Abstract
Background/Aim: One of the most studied bacterial resistance mechanisms is the resistance related to multidrug efflux pumps. In our study the pump activity of the Escherichia coli K-12 AG100 strain expressing the AcrAB-TolC pump system was investigated at pH 7 and pH 5 in the presence of the efflux pump inhibitor (EPI) promethazine (PMZ). Materials and Methods: The EPI activity was assessed by real-time fluorimetry. The influence of PMZ treatment on the relative expression of the pump genes acrA, acrB and their regulators marA, marB, marR, the stress genes soxS, rob, as well as the bacterial growth control genes ftsI, and sdiA were determined by RT-qPCR. Results: The EPI activity of PMZ was more effective at neutral pH. The PMZ treatment induced a significant stress response in the bacterium at acidic pH by the up-regulation of genes. Conclusion: The genetic system that regulates the activity of the main efflux pump is pH-dependent.
Keywords: Multidrug resistance, efflux pump, promethazine, Escherichia coli K-12 AG100, efflux pump genes, pH-dependent
Bacterial infections that are resistant to two or more classes of antibiotics are deemed multidrug resistant (MDR). The frequency of MDR clinical isolates is global and renders commonly available antibiotics useless. Among the many ways by which the MDR phenotype arises, the over-expression of MDR transporters promotes the expelling of antibiotics and other toxins from the bacterium prior to their reaching their targets (1).
The main efflux pump of E. coli K-12 AG100 is the AcrAB-TolC system which is either transiently over-expressed in the MDR isolate or permanently over-expressed as a consequence of mutations of the regulatory genes that encode the pump (2).
The AcrAB-TolC efflux pump system consists of three proteins, the AcrB transporter and two fusion proteins that attach the AcrB to the plasma membrane. The AcrB transporter recognizes and binds the toxic agent (antibiotic) and releases it to the TolC channel to which it is connected. The AcrB is attached to the plasma membrane via the two fusion proteins whose peristaltic action drives water through the transporter and TolC channel, ultimately releasing the agent to the environment at the junction of TolC to the outer membrane, which consists partly of lipopolysaccharides (LPS). Transport of drugs from the cell to the outside by this system is coupled to proton motive force (PMF) from the periplasm to the cytoplasm. Proton binding and release takes place in the transmembrane domain. The energy transduction and substrate transport seem to be spatially separated (3-5). The activity of the AcrAB-TolC pump in E. coli K-12 AG100 is inhibited by all phenothiazines studied to date. Whether inhibition is direct or indirect is not fully understood. Nevertheless, because the inhibition of the over-expressed efflux pump renders the MDR bacterium susceptible to antibiotics to which it was initially resistant, phenothiazines may have a clinical use as adjuvants. Such examples are PMZ that reduced the frequency of pyelonephritis in children treated with gentamicin, and thioridazine that was effective against XDR-Mycobacterium tuberculosis in combination with antibiotics to which the isolate was initially resistant (6). The activity of the AcrAB-TolC system at neutral pH depends on metabolic energy whereas at pH of 5 no metabolic energy is needed. Moreover, the effect of antibiotics at acidic pH is less pronounced. The effect of pH on the growth of bacteria is well known regarding the stress promoted at acidic conditions (7). The expression of the AcrAB-TolC efflux pump system is controlled by transcriptional regulators, such as MarA, MarB, MarR and the stress proteins SoxS and Rob (8-11). In addition, the bacterial growth is controlled by SdiA and its over-expression results in the over-expression of the AcrAB-TolC system (12).
The bacterial cell division is controlled by the transpeptidase FtsI required for synthesis of peptidoglycan (13). The role of phenothiazines as EPIs of the AcrAB-TolC pump has been studied at neutral pH under conditions that permit the phenothiazine to affect the activity of the pump, however, these compounds have not been studied at acidic pH (3,14). It is the intent of the study to describe the expression of genes that are known to be affected at neutral and acidic pH and determine the effect of PMZ on the expression of these genes.
