Abstract
The Drosophila nasuta species complex contains over a dozen recently diverged species that are distributed widely across South-East Asia, and which shows varying degrees of pre- and postzygotic isolation. Here, we assemble a high-quality genome for D. albomicans using single-molecule sequencing and chromatin conformation capture, and draft genomes for 11 additional species and 67 individuals across the clade, to infer the species phylogeny and patterns of genetic diversity in this group. Our assembly recovers entire chromosomes, and we date the origin of this radiation ∼2 Ma. Despite low levels of overall differentiation, most species or subspecies show clear clustering into their designated taxonomic groups using population genetics and phylogenetic methods. Local evolutionary history is heterogeneous across the genome, and differs between the autosomes and the X chromosome for species in the sulfurigaster subgroup, likely due to autosomal introgression. Our study establishes the nasuta species complex as a promising model system to further characterize the evolution of pre- and postzygotic isolation in this clade.
Keywords: speciation, Drosophila, introgression
Introduction
Species radiations are responsible for most of today’s biodiversity and are a prime study system to learn about the factors resulting in the origin of new species. Recent work in diverse species groups, ranging from humans, birds, fish to mosquitos, butterflies, and other insects has highlighted that genealogical relationships among closely related species can be complex and can vary across the genome and among individuals (Martin et al. 2013; Brawand et al. 2014; Fontaine et al. 2015; Lamichhaney et al. 2015; Dannemann and Racimo 2018). Recently diverged species often have incomplete reproductive barriers and may hybridize. Ancestral polymorphism predating lineage splitting may also be sorted stochastically among descendant lineages (i.e., incomplete lineage sorting). Phylogenetic heterogeneity can be caused both by hybridization and introgression and by incomplete lineage sorting in ancestral populations, causing some parts of the genome to have genealogies that are discordant with the species tree.
Genome-wide studies have revealed that certain genomic regions such as sex chromosomes can have distinct phylogenetic histories, possibly reflecting systematic differences in the extent of interspecific gene flow across the genome (Fontaine et al. 2015; Wong Miller et al. 2017; Fuller et al. 2018). Introgression can transfer beneficial alleles between closely related species, but interspecific gene flow can also be counteracted by natural selection at particular “barrier loci” (Dannemann and Racimo 2018). Thus, the landscape of genomic divergence contains information on the evolutionary forces that contribute to the origin of new species (Martin and Jiggins 2017). Here, we characterize the evolutionary history of the rapidly radiating nasuta subgroup of the immigrans species group of Drosophila. The nasuta group consists of more than a dozen closely related species or subspecies that are widely distributed across South-East Asia (Wilson et al. 1969; Kitagawa et al. 1982; fig. 1), and that show varying degrees of pre- and postzygotic isolation. Members from different species or subspecies often produce viable and sometimes fertile hybrids (Kitagawa et al. 1982), but show differences in their behavior and morphology (Spieth 1969; Wilson et al. 1969; Kitagawa et al. 1982).
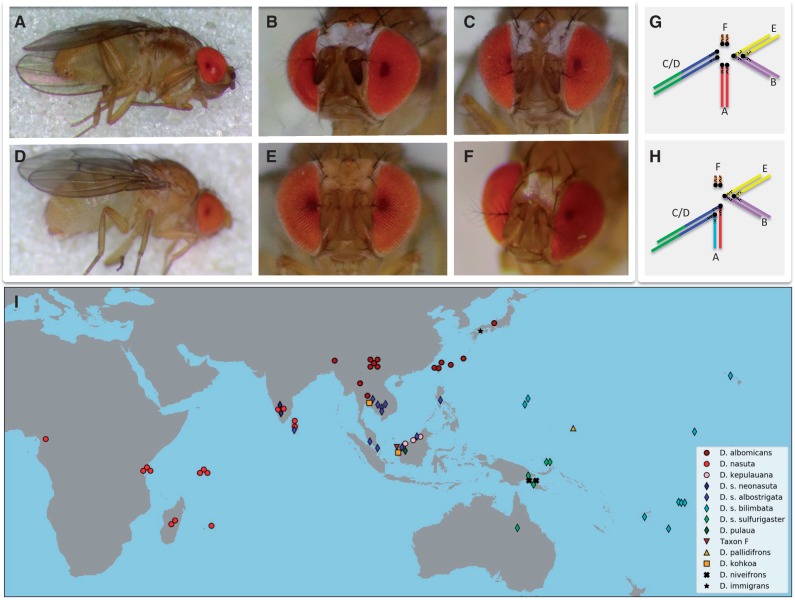
Fig. 1.
