Abstract
Genomic data can be a powerful tool for inferring ecology, behavior, and conservation needs of highly elusive species, particularly, when other sources of information are hard to come by. Here, we focus on the Dryas monkey (Cercopithecus dryas), an endangered primate endemic to the Congo Basin with cryptic behavior and possibly <250 remaining adult individuals. Using whole-genome sequencing data, we show that the Dryas monkey represents a sister lineage to the vervets (Chlorocebus sp.) and has diverged from them ∼1.4 Ma with additional bidirectional gene flow ∼750,000–∼500,000 years ago that has likely involved the crossing of the Congo River. Together with evidence of gene flow across the Congo River in bonobos and okapis, our results suggest that the fluvial topology of the Congo River might have been more dynamic than previously recognized. Despite the presence of several homozygous loss-of-function mutations in genes associated with sperm mobility and immunity, we find high genetic diversity and low levels of inbreeding and genetic load in the studied Dryas monkey individual. This suggests that the current population carries sufficient genetic variability for long-term survival and might be larger than currently recognized. We thus provide an example of how genomic data can directly improve our understanding of highly elusive species.
Keywords: genomics, conservation, introgression, guenons, genetic diversity, inbreeding
Introduction
The Dryas monkey (Cercopithecus dryas) is a little-known species of guenon endemic to the Congo Basin, previously only recorded from a single location (fig. 1). The recent discovery of a second geographically distinct population in the upper basins of the Lomami and Lualaba rivers has led to the elevation of its conservation status from critically endangered to endangered. However little is known about its population size, distribution range, behavior, ecology, and evolutionary history (Hart et al. 2019). Based on pelage coloration, the Dryas monkey was first classified as the Central African representative of the Diana monkey (Cercopithecus diana) (Schwarz 1932). However, later examinations of the few available specimens suggested that the Dryas monkey should be classified as a unique Cercopithecus species (Colyn, Gautier-Hion, and van den Audenaerde 1991). More recently, Guschanski et al. (2013) described the mitochondrial genome sequence of the Dryas monkey type specimen, preserved at the Royal Museum for Central Africa (Tervuren, Belgium), providing the first genetic data for this species. The mitochondrial genome-based phylogeny placed the Dryas monkey within the vervet genus Chlorocebus, supporting previously suggested grouping based on similarities in feeding behavior, locomotion, and cranial morphology (Kuroda et al. 1985; Butynski 2013). The vervets consist of six recognized species: Chl. sabaeus, aethiops, tantalus, djamdjamensis, pygerythrus, and cynosuros (Zinner et al. 2013). The pygerythrus species is further divided into the subspecies Chl. p. pygerythrus and Chl. p. hilgerti, from here on referred to by their subspecies name. They are common in savannahs and riverine forests throughout sub-Saharan Africa, as well as on several Caribbean islands, where they were introduced during the colonial times (fig. 1A). However, the Dryas monkey is geographically isolated from all vervets and has numerous highly distinct morphological characteristics (Zinner et al. 2013). As vervets are characterized by a dynamic demographic history with extensive hybridization (Svardal et al. 2017), including female-mediated gene flow, transfer of mitochondrial haplotypes between species can result in discordance between the mitochondrial tree and the true species evolutionary history. Thus, the phylogenetic placement of the Dryas monkey remains uncertain.
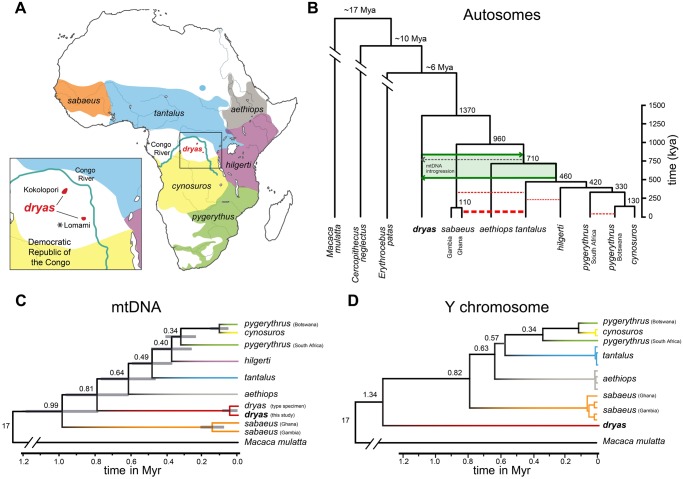
Fig. 1.
Phylogeny of the Dryas monkey and the vervets. (A) Distribution ranges of vervets. Cercopithecus dryas is currently known from only two isolated populations: The Kokolopori–Wamba and the upper basins of the Lomami and Lualaba rivers. The sequenced Dryas monkey individual was sampled from the Lomami population as indicated by the asterisk. (B) The autosomal consensus phylogeny and MCMC dating of the Dryas monkey and the vervets in comparison to two other guenon genomes, using the rhesus macaque (Macaca mulatta) as outgroup. The tree topology is supported by multiple analyses (see Materials and Methods and supplementary figs. S2 and S3, Supplementary Material online). Red dotted lines depict previously reported admixture events between the different vervet species (Svardal et al. 2017), with the line width corresponding to the relative strength of admixture. Green arrows show the best-supported timing of admixture between the Dryas monkey and the vervets. Chlorocebus sabaeus individuals from Ghana have a higher proportion of shared derived alleles with the Dryas monkey than Chl. sabaeus individuals from Gambia, which is the result of a strong secondary admixture pulse between Chl. tantalus and the Ghana Chl. sabaeus population (Svardal et al. 2017). Numbers at the nodes represent estimated mean divergence times (see supplementary fig. S3, Supplementary Material online). (C) Mitochondrial phylogeny and divergence times, provided as numbers at the nodes. Gray blocks show the 95% confidence intervals as obtained with MCMCTree. (D) UPGMA Y-chromosomal phylogeny, calibrated using the rhesus macaque Y-chromosome as outgroup. Number at the nodes represent estimated divergence times.
With only two known populations and possibly fewer than 250 adult individuals, the Dryas monkey is of significant conservation concern (Hart et al. 2019). The goal of this study was thus 2-fold: First, reconstruct the evolutionary and demographic history of the Dryas monkey and second, assess the long-term genetic viability of this species. To this end, we sequenced the genome of a male Dryas monkey at high coverage (33×), which represents the first genome-wide information available for this cryptic and little-known species.