Materials and Methods
Reagents and media. Promethazine (EGIS), ethidium bromide (EB) and Luria-Bertani (LB) broth were purchased from Sigma-Aldrich Chemie GmbH (Steinheim, Germany). Mueller Hinton (MH) broth was purchased from Scharlau Chemie S. A. (Barcelona, Spain). Sterile glucose solution (40%) was applied in the experiments (PannonPharma, Hungary). The pH was adjusted to 5.0 and 7.0. In the case of PMZ the solvent for the stock solution was distilled water. Phenothiazines are affected by light, thus, PMZ was protected from it (15). The optical properties of the broth with and without PMZ were not altered.
Bacterial strain. The wild-type Escherichia coli K-12 AG100 strain [argE3 thi-1 rpsL xyl mtl Δ(gal-uvrB) supE44] expresses the AcrAB-TolC efflux pump at its basal level (16,17). This strain was kindly provided by Prof. Dr. Hiroshi Nikaido (Department of Molecular and Cell Biology and Chemistry, University of California, Berkeley, CA, USA).
Determination of minimum inhibitory concentrations by microdilution method. The minimum inhibitory concentration (MIC) of PMZ was determined according to Clinical and Laboratory Standard Institute (CLSI) guidelines using MH media at pH 5 and pH 7. Briefly, the bacterial strain was separately cultured in broth of pH 5 and pH 7 overnight at 37˚C. On the following day, the determination of MIC was carried out using broth dilution method in 96-well plates and the plates were incubated for 18 h at 37˚C.
Determination of growth curves at pH 5 and pH 7. The growth curves of the E. coli K-12 AG100 strain were determined by measuring the optical density (OD600) at pH 5 and pH 7 in LB broth with and without 25 μg/ml of PMZ. The bacterial cultures were incubated for further 24 h at 37˚C with shaking (220 rpm) and the optical density was monitored at 600 nm. In addition, we determined the colony forming units (CFUs) at various time points (0, 2, 5, 8, and 24 h) on LB agar plates.
Phenothiazines promote the elongation and subsequent filamention of the bacterium (18). This morphological response affects the interpretation of the results that define growth and optical properties of the culture. From the MIC data obtained at pH 5 and pH 7, the concentration of PMZ that produced no obvious effect on the viability, elongation or filamentation of E. coli K-12 AG100 at both acidic and neutral pH was determined.
Real-time EB accumulation assay. The effect of PMZ on the real-time accumulation of EB in the presence and absence of glucose (0.4%) was assessed by an automated EB method as described previously (19), using a LightCycler real-time thermocycler (LightCycler 1.5; Roche, Indianapolis, IN, USA). The final concentrations of PMZ and EB were 25 μg/ml and 1 μg/ml, respectively. The capillaries were placed into a carousel (Roche) and the fluorescence was monitored at the FL-2 channel every minute on a real-time basis. From the real-time data, the activity of the compound, namely the relative fluorescence index (RFI) of the last time point (minute 30) of the EB accumulation assay was calculated according to the formula:
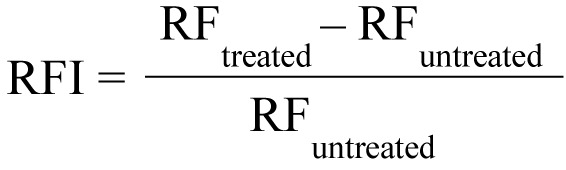
where RFtreated is the relative fluorescence at the last time point of the EB retention curve in the presence of an inhibitor, and RFuntreated is the relative fluorescence at the last time point of the EB retention curve of the untreated solvent control. The solvent was distilled water in the case of PMZ.
Total RNA isolation. The E. coli K-12 AG100 strain was cultured overnight in LB broth at pH 5 and pH 7 at 37˚C with shaking (OD600: 0.6). Bacterial suspensions were prepared with and without PMZ (25 μg/ml) in 3.5 ml of LB medium at pH 5 and pH 7 and were incubated at 37˚C with shaking. The total RNA was isolated at various time points (0, 1, 2, 4, 8, and 18 h). The RNA preparation was carried out in an RNase-free environment using the NucleoSpin RNA kit (Macherey Nagel, Germany), according to the manufacturer’s instructions. Purified RNA was stored in RNase-free water in nuclease-free collection tubes and was maintained at –20˚C until quantification was performed. The concentration of the extracted RNA templates was assessed by spectrophotometry SmartSpec™ Plus at 260 nm (Bio-Rad, Hercules, CA, USA).
