Abstract
Background/Aim: Microtubule-associated scaffold protein 1 (MTUS1) acts as tumor suppressor in several cancer types. This study assessed the relationship between clinicopathological characteristics and expression of microRNA candidates based on MTUS1 expression in gallbladder cancer (GBC). Materials and Methods: MTUS1 expression was evaluated by immunohistochemical staining of tissue microarrays from 109 cases of GBC. The association of MTUS1 expression with clinicopathological factors was explored. Two microRNA candidates (miR-19a-3p, and miR-19b-3p), which were identified by a literature review and computational analysis, were assessed in GBC tissue samples by quantitative real-time polymerase chain reaction. Results: Low MTUS1 expression in GBC was associated with high histological grade, perineural invasion, lymphovascular invasion, high T-stage, advanced TNM stage, poorer disease-free survival, and poorer cancer-specific survival. No statistical association between MTUS1 expression and expression of microRNA candidates was observed. Conclusion: MTUS1 may act as tumor suppressor and might be a potential biomarker for predicting prognosis in GBC.
Keywords: MTUS1 protein, gallbladder cancer, immunohistochemistry, prognosis, microRNA, real-time polymerase chain reaction
Although gallbladder cancer (GBC) is rare, it is the most common malignant tumor of the biliary tract (1-4). No specific symptoms or reliable diagnostic markers are established for GBC. In addition, GBC is diagnosed late; moreover, systemic chemotherapy and radiation therapy do not produce significant improvement in survival or quality of life for patients with GBC (1-3). Thus, patients with GBC do not have a favorable clinical outcome (1-3). Therefore, it is essential to evaluate new biomarkers for predicting the prognosis of patients with GBC.
Microtubule-associated scaffold protein 1 (MTUS1), also known as angiotensin II receptor-interacting protein (ATIP), is located on chromosome 8p22 and contains 17 exons (5,6). MTUS1 acts as a tumor-suppressor gene, exhibiting low expression in a variety of malignancies, including breast, colorectal, head and neck, ovary, stomach, salivary gland, and urinary bladder cancer (7-18).
MicroRNAs (miRNAs) are short, noncoding RNAs that regulate gene expression by binding target mRNA at the post-transcriptional level. As miRNA pairing targets mRNAs in an imperfect manner, a single miRNA can target multiple genes; and one gene may be affected by multiple miRNAs (1,19,20). Recently, Ge et al. demonstrated that miR-19a and miR-19b co-regulate MTUS1 in lung cancer (14). Kara et al. reported decreased MTUS1 expression and elevated miR-183-5p expression in advanced stages of breast cancer (10). Additionally, several miRNAs including miR-135b-5p, miR-373-3p, miR-183-5p, miR-142-5p, miR-200c-3p, and miR-19a-3p, may play important roles in colorectal carcinoma through MTUS1 (21).
The role of MTUS1 as a tumor suppressor has been reported, however, the expression of MTUS1 and its clinicopathological significance in GBC has not yet been evaluated. Thus, this study aimed to assess the clinicopathological parameters in association with MTUS1 expression and the correlation between MTUS1 expression and expression of candidate miRNAs.
Materials and Methods
Patients and tissue specimens. Patients who underwent surgery for primary GBC at the Hanyang University Medical Center, Seoul, between January 2001 and December 2015, were included in this study. Adequate quantities of formalin-fixed, paraffin-embedded (FFPE) tissues were collected from 109 patient samples. All patients had undergone radical cholecystectomy, or pancreaticoduodenectomy with/without lymph node dissection or with/without adjuvant chemoradiation therapy. Clinicopathological factors including TNM stage, tumor size, histological type, gross type, histologic grade, lympho-vascular invasion, perineural invasion, and clinical outcomes were assessed by reviewing electronic medical records from intranet resources at the Hanyang University Medical Center, Seoul. Tumor stage was based on the TNM classification described in the seventh edition of the American Joint Committee on Cancer (AJCC) (22).
Tissue microarray (TMA) construction, immunohistochemical staining and scoring. For TMA construction, hematoxylin and eosin (HE)-stained slides were reviewed to select the cellular portion of the malignant tumors. A 3.0-mm sized tumor core was taken from the FFPE blocks of each sample.