Results and Discussion
The Dynamic Demographic History of the Dryas Monkey and the Vervets
We present the first genome sequence of the Dryas monkey, and show that it is a sister lineage to all vervets (fig. 1B andsupplementary figs. S2 and S3, Supplementary Material online). multiple phylogenomic approaches (MSC, maximum likelihood, and Bayesian) unambiguously support the same tree topology, thus contradicting the suggested placement within the Chlorocebus genus as inferred from the mitochondrial data (fig. 1C; Guschanski et al. 2013). After analyzing 3,602 gene trees from autosomal genomic windows, we obtained a multispecies coalescent tree with maximum support values (lpp = 1.0) for all nodes (supplementary fig. S2A, Supplementary Material online). Our topology is consistent with the vervet phylogeny previously reported by Warren et al. (2015). Although a majority-rule consensus tree (supplementary fig. S2B, Supplementary Material online) and network analyses at different threshold values showed some poorly resolved nodes within the vervet clade (supplementary fig. S2C, Supplementary Material online), the position of Dryas monkey as sister lineage to all vervets remains unambiguous (supplementary fig. S2, Supplementary Material online). By using an approximate likelihood MCMC method on all autosomes, we estimate that the Dryas monkey and vervets diverged ∼1.4 Ma, whereas the estimates of divergence times to both the genus Cercopithecus as exemplified by the De Brazza's monkey, Cer. neglectus (∼10.2 Ma) and the genus Erythrocebus represented by the patas monkey, E. patas (∼5.8 Ma) are much older (fig. 1B and supplementary fig. S3, Supplementary Material online). Artificial hybrid PSMC (hPSMC) analysis, which is based on mutation rate rather than calibration nodes, suggests a divergence with ongoing gene flow between the Dryas monkey and the vervets starting at ∼2 Ma with the final separation of the two lineages ∼1.5 Ma (supplementary fig. S5, Supplementary Material online). Both the MCMC and hPSMC analyses suggest that the radiation within the vervets started ca. 960 ka (fig. 1B and supplementary fig. S6, Supplementary Material online). Although our inferred divergence times are sensitive to both node calibration used in the MCMC and the mutation rate used for the hPSMC, they unambiguously support the sister relationship between the Dryas monkey and the Chlorocebus genus. Based on our analyses with only two representatives of the genus Cercopithecus (the Dryas monkey and de Brazza’s monkey), this genus appears paraphyletic. More guenon genomes will be needed to disentangle their complex evolutionary history, which may call for reconsideration of the taxonomic classification of the Dryas monkey.
The divergence times within the vervets inferred by us are more ancient than reported by Warren et al. (2015). This is likely explained by the use of a more ancient date for the divergence between the rhesus macaque and vervets as calibration node (14.5–19.6 Ma in this study vs. 12 Ma in Warren et al. 2015), the mapping against an outgroup (Macaca mulatta) in contrast to Warren et al. (2015) who used an ingroup reference (Chl. sabaeus), which likely artificially increased the similarity to Chl. sabaeus thus lowering the estimated divergence time, and the used methods (MCMC and hPSMC in this study vs. pairwise differences in Warren et al. 2015). The Y-chromosome-based phylogeny shows the same topology and similar divergence time estimates as the autosomal tree (fig. 1D). This stands in stark contrast to the inferences based on mitochondrial sequences, which show that all four Dryas monkey individuals with available mtDNA sequence data are nested within the vervet genus Chlorocebus (fig. 1C and supplementary figs. S7 and S8, Supplementary Material online).
The discrepancies in tree topologies derived from genomic regions with different inheritance modes (autosomal and mitochondrial) and the known history of introgression among the vervets (Svardal et al. 2017) suggest a possible role of gene flow in shaping the evolutionary history of the Dryas monkey. Therefore, we explored if ancient admixture events can resolve the observed phylogenetic discordance between the autosomal and mitochondrial data. We found that Chl. sabaeus individuals from Gambia share significantly fewer derived alleles with the Dryas monkey (D-statistic) than all other vervets (fig. 2A). As derived alleles should be approximately equally frequent in all species under the scenario of incomplete lineage sorting without additional gene flow (Green et al. 2010), such a pattern strongly suggests that alleles were exchanged between the Dryas monkey and the vervets excluding Chl. sabaeus that has split off from the others ∼960 ka (fig. 1B). Chlorocebus aethiops individuals share significantly fewer derived alleles with the Dryas monkey than Chl. tantalus, hilgerti, cynosuros, and pygerythrus (fig. 2A), suggesting that gene flow likely occurred for an extended period of time, at least until after the separation of Chl. aethiops from the common ancestor of the other vervets (∼710 ka, fig. 1B). As Chl. tantalus, hilgerti, cynosuros, and pygerythrus vervets share a similar amount of derived alleles with the Dryas monkey, gene flow most likely ended before this group radiated (∼460 ka, fig. 1B). The small observed differences in the D-statistic between Chl. tantalus, hilgerti, cynosuros, and pygerythrus (±12%, fig. 2A) could be the result of drift and selection due to population size differences among these vervet species or hint at ancient substructure within these species (Slatkin and Pollack 2008; Svardal et al. 2017). The inferred history of gene flow is concordant with the mitochondrial phylogeny, which suggests that the Dryas monkey mitochondrial genome was introgressed from the common ancestor of all non-sabaeus vervets ∼810 ka (fig. 1B and C). Gene flow between the common ancestor of the non-sabaeus vervets and the Dryas monkey is also supported by TreeMix analyses (supplementary fig. S9, Supplementary Material online), but we note that these inferences have to be interpreted with caution, as this model-based approach relies on accurate allele frequency estimates, which are absent for the Dryas monkey population, as it is represented by a single individual.
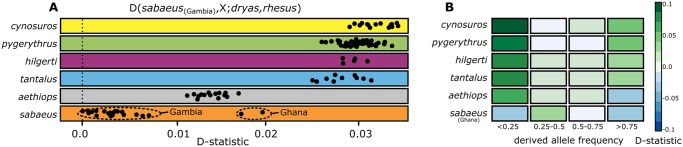
Fig. 2.
Genome-wide statistics support gene flow between the Dryas monkey and the vervets. (A) Obtained D-statistics for individual comparisons. Each dot depicts a comparison of a single vervet genome (X) with the Chl. sabaeus individual that showed the least amount of shared derived alleles with the Dryas monkey in all individual comparisons. (B) D-statistics stratified by derived allele frequency for each species, using all Chl. sabaeus Gambia individuals as ingroup.
In contrast to the Chl. sabaeus individuals from Gambia, Chl. sabaeus vervets from Ghana also carry an excess of shared derived alleles with the Dryas monkey (fig. 2A). The Ghanaian Chl. sabaeus population recently hybridized with Chl. tantalus and thus a large proportion of their genome (∼15%) is of recent Chl. tantalus ancestry (fig. 1B) (Svardal et al. 2017). As Chl. tantalus individuals carry many shared derived alleles with the Dryas monkey, this secondary introgression event likely led to the introduction of Dryas monkey alleles into the Ghanaian Chl. sabaeus, explaining the high D-statistics in this population.
Next, we obtained approximations of the directionality of gene flow using frequency-stratified D-statistics as in de Manuel et al. (2016). We found that the vervet populations carry derived alleles shared with the Dryas monkey at either low- or high frequency, but few such alleles are found at intermediate frequencies (fig. 2B). High-frequency alleles in the donor population are more likely to be introgressed and are subsequently present at low frequency in the recipient population (Kuhlwilm et al. 2016). Therefore, our observation is consistent with bidirectional gene flow between the Dryas monkey and the non-sabaeus vervets. The general direction of the gene flow appears to have been dominated by the introgression from the Dryas monkey into the non-sabaeus vervets, as we observe an overall higher proportion of low-frequency shared derived alleles in the vervets (fig. 2B). This difference is particularly pronounced in Chl. aethiops, suggesting that the gene flow was primarily from the Dryas monkey into the vervets before Chl. aethiops separated from the common ancestor of Chl. tantalus, hilgerti, cynosuros, and pygerythrus. After this split, gene flow likely became more bidirectional with increased proportion of introgression events into the Dryas monkey, as evidenced by the presence of high-frequency putatively introgressed alleles in Chl. tantalus, hilgerti, cynosuros, and pygerythrus. An alternative explanation is that the observed allele frequencies are driven by selection, as introgressed alleles might be on an average selected against Schumer et al. (2018). It is also noteworthy that all four Dryas monkey individuals for which mitochondrial sequence data are available carry the vervet-like mitochondrial haplotype (supplementary fig. S8, Supplementary Material online), which must have been introduced into the population through female-mediated gene flow. Given that the three Dryas monkeys in this study come from a different population than the Dryas monkey museum type specimen, this vervet-like haplotype is most likely fixed (or present at high frequency) in the Cercopithecus dryas population.
