Abstract
Background & Aims:
Precursors of pancreatic cancer arise in the ductal epithelium; markers exfoliated into pancreatic juice might be used to detect high-grade dysplasia (HGD) and cancer. Specific methylated DNA sequences in pancreatic tissue have been associated with adenocarcinoma. We analyzed these methylated DNA markers (MDMs) in pancreatic juice samples from patients with pancreatic ductal adenocarcinomas (PDACs) or intraductal papillary mucinous neoplasms (IPMNs) with HGD (cases), and assessed their ability to discriminate these patients from individuals without dysplasia or with IPMNs with low-grade dysplasia (IPMN-LGD) (controls).
Methods:
We obtained pancreatic juice samples from 38 patients (35 with biopsy-proven PDAC or pancreatic cystic lesions with invasive cancer and 3 with HGD) and 73 controls (32 with normal pancreas and 41 with benign disease), collected endoscopically from the duodenum after secretin administration from February 2015 through November 2016 at 3 medical centers. Samples were analyzed for the presence of 14 MDMs (in the genes NDRG4, BMP3, TBX15, C13orf18, PRKCB, CLEC11A, CD1D, ELMO1, IGF2BP1, RYR2, ADCY1, FER1L4, and EMX1, LRRC4), by quantitative allele-specific real-time target and signal amplification. We performed area under the receiver operating characteristic (AUROC) curve analyses to determine the ability of each marker, and panels of markers, to distinguish patients with of HGD and cancer from controls. MDMs were combined to form a panel for detection using recursive partition trees.
Results:
We identified a group of 3 MDMs (at C13orf18, FER1L4, and BMP3) in pancreatic juice that distinguished cases from controls with an AUROC value of 0.90 (95% CI, 0.83–0.97). Using a specificity cut-off value of 86%, this group of MDMs distinguished patients with any stage of pancreatic cancer from controls with 83% sensitivity (95% CI, 66%–93%) and identified patients with stage I or II PDAC or IPMN with HGD with 80% sensitivity (95% CI, 56%–95%).
Conclusions:
We identified a group of 3 MDMs in pancreatic juice that identifies patients with pancreatic cancer with an AUROC value of 0.90, including patients with early-stage disease or advanced precancer. These DNA methylation patterns might be included in algorithms for early detection of pancreatic cancer, especially in high-risk cohorts. Further optimization and clinical studies are needed.
Keywords: secretin, prognostic, chemistry, neoplasm
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is currently the third leading cause of cancer death in the United States and is projected to be the second leading cause by 2030 (1). The extremely low 5-year survival rate has changed little over recent decades and remains at under 10% (2). Fewer than 10% of PDACs are detected at a localized stage, and the 5-year survival in this subgroup is 31.5% overall (2). However, for small tumors (<1 cm) confined to the pancreas in pre-symptomatic patients, the 5-year survival may be as high as 75% (3). In addition to PDAC, a small subset of pancreatic cancer arises from malignant transformation of pancreatic cystic lesions, most commonly intraductal papillary mucinous neoplasms (IPMN). To improve patient outcomes, a test that accurately detects pre-symptomatic early stage PDAC or advanced precancer in pancreatic cystic lesions is urgently needed.
We use the term pancreatic cancer for PDAC and malignancy arising from intraductal papillary mucinous neoplasm (IPMN). The cell of origin for both is thought to be in the ductal epithelium which is bathed by pancreatic juice. It is biologically plausible that assay of an informative biomarker exfoliated into pancreatic juice could lead to an accurate approach to early detection of pancreatic cancer and its high grade precursor lesions. Such an approach could complement imaging for surveillance of patients at high risk for PDAC and for the diagnostic evaluation and surveillance of IPMN.
Currently there are no clinically available biomarkers for detection of PDAC from pancreatic juice. Candidate biomarkers have been tested in early phase studies by our group and others to address this need (4–6). Aberrant methylation has been described in both flat and cystic precursor lesions of PDAC (7, 8). Recent studies have explored this class of markers in pancreas cancer cell lines as well as in pancreatic juice cell-free DNA yielding promising early results (9, 10). Using a whole methylome interrogation approach followed by technical and biological validation of best candidate markers, our group at Mayo Clinic has identified novel methylated DNA markers (MDMs) that are strongly associated with PDAC in tissue (4). In a non-optimized pilot study using archival samples, we have shown that selected MDMs from this discovery effort when assayed from pancreatic juice can discriminate patients with PDAC from those with chronic pancreatitis or a normal pancreas. We also recently identified, and validated in tissue, MDMs that distinguish cases with high-grade precursor lesions (IPMN-HGD, PanIN-3) and PDAC from controls with normal pancreas or low-grade precursors (IPMN-LGD, PanIN-1, and PanIN-2) (11). Based on these findings in tissue and preliminary results in pancreatic juice, the present study was undertaken to further explore the diagnostic performance of this marker class in a multicenter referral setting. The aims of this blinded case-control study were to derive a candidate MDM panel and assess its accuracy in pancreatic juice for detection of PDAC and advanced precancer.
MATERIALS AND METHODS
Study overview and patient charecteristics
This was a multicenter case-control study enrolling patients from February 2015 to Novemeber 2016 at 3 Mayo Clinic participating sites in Rochester, MN, Scottsdale, AZ and Jacksonville, FL. The study was reviewed and approved by our institutional review board. Cases included adult patients with biopsy-proven PDAC and pancreatic cystic lesions harboring invasive cancer or high-grade dysplasia (HGD). Controls included patients with no clinical history of pancreatic disease and no imaging evidence of morphologic abnormality in the pancreas as well as those with evidence of benign pancreatic diseases. This latter group termed ‘disease controls’ included subjects with fatty pancreas, personal history of acute pancreatitis, chronic pancreatitis, pancreatic cystic lesions without any evidence of advanced neoplasia (i.e. no dysplasia or low grade dysplasia (LGD) only) and patients indeterminate for chronic pancreatitis by endoscopic ultrasound. Patients with prior history of GI malignancy, contraindication to endoscopy or intravenous secretin administration, chemotherapy or radiation for PDAC or surgical resection of target lesion prior to pancreatic juice collection and those unable to provide informed consent were excluded. All women of child-bearing age underwent a pregnancy test and were only eligible for secretin-stimulated endoscopy if negative. Clinical and demographic data, radiologic and endosonographic features, and surgical pathology for study subjects was entered by study coordinators at each center under supervision of site-specific principal investigators using pre-designed study forms. Data from the study forms were subsequently uploaded in the study database, and the final dataset was verified by expert review of electronic medical records. Pancreatic juice was collected and processed as per study protocol (Figure 1). Candidate MDMs were selected using pre-defined filtration criteria. From 1 ml of thawed pancreatic juice, DNA was extracted bisulfite converted and QuARTs (quantitative allele-specific real-time target and signal amplification) assays were performed (for details of sample collection, processing and assay methods please see Supplemental Material).
Figure 1:
Pancreatic juice collection. A. The suction catheter is passed through the endoscope. B. The catheter is advanced till the tip is visible in the duodenal lumen. C. Intravenous secretin administered leading to pancreatic juice secretion. D. The suction catheter tip is immersed in pancreatic juice that pools in the duodenal lumen and suction is applied. E. A specimen collection trap is connected to the suction catheter and for collecting the pancreatic juice. F. After collection the trap is detached, capped and transported to the research laboratory for processing and storage.
Sample size estimate and statistical analysis plan
Continuous variables were summarized as a median with corresponding 25th and 75th percentiles and comparisons between subgroups was based on the Wilcoxon rand sum test. Categorical variables were summarized as a percent of group totals and comparisons between subgroups was based on Fisher’s exact test for proportions. Individual MDMs were normalized to ACTB and stratified by the presence or absence of the protease inhibitor at collection via non-linear calibration curves. The MDMs were then scaled by their respective sample standard deviations for display purposes. For the primary analysis, the area under (AUC) the Receiver Operating Characteristic (ROC) curve was used to summarize the global discrimination of HGD/cancer from controls for each MDM individually as well as a panel of multiple MDMs. MDMs were combined to form a panel by using recursive partitioning trees (rPart). The entire modeling process was cross-validated in silico by creating 500 randomly selected training sets from the original data that were then applied to the unselected samples from the test set. All results were averaged across the 500 iterations. For the secondary analysis, the above analyses were repeated after excluding cystic lesions and IPMNs. The association clinical factors with the entire panel of MDMs were evaluated by comparing the average MDM intensity (“panel intensity”) per individual across strata using the Wilcoxon Rank Sums Test or Kruskal Wallis Test where appropriate. The association of clinical factors with the diagnostic accuracy of the prediction model was assessed by comparing the AUC across strata. This study was powered to address hypotheses of the primary analysis. With 38 cases and 73 controls, the minimum detectable AUC relative to a null AUC value of 0.7 for each individual MDM was 0.83 using a one-sided significance test of 0.05 with 80% power. With a Bonferroni corrected significance level of 0.05/12, the minimum detectable AUC was slightly higher at 0.87.
RESULTS
Patient and Lesion Characteristics
Pancreatic juice was collected from a total of 135 patients. Six patients found to have a pancreatic neoplasm that was not PDAC (3 neuroendocrine, 2 adenosquamous carcinoma and 1 B-cell lymphoma) were excluded. Of the remaining 129 subjects, 18 (5 cases and 13 controls), did not meet study quality control criteria (recovered DNA fell below pre-determined cutoff of BTACT<100) and were also excluded. Demographic characteristics of the remaining 111 patients (38 cases, 73 controls) are summarized in Table 1. Of the 73 controls, 32 had a normal pancreas (no clinical history of pancreatic disease and normal pancreas on imaging) and 41 were disease controls that included 17 patients with CP, 2 with surgically resected IPMNs harboring low or intermediate grade dysplasia, 3 with small cysts presumed IPMN without high-risk or worrisome features, 4 with fatty pancreas, and 15 with imaging findings indeterminate for CP. The 38 cases included 3 patients with IPMN-HGD and 35 with PDAC; there were 5, 15, 6 and 9 patients in stages 1-4 respectively. Cases were significantly older than controls and had a higher frequency of diabetes mellitus. There were no significant differences between the study groups with regard to sex, race, tobacco use, alcohol use, or family history of pancreatic cancer.
Table 1:
Demographic features of cases and controls
| Case (N=38) | Control (N=73) | p value | |
|---|---|---|---|
| Age | 0.005 | ||
| Median (Q1, Q3) | 67.7 (60.2, 72.4) | 60.7 (42.8, 72.2) | |
| Sex | 0.351 | ||
| Men, n (%) | 22 (57.9%) | 34 (46.6%) | |
| Race | 0.381 | ||
| Black, n (%) | 0 (0.0%) | 2 (2.7%) | |
| Caucasian | 35 (92.1%) | 66 (90.4%) | |
| Other | 1 (2.6%) | 0 (0.0%) | |
| Unknown | 2 (5.3%) | 5 (6.8%) | |
| Alcohol use | 0.570 | ||
| No | 18 (48.6%) | 27 (38.0%) | |
| Yes, but not currently | 1 (2.7%) | 3 (4.2%) | |
| Yes, currently less than 3 times per week | 10 (27.0%) | 28 (39.4%) | |
| Yes, currently 3 or more times per week | 8 (21.6%) | 13 (18.3%) | |
| Tobacco use | 0.429 | ||
| Yes, but not in the last 3 months | 12 (31.6%) | 22 (30.1%) | |
| Yes, currently or quit in the last 3 months | 5 (13.2%) | 17 (23.3%) | |
| Never | 21 (55.3%) | 34 (46.6%) | |
| History of Diabetes mellitus | 0.007 | ||
| Yes | 16 (42.1%) | 11 (15.1%) | |
| Family history of pancreatic cancer | 0.309 | ||
| Yes | 5 (13.5%) | 4 (5.7%) |
Performance of Individual and Combined Methylated DNA Markers
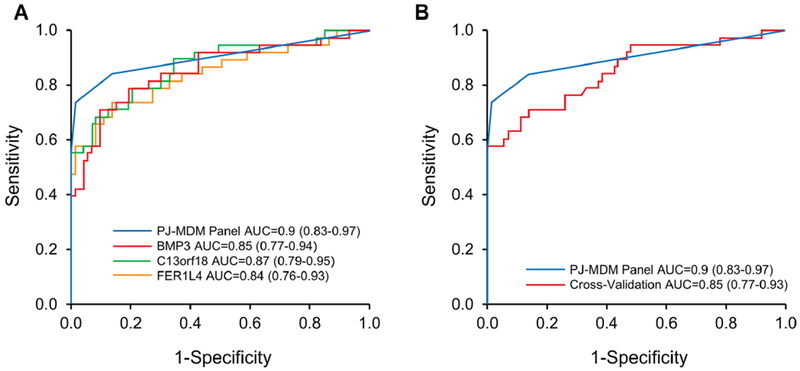
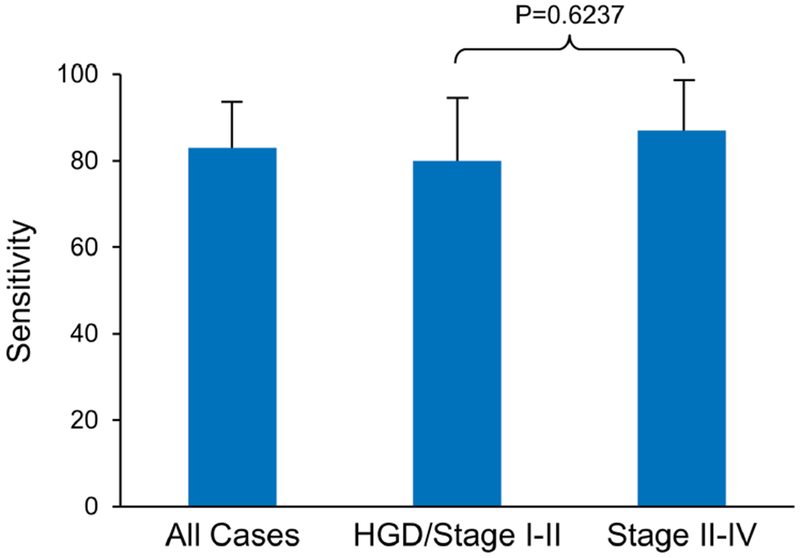