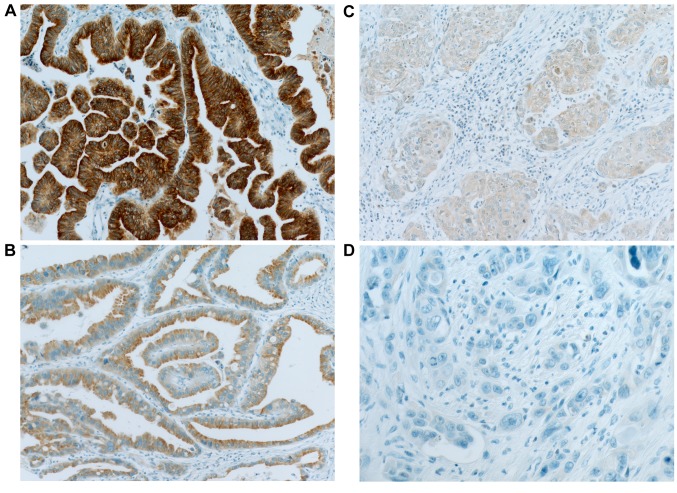
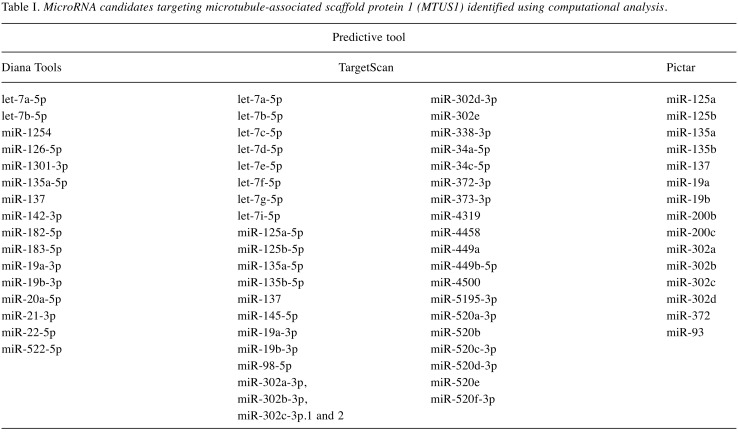
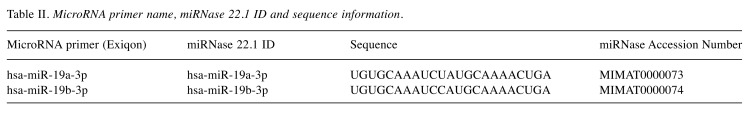
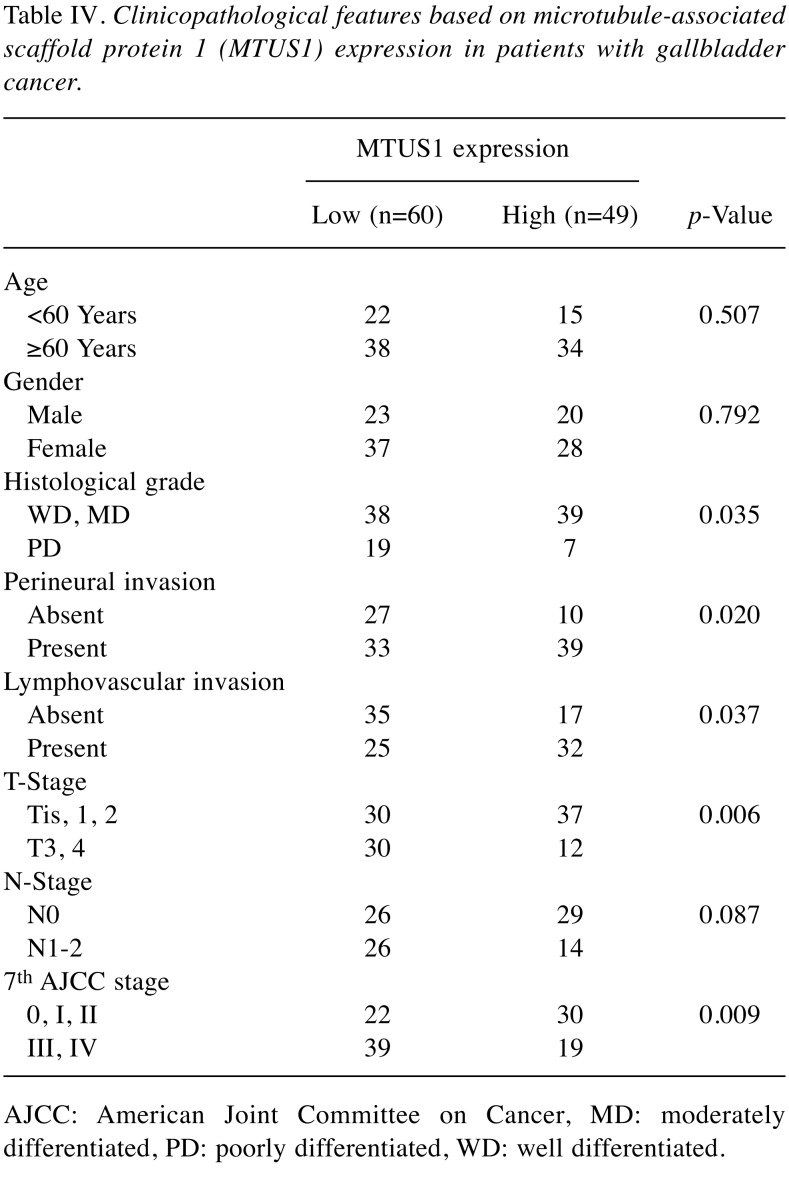
Each TMA block was divided into 4-mm thick sections. Immunohistochemical (IHC) staining of the TMA sections was carried out with a fully automated slide preparation Benchmark XT System (Ventana Medical Systems Inc., Tucson, AZ, USA). Primary antibodies against MTUS1 (1:100; Aviva, San Diego, CA, USA) were used according to the manufacturer’s instructions. MTUS1 expression was assessed using the H-score (23). Tumor cells were scored for either membranous or cytoplasmic expression of MTUS1. Strong positivity was scored as 3+, intermediate positivity as 2+, weak positivity as 1+, and no reactivity as 0 (Figure 1). The proportion of tumor cells at each staining intensity was assessed by the eyeball estimation method. Each sample was scored using the following formula: H-score=[3×(% cells 3+)+2×(% cells 2+)+1×(% cells 3+)] (23). Patients were divided as low expression group (H-score <55) or high expression group (H-score ≥55) for MTUS1 protein expression using the receiver operating characteristics (ROC) curve. All sample assessments were blinded to clinicopathological factors and clinical outcome.
Figure 1. Representative images of immunohistochemical staining of microtubule-associated scaffold protein 1 in gallbladder carcinoma. A: Strong (3+); (B: intermediate (+2); C: weak (+1); and D: negative (0) staining. Original magnification, ×400.
Selection of candidate microRNAs, RNA extraction, and quantitative real-time polymerase chain reaction (PCR). As mentioned above, miRNA candidates targeting MTUS1 were identified from reviewing the literature. We used computational analysis of the retrieved miRNA candidates using Diana Tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php), Target Scan (www.
targetscan.org), and Pictar (https://pictar.mdc-berlin.de/) and confirmed four miRNAs, miR-135a (-5p), miR-137, miR-19a (-3p), and miR-19b (-3p), as miRNA candidates targeting MTUS1. Suggested miRNAs for targeting MTUS1 are listed in Table I. Of these, miR-19a-3p, and miR-19b-3p were experimentally proven to target MTUS1 in lung cancer directly (14), thus suggesting that these two miRNAs may potentially target MTUS1 in GBC.