The putatively introgressed alleles in the Chl. sabaeus population from Ghana are found at intermediate frequency (>0.25 and <0.50) (fig. 2B), which is in agreement with the indirect introduction of these alleles through recent introgression from Chl. tantalus. As the Chl. tantalus population carries derived alleles at high- and low frequency (fig. 2B), the Ghanaian Chl. sabaeus population received a mixture of both high- and low-frequency alleles, resulting in the observed intermediate frequency.
Using approaches that are relatively insensitive to demographic processes (e.g., genetic drift and changes in effective population size), we obtained strong support for the presence of gene flow between the Dryas monkey and the vervets, but we caution that they may incorrectly infer gene flow in situations with ancestral subdivision (Slatkin and Pollack 2008). However, such ancestral population structure would have to persist over an extended period of time, encompassing multiple speciation events. Furthermore, our inferences of gene flow are supported by the discordance between the nuclear and mitochondrial phylogenies. Thus, gene flow seems to be the most parsimonious explanation.
Identifying Introgressed Regions and Inferring Their Functional Significance
Using Dxy and d statistic (Martin et al. 2015) we identified putatively introgressed regions in sliding windows of 10,000 bp for each individual. As expected under the scenario of secondary gene flow, windows containing an excess of shared derived alleles with the Dryas monkey have low genetic divergence (Dxy) to the Dryas monkey and high genetic divergence to Chl. sabaeus (supplementary fig. S10, Supplementary Material online) (de Manuel et al. 2016). Summing over all putatively introgressed windows, we roughly estimate that 0.4–0.9% in Gambia Chl. sabaeus, 1.6–2.4% in Chl. aethiops, and 2.7–4.8% of the genome in the other vervets show as signature of introgression with the Dryas monkey. However, we note that we can only identify the excess of Dryas monkey ancestry over that present in the gambian Chl. sabaeus population. These estimates are thus likely a lower bound, as the Chl. sabaeus population might carry some Dryas monkey ancestry due to secondary gene flow with other vervet species (fig. 1B;Svardal et al. 2017). We estimate that putatively introgressed haplotypes average <10,000 bp and identify putatively introgressed blocks of up to 180 kb (supplementary fig. S11, Supplementary Material online). However, we note that we cannot estimate the length of introgressed blocks below the length of 10,000 bp, as such short windows do not provide sufficient number of informative sites. The similar length of putatively introgressed haplotypes in all non-sabaeus vervet species strongly supports that gene flow occurred in their common ancestor. The putatively introgressed haplotypes into the Ghanaian and (to a lesser extent) Gambian Chl. sabaeus populations were later introduced during secondary gene flow with Chl. tantalus, as an independent recent gene flow event from the Dryas monkey into these Chl. sabaeus individuals would have resulted in significantly longer haplotypes (Liang and Nielsen 2014). However, we caution that our ability to accurately identify the haplotype lengths is low given the short length of the introgressed haplotypes and a single available Dryas monkey genome.
Interestingly, the proposed gene flow between bonobos (Pan paniscus) and non-western chimpanzees (Pan troglodytes), which have overlapping distribution range with the Dryas monkey and the vervets, respectively, possibly occurred around a similar time period, (∼500,000 years ago, de Manuel et al. 2016). It is noteworthy that the gene flow that occurred between the Dryas monkey and the vervets most likely involved the crossing of the Congo River (fig. 1), previously thought to be an impenetrable barrier for mammals (Colyn 1987; Colyn, Gautier-Hion and Verheyen 1991; Colyn and Deleporte 2004; Eriksson et al. 2004). A similar scenario of cross-Congo River gene flow was proposed for the okapi populations, and bonobos-chimpanzees (Stanton et al. 2014; de Manuel et al. 2016), suggesting that the fluvial topology of the Congo River and the geology within the Congo basin might have been more dynamic 750,000–500,000 years ago than previously recognized (Beadle 1981; Stankiewicz and de Wit 2006).
Having identified introgressed regions, we explored if they may carry functional significance. First, we used Twisst to estimate the most likely topology along short sliding windows (50 SNPs), where multiple consecutive windows showing a contrasting topology to the majority topology can indicate adaptive introgression (Martin and Van Belleghem 2017). Although this analysis supported our inferred species topology with additionally high support of introgression between the Dryas monkey and the vervets, we did not detect strong signals of long introgressed blocks, likely due to the very ancient timing of gene flow (supplementary fig. S11, Supplementary Material online). As Twisst does not distinguish between incomplete lineage sorting and introgression for very short windows, we focused on introgressed regions identified with d/Dxy statistics. We find that genes previously identified to be under strong selection within vervets (top 10% of genes under the strongest selective pressure; Svardal et al. 2017) are less often introgressed with the Dryas monkey (average introgression frequency 0.021) than genes that did not experience strong selection (bottom 10%; average allele frequency 0.029). This may indicate weak selection against introgressed genes on average, which may be deleterious in the nonhost background, a pattern also observed for Neanderthal genes in Homo sapiens and bonobo genes in the chimpanzee genetic background (Sankararaman et al. 2014; Schumer et al. 2018). However, we cannot exclude that the difference in these gene frequencies is due to stochastic events or drift. To identify genes with adaptive functions, we focused on 109 putatively introgressed genes that are fixed in all non-sabaeus vervets. Gene ontology analysis did not reveal significant enrichment for any functional category (FDR = 1). Vervets are the natural host of the simian immunodeficiency virus (SIV) and the genes under strongest selection in the vervets are related to immunity against this virus (Svardal et al. 2017). We find POU2F1, AEBP2, and PDCD6IP among fixed putatively introgressed genes in all non-sabaeus individuals. POU2F1 is a member of the pathway involved in the formation of the HIV-1 elongation complex in humans (Sturm et al. 1993), AEBP2 is a RNA polymerase II repressor (Kim et al. 2009) known to interact with viral transcription (Zhou and Rana 2002; Debaisieux et al. 2012), and PDCD6IP is involved in virus budding of the human immunodeficiency and other lentiviruses (Strack et al. 2003; von Schwedler et al. 2003). However, the SIV resistance-related genes that experienced the strongest selection in vervets (e.g., RANBP3, NFIX, CD68, FXR2, and KDM6B) do not show a signal of introgression between vervets and the Dryas monkey. Therefore, while adaptive importance can be plausible for some of the introgressed loci, it does not appear to be a strong driver for retaining particular gene classes.
