; hence this approach could not provide insight about our strongest signal rs6528024 near GPM6B. Extended results are included in the Supplementary Table 9.
Data availability.
We have provided summary statistics for the top 10,000 SNPs (Supplementary Data Set, Supplementary Readme). Full GWAS summary statistics for the 23andMe dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Interested investigators should email dataset-request@23andme.com for more information.
URLs.
EPACTS http://genome.sph.umich.edu/wiki/EPACTS, Genes For Good https://genesforgood.sph.umich.edu/about_study, GTEx database (http://broadinstitute.org/gtex/, LD Score software https://github.com/bulik/ldsc/, LDHub http://ldsc.broadinstitute.org/, S-PrediXcan https://github.com/hakyimlab/S-PrediXcan-Working, The NHGRI GWAS Catalog http://www.genome.gov/gwastudies/, Regulome DB database http://www.regulomedb.org/, PhenX toolkit https://www.phenxtoolkit.org, PredictDB Data Repository http://predictdb.hakyimlab.org/
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the research participants and employees of 23andMe for making this work possible. J.M.’s contributions were partially supported by the Peter Boris Chair in Addictions Research. S.S-R was supported by the Frontiers of Innovation Scholars Program (FISP), the Interdisciplinary Research Fellowship in NeuroAIDS (IRFN; MH081482) and a pilot award from DA037844.
Footnotes
COMPETING FINANCIAL INTERESTS
Members of the 23andMe Research Team are employees of 23andMe Inc. The opinions and assertions expressed herein are those of the authors; specifically, with respect to JCG, they do not reflect the official policy or position of the Uniformed Services University or the Department of Defense.
METHODS
Methods, along with Supplemental Information with 10 additional tables, are available in the online version of the paper.
REFERENCES
- 1.Bari A & Robbins TW Prog. Neurobiol 108, 44–79 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Hamilton KR et al. Personal. Disord 6, 182–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson JNS & MacKillop J Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 316–325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amlung M, Vedelago L, Acker J, Balodis I & MacKillop J Addict. Abingdon Engl 112, 51–62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClelland J et al. Neurosci. Biobehav. Rev 71, 506–528 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Insel T et al. Am. J. Psychiatry 167, 748–751 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Kirby KN, Petry NM & Bickel WK J. Exp. Psychol. Gen 128, 78–87 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Anokhin AP, Grant JD, Mulligan RC & Heath AC Biol. Psychiatry 77, 887–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Lee H, Goddard ME & Visscher PM Am. J. Hum. Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fjorback AW, Müller HK & Wiborg OJ Mol. Neurosci. MN 37, 191–200 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Dere E et al. Behav. Brain Res 277, 254–263 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Darna M et al. Behav. Brain Res 293, 134–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalley JW & Roiser JP Neuroscience 215, 42–58 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Schweighofer N et al. J. Neurosci 28, 4528–4532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchsova B, Alvarez Juliá A, Rizavi HS, Frasch AC & Pandey GN Neuroscience 299, 1–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacKillop JJ Exp. Anal. Behav 99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamazon ER et al. (2017). Nat Genet 47, 1091–8 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan B et al. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patros CHG et al. Clin. Psychol. Rev 43, 162–174 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Gottesman II & Gould TD Am. J. Psychiatry 160, 636–645 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Durand EY, Do CB, Mountain JL, Macpherson JM 10.1101/010512 (2014). [DOI]
- 22.Eriksson N et al. PLoS Genet 6, e1000993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyde CL et al. Nat. Genet 48, 1031–1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo M-T et al. Nat. Genet 49, 152–156 (2016).27918536 [Google Scholar]
- 25.Henn BM et al. PLoS ONE 7, e34267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browning SR & Browning BL Am. J. Hum. Genet 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchsberger C, Abecasis GR & Hinds DA Bioinformatics 31, 782–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng X et al. Pharmacogenomics J 14, 192–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray JC, Amlung MT, Palmer AA & MacKillop JJ Exp. Anal. Behav 106, 156–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders JB, Aasland OG, Babor TF, de la Fuente JR & Grant M Addict. Abingdon Engl 88, 791–804 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL & Kraft P Am. J. Hum. Genet 96, 329–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durbin RM et al. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Li Y & Abecasis GR Bioinforma. Oxf. Engl 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Lee SH, Goddard ME & Visscher PM Methods Mol. Biol. Clifton NJ 1019, 215–236 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Bulik-Sullivan B et al. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International HapMap 3 Consortium et al. Nature 467, 52–58 (2010).20811451 [Google Scholar]
- 37.Price AL, Zaitlen NA, Reich D & Patterson N Nat. Rev. Genet 11, 459–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y & Hochberg MT J. R. Stat. Soc 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have provided summary statistics for the top 10,000 SNPs (Supplementary Data Set, Supplementary Readme). Full GWAS summary statistics for the 23andMe dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Interested investigators should email dataset-request@23andme.com for more information.