Abstract
Eukaryotic cells are divided into the nucleus and the cytosol, and, to enter the nucleus, proteins typically possess short signal sequences, known as nuclear localization signals (NLSs). Although NLSs have long been considered as features unique to eukaryotic proteins, we show here that similar or identical protein segments are present in ribosomal proteins from the Archaea. Specifically, the ribosomal proteins uL3, uL15, uL18, and uS12 possess NLS-type motifs that are conserved across all major branches of the Archaea, including the most ancient groups Microarchaeota and Diapherotrites, pointing to the ancient origin of NLS-type motifs in the Archaea. Furthermore, by using fluorescence microscopy, we show that the archaeal NLS-type motifs can functionally substitute eukaryotic NLSs and direct the transport of ribosomal proteins into the nuclei of human cells. Collectively, these findings illustrate that the origin of NLSs preceded the origin of the cell nucleus, suggesting that the initial function of NLSs was not related to intracellular trafficking, but possibly was to improve recognition of nucleic acids by cellular proteins. Overall, our study reveals rare evolutionary intermediates among archaeal cells that can help elucidate the sequence of events that led to the origin of the eukaryotic cell.
Keywords: ribosomal proteins, translation, origin of the nucleus, nuclear localization signals
Introduction
Our understanding of past evolutionary events mainly relies on the discovery of transitional forms. Classical examples of these transitional forms include the fossilized bird-like dinosaur Archaeopteryx lithographica and the crawling fish Tiktaalik roseae, whose bone structures illuminated many aspects of how, between the Late Jurassic and the Late Devonian, the fins of ancestral fish were transformed into the legs of terrestrial animals and subsequently into the feathered wings of birds (Chiappe 1999; Norell and Xu 2005; Daeschler et al. 2006). However, the further we delve into the past, the more we find ourselves limited in our archeological record, so that the most ancient events in history of life on Earth are known in sketchy outline or remain enigmatic.
One such enigmatic event is related to the question how the cell nucleus originated. Fossil records indicate that, between 1.7 and 2.7 billion years ago (Bya), a group of ancestral prokaryotic cells were transformed into what we now refer to as eukaryotes: they acquired a DNA-storage compartment—the nucleus—that was separated from the cytoplasm by a nuclear membrane equipped with selectively penetrable channels, the nuclear pores (Brocks et al. 1999; Rasmussen et al. 2008). To help them pass via the nuclear pores from the cytoplasm into the nucleus, cellular proteins have evolved specialized signal sequences, the nuclear localization signals (NLSs) (Dingwall et al. 1982). A typical NLS is a short and surface-exposed stretch of basic residues that is recognized by specialized receptors, karyopherins (Gorlich et al. 1994). Karyopherins transfer NLS-containing proteins across the nuclear pores (Gorlich et al. 1994). Although the nuclear–cytoplasmic trafficking machinery includes more than 100 different components, including karyopherins, nuclear pore components, and other proteins, the order in which these proteins evolved and the factors that drove the transition from prokaryotic to eukaryotic cell structures are currently unknown. In the absence of evidence of a single transitional form between prokaryotes and eukaryotes, there are at least twelve alternative hypotheses of how the system of nuclear–cytoplasmic trafficking might have emerged (Martin et al. 2015).
Although the origin of the nucleus remains a matter of debate, we have some general understanding of the preceding events based on the available fossil record and molecular clock analyses. According to the existing evidence, life originated on Earth between 3.9 and 4.3 Bya (Dodd et al. 2017); and about 2–2.7 Bya the life forms were split into two major lineages—the lineage that gave rise to modern Bacteria, and the lineage that gave rise to modern Archaea and Eukarya (Eme et al. 2018). Before Eukarya have emerged as a separate branch on the tree of life, their ancestors belong to the same branch of life as modern Archaea (Eme et al. 2018). It is therefore not surprising that presently living Archaea are studied to gain insights into the origin of eukaryotes.
Also, previous studies have shown that many aspects of the early evolution of life on Earth can be understood through analysis of the macromolecules of the ancient origin of a living cell, such as ribosomes. Because of the ancient origin of ribosomes, their structure has been used as a living molecular fossil to gain an understanding of such evolutionary enigmas as the origin of catalytic RNA (Krupkin et al. 2011; Noller 2012), the evolution of protein folding (Klein et al. 2004; Hsiao et al. 2009), the origin of the genetic code (Johnson and Wang 2010; Hartman and Smith 2014), and the reason for the stereospecific structure of proteins (Melnikov et al. 2019), among others (Bokov and Steinberg 2009; Petrov et al. 2015; Melnikov, Manakongtreecheep, et al. 2018). Additionally, eukaryotic ribosomes have been shown to be adapted to the nuclear–cytoplasmic separation of eukaryotic cells because eukaryotic ribosomal proteins, unlike their bacterial counterparts, have evolved NLSs that allow ribosomal proteins to enter the cell nucleus where they are subsequently incorporated into nascent ribosomes (Melnikov et al. 2015). This finding suggested that further investigation into ribosomes’ structures may lead to a better understanding of the origin of the nuclear–cytoplasmic trafficking (Melnikov et al. 2015).
In this study we have analyzed ribosome structures from the three domains of life to investigate origins of NLSs in eukaryotic proteins. By comparing homologous ribosomal proteins from each of the three domains of life, we found that protein segments that were described as NLSs in eukaryotic ribosomal proteins were also present in homologous proteins from Archaea. This finding indicates that at least some NLSs evolved in proteins considerably earlier than the event when cells separated into the nucleus and the cytoplasm. This finding reveals a group of rare evolutionary intermediates—NLS-type motifs in archaeal ribosomal proteins—which may shed light on the sequence of events that eventually resulted in the origin of the nuclear–cytoplasmic separation of eukaryotic cells.
Results
Archaeal Ribosomal Proteins Possess NLS-Motifs
Seeking to better understand when NLS-motifs might have emerged in ribosomal proteins, we assessed their conservation among ribosomal proteins from the three domains of life. To date, NLS-motifs have been characterized in ten ribosomal proteins from several eukaryotic species (table 1). We assessed the conservation of these NLS-motifs by using multiple sequence alignments of eukaryotic ribosomal proteins (from 482 species) and their homologs in the Bacteria (2,951 species) and Archaea (402 species) (supplementary data 1, Supplementary Material online).
