Abstract
Background/Aim: The difficulty of early diagnosis of colitis associated colorectal cancer (CACRC) due to colonic mucosal changes in long-standing ulcerative colitis (UC) patients is often experienced in daily clinical practice. Noninvasive objective monitoring for cancer development is advantageous for optimizing treatment strategies in UC patients. We aimed to examine the epigenetic alterations occurring in CACRC, focusing on DNA hypermethylation of CpG islands. Materials and Methods: The level of DNA methylation in CpG cites was compared between CACRC and the counterpart non-tumorous mucosa using Infinium HumanMethylation 450K BeadChip. Results: Our subjects included 3 males and 3 females (median age, 49.5 years). The 450K CpG site DNA methylation microarray revealed that the difference in β value (level of hypermethylation) was the highest for corcicotropin releasing hormone receptor 2 (CRHR2) between CACRC and counterpart non-tumorous mucosa. Conclusion: Detection of hypermethylation of CRHR2 may be promising for cancer screening in UC patients.
Keywords: Ulcerative colitis, colitis associated colorectal cancer, epigenetic alteration, hypermethylation, corticotropin releasing hormone receptor 2, carcinogenesis
Colorectal cancer (CRC) is one of the representative causative factors for cancer-related mortality worldwide (1). CRC is considered to occur through multistep genetic or epigenetic alterations along with a pathological change (i.e., adenoma-carcinoma sequence or serrated pathway) (1-6). The representative genetic pathways of sporadic CRC are: 1) the chromosome instability pathway resembling the pathway of hereditary CRC occurring in familial adenomatous polyposis due to mutations in KRAS, APC, and p53, and 2) the microsatellite instability pathway resembling the pathway of Lynch syndrome CRC due to alterations in mismatch repair genes (mainly methylation of MLH1) (7).
Ulcerative colitis (UC) is defined as an idiopathic inflammatory disorder mainly involving the colonic mucosa and submucosa according to the report of the World Health Organisation (WHO) Council for International Organization of Medical Sciences, and is one of the main inflammatory bowel diseases (IBDs) (8). The etiologies for UC are unclear, however, UC development and acute UC exacerbation have been associated with both physical and mental stress (9-11). Colorectal epithelium in UC patients with long disease duration has an increased risk of developing CRC, and IBD-related CRC is responsible for 10-15% of the annual deaths in IBD patients (12). It is therefore of high importance to detect malignancy from UC at an earlier stage of the disease (13-15). Mutation analyses of genes have suggested that tumorigenesis derived from UC patients differ from that derived from patients without UC (13,14,16). The protein expression of p53 in dysplastic crypts of the mucosa involves potential to serve as a reliable biomarker in the early stages of UC-related tumorigenesis (13). Tanaka, et al. reported that mutational rates in APC and KRAS differ between patients with CACRC and those with sporadic CRC (14).
As the role of genetic alterations in colorectal carcinogenesis has been fully investigated, CRC also provides an excellent model for the clarification of epigenetic mechanisms involved in carcinogenesis (17). DNA methylation is the most widely appreciated epigenetic modification (18,19). DNA hypermethylation of CpG islands alters the expression of genes in tumor cells and exerts an essential role in carcinogenesis (20,21). In general, DNA methylation of cancer-related gene promoters starts early in the process of tumorigenesis, affecting various types of CRCs to various degrees (22). Promoter hypermethylation at CpG islands and global hypomethylation can be observed in tumor cells (17,23). Regulation of transcript expression by DNA methylation involves genes relevant to colon tumorigenesis and may account for differences in clinical findings and outcomes between CACRC and sporadic CRC. Different mechanisms of carcinogenesis involving epigenetic alterations is suggested to account for CACRC and sporadic CRC. Advancements leading to the better understanding of the tumor biology can be expected to offer reliable biomarkers that will aid future diagnosis, risk stratification, and treatment strategies for patients with CRC (17). The difficulty in the early diagnosis of CACRC due to colonic mucosal changes in long-standing UC patients is often experienced in daily clinical practice. Noninvasive objective monitoring for cancer development is helpful and advantageous for optimizing treatment strategies in UC patients (24). Abnormal hypermethylation at specific DNA sequences can serve as biomarkers for predicting diagnosis, prognosis or treatment efficacy (25).
In this study, we aimed to examine epigenetic alterations occurring in CACRC focusing on DNA hypermethylation of CpG islands, compared with counterpart colonic non-tumorous mucosa.