Relative gene expression analyses by real-time reverse transcriptase quantitative polymerase chain (RT-qPCR) reaction. The relative gene expression levels were determined at pH 5 and pH 7 in the presence and absence of PMZ. The E. coli K-12 AG100 strain was cultured in LB at pH 5 and pH 7 and total RNA was isolated at various time points (after 0, 1, 2, 4, 8, and 18 h). The relative expression levels of the efflux pump genes, their regulators, stress genes and genes involved in cell wall biosynthesis and quorum sensing were determined by RT-qPCR (20,21) (Table I), using CFX96 Touch real-time PCR detection system (BioRad), strictly following the manufacturer’s recommendations for the SensiFAST™ SYBR No-ROX One-Step Kit (Bioline GmbH, Luckenwalde, Germany). Briefly, each well of the 96-well microtiter plate contained 20 μl as follows: 10 μl of the 2x SensiFAST™ SYBR No-ROX One-Step Mix, 0.2 μl Reverse Transcriptase, 0.4 μl RiboSafe RNase Inhibitor, 5.4 μl Diethylpyrocarbonate (DEPC)-treated water, 500 nM of each primer and approximately 20 ng of total RNA in RNase-free water. Thermal cycling was initiated with a denaturation step of 5 min at 95˚C, followed by 40 cycles each of 10 s at 95˚C, 30 s at 57˚C and 20 s at 72˚C. The relative quantities of the mRNA of each gene of interest were determined by the use of the ΔΔCT method. Gene transcript levels were normalized against the E. coli housekeeping gene GAPDH measured in the same sample. The equation 2–ΔΔCT allows the relative quantification of differences in each gene’s expression level between two samples, the sample of interest and a calibrator or reference sample. The relative gene expression analysis was calculated, according to the following formulas:
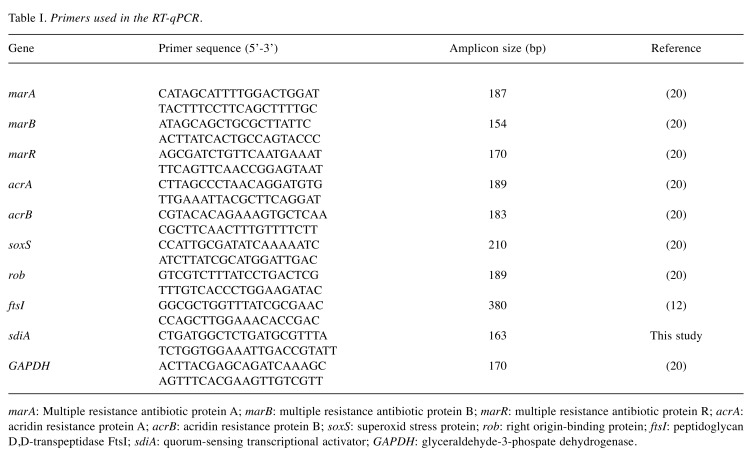
Table I. Primers used in the RT-qPCR.
marA: Multiple resistance antibiotic protein A; marB: multiple resistance antibiotic protein B; marR: multiple resistance antibiotic protein R; acrA: acridin resistance protein A; acrB: acridin resistance protein B; soxS: superoxid stress protein; rob: right origin-binding protein; ftsI: peptidoglycan D,D-transpeptidase FtsI; sdiA: quorum-sensing transcriptional activator; GAPDH: glyceraldehyde-3-phospate dehydrogenase.
ΔCT=CT (gene of interest) – CT (reference gene)
ΔΔCT=ΔCT (PMZ treated) – ΔCT (PMZ untreated)
Results
The MIC of PMZ was determined using MH broth at pH 5 and pH 7 on E. coli K-12 AG100 strain expressing the AcrAB-TolC efflux pump system. The MIC of PMZ was 200 μg/ml at pH 5 and 7.
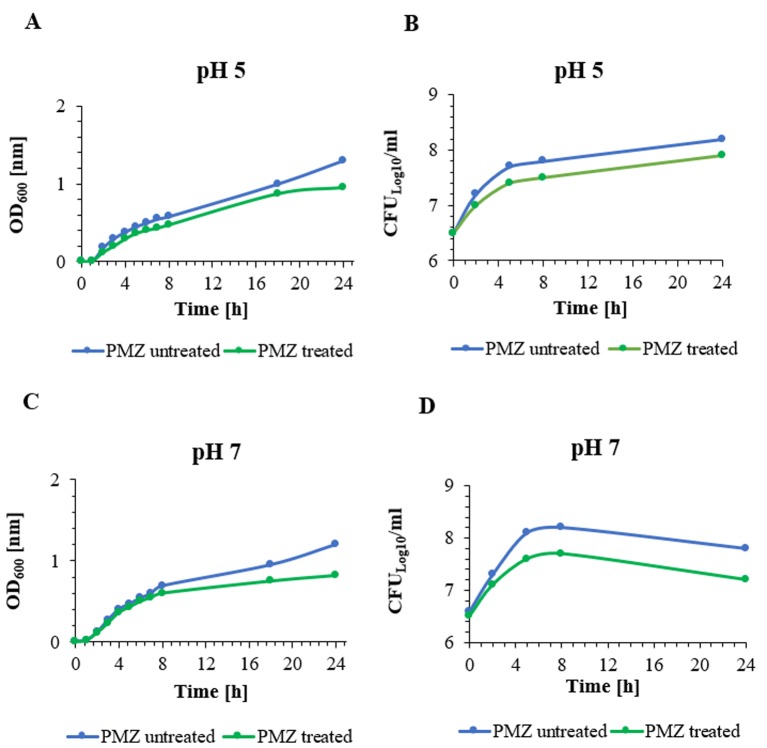
The growth of the E. coli K-12 AG100 strain at pH 5 and pH 7 in LB broth with and without the lowest concentration of PMZ that had a nominal effect on growth for 24 h is shown in Figure 1. Briefly, the growth of PMZ treated bacterial culture was slower at both pH, although the effect on growth was greater at pH 7.
Figure 1. The growth curves of E. coli K-12 AG100 strain were determined at pH 5 (A, B) and pH 7 (C, D) in LB broth in the presence and absence of 25 μg/ml of PMZ. Graphs A and C show the growth curves by measuring the optical density (OD600) at pH 5 and pH 7, respectively. Graphs B and D show the growth curves by counting the CFUs at pH 5 and pH 7, respectively. The growth of PMZ treated bacterial culture was slower at pH 5 compared to pH 7. The growth of bacterial culture was more rapid at pH 7, moreover after 8 h of culturing the declination phase of the bacterial culture could be detected at pH 7 (Graph D).
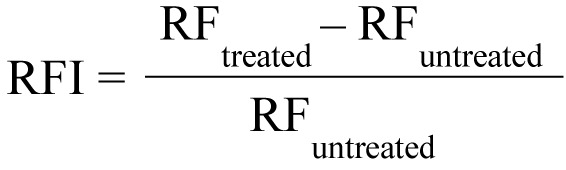
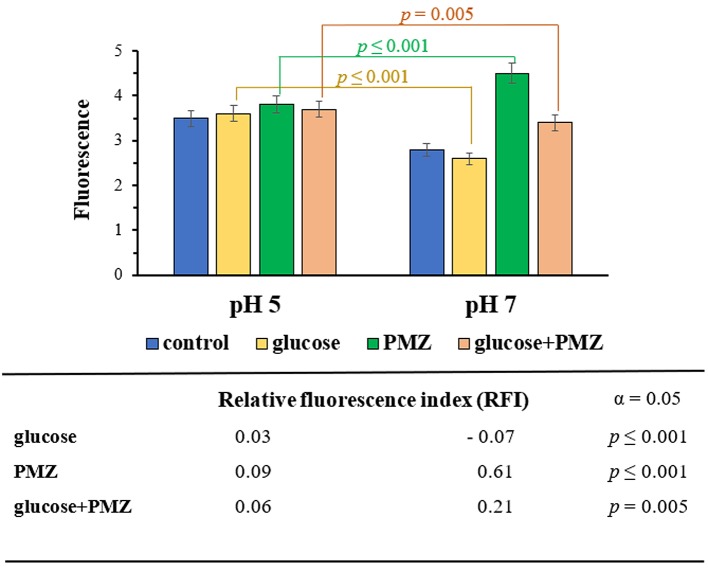
The effect of PMZ on the real-time accumulation of EB was assessed using an automated EB method in the presence and absence of 0.4% glucose. The real-time accumulation curves demonstrated a higher intracellular EB concentration without glucose at pH 7 compared to the EB accumulation at pH 5. The intracellular concentration of EB increased in the presence of PMZ at neutral pH, however the PMZ treated sample exhibited lower EB accumulation at acidic pH. In the case of PMZ treated sample the intracellular EB accumulation was significantly higher at pH 7 compared to pH 5. The efflux pump inhibitor PMZ could exert a more potent EPI effect at neutral pH (Figure 2).
Figure 2. Accumulation of EB at pH 5 and pH 7 by E. coli K-12 AG100 in the presence and absence of glucose 0.4%, with and without 25 μg/ml of PMZ. The real-time accumulation curves demonstrated a higher intracellular EB concentration without glucose at pH 7 compared to pH 5. The intracellular concentration of EB was significantly higher in the presence of PMZ at pH 7, in addition the PMZ treated sample exhibited lower EB accumulation at pH 5. The calculation of statistical significance and p-value was based on the relative fluorescence index (RFI) of the given sample. The correlation is significant: p≤0.001 and p=0.005.
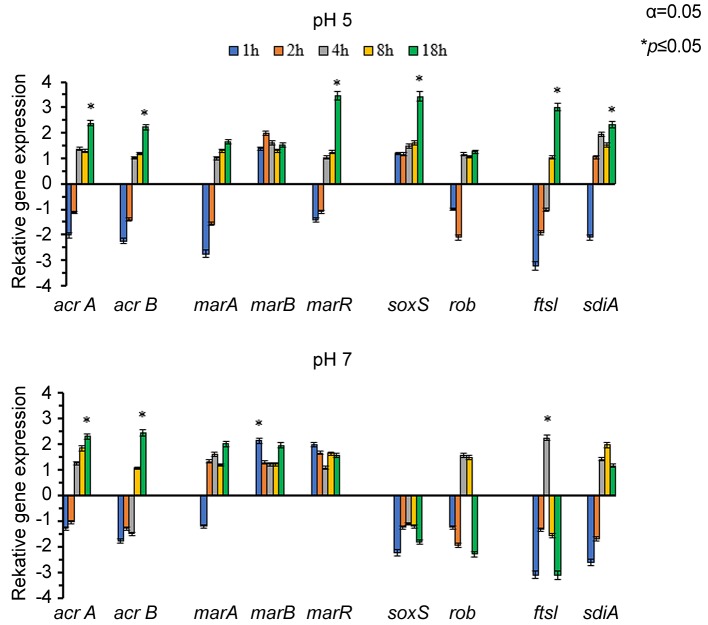
The influence of PMZ treatment was examined on the relative expression of the efflux pump genes (acrA, acrB) and their regulators at neutral and acidic pH. The RT-qPCR results showed that the external environment can influence the activity of efflux pumps. In the case of acidic pH every gene except for soxS exhibited a decreased gene expression pattern in the first 1-2 h. After this period of time the gene expression levels started to increase. Increase in gene expression was detected in the cases of the efflux pump genes acrA and B, as well as in marR regulator, soxS stress gene and QS regulator sdiA after the 18th hour (Figure 3). At neutral pH almost all genes except for marB and marR exhibited a decreased expression pattern in the first 1-2 h. Significant gene expression could be observed in the expression levels of acrA, acrB, and marA genes in the 18th hour, of marB in the 1st hour and of ftsI after 4 h. Initially the efflux pump genes acrA and acrB were down-regulated, but at the end of the culturing period (18th hour) both genes were up-regulated (Figure 3).
Figure 3. Relative expression of genes involved in stress response on E. coli K-12 AG100 strain in the presence of 25 μg/ml of PMZ at pH 5 and pH 7 at different time points (1-18 h). At the beginning of the culturing period most of the studied genes showed a decreased expression pattern. Increase in gene expression level was detected in the cases of the acrA and B, marR, soxS, sdiA genes after the 18th hour at pH 5. Significant gene expression could be observed in the expression levels of acrA, acrB, and marA genes in the 18th hour, of marB in the 1st hour and of ftsI after 4 h. Initially the efflux pump genes acrA and acrB were down-regulated, but at the end of the culturing period (18th hour) both genes were up-regulated at pH 7. The significant correlation is: p≤0.05.
Discussion
The virulence and adaptation of bacteria to the environmental conditions depend on the stress response induced by different factors, such as reduced nutrient source and starvation, pH, low and high osmolarity (22). The survival and colonization of enteric bacteria depend on extreme pH tolerance (23). Before the pathogenic bacteria reach the alkaline pH of the small intestine they must survive the acidic pH of the stomach (24). According to our results, the pH can influence the accumulation and efflux of the RND pump substrate EB. Applying resistance modifiers in our experimental model, the consideration of different pH received special attention in order to mimic the physiological conditions in the gastrointestinal tract (25).
In order to restore survival of E. coli K-12 AG100 and maintain the conditions of cell growth, the acidic stress and PMZ treatment have to be overcome by different cellular mechanisms, such as proton pumps and induce a buffering effect through increasing the concentration of intracellular alkaline compounds (26). In addition, E. coli K-12 AG100 has two major energy sources, and the ATP synthesis. The latter is generated via the respiratory chain and is used mainly for ATP synthesis and various membrane transports (27). At acidic pH, the ATP supply is crucial for the survival of E. coli. At pH 7, E. coli cells are able to maintain a constant internal pH over a range of external pH values from 6.7 to 7.9 (28).
In the EB accumulation assay the EPI activity of PMZ was less effective at acidic pH compared to neutral pH. From this, it can be concluded that the EPI activity of PMZ is pH-dependent because the proton motive force provides a more pronounced energy supply for the AcrB pump at pH 5. At acidic pH the PMF is higher compared to the neutral pH, for this reason the EB that accumulated in the presence of PMZ was lower at pH 5. It has been demonstrated by Mulkidjanian and coworkers that protons generated through metabolic pathways are transported as hydronium ions via channels to the surface of the cell where they are distributed and bound to reactive groups of LPS. These hydronium ions are then transported to the periplasm where they drive the efflux system, thus promoting the rapid extrusion of the substrate (29).
In addition, the acidic pH and PMZ treatment induced a significant stress response in E. coli, and this fact was confirmed by the up-regulation of marB, marR, acrA, acrB, soxS, ftsI, and sdiA genes at acidic pH compared to the neutral pH. Interestingly, soxS exhibited a continuous increase at pH 5, however, at pH 7 the soxS gene was down-regulated until the end of the culturing period. It has to be emphasized that the presented experimental setting applied PMZ at a very low concentration, for which at pH 7 it does not affect the stress genes in the wild type E. coli. For this reason, the rob and ftsI genes required for the initiation of replication and cell wall synthesis, respectively, have the highest expression rate in the 4th hour of culture. In addition, the expressions of ftsI and sdiA were higher at acidic pH compared to neutral pH. The over-expression of the efflux pump genes acrA and B at pH 5 and pH 7 indicates the continuous removal of toxic substances by efflux pumps. Finally, it can be concluded that the acidic pH and PMZ treatment induced a stress response in the bacterium.
Taking our results together we were able to demonstrate that efflux pump inhibitors are promising therapeutic options that may help overcome bacterial multidrug resistance, as well as improve the efficacy of combined antibacterial chemotherapy using conventional antibiotics and EPIs. EPI compounds can influence virulence factors (30), and should be further studied using different bacterial model systems imitating the environmental conditions present in the host organism.
Conflicts of Interest
None declared.
Authors’ Contributions
GS conceived and designed the study. MN and AK performed the laboratory work. MN, AK and GS wrote the article. JM and LA revised the manuscript critically. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by the European Union and the State of Hungary, co-financed by the European Social Fund in the framework of TÁMOP 4.2.4. A/2-11-1-2012-0001 ‘National Excellence Program’ and EFOP 3.6.3-VEKOP-16-2017-00009 and the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. Annamária Kincses was supported by the ÚNKP-18-3 New National Excellence Program of the Ministry of Human Capacities of Hungary.
References
- 1.Weston N, Sharma P, Ricci V, Piddock LJV. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol. 2018;169(7-8):425–431. doi: 10.1016/j.resmic.2017.10.005. PMID: 29128373. DOI: 10.1016/j.resmic.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794(5):769–781. doi: 10.1016/j.bbapap.2008.10.004. PMID: 19026770. DOI: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453(2):254–267. doi: 10.1016/j.bbrc.2014.05.090. PMID: 24878531. DOI: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Fan G, Hryc CF, Blaza JN, Serysheva II, Schmid MF, Chiu W, Luisi BF, Du D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife. 2017:e24905. doi: 10.7554/eLife.24905. PMID: 28355133. DOI: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794(5):782–793. doi: 10.1016/j.bbapap.2008.12.015. PMID: 19166984. DOI: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Varga B, Csonka A, Csonka A, Molnar J, Amaral L, Spengler G. Possible biological and clinical applications of phenothiazines. Anticancer Res. 2017;37(11):5983–5993. doi: 10.21873/anticanres.12045. PMID: 29061777. DOI: 10.21873/anticanres.12045. [DOI] [PubMed] [Google Scholar]
- 7.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187(1):304–319. doi: 10.1128/JB.187.1.304-319.2005. PMID: 15601715. DOI: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duval V, Lister IM. MarA, SoxS and Rob of Escherichia coli- global regulators of multidrug resistance, virulence and stress response. Int J Biotechnol Wellness Ind. 2013;2(3):101–124. doi: 10.6000/1927-3037.2013.02.03.2. PMID: 24860636. DOI: 10.6000/1927-3037.2013.02.03.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178(1):306–308. doi: 10.1128/jb.178.1.306-308.1996. PMID: 8550435. DOI: 10.1128/jb.178.1.306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A, Fuentes SM, Grove A. Redox-sensitive MarR homologue BifR from Burkholderia thailandensis regulates biofilm formation. Biochemistry. 2017;56(17):2315–2327. doi: 10.1021/acs.biochem.7b00103. PMID: 28406615. DOI: 10.1021/acs.biochem.7b00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grove A. MarR family transcription factors. Curr Biol. 2013;23(4):R142–R143. doi: 10.1016/j.cub.2013.01.013. PMID: 23428319. DOI: 10.1016/j.cub.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Tavio MM, Aquili VD, Poveda JB, Antunes NT, Sanchez-Cespedes J, Vila J. Quorum-sensing regulator sdiA and marA overexpression is involved in in vitro-selected multidrug resistance of Escherichia coli. J Antimicrob Chemother. 2010;65(6):1178–1186. doi: 10.1093/jac/dkq112. PMID: 20395215. DOI:10.1093/jac/dkq112. [DOI] [PubMed] [Google Scholar]
- 13.Askoura M, Mottawea W, Abujamel T, Taher I. Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J Med. 2011;6 doi: 10.3402/ljm.v6i0.5870. PMID: 21594004. DOI: 10.3402/ljm.v6i0.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pages JM, Sandrine AF, Mahamoud A, Bolla JM, Davin-Regli A, Chevalier J, Garnotel E. Efflux pumps of Gram-negative bacteria, a new target for new molecules. Curr Top Med Chem. 2010;10(18):1848–1857. doi: 10.2174/156802610793176620. PMID: 20615189. DOI: 10.2174/156802610793176620. [DOI] [PubMed] [Google Scholar]
- 15.Pascu ML, Danko B, Martins A, Jedlinszki N, Alexandru T, Nastasa V, Boni M, Militaru A, Andrei IR, Staicu A, Hunyadi A, Fanning S, Amaral L. Exposure of chlorpromazine to 266 nm laser beam generates new species with antibacterial properties: Contributions to development of a new process for drug discovery. PLoS One. 2013;8(2):e55767. doi: 10.1371/journal.pone.0055767. PMID: 23405212. DOI: 10.1371/journal.pone.0055767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins A, Machado L, Costa S, Cerca P, Spengler G, Viveiros M, Amaral L. Role of calcium in the efflux system of Escherichia coli. Int J Antimicrob Agents. 2011;37(5):410–414. doi: 10.1016/j.ijantimicag.2011.01.010. PMID: 21419607. DOI: 10.1016/j.ijantimicag.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Sugawara E, Nikaido H. Properties of AdeABC and AdeIJK efflux systems of Acinetobacter baumannii compared with those of the AcrAB-TolC system of Escherichia coli. Antimicrob Agents Chemother. 2014;58(12):7250–7257. doi: 10.1128/AAC.03728-14. PMID: 25246403. DOI: 10.1128/AAC.03728-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaral L, Lorian V. Effects of chlorpromazine on the cell envelope proteins of Escherichia coli. Antimicrob Agents Chemother. 1991;35(9):1923–1924. doi: 10.1128/aac.35.9.1923. PMID: 1952868. DOI: 10.1128/aac.35.9.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viveiros M, Martins A, Paixao L, Rodrigues L, Martins M, Couto I, Fahnrich E, Kern WV, Amaral L. Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int J Antimicrob Agents. 2008;31(5):458–462. doi: 10.1016/j.ijantimicag.2007.12.015. PMID: 18343640. DOI: 10.1016/j.ijantimicag.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pages JM, Amaral L. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One. 2007;2(4):e365. doi: 10.1371/journal.pone.0000365. PMID: 17426813. DOI: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spengler G, Rodrigues L, Martins A, Martins M, McCusker M, Cerca P, Machado L, Costa SS, Ntokou E, Couto I, Viveiros M, Fanning S, Molnar J, Amaral L. Genetic response of Salmonella enterica serotype Enteritidis to thioridazine rendering the organism resistant to the agent. Int J Antimicrob Agents. 2012;39(1):16–21. doi: 10.1016/j.ijantimicag.2011.08.013. PMID: 21982147. DOI: 10.1016/j.ijantimicag.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Machado L, Spengler G, Evaristo M, Handzlik J, Molnar J, Viveiros M, Kiec-Kononowicz K, Amaral L. Biological activity of twenty-three hydantoin derivatives on intrinsic efflux pump system of Salmonella enterica serovar Enteritidis NCTC 13349. In Vivo. 2011;25(5):769–772. PMID: 21753132. [PubMed] [Google Scholar]
- 23.Pienaar JA, Singh A, Barnard TG. Acid-happy: Survival and recovery of enteropathogenic Escherichia coli (EPEC) in simulated gastric fluid. Microb Pathog. 2019;128:396–404. doi: 10.1016/j.micpath.2019.01.022. PMID: 30660737. DOI: 10.1016/j.micpath.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 24.Cotter PD, Hill C. Surviving the acid test: Responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev. 2003;67(3):429–453. doi: 10.1128/MMBR.67.3.429-453.2003. PMID: 12966143. DOI: 10.1128/mmbr.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amaral L, Cerca P, Spengler G, Machado L, Martins A, Couto I, Viveiros M, Fanning S, Pages JM. Ethidium bromide efflux by Salmonella: Modulation by metabolic energy, pH, ions and phenothiazines. Int J Antimicrob Agents. 2011;38(2):140–145. doi: 10.1016/j.ijantimicag.2011.03.014. PMID: 21565465. DOI: 10.1016/j.ijantimicag.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Shabayek S, Spellerberg B. Acid stress response mechanisms of group B Streptococci. Front Cell Infect Microbiol. 2017;7:395. doi: 10.3389/fcimb.2017.00395. PMID: 28936424. DOI: 10.3389/fcimb.2017.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Fukamachi T, Saito H, Kobayashi H. ATP requirement for acidic resistance in Escherichia coli. J Bacteriol. 2011;193(12):3072–3077. doi: 10.1128/JB.00091-11. PMID: 21478347. DOI: 10.1128/JB.00091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padan E, Zilberstein D, Rottenberg H. The proton electrochemical gradient in Escherichia coli cells. Eur J Biochem. 1976;63(2):533–541. doi: 10.1111/j.1432-1033.1976.tb10257.x. PMID: 4325. DOI: 10.1111/j.1432-1033.1976.tb10257.x. [DOI] [PubMed] [Google Scholar]
- 29.Mulkidjanian AY, Heberle J, Cherepanov DA. Protons @ interfaces: Implications for biological energy conversion. Biochim Biophys Acta. 2006;1757(8):913–930. doi: 10.1016/j.bbabio.2006.02.015. PMID: 16624250. DOI: 10.1016/j.bbabio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Blanco P, Sanz-Garcia F, Hernando-Amado S, Martinez JL, Alcalde-Rico M. The development of efflux pump inhibitors to treat gram-negative infections. Expert Opin Drug Discov. 2018;13(10):919–931. doi: 10.1080/17460441.2018.1514386. PMID: 30198793. DOI: 10.1080/17460441.2018.1514386. [DOI] [PubMed] [Google Scholar]