Table 1.
Previously Determined NLSs in Eukaryotic Ribosomal Proteins.
| Protein | Old Name | Organism | Sequence | Reference | Tag Used to Monitor the Nucleolar Accumulation | Presence in Archaea |
|---|---|---|---|---|---|---|
| uS3 | S3 | S. cerevisiae | 3-KKRK-7 | [Koch et al. 2012] | eGFP at the C-terminus | − |
| H. sapiens | (S. cerevisiae) | |||||
| uS4 | S9 | H. sapiens | 171-GRVKRKNAKKGQ-182 | [Lindstrom 2012] | eGFP at the N- and C-termini | − |
| uS8 | S22 | S. cerevisiae | 20-GKRQVLIRP-28 | [Timmers et al. 1999] | β-galactosidase at the N-terminus | − |
| uS12 | S23 | H. sapiens | 2-GKCRGLRTARKLRSHRRDQKWHDKQYKKAHLGTALKANPF-41 | [Melnikov et al. 2015] | eGFP at the C-terminus | + |
| uL3 | L3 | S. cerevisiae | 2-SHRKYEAPRHGHLGFLPRKRA-21 | [Moreland et al. 1985] | β-galactosidase at the C-terminus | + |
| uL13 | L13a | H. sapiens | 59-RRK-61 | [Das et al. 2013] | HA-tag, position is not specified | − |
| uL15 | L29 | S. cerevisiae | 6-KTRKHRG-13 | [Underwood and Fried 1990] | β-galactosidase at the C-terminus | + |
| 23-KHRKHPG-29 | ||||||
| uL18 | L5 | H. sapiens | 21-RRRREGKTDYYARKRLV-37 | [Rosorius et al. 2000] | eGFP at the C-terminus | + |
| X. laevis | 255-KKPKKEVKKKR-265 | |||||
| uL23 | L23a, L25 | S. cerevisiae, | 11-KKAVVKG-17 | [Schaap et al. 1991] | β-galactosidase at the C-terminus | − |
| H. sapiens | 18-TNGKKALKVRT-28 | (S. cerevisiae) | ||||
| uL29 | L35 | H. sapiens | 71-KGKKYQPKDLRAKKTRALRRALTKF-95 | [Chen et al. 2008] | GST at the C-terminus | − |
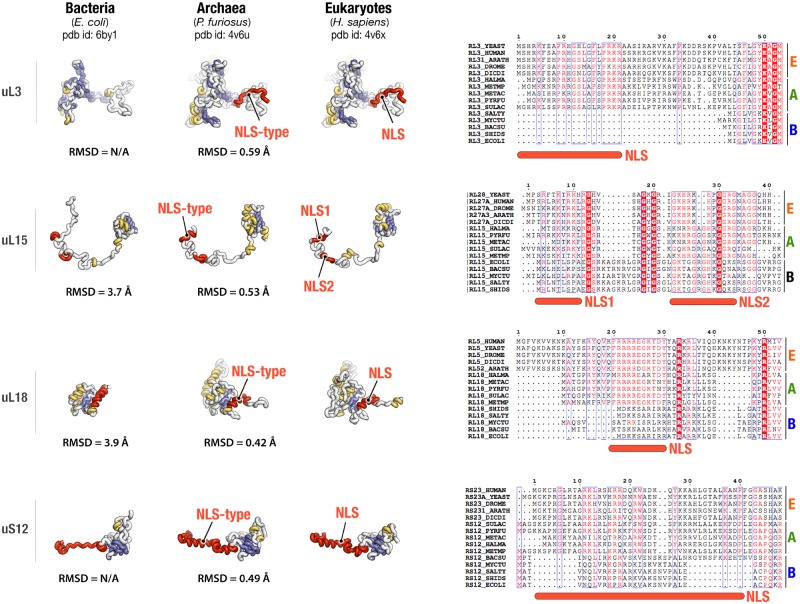
We found that six NLS-motifs—including those in ribosomal proteins uS3, uS4, uS8, uL13, uL23, and uL29—are highly conserved among the Eukarya but absent in Bacteria and Archaea (table 1). This finding was consistent with studies of other proteins, such as histones, illustrating that NLSs are found only in eukaryotic ribosomal proteins (Jenkinson and Chong 2003; Henneman et al. 2018). However, four proteins—uL3, uL15, uL18, and uS12—were found to have NLS-type motifs not only in the Eukarya but also in the Archaea (fig. 1, supplementary data 1, Supplementary Material online).
Fig. 1.
Archaeal ribosomal proteins have segments that are similar or identical to NLSs. This figure compares the crystal structures and sequences of homologous ribosomal proteins from Archaea, Bacteria, and Eukarya. NLSs in eukaryotic proteins and corresponding segments in homologous archaeal proteins are highlighted in red. This figure illustrates that NLSs of eukaryotic proteins are absent in homologous bacterial ribosomal proteins, but present in homologous archaeal ribosomal proteins. To compare structural similarity between eukaryotic NLSs and corresponding protein segments in bacterial and archaeal proteins, the RMSD values are shown for superposition of Cα-atoms of NLS segment in eukaryotic protein and a corresponding protein segment in archaeal and bacterial homologs.
The ribosomal structure has been determined for several archaeal species, including Haloarcula marismortui (Ban et al. 2000) and Pyrococcus furiosus (Armache et al. 2013), so we therefore next investigated whether archaeal and eukaryotic NLS-type motifs have conserved structures. The NLS-type motifs in proteins uL3, uL15, uL18, and uS12 turned out to have conserved secondary and tertiary structures between Eukarya and Archaea (fig. 1). Thus, our analysis revealed that NLS-motifs are not limited to eukaryotic proteins but can also be found in their archaeal homologs.
NLS-Type Motifs Are Conserved in All Groups of Archaea, Including the Most Ancient Archaeal Branches
Having found NLSs-type segments in archaeal ribosomal proteins, we next asked whether these motifs are preserved in all branches of the Archaea or just in a subset of archaeal species. To answer this, we analyzed the conservation of NLS-motifs in the archaeal proteins uL3, uL15, uL18, and uS12 (fig. 2, supplementary data 2, Supplementary Material online). To date, archaeal species have been divided into four large lineages, including DPANN (a superphylum named after Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaea lineages), TACK (a superphylum named after Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota lineages), Euryarchaeota, and Asgard superphyla (fig. 2) (Eme et al. 2018). We anticipated that NLS-type motifs would be present only in the most recently evolved and eukaryote-like branches of archaeal species, such as Asgard. Indeed, we found that sequence similarity between eukaryotic NLSs and archaeal NLS-type motifs increases with the transition from ancient archaeal branches (DPANN) to more recently emerged branches (Asgard) (supplementary data 2, Supplementary Material online). However, even in the DPANN superphylum the sequence similarity remains above 50% for each of the four NLS-type motifs (fig. 2), with some DPANN species (e.g., Mancarchaeum acidiphilum, Nanoarchaeota archaeon, and Aenigmarchaeota archaeon) having only a single substitution in their NLS-type motifs (typically lysine-to-arginine) compared with eukaryotic ribosomal proteins (supplementary data 2, Supplementary Material online). Thus, contrary to our expectations, we found that NLS-type motifs are conserved across all the archaeal branches, including the most ancient superphylum, DPANN.
Fig. 2.
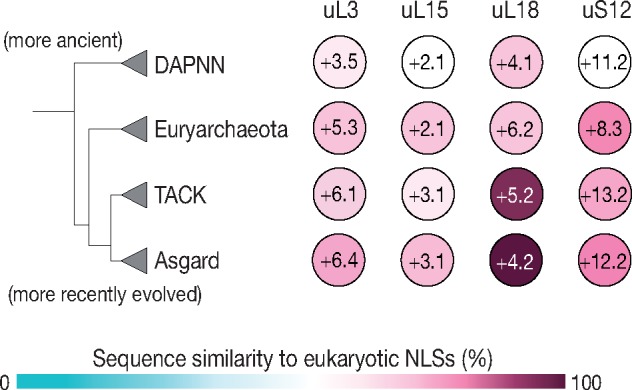
The NLS-type motifs are conserved in ribosomal proteins from all known branches of the Archaea. A phylogenetic tree of life, as according to Eme et al. (2018), showing the major branches of archaeal species and a diagram illustrating high sequence conservation among NLS-type motifs in archaeal ribosomal proteins of the major groups of archaeal species, from the most ancient branches (DAPNN superphylum) to the most recently emerged eukaryote-like branch (Asgard superphylum). The numbers in each circle (e.g., +3.5 in uL3 from DAPNN) show an average net charge of an NLS-type segment in a corresponding clade of Archaea.
Notably, many archaeal NLS-type motifs have precisely same sets of hydrophobic and basic residues as the NLS-motifs of eukaryotic ribosomal proteins, which is particularly common among species from the eukaryote-like Asgard superphylum (supplementary data 2, Supplementary Material online). For instance, the NLS of the ribosomal protein uL18 in the eukaryote Saccharomyces cerevisiae has a set of hydrophobic and basic residues identical to those in 89 archaeal species, including species from Pyrococcus (Euryarchaeota), Acidianus (TACK), and Lokiarchaeum (Asgard) genera (supplementary data 2, Supplementary Material online). Similarly, an identical set of hydrophobic and basic residues can be found in the NLS of S. cerevisiae protein uL3 and in the corresponding NLS-type sequences from 27 archaeal species (supplementary data 2, Supplementary Material online). Furthermore, in three of these archaeal species uL3 has a preserved adjacent serine residue (Ser24 in S. cerevisiae), phosphorylation of which in yeast is thought to regulate uL3 intracellular transport and ribosome biogenesis (Chi et al. 2007). Overall, this analysis revealed that even the most ancient lineages of archaeal species carry highly conserved protein segments that closely resemble eukaryotic NLSs.
NLS-Type Motifs Have Evolved Independently of Changes in Ribosomal RNA
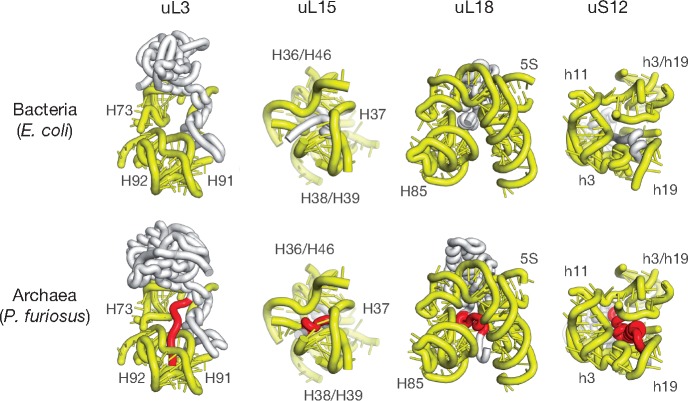
Finding NLS-type motifs in archaeal proteins was surprising as it raised the question: why do organisms that lack a cell nucleus have conserved NLS-type motifs? What advantage would be conferred by having these signal sequences in archaeal cells? Many segments in ribosomal proteins have previously been shown to have coevolved with novel segments in rRNA (Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011; Melnikov et al. 2012), so we sought to determine whether the emergence of NLSs in ribosomal proteins was somehow related to the evolution of ribosomal RNA. To answer this question, we analyzed NLSs’ contacts with rRNA (fig. 3).
Fig. 3.
NLS-type motifs have emerged independently of changes in ribosomal RNA. The panels show the interior of bacterial (E. coli, PDB ID: 6by1) and archaeal (P. furiosus, PDB ID: 4v6u) ribosomes where the NLS-type motifs of ribosomal proteins (highlighted in red) and corresponding segments in bacterial ribosomal proteins interact with rRNA. The figure illustrates that Archaea/Eukarya-specific NLS-type motifs are buried in the ribosome interior where they bind highly conserved rRNA segments. In the archaeal ribosome structure, the NLS-type motifs bind rRNA helical junctions, suggesting that the NLS-motifs may help to recognize rRNA or govern rRNA-folding during ribosome biogenesis. Overall, this figure illustrates that the rRNA structure before and after the emergence of NLS-type motifs remains conserved, illustrating that NLS-type motifs in archaeal ribosomal proteins have evolved independently of changes in rRNA.
Previously, we showed that NLSs in ribosomal proteins are buried within the ribosome interior, and explained how these signals become inactivated during ribosome biogenesis to prevent nuclear import of mature ribosomes (Melnikov et al. 2015). We also showed that, within the ribosomal structure, NLSs of ribosomal proteins interact with helical junctions, suggesting that NLSs may facilitate rRNA-folding during ribosome biogenesis (Melnikov et al. 2015). Here, comparing ribosome structures from the archaeon P. furiosus and the bacterium E. coli, we found that the NLS-type motifs of archaeal ribosomal proteins bind conserved rRNA segments that have overall conserved secondary and tertiary structure in Archaea and Bacteria (fig. 3). For instance, in archaeal ribosomes, the NLS-type segment of protein uL3 interacts with 23S rRNA helices H99 and H100, and these helices show only local conformational changes compared with the corresponding 23S rRNA segments in bacterial ribosomes (fig. 3). Furthermore, these local rearrangements may be caused in part by structural divergence of the adjacent helices H1 and H98 (fig. 3).
Similarly, the NLS-type motif of protein uS12 binds 16S rRNA segments that have conserved secondary and tertiary structures between Archaea and Bacteria, even though these rRNA segments may differ in sequence (e.g., nucleotides 23, 303, 304, 503, 504, and 906 between T. thermophilus and P. furiosus 23S rRNA, according to the E. coli numbering). Similar structural conservation with a moderate degree of sequence variability can be observed in the rRNA that contacts the NLS-type segment of uL18 (fig. 3). The NLS-type segment of uL18, together with another Archaea/Eukaryote-specific protein segment of uL18, binds 5S rRNA and mediates its interaction with 23S rRNA in the central protuberance region of the large ribosomal subunit. Compared with bacterial 5S rRNA, archaeal 5S rRNA shows sequence variations and local base-pairing change in the vicinity of the NLS-type segment, but retains its overall secondary and tertiary structure (fig. 3). Similarly, the NLS-type segment of protein uL15 binds 23S helical junctions H36/H46 and H38/H39 that have conserved tertiary structure in bacterial and archaeal 23S rRNA, even though in the vicinity of this NLS-type segment there are additional Archaea/Eukaryote-specific protein features, including extensions in proteins uL4 and eL18 that are missing in bacteria. Overall, this structural analysis indicates that NLSs emerged in ribosomal proteins independently of large structural changes in rRNA (such as expansion segments) and was accompanied with relatively minor local conformation and/or sequence variations in 5S, 16S, and 23S rRNA.
Archaeal NLS-Type Motifs Can Substitute Eukaryotic NLSs to Direct Intracellular Transport of Ribosomal Proteins in Eukaryotic Cells
Finally, we explored whether archaeal NLS-type motifs can functionally substitute NLSs of eukaryotic ribosomal proteins. NLSs in ribosomal proteins were described as proteins segments comprising ∼10–20 residues, but none of these residues were studied by point mutagenesis to elucidate their individual contribution to NLS activity (Table 1, Supplementary Material online). However, studies of NLSs in other eukaryotic proteins revealed that, commonly, NLS activity relies on patches of basic and hydrophobic residues that mediate NLS interactions with karyopherines (Rout et al. 1997; Lee et al. 2006). Are these functional NLS residues present in the archaeal NLS-type motifs?
As noted above, many archaeal species have identical sequences of the NLS-type segments in proteins uL3 and uL18 compared with NLS-segments in homologous eukaryotic proteins, meaning that many archaeal species have fully intact NLSs segments in proteins uL3 and uL18 (supplementary data 2, Supplementary Material online). In ribosomal protein uL15, Archaea have only one of the two NLS-segments that are required for the nuclear import of eukaryotic uL15, indicating that the complete NLS in eukaryotic uL15 was possibly formed only after the Archaea/Eukarya separation. However, in ribosomal protein uS12, the 3D-structure of the NLS-containing segment is conserved between Archaea and Eukarya (fig. 1), but the patches of basic and hydrophobic residues are not fully preserved in any of the archaeal lineages (supplementary data 2, Supplementary Material online).
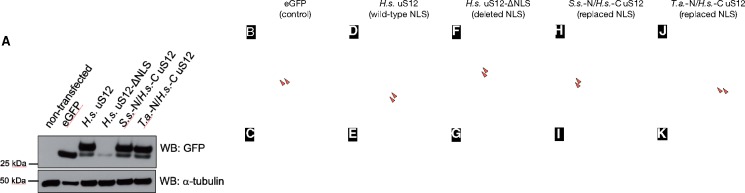
To test if this NLS-like motif in archaeal uS12 can fulfil the NLS function, we have replaced the NLS in human uS12 with the corresponding NLS-type segment of archaeal uS12 from Sulfolobus solfataricus or Thermoplasma acidophilum. We then expressed these chimeric proteins as eGFP-fusions in human cell lines and tested if these proteins can be efficiently delivered to the nucleoli, a site of ribosome biogenesis in the cell nucleus (fig. 4A–K). Notably, as overexpressed ribosomal proteins are being rapidly degraded in eukaryotic cells (Lam et al. 2007; Sung et al. 2016), we observed that all our expression constructs (including wild-type human uS12-eGFP fusion) have generated not only uS12-eGFP fusion but also some residual quantities of eGFP, which possibly represents an intermediate product of uS12-eGFP degradation (fig. 4A). We then observed that deletion of NLS-segment in uS12 resulted in loss of protein stability and loss of fluorescent signal from human nucleoli (fig. 4A, G, and K). However, uS12 chimeras with archaeal NLS-type motifs had similar expression levels and nucleoli localization as the wild-type human uS12, illustrating that the NLS-type motif of archaeal ribosomal protein uS12 is not only similar in sequence and structure to eukaryotic NLSs, but can also complements the activity of eukaryotic NLSs (fig. 4A and H–K). Overall, these observations suggest that—in some cellular proteins—protein segments, which can fulfil biological activity of eukaryotic NLSs, preexisted the origin of the nucleus and the split of archaeal and eukaryotic lineages of life.
Fig. 4.