Materials and Methods
Diagnosis and treatment of UC. Diagnosis for UC was based on clinical, endoscopic, and histopathological findings based on the diagnostic criteria of the Research Committee on Inflammatory Bowel Disease in Japan. The severity of inflammation in UC patients was graded as mild, moderate or severe (26). Treatment strategies for UC were in compliance with the current Japanese guidelines (27).
Clinical samples. Cancer tissue samples and counter background colon epithelium (paraffin-embedded tissue sections) were obtained from the surgical specimens of 7 UC patients with CACRC between July 2011 and February 2013. One case with insufficient material was excluded and thus a total of 6 cases were analyzed in the current analysis. All analyzed patients were treated with total colectomy. DNA was extracted by the standard procedure involving digestion with proteinase K and phenol chloroform extraction. All samples were fixed in formalin and stored at 4˚C until use.
450K CpG site DNA methylation microarray. Bisulfite-converted DNA was used for hybridization on Infinium HumanMethylation 450K BeadChip, which contains all the reagents required for the DNA methylation analysis, following the Illumina Infinium HD Methylation protocol (28,29). In brief, Infinium HumanMethylation450 BeadChip includes 485,764 cytosine positions of the human genome. Of these cytosine sites, 482,421 positions (99.3%) are CpG dinucleotides, while 3,343 sites (0.7%) are CNG targets. According to their associated RNA transcripts, 361,766 CpGs (74.4%) correspond to classic coding messenger RNA genes and 4,168 (0.85%) correspond to non-coding RNAs (microRNAs and long non-coding RNAs). CpG contents were: 30.9% in CpG islands, 23% in CpG shores, 9.7% in CpG shelves and 36.3% in isolated CpG (open sea). We thoroughly investigated the methylation of CpG cites using Infinium HumanMethylation 450K BeadChip.
The level of DNA methylation in the CpG cites was compared between CACRC and the counterpart non-tumorous mucosa. The level of DNA methylation in the CpG cites (β value) was calculated using the following formula: Max (signal B, 0) divided by {Max (signal A, 0) + Max (signal B, 0) + 100}. Signal A indicates signal intensity of detection of non-methylated DNA, while signal B indicates signal intensity of detection of methylated DNA. When β value is close to 0, the proportion of non-methylated DNA is considered to be high. When β value is close to 1, the proportion of methylated DNA is considered to be high. When comparing the level of DNA methylation between two groups, and a ≥0.15 difference in the β values between the two groups is observed, there appears to be a difference in the methylation level between the two groups.
Traditional hierarchical clustering algorithm. The traditional hierarchical clustering algorithm was performed before methylation analysis using heat map (30). Clustering of genes was performed for different levels of methylation by analysis of variance (ANOVA).
All analyzed subjects provided written informed consent for this study. The study protocol was acknowledged by the institutional review board in Hyogo College of Medicine Hospital (approval no. 0246) and declaration of Helsinki was strictly followed in order to guarantee the right of the study subjects. Personal information (clinical data or genetic information) were protected during data collection.
Statistical considerations. In the analysis of continuous parameters, we employed Student’s t-test or ANOVA to assess group differences, as appropriate. The correction method for multiple comparison was not used. We set the threshold for statistical significance at p<0.05. SPSS was employed for statistical analyses.
Results
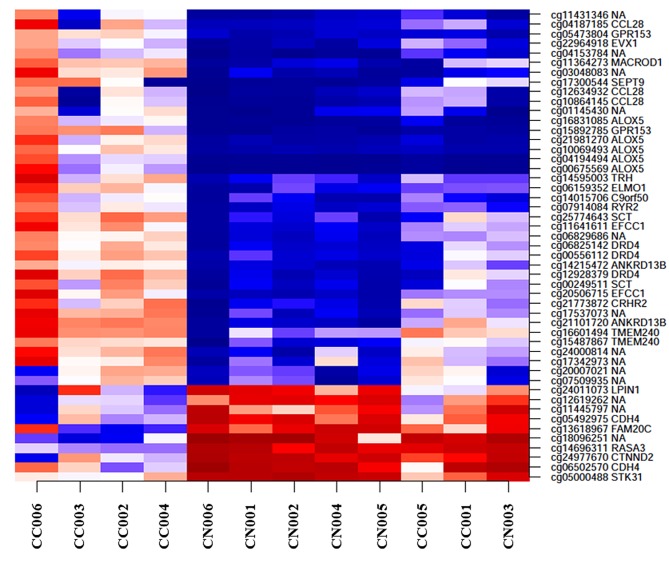
Baseline characteristics of our UC-related CC cases are shown in Table I. They included 3 males and 3 females with a median age of 49.5 years. All cases had a long-term UC duration of 10 years or more, ranging from 17 to 37 years. The pathological findings of CACRC were varied, including well, moderate or poorly differentiated adenocarcinoma, signet cell ring carcinoma, and mucinous adenocarcinoma. Tumor locations of CACRC were also varied, including rectum, sigmoid colon and transverse colon. In terms of disease inflammatory activity of UC, there were 3 cases with mild activity and 3 cases with severe activity.