Table I. MicroRNA candidates targeting microtubule-associated scaffold protein 1 (MTUS1) identified using computational analysis.
HE-stained slides were reviewed to assess tumor areas with minimal necrosis and non-neoplastic gallbladder tissue. HE-stained slides and FFPE blocks were aligned to remove the non-tumoral areas. Three or four sections of 10-μm thickness were obtained from each of the trimmed FFPE blocks. Total RNA was extracted from FFPE tumor specimens using miRNeasy FFPE kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The RNA concentration was determined using a NanoDrop 2000 instrument (NanoDrop Technologies, Waltham, MA, USA).
Using a universal cDNA synthesis Kit (Exiqon, Hilden, Germany), complementary DNA was synthesized from RNA following the manufacturer’s instructions. The expression level of miR-19a-3p and miR-19b-3p was measured by quantitative real-time PCR using a specific primer set (microRNA LNATM PCR primer set; Exiqon) and ExiLENT SYBR Green Master mix (Exiqon). Primers used in the study are listed in Table II. Amplification was performed on a CFX96 thermocycler (Bio-Rad, Hercules, CA, USA). PCR conditions were as follows: 15 min at 95˚C, followed by 45 cycles of 10 s at 95˚C and 1 min at 60˚C. Melting curve analysis was carried out after the PCR cycle. The expression levels of the two miRNAs were normalized using U6 small nuclear RNA (RNU6B). The relative expression was estimated with the 2−ΔΔCt method. Patients were classified into groups with high or low expression based on the median miRNA expression level.
Table II. MicroRNA primer name, miRNase 22.1 ID and sequence information.
Statistical analysis. All statistical analyses were performed using version 23.0 of the Statistical Package for the Social Sciences (IBM, Armonk, NY, USA). The chi-squared test and Student’s t-test were used to assess correlations between MTUS1 protein expression and clinicopathological characteristics. Kaplan-Meier curves were used to present the clinical outcomes, including disease-free survival (DFS) and cancer-specific survival (CSS). For all statistical analyses, values of p<0.05 were considered statistically significant.