Archaeal NLS-type motifs can functionally substitute NLS-signals of the eukaryotic ribosomal protein uS12 in human cells. (A) Western blot of HEK293T cells extracts to evaluate expression of uS12-eGFP fusions: the labels correspond to eGFP and uS12-eGFP fusions where H.s. stands for Human sapiens, S.s.—Sulfolobus solfataricus, and T.a.—Thermoplasma acidophilum. (B–K) The panels show microscopic snapshots of eGFP fluorescence (green, top panels) and fluorescence of the DNA-staining agent DAPI (blue, bottom panels) in the human cell line HEK293T. Red arrows point to the location of human cell nucleoli. Cells are expressing the following eGFP fusions: (B, C) eGFP alone (as a negative control) shows both cytoplasmic and nuclear localization; (D, E) eGFP fusion with human uS12 accumulates in nucleoli; (F, G) eGFP fusion with human uS12 lacking the NLS-containing N-terminus (residues 1–41) is no longer localized in cell nucleoli; (H, I) eGFP fusion with human uS12 in which the N-terminus (residues 1–41) is replaced by the N-terminus of uS12 from the archaeon S. solfataricus; (J, K) eGFP fusion with human uS12 in which the N-terminus (residues 1–41) is replaced by the N-terminus of uS12 from the archaeon T. acidophilum. The panels H–K illustrate that N-terminal segments of the archaeal ribosomal protein uS12 (either from S. solfataricus or T. acidophilum) are able to functionally substitute the NLS-containing N-terminal segment of human uS12, to direct uS12 accumulation in human cell nucleoli.
Discussion
In this study, we have identified a group of rare evolutionary intermediates that can shed light on the sequence of events that led to the emergence of the nuclear–cytoplasmic transport system. Our finding of NLS-type sequences in the ribosomal proteins uS12, uL3, uL15, and uL18 suggests that at least some cellular proteins evolved NLS-motifs long before the nucleus emerged, indicating that the initial function of NLSs could have been distinct from directing protein transport. Why the NLS-motifs emerged in ribosomal proteins in the Archaea—in species that have neither cell nucleus nor karyopherin proteins to recognize the NLS-type motifs—remains unclear (Koonin and Aravind 2009; Field and Rout 2019), as does what advantage might have been conferred by having these signals in the absence of nuclear–cytoplasmic transport. Furthermore, the initial function of NLS-type sequences in cells in the absence of the nuclear envelope and the nuclear–cytoplasmic trafficking system is unknown.
These questions resonate with several classical studies in evolutionary biology, such as studies on the origin of birds (Dial et al. 2006). In the origin of birds theory, there is a famous question “of what use is half a wing?” This refers to the fact that it took multiple millions of years to transform the limbs of ancestral reptiles into the wings of ancestral birds. Yet, for most of this time, the transitional forms of a wing could not support flight. Thus, the advantage of flight could not drive the positive selection of species with “germinal,” “premature” wings. Thus, what was the advantage of “half a wing”? Studies of modern birds suggest that wings could initially have emerged to facilitate gliding or accelerated climbing—two forms of motion related to flying that are preserved in the behavior of modern birds (Padian and Chiappe 1998; Zhou 2004; Dial et al. 2008).
Similar to the origin of wings, it is possible that NLSs could initially have emerged to fulfil similar, yet not identical, or incomplete functions of modern NLSs. In this regard, it is important to note that NLSs are known to fulfil three biological activities: 1) they participate in the recognition of nucleic acids, because most known NLSs reside within DNA- or RNA-binding protein domains (LaCasse and Lefebvre 1995); 2) they mediate protein transport across nuclear pores by recruiting karyopherins (Dingwall et al. 1982; Gorlich et al. 1994); and 3) karyopherin-binding to NLSs shields their electrostatic charges and prevents nonspecific interactions of nucleic acid-binding domains(Jakel et al. 2002). In other words, karyopherins that bind NLSs not only fulfil the role of transport factors but also serve as chaperones for highly charged proteins in eukaryotic cells (Jakel et al. 2002).
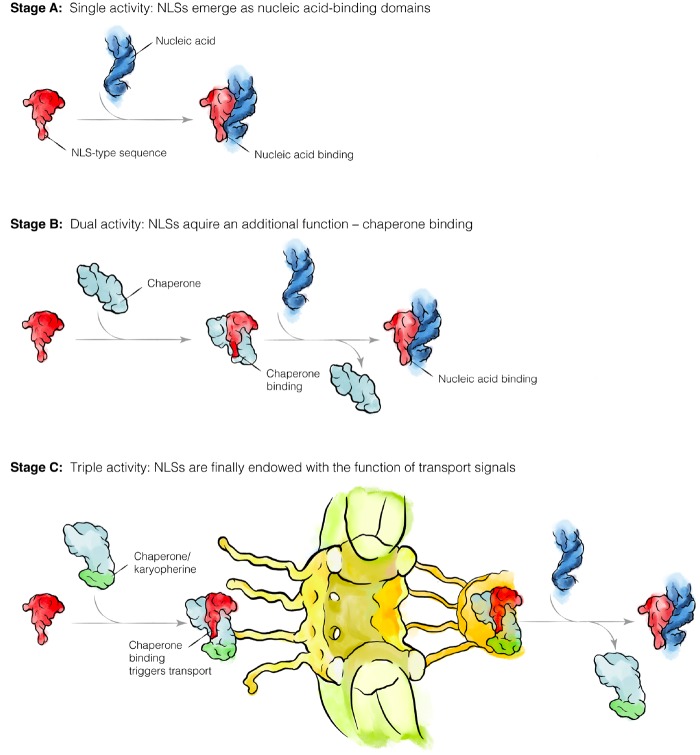
Based on our findings, it is tempting to suggest an order in which these three biological activities of NLSs may have evolved. High conservation of NLS-type motifs in archaeal ribosomal proteins and their abundant contacts with rRNA suggest that, at least in some proteins, the NLS-type sequences could have evolved as nucleic acid-binding domains that were required to increase the specificity of interactions in the increasingly complex macromolecular content of evolving cells (fig. 5A). Later, these cells could have evolved a DNA/RNA-mimicking chaperone that would recognize NLS-type sequences in the way chaperones for hydrophobic proteins recognize hydrophobic protein segments to prevent nonspecific interactions and protein aggregation (fig. 5B). Only later, when this chaperone for charged proteins had evolved the ability to bind to the nuclear pores and direct protein transport to the nucleus, did the NLSs eventually become signals of intracellular transport (fig. 5C). If this hypothetical order of events is correct, then the system of nuclear–cytoplasmic trafficking could originally have evolved as a system of chaperones for highly charged proteins, to prevent nucleic acid-binding domains from nonspecific interactions in the increasingly complex environment of ancestral eukaryotic cells.
Fig. 5.
