Abstract
Background/Aim: Early detection of gastrointestinal cancer may reduce mortality. Recently, Caenorhabditis elegans has been reported to be capable of differentiating patients with cancers from healthy persons by the smell of urine. This novel technique using C. elegans olfaction has been named as Nematode-NOSE (N-NOSE). Materials and Methods: We collected 180 urine samples from patients with gastrointestinal cancer and 76 samples from healthy subjects. N-NOSE test was performed using these samples and N-NOSE index was obtained. Quantification of the olfactory behavior of C. elegans was performed as established in past studies. By receiver operating characteristic (ROC) analysis, we examined the diagnostic capability of N-NOSE. Results: ROC analysis revealed that N-NOSE showed an area under the curve value of more than 0.80, even in early-stage cancers. Conclusion: C. elegans olfaction enabled the detection of gastrointestinal cancers from urine with high sensitivity, which can provide the basis for the development of N-NOSE as a gastrointestinal cancer screening test.
Keywords: Gastrointestinal cancer, early-stage, screening tests, NNOSE, C elegans.
Gastrointestinal cancer causes various complicating diseases and has high mortality rates. Gastrointestinal cancers account for about 18% of new cases of invasive cancers and for about 26% of cancer-related deaths (1). The survival rate of patients with cancer depends on the stage of cancer. For example, in patients with early-stage colorectal cancer, five-year survival rate is approximately 90%, whereas the survival rate is decreased to less than 20% in patients with late-stage colorectal cancer (1).
To decrease cancer-related mortality, screening tests for early detection of gastrointestinal cancer are essential. Various types of screening tests are available to detect early-stage colorectal cancer (2-5). Although previous studies have demonstrated that conventional screening tests could decrease incidence and mortality, these tests need to be improved, in terms of ineffectiveness, invasiveness, and expensiveness. For example, colonoscopic tests achieve more than 80% sensitivity though they are relatively expensive and invasive. Also, fecal DNA and occult blood test are relatively low-cost, without requiring invasive procedures, though their sensitivity is less than 65% (2-5). Regarding biochemical methods, detection of carcinoembryonic antigen (CEA) was one of the first oncofetal antigens to be used clinically (6), though its sensitivity is less than 10% at an early-stage cancer (7). Another biomarker, carbohydrate antigen (CA) 19-9, can help in the diagnosis of gastrointestinal cancer (8,9). However, detection of early-stage cancers is still difficult.
Novel diagnostic approaches are required to increase detection of early-stage cancers. For this purpose, some biological methods that use the olfactory systems of animals have been invented. Especially, the nematode, Caenorhabditis elegans (C. elegans), which contains over 1000 candidate G-protein coupled olfactory receptors has a very sensitive olfactory system (10-12), and has the innate ability of sensing odors in the urine of patients with cancer. A previous study has indicated that C. elegans exerts a urine-specific behavior; it is attracted by the urine of cancer patients, even if cancer is at an early-stage and repelled by the urine of healthy subjects, and this behavior is due to their olfactory system (13). This olfactory behavior of C. elegans is supported by several previous studies, which have indicated that urine from patients with cancer contains volatile organic compounds (VOCs) produced specifically in response to cancers (14-16). We named this novel method of detection of cancer from urine by C. elegans olfaction as Nematode-NOSE (N-NOSE).
In this study, we evaluated the potential of N-NOSE to be used as a gastrointestinal cancer screening test, since a previous study has suggested the possibility that the olfaction of C. elegans can be utilized to detect cancer from urine (13). The present study examined the diagnostic performance of N-NOSE using 180 samples from cancer patients and 76 samples from healthy subjects. Using receiver operating characteristics (ROC) analysis, we compared the diagnostic ability between N-NOSE and classic tumor markers, CEA and CA19-9. The present study may provide the basis for developing N-NOSE as a novel cancer screening test.