Ethics statement. The study protocol was reviewed and approved by the Institutional Review Board of Hanyang University Hospital (IRB file no. 2018-08-031-002). Informed consent was waived due to the retrospective nature of this work.
Results
Patient characteristics. The clinicopathological features of patients with GBC are summarized in Table III. The patients’ age ranged from 28 to 90 years, with a mean of 63.4 years. The median follow-up period for the patients in this study was 48.4 months (range, 0.5 to 242.2 months). We included 42 male patients and 67 female patients. Among the 109 GBC cases, the most common histological type was adenocarcinoma (103 cases, 94.5%). T-Stage was T2 or more in 80.7%. Of the 109 patients, 46 (42.2%) had lymph node metastasis and 10 (9.2%) had distant metastasis. Forty-five patients (41.3%) exhibited a relapse, and 42 patients (38.5%) died due to GBC.
Table III. Histological and clinical characteristics of patients with gallbladder cancer (GBC) (n=109).
AJCC: American Joint Committee on Cancer. *Histological grade was not applied to adenosquamous carcinoma.
MTUS1 expression and correlation with clinicopathological factors in patients with GBC. We divided patients into two groups based on the MTUS1 expression: High and low expression groups. The correlation between MTUS1 expression and clinicopathological characteristics was evaluated to assess the clinical and pathological significance (Table IV). Low MTUS1 expression was correlated with aggressive tumor parameters, such as higher histological grade (p=0.035), perineural invasion (p=0.02), lymphovascular invasion (p=0.037), higher T-stage (p=0.006), and higher AJCC stage (p=0.009). Age, gender, and N-stage were not associated with the expression of MTUS1.
Table IV. Clinicopathological features based on microtubule-associated scaffold protein 1 (MTUS1) expression in patients with gallbladder cancer.
AJCC: American Joint Committee on Cancer, MD: moderately differentiated, PD: poorly differentiated, WD: well differentiated.
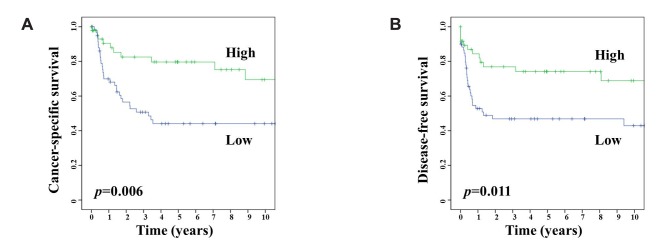
Survival analysis and MTUS1 expression in patients with GBC. We evaluated the correlation between the clinical outcome of patients with GBC and the expression of MTUS1. The Kaplan-Meier survival curves stratified by MTUS1 expression demonstrated that high MTUS1 expression was associated with significantly good clinical outcomes, both in DFS (log-rank test, p=0.011), and CSS (log-rank test, p=0.006) (Figure 2).
Figure 2. Kaplan-Meier analysis for cancer-specific (A) and disease-free (B) survival of patients with gallbladder carcinoma based on expression of microtubule-associated scaffold protein 1 (MTUS1).
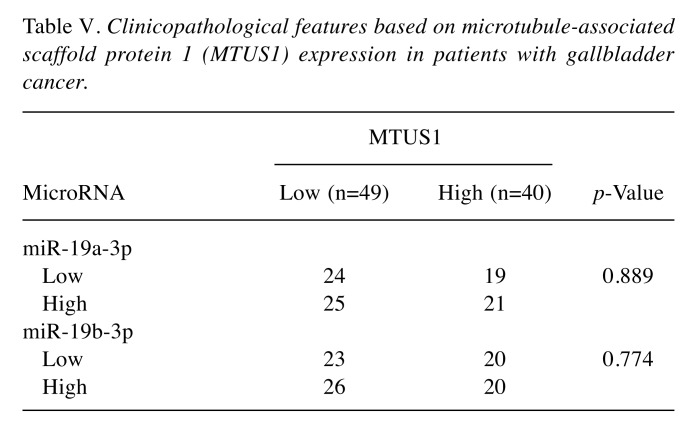
Correlation between MTUS1 expression and miRNA expression. We examined the association between MTUS1 expression and expression of two candidate miRNAs (miR-19a-3p, and miR-19b-3p). In 20 cases, tumor size was too small to extract RNA from FFPE tissue. Based on the median value of expression of miRNA, patients were classified into high and low miRNA expression groups. However, we failed to demonstrate a significant relationship between MTUS1 expression and expression of these two miRNAs (Table V).
Table V. Clinicopathological features based on microtubule-associated scaffold protein 1 (MTUS1) expression in patients with gallbladder cancer.
Discussion
In this study, we used TMA blocks of tumor samples from patients with GBC to perform MTUS1 IHC staining. Patients were classified into two groups using the ROC curve: high and low expression groups. Low MTUS1 expression was significantly associated with poor prognostic factors, including high histological grade, perineural invasion, lymphovascular invasion, high T-stage, and high AJCC stage. Additionally, low MTUS1 expression was significantly correlated with poor clinical outcomes in terms of DFS and CSS. RNAs were extracted from representative sections of FFPE tissues of tumors to explore correlation between expression of MTUS1 and candidate miRNAs. However, we failed to identify any association between expression of candidate miRNAs and MTUS1 expression in patients with GBC.
Biliary tract malignancy including GBC is aggressive and associated with poor clinical outcomes, as it is typically diagnosed at advanced stage and chemotherapy generally has no significant effect (24,25). Radiation therapy appears to have an important role in GBC therapy in improving DFS and overall survival, but requires further study (24-26). Recent study suggested that high proportion (more than 50%) of neoplastic spindle cells were an independent prognostic factor for patients with GBC (27). In addition, GBC generally has several genomic alterations: Adenomatous polyposis coli (APC); AT-rich interactive domain 1A; B-Raf proto-oncogene (BRAF); cyclin-dependent kinase inhibitor 2A and cyclin-dependent kinase 4 inhibitor B; catenin beta 1; human epidermal growth factor receptor (HER) family genes; isocitrate dehydrogenase 1 and 2; lysine (K)-specific methyltransferase 2C RAS family; phosphatidylinositol-3-kinase/AKT and the mammalian target of rapamycin pathway genes; polybromo 1; SMAD family member 4; and tumor protein p53 (3,4,28). Of these, targeted therapeutic clinical trials for targeting epidermal growth factor receptor, BRAF, vascular endothelial growth factor receptor, MET proto-oncogene, mitogen-activated protein kinase kinase 1/2, KIT, platelet-derived growth factor receptor A, and poly ADP ribose polymerase for GBC are ongoing. Nevertheless, GBC remains aggressive, needing new biomarkers.
MTUS1 is a tumor-suppressor gene as was shown in several studies (7-18). Thus, the expression of MTUS1 protein and mRNA in tumor tissues was significantly lower than that in normal tissues of several human tumor types, including breast cancer, gastric carcinoma, head and neck squamous cell carcinoma, salivary adenoid cystic carcinoma, and urinary bladder cancer (8,11,13,16,17). In bladder carcinoma, the expression of MTUS1 protein was reduced in papillary and advanced bladder carcinomas (7). Low expression of MTUS1 was correlated with adverse clinical outcome. Moreover, significant correlation of MTUS1 expression and histological subtype was present: expression was positive in all micropapillary types and aberrant nuclear expression was observed in a subset of the plasmacytoid variants. MTUS1 expression in bladder cancer cell lines led to significant inhibition of cell viability. In breast cancer, low levels of MTUS1 were associated with high grade, and distant metastasis (11). Moreover, expression of MTUS1 was significantly lower in triple-negative breast cancer (11). Silencing of MTUS1 expression promoted breast cancer cell proliferation, a result further supported in experiments using xenograft models (11,29). In colon cancer, Zuern et al. found that silencing of MTUS1 significantly promoted cellular proliferation (12). Li et al. showed that overexpression of MTUS1 significantly decreased both the proliferative and metastatic potential of stomach cancer cell line, an observation further supported in experiments with mouse tumor xenograft models, showing that overexpression of MTUS1 inhibited both tumor growth and metastasis (16). Zhao and Ding et al. showed that down-regulation of MTUS1 was frequent in salivary adenoid cystic carcinoma, and was associated with distant metastasis and poor clinical outcomes, including DFS and overall survival. Additionally, they identified that restoration of expression of ATIP3a, the major isoform of MTUS1, promoted antiproliferative activity and limited the migration and invasion of salivary adenoid cystic carcinoma cell lines (17). In oral tongue squamous cell carcinoma, low expression of MTUS1 correlated with short overall survival (18). Moreover, functional in vitro studies showed that restoration of ATIP1 expression led to G1 arrest, apoptosis and reduction of cell proliferation in oral tongue squamous cell carcinoma cell lines. Similar to earlier studies with different types of cancer, in our study, MTUS1 appeared to plays a tumor-suppressive role in GBC.
Aberrant expression of miRNAs is associated with tumor cell carcinogenesis via binding to mRNA of genes (30,31). As pointed out, several miRNAs were identified as targeting MTUS1 in different types of cancer. MTUS1 mRNA expression in colorectal carcinoma was lower compared to normal colon tissue (21). Several miRNAs, including miR-135b-5b, miR-373-3p, miR-183-5p, miR-142-5p, miR-200c-3p, and miR-19a-3p were overexpressed in colorectal cancer. However, experimental validation was not carried out in their study to assess the relationship between miRNAs and MTUS1 expression. In breast cancer, MTUS1 expression was reduced compared to fibroadenoma, and 15 miRNA candidates for MTUS1 targeting were identified using computational prediction (19). Of these, miR-183-5p was highly expressed in breast cancer. The MTUS1-miR-183-5p axis may play an important role in breast cancer, however, experiments were not conducted to confirm the role of the MTUS1-miR-183-5p axis. Finally, MTUS1 protein and mRNA were significantly lower in patients with lung cancer compared to healthy controls Additionally, in the study, miR-19a/miR-19b inhibited MTUS1 expression, stimulating cell proliferation and migration of lung cancer cell lines (14). However, our results showed that miR-19a-3p, and miR-19b-3p were not associated with MTUS1 expression in patients with GBC.
This is the first study, to our knowledge, to investigate the role of MTUS1 in GBC. Our data support the inverse correlation of MTUS1 expression with poor prognostic factors and unfavorable clinical outcomes and confirm its role as a tumor suppressor in GBC. However, the number of samples in this study was limited. Thus, additional studies with a greater number of patients and experiments to identify miRNAs targeting MTUS1 in GBC are required.
Conflicts of Interest
All Authors declare no potential conflicts of interest.
Authors’ Contributions
Conceptualization: J Sim and K Jang. Methodology: S Bang. Formal analysis: J Sim. Data curation: Y Kim, H Kim, S Jee, S Park, SJ Shin., Investigation: J Sim, K Jang. Writing and original draft preparation: J Sim. Writing, review and editing: K Jang. Approval of final article. All Authors.
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korean government (Ministry of Science, ICT & Future Planning) (NRF-2015R1C1A1A01056091), and Basic Science Research Program through the National Research Foundation of Korea(NRF), funded by the Ministry of Education (NRF-2018R1D1A1B07048798).
References
- 1.Chandra V, Kim JJ, Mittal B, Rai R. MicroRNA aberrations: An emerging field for gallbladder cancer management. World J Gastroenterol. 2016;22(5):1787–1799. doi: 10.3748/wjg.v22.i5.1787. PMID: 26855538. DOI: 10.3748/wjg.v22.i5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 2017;23(22):3978–3998. doi: 10.3748/wjg.v23.i22.3978. PMID: 28652652. DOI: 10.3748/wjg.v23.i22.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sicklick JK, Fanta PT, Shimabukuro K, Kurzrock R. Genomics of gallbladder cancer: The case for biomarker-driven clinical trial design. Cancer Metastasis Rev. 2016;35(2):263–275. doi: 10.1007/s10555-016-9602-8. PMID: 26857926. DOI: 10.1007/s10555-016-9602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montalvo-Jave EE, Rahnemai-Azar AA, Papaconstantinou D, Deloiza ME, Tsilimigras DI, Moris D, Mendoza-Barrera GE, Weber SM, Pawlik TM. Molecular pathways and potential biomarkers in gallbladder cancer: A comprehensive review. Surg Oncol. 2019;31:83–89. doi: 10.1016/j.suronc.2019.09.006. PMID: 31541911. DOI: 10.1016/j.suronc.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Seibold S, Rudroff C, Weber M, Galle J, Wanner C, Marx M. Identification of a new tumor-suppressor gene located at chromosome 8p21.3-22. FASEB J. 2003;17(9):1180–1182. doi: 10.1096/fj.02-0934fje. PMID: 12692079. DOI: 10.1096/fj.02-0934fje. [DOI] [PubMed] [Google Scholar]