A hypothetical order of NLS evolution in eukaryotic proteins. In the modern eukaryotic cell, NLSs fulfil three biological activities: 1) they serve as signal peptides to direct protein transport into the nucleus; 2) during this protein transport, they recruit trafficking factors (karyopherins) that shield high positive charge of nucleic acid-binding domains (which prevents nonspecific interactions of NLS-containing proteins with other molecules in a cell); and 3) typically, NLSs reside within DNA- or RNA-binding domains of proteins and, therefore, NLSs mediate specific recognition of nucleic acids. Our model suggests that NLSs may have originally emerged as nucleic acid-binding domains in a cell lacking the nuclear–cytoplasmic separation (Stage A). Later, a chaperone emerged that shielded the highly positively charged nucleic acid-binding domains (Stage B). Finally, when cells got separated into the nucleus and the cytoplasm, this chaperone turned into a karyopherin when it acquired the additional capacity to mediate the long-distance trafficking of proteins across nuclear pores (Stage C). Thus, NLSs could initially emerge to help cellular proteins to recognize nucleic acids, and only later NLSs were endowed with an additional function of trafficking signals.
Noteworthy, we showed earlier that many ribosomal proteins undergo local structural remodeling in Archaea and Eukarya compared with Bacteria. We termed the locally altered protein segments as “AE”-segments, where “AE” indicates conservation of their 3D structures in Eukarya and Archaea, but not in Bacteria. Curiously, all the four NLS-type motifs described in this study precisely overlap with the N-terminal AE-segments in ribosomal proteins uS12, uL3, uL15, and uL18 (fig. 1). Given that NLSs have been experimentally identified only in about a quarter of ribosomal proteins, it is tempting to suggest that some of the AE segments may point to the location of NLS and NLS-type motifs in eukaryotic and archaeal ribosomal proteins for which location of NLSs is currently unknown.
Our study leaves unanswered the question: why do the nonglobular extensions of ribosomal proteins have different structures in Bacteria and Archaea, even though these extensions bind invariable, universally conserved rRNA segments? Previously, it was suggested that these structural differences stemmed from an independent evolutionary origin for nonglobular segments in bacterial and eukaryotic ribosomal proteins (Klein et al. 2004). Another possible reason may be related to differences in ribosome biogenesis pathways between Bacteria (in which ribosome biogenesis largely occurs via self-assembly and with the help of protein-guided rRNA modification machinery) and Archaea/Eukarya (in which ribosome biogenesis requires archaea/eukaryote-specific biogenesis factors and RNA-guided machinery for rRNA modification) (Hage and Tollervey 2004; Blombach et al. 2011; Yip et al. 2013; de la Cruz et al. 2015).
We also cannot exclude the possibility that these differences may be related not to the problem of ribosome evolution and biogenesis but to a more general problem of specific macromolecular interactions in a cell. When life first emerged on our planet, the earliest life forms were likely made of a very limited number of molecules. Therefore, it was relatively easy for these molecules to find one specific partner among all the other molecules in a living cell. However, as cells grew in size and complexity, it is possible, even probable, that the old rules of specific interactions between cellular molecules had to be gradually redefined in order to help cellular proteins and nucleic acids to find their specific partners more easily in the complex environment of a complex cell. Given that all the NLS-type sequences identified in this study create a new RNA/protein interface, it is possible that evolving these sequences gave ribosomal proteins the advantage of more specific recognition of nucleic acid and thereby gave cells a chance to further expand diversity of their proteomes and nucleic acids without a risk of nonspecific interactions. In other words, it is possible that NLS-type sequences could originally emerge to increase specific molecular interactions and only later were repurposed into the signals of intracellular trafficking.
Materials and Methods
Comparison of Protein Sequences
The sequences of ribosomal proteins were retrieved from the Uniprot protein databank (https://www.uniprot.org; last accessed September 16, 2019) by using a script described in Melnikov, van den Elzen, et al. (2018). For species containing numerous copies of ribosomal proteins’ genes (for instance, S. cerevisiae or Arabidopsis thaliana) only one isoform (isoform A or isoform 1) was used per species. For bacterial species in which several ribosomal proteins are encoded by two genes (corresponding to Zn-coordinating and Zn-free isoforms of ribosomal proteins), we only used the genes coding for Zn-coordinating isoforms. All the retrieved sequences are listed in supplementary data 1, Supplementary Material online. To create multiple sequence alignments, we used the precompiled package of MAFFT (MAFFT-7.427) with default settings (Nakamura et al. 2018).
To assess the conservation of NLS-type motifs in archaeal proteins, we calculated consensus sequences of NLS-type motifs for each of the four ribosomal proteins (uS12, uL3, uL15, and uL18) in each of the major archaeal branches (DAPNN, Euryarchaeota, TACK, and Asgard) by using multiple sequence alignments (MAFFT with default settings) of archaeal protein sequences listed in supplementary data 1, Supplementary Material online. These consensus sequences were then compared with the corresponding consensus sequences of the eukaryotic NLSs. The NLS-consensus sequences were defined as follows: 11-RKLxxxRRxxRWxxxx-YKKRxxxxxxKxxP−40 for protein uS12 (residue numbers indicate their position in uS12 from S. cerevisiae, “x” designates any amino acid); 3-HRKxxxPRHxxxxxxPRKR−21 for protein uL3; 4-RxxKxRKxR−12 for protein uL15; and 20-FRRRRxxKxxY−30 for protein uL18. To estimate an average net charge of the NLS-type segments, the net charge was calculated for each NLS segment (as they are defined in table 1) from each archaeal species shown in supplementary data 1, Supplementary Material online by using the Prot pi Protein Tool of Zurich University of Applied Sciences (https://www.protpi.ch/Calculator/ProteinTool; last accessed September 16, 2019) with default values (pH 7.4), and then average net-charge values were shown for proteins uL3, uL15, uL18, and uS12 from each of the archaeal clades in fig. 2. Sequence similarity shown in fig. 2 was calculated according to the AMAS definitions for the hierarchical analysis of residue conservation (Livingstone and Barton 1993) using Jalview (Troshin et al. 2011) (supplementary data 2, Supplementary Material online).