Materials and Methods
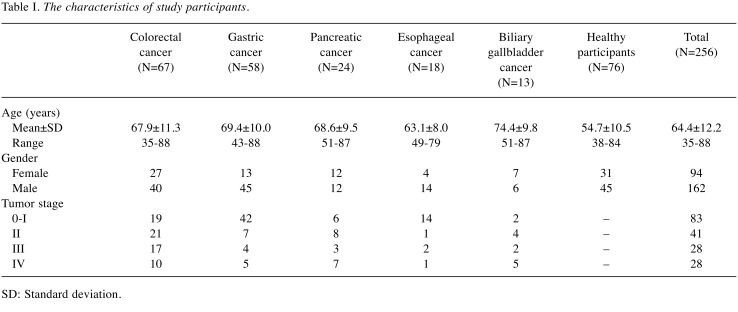
Study populations and ethics. Patients diagnosed at the Nanpuh Hospital (Kagoshima, Japan) with colorectal cancer (n=67), gastric cancer (n=58), pancreatic cancer (n=24), biliary tract cancer (n=9), gallbladder cancer (n=4), and esophageal cancer (n=18) from June 2016 to March 2018 were recruited. To identify healthy individuals, participants were examined using the positron emission tomography (PET) scanning test, blood tests, and cancer biomarker tests, and interviewed by a medical doctor. We excluded individuals who had already undergone treatment for other diseases, had undergone PET showing some abnormalities, and had a history of cancer. The remaining individuals were defined as healthy individuals (n=76). Urine and blood samples from the participants were stored at -80˚C. The PET scanning test was performed using a Discovery ST Elite PET scanner (GE Healthcare, Chicago, IL, USA). The characteristics of all participants are summarized in Table I. Cancer staging was based on a routine histopathological analysis and clinical assessment, according to the tumor-node-metastasis (TNM) classification.
Table I. The characteristics of study participants.
SD: Standard deviation.
The research was planned in accordance with the Declaration of Helsinki and approved by the hospital ethics committee. Informed consent was obtained in writing. The first author guarantees the accuracy and completeness of the data and analysis, and of the study’s fidelity in regard to technical and biostatistics protocols.
Measurement method of N-NOSE. The behavior of the nematode C. elegans towards odors has been analyzed by a number of researchers, and the method to quantify the olfactory behavior has been established (11,12,17-20). In the present study, the culture of animals and chemotaxis assay were performed according to previous C. elegans studies.
C. elegans was cultured on Nematode Growth Media (NGM) agar plate seeded with Escherichia coli as a food source, according to standard methods (11,12,17-20). The method for measuring N-NOSE, a biological cancer test using C. elegans, was according to previous C. elegans olfaction studies (11,17-20) and as described in Hirotsu et al (13). Briefly, 1 μl of diluted urine sample was spotted at one side of the agar plate (9 cm). Then, approximately 50 worms (N2: wild-type strain) were placed in the center of the agar plate. After 30 min, the worms that were present on the half surface of the sample side (A) and on the half surface of the other side (B) were counted. The value of the chemotaxis index was calculated using the following equation: Index = (A-B)/(A+B). The value (-1~0) means repulsion to urine sample; the value (0~1) means attraction to urine sample. Using this value of chemotaxis index, past C. elegans studies quantified the olfactory behavior to various odor samples and concentrations (11,17-20). The average of chemotaxis indices of more than 10 assay plates were used for ROC analysis. Hirotsu et al. have reported that the chemotaxis index is positive in patients' urine samples diluted by 10-1000-fold by water, which is not observed in urine samples from healthy volunteers (13). Therefore, the olfactory behavior against 10-1000-fold diluted urine was investigated.
Collection of urine/blood. Before surgery or chemotherapy, we collected urine and blood samples from participants. Urine samples were stored frozen until analysis using N-NOSE. We measured concentrations of CEA and CA19-9 using an electro-chemiluminescence immunoassay (ECLIA) with LUMIPULSE G1200® (Fujire bio, Inc., Tokyo, Japan) according to the manufacturer’s instructions. Concentrations of squamous cell carcinoma (SCC)-associated antigen in serum was determined by an ECLIA using Elecsys® (Roche Diagnostics K.K., Tokyo, Japan).
Statistical analysis. The area under the curve (AUC) values were calculated using Receiver Operating Characteristics (ROC) analysis with SPSS Version 25 (IBM Co., Armonk, NY, USA). A value of p<0.05 was considered as significant.