Table I. Baseline characteristics (n=6).
R: Rectum; S: sigmoid colon; D: descending colon; Tr: transverse colon; Rs: recto-sigmoid colon.
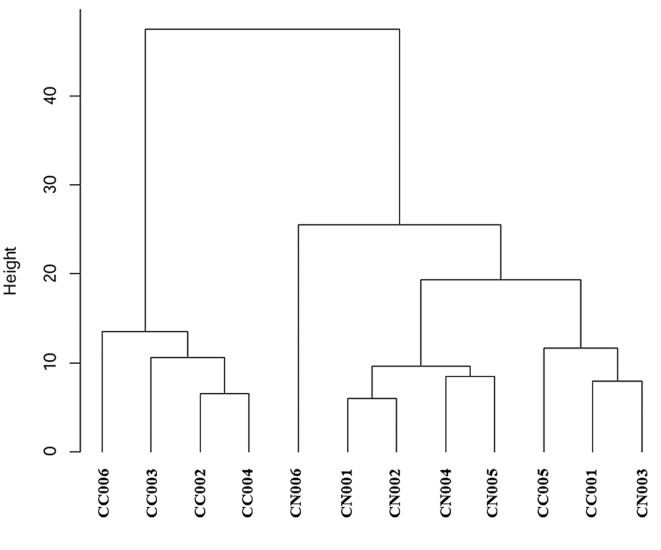
Our hierarchical clustering algorithm and methylation analysis using heat map are shown in Figure 1 and Figure 2. Our case numbers are presented is CC001 to CC006 in CACRC patients and as CN001 to CN006 for counterpart colonic non-tumorous mucosa. In the heat map, the left four CACRCs (CC006, CC003, CC002 and CC004) clearly showed different methylation patterns from non-tumorous mucosa. The CC001 and CC005 on the right also showed different methylation patterns.
Figure 1. Our hierarchical clustering algorithm. Our case numbers are presented as CC001 to CC006 in CACRC patients and as CN001 to CN006 for counterpart colonic non-tumorous mucosa.
Figure 2. Heat map comparing the degree of methylation in CACRC and background non-tumorous mucosa. Our case numbers are presented as CC001 to CC006 in CACRC patients and as CN001 to CN006 in counterpart colonic non-tumorous mucosa. The horizontal axis shows each sample number and the vertical axis shows the location of methylation and the corresponding gene. Clustering of genes was performed for different levels of methylation by ANOVA analysis. The higher the intensity of red, the higher the methylation, whereas the higher the intensity of the blue colour, the lower the methylation. The left 4 CACRCs (CC006, CC003, CC002 and CC004) clearly show different methylation patterns from non-tumorous mucosa. The CC001 and CC005 on the right also show different methylation patterns. NA: Not applicable.
The 450K CpG site DNA methylation microarray revealed that when comparing the levels of DNA methylation between CACRC and the counterpart non-tumorous mucosa, the highest difference in β value was found in corcicotropin releasing hormone receptor 2 (CRHR2). There were 4 genes with differences in β values between CACRC and counterpart non-tumorous mucosa >0.5: CRHR2 gene on chromosome 7 (difference=0.55729602, p=0.002023504), Ankyrin Repeat Domain 13B gene on chromosome 17 (difference= 0.535918997, p=0.008445845), Crystallin Gamma D gene on chromosome 2 (difference=0.51510056, p=0.013923275) and SLIT-ROBO Rho GTPase Activating Protein 3 gene on chromosome 3 (difference=0.501726707, p=0.024044769). These four genes showed hypermethylation.
Discussion
In this study, CACRCs clearly showed different methylation patterns from non-tumorous mucosa. We focused on the CRHR2 gene on chromosome 7 that was specifically methylated in CACRC patients, and a higher frequency of hypermethylation of CRHR2 was identified in CC compared with non-tumorous mucosa. This is the major finding of the current study. To identify genes linked to CACRC in UC patients is clinically important because they may determine clinical outcome and assist in the early diagnosis of CACRC. An increase or a decrease in DNA hypermethylation can contribute to or be a marker for cancer development and tumor progression (25). Moriyama, et al., have reported that hypermethylation of p14 seems to be linked to an early stage of dysplasia in UC patients (31). However, to the best of our knowledge, there have been few reports on the role of hypermethylation of CRHR2 on the development of CACRC in UC patients (32).
Members of the CRH family, which consists of CRH, urocortin I, II and III, and CRHR 1 and 2, are widely expressed in the brain, peripheral tissues, intestine and cardiovascular system (33,34). Urocortin is a member of the CRH neuropeptide family (34). CRH is one of the major modulators of various stress-related behavioral, autonomic, and visceral changes (33). CRH binds to two known receptors, CRHR1 and CRHR2 (35). A previous study reported that CRHR2 exacerbates chronic cardiac dysfunction (36). On the other hand, a functional alteration in the epithelial intestinal barrier recognized in IBD is considered to be a consequence of stress. It has been proposed that CRHR2, the principal neuromediator of stress, acts on cholinergic nerves and results in stress-mediated dysfunction of the epithelial barrier (32). Chatzaki, et al. have reported that CRHR2 expression in the colonic epithelial cells was down-regulated both in patients with moderately active UC and those in remission (37). CRHR1 has a pro-inflammatory and therefore a pro-tumorigenesis effect in terms of colitis associated cancer (38). Inhibition of the expression of CRHR2 correlates with tumor expansion, epithelial-mesenchymal transition, distant metastasis risk and poor survival in experimental CRC models and in CRC patients (39). Therefore, hypermethylation of CRHR2 may be associated with cancer development in UC patients through inactivation of tumor suppressor genes (39). These may support the importance of CRHR2 in CACRC carcinogenesis.
Angiogenesis indicates the neovascularization in existing vessels, and also reflects an underlying cause of many diseases such as cancer and inflammation. In the examinations of prostate cancer, although urocortin expression did not differ between prostate cancer and non-tumorous tissues, CRHR2 expression was identified in the vascular endothelial cells of normal prostate tissue, but not detected in prostate cancer tissue, and therefore a decrease in CRHR2 expression in the prostate could be associated with tumor progression and increased neovascularization (40,41). These results are in accordance with our current data.
Assaying molecular markers in stool samples is a promising noninvasive method for screening CRC (42-46). A recent systematic review demonstrated the eligibility and feasibility of DNA methylation as a noninvasive biomarker in stool samples for the diagnosis of CRC and adenoma (44). Potential expanded applications of stool DNA testing involve identification of cancers and high-grade dysplasia (HGD) in IBD patients (42,47). In UC patients undergoing surveillance colonoscopy, when observation is insufficient due to poor pretreatment, if the stool examination confirms the hypermethylation of the CRHR2 gene, re-examination of areas that have been poorly observed may be required to detect neoplasia. If hypermethylation of the CRHR2 gene is confirmed in UC patients by a stool test, the patients should be subjected to colorectal cancer screening by colonoscopy. While in this study, UC patients with HGD were not included, however, if hypermethylation of the CRHR2 gene occurs frequently in UC patients with HGD, it may be a useful marker for the development of HGD. It is widely accepted that total colectomy should be indicated for UC patients with HGD or histologically severe lesions (48,49). Also, the prognostic impact of the hypermethylation of CRHR2 in UC patients should be further investigated.
The major limitation of the current study was its small sample size, and that the study data were derived from Japanese UC patient data. Additional investigations on different races are needed to further verify these findings and extend the application to different races. In addition, the direct association between hypermethylation of CpG islands and CACRC development remains unclear due to the cross-sectional nature of our study. Caution must be therefore exercised for the interpretation of our study data. However, our DNA methylation microarray demonstrated that hypermethylation of CRHR2 in UC patients can be a useful marker for the development of CACRC. In conclusion, detection of hypermethylation of CRHR2 may be promising for cancer screening in UC patients.
Conflicts of Interest
The Authors declare that they have no conflicts of interest regarding this study.
Authors’ Contributions
Study conception and design: NM; data acquisition: NM, MK, MU, HI, JS, KK, MY, AB, TY, MI, NT; data analysis and interpretation: NM, YN, YO, IM, SH; drafting the article: MK, NM, HN.
Acknowledgements
The Authors would like to thank Aya Yano for her technical support. This work was supported in part by a Grant-in Aid from the Japanese Ministry of Education, Science, Sports and Culture of Japan (23591974).
References
- 1.De Palma FDE, D'Argenio V, Pol J, Kroemer G, Maiuri MC, Salvatore F. The molecular hallmarks of the serrated pathway in colorectal cancer. Cancers (Basel) 2019;11(7):1017. doi: 10.3390/cancers11071017. PMID: 31330830. DOI: 10.3390/cancers11071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. PMID: 2841597. DOI: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 3.Nassif NT, Lobo GP, Wu X, Henderson CJ, Morrison CD, Eng C, Jalaludin B, Segelov E. Pten mutations are common in sporadic microsatellite stable colorectal cancer. Oncogene. 2004;23(2):617–628. doi: 10.1038/sj.onc.1207059. PMID: 14724591. DOI: 10.1038/sj.onc.1207059. [DOI] [PubMed] [Google Scholar]
- 4.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. Apc mutations occur early during colorectal tumorigenesis. Nature. 1992;359(6392):235–237. doi: 10.1038/359235a0. PMID: 1528264. DOI: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. PMID: 23539594. DOI: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119(3):854–865. doi: 10.1053/gast.2000.16507. PMID: 10982779. DOI: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 7.Raskov H, Pommergaard HC, Burcharth J, Rosenberg J. Colorectal carcinogenesis – update and perspectives. World J Gastroenterol. 2014;20(48):18151–18164. doi: 10.3748/wjg.v20.i48.18151. PMID: 25561783. DOI: 10.3748/wjg.v20.i48.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornbluth A, Sachar DB. Ulcerative colitis practice guidelines in adults: American College of Gastroenterology, Practice Parameters Committee. Am J Gastroenterol. 2010;105:501–523. doi: 10.1038/ajg.2009.727. PMID: 20068560. DOI: 10.1038/ajg.2009.727. [DOI] [PubMed] [Google Scholar]
- 9.Mawdsley JE, Rampton DS. Psychological stress in IBD: new insights into pathogenic and therapeutic implications. Gut. 2005;54:1481–1491. doi: 10.1136/gut.2005.064261. PMID: 16162953. DOI: 10.1136/gut.2005.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mawdsley JE, Macey MG, Feakins RM, Langmead L, Rampton DS. The effect of acute psychologic stress on systemic and rectal mucosal measures of inflammation in ulcerative colitis. Gastroenterology. 2006;131:410–419. doi: 10.1053/j.gastro.2006.05.017. PMID: 16890594. DOI: 10.1053/j.gastro.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Langhorst J, Cobelens PM, Kavelaars A, Heijnen CJ, Benson S, Rifaie N, Dobos GJ, Schedlowski M, Elsenbruch S. Stressrelated peripheral neuroendocrine-immune interactions in women with ulcerative colitis. Psychoneuroendocrinology. 2007;32:1086–1096. doi: 10.1016/j.psyneuen.2007.09.003. PMID: 17933470. DOI: 10.1016/j.psyneuen.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Keller DS, Windsor A, Cohen R, Chand M. Colorectal cancer in inflammatory bowel disease: review of the evidence. Tech Coloproctol. 2019;23(1):3–13. doi: 10.1007/s10151-019-1926-2. PMID: 30701345. DOI: 10.1007/s10151-019-1926-2. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K, Tomita H, Shimizu M, Tanaka T, Suzui N, Miyazaki T, Hara A. p53 Expression as a diagnostic biomarker in ulcerative colitis-associated cancer. Int J Mol Sci. 2017;18(6):E1284. doi: 10.3390/ijms18061284. PMID: 28621756. DOI: 10.3390/ijms18061284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka T, Kobunai T, Yamamoto Y, Emoto S, Murono K, Kaneko M, Sasaki K, Otani K, Nishikawa T, Kawai K, Hata K, Nozawa H, Watanabe T. Colitic cancer develops through mutational alteration distinct from that in sporadic colorectal cancer: A comparative analysis of mutational rates at each step. Cancer Genomics Proteomics. 