Comparison of Ribosome Structures
The ribosome structures were retrieved from the protein databank (https://www.rcsb.org; last accessed September 16, 2019) and were visualized and inspected by using PyMOL Molecular Graphics System (Version 2.0 Schrödinger, LLC.). Homologous ribosomal proteins were aligned by using “align” command to superpose Cα-atoms of conserved globular domains of homologous proteins or “pair_fit” command to superpose phosphate atoms of conserved rRNA segments in bacterial and archaeal ribosomes. To measure RMSD values between NLSs in eukaryotic ribosomal proteins and corresponding segments in bacterial and archaeal ribosomal proteins, we used “pair_fit” command in the PyMOL Molecular Graphics System (Version 2.0 Schrödinger, LLC.) to align corresponding Cα-atoms as defined in the multiple sequence alignment in fig. 1.
Cloning of Ribosomal Proteins
The cDNA of human protein uS12 (NCBI Gene ID: 6228 and Uniport ID: P62266) and its N-terminally truncated mutant (deleted residues 1–41) were cloned into pEGFP-N1 vector (Promega) between KpnI and XhoI sites as described in (Melnikov et al. 2015). To create chimeras between human uS12 and homologous proteins from T. acidophilum and S. solfataricus, the genomic sequence that corresponds to residues 1–41 of human uS12 was replaced by codon-optimized sequences from T. acidophilum (NCBI Gene ID: 1455748) and S. solfataricus (NCBI Gene ID: 38467843) corresponding to residues 1–37 of uS12 from T. acidophilum (Uniprot ID: Q9HLY2) or residues 1–42 from of uS12 from S. solfataricus (Uniport ID: P39573). The codon optimization for protein expression in human cells was done by using the codon optimization algorithm from the Integrated DNA Technologies (https://www.idtdna.com; last accessed September 16, 2019). The DNA fragments corresponding to the N-termini of archaeal uS12 were purchased from GenScript (https://www.genscript.com; last accessed September 16, 2019) and cloned into the H. sapiens uS12-pEGFP-N1 construct by using In-Fusion HD Cloning Plus (Takara).
Cell Lines, Transfection, Confocal Microscopy, and Western Blot Analysis
Nucleolar accumulation of eGFP-fused human ribosomal protein uS12 and its variants were examined in the human cell line HEK293T (ATCC, CCL11268). HEK293T cells were maintained in DMEM (Gibco) supplemented with 10% FBS (Gibco). Cells were plated on poly-l-lysine-coated glass cover slips at 70–80% confluence, and transfected with the respective plasmids encoding uS12 and its variants using Lipofectamine 2000 (Invitrogen), according to the manufacturers’ protocol.
Confocal fluorescent images were obtained by a Zeiss LSM 880 Airyscan NLO/FCS confocal microscope 48 h after transfection. Prior to imaging, cell samples were fixed with 4% paraformaldehyde, permeabilized with 0.1% triton X-100 and mounted using with ProLong® Diamond Antifade Mountant (Invitrogen) with DAPI. For each eGFP-uS12 variant (and also for eGFP protein as a negative control), the snapshots were taken for six nonoverlapping fields at 63x—for better data redundancy (supplementary fig. S1, Supplementary Material online). For each genetic construct, experiments were repeated three times.
To carry out the Western blot analysis, HEK293T cells were transfected with the constructs encoding eGFP-fused human ribosomal protein uS12 and its variants using Fugene HD (Promega). The cells were lysed in radioimmunoprecipitation (RIPA) lysis buffer, supplemented with 1x Halt TM protease inhibitor cocktail (Thermo Scientific), for 15 min on ice. Lysates were cleared at 15,000 × g for 15 min at 4 °C. Protein concentrations were quantified with Quick Start TM Bradford 1x dye reagent (Bio-Rad). Samples were resolved on 10% SDS-PAGE gels. Immunoblots were incubated with anti-GFP (Cell Signaling Technology, #2956), or anti-α-tubulin (Cell Signaling Technology, #3873) overnight at 4 °C, followed by a secondary horseradish peroxidase HRP-conjugated antibody for 1 h at room temperature.
Supplementary Material
Acknowledgments
We thank members of Dieter Söll, Michael Rout, and Marat Yusupov laboratories for valuable discussions at the early stage of this project. We also thank Richard Prum and Günter Wagner (Department of Ecology and Evolutionary Biology at Yale University) for critical feedback, stimulating discussions and inspirational comments during preparation of the manuscript, Adam Bodley for editing, and Vladimir Roudko (Icahn School of Medicine at Mount Sinai), and anonymous reviewers for thorough reading, thoughtful comments, and valuable suggestions that greatly improved our work. This work was supported by grants from the European Molecular Biology Organization (ASTF 434-2014 to S.M.) and from the National Institute of General Medical Sciences (R35GM122560 to D.S.).