- 6.Di Benedetto M, Bieche I, Deshayes F, Vacher S, Nouet S, Collura V, Seitz I, Louis S, Pineau P, Amsellem-Ouazana D, Couraud PO, Strosberg AD, Stoppa-Lyonnet D, Lidereau R, Nahmias C. Structural organization and expression of human MTUS1, a candidate 8p22 tumor-suppressor gene encoding a family of angiotensin II AT2 receptor-interacting proteins, ATIP. Gene. 2006;380(2):127–136. doi: 10.1016/j.gene.2006.05.021. PMID: 16887298. DOI: 10.1016/j.gene.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Rogler A, Hoja S, Giedl J, Ekici AB, Wach S, Taubert H, Goebell PJ, Wullich B, Stockle M, Lehmann J, Petsch S, Hartmann A, Stoehr R. Loss of MTUS1/ATIP expression is associated with adverse outcome in advanced bladder carcinomas: Data from a retrospective study. BMC Cancer. 2014;14(214) doi: 10.1186/1471-2407-14-214. PMID: 24650297. DOI: 10.1186/1471-2407-14-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao J, Chen JX, Zhu YP, Zhou LY, Shu QA, Chen LW. Reduced expression of MTUS1 mRNA is correlated with poor prognosis in bladder cancer. Oncol Lett. 2012;4(1):113–118. doi: 10.3892/ol.2012.673. PMID: 22807972. DOI: 10.3892/ol.2012.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozgeyik I, Yumrutas O, Bozgeyik E. MTUS1, a gene encoding angiotensin-II type 2 (AT2) receptor-interacting proteins, in health and disease, with special emphasis on its role in carcinogenesis. Gene. 2017;626:54–63. doi: 10.1016/j.gene.2017.05.019. PMID: 28499941. DOI: 10.1016/j.gene.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Kara M, Kaplan M, Bozgeyik I, Ozcan O, Celik OI, Bozgeyik E, Yumrutas O. MTUS1 tumor suppressor and its miRNA regulators in fibroadenoma and breast cancer. Gene. 2016;587(2):173–177. doi: 10.1016/j.gene.2016.05.006. PMID: 27155522. DOI: 10.1016/j.gene.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues-Ferreira S, Di Tommaso A, Dimitrov A, Cazaubon S, Gruel N, Colasson H, Nicolas A, Chaverot N, Molinie V, Reyal F, Sigal-Zafrani B, Terris B, Delattre O, Radvanyi F, Perez F, Vincent-Salomon A, Nahmias C. 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One. 2009;4(10):e7239. doi: 10.1371/journal.pone.0007239. PMID: 19794912. DOI: 10.1371/journal.pone.0007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuern C, Heimrich J, Kaufmann R, Richter KK, Settmacher U, Wanner C, Galle J, Seibold S. Down-regulation of MTUS1 in human colon tumors. Oncol Rep. 2010;23(1):183–189. PMID: 19956880. [PubMed] [Google Scholar]
- 13.Mahjabeen I, Kayani MA. Loss of mitochondrial tumor suppressor genes expression is associated with unfavorable clinical outcome in head and neck squamous cell carcinoma: Data from retrospective study. PLoS One. 2016;11(1):e0146948. doi: 10.1371/journal.pone.0146948. PMID: 26785117. DOI: 10.1371/journal.pone.0146948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y, Liu S, Zhang X, Chen G, Liang H, Yu M, Liao Z, Zhou Y, Zhang CY, Wang T, Wang C, Zhang J, Chen X. Oncogenic miR-19a and miR-19b co-regulate tumor suppressor MTUS1 to promote cell proliferation and migration in lung cancer. Protein Cell. 2017;8(6):455–466. doi: 10.1007/s13238-017-0393-7. PMID: 28364280. DOI: 10.1007/s13238-017-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Ju B, Tian J, Liu F, Yu H, Xiao H, Liu X, Liu W, Yao Z, Hao Q. Ovarian cancer stem cell-specific gene expression profiling and targeted drug prescreening. Oncol Rep. 2014;31(3):1235–1248. doi: 10.3892/or.2014.2976. PMID: 24424387. DOI: 10.3892/or.2014.2976. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Liu H, Yu T, Dong Z, Tang L, Sun X. Loss of MTUS1 in gastric cancer promotes tumor growth and metastasis. Neoplasma. 2014;61(2):128–135. doi: 10.4149/neo_2014_018. PMID: 24299308. DOI: 10.4149/neo_2014_018. [DOI] [PubMed] [Google Scholar]
- 17.Zhao T, Ding X, Chang B, Zhou X, Wang A. MTUS1/ATIP3a down-regulation is associated with enhanced migration, invasion and poor prognosis in salivary adenoid cystic carcinoma. BMC Cancer. 2015;15(203) doi: 10.1186/s12885-015-1209-x. PMID: 25885343. DOI: 10.1186/s12885-015-1209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding X, Zhang N, Cai Y, Li S, Zheng C, Jin Y, Yu T, Wang A, Zhou X. Down-regulation of tumor suppressor MTUS1/ATIP is associated with enhanced proliferation, poor differentiation and poor prognosis in oral tongue squamous cell carcinoma. Mol Oncol. 2012;6(1):73–80. doi: 10.1016/j.molonc.2011.11.002. PMID: 22153618. DOI: 10.1016/j.molonc.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonardi GC, Candido S, Carbone M, Colaianni V, Garozzo SF, Cina D, Libra M. microRNAs and thyroid cancer: Biological and clinical significance (Review) Int J Mol Med. 2012;30(5):991–999. doi: 10.3892/ijmm.2012.1089. PMID: 22895530. DOI: 10.3892/ijmm.2012.1089. [DOI] [PubMed] [Google Scholar]