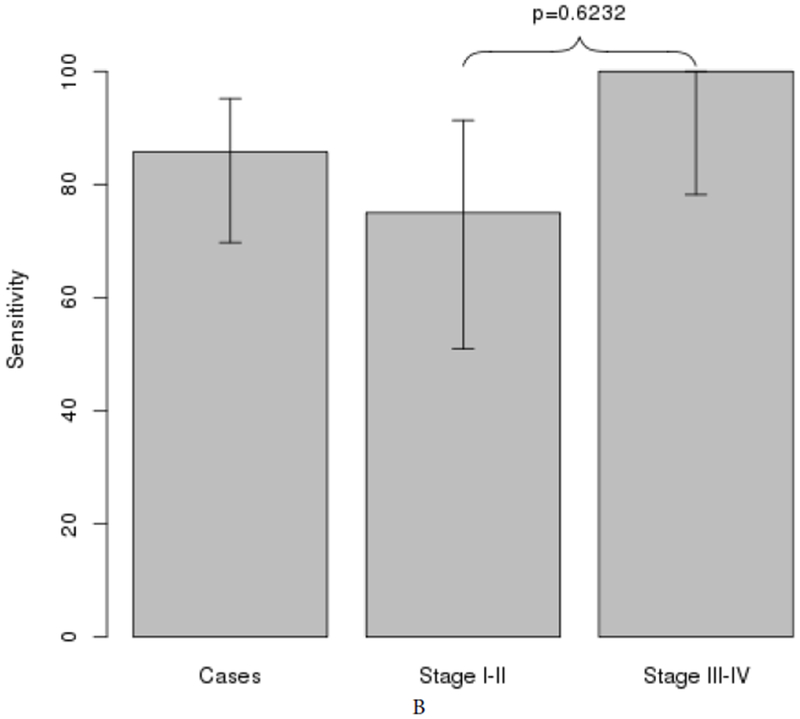
AUCs for individual MDMs ranged from 0.6 to 0.87 (Supplementary Table 1). Individual markers demonstrated high fold-change and clear discrimination between cases and both normal and disease controls (Figure 2). A panel of 3 MDMs (C13orf18, FER1L4, and BMP3), identified using rPart, achieved an AUC of 0.9 (95% CI: 0.83-0.97) (Figure 3A); the cross validated AUC of the panel was 0.85 (95% CI: 0.77-0.93) (Figure 3B). The sensitivities of the panel at specificity cut-offs of 99% and 86% were 74% (95% CI: 57-87%) and 83% (95% CI: 66-93%) for all stages of PDAC. For early stage disease (Stages 1 and 2), at the same specificity cut-offs, the sensitivities were 70% (95% CI: 46-88%) and 80% (95% CI: 56-95%), respectively (Figure 4). The panel detected all cases with IPMN-HGD. Specificity was not statistically different between normal and disease controls for any of the individual MDMs in the 3-marker panel. At the overall specificity cutoff of 86% (95% CI: 76-93%), sensitivity was 88% (95% CI: 71-97%) with normal controls and 85% (95% CI: 69-94) with disease controls (p=0.9961). Eliminating the IPMN-HGD cases (n=3) and cyst without advanced neoplasia from controls (n=5) resulted in an improved best fit AUC of 0.95 (0.91-0.99) for discriminating PDAC cases from controls (Figure 4A). In addition to the 3 MDMs in the panel described above, the tree prediction model identified methylated CD1D as an additional MDM for inclusion in the panel when cystic cases and controls are eliminated. The cross-validated AUC for this 4-marker panel however remained similar at 0.83 (0.74-0.92) (Figure 5A). The sensitivities of the panel at specificity cut-offs of 92% (95% CI: 82-97%) was 89% (95% CI: 73-97%) for all stages of PDAC combined and 80% (95% CI: 56-94%) and 100% (95% CI: 78-100%) for early and late stage disease respectively (Figure 5B). At a specificity of 90%, pancreatic juice KRAS had 68% sensitivity for detecting PDAC. The AUC for KRAS was 0.62 (95% CI: 0.51-0.73); this was significantly lower than the AUC of 0.90 (95% CI: 0.83-0.97) with MDM panel (p<0.0001). Combining KRAS with MDMs did not improve performance of MDM panel.
Figure 2:
Distributions of selected methylated DNA markers in pancreatic juice from controls (healthy controls (N-Control) n=32, disease controls (Dx-Control) n=41) and cases (pancreatic ductal adenocarcinoma and intraductal papillary mucinous neoplasms with high-grade dysplasia, n=38). (A) C13orf18, (B) FER1L4, and (C) BMP3
Figure 3:
Methylated DNA markers (MDM) assayed from pancreatic juice discriminate pancreatic cancer and IPMN with high grade dysplasia from healthy and diseased controls. ROC curves plotted (Numerical areas under the curve (AUCs) with 95% confidence shown within the figures). A. AUCs of 3-MDM panel and individual MDMs within panel. B. AUCs of MDM panel: best-fit model in comparison to the cross-validated model.
Figure 4:
Sensitivity at 85% specificity for all cases (PDAC/IPMN-HGD) across all stages combined and when stratified by early or late stage
Figure 5:
Subgroup analysis after removing cystic lesions from control group and IPMN-HGD from case group A. AUCs of 4-MDM panel, best-fit and cross-validation. B. Sensitivity and specificity of the 4-MDM panel across all stages and when stratified by early or late stage PDAC.
Covariate Analysis
The AUCs for the 3-MDM panel was not significantly affected by age, sex, or presence of diabetes (Table 2). Tumor site also did not affect detection rates; stratifying by tumor location, the 3-MDM panel detected 74% (95% CI: 54-89%), 71% (95% CI: 29-96%) and 67% (95% CI: 10-99%) of cases located in the head, body and tail respectively (p=0.99).
Table 2:
Area under the curve (with 95% confidence interval) was not influenced by baseline differences in clinical covariates, but was significantly higher in samples collected without protease inhibitor
| Covariate | No | Yes | P value |
|---|---|---|---|
| Age>65 | 0.91 (0.82-1.00) | 0.89 (0.8-0.99) | 0.7871 |
| Male | 0.93 (0.84-1.00) | 0.88 (0.78-0.98) | 0.5175 |
| Diabetes | 0.86 (0.76-0.96) | 0.96 (0.88-1.00) | 0.1264 |
| Inhibitor | 1.00 (1.00-1.00) | 0.89 (0.82-0.96) | 0.0036 |
Pancreatic juice was collected without proteinase inhibitor in 4 cases and 15 controls. In this subset, the AUC for the 3 MDM panel for distinguishing between cases and controls was 1 (95% CI 1-1) and significantly better than the AUC in samples collected with proteinase inhibitor (0.89 (0.82-0.96)), p=0.0036 (Table 2). Furthermore, technical failures were 0% (0/19) in the subset collected without inhibitor versus 14% (18/129) in those with inhibitor (p=0.1297). Median DNA yield was roughly five times higher in pancreatic juice samples without inhibitor compared to those collected with inhibitor (3943 (654-8242) ng ACTB/ml vs.779 (212-3360) ng/ml, p=0.0085). A MDM panel intensity analysis showed no significant differences in cases based on tumor size, location, prior fine needle aspiration, stent placement or inhibitor use in either cases or controls (Supplemental Figures A–E).
DISCUSSION
In this prospective study using previously discovered pancreatic juice-MDMs, we derive a panel of selected MDMs in pancreatic juice discriminates PDAC and IPMN-HGD cases from controls with normal pancreas, IPMN-LGD and non-neoplastic pancreatic diseases. This 3 MDM panel has moderately high sensitivity and specificity for detecting PDAC across all stages of the disease. Findings corroborate our previous preliminary report on the use of MDMs in pancreatic juice for detection of pancreatic neoplasia (4). As endoscopic collection of pancreatic juice is safe and less invasive than biopsy, (12) and as the majority of surveillance-detected PDAC cases are resectable and associated with significantly improved survival (13), this approach is clinically appealing, particularly for high risk subsets. Furthermore, it is biologically plausible that exfoliated cells and cell-free DNA from epithelial precursor lesions and early stage PDAC may appear in pancreatic juice before such lesions are visible on abdominal imaging or detectable using a blood-based test. Pancreatic juice testing could prove to be a helpful adjunct to imaging for early detection in high risk subsets and in patients with a pancreatic solid or cystic lesion where the fine needle cytology is indeterminate.
The majority of previous efforts for molecular analysis of pancreatic juice for PDAC detection have focused on genetic mutation testing. A recent study demonstrated a high frequency of detectable pancreatic juice KRAS mutations in patients with PDAC (73%). However KRAS mutations were also present in 19% of healthy controls and in 50% of patients without PDAC who had a known risk factor for pancreatic malignancy (14). A meta-analysis summarizing data from 16 studies on pancreatic juice KRAS reported a pooled sensitivity and specificity of 59% and 87% respectively (15) for diagnosing PDAC. The clinical application of pancreatic juice KRAS mutation testing for early detection of PDAC is limited owing to the knowledge that KRAS mutation is an early event in pancreatic oncogenesis and mutations are prevalent in early precursor lesions. Mutant TP53 although more specific for advanced neoplasia, has limited diagnostic utility given low prevalence in PDAC (16). In a recently published study, the combination of mutant TP53 and SMAD4 assayed in pancreatic juice yielded an AUC of 0.82 for discriminating PDAC from normal controls (6). Guanine nucleotide-binding protein α-stimulating (GNAS) mutations had lower sensitivity (38%) and specificity (83%) compared to mutant KRAS, TP53 and SMAD4 (6).
Earlier attempts to identify epigenetic markers of pancreatic neoplasia in pancreatic juice have targeted promoter methylation of known genes associated with PDAC (17) (18). More recently, mucin expression in pancreatic juice was regulated by DNA methylation and reported to have sensitivity and specificity of 80% and 87% respectively for PDAC (19). Our candidate marker selection was based on a prior discovery effort using whole methylome sequencing (4, 11). Our previously published pilot data used non-optimized first-pass primer designs on a limited set of candidate markers; when these were tested on archival pancreatic juice samples, we demonstrated AUCs ranging from 0.75-0.92 for discriminating PDAC from normal pancreas (4). In the present study utilizing an expanded candidate marker panel, we further establish the potential feasibility of pancreatic juice-MDMs for detection of advanced pancreatic neoplasia. Importantly, early stage PDAC and IPMN-HGD were detected as well as later stage PDAC. Moreover, the demonstrated ability of the pancreatic juice-MDM panel to discriminate PDAC from a disease control group comprising pancreatic cystic lesions with low-grade dysplasia and chronic pancreatitis, suggests a potential future role for pancreatic juice-MDM testing to help guide surveillance of patients with such conditions.