References
- Armache JP, Anger AM, Marquez V, Franckenberg S, Frohlich T, Villa E, Berninghausen O, Thomm M, Arnold GJ, Beckmann R.. 2013. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 41(2):1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA.. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289(5481):905–920. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M.. 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334(6062):1524–1529. [DOI] [PubMed] [Google Scholar]
- Blombach F, Brouns SJ, van der Oost J.. 2011. Assembling the archaeal ribosome: roles for translation-factor-related GTPases. Biochem Soc Trans. 39(1):45–50. [DOI] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV.. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457(7232):977–980. [DOI] [PubMed] [Google Scholar]
- Brocks JJ, Logan GA, Buick R, Summons RE.. 1999. Archean molecular fossils and the early rise of eukaryotes. Science 285(5430):1033–1036. [DOI] [PubMed] [Google Scholar]
- Chen IJ, Wang IA, Tai LR, Lin A.. 2008. The role of expansion segment of human ribosomal protein L35 in nuclear entry, translation activity, and endoplasmic reticulum docking. Biochem Cell Biol 86:271–217. [DOI] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF.. 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 104(7):2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappe P. 1999. The wing of Archaeopteryx as a primary thrust generator. Nature 399:60–62. [Google Scholar]
- Daeschler EB, Shubin NH, Jenkins FA Jr.. 2006. A Devonian tetrapod-like fish and the evolution of the tetrapod body plan. Nature 440(7085):757–763. [DOI] [PubMed] [Google Scholar]
- Das P, Basu A, Biswas A, Poddar D, Andrews J, Barik S, Komar AA, Mazumder B.. 2013. Insights into the mechanism of ribosomal incorporation of mammalian L13a protein during ribosome biogenesis. Mol Cell Biol. 33:2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K, Woolford JL Jr.. 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem. 84:93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dial KP, Jackson BE, Segre P.. 2008. A fundamental avian wing-stroke provides a new perspective on the evolution of flight. Nature 451(7181):985–989. [DOI] [PubMed] [Google Scholar]
- Dial KP, Randall RJ, Dial TR.. 2006. What use is half a wing in the ecology and evolution of birds? BioScience 56(5):437–445. [Google Scholar]
- Dingwall C, Sharnick SV, Laskey RA.. 1982. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 30(2):449–458. [DOI] [PubMed] [Google Scholar]
- Dodd MS, Papineau D, Grenne T, Slack JF, Rittner M, Pirajno F, O’Neil J, Little CTS.. 2017. Evidence for early life in Earth's oldest hydrothermal vent precipitates. Nature 543(7643):60–64. [DOI] [PubMed] [Google Scholar]
- Eme L, Spang A, Lombard J, Stairs CW, Ettema T.. 2018. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 16(2):120.. [DOI] [PubMed] [Google Scholar]
- Field MC, Rout MP.. 2019. Pore timing: the evolutionary origins of the nucleus and nuclear pore complex. F1000 Res. 8:369.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E.. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79(5):767–778. [DOI] [PubMed] [Google Scholar]
- Hage AE, Tollervey D.. 2004. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 1(1):10–15. [PubMed] [Google Scholar]
- Hartman H, Smith TF.. 2014. The evolution of the ribosome and the genetic code. Life 4(2):227–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneman B, van Emmerik C, van Ingen H, Dame RT.. 2018. Structure and function of archaeal histones. PLoS Genet. 14(9):e1007582.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C, Mohan S, Kalahar BK, Williams LD.. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 26(11):2415–2425. [DOI] [PubMed] [Google Scholar]
- Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D.. 2002. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21(3):377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson ER, Chong JP.. 2003. Initiation of archaeal DNA replication. Biochem Soc Trans. 31(Pt 3):669–673. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Wang L.. 2010. Imprints of the genetic code in the ribosome. Proc Natl Acad Sci U S A. 107(18):8298–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padian K, Chiappe LM.. 1998. The origin and early evolution of birds. Biol Rev. 73(1):1–42. [Google Scholar]
- Klein DJ, Moore PB, Steitz TA.. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 340(1):141–177. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N.. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334(6058):941–948. [DOI] [PubMed] [Google Scholar]
- Koch B, Mitterer V, Niederhauser J, Stanborough T, Murat G, Rechberger G, Bergler H, Kressler D, Pertschy B.. 2012. Yar1 protects the ribosomal protein Rps3 from aggregation. J Biol Chem. 287:21806–21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L.. 2009. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 8(13):1984–1985. [DOI] [PubMed] [Google Scholar]
- Krupkin M, Matzov D, Tang H, Metz M, Kalaora R, Belousoff MJ, Zimmerman E, Bashan A, Yonath A.. 2011. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond, B Biol Sci. 366(1580):2972–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse EC, Lefebvre YA.. 1995. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 23(10):1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS.. 2007. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 17(9):749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM.. 2006. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126(3):543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS. 2012. Elucidation of motifs in ribosomal protein S9 that mediate its nucleolar localization and binding to NPM1/nucleophosmin. PLoS One 7:e52476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone CD, Barton GJ.. 1993. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 9(6):745–756. [DOI] [PubMed] [Google Scholar]
- Norell MA, Xu X.. 2005. Feathered dinosaurs. Annu Rev Earth Planet Sci. 33(1):277–299. [Google Scholar]
- Martin WF, Garg S, Zimorski V.. 2015. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond. B Biol Sci. 370(1678):20140330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M.. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 19(6):560–567. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Yusupova G, Yusupov M.. 2015. Insights into the origin of the nuclear localization signals in conserved ribosomal proteins. Nat Commun. 6:7382.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Manakongtreecheep K, Soll D.. 2018. Revising the structural diversity of ribosomal proteins across the three domains of life. Mol Biol Evol. 35(7):1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov SV, Khabibullina NF, Mairhofer E, Vargas-Rodriguez O, Reynolds NM, Micura R, Soll D, Polikanov YS.. 2019. Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site. Nucleic Acids Res. 47(4):2089–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov SV, van den Elzen A, Stevens DL, Thoreen CC, Soll D.. 2018. Loss of protein synthesis quality control in host-restricted organisms. Proc Natl Acad Sci U S A. 115(49):E11505–E11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland RB, Nam HG, Hereford LM, Fried HM.. 1985. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc Natl Acad Sci U S A 82:6561–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yamada KD, Tomii K, Katoh K.. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34(14):2490–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF. 2012. Evolution of protein synthesis from an RNA world. Cold Spring Harb Perspect Biol. 4(4):a003681.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A. 112(50):15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N.. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331(6018):730–736. [DOI] [PubMed] [Google Scholar]
- Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR.. 2008. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455(7216):1101–1104. [DOI] [PubMed] [Google Scholar]
- Rosorius O, Fries B, Stauber RH, Hirschmann N, Bevec D, Hauber J.. 2000. Human ribosomal protein L5 contains defined nuclear localization and export signals. J Biol Chem. 275:12061–12068. [DOI] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD.. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell 89(5):715–725. [DOI] [PubMed] [Google Scholar]
- Schaap PJ, van't Riet J, Woldringh CL, Raue HA.. 1991. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J Mol Biol. 221:225–237. [DOI] [PubMed] [Google Scholar]
- Sung MK, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ.. 2016. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol Biol Cell. 27(17):2642–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers AC, Stuger R, Schaap PJ, van 't Riet J, Raue HA.. 1999. Nuclear and nucleolar localization of Saccharomyces cerevisiae ribosomal proteins S22 and S25. FEBS Lett. 452:335–340. [DOI] [PubMed] [Google Scholar]
- Troshin PV, Procter JB, Barton GJ.. 2011. Java bioinformatics analysis web services for multiple sequence alignment–JABAWS: MSA. Bioinformatics 27(14):2001–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MR, Fried HM.. 1990. Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J. 9:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip WS, Vincent NG, Baserga SJ.. 2013. Ribonucleoproteins in archaeal pre-rRNA processing and modification. Archaea 2013:614735.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. 2004. The origin and early evolution of birds: discoveries, disputes, and perspectives from fossil evidence. Naturwissenschaften 91(10):455–471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.