Results
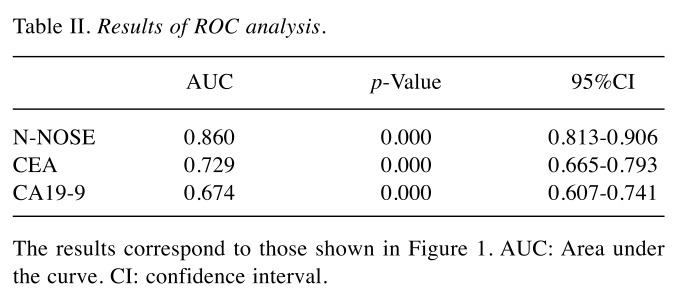
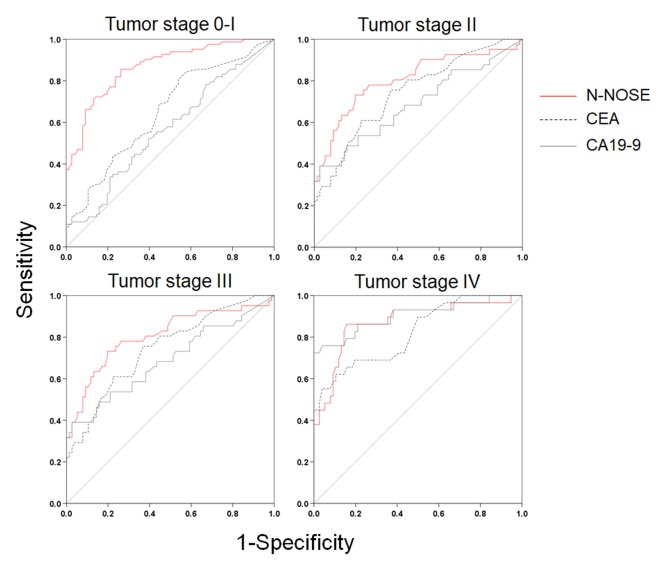
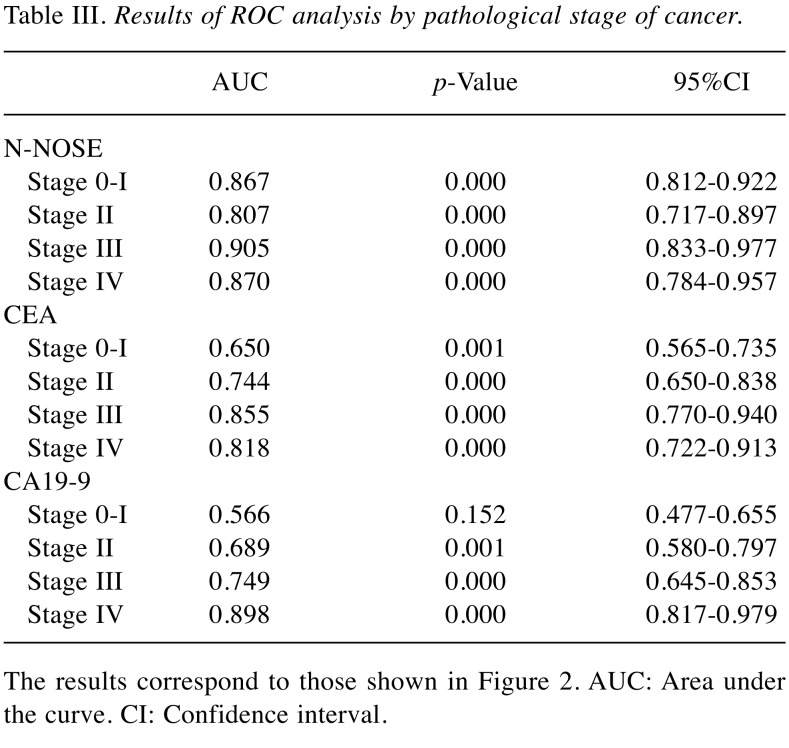
To evaluate the performance of N-NOSE in the detection of several gastrointestinal cancers, we performed ROC analysis. The baseline clinical characteristics of the 180 subjects are shown in Table I. The mean age was 64.4±12.2 years, with 162 (63.3%) men and 94 (36.7%) women. N-NOSE had a higher AUC value than that of CEA and CA19-9 (Figure 1 and Table II). The diagnostic ability of N-NOSE was examined for the various pathological stages. In stage 0-I cancer, the AUC value of N-NOSE was higher than that of CEA and CA19-9 (Figure 2 and Table III). AUC values were also compared with regard to cancer types. N-NOSE scored higher than tumor markers in detecting colorectal cancer, gastric cancer, pancreatic cancer, esophageal cancer, and biliary gallbladder cancer (Figure 3 and Table IV). In stage 0-II cancer, we examined the diagnostic ability of N-NOSE in colorectal cancer, gastric cancer, pancreatic cancer, esophageal cancer, and biliary gallbladder cancer (Figure 4 and Table V). These results suggested that N-NOSE performed better in detecting early stage cancers of gastrointestinal organs.