Our study has several limitations. Firstly, the addition of a protease inhibitor to pancreatic juice samples, previously a standard element of our collection protocol was recently discontinued after observations of an adverse impact on DNA recovery. Interestingly, in the small subset of samples obtained in this study without addition of inhibitor, MDM testing yielded significantly higher DNA concentration and superior discrimination. Prospective studies are underway using refined sample collection and assay techniques. Secondly, the small number of cases with HGD limits our ability to confidently report pancreatic juice-MDM performance in precancer lesions for discriminating between LGD and HGD. Pancreatic cancer precursors can be cystic or flat. We have demonstrated in pancreatic cyst fluid the ability of MDMs to distinguish between IPMN-LGD and IPMN-HGD and studies exploring the role of pancreatic juice-MDM in pancreatic cysts are currently underway. Most precursors (i.e., Pan-IN lesions) of PDAC cannot be reliably visualized using currently available imaging techniques and require longitudinal studies to evaluate test performance. Thirdly, duodenal collection of pancreatic juice has been considered a limitation for pancreatic juice biomarker studies (20). There are multiple potential confounding factors from duodenal contents including exfoliated dysplastic cells from the duodenum and more proximal GI tract, exfoliated normal epithelial or inflammatory cells within the duodenal lumen, and variably active DNase activity that could compromise target sequence integrity. In our earlier tissue discovery steps for pancreatic neoplasia and based on the large database we have on methyl-sequencing of normal and neoplastic GI tissues, aberrantly methylated sequences selected as target markers for pancreatic juice assays are not present in normal GI epithelium or in inflammatory cells. Some, but not all, of the targeted methylated DNA markers we use in this study are found in Barrett’s esophagus and esophageal cancer (21) and in gastric cancer (22), but these should be apparent in most instances on esophagogastroscopy. We have attempted to mitigate such confounding in several ways: all pooled gastric and duodenal fluids are routinely removed by aspiration prior to secreting stimulation and pancreatic juice collection to minimize contamination with non-pancreatic epithelial cells, markers targeted were selected to minimize non-specific expression in normal epithelium and inflammatory cells, and collection of pancreatic juice occurs only during clear efflux from the papilla following secretin stimulation which enhances pancreatic juice purity.
In conclusion, this study supports a potential role for pancreatic juice MDM assays in .the early detection of pancreatic cancer and its advanced precursors. Technical optimization promises to further improve accuracy of pancreatic juice testing. Prospective studies in high risk patient populations using optimized pancreatic juice methods and validating marker cut-offs for the MDM panel described here are indicated to further evaluate clinical application.
Supplementary Material
Need to Know.
Background:
Differences in methylation patterns in DNA from in pancreatic tissue have been associated with adenocarcinoma. We analyzed these methylated DNA markers (MDMs) in pancreatic juice samples from patients with pancreatic ductal adenocarcinomas (PDACs) or intraductal papillary mucinous neoplasms (IPMNs) with HGD, and assessed their ability to discriminate these patients from individuals without dysplasia or with IPMNs with low-grade dysplasia (IPMN-LGD) (controls).
Findings:
We identified a group of 3 MDMs (at C13orf18, FER1L4, and BMP3) in pancreatic juice that distinguish patients with pancreatic cancer from controls with an AUROC value of 0.90, including patients with early-stage disease or advanced precancer.
Implications for Patient Care:
Methylation patterns at 3 loci can be used as biomarkers that might be included in algorithms for early detection of pancreatic cancer, especially in high-risk cohorts.
Acknowledgments
Conflict of Interest Statement and Disclosures: Mayo Clinic has licensed intellectual property to Exact Sciences on molecular markers and sample processing techniques for multiple cancers and precancers, including pancreatic cancer. As co-inventors on licensed technologies, several co-authors (DAA, JBK, WRT, SM, DWM, and TCY) could share potential future royalties to Mayo Clinic from Exact Sciences in accordance with institutional policy and oversight. Drs. Lidgard and Alawi are Exact Sciences employees. Exact Sciences provided assay materials and partial funding but had no role in the protocol design, study execution, or analysis of data. The other authors of this manuscript have no conflicts of interest to declare.