Morphology, karyotype, and distribution of species in the nasuta subgroup. (A–F) Male flies of the nasuta subgroup differ with regards to their morphology. (A and B) Drosophila albomicans; (C) D. s. albostrigata; (D and E) D. pulaua; (F) D. niveifrons. (G and H) Karyotypes of members of the nasuta group. Muller elements A–F are color-coded. (G) All species (apart from D. albomicans) have a telocentric X chromosome (Muller A), a metacentric autosome (Muller B/E fusion), and a large telocentric autosome (Muller C/D fusion), and the small dot chromosome (Muller F). (H) In D. albomicans, a neo-sex chromosome formed by the fusion of Muller C/D to both the X and Y chromosome. (I) Sampling locations of species and strains investigated. Note that for flies with overlapping sampling locations, the markers where slightly shifted on the map for visualization.
Females of the nasuta species complex are indistinguishable from their external morphology. However the males can be differentiated into phenotypic groups based on markings on the frons and thorax (Wilson et al. 1969; Kitagawa et al. 1982; see fig. 1A–F). The first category includes species where males have a continuous silver patch on their frons and dark bands on their thorax (i.e., D. nasuta, D. albomicans, D. kepulauana, and D. kohkoa). Species in the second category have prominent whitish orbits along the edges of their compound eyes and slightly dark thoracic bands; these include all subspecies of the D. sulfurigaster sp. group. Drosophila s. albostrigata and D. s. neonasuta have broader bands than D. s. sulfurigaster and D. s. bilimbata. Drosophila pulaua males have very pale white bands. The third category contains species without whitish patterns (D. pallidifrons, Taxon-F). The darkness of the bands on the mesopleuron on the thorax is correlated with the coloration of the frons, with flies with more bright areas on the frons showing more dark bands on the thorax. Drosophila niveifrons males have an X-shaped silver patch on their forehead and no coloration on their thorax.
Species in this group also display clear differences in mating behavior (Spieth 1969), and both acoustic and visual signals appear important during courtship display (Spieth 1969). Courtship songs, caused by wing vibration of the courting males, are often species-specific and contribute to prezygotic isolation between closely related species (Gleason and Ritchie 1998). Indeed, species or species groups in the nasuta species clade differ in male song, both with regards to quantitative and qualitative song parameters (Shao et al. 1997; Nalley MJ and Bachtrog D, unpublished data). Visual stimuli have also diverged among species in this group. During courtship, males in this species group show species-specific patterns of wing displays, circling of the females, and frontal displays of the males (Spieth 1969; Kitagawa et al. 1982).
Patterns of hybrid viability and sterility are complex within the nasuta species group (Kitagawa et al. 1982). In general, flies with similar frons patterns often produce viable and fertile hybrids (but D. kohkoa, for example, is clearly more reproductively isolated from other species with continuous white frons) and other crosses also sometimes produce viable offspring (in particular, D. albomicans females produce viable, but often sterile crosses with several species; Kitagawa et al. 1982).
Thus, levels of both pre- and postzygotic isolation differ among members of this species group, making it an ideal system to study the evolution of sexual isolation. Drosophila albomicans is of special interest in this clade, because of its recently formed neo-sex chromosomes: chromosomal fusions between an autosome and both the X and Y have created a neo-sex chromosome roughly ∼100,000 years ago (fig. 1G and H). Neo-sex chromosomes of Drosophila have served as a powerful tool to study the evolutionary forces driving sex chromosome differentiation (Bachtrog and Charlesworth 2002; Zhou and Bachtrog 2015; Mahajan et al. 2018).