- 20.Weber C. MicroRNAs: From basic mechanisms to clinical application in cardiovascular medicine. Arterioscler Thromb Vasc Biol. 2013;33(2):168–169. doi: 10.1161/ATVBAHA.112.300920. PMID: 23325472. DOI: 10.1161/atvbaha.112.300920. [DOI] [PubMed] [Google Scholar]
- 21.Ozcan O, Kara M, Yumrutas O, Bozgeyik E, Bozgeyik I, Celik OI. MTUS1 and its targeting miRNAs in colorectal carcinoma: Significant associations. Tumour Biol. 2016;37(5):6637–6645. doi: 10.1007/s13277-015-4550-4. PMID: 26643896. DOI: 10.1007/s13277-015-4550-4. [DOI] [PubMed] [Google Scholar]
- 22.Compton CC, Byrd DR, Garcia-Aguilar J, Kurtzman SH, Olawaiye A, Washington MK. AJCC cancer staging atlas: A companion to the seventh editions of the AJCC cancer staging manual and handbook. Springer Science & Business Media. 2012 [Google Scholar]
- 23.Choudhury KR, Yagle KJ, Swanson PE, Krohn KA, Rajendran JG. A robust automated measure of average antibody staining in immunohistochemistry images. J Histochem Cytochem. 2010;58(2):95–107. doi: 10.1369/jhc.2009.953554. PMID: 19687472. DOI: 10.1369/jhc.2009.953554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh Rde W, Alonzo M, Bajaj S, Baker M, Elton E, Farrell TA, Gore RM, Hall C, Nowak J, Roy H, Shaikh A, Talamonti MS. Comprehensive review of the diagnosis and treatment of biliary tract cancer 2012. Part I: Diagnosis-clinical staging and pathology. J Surg Oncol. 2012;106(3):332–338. doi: 10.1002/jso.23028. PMID: 22488652. DOI: 10.1002/jso.23028. [DOI] [PubMed] [Google Scholar]
- 25.Marsh Rde W, Alonzo M, Bajaj S, Baker M, Elton E, Farrell TA, Gore RM, Hall C, Nowak J, Roy H, Shaikh A, Talamonti MS. Comprehensive review of the diagnosis and treatment of biliary tract cancer 2012. Part II: Multidisciplinary management. J Surg Oncol. 2012;106(3):339–345. doi: 10.1002/jso.23027. PMID: 22488601. DOI: 10.1002/jso.23027. [DOI] [PubMed] [Google Scholar]
- 26.Bisello S, Buwenge M, Palloni A, Autorino R, Cellini F, Macchia G, Deodato F, Cilla S, Brandi G, Tagliaferri L, Cammelli S, Valentini V, Morganti AG, Mattiucci GC. Radiotherapy or chemoradiation in unresectable biliary cancer: A retrospective study. Anticancer Res. 2019;39(6):3095–3100. doi: 10.21873/anticanres.13445. PMID: 31177154. DOI: 10.21873/anticanres.13445. [DOI] [PubMed] [Google Scholar]
- 27.Midorikawa R, Hisaka T, Sakai H, Nomura Y, Goto Y, Sato T, Kawahara R, Ishikawa H, Fujita F, Yasunaga M, Tanigawa M, Naito Y, Akiba J, Yano H, Tanaka H, Akagi Y, Okuda K. Clinical and prognostic significance of neoplastic spindle cells in gallbladder cancer. Anticancer Res. 2019;39(8):4561–4568. doi: 10.21873/anticanres.13634. PMID: 31366560. DOI: 10.21873/anticanres.13634. [DOI] [PubMed] [Google Scholar]
- 28.Mishra SK, Kumari N, Krishnani N. Molecular pathogenesis of gallbladder cancer: An update. Mutat Res. 2019;111674:816–818. doi: 10.1016/j.mrfmmm.2019.111674. PMID: 31330366. DOI: 10.1016/j.mrfmmm.2019.111674. [DOI] [PubMed] [Google Scholar]
- 29.Molina A, Velot L, Ghouinem L, Abdelkarim M, Bouchet BP, Luissint AC, Bouhlel I, Morel M, Sapharikas E, Di Tommaso A, Honore S, Braguer D, Gruel N, Vincent-Salomon A, Delattre O, Sigal-Zafrani B, Andre F, Terris B, Akhmanova A, Di Benedetto M, Nahmias C, Rodrigues-Ferreira S. ATIP3, a novel prognostic marker of breast cancer patient survival, limits cancer cell migration and slows metastatic progression by regulating microtubule dynamics. Cancer Res. 2013;73(9):2905–2915. doi: 10.1158/0008-5472.CAN-12-3565. PMID: 23396587. DOI: 10.1158/0008-5472.Can-12-3565. [DOI] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. PMID: 14744438. DOI: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 31.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. PMID: 24079833. DOI: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]