Funding Sources:
Funding was provided by the Carol M. Gatton Foundation (to DAA). Exact Sciences (Madison WI) provided funds for sequencing and critical assay reagents. Dr. Majumder was supported by a career enhancement award funded by Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFRENCES
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–21. [DOI] [PubMed] [Google Scholar]
- 2.Howlader NNA, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975-2014, National Cancer Institute; Bethesda, MD, http://seer.cancer.gov/csr/1975_2014/, based on November 2016 SEER data submission, posted to the SEER web site, April 2017. [Google Scholar]
- 3.Kenner BJ, Chari ST, Maitra A, Srivastava S, Cleeter DF, Go VL, et al. Early Detection of Pancreatic Cancer-a Defined Future Using Lessons From Other Cancers: A White Paper. Pancreas. 2016;45(8):1073–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kisiel JB, Raimondo M, Taylor WR, Yab TC, Mahoney DW, Sun Z, et al. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(19):4473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Raimondo M, Guha S, Chen J, Diao L, Dong X, et al. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. Journal of Cancer. 2014;5(8):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J, Sadakari Y, Shindo K, Suenaga M, Brant A, Almario JAN, et al. Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut. 2017;66(9):1677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato N, Ueki T, Fukushima N, Iacobuzio-Donahue CA, Yeo CJ, Cameron JL, et al. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2002;123(1):365–72. [DOI] [PubMed] [Google Scholar]
- 8.Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2008;21(3):238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi JM, Guzzetta AA, Bailey VJ, Downing SR, Van Neste L, Chiappinelli KB, et al. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin Cancer Res. 2013;19(23):6544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksen SD, Madsen PH, Larsen AC, Johansen MB, Drewes AM, Pedersen IS, et al. Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma. Clinical epigenetics. 2016;8:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majumder S, Taylor WR, Yab TC, Cao X, Foote PH, Berger CK, et al. 596 Detection of Pancreatic High-Grade Dysplasia and Cancer using Novel Methylated DNA Markers: Discovery and Tissue Validation. Gastroenterology.150(4):S120–S1. [Google Scholar]
- 12.Hart PA, Topazian M, Raimondo M, Cruz-Monserrate Z, Fisher WE, Lesinski GB, et al. Endoscopic Pancreas Fluid Collection: Methods and Relevance for Clinical Care and Translational Science. The American journal of gastroenterology. 2016;111(9):1258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canto MI, Almario JA, Schulick RD, Yeo CJ, Klein A, Blackford A, et al. Risk of Neoplastic Progression in Individuals at High Risk for Pancreatic Cancer Undergoing Long-term Surveillance. Gastroenterology. 2018;155(3):740–51.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Li S, Li J, Wang F, Chen K, Zheng Y, et al. A meta-analysis of the diagnostic value of detecting K-ras mutation in pancreatic juice as a molecular marker for pancreatic cancer. Pancreatology. 2016;16(4):605–14. [DOI] [PubMed] [Google Scholar]
- 16.Kanda M, Sadakari Y, Borges M, Topazian M, Farrell J, Syngal S, et al. Mutant TP53 in duodenal samples of pancreatic juice from patients with pancreatic cancer or high-grade dysplasia. Clin Gastroenterol Hepatol. 2013;11(6):719–30.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan L, McFaul C, Howes N, Leslie J, Lancaster G, Wong T, et al. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology. 2005;128(7):2124–30. [DOI] [PubMed] [Google Scholar]
- 18.Matsubayashi H, Canto M, Sato N, Klein A, Abe T, Yamashita K, et al. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer research. 2006;66(2):1208–17. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama S, Kitamoto S, Higashi M, Goto Y, Hara T, Ikebe D, et al. Diagnosis of pancreatic neoplasms using a novel method of DNA methylation analysis of mucin expression in pancreatic juice. PLoS One. 2014;9(4):e93760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadakari Y, Kanda M, Maitani K, Borges M, Canto MI, Goggins M. Mutant KRAS and GNAS DNA Concentrations in Secretin-Stimulated Pancreatic Fluid Collected from the Pancreatic Duct and the Duodenal Lumen. Clinical and translational gastroenterology. 2014;5:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer PG, Taylor WR, Johnson ML, Lansing RL, Maixner KA, Yab TC, et al. Highly Discriminant Methylated DNA Markers for the Non-endoscopic Detection of Barrett’s Esophagus. The American journal of gastroenterology. 2018;113(8):1156–66. [DOI] [PubMed] [Google Scholar]
- 22.Anderson BW, Suh YS, Choi B, Lee HJ, Yab TC, Taylor WR, et al. Detection of Gastric Cancer with Novel Methylated DNA Markers: Discovery, Tissue Validation, and Pilot Testing in Plasma. Clin Cancer Res. 2018;24(22):5724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.