Genomic View on Conservation of the Endangered Dryas Monkey
The Dryas monkey is considered the only representative of the Dryas species group (Grubb et al. 2003) and is listed as endangered in the IUCN Red List due to its small population size of ca. 250 adult individuals (Hart et al. 2019). We therefore used demographic modeling and genome-wide measures of heterozygosity and inbreeding to assess the long- and short-term population history of the Dryas monkey. Pairwise Sequential Markovian Coalescent (PSMC) analysis of the Dryas monkey genome revealed a dynamic evolutionary history, with a marked increase in effective population size starting ca. 600,000 years ago, followed by continuous decline in the last ∼150,000 years (fig. 3A). To eliminate the possibility that our PSMC inferences are driven by the increased heterozygosity due to gene flow, we removed all putatively introgressed regions and repeated the PSMC analysis, which produced a highly similar result (fig. 3A). We therefore suggest that the population size increase in the Dryas monkey and the associated likely range expansion facilitated secondary contact between the Dryas monkey and the vervets.
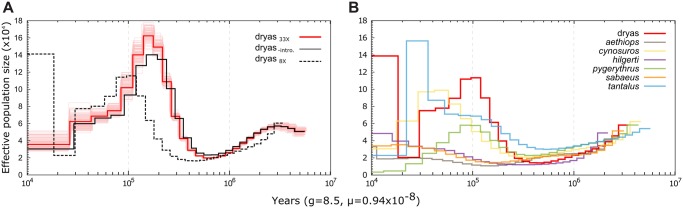
Fig. 3.
Demographic history of the Dryas monkey and the vervets. (A) PSMC analyses for the repeat-filtered Dryas monkey genome (red), after removing putatively introgressed regions (black), and after down-sampling to 8× coverage (dotted line). As the curve is strongly shifted at lower coverage, the down-sampled genome was used for between-species comparison in (B). (B) PSMC analysis of medium-coverage genomes (7.4–9.8×) for all vervets and the down-sampled Dryas monkey genome (8×).
As previously reported, low genomic coverage shifts the PSMC trajectory and makes inference less reliable, particularly, for more recent time periods (Nadachowska-Brzyska et al. 2016). Therefore, to allow for demographic comparisons to the vervets, we reran the PSMC on the Dryas monkey genome down-sampled to similar coverage as the genomic data available for the vervets. We indeed find a strong shift of the PSMC trajectory as a result of reduced coverage to more recent times and lower effective population size (fig. 3A). Nonetheless, bias related to coverage affect the different vervet genomes in a similar manner (Nadachowska-Brzyska et al. 2016), thus relative comparison between the species suggest that 100,000–300,000 years ago the Dryas monkey population was the largest among all vervets (fig. 3B).
The genetic diversity of the Dryas monkey (measured as between chromosome–pair differences), a proxy for the adaptive potential of a population (Lande and Shannon 1996), is high compared with that of the much more abundant vervets (fig. 4A). The Dryas monkey individual also shows no signs of excessive recent inbreeding, which would manifest itself in a high fraction of the genome contained in long tracts of homozygosity (>2.5 Mb) (fig. 4B).
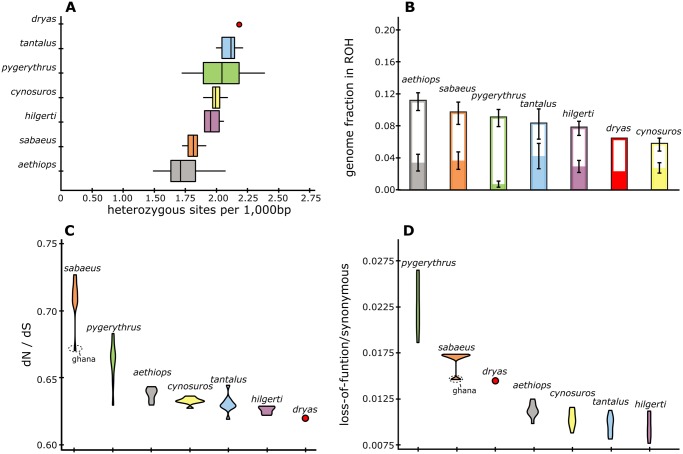
Fig. 4.
Genome-wide measures of genetic diversity and genetic load in the Dryas monkey. (A) Average number of heterozygous sites per 1,000 base pairs. (B) Fraction of the genome in runs of homozygosity (ROH) >100 kb (open bars) and fraction of the genome in ROH >2.5 Mb (solid bars). (C) Ratio of missense to synonymous mutations, excluding all missense mutations within genes containing one or more LOF mutations. (D) Ratio of LOF to synonymous mutations, counting genes with more than one LOF mutation only once.
To estimate genetic load, we identified all genes in the Dryas monkey genome containing one or more loss-of-function (LoF) mutations and identified all missense mutations in genes other than those already containing LoF mutation(s) (as such mutations likely behave neutrally). We find multiple genes in the Dryas monkey containing a homozygous LoF mutation associated with a disease phenotype in humans (n = 27), including in SEPT12, associated with reduced sperm mobility (Kuo et al. 2015) and in SLAMF9, associated with reduced immunity to tapeworm infections (Cárdenas et al. 2014). However, genome-wide measures of genetic load, measured as the ratio between LoF or missense and synonymous mutations (dN/dS), do not show an increased genomic burden of deleterious mutations in the Dryas monkey compared with the much more abundant and widely distributed vervets (fig. 4C and D). The demographic history and the genome-wide measures of genetic diversity and genetic load of the Dryas monkey thus suggest that the population of this endangered primate might be larger than currently recognized and has good chances for long-term survival, if appropriate conservation measures are implemented.
Materials and Methods
Tissue Sample Collection and Whole-Genome Sequencing
Three Dryas monkey tissue samples were obtained from individuals from the Lomami National Park buffer zone in the Democratic Republic of Congo (supplementary table S1, Supplementary Material online and fig. 1A) and exported to Florida Atlantic University, United States under approved country-specific permits, where DNA was extracted using the Qiaqen DNeasy Blood & Tissue kit, following the manufactures protocol. For library preparation and sequencing the DNA extracts of two individuals were sent to Uppsala University, Sweden. The Illumina TruSeq DNA PCR-free kit was used for standard library preparation and the sample showing the highest DNA extract concentration (Field ID JH024, FAU Primatology Lab ID: DRD14M418) was sequenced on one lane of the Illumina HiseqX platform (2× 150 bp). In addition, we obtained previously published FASTQ data for all currently recognized vervet species from mainland Africa (Warren et al. 2015; Svardal et al. 2017) and divided them into two subsets: 23 Chl. sabaeus, 16 Chl. aethiops, 11 Chl. tantalus, 6 Chl. hilgerti, 16 Chl. cynosuros, and 51 Chl. pygerythrus individuals sequenced on the Hiseq2000 platform at 1.9–6.8× coverage (the low-coverage data set), and one individual of each vervet species sequenced on the Genome-Analyzer II platform at 7.4–9.8× coverage (the medium-coverage data set). We also obtained publicly available de novo assembled genomes of two additional guenon species, the patas monkey (Erythrocebus patas, GenBank: GCA_004027335.1) and the De Brazza’s monkey (Cercopithecus neglectus, GenBank: GCA_004027615.1), and previously published high-coverage genomes (27× and 31×) of two rhesus macaques (Macaca mulatta) to be used as outgroup (Xue et al. 2016).
Mapping, Variant Detection, and Filtering
All FASTQ data were adapter and quality trimmed using Trimmomatic on recommended settings (Bolger et al. 2014) and initially mapped against the Chlorocebus sabaeus reference genome (ChlSab1.1) (Warren et al. 2015) using bwa-mem (Li 2013) with default settings. The publicly available de novo patas and De Brazza’s monkey assemblies were first converted into 100 base pair nonoverlapping FASTQ-reads and then mapped as above. For some analyses, we noted significant reference bias arising from the ingroup position of Chlorocebus sabaeus reference with respect to the set of species studied here. As a result, FASTQ reads showing an alternative allele to the reference obtained lower mapping scores than reads carrying the references allele. This bias increases with genetic distance to the reference, and thus to avoid potential errors in our inferences, we additionally mapped all samples against the closest available outgroup reference (Macaca mulatta, Mmul_10) as above. Although mapping to a distant outgroup reference results in underrepresentation of fast evolving sites, demographic inference will not be biased toward any of the studied taxa. Thus, all our subsequent analyses are based on the mappings to Macaca mulatta, unless stated otherwise. Samtools was used to filter out reads below a mapping quality of 30 (phred-scale) (Li et al. 2009). Next, reads were realigned around indels using GATK IndelRealigner (McKenna et al. 2010; DePristo et al. 2011) and duplicates marked using Picard2.10.3 (https://broadinstitute.github.io/picard/). After these filtering steps, we obtained a genome-wide coverage of 33× for the Dryas monkey. Next, we called single nucleotide polymorphisms (SNPs) with GATK UnifiedCaller outputting all sites (McKenna et al. 2010; DePristo et al. 2011). Raw variant calls were then hard filtered following the GATK best practices (Van der Auwera et al. 2013). Additionally, we removed all sites below quality 30 (phred-scale), those with more than three times average genome-wide coverage across the data set, sites for which >75% of samples had a nonreference allele in a heterozygous state, indels, and sites within highly repetitive regions as identified from the repeatmask-track for the Mmul_10 reference using VCFtools (Danecek et al. 2011).
Cytochrome B Sequencing
We amplified the cytochrome B sequence (1,140 bp) of the mitochondrial genome for all three Dryas monkey samples (supplementary table S1, Supplementary Material online) in five overlapping fragments (Haus et al. 2013). PCR reactions were run in 25 µl final volume containing 1 U GoTaq G2 Green Master Mix, 0.33 µM of each primer (forward and reverse, supplementary table S1, Supplementary Material online), 2 µl of genomic DNA, and 6.5 µl ddH20. The cycling conditions followed the procedure recommended by Haus et al. (2013) with a few modifications to the annealing temperatures of each primer, 94 °C for 2 min, followed by 40 cycles of 94 °C for 1 min, primer-specific annealing temperature (supplementary table S2, Supplementary Material online) for 1 min, 72 °C for 1 min, and 72 °C for 5 min. All PCR products were checked on 2% agarose gels and then cleaned with the Wizard SV Gel and PCR Clean-up System from Promega (Madison, WI) and sent off for Sanger-sequencing along with the amplification primers to Molecular Cloning Laboratories (San Francisco, CA). Sequence chromatograms were inspected by eye for accurate base calls and assembled using Geneious R11 11.0.5.
Autosomal Phylogeny and Divergence Dating
First, we used the vervet and the Dryas monkey SNP-calls to assess the genetic similarity between all individuals by applying a clustering algorithm. The R package ape was used to calculate pairwise distances among all individuals using the Tamura and Nei (1993) model, which allows for different rates of transitions and transversions, heterogeneous base frequencies, and between-site variation of the substitution rate (Tamura and Nei 1993; Paradis et al. 2004). We then used the R package phangorn to construct a UPGMA phylogeny from the resulting pairwise distance matrix (Schliep 2011).
Next, we reconstructed the phylogeny of the Dryas monkey in relation to the De Brazza’s monkey, patas monkey, and the vervets using genome-wide phylogenetic methods. First, we generated haploidized sequences for one individual per species using ANGSD, by randomly selecting a single high-quality base call (BaseQuality ≥30, read MapQuality ≥30, max depth below three times the genome-wide coverage) at each site in the reference genome, excluding sex chromosomes and sites within repetitive regions (Korneliussen et al. 2014). We then concatenated autosomal sequences into a multispecies alignment file. Next, nonoverlapping 350-kb genomic windows were extracted from the alignment and filtered for missing sites using PHAST v1.4 (Hubisz et al. 2011). After filtering, we excluded all windows with a length <200 kb resulting in a final data set consisting of 3,602 genomic windows (mean sequence length of 298 kb ±SD 18.5 kb). Individual approximately maximum-likelihood gene trees were then generated for the set of windows using FastTree2 v2.1.10 (Price et al. 2010) and the GTR model of sequence evolution. Next, we constructed a coalescent species tree from the obtained gene trees with ASTRAL v5.6.2 (Zhang et al. 2018) on default parameters. ASTRAL estimates branch length in coalescent units and uses local posterior probabilities to compute branch support for the species tree topology, which gives a more reliable measure of support than the standard multilocus bootstrapping (Sayyari and Mirarab 2016). Additionally, we obtained an extended majority-rule consensus tree using the CONSENSE algorithm in PHYLIP v3.695 (Felsenstein 2005). Finally, to explore phylogenetic conflict among the different gene trees, we created consensus networks with SplitsTree v4 using different threshold values (15%, 20%, and 25%) (Huson and Bryant 2006).
After obtaining the consensus phylogeny, we used a Monte–Carlo–Markov-Chain (MCMC)-based method to estimate divergence times between the different species. For large alignments, calculation of the likelihood function during the MCMC is computationally expensive, and we therefore estimated divergence times using an approximate method as implemented in the software MCMC-Tree that significantly improves the speed of the MCMC calculations (Yang 2007; Reis and Yang 2011). For this analysis, we used the independent molecular clock (the rates follow a log-normal distribution) and the JC69 evolutionary model with the following parameters as recommended by Reis and Yang (2011): alpha = 0, ncatG = 5, cleandata = 1, BDparas = 1 1 0.1, kappa_gamma = 6 2, alpha_gamma = 1 1, rgene_gamma = 2 20 1, sigma2_gamma = 1 10 1, finetune = 1: .1 .1 .1 .1 .1 .1, print = 1, burnin = 2,000, sampfreq = 10, nsample = 20,000. The divergence times were calibrated using a soft bound of 14.5–19.6 Ma for the most recent common ancestor between the rhesus macaque (tribe Papionini) and the guenons (tribe Cercopithecini), as recently estimated in Reis et al. (2018). We repeated the MCMC run twice and checked for run convergence in R (R2 = 0.991).