2017;14(5):341–348. doi: 10.21873/cgp.20044. PMID: 28871001. DOI: 10.21873/cgp.20044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanaoka M, Ishikawa T, Ishiguro M, Tokura M, Yamauchi S, Kikuchi A, Uetake H, Yasuno M, Kawano T. Expression of ATF6 as a marker of pre-cancerous atypical change in ulcerative colitis-associated colorectal cancer: a potential role in the management of dysplasia. J Gastroenterol. 2018;53(5):631–641. doi: 10.1007/s00535-017-1387-1. PMID: 28884228. DOI: 10.1007/s00535-017-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita M, Matsubara N, Matsuda I, Maejima K, Oosawa A, Yamano T, Fujimoto A, Furuta M, Nakano K, Oku-Sasaki A, Tanaka H, Shiraishi Y, Mateos RN, Nakai K, Miyano S, Tomita N, Hirota S, Ikeuchi H, Nakagawa H. Genomic landscape of colitis-associated cancer indicates the impact of chronic inflammation and its stratification by mutations in the Wnt signaling. Oncotarget. 2017;9(1):969–981. doi: 10.18632/oncotarget.22867. PMID: 29416670. DOI: 10.18632/oncotarget.22867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsubara N. Epigenetic regulation and colorectal cancer. Dis Colon Rectum. 2012;55(1):96–104. doi: 10.1097/DCR.0b013e318233a1ef. PMID: 22156874. DOI: 10.1097/DCR.0b013e318233a1ef. [DOI] [PubMed] [Google Scholar]
- 18.Nowacka-Zawisza M, Wiśnik E. DNA methylation and histone modifications as epigenetic regulation in prostate cancer (Review) Oncol Rep. 2017;38(5):2587–2596. doi: 10.3892/or.2017.5972. PMID: 29048620. DOI: 10.3892/or.2017.5972. [DOI] [PubMed] [Google Scholar]
- 19.Barchitta M, Maugeri A, Destri GL, Basile G, Agodi A. Epigenetic biomarkers in colorectal cancer patients receiving adjuvant or neoadjuvant therapy: A systematic review of epidemiological studies. Int J Mol Sci. 2019;20(15):3842–3842. doi: 10.3390/ijms20153842. PMID: 31390840. DOI: 10.3390/ijms20153842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiuri AR, Savant SS, Podicheti R, Rusch DB, O'Hagan HM. DNA methyltransferase inhibition reduces inflammation-induced colon tumorigenesis. Epigenetics. 2019;26:1–15. doi: 10.1080/15592294.2019.1634986. PMID: 31240997. DOI: 10.1080/15592294.2019.1634986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90(24):11995–11999. doi: 10.1073/pnas.90.24.11995. PMID: 7505451. DOI: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pekow J, Hernandez K, Meckel K, Deng Z, Haider HI, Khalil A, Zhang C, Talisila N, Siva S, Jasmine F, Li YC, Rubin DT, Hyman N, Bissonnette M, Weber C, Kibriya MG. IBD-associated colon cancers differ in DNA methylation and gene expression profiles compared with sporadic colon cancers. J Crohns Colitis. 2019;13(7):884–893. doi: 10.1093/ecco-jcc/jjz014. PMID: 30753380. DOI: 10.1093/ecco-jcc/jjz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Grieu F, Watanabe G, Iacopetta B. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94(4):593–598. doi: 10.1038/sj.bjc.6602940. PMID: 16421593. DOI: 10.1038/sj.bjc.6602940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura S, Imaeda H, Nishikawa H, Iimuro M, Matsuura M, Oka H, Oku J, Miyazaki T, Honda H, Watanabe K, Nakase H, Andoh A. Usefulness of fecal calprotectin by monoclonal antibody testing in adult Japanese with inflammatory bowel diseases: a prospective multicenter study. Intest Res. 2018;16(4):554–562. doi: 10.5217/ir.2018.00027. PMID: 30301337. DOI: 10.5217/ir.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehrlich M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics. 2019;8:1–23. doi: 10.1080/15592294.2019.1638701. PMID: 31284823. DOI: 10.1080/15592294.2019.1638701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh AJ, Bryant RV, Travis SP. Current best practice for disease activity assessment in IBD. Nat Rev Gastroenterol Hepatol. 2016;13:567–579. doi: 10.1038/nrgastro.2016.128. PMID: 27580684. DOI: 10.1038/nrgastro.2016.128. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka K, Kobayashi T, Ueno F, Matsui T, Hirai F, Inoue N, Kato J, Kobayashi K, Kobayashi K, Koganei K, Kunisaki R, Motoya S, Nagahori M, Nakase H, Omata F, Saruta M, Watanabe T, Tanaka T, Kanai T, Noguchi Y, Takahashi KI, Watanabe K, Hibi T, Suzuki Y, Watanabe M, Sugano K, Shimosegawa T. Evidence-based clinical practice guidelines for inflammatory bowel disease. J Gastroenterol. 2018;53(3):305–353. doi: 10.1007/s00535-018-1439-1. PMID: 29429045. DOI: 10.1007/s00535-018-1439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Kaneda M, Futagawa M, Takeshita M, Kim S, Nakama M, Kawashita N, Tatsumi-Miyajima J. Genetic and genomic basis of the mismatch repair system involved in Lynch syndrome. Int J Clin Oncol. 2019;24(9):999–1011. doi: 10.1007/s10147-019-01494-y. PMID: 31273487. DOI: 10.1007/s10147-019-01494-y. [DOI] [PubMed] [Google Scholar]
- 29.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. PMID: 21593595. DOI: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 30.Hunter DJ. Essentials of Discrete Mathematics. 3. Burlington: Jones & Bartlett Learning, LLC; 2015. Thinking through applications. 2015. Available at http://www.webassign.net/features/textbooks/hunterdm3/details.html?l=publisher.
- 31.Moriyama T, Matsumoto T, Nakamura S, Jo Y, Mibu R, Yao T, Iida M. Hypermethylation of p14 (ARF) may be predictive of colitic cancer in patients with ulcerative colitis. Dis Colon Rectum. 2007;50(9):1384–1392. doi: 10.1007/10350-007-0302-x. PMID: 17665255. DOI: 10.1007/10350-007-0302-x. [DOI] [PubMed] [Google Scholar]
- 32.Pelissier-Rota M, Chartier NT, Bonaz B, Jacquier-Sarlin MR. A crosstalk between muscarinic and CRF2 receptors regulates cellular adhesion properties of human colon cancer cells. Biochim Biophys Acta Mol Cell Res. 2017;1864(7):1246–1259. doi: 10.1016/j.bbamcr.2017.04.008. PMID: 28432022. DOI: 10.1016/j.bbamcr.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Im E. Corticotropin-releasing hormone and its biological diversity toward angiogenesis. Intest Res. 2014;12(2):96–102. doi: 10.5217/ir.2014.12.2.96. PMID: 25349575. DOI: 10.5217/ir.2014.12.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muramatsu Y, Fukushima K, Iino K, Totsune K, Takahashi K, Suzuki T, Hirasawa G, Takeyama J, Ito M, Nose M, Tashiro A, Hongo M, Oki Y, Nagura H, Sasano H. Urocortin and corticotropin-releasing factor receptor expression in the human colonic mucosa. Peptides. 2000;21(12):1799–1809. doi: 10.1016/s0196-9781(00)00335-1. PMID: 11150640. DOI: 10.1016/s0196-9781(00)00335-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Li S. Corticotropin-releasing factor family and its receptors: tumor therapeutic targets. Biochem Biophys Res Commun. 2007;362(4):785–788. doi: 10.1016/j.bbrc.2007.08.014. PMID: 17822675. DOI: 10.1016/j.bbrc.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Tsuda T, Takefuji M, Wettschureck N, Kotani K, Morimoto R, Okumura T, Kaur H, Eguchi S, Sakaguchi T, Ishihama S, Kikuchi R, Unno K, Matsushita K, Ishikawa S, Offermanns S, Murohara T. Corticotropin releasing hormone receptor 2 exacerbates chronic cardiac dysfunction. J Exp Med. 2017;214(7):1877–1888. doi: 10.1084/jem.20161924. PMID: 28550160. DOI: 10.1084/jem.20161924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatzaki E, Anton PA, Million M, Lambropoulou M, Constantinidis T, Kolios G, Taché Y, Grigoriadis DE. Corticotropin-releasing factor receptor subtype 2 in human colonic mucosa: Down-regulation in ulcerative colitis. World J Gastroenterol. 2013;19(9):1416–1423. doi: 10.3748/wjg.v19.i9.1416. PMID: 23539366. DOI: 10.3748/wjg.v19.i9.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Fang X, Yuan J, Sun Z, Li C, Li R, Li L, Zhu C, Wan R, Guo R, Jin L, Li S. The role of corticotropin-releasing hormone receptor 1 in the development of colitis-associated cancer in mouse model. Endocr Relat Cancer. 