Figure 1. ROC curves indicating the diagnostic ability of N-NOSE. Area under the ROC curve for N-NOSE (red line), CEA (black dotted line), and CA19-9 (grey line).
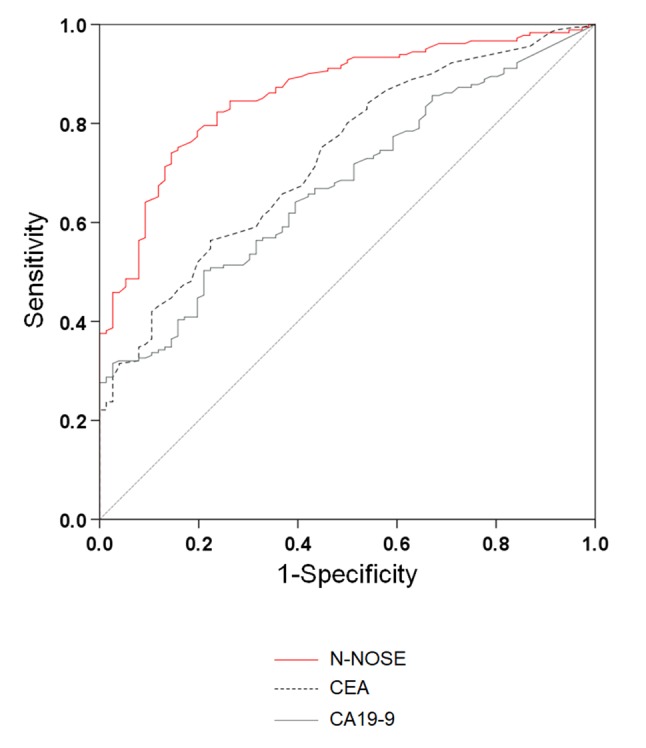
Table II. Results of ROC analysis.
The results correspond to those shown in Figure 1. AUC: Area under the curve. CI: confidence interval.
Figure 2. ROC curves of N-NOSE by pathological stage. Area under the ROC curve for N-NOSE (red line), CEA (black dotted line), and CA19-9 (grey line) in stage 0–I, stage II, stage III, and stage IV.
Table III. Results of ROC analysis by pathological stage of cancer.
The results correspond to those shown in Figure 2. AUC: Area under the curve. CI: Confidence interval.
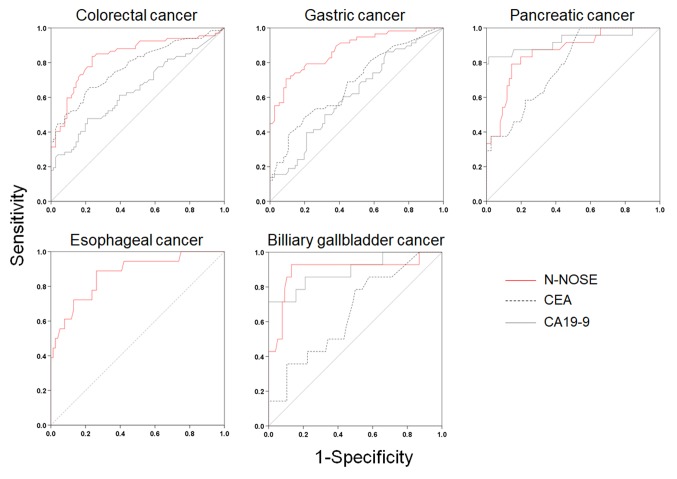
Figure 3. ROC curves of N-NOSE by types of cancer. Area under the ROC curve for N-NOSE (red line), CEA (black dotted line), and CA19-9 (grey line) in colorectal, gastric, pancreatic, esophageal, and biliary gallbladder cancer.
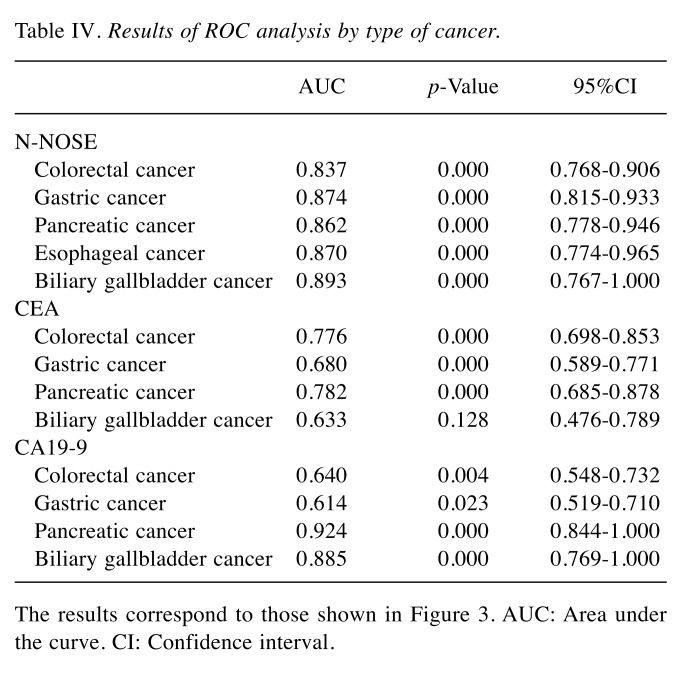
Table IV. Results of ROC analysis by type of cancer.
The results correspond to those shown in Figure 3. AUC: Area under the curve. CI: Confidence interval.
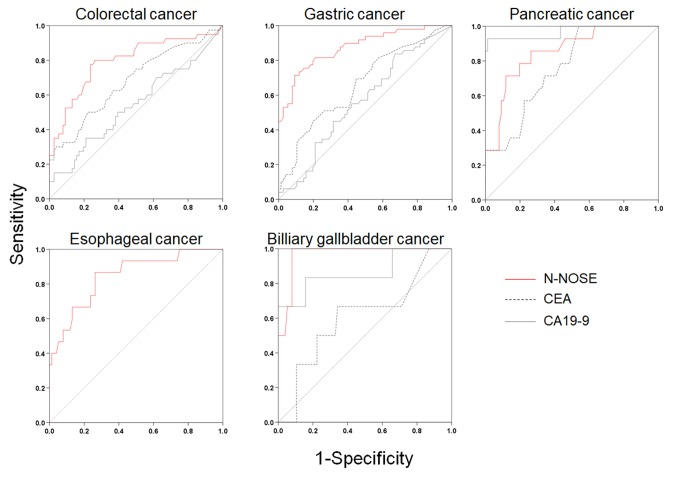
Figure 4. ROC curves of N-NOSE by types of cancer in stage 0–II. Area under the ROC curve for N-NOSE (red line), CEA (black dotted line), and CA19-9 (grey line) in colorectal, gastric, pancreatic, esophageal, and biliary gallbladder cancer.
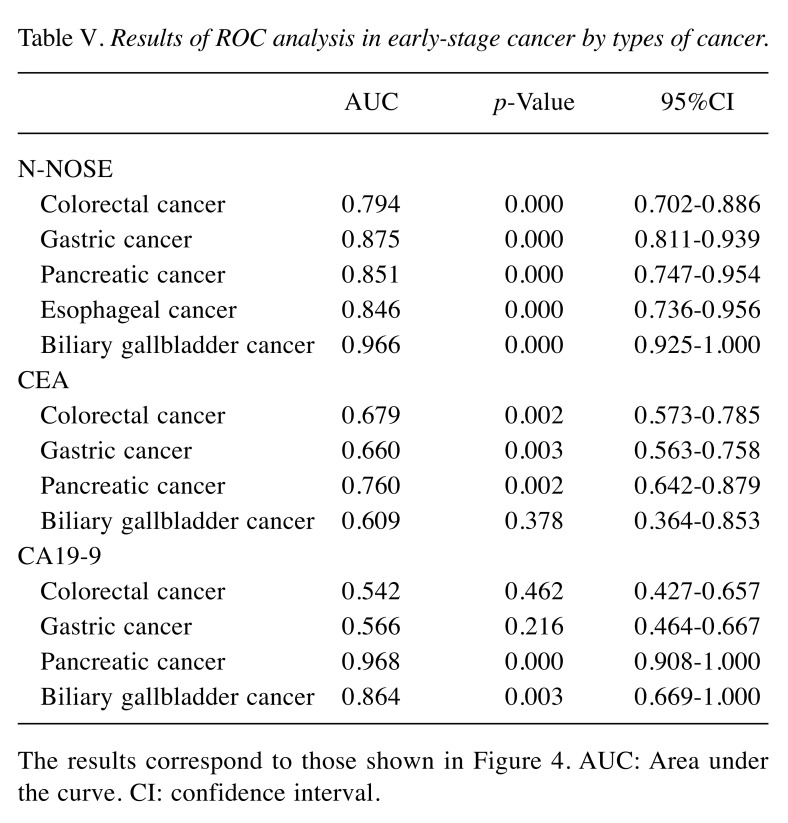
Table V. Results of ROC analysis in early-stage cancer by types of cancer.
The results correspond to those shown in Figure 4. AUC: Area under the curve. CI: confidence interval.
Discussion
We investigated the potential of N-NOSE to be a screening test for digestive cancer. N-NOSE had higher AUC value than CEA and CA19-9. The comparison of the AUC values with regard to pathological stages indicated that N-NOSE had higher AUC value than CEA and CA19-9. Regarding cancer types, the AUC values of N-NOSE were approximately 0.85 in all cancer types, higher than CEA and CA19-9. These results indicated that N-NOSE is a useful surrogate marker for detecting gastrointestinal cancers.
Our results indicated the possibility that N-NOSE could be used as a gastrointestinal cancer screening test. We found that N-NOSE has the diagnostic ability to detect gastrointestinal cancers from urine samples. Consistent with a previous study (13), our study found N-NOSE to have higher AUC values, regardless of stage and type of cancer (Figures 2 and 3, Tables III and IV). Especially, the value of AUC for early-stage cancer remained more than 0.85 in N-NOSE, whereas the value fell below 0.70 in the classic tumor markers in early-stage cancer (Figure 2 and Table III). To examine the performance of N-NOSE in early-stage cancers in more detail, we performed ROC analysis and compared the AUC values in early-stage cancer (i.e. samples from patients in stages 0-II of cancer) by the types of cancer. Even in early-stage colorectal, gastric, pancreatic, esophageal, and biliary gallbladder cancers, the AUC values remained above 0.790 in N-NOSE, whereas the AUC values of CEA were below 0.760 (Figure 4 and Table V). Regarding CA19-9, the AUC values were higher than 0.85 in pancreatic and biliary gallbladder cancers. This may suggest that the combined use of N-NOSE and CA19-9 could lead to a more effective and early detection. The follow-up ROC analysis in early-stage cancer based on types showed the possibility that the sole use of N-NOSE can lead to early detection of cancers occurring in the gastrointestinal system; this ability of N-NOSE would be essential to cancer screening. Early detection of cancer helps to diminish cancer-related mortality, since the five-year survival rates are largely different between early- and late-stage cancers (1). Due to the high diagnostic performance of early-stage and of several types of digestive cancer, N-NOSE would be suitable as a screening test. Furthermore, N-NOSE is less costly and non-invasive. The method used in N-NOSE is based on the C. elegans chemotaxis assay, which has been established in previous C. elegans studies (11,17-20); the assay can be performed using basic laboratory instruments and requires less than 100 μl of urine samples. In addition, it is non-invasive compared to the sampling of blood serum and endoscopic examination.
There were some false positive responses, where C. elegans was attracted to the urine samples from healthy subjects or repelled to the samples from cancer patients. The attraction to a healthy false response may be related with the age of participants. In a previous study, the mean of age in healthy subjects was 47 and 207 out of 218 samples showed the negative chemotaxis index (i.e., the specificity was 95.0%) (13), whereas in the present study the mean of age was 55 and 67 out of 76 samples showed negative chemotaxis index (i.e., the specificity was 88.2%, if the cut-off of chemotaxis index is 0). Although the previous study indicated a non-significant correlation between age and the result of N-NOSE (13), our follow-up analysis suggested a correlation between age and detection of healthy subjects by comparing the different populations (i.e., comparison of the specificity between 218 healthy subjects of the previous study and 76 subjects of the present study). The repulsion to cancer, which is a false response, may be related to the dose-dependent olfactory behavior of C. elegans. Previous studies have revealed that C. elegans shows aversive behavior to high-concentration odors, even if the odor is attractive at a low-concentration (17,20). Similarly, the olfactory behavior of C. elegans to urine samples could depend on the concentration (13). Thus, we could suggest the possibility that the ratio of the cancer-specific odorants was relatively high in the urine from cancer patients, resulted in the aversive olfactory behavior.
This study has some limitations. First, to rigorously establish N-NOSE as a cancer screening test, several additional studies are needed. For example, Pepe et al. have suggested guidelines to develop a biomarker-based screening tool, in which five consecutive phases are used to evaluate and examine the screening tool (21). Otherwise, comprehensive research, using an enormous number of samples, might be needed to evaluate and examine the outcome. For instance, fecal occult blood test has been recommended in Europe and the USA, and a large number of samples have been tested (2,3). Such comprehensive research would enable the evaluation of outcomes and provide an appropriate cut-off value for N-NOSE. Second, in esophageal cancer, the diagnostic ability of N-NOSE was not compared with that of classic tumor markers. All subjects of this study were Japanese; the Japanese population has a high prevalence of esophageal squamous cell carcinoma (22,23). Thus, another tumor marker, such as squamous cell carcinoma (SCC)-associated antigen, should be measured and compared with N-NOSE, rather than CEA and CA19-9, though no measurement of the SCC-associated antigen was performed in this study. To evaluate the performance of N-NOSE in esophageal cancer in more detail, such comparison between N-NOSE and SCC-associated antigen is needed.
In conclusion, the present study evaluated the potential of N-NOSE as a gastrointestinal cancer screening test. We demonstrated that the olfactory system of C. elegans can detect gastrointestinal cancers, regardless of the type and stage. Establishment and practical realization of this novel technique in cancer screening may help in early detection of various cancer types, and eventually a reduction in cancer-related mortality.
Conflicts of Interest
The Authors have no conflicts of interest to declare regarding this study.
Authors’ Contributions
HK, TH, and TU designed the conception of the study. TH and TU experimentally designed the N-NOSE test. TN, KT, SS, KTsu, YB, SF and JF collected urine and serum samples from participants. HK and TN evaluated clinical data. HK, TU, and TH interpreted the results. HK, TU, and TH wrote the manuscript.
Acknowledgements
The Authors appreciate the support of the Division of Clinical Laboratory and Clinical Application of Nanpuh Hospital.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. PMID: 30620402. DOI: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda T, Ono A, Kakugawa Y, Matsumoto M, Saito Y. Impact of screening colonoscopy on outcomes in colorectal cancer. Jpn J Clin Oncol. 2015;45:900–905. doi: 10.1093/jjco/hyv117. PMID: 26254299. DOI: 10.1093/jjco/hyv117. [DOI] [PubMed] [Google Scholar]
- 3.Hoff G, Telemark S, Dominitz J. Contrasting US and European approaches to colorectal cancer screening: Which is best? Contrasting US and European approaches to colorectal cancer screening: which is best. Gut. 2010;59:407–414. doi: 10.1136/gut.2009.192948. PMID: 20207645. DOI: 10.1136/gut.2009.192948. [DOI] [PubMed] [Google Scholar]
- 4.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME, Colorectal Cancer Study Group Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. PMID: 15616205. DOI: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 5.Stoop EM, de Haan MC, de Wijkerslooth TR, Bossuyt PM, van Ballegooijen M, Nio CY, van de Vijver MJ, Biermann K, Thomeer M, van Leerdam ME, Fockens P, Stoker J, Kuipers EJ, Dekker E. Participation and yield of colonoscopy versus non-cathartic CT colonography in population-based screening for colorectal cancer: a randomised controlled trial. Lancet Oncol. 2012;13:55–64. doi: 10.1016/S1470-2045(11)70283-2. PMID: 22088831. DOI: 10.1016/S1470-2045(11)70283-2. [DOI] [PubMed] [Google Scholar]