We also estimated the divergence times between the Dryas monkey and the vervets using Pairwise Sequential Markovian Coalescent (PSMC) modeling specifically suited for low-coverage genomes (Cahill et al. 2016). PSMC plots on artificial hybrid genomes (hPSMC) show a rapid increase in effective population size at the time when the two parental lineages diverge (Cahill et al. 2016). Haploidized genomic sequences of the Dryas monkey and the vervets were merged using seqtk mergefa (https://github.com/lh3/seqtk.git) and we then ran hPSMC on artificial hybrids of all pairwise combination between Dryas monkey and the vervets (using four different vervet genomes per species) assuming a generation time of 8.5 years (Warren et al. 2015) and a mutation rate per generation of 0.94×10−8 (Pfeifer 2017).
Mitochondrial Phylogeny
We de novo assembled the mitochondrial genomes for the Dryas monkey, the vervets, and rhesus macaque with NOVOplasty (Dierckxsens et al. 2016) with recommended settings, using a K-mer size of 39 and the Chlorocebus sabaeus (ChlSab1.1) mitochondrial reference genome as seed sequence. We also included the previously published mitochondrial genome sequence of the Dryas monkey type specimen (Guschanski et al. 2013). Mitochondrial genomes were aligned using Clustal Omega with default settings (Sievers et al. 2011). We then obtained a maximum-likelihood phylogeny in MEGAX, using the Tamura–Nei model with uniform rates among sites and running 1,000 bootstrap replicates (Kumar et al. 2018). Next, we partitioned the protein-coding genes in the alignment into 1st, 2nd, and 3rd coding position, using the ChlSab1.1 annotation in Geneious 10.1.2 (https://www.geneious.com; last accessed September 24, 2019). We estimated divergence times with MCMC-tree (which allows for different evolutionary rates of the partitions) using the same parameters as for the autosomal divergence estimates, however with the likelihood (instead of approximate likelihood) function. Finally, we also aligned the cytochrome B sequences of two additional Dryas monkey individuals to all other mitochondrial genomes and reran the maximum-likelihood estimates as above, this time only for the cytochrome-B sequence region.
Y-Chromosome Phylogeny
The mammalian Y-chromosome sequence is enriched for repeats and palindromes, and thus accurate assembly from short-read data is challenging (Tomaszkiewicz et al. 2017; Kuderna et al. 2019). We therefore obtained partial Y-chromosome consensus sequences using the filtered SNP calls. First, we identified all male individuals in our low-coverage data set using the ratio of X-chromosome to autosomal coverage (supplementary fig. S1, Supplementary Material online). Next, GATK FastaAlternateReferenceMaker was used to obtain a Y-chromosome consensus sequence for each male individual using the filtered variant calls as input. We masked all sites for which at least one individual showed a heterozygous call, as these represent SNP-calling errors. Additionally, we masked all repetitive regions and all sites for which one or more female individuals also showed a variant call, as these regions are likely enriched for SNP-errors due to mismappings. Given the scarcity of the retained genomic data (only 4% of the Y-chromosome was retained), we could not use any model-based phylogenetic approaches and instead constructed a UPGMA tree, as was done for the autosomal clustering, only including the filtered sites called in all male individuals. The tree was time calibrated using relative divergence to the rhesus Y-chromosome, and a rhesus macaque–guenon divergence time of 17 My (Reis et al. 2018), assuming a uniform mutation rate.
Gene Flow
We performed a model-free test of unbalanced allele sharing between the vervet individuals and the Dryas monkey (D-statistic) (Green et al. 2010) using all autosomal biallelic sites called in all individuals. First, we calculated D-statistics for all individual combinations as H1, H2, H3, H4, with the Dryas monkey as H3 and the rhesus macaque as the representative of the ancestral variant H4 (we excluded sites for which the two rhesus macaque genomes were not identical). Next, we calculated frequency-stratified D-statistics by grouping all individuals by species and calculating the frequency of shared derived alleles within each species (e.g., for each genomic site where the Dryas monkey carries the derived allele, we calculated the frequency of the derived allele in each of the vervet species), again using the Dryas monkey genome as H3 and rhesus as H4. Genome-wide D-statistic on population level were then calculated for four different bins of derived allele frequencies (<0.25, 0.25–0.5, 0.5–0.75, and >0.75).
A model-based estimate of introgression was obtained by constructing a maximum-likelihood (ML) tree using TreeMix v. 1.12 (Pickrell and Pritchard 2012), accounting for linkage disequilibrium (LD) by grouping sites in blocks of 1,000 SNPs (-k 1,000). Based on the previous phylogenetic inferences, the Dryas monkey was set as root. Standard errors (-SE) and bootstrap replicates (-bootstrap) were used to evaluate the confidence in the inferred tree topology and the weight of migration events. After constructing a maximum-likelihood tree, migration events were added (−m) and iterated 50 times for each value of “m” (1–10) to check for convergence in terms of the likelihood of the model as well as the explained variance following each addition of a migration event. The inferred maximum-likelihood trees were visualized with the in-build TreeMix R script plotting functions.
To identify putatively introgressed regions in all vervet individuals, we performed a screen for such segments following a strategy outlined in Martin et al. (2015). As this method is sensitive to the number of informative sites within windows, we used the mappings against the Chlorocebus sabaeus reference to maximize the number of informative sites (at the cost of reference bias). Briefly, in sliding windows of 10 kb, we calculated d statistics (which is related to D-statistic but not subject to the same biases as D when calculated in sliding windows; Martin et al. 2015) and Dxy using all Chl. sabaeus individuals from Gambia as ingroup (H1) (as these showed the least amount of shared derived alleles with the Dryas monkey, see below). Genetic distance (Dxy) was calculated for each window to the Dryas monkey (Dxydryas-X), and to Chl. sabaeus (Dxysabaeus(Gambia)-X), where x refers to the focal individual. Next, we calculated the ratio between Dxydryas-X/Dxysabaeus(Gambia)-X for each window. As Dxy is a measure of sequence divergence, introgressed windows between the Dryas monkey and non-sabaeus vervets are expected to have a relatively low Dxydryas-X and relatively high Dxysabaeus(Gambia)-X. Next, windows showing an excess of shared derived alleles with the Dryas monkey (d value >2 times SD) and an unusual low genetic divergence toward the Dryas monkey (the ratio of DxyDryas-X/Dxysabaeus(Gambia)-X < the genome-wide average minus two times the SD) were flagged as putatively introgressed. As regions with high d statistics tend to cluster in regions of low absolute divergence (Dxy) (Martin et al. 2015), some of our identified introgressed regions might be false positives.
Additionally, we used a topology weighting method, Twisst, for quantifying the relationships between the different taxa and visualizing how these relationships change across the genome (Martin and Van Belleghem 2017). Twisst estimates the topology across the genome in sliding windows, where consecutive blocks of contrasting topologies to the majority topology are an indication of adaptive introgression. For this analysis, we first computationally phased the SNP-calls using SHAPEIT (-phase–input-vcf beagle_out.vcf–window 0.1) (Delaneau et al. 2013) and subsequently obtained topologies in sliding windows of 50 SNPs using PhyML grouping of all samples by species, following the Twisst guidelines (https://github.com/simonhmartin/twisst.git). As the number of topologies increases exponentially with increasing number of species, running Twisst for many species becomes computationally unfeasible. We thus ran Twisst on the complete data set of inferred PhyML trees from the rhesus macaque, the Dryas monkey, Chl. sabaeus, aethiops and cynosuros samples (excluding Chl. pygerythrus, hilgerti, and tantalus as we did not find strong differences in introgression proportions between these and Chl. cynosuros) using the complete method.
Selection and Gene Ontology Enrichment of Introgressed Genes
Using the Chlorocebus sabaeus genome annotation (Warren et al. 2015), we obtained for each individual all genes within putatively introgressed windows as identified from d and Dxy statistics. A gene ontology enrichment was run for all putatively introgressed genes fixed in all non-sabaeus individuals in Blast2GO using Fisher’s exact test (Gotz et al. 2008). Next, for all genes, we obtained previously calculated selection score coefficients (XP-CLR selection scores) from Svardal et al. (2017). These selection scores are calculated for the different vervet populations and are based on a multilocus test of allele frequency differentiation, identifying regions in the genome where the change in allele frequency at the locus within the vervets occurred too quickly to be explained by drift using the XP-CLR method from Chen et al. (2010). Candidate genes for adaptive introgression were then identified as those with high gene-selection scores (top 10% of XP-CLR scores).
Demographic History
To infer long-term demographic history of the studied species, we used a pairwise sequentially Markovian coalescent model (PSMC) (Li and Durbin 2011). As PSMC analysis is sensitive to the number of present genomic sites (Nadachowska-Brzyska et al. 2016), we used the medium-coverage vervets (7.4–9.8× coverage) and the Dryas monkey genome mapped to the Chlorocebus sabeus reference. We excluded sex chromosomes, repetitive regions and all sites for which read depth was less than five and higher than two times the genome-wide average. We scaled the PSMC output using a generation time of 8.5 years (Warren et al. 2015) and a mutation rate of 0.94×10−8 per site per generation (Pfeifer 2017), as above. Bootstrap replicates (n = 100) were performed for the high-coverage Dryas monkey genome by splitting all chromosomal sequences into smaller segments using the splitfa implementation in the PSMC software and then randomly sampling with replacement from these fragments (Li and Durbin 2011). Our Dryas monkey genome coverage (33×) differed strongly from that of the medium-coverage vervets. As limited coverage is known to biases PSMC results (Nadachowska-Brzyska et al. 2016), we down-sampled the Dryas monkey genome to a similar coverage (8×) as the medium-coverage vervet genomes by randomly removing 75% of reads (samtools view –s 0.25) and repeated the PSMC analysis, allowing for relative comparisons between species. Demographic estimates from PSMC can also be biased by admixture events between divergent populations, giving a false signal of population size change (Hawks 2017). We thus removed all putatively introgressed regions (see above) from the Dryas monkey genome and reran the PSMC analysis.
Heterozygosity and Inbreeding
We measured genome-wide autosomal heterozygosity for all individuals with average genome coverage >3× using realSFS as implemented in ANGSD, considering only uniquely mapping reads (-uniqueOnly 1) and bases with quality score >19 (-minQ 20) (Fumagalli et al. 2013; Korneliussen et al. 2014). ANGSD uses genotype-likelihoods, rather than variant calls, allowing for the incorporation of statistical uncertainty in low-coverage data and shows high accuracy in estimating heterozygosity for genomes >3× coverage (Fumagalli 2013; van der Valk et al. 2019). Next, we used PLINK1.9 (Purcell et al. 2007) to identify stretches of the genome in complete homozygosity (runs of homozygosity: ROH) for all individuals with average genome coverage >3×. To this end, we ran sliding windows of 50 SNPs on the VCF files of all included genomes, requiring at least one SNP per 50 kb. In each individual genome, we allowed for a maximum of one heterozygous and five missing calls per window before we considered the ROH to be broken.
Genetic Load
To estimate genetic load in the Dryas monkey and vervets, we used the mappings and genome annotation of the Chlorocebus sabaeus. The variant effect predictor tool (McLaren et al. 2016) was used to identify loss-of-function mutations (transcript ablation, splice donor variant, splice acceptor variant, stop gained, frameshift variant, inframe insertion, inframe deletion, and splice region variant), missense, and synonymous mutations on the filtered SNP calls. As an indication of mutational load, for each individual, we counted the number of genes containing one or more loss-of-function and the total number of missense mutations divided by the number of synonymous mutations (dN/dS) (Fay et al. 2001). We excluded all missense mutations within genes containing a loss-of-function mutation, as these are expected to behave neutrally. Dividing by the number of synonymous mutations mitigates species-specific biases, such as mapping bias due to the fact that the reference genome was derived from a Chl. sabaeus individual, coverage differences, and mutation rate (Xue et al. 2015).
Supplementary Material
Acknowledgments
We thank the DRC government, ICCN, and the Lukuru foundation for facilitating sample collection and the 200 Mammals Consortium for providing assemblies of Cercopithecus neglectus and Erythrocebus patas. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The Margot Marsh Biodiversity Fund, Yale Peabody Museum of Natural History, and Florida Atlantic University provided support to KMD for sample export/import and molecular work at the Primatology Lab at FAU. The authors acknowledge support from the US Fish and Wildlife Service and Full Circle Foundation for field work and the Uppsala Multidisciplinary Centre for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. This work was supported by the Swedish research council FORMAS (2016-00835) to K.G.
Whole-genome sequence data generated in this study are available in the European nucleotide archive under accession number PRJEB32105. Cytochrome B sequences are available in the GenBank under accession number MN450179-MN450181.
References
- Beadle L. 1981. The inland waters of tropical Africa: an introduction to tropical limnology. 2nd ed. London/New York: Longman. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butynski TM. 2013. Cercopithecus dryas dryad monkey (Salongo monkey) In: Butynski T, Kingdon J, Kalina J, editors. Mammals of Africa: volume II: primates. London: Bloomsbury Publishing; p. 306–309. [Google Scholar]
- Cahill JA, et al. 2016. Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos Trans R Soc Lond B Biol Sci. 371:1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas G, Fragoso G, Rosetti M, Uribe-Figueroa L, Rangel-Escareño C, Saenz B, Hernández M, Sciutto E, Fleury A.. 2014. Neurocysticercosis: the effectiveness of the cysticidal treatment could be influenced by the host immunity. Med Microbiol Immunol. 203(6):373–381. [DOI] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D.. 2010. Population differentiation as a test for selective sweeps. Genome Res. 20(3):393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colyn M. 1987. Les primates des forêts ombrophiles de la cuvette du Zaïre: interprétations zoogéographiques des modèles de distribution. Rev Zool Afr. 101:183–196. [Google Scholar]
- Colyn M, Deleporte P.. 2004. Biogeographic analysis of central African forest guenons. Guenons 1:61–78. [Google Scholar]
- Colyn M, Gautier-Hion A, vans den Audenaerde T.. 1991. Cercopithecus dryas Schwarz 1932 and C. salongo Thys van den Audenaerde 1977 are the same species with an age-related coat pattern. Folia Primatol. 56(3):167–170. [DOI] [PubMed] [Google Scholar]
- Colyn M, Gautier-Hion A, Verheyen W.. 1991. A re-appraisal of palaeoenvironmental history in Central Africa: evidence for a major fluvial refuge in the Zaire Basin. J Biogeogr. 18(4):403. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaisieux S, Rayne F, Yezid H, Beaumelle B.. 2012. The ins and outs of HIV-1 tat. Traffic 13(3):355–363. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Howie B, Cox AJ, Zagury J-F, Marchini J.. 2013. Haplotype estimation using sequencing reads. Am J Hum Genet. 93(4):687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, Kuhlwilm M, Frandsen P, Sousa VC, Desai T, Prado-Martinez J, Hernandez-Rodriguez J, Dupanloup I, Lao O, Hallast P, et al. 2016. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science 354(6311):477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Hohmann G, Boesch C, Vigilant L.. 2004. Rivers influence the population genetic structure of bonobos (Pan paniscus). Mol Ecol. 13(11):3425–3435. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI.. 2001. Positive and negative selection on the human genome. Genetics 158(3):1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package). Seattle: University of Washington. [Google Scholar]
- Fumagalli M. 2013. Assessing the effect of sequencing depth and sample size in population genetics inferences. PLoS One 8(11):e79667.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Korneliussen TS, Linderoth T, Huerta-Sánchez E, Albrechtsen A, Nielsen R.. 2013. Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195(3):979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10):3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MHY, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb P, Butynski TM, Oates JF, Bearder SK, Disotell TR, Groves CP, Struhsaker TT.. 2003. Assessment of the diversity of African primates. Int J Primatol. 24(6):1301–1357. [Google Scholar]
- Guschanski K, Krause J, Sawyer S, Valente LM, Bailey S, Finstermeier K, Sabin R, Gilissen E, Sonet G, Nagy ZT, et al. 2013. Next-generation museomics disentangles one of the largest primate radiations. Syst Biol. 62(4):539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart JA, et al. 2019. Cercopithecus dryas. The IUCN red list of threatened species. Cambridge.
