Abstract
Background/Aim: Intestinal damage induced by total body irradiation (TBI) reduces leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5)-expressing stem cells, goblet, and Paneth cells, breaching the epithelial lining, and facilitating bacterial translocation, sepsis, and death. Materials and Methods: Survival was measured after TBI in animals that received wild-type or recombinant bacteria producing interleukin-22 (IL-22). Changes in survival due to microbially delivered IL-22 were measured. Lactobacillus reuteri producing IL-22, or Escherichia coli-IL-22 were compared to determine which delivery system is better. Results: C57BL/6 mice receiving IL-22 probiotics at 24 h after 9.25 Gy TBI, demonstrated green fluorescent protein-positive bacteria in the intestine, doubled the number of Lgr5+ intestinal stem cells, and increased 30-day survival. Bacteria were localized to the jejunum, ileum, and colon. Conclusion: Second-generation probiotics appear to be valuable for mitigation of TBI, and radiation protection during therapeutic total abdominal irradiation.
Keywords: Lactobacillus reuteri, E coli, interleukin-22, total body irradiation, radiation mitigators
Radiation mitigators that are delivered 24 h or later after total body irradiation (TBI) are needed as countermeasures against a radiation terrorist event or nuclear reactor accidents (1,2). Ionizing irradiation damages the intestine and leads to the gastrointestinal syndrome (3-6). TBI at levels that exceed the dose that can be rescued by bone marrow transplantation (hematopoietic syndrome) kills intestinal crypt cells, damages villi, and results in a systemic increase in gut bacteria, which leads to sepsis, and, ultimately, death (4-6). A distinct gastrointestinal syndrome is observed after TBI at higher doses, and kills mice 5-10 days after exposure to doses in excess of 12 Gy. Animals receiving 15 Gy total abdominal irradiation die within 10 days associated with shrinkage of antimicrobial intestinal Paneth cells that, naturally, produce defensins, lysozyme and other antimicrobial factors (7-16). Loss of intestinal crypt cells, principally leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5+) stem cells (17), is also detected after doses that produce death from gastrointestinal syndrome (4-6). Following irradiation, there is rapid depletion of intestinal immunocytes, prominently, γδT-cells and helper T-cells, which are known to produce interleukin-22 (IL-22) and other anti-inflammatory cytokines (18-28). IL-22 produced by anti-inflammatory immunocytes helps to stabilize both intestinal Paneth cells and Lgr5+ intestinal stem cells (17). Other differentiated cell populations within intestinal villi include goblet cells that produce mucin (6), which protects the intestinal epithelium, has a barrier function and prevents entry of bacteria. In addition to production of anti-microbial defensins and lysozyme, Paneth cells are critical for support of Lgr5+ stem cells. The search for radiation protector and radiation mitigator drugs (the latter showing effectiveness after irradiation exposure) has led to discoveries of multiple classes of therapeutic agents (1,2). These include drugs that block distinct radiation-induced cell death pathways, including apoptosis, necroptosis, ferroptosis, and parthanatos (29). Other classes of radiation mitigators include anti-inflammatory cytokines which delivered at various stages before and after irradiation lead to preservation and regeneration of intestinal epithelium. Recent evidence has demonstrated that the anti-inflammatory cytokine IL-22, is likely a common target in radiation mitigation (6) as it ameliorates intestinal radiation damage.
In this work, we tested the hypothesis that microbial-mediated delivery of recombinant IL-22 to the intestine promotes the survival of mice exposed to TBI.
Materials and Methods
Mice and animal care. Female C57BL/6NTac (Taconic Biosciences, Wayne, PA, USA) and C57BL/6 Lgr5+ green fluorescent protein tagged (5,6) mice were housed five per cage and fed standard laboratory chow and deionized drinking water according to University of Pittsburgh Institutional Animal Care and Use Committee regulations.
Irradiation. TBI was delivered with a cesium-137 gamma cell irradiator with filters removed at a dose rate of 310 cGy/min; beam flatness, homogeneity, and analysis of spectrum of gamma ray precise dosimetry were carried out according to published methods (2). Mice were irradiated in a plexiglass pie plate of 12 sections. Five mice were placed in each pie plate. The mice were not placed in adjacent sections, but there were one or two empty sections between mice so that the mice would not shield the other mice in the pie plate. Irradiation dose was verified using thermoluminescent readings according to published methods (2).
Bacterial strains and growth conditions. Escherichia coli DH5α (Thermo Fisher Scientific, Waltham, MA, USA) was used for subcloning and plasmid amplification. E. coli BL21 (New England Biolabs, Ipswich, MA, USA) was used as the expression host. E. coli was grown in Luria-Bertani broth at 37˚C with 200 rpm shaking. Ampicillin (100 μg/ml) was added when needed for selection. Lactobacillus reuteri (American Type Culture Collection, Manassas, VA USA) strains were cultured at 37˚C under static conditions in de Man Rogosa Sharpe medium (Sigma-Aldrich Co, St. Louis, MO, USA).
Selection of E. coli bacterial transformants was carried out using the metabolic marker construct ampicillin. Successful transformants were assayed for both intracellular and secreted (into the culture broth) of IL-22. Green fluorescent protein (GFP) positivity was determined by both observation of green color in bacterial transformants and also by PCR assay for the transgene for GFP.
Plasmid construction, expression, and plasmid transfection reagents. Plasmid pRSET-EmGFP purchased from Thermo Fisher Scientific was used as E. coli BL21 expression vector.
The gene coding for IL-22 (GeneBank: NM_016971.2) was amplified from a cDNA library of C57BL/6 mouse bone marrow stromal cells. IL-22 was fused to the GFP protein gene on vector pRSET-EmGFP between BamHI and NcoI restriction sites. The IL-22 gene was amplified by the primers: IL22-forward: 5’-GGT GGT GGA TCC ATG GCT GTC CTG CAG AAA TC; IL22-reverse: 5’-GGT GGT CCA TGG GAC GCA AGC ATT TCT CAG AG.
The plasmids were first transformed into E. coli DH5α and the expression cassettes were verified by DNA sequencing. The plasmid pRSETEmGFP/IL22 was transformed into E. coli BL21 by heating transformation generating an E. coli BL21 strain expressing IL-22.
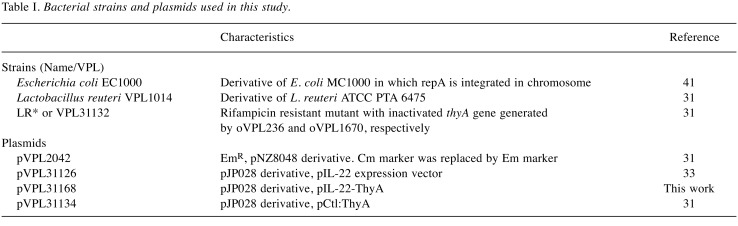
Construction of LRΔthyA:rpoB(H488R)/pIL-22-thyA. By single-stranded DNA recombineering, we inactivated thyA in a rifampicin-resistant derivative of L. reuteri VPL1014 [LR:rpoB(H488R)] to yield LRΔthyA (Rif®), as described previously (30,31) (Table I and Table II). The gene providing chloramphenicol resistance in the vector pVPL31126 was replaced with the thyA gene derived from L. reuteri VPL1014 via blunt-end ligation (T4 DNA ligase: Thermo Fisher Scientific) and transformed into LR* by electroporation to construct LR*/pIL-22-thyA, as described previously (31-33). LR* harboring the previously constructed vector pCtl-thyA served as an empty vector control (31). E. coli EC1000 was used as an intermediate cloning host.
Table I. Bacterial strains and plasmids used in this study.
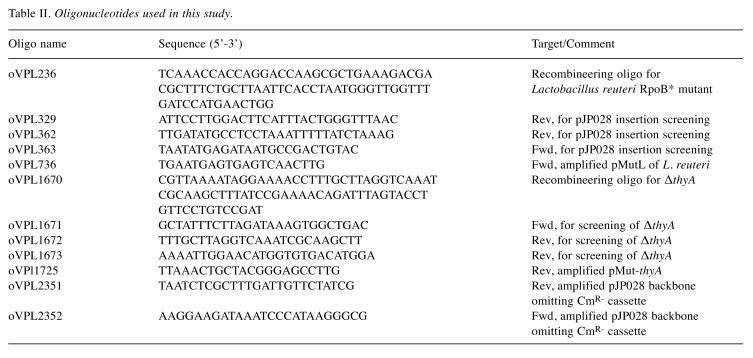
Table II. Oligonucleotides used in this study.
ELISA assays. IL-22 ELISA kit (Thermo Fisher Scientific) was used to quantify IL-22 levels following the company’s instructions. Lysates of transformed bacterial strains (lysed by sonication) and culture medium were used to determine IL-22 levels.
IL-22 biological activity assay. The activity of both E. coli IL-22 and Lactobacillus-IL-22 was determined by measuring secretion of IL-10 in the Colo 205 (ATCC CCL-222; American Type Culture Collection) human colon carcinoma cell line. IL-10 production is positively regulated by IL-22. Colo 205 cells were maintained in RPMI-1640 supplemented with 10% fetal bovine serum, 1% pen-strep and 1% L-glutamine. Cells were seeded in 6-well plates at a density of 1×106 cells/well several hours before adding IL-22. At 24 h after addition of IL-22 (500 μl of IL-22 medium into 2 ml of Colo 205 medium), the amount of IL-10 in the cell-free medium from the Colo 205 cell was determined by IL-10 ELISA kit (Thermo Fisher Scientific). A standard curve was established to quantitate the IL-22 in the culture medium using positive control IL-22 (Peprotech, Rocky Hill, NJ, USA) at levels ranging from 1 pg/ml to 1 μg/ml.
Dose-response curve of number of gavaged L. reuteri, or E. coli producing IL-22. Mice were irradiated to 9.25 Gy TBI, which took 13-14 min, and 24 h later were then gavaged with 100 μl saline solution containing L. reuteri-IL-22, or E. coli-IL-22. All groups of mice received 100 μl saline, which contained different numbers of bacteria (106, 107, 108 and 109 bacteria; n=10 animals per group).
Assay for transgene IL-22, and GFP transgene by PCR. Bacterial strains were evaluated by recombinant DNA reagents in assays for transgene insertion using standard procedures for PCR. Primers to detect transgene IL-22, and GFP were obtained from (Integrated DNA Technologies, Austin, TX, USA) and have been described above.
Assays for intestinal paneth, goblet, and Lgr5+ stem cells. These assays were carried out using antibodies and assay conditions according to published methods (5,6).
Administration of small molecule radiation mitigator, IL-22 protein, L. reuteri lacking IL-22, and fecal microbiome. As a control for our probiotic mitigators, the GS-nitroxide, JP4-039 (1), was prepared and administered according to published methods, i.m. at 20 mg/kg in 50 μl of 30% cyclodextrin aqueous (34). Cyclodextrin alone has no radiomitigative effect (34). IL-22 protein was administered i.p. at 0.1 mg/kg. L. reuteri lacking IL-22 (109) were administered by gavage in 100 μl saline. Fecal microbiome transplant was carried out by delivery by gavage in 100 μl saline of 109 bacteria from the feces of 30-day survivors of 9.25 Gy TBI of C57BL/6 female mice.
Immunostaining and imaging. Imaging of intestinal barrier function was carried out using multicolor staining techniques according to published methods (35). In particular, imaging of GFP+ bacteria location relative to breeches in intestinal keratin barrier were carried out according to published methods (35).
Statistical analysis. In vivo survival curves were analyzed using a log-rank test. Comparisons between two groups were evaluated using Student’s t-test (34).
Results
Oral administration of recombinant L. reuteri producing interleukin-22 improves survival after TBI. In our study, we used a derivative of a previously constructed L. reuteri strain that produces IL-22. To ensure plasmid stability without the need for antibiotic selection, we deleted the essential gene thyA, which encodes thymidylate synthase, and cloned this on the plasmid backbone encoding IL-22 (36). The plasmid will never be lost in the absence of antibiotic selection because it provides ThyA in trans (32).
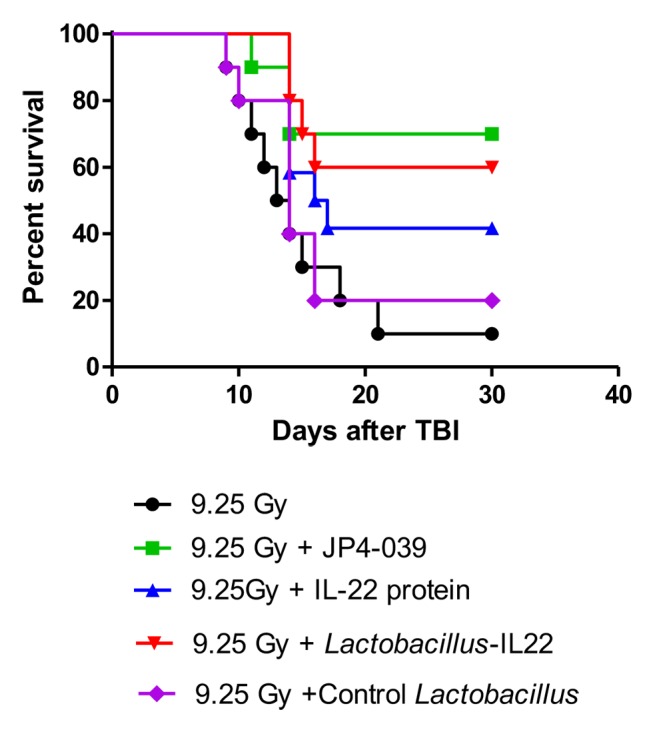
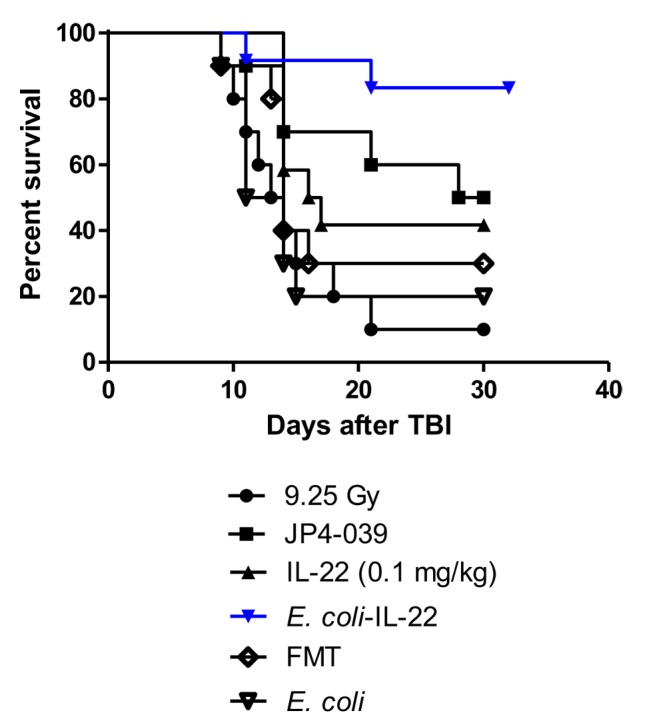
C57BL/6NTac mice were irradiated to 9.25 Gy TBI and gavaged with L. reuteri-expressing IL-22 or L. reuteri harboring the empty vector control. Additional controls included animals subjected to i.p. injection of recombinant IL-22 protein or the radiation mitigator, JP4-039. Compared to the animals that only received TBI, we observed increased survival at 30 days after TBI in mice administered Lactobacillus-IL-22 (60%), IL-22 protein (40%), or JP4-039 (70%) (Figure 1). There was no significant increase in survival following gavage of wild-type L. reuteri compared to control 9.25 Gy TBI (10%). Thus, probiotic-mediated delivery of IL-22 increased the survival of animals exposed to TBI at levels that are comparable to those induced by the radiation-mitigation compound JP4-039.
Figure 1. Improved survival of mice treated with total body irradiation (TBI) and Lactobacillus reuteri-IL-22 (n=10). Groups of 10 mice received 9.25 Gy TBI then 24 h later gavage of 100 μl of saline containing 109 Lactobacillus reuteri-interleukin (IL-22), or IL-22 protein delivered intraperitoneally at 20 mg/kg in 100 μl, or 100 μl cyclodextrin containing 20 mg/kg of the radiation mitigator JP4-039. Significant increase in survival was seen in irradiated mice treated with JP4-039 (p=0.0079), IL-22 protein (p=0.0428) or Lactobacillus-IL-22 (p=0.0014) but not control Lactobacillus (p=0.5021) compared to control irradiated mice.
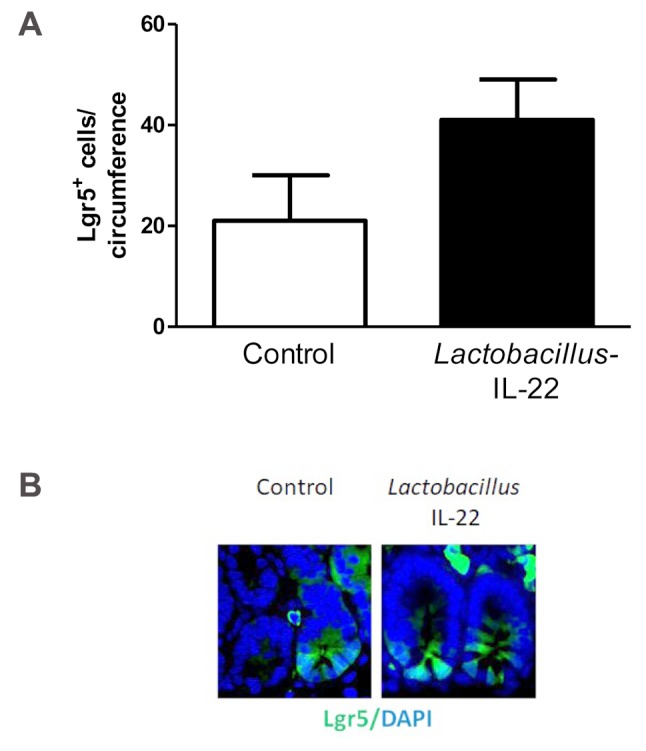
Histopathological analysis of Lgr5+ intestinal stem cells. To better understand what the biological effect of L. reuteri-mediated delivery of IL-22 was we analyzed the intestinal crypt cells (5-6). Gavage of Lactobacillus-IL-22 at 24 h after irradiation was associated with increased numbers of Lgr5+ crypt cells compared to the numbers detected in control irradiated mice (Figure 2). There were also increased numbers of intestinal Paneth cells and intestinal goblet cells, and increased intestinal villus length at 2, 5, and 7 days after TBI in the Lactobacillus-IL-22 treated mice, which demonstrates that recombinant IL-22 delivered by L. reuteri is biologically active. Control L. reuteri had no protective effect.
Figure 2. Lactobacillus reuteri-interleukin (IL-22) gavage at 24 h after total body irradiation (TBI) rescues and preserves critical leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5+) cells in ileum of Lgr5+ green fluorescent protein (GFP)+ mice at day 7. A: Groups of 10 mice received 9.25 Gy TBI, then 24 h later gavage of 100 μl of saline containing 109 Lactobacillus reuteri-IL-22. At day 7, mice were sacrificed, ileum removed and fixed, as described in prior studies (5, 6), then 20 cross-sections of ileum were scored for number of Lgr5+ GFP+ intestinal stem cells. Results are the mean ± SEM. *Significantly different at p=0.0357. B: Photographs of Lgr5+ cells in ileum from control irradiated mice and irradiated mice treated with Lactobacillus- IL-22. Original magnification, ×1000.
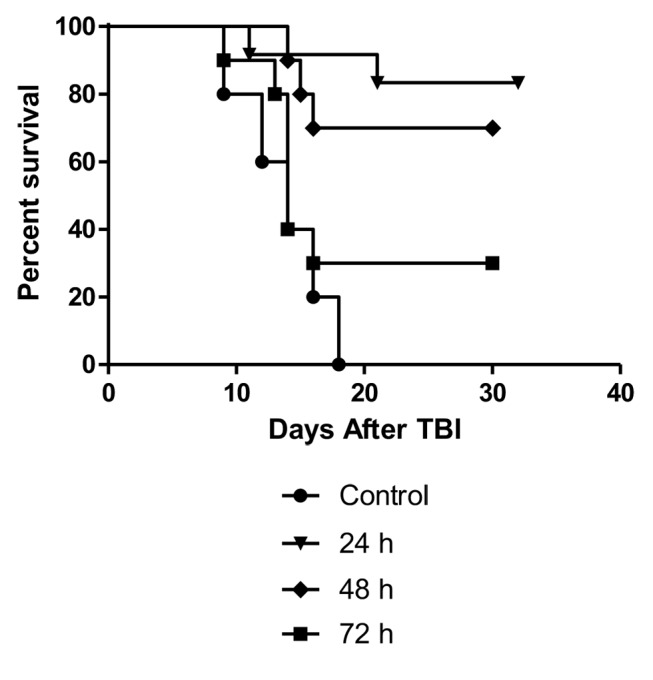
Optimization of time of delivery of Lactobacillus-IL-22. Having established that recombinant L. reuteri can rescue mice from TBI-induced death, we aimed to identify to what extent delivery of Lactobacillus-L-22 was effective at times later than 24 h after TBI. Administration of Lactobacillus-IL-22 was effective at all time points with a linear correlate tested after TBI, but was most effective at mitigation when delivered at 24 h (Figure 3). While mitigators should be given as soon after TBI exposure as possible, these data suggest that mitigation can still be achieved by Lactobacillus-IL-22 if delivered after 24 h.
Figure 3. Significant mitigation of 9.25 Gy total body irradiation (TBI) delivered to C57BL/6 female mice by gavage of Lactobacillus reuteriinterleukin- 22-green fluorescent protein fusion protein (IL-22-GFP+) (109 bacteria in 100 μl saline) administered at 24, 48, or 72 h after TBI (n=12 animals per group). Compared to control irradiated mice, mice gavaged with Lactobacillus-IL-12-GFP at 24, 48 or 72 h after irradiation had increased survival (p=0.0001, 0.0047 and 0.0448, respectively.
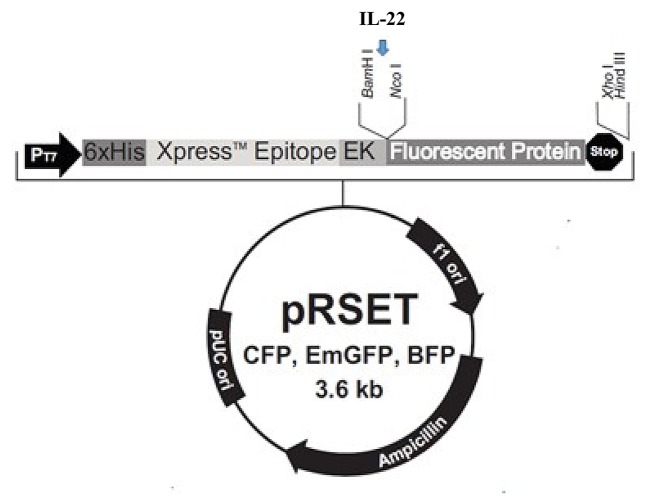
Radiomitigation by E. coli-IL-22. To determine whether our findings were specific for recombinant L. reuteri, E. coli was engineered to produce recombinant IL-22 and a fusion of IL-22 and GFP (Figure 4-Figure 6). E. coli was also transformed with the pRSET, as empty plasmid. Irradiated C57BL/6NTac mice were administered E. coli-IL-22 by gavage at 24 h post-irradiation and followed for death. Mice administered E. coli-IL-22 had increased (85%) survival after TBI compared to control mice to TBI alone (10%) (Figure 7). Thus, TBI irradiation mitigation can be achieved by derivatives of the gut symbiont L. reuteri, as well as the laboratory-adapted E. coli K12.
Figure 4. pRSET-EmGFP plasmid map used to transform Escherichia coli. The mouse interleukin (IL-22) construct was inserted into the vector between the BamHI and NcoI restriction sites (blue arrow) forming pRSET-IL-22-EmGFP.
Figure 6. Gene expression for interleukin-22 (IL-22) in clones of Escherichia coli-IL-22, as demonstrated by polymerase chain reaction. Five representative clones were positive for IL-22. N.C.: Negative control.
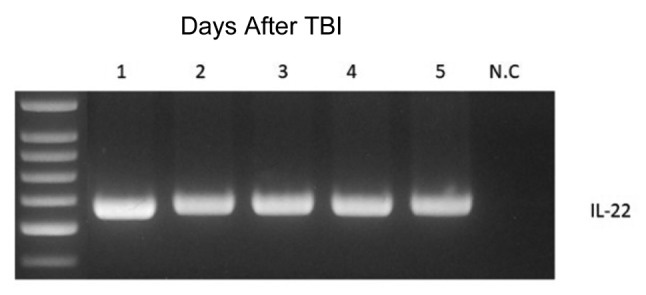
Figure 7. Gavage of Escherichia coli-interleukin-22-green fluorescent protein (IL-22-GFP+) increases survival of mice irradiated to 9.25 Gy. C57BL/6NTac mice were irradiated to 9.25 Gy total body irradiation (TBI). At 24 hours after irradiation, mice were gavaged with100 μl saline containing 109 Escherichia coli-interleukin IL-22-green fluorescent protein (IL-22-GFP+), or control Escherichia coli. Other mice were given a fecal microbiome transplant (FMT) of 109 bacteria from 30-day survivors of 9.25 Gy TBI, IL-22 protein dissolved in phosphate-buffered saline administered as an intraperitoneal injection or radiation mitigator JP4-039 dissolved in 30% (2-hydroxypropyl)-β- cyclodextrin and injected intramuscularly. Compared to control irradiated mice, administration of E.coli-IL-22 (p=0.0004), JP4-039 (p=0.0195), or IL-22 protein (p=0.0428) increased survival. FMT, and control E. coli did not increase survival (p=0.6876 and 0.8851, respectively).
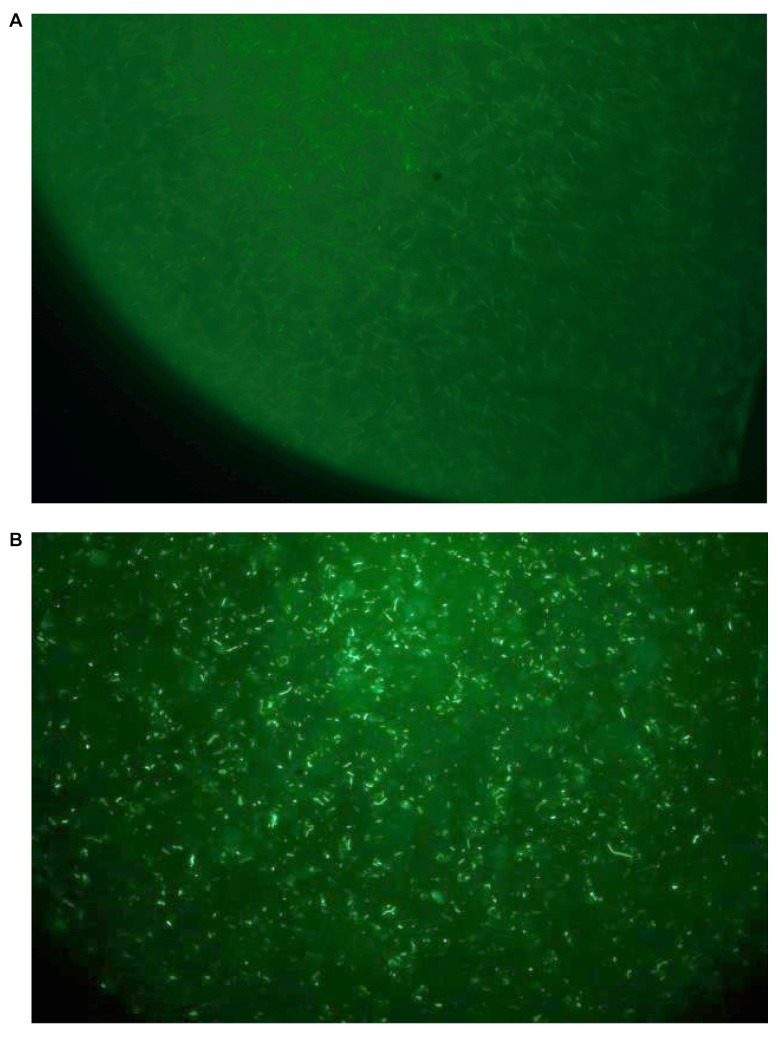
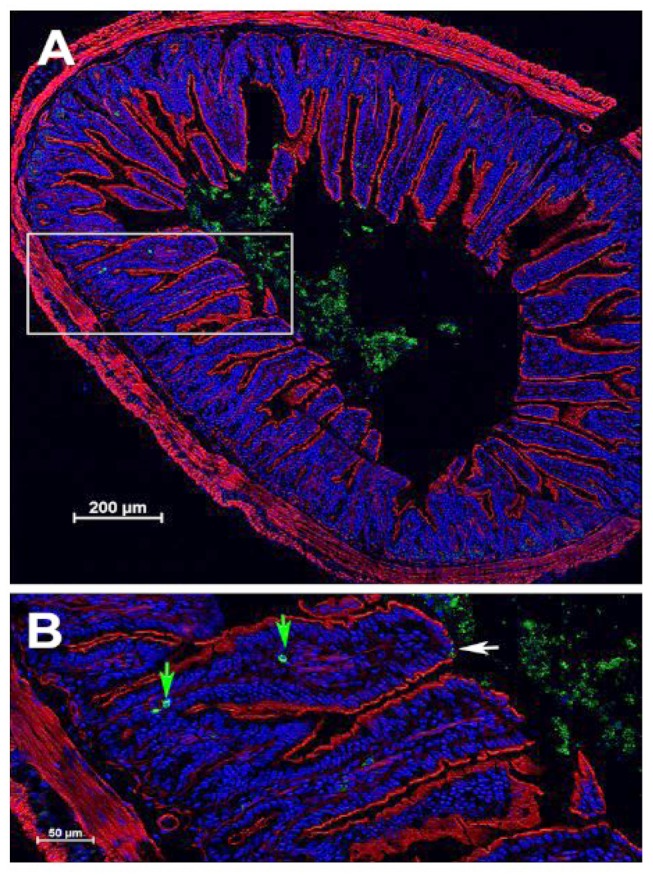
Using the fluorescent derivative of E. coli producing IL-22 (Figure 8), we demonstrated that bacteria were localized to the intestine, specifically, the jejunum, ileum, and colon. The data confirm presence of bacteria at the small intestinal villi. As shown in Figure 8 and Figure 9, E. coli-IL-22-GFP+ were observed in the jejunum and ileum 24 h after gavage and colon. Recombinant E. coli were cleared from the colon by day 5 after gavage.
Figure 8. Green fluorescent protein (GFP) expression in Escherichia coli-interleukin-22-green fluorescent protein (IL-22-GFP+) cells. A: Control Escherichia coli. B: IL-22-secreting E. coli clone. Original magnification, ×1000.
Figure 9. Histological localization of green fluorescent protein (GFP) to the ileum at 4 h after gavage of 109 Escherichia coli-interleukin-22- (IL-22)-GFP) cells at 24 h after total body irradiation (TBI) in C57BL/6J mice. A: Cross section of ileum. B: Inset from A showing localization of E. coli-IL-22-GFP+ fusion protein to the epithelial surface of the ileum. White arrow: Epithelial break (absence of red actin staining); green arrows: bacteria in villi near macrophages.
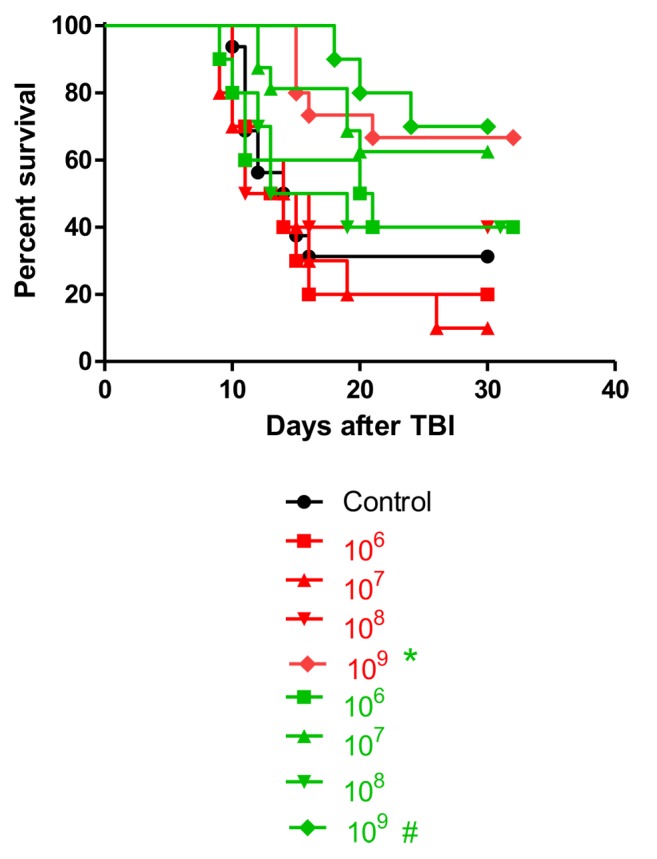
Dose-response of bacteria administered for effective radiation mitigation. To better understand the dynamics of the irradiation mitigation by recombinant L. reuteri and E. coli, we conducted a dose–response experiment. C57BL/6NTac mice were irradiated to 9.25 Gy and 24 h later gavaged with Lactobacillus-IL-22 or E. coli-IL-22-GFP. The number of bacteria gavaged ranged from 106 to 109 cells (Figure 10). Radiation mitigation was optimal for 109 Lactobacillus-IL-22. E. coli-IL-22 bacteria also provided the best mitigation when 109 cells were gavaged. The number of Lactobacillus-IL-22 bacteria was correlated with improved Lgr5+ cell numbers and survival. In contrast, there was not a linear effect of the number of E. coli-IL-22 with improved survival.
Figure 10. Survival of C57BL/6 mice gavaged at 24 h after 9.25 Gy total body irradiation (TBI) with different numbers of Lactobacillus reuteri-IL-22 (red), or Escherichia coli-IL-22 (green) (n=10 animals per group). Significant increase in survival compared with control irradiation only at *p=0.0139, and #p=0.303.
E. coli-IL-22 secretes IL-22 in vitro. Perhaps the difference in the dose response between recombinant L. reuteri and E. coli was attributed to differences in IL-22 production. To test this, we quantitated in vitro production of IL-22. Bacteria were grown in standard culture broth and both the bacteria cell pellets, and culture medium were assayed for IL-22 by ELISA assay. Figure 11A shows that E. coli-IL-22 contained IL-22 in the culture medium. Since in vitro growth in conventional medium does not promote bacteriophage-mediated lysis of L. reuteri, we observed the expected finding that limited IL-22 was present in the supernatant. Previously, it has been demonstrated that wild-type L. reuteri does not produce IL-22 (33), and, therefore, this control was excluded. Figure 11B shows that cell lysates from both E. coli and Lactobacillus-IL-22 contained detectable amounts of IL-22. As shown in Figure 11C, the lysates from E. coli-IL-22 and Lactobacillus-IL-22 bacteria contained IL-22, showing induction of IL-10, while the control bacteria lacking the plasmid encoding IL-22 did not. The medium in which the bacteria were grown was assayed by IL-22 ELISA and itself was negative. The medium in which the E. coli-IL-22 were grown was positive for IL-22 (Figure 11B), but that containing Lactobacillus-IL-22 or E. coli-GFP medium did not contain detectable IL-22.
Figure 11. Secretion of interleukin-22 (IL-22) by Escherichia coli cells transformed with plasmid pRSET containing the IL-22-green fluorescent protein transgene (pRSET-IL-22-GFP) plasmid. A: Level of IL-22 detected in culture medium collected at 48 h. B: Level of IL-22 in cell lysates from E. coli-IL-22 and Lactobacillus reuteri-IL-22. C: Biological activity of IL-22 produced by Lactobacillus-IL-22 using the induction of IL-10 in Colo205 cells. Significantly different at **p<0.01 versus E. coli-GFP; #p<0.05 versus Colo205; and ^p<0.01 versus control Lactobacillus.
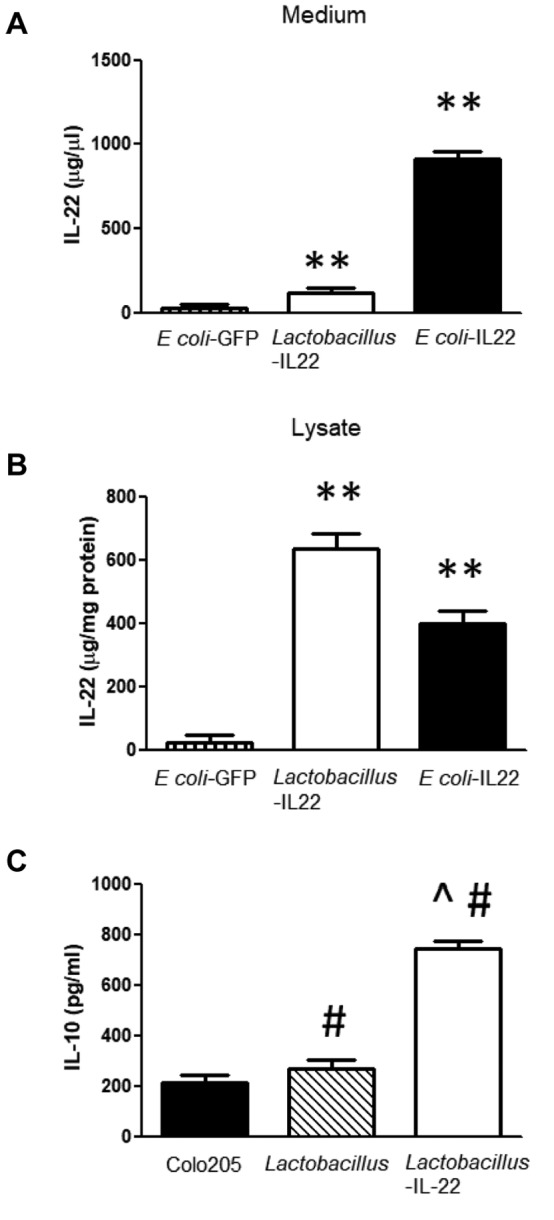
We next tested the in vitro biological activity of IL-22 produced by Lactobacillus-IL-22. As shown in Figure 11C, lysates from L. reuteri-IL-22 induced production of IL-10 in the target colon cancer cell line in vitro. This demonstrates that the IL-22 produced by recombinant L. reuteri was biologically active.
Fecal microbiome transplant does not mitigate TBI. Many microbiologists and gastroenterologists believe that fecal microbiome transplant, which delivers communities of bacteria, is superior to delivery of any one probiotic. There was no significant mitigation by delivery by gavage 109 bacteria in fecal microbiome transplant from 30-day survivors of 9.25 Gy TBI (Figure 7). Fecal microbiome transplant of 109 bacteria from pre-irradiated control mice also produced no mitigation after 9.25 Gy TBI (data not shown).
Discussion
The present study demonstrates that second-generation probiotics, specifically, L. reuteri or E. coli harboring the transgene for IL-22, can be used as radiation mitigators. Mice receiving 9.25 Gy TBI were gavaged at 24 h after irradiation with Lactobacillus-IL-22 or E. coli-IL-22. Animals that received control bacteria not harboring IL-22 showed no significant improvement in survival, while survival in those that received the second-generation probiotics producing IL-22 had increased by up to 85% at 30 days after TBI. The results establish the effectiveness of two different platforms for delivery of IL-22. L. reuteri harbor the IL-22 transgene and releases IL-22 into the intestine upon lysis of the bacteria. In contrast, E. coli secrete IL-22. Intraperitoneal delivery of IL-22 protein was not as effective a radiation mitigator as was delivery of Lactobacillus-IL-22 by gavage (17-18). These two second-generation probiotics also present opportunities for further studies of the effect of systemic compared to intraluminal delivery of IL-22.
We observed that E. coli-GFP localized to the site of intestinal lumen breakdown caused by irradiation damage. While the data do not conclusively demonstrate that the effect of second-generation probiotic bacteria directly ameliorates damage to the intestine, they do correlate with both increased numbers of intestinal stem cells at day 5 after TBI and with improved survival at day 30 after TBI.
The present results extend the scope of radiation mitigators, defined as agents that increase survival following exposure to TBI. Whether a mitigator ameliorates specific cellular, tissue, or organ related toxicity is different for each category of agents (36-39). Successful radiation mitigation is defined as improved survival only after administration of an agent after irradiation, but prior to presentation of symptoms or signs of irradiation damage. Radiation mitigators are currently confined to categories of small molecule antioxidants, including GS-nitroxides (1), Manganese superoxide dismutase-mimic molecules (36), and free radical scavenger molecules (36). Another category of mitigators includes cytokines granulocyte colony-stimulating factor, granulocyte macrophage colony-stimulating factor, epidermal growth factor, fibroblast growth factor, interleukin-10, and interleukin-11 (36,39). These all work by systemic effects on inflammation or on targets of apoptosis. Target organs for design of new radiation mitigation have focused on bone marrow and small intestine (39), organs which contain the greatest self-renewing stem cell populations which are rapidly depleted after exposure to ionizing irradiation (1). Second-generation probiotic administration would provide an advantageous alternative to these existing mitigators, and may be provided in tandem with these other mitigators.
TBI exposure leads to dose-dependent syndromes. In mice, the hematopoietic syndrome is defined as a dose of irradiation that results in bone marrow failure leading to death in 10 to 14 days following irradiation. At our animal facility C57BL/6J mice display hematopoietic syndrome at a dose of 9.0-9.5 Gy. We used a TBI dose to simulate the dose received from a fission bomb to individuals who will survive to day 30 rather than 12 Gy, which may be lethal by days 5-10. Doses above this level lead to the gastrointestinal syndrome in the C57BL/6 mouse strain doses usually above 12 Gy (39).
Intestinal irradiation damage is observed over a wide range of TBI doses, and is associated with the breakdown of the protective mucosal barrier, entry of intestinal bacteria into the villi, reduction in number of Lgr5+ intestinal stem cells, and depletion of differentiated intestinal stem cells including Paneth cells, which produce antibacterial defensins, and goblet cells that produce the protective mucin barrier lining the intestine (5,6). While recent interest has focused on discovery of mitigators for radiation intestinal damage, administration of new radiation mitigators targeted to the intestine has also been tested, but by systemic delivery, whether intravenous, intramuscular, or intraperitoneal (5, 6, 36-39).
The present report represents the first presentation of a novel approach to radiation mitigation, namely enteric (gavage) delivery of second-generation probiotics to deliver a therapeutic cytokine IL-22 locally to the intestinal mucosa. It presents evidence of successful radiation mitigation with two different platforms for enteric delivery of IL-22, with one involving cytokine secretion by Escherichia coli, the other involving the lysis of bacteria and release of cytokine in L. reuteri. It is known that bacteriophage lyse L. reuteri during gastrointestinal transit resulting in release of IL-22 (31). L. reuteri is known to target the jejunum and ilium, of the small intestine (40). Release of IL-22 at this site is beneficial since the intestinal villi at this location are known to be involved in the gastrointestinal syndrome. E. coli is usually prominent in the colon (40), but our results show that release of IL-22 by E. coli increases survival. Both second-generation probiotic platforms were successful as radiation mitigators when delivered 24 h after TBI. Furthermore, our studies with Lactobacillus-IL-22 demonstrated effective radiation mitigation when delivered by the enteric route as late as 72 h after TBI. Symptoms of the gastrointestinal syndrome usually begin at 24-72 h, so this data are exciting.
The present results alone open an opportunity for studies of the combination of enteric delivered second-generation probiotics with other systemic radiation mitigators to further ameliorate the effects of TBI including the further increase of survival. Further studies of combinations of enteric and systemic radiation mitigators will be required to develop an optimal protocol for management of individuals exposed to TBI.
Conflicts of Interest
The Authors have no conflicts of interest.
Authors’ Contributions
Xichen Zhang designed and characterized the E. coli-IL-22 bacteria. Renee Fisher performed the in vivo experiments. Wen Hou performed the IL-22 ELISA. Donna Shields grew bacteria and cells in vitro. Michael W. Epperly helped design, perform and analyze in vitro and in vivo experiments. Hong Wang performed the statistical analysis. Liang Wei aided in the staining of the intestines for Lgr5. Brian J. Leibowitz aided in the staining of the intestines for Lgr5. Jian Yu aided in the experimental design and data analysis. Jan-Peter van Pijkeren conceptualized the design of L. reuteri to deliver IL-22, and the resultant strains were constructed by Laura M. Alexander. Simon Watkins demonstrated localization of GFP+ bacteria in intestine. Peter Wipf designed and supplied JP4-039. Joel S. Greenberger aided in design of Lactobacillus-IL-22 bacteria, experimental protocols and analysis of experimental data.
Acknowledgements
Supported by grant from the NIH/NIAID U19A168021, and the UPCI-Hillman Animal Research Core Facility award P30CA047904.
Figure 5. Gene sequence of interleukin-22-green fluorescent protein (IL-22–GFP) fusion protein inserted into pRSET plasmid. IL-22–GFP fusion protein. Blue: Cytokine sequence; green: GFP sequence.
References
- 1.Rwigema J-CM, Beck B, Wang W, Doemling A, Epperly MW, Shields D, Franicola D, Dixon T, Frantz M-C, Wipf P, Tyurina Y, Kagan VE, Wang H, Greenberger JS. Two strategies for the development of mitochondrial-targeted small molecule radiation damage mitigators. Int J Radiat Oncol Biol Phys. 2011;80(3):860–868. doi: 10.1016/j.ijrobp.2011.01.059. PMID: 21493014. DOI: 10.1016/j.ijrobp.2011.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinman J, Epperly M, Hou W, Willis J, Wang H, Fisher R, Liu B, Bahar I, McCaw T, Kagan V, Bayir H, Yu J, Wipf P, Li S, Huq MS, Greenberger JS. Improved total-body irradiation survival by delivery of two radiation mitigators that target distinct cell death pathways. Radiat Res. 2018;189(1):68–83. doi: 10.1667/RR14787.1. PMID: 29140165. DOI: 10.1667/RR14787.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano J, Sato T, Ichinose S, Kajita M, Onai N, Shimizu S, Ohteki T. Intrinsic autophagy is required for the maintenance of intestinal stem cells and for irradiation-induced intestinal regeneration. Cell Rep. 2017;20(5):1050–1060. doi: 10.1016/j.celrep.2017.07.019. PMID: 28768191. DOI: 10.1016/j.celrep.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wei L, Cramer JM, Leibowitz BJ, Judge C, Epperly M, Greenberger J, Wang F, Li L, Martin MG, Lagasse E, Zhang L, Yu J. Pharmacologically blocking p53-dependent apoptosis protects intestinal stem cells and mice from radiation. Scientific Rep. 2015;5:8566. doi: 10.1038/srep08566. PMID: 25858503. DOI: 10.1038/srep08566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei L, Leibowitz BJ, Wang X, Epperly M, Greenberger J, Zhang L, Yu J. Inhibition of CDK4/6 protects against radiation-induced intestinal injury in mice. J Clin Invest. 2016;126(11):4076–4086. doi: 10.1172/JCI88410. PMID: 27701148. DOI: 10.1172/JCI88410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei L, Leibowitz BJ, Epperly M, Bi C, Li A, Steinman J, Wipf P, Zhang L, Greenberger JS, Yu J. The GS-nitroxide JP4-039 improves intestinal barrier and stem cell recovery in irradiated mice. Scientific Rep. 2018;8:2072. doi: 10.1038/s41598-018-20370-9. PMID: 29391546. DOI: 10.1038/s41598-018-20370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Huang C, Wang J, Zhou H, Lu Y, Lou L, Zhang J, Tian L, Wang X, Cao Z, Zeng Y. Dysbiosis of intestinal microbiota and decrease in Paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS One. 2017;12(4):e0176583. doi: 10.1371/journal.pone.0176583. PMID: 28441432. DOI: 10.1371/journal.pone.0176583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riba A, Olier M, Lacroix-Lamande S, Lencina C, Bacquie V, Harkat C, Gillet M, Baron M, Sommer C, Mallet V, Salvador-Cartier C, Laurent F, Theodorou V, Menard S. Paneth cell defects induce microbiota dysbiosis in mice and promote visceral hypersensitivity. Gastroenterology. 2017;153:1594–1606. doi: 10.1053/j.gastro.2017.08.044. PMID: 28865734. DOI: 10.1053/j.gastro.2017.08.044. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Tian F, Wang P, Zheng H, Zhang Y, Tian H, Zhang L, Gao X, Wang X. Gut microbiota as a modulator of Paneth cells during parenteral nutrition in mice. J Parenter Enteral Nutr. 2018;42(8):1280–1287. doi: 10.1002/jpen.1162. PMID: 29701912. DOI: 10.1002/jpen.1162. [DOI] [PubMed] [Google Scholar]
- 10.Guo X, Tang R, Yang S, Lu Y, Luo J, Liu Z. Rutin and its combination with inulin attenuate gut dysbiosis, the inflammatory status and endoplasmic reticulum stress in Paneth cells of obese mice induced by high-fat diet. Front Microbiol. 2018;9:2651. doi: 10.3389/fmicb.2018.02651. PMID: 30455677. DOI: 10.3398/fmicb.2018.02651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gassier N. Paneth cells in intestinal physiology and pathophysiology. World J Gastrointest Pathophysiol. 2017;8(4):150–160. doi: 10.4291/wjgp.v8.i4.150. PMID: 29184701. DOI: 10.4291/wjgp.v8.i4.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dayton TL, Clevers H. Beyond growth signaling: Paneth cells metabolically support ISCs. Cell Res. 2017;17:851–852. doi: 10.1038/cr.2017.59. PMID: 28429766. DOI: 10.1038/cr.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langlands AJ, Almet AA, Appleton PL, Newton IP, Osborne JM, Nathke IS. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLoS Biol. 2016;14(6):e1002491. doi: 10.1371/journal.pbio.1002491. PMID: 27348469.DOI: 10.1371/journal.pbio.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, Hooper LV. Paneth cells secrete lysosome via secretory autophagy during bacterial infection of the intestine. Science. 2017;357(6355):1047–1052. doi: 10.1126/science.aal4677. PMID: 28751470. DOI: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogala AR, Schoenborn AA, Fee BE, Cantillana VA, Joyce MJ, Gharaibeh RZ, Roy S, Fodor AA, Sartor RB, Taylor GA, Gulati AS. Environmental factors regulate Paneth cell phenotype and host susceptibility to intestinal inflammation in Irgm1-deficient mice. Dis Model Mech. 2018;11(2):ppi: dmm031070. doi: 10.1242/dmm.031070. PMID: 29361512. DOI: 10.1242/dmm.031070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu S, Tong K, Zhao Y, Balasubramanian I, Yap GS, Ferraris RP, Bonder EM, Verzi MP, Gao N. Paneth cells multipotency induced by notch activation following injury. Cell Stem Cell. 2018;23(1):46–59. doi: 10.1016/j.stem.2018.05.002. PMID: 29887318. DOI: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zha J-M, Li H-S, Lin Q, Kuo W-T, Jiang Z-H, Tsai P-Y, Ding N, Wu J, Xu S-F, Wang Y-T, Pan J, Zhou X-M, Chen K, Tao M, Odenwald MA, Tamura A, Tsukita S, Turner JR, He W-Q. Interleukin 22 expands transit-amplifying cells while depleting Lgr5+ stem cells via inhibition of Wnt and notch signaling. Cell Mol Gastroeneterol Hepatol. 2018;7(2):255–274. doi: 10.1016/j.jcmgh.2018.09.006. PMID: 30686779. DOI: 10.1016/j.jcmgh.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas LS, Targan SR, Tsuda M, Yu QT, Salumbides BC, Haritunians T, Mengesha E, McGovern DPB, Michelsen KS. The TNF family member TL1A induces IL-22 secretion in committed human TH17 cells via IL-9 induction. J Leuk Biol. 2017;101:727. doi: 10.1189/jlb.3A0316-129R. PMID: 27733581. DOI: 10.1189/jlb.3A0316-129R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K, Ruan S, Tian Y, Zhao D, Chen C, Pan B, Yan Z, Yin L, Zhu S, Xu K. IL-22 promoted CD3+ T-cell infiltration by IL-22R induced STAT3 phosphorylation in murine acute graft versus host disease target organs after allogeneic bone marrow transplantation. Int Immunopharmacol. 2016;39:383–388. doi: 10.1016/j.intimp.2016.08.012. PMID: 27551984. DOI: 10.1016/j.intimp.2016/08.012. [DOI] [PubMed] [Google Scholar]
- 20.Pan B, Xia F, Wu Y, Zhang F, Lu Z, Fu R, Shang L, Li L, Sun Z, Zeng L, Xu K. Recipient-derived IL-22 alleviates murine acute graft-versus-host disease in association with reduced activation of antigen-presenting cells. Cytokine. 2018;111:33–40. doi: 10.1016/j.cyto.2018.08.010. PMID: 30114627. DOI: 10.1016/j.cyto.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kulper JW, Tschurtschenthaler M, Savelijeva S, Bhattacharyya J, Hasier R, Bartsch K, Luzius A, Jentzsch M, Falk-Paulsen M, Stengl ST, Wetz L, Schwarzer R, Rabe B, Barchet W, Krautwald S, Hartmann G, Pasparaki M, Blumberg RS, Schreiber S, Kaser A, Rosenstiel P. ATG16L1 orchestrates Interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018;215(11):2868–2886. doi: 10.1084/jem.20171029. PMID: 30254094. DOI: 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gronke K, Hernandez PP, Zimmerman J, Klose CN, Kofoed-Branzk M, Guendel F, Witkowski M, Tizian C, Amann L, Schumacher F, Glatt H, Triantafyllopoulou A, Diefenbach A. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019;566:249–255. doi: 10.1038/s41586-019-0899-7. PMID: 30700914. DOI: 10.1038/s41586-019-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bootz F, Ziffels B, Neri D. Antibody-based targeted delivery of interleukin-22 promotes rapid clinical recovery in mice with DSS-induced colitis. Inflamm Bowel Dis. 2016;22(9):2098. doi: 10.1097/MIB.0000000000000851. PMID: 27482975. DOI: 10.107/MIB.0000000000000851. [DOI] [PubMed] [Google Scholar]
- 24.Hammer AM, Morris NL, Cannon AR, Khan OM, Gagnon RC, Movichan NV, Langeveid I van, Li X, Gao B, Choudhry MA. Interleukin-22 prevents microbial dysbiosis and promotes intestinal barrier regeneration following acute injury. Shock. 2017;48(6):657–665. doi: 10.1097/SHK.0000000000000900. PMID: 28498296. DOI: 10.1097/SHK.0000000000000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyerich K, Dimartino V, Cavani A. IL-17 and IL-22 in immunity: driving protection and pathology. Eur J Immunol. 2017;47:607–614. doi: 10.1002/eji.201646723. PMID: 28295238. DOI: 10.1002/eji.201646723. [DOI] [PubMed] [Google Scholar]
- 26.Hu M, Lin H, Yang L, Cheng Y, Zhang H. Interleukin-22 restored mitochondrial damage and impaired glucose-stimulated insulin secretion through down-regulation of uncoupling protein-2 in INS-1 cells. J Biochem. 2017;161(5):433–439. doi: 10.1093/jb/mvw084. PMID: 28069865. DOI: 10.1093/jb/mvw084. [DOI] [PubMed] [Google Scholar]
- 27.Khawar MB, Azam F, Sheikh N, Mujeeb KA. How does interleukin-22 mediate liver regeneration and prevent injury and fibrosis. J Immunol Res. 2016;2016:2148129. doi: 10.1155/2016/2148129. PMID: 28050571. DOI: 10.1155/2016/2148129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valeri M, Raffatellu M. Cytokines IL-17 and IL-22 in the host response to infection. Pathog Dis. 2016;74(9):1. doi: 10.1093/femspd/ftw111. PMID: 27915228. DOI: 10.1093/femspd/ftw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. PMID: 24452471. DOI: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 30.van Pijkeren J-P, Britton RA. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012;40:e76. doi: 10.1093/nar/gks147. PMID: 22328729. DOI: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander LM, Oh J-H, Stapleton DS, Schueler KL, Keller MP, Attie AD, van Pijkeren J-P. Exploiting prophage-mediated lysis for biotherapeutic release by Lactobacillus reuteri. Appl Environ Microbiol. 2019;85:e02335-18. doi: 10.1128/AEM.02335-18. PMID: 30683744. DOI: 10.1128/AEM.02335-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu X, Xu J-G. Development of a chromosome-plasmid balanced lethal system for Lactobacillus acidophilus with thyA gene as selective marker. Microbiol Immunol. 2000;44:551–556. doi: 10.1111/j.1348-0421.2000.tb02533.x. PMID: 10981827. DOI: 10.1111/j.1348-0421.2000.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 33.Hendrikx T, Duan Y, Wang Y, Oh J-H, Alexander LM, Huang W, Starkel P, Ho SB, Gao B, Fiehn O, Emond P, Sokol H, van Pijkeren J-P, Schnabl B. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–1515. doi: 10.1136/gutjnl-2018-317232. PMID: 30448775. DOI: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epperly MW, Wipf P, Fisher R, Franicola D, Beumer J, Li S, Brand R, Falo L, Greenberger JS. Evaluations of different formulations and routes for the delivery of the ionizing radiation mitigation GS-nitroxide (JP4-039) In Vivo. 2018;32:1009–1023. doi: 10.21873/invivo.11341. PMID: 30150422. DOI: 10.3389/fphar.2018.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyurina YY, St. Croix C, Watkins S, Epperly M, Anthonymuthu T, Kisin E, Vlassova II, Krysko O, Krysko D, Kapralov A, Dar H, Tyurin VA, Popova EN, Bolevich SB, Timashev PS, Kellum JA, Wenzel SE, Mallampalli R, Greenberger JS, Bayir H, Shvedova AA, Kagan VE. Redox (phospho)lipidomics of signaling in inflammation and programmed cell death. J Leukocyte Biol. 2019;106:57–81. doi: 10.1002/JLB.3MIR0119-004RR. PMID: 31071242. DOI: 10.1002/JLB.3MIR0119-004RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenberger JS. Radioprotection. In Vivo. 2009;23:323–336. PMID: 19414422. [PMC free article] [PubMed] [Google Scholar]
- 37.Greenberger JS, Kagan VE, Pearce L, Boriseniao G, Tyurina Y, Epperly MW. Modulation of redox signal transduction pathways in the treatment of cancer. Antioxidants Redox Signaling. 2001;3(3):347–359. doi: 10.1089/15230860152409004. PMID: 11491649. DOI: 10.1089/15230860152409004. [DOI] [PubMed] [Google Scholar]
- 38.Greenberger JS, Epperly MW. Radioprotective antioxidant gene therapy: potential mechanisms of action. Gene Ther Mol Biol. 2004;8:31–44. [Google Scholar]
- 39.Stone HB, Coleman CN, Moulder JE, Ang KK, Anscher MS, Barcellos-Hoff MH, Dynan WS, Fike JR, Grdina DJ, Greenberger JS, Hauer-Jensen M, Hill RP, Kolesnick RN, MacVittie TJ, Marks C, McBride WH, Metting N, Pellmar T, Purucker M, Robbins MEC, Schiestl RH, Seed TM, Tomaszewski J, Travis EL, Wallner PE, Wolpert M, Zaharevitz D. Models for evaluating agents intended for the prophylaxis, mitigation and treatment of radiation injuries. Report of an NCI Workshop, December 3-4, 2003. Radiat Res. 2004;162:711–718. doi: 10.1667/rr3276. PMID: 15548121. DOI: 10.1667/rr3276. [DOI] [PubMed] [Google Scholar]
- 40.Meisel M, Hinterleitner R, Pacis A, Chen L, Earley ZM, Mayassi T, Pierre JF, Ernest JD, Galipeau HJ, Thuille N, Bouziat R, Buscarlet M, Ringus DL, Wang Y, Li Y, Dinh V, Kim SM, McDonald BD, Zurenski MA, Musch MW, Furtado GC, Lira SA, Baier G, Change EB, Eren AM, Weber CR, Busque L, Godley LA, Verdu EF, Barreiro LB, Jabri B. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 2018;447:20–30. doi: 10.1038/s41586-018-0125-z. PMID: 29769727. DOI: 10.1038/s41586-018-0125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. A general system for generating unlabeled gene replacements in bacterial chromosomes. Mol Gen Genet MGG. 1996;253:217–224. doi: 10.1007/s004380050315. PMID: 9003306. DOI:10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]