- 6.Gold BYP, Freedman SO. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J Exp Med. 1965;121:439–462. doi: 10.1084/jem.121.3.439. PMID: 14270243. DOI: 10.1084/jem.121.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castaldi F, Marino M, Beneduce L, Belluco C, De Marchi F, Mammano E, Nitti D, Lise M, Fassina G. Detection of circulating CEA-IgM complexes in early stage gastric cancer. Int J Biol Markers. 2005;20:204–208. doi: 10.1177/172460080502000402. PMID: 16398401. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: A meta-analysis. Int J Clin Exp Med. 2015;8:11683–11691. PMID: 26380005. [PMC free article] [PubMed] [Google Scholar]
- 9.Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G, Guo M, Lian X, Fan D, Zhang H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:1–6. doi: 10.1186/s12885-017-3738-y. PMID: 29121872. DOI: 10.1186/s12885-017-3738-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson H, Thomas JH. The putative chemoreceptor families of C. elegans. WormBook. 2006;2006:1–12. doi: 10.1895/wormbook.1.66.1. PMID: 18050473. DOI: 10.1895/wormbook.1.66.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. PMID: 8348618. DOI: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 12.Sengupta P, Chou JH, Bargmann CI. Odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. PMID: 8601313. DOI: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 13.Hirotsu T, Sonoda H, Uozumi T, Shinden Y, Mimori K, Maehara Y, Ueda N, Hamakawa M. A highly accurate inclusive cancer screening test using Caenorhabditis elegans scent detection. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0118699. PMID: 25760772. DOI: 10.1371/journal.pone.0118699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanai Y, Shimono K, Matsumura K, Vachani A, Albelda S, Yamazaki K, Beauchamp Gk, Oka H. Urinary volatile compounds as biomarkers for lung cancer. Biosci Biotechnol Biochem. 2012;76:679–684. doi: 10.1271/bbb.110760. PMID: 22484930. DOI: 10.1271/bbb.110760. [DOI] [PubMed] [Google Scholar]
- 15.Shirasu M, Touhara K. The scent of disease: Volatile organic compounds of the human body related to disease and disorder. J Biochem. 2011;150:257–266. doi: 10.1093/jb/mvr090. PMID: 21771869. DOI: 10.1093/jb/mvr090. [DOI] [PubMed] [Google Scholar]
- 16.Grayson K, Gregory E, Khan G, Guinn BA. Urine biomarkers for the early detection of ovarian cancer – are we there yet. Biomark Cancer. 2019;11:1179299X1983097. doi: 10.1177/1179299X19830977. PMID: 30833816. DOI: 10.1177/1179299X19830977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida K, Hirotsu T, Tagawa T, Oda S, Wakabayashi T, Iino Y, Ishihara T. Odour concentration-dependent olfactory preference change in C. elegans. Nat Commun. 2012;3:711–739. doi: 10.1038/ncomms1750. PMID: 22415830. DOI: 10.1038/ncomms1750. [DOI] [PubMed] [Google Scholar]
- 18.Cho CE, Brueggemann C, L’Etoile ND, Bargmann CI. Parallel encoding of sensory history and behavioral preference during Caenorhabditis elegans olfactory learning. Elife. 2016;5:1–20. doi: 10.7554/eLife.14000. PMID: 27383131. DOI: 10.7554/eLife.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worthy SE, Rojas GL, Taylor CJ, Glater EE. Identification of odor blend used by Caenorhabditis elegans for pathogen recognition. Chem Senses. 2018;43:169–180. doi: 10.1093/chemse/bjy001. PMID: 29373666. DOI: 10.1093/chemse/bjy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi G, Uozumi T, Kiriyama K, Kamizaki T, Hirotsu T. Screening of odor-receptor pairs in Caenorhabditis elegans reveals different receptors for high and low odor concentrations. Sci Signal. 2014;7 doi: 10.1126/scisignal.2005136. PMID: 24782565. DOI: 10.1126/scisignal.2005136. [DOI] [PubMed] [Google Scholar]
- 21.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson M Lou, Thornquist M, Winget M, Yasui Y. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. PMID: 11459866. DOI: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 22.Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg. 2018;41:210–215. doi: 10.1016/j.asjsur.2016.10.005. PMID: 27986415. DOI: 10.1016/j.asjsur.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Pickens A, Orringer MB. Geographical distribution and racial disparity in esophageal cancer. Ann Thorac Surg. 2003;76:1367–1369. doi: 10.1016/s0003-4975(03)01202-5. PMID: 14530066. DOI: 10.1016/s0003-4975(03)01202-5. [DOI] [PubMed] [Google Scholar]