2014;21(4):639–651. doi: 10.1530/ERC-14-0239. PMID: 25015995. DOI: 10.1530/ERC-14-0239. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez JA, Huerta-Yepez S, Law IK, Baay-Guzman GJ, Tirado-Rodriguez B, Hoffman JM, Iliopoulos D, Hommes DW, Verspaget HW, Chang L, Pothoulakis C, Baritaki S. Diminished expression of CRHR2 in human colon cancer promotes tumor growth and EMT via persistent IL-6/Stat3 signaling. Cell Mol Gastroenterol Hepatol. 2015;1(6):610–630. doi: 10.1016/j.jcmgh.2015.08.001. PMID: 26495412. DOI: 10.1016/j.jcmgh.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tezval H, Jurk S, Atschekzei F, Becker JU, Jahn O, Serth J, Kuczyk MA. Urocortin and corticotropin releasing factor receptor 2 in human renal cell carcinoma: disruption of an endogenous inhibitor of angiogenesis and proliferation. World J Urol. 2009;27:825–830. doi: 10.1007/s00345-009-0417-x. PMID: 19437022. DOI: 10.1007/s00345-009-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tezval H, Jurk S, Atschekzei F, Serth J, Kuczyk MA, Merseburger AS. The involvement of altered corticotropin releasing factor receptor 2 expression in prostate cancer due to alteration of antiangiogenic signaling pathways. Prostate. 2009;69:443–448. doi: 10.1002/pros.20892. PMID: 19058138. DOI: 10.1002/pros.20892. [DOI] [PubMed] [Google Scholar]
- 42.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192–206. doi: 10.1053/j.gastro.2004.10.041. PMID: 15633136. DOI: 10.1053/j.gastro.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 43.Müller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Mühlthaler M, Ofner D, Margreiter R, Widschwendter M. Methylation changes in faecal DNA: a marker for colorectal cancer screening. Lancet. 2004;363:1283–1285. doi: 10.1016/S0140-6736(04)16002-9. PMID: 15094274. DOI: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 44.Liu R, Su X, Long Y, Zhou D, Zhang X, Ye Z, Ma J, Tang T, Wang F, He C. A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma. Mutat Res. 2019;779:45–57. doi: 10.1016/j.mrrev.2019.01.003. PMID: 31097151. DOI: 10.1016/j.mrrev.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Mojtabanezhad Shariatpanahi A, Yassi M, Nouraie M, Sahebkar A, Varshoee Tabrizi F, Kerachian MA. The importance of stool DNA methylation in colorectal cancer diagnosis: A meta-analysis. PLoS One. 2018;13(7):e0200735. doi: 10.1371/journal.pone.0200735. PMID: 30024936. DOI: 10.1371/journal.pone.0200735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagasaka T, Tanaka N, Cullings HM, Sun DS, Sasamoto H, Uchida T, Koi M, Nishida N, Naomoto Y, Boland CR, Matsubara N, Goel A. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009;101(18):1244–1258. doi: 10.1093/jnci/djp265. PMID: 19700653. DOI: 10.1093/jnci/djp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kisiel JB, Klepp P, Allawi HT, Taylor WR, Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann S, Mahoney DW, Roseth A, Ahlquist DA. Analysis of DNA methylation at specific loci in stool samples detects colorectal cancer and high-grade dysplasia in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol. 2019;17(5):914–921.e5. doi: 10.1016/j.cgh.2018.05.004. PMID: 29775793. DOI: 10.1016/j.cgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itzkowitz SH, Harpaz N. Diagnosis and management of dysplasia in patients with inflammatory bowel diseases. Gastroenterology. 2004;126:1634–1648. doi: 10.1053/j.gastro.2004.03.025. PMID: 15168373. DOI: 10.1053/j.gastro.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 49.Yamasaki K, Matsui T, Hisabe T, Yano Y, Hirai F, Morokuma T, Iwao Y, Matsumoto T, Ohi H, Andoh A, Esaki M, Aoyagi K, Sugita A, Nakase H, Fujiya M, Higashi D, Futami K. Retrospective analysis of growth speed of 54 lesions of colitis-associated colorectal neoplasia. Anticancer Res. 2016;36(7):3731–3740. PMID: 27354647. [PubMed] [Google Scholar]