- Haus T, Akom E, Agwanda B, Hofreiter M, Roos C, Zinner D.. 2013. Mitochondrial diversity and distribution of African green monkeys (Chlorocebus Gray, 1870). Am J Primatol. 75(4):350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawks J. 2017. Introgression makes waves in inferred histories of effective population size. Hum Biol. 89(1):67–80. [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A.. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinformatics. 12(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267. [DOI] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J.. 2009. AEBP2 as a potential targeting protein for polycomb repression complex PRC2. Nucleic Acids Res. 37(9):2940–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15(1):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuderna LFK, Lizano E, Julià E, Gomez-Garrido J, Serres-Armero A, Kuhlwilm M, Alandes RA, Alvarez-Estape M, Juan D, Simon H, et al. 2019. Selective single molecule sequencing and assembly of a human Y chromosome of African origin. Nat Commun. 10(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, et al. 2016. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530(7591):429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y-C, Shen Y-R, Chen H-I, Lin Y-H, Wang Y-Y, Chen Y-R, Wang C-Y, Kuo P-L.. 2015. SEPT12 orchestrates the formation of mammalian sperm annulus by organizing core octameric complexes with other SEPT proteins. J Cell Sci. 128(5):923–934. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Kano T, Muhindo K.. 1985. Further information on the new monkey species, Cercopithecus salongo Thys van den Audenaerde, 1977. Primates 26(3):325–333. [Google Scholar]
- Lande R, Shannon S.. 1996. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution 50(1):434. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v1 [q-bio.GN].
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Nielsen R.. 2014. The lengths of admixture tracts. Genetics 197(3):953–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Van Belleghem SM.. 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics 206(1):429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F.. 2016. The Ensembl variant effect predictor. Genome Biol. 17(1):122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H.. 2016. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol Ecol. 25(5):1058–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Pfeifer SP. 2017. Direct estimate of the spontaneous germ line mutation rate in African green monkeys. Evolution 71(12):2858–2870. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M. D, Yang Z.. 2011. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 28(7):2161–2172. [DOI] [PubMed] [Google Scholar]
- Reis MD, Gunnell GF, Barba-Montoya J, Wilkins A, Yang Z, Yoder AD.. 2018. Using phylogenomic data to explore the effects of relaxed clocks and calibration strategies on divergence time estimation: primates as a test case. Syst Biol. 67(4):594–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D.. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507(7492):354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyari E, Mirarab S.. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol Biol Evol. 33(7):1654–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27(4):592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360(6389):656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E. 1932. Der Vertreter der Diana-Meerkatze in Zentral-Afrika. Rev Zool Bot Afr. 21:251–254. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(1):539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Pollack JL.. 2008. Subdivision in an ancestral species creates asymmetry in gene trees. Mol Biol Evol. 25(10):2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz J, de Wit MJ.. 2006. A proposed drainage evolution model for Central Africa—did the Congo flow east? J Afr Earth Sci. 44(1):75–84. [Google Scholar]
- Stanton DWG, Hart J, Galbusera P, Helsen P, Shephard J, Kümpel NF, Wang J, Ewen JG, Bruford MW.. 2014. Distinct and diverse: range-wide phylogeography reveals ancient lineages and high genetic variation in the endangered Okapi (Okapia johnstoni). PLoS One 9(7):e101081.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Göttlinger HG.. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114(6):689–699. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Cassady JL, Das G, Romo A, Evans GA.. 1993. Chromosomal structure and expression of the human OTF1 locus encoding the Oct-1 protein. Genomics 16(2):333–341. [DOI] [PubMed] [Google Scholar]
- Svardal H, Jasinska AJ, Apetrei C, Coppola G, Huang Y, Schmitt CA, Jacquelin B, Ramensky V, Müller-Trutwin M, Antonio M, et al. 2017. Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat Genet. 49(12):1705–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M.. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526. [DOI] [PubMed] [Google Scholar]
- Tomaszkiewicz M, Medvedev P, Makova KD.. 2017. Y and W chromosome assemblies: approaches and discoveries. Trends Genet. 33(4):266–282. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, et al. 2013. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma 43(SUPL.43):11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk T, Díez-del-Molino D, Marques-Bonet T, Guschanski K, Dalén L.. 2019. Historical genomes reveal the genomic consequences of recent population decline in eastern gorillas. Curr Biol. 29(1):165–170.e6. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung H-Y, Morita E, Wang HE, Davis T, He G-P, Cimbora DM, et al. 2003. The protein network of HIV budding. Cell 114(6):701–713. [DOI] [PubMed] [Google Scholar]
- Warren WC, Jasinska AJ, García-Pérez R, Svardal H, Tomlinson C, Rocchi M, Archidiacono N, Capozzi O, Minx P, Montague MJ, et al. 2015. The genome of the vervet (Chlorocebus æthiops sabæus). Genome Res. 25(12):1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Raveendran M, Harris RA, Fawcett GL, Liu X, White S, Dahdouli M, Rio Deiros D, Below JE, Salerno W, et al. 2016. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome Res. 26(12):1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, Szpak M, Frandsen P, Chen Y, Yngvadottir B, Cooper DN, et al. 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348(6231):242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S.. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(S6):153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Rana TM.. 2002. A bimolecular mechanism of HIV-1 Tat protein interaction with RNA polymerase II transcription elongation complexes. J Mol Biol. 320(5):925–942. [DOI] [PubMed] [Google Scholar]
- Zinner D, et al. 2013. Family Cercopithecidae (old world monkeys) In: Mittermeier RA, Rylands AB, Wilson DE, editors. Handbook of the mammals of the world: primates. Lynx Edicions; p. 550–753. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.