Despite its general promise as a model system for speciation genomics, and detailed morphological, behavioral, and genetic investigations, little is known about the phylogenetic relationship among members of this group, or general patterns of sequence differentiation, and the correct species branching order has remained controversial and unresolved (Yu et al. 1999; Nagaraja et al. 2004). Here, we utilize whole genome sequencing to study patterns of genomic differentiation in the nasuta species complex. We assemble a high-quality genome of D. albomicans using single-molecule sequencing and chromatin conformation capture, and draft genomes for 11 additional species, and obtain genome-wide polymorphism data for a total of 67 strains of the nasuta group (supplementary table S1, Supplementary Material online and fig. 1I). This comprehensive data set allowed us to clarify species phylogenetic relationships, and describe overall patterns of differentiation and divergence among species in this group. As expected for such a recently diverged species group, patterns of genomic differentiation are highly heterogeneous across the genome. Detailed knowledge of background levels of genomic differentiation will provide a foundation for future studies on the genetic basis of pre- and postzygotic isolation in this clade.
Results
Assembly of D. albomicans Genome and Annotation
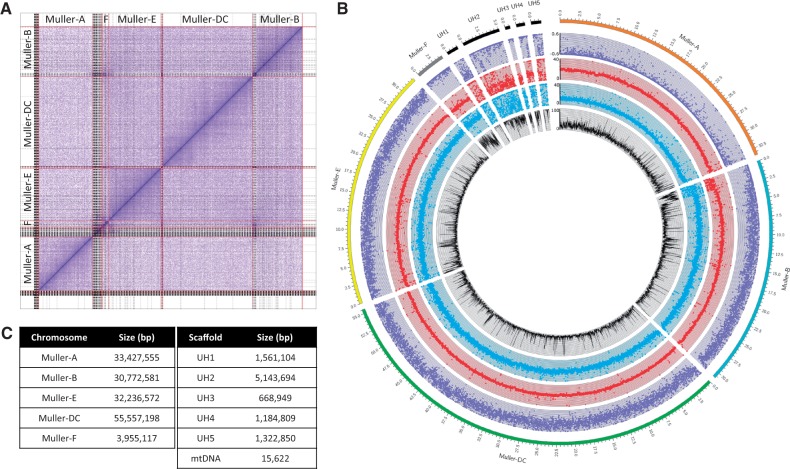
Drosophila albomicans is a species of particular interest in this clade, due to its recently formed neo-sex chromosomes (fig. 1H; Zhou et al. 2012). We used a combination of single-molecule long sequencing reads, Illumina reads, and chromatin conformation capture to create a chromosome level genome assembly of D. albomicans (supplementary fig. S1, Supplementary Material online). Our final assembly is 165.8 Mb in size, with an N50 of 33.4 Mb (fig. 2), and with all of the major chromosomes being contained within a single scaffold (fig. 2A). We verified X-linked scaffolds on the basis of significant differences in read depth between males (XY) and females (XX) (fig. 2B). As expected, Muller element A shows half the coverage in males relative to females; Muller CD (the neo-sex chromosome), on the other hand, shows similar levels of genomic coverage in both sexes. This means that most reads from the neo-Y in males still fully map to the neo-X, indicating low levels of differentiation between the neo-sex chromosomes. Our final genome annotation contained 12,387 genes, and the repeat content is ∼21%. We examined the genome for completeness using BUSCO scores (Simão et al. 2015), and found that 98% of core eukaryotic genes were present in our reference genome (supplementary table S2, Supplementary Material online). This assembly is a significant improvement over a previous one based on Illumina reads (supplementary fig. S2, Supplementary Material online; Zhou et al. 2012).
Fig. 2.
Assembly of Drosophila albomicans genome. (A) Hi-C scaffolding of contigs. Gray lines denote PacBio contigs, and red lines indicate different chromosomes. (B) Coverage analysis of chromosomes (Muller elements). Genomic reads from D. albomicans 15112-1751.03 males and females were mapped to the genome (20× coverage each); each point represents the mean coverage in nonoverlapping 10-kb windows (blue: male coverage, red: female coverage, purple: log2(male/female) coverage). The black line shows the mean repeat content (% repeat-masked bp along 10-kb windows). Unmapped scaffolds are highly repeat-rich and presumably correspond to pericentromeric regions. (C) Assembled size of the different chromosomes, and of various unmapped scaffolds (UH1-5), and the mitochondrial DNA.
Clustering of Species and Population
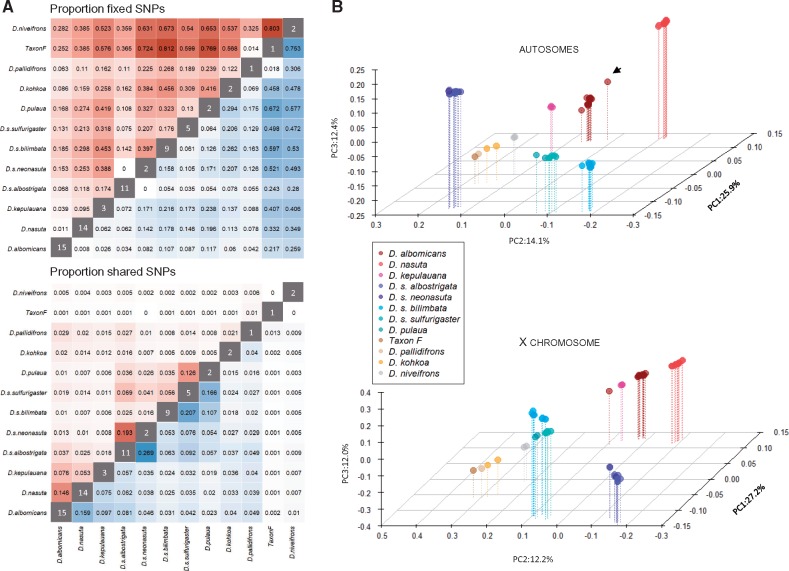
We resequenced 67 individuals from 11 species across the nasuta species group (median sequence coverage per fly 24-fold; supplementary fig. S3, Supplementary Material online). Reads were aligned to the genome assembly generated for D. albomicans, and stringent variant calling revealed ∼17.6 million variable sites within or between populations. We found considerable levels of genetic diversity (average pairwise diversity π) within each species, in the range 0.18–0.61% (supplementary table S3, Supplementary Material online), similar to that reported in other Drosophila populations. Genetic diversity within species does not appear to be determined by geographic range: D. s. bilimbata is widely scattered on many islands in the Pacific Ocean but has the lowest level of genetic diversity (0.18%), while D. albomicans has the highest level of diversity (0.61%) yet a more limited distribution than its close sister D. nasuta (Spieth 1969; Wilson et al. 1969; Kitagawa et al. 1982). Pairwise diversity between D. nasuta strains increases with geographic distance (isolation by distance; supplementary fig. S4, Supplementary Material online). We compared the proportions of shared and fixed SNPs between species in the nasuta group (fig. 3A andsupplementary fig. S5, Supplementary Material online). Extensive sharing of genetic variation and few fixed differences among populations was evident, particularly among subspecies of the sulfurigaster group and D. pulaua, and between D. albomicans, D. nasuta, and D. kepulauana (fig. 3A), indicative of their recent divergence time. Drosophila niveifrons appeared most divergent from all other species (fig. 3A). Principle component analysis (PCA) revealed similar patterns of clustering between species, with flies from the sulfurigaster group and D. pulaua consistently forming a cluster, and D. albomicans, D. nasuta, and D. kepulauana clustering (fig. 3B). Interestingly, one strain of D. albomicans (E-10815_SHL48) clusters more closely with D. nasuta on the autosome compared with other D. albomicans strains; admixture analysis reveals that this strain indeed has some D. nasuta ancestry (Cheng et al. 2017; supplementary fig. S6, Supplementary Material online). We also find clusters of D. pallidifrons and D. kohkoa flies. Drosophila s. albostrigata and D. s. neonasuta consistently overlap in the PCA analysis (fig. 3B), and also do not separate in the structure analysis (supplementary fig. S6, Supplementary Material online), indicating that they are genetically indistinguishable from each other. Drosophila s. sulfurigaster and D. pulaua also fail to clearly separate in the structure plots, especially for autosomes (supplementary fig. S6, Supplementary Material online). Drosophila s. bilimbata individuals form their own group, but D. s. bilimbata strain 1821.03 shows high levels of D. s. sulfurigaster/D. pulaua ancestry on the autosomes (supplementary fig. S6, Supplementary Material online).
Fig. 3.
Patterns of genome-wide differentiation in the nasuta group. (A) Proportion of fixed (top) and shared (bottom) SNPs in the nasuta group between the X chromosomes (red) and autosomes (blue). Darker shading indicates larger values. The values in the diagonal indicate the sample size. (B) Principle component analysis of autosomal (top) and X-linked (bottom) SNPs in the nasuta species group. The black arrow indicates Drosophila albomicans E-10815_SHL48.
Phylogenomic Clustering of Species
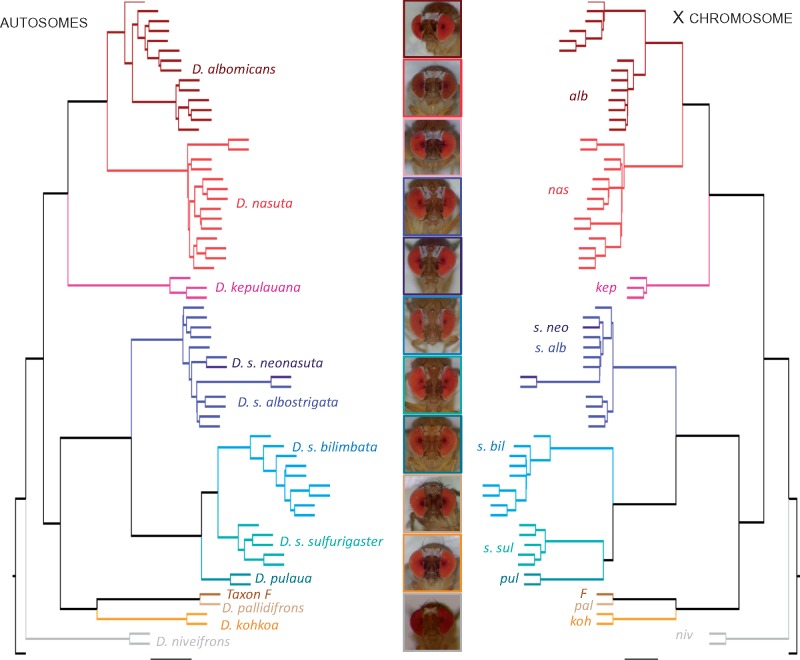
We inferred phylogenetic relationships among species using nonoverlapping 500-kb genomic windows (Stamatakis 2014), and inferred consensus trees separately for the X chromosome and the autosomes (Mirarab et al. 2014; Zhang et al. 2018). Our species tree topology is overall consistent with groupings based on PCA, identifying the same major clades (fig. 4). In particular, D. albomicans and D. nasuta are sister taxa, and group with D. kepulauana (the nasuta subclade). Likewise, all the different sulfurigaster subspecies form a cluster together with D. pulaua, with D. s. neonasuta and D. s. albostrigata being intermingled in the tree (in agreement with the clustering analysis above), and with D. s. sulfurigaster, D. s. bilimbata and D. pulaua forming a separate group. Interestingly, however, the topology of the consensus tree for this subgroup differs for the X and the autosomes: for the autosomes, D. s. sulfurigaster, and D. s. bilimbata cluster and D. pulaua is the outgroup, while the X topology places D. s. bilimbata as the outgroup (fig. 4). Drosophila pallidifrons is most closely related to Taxon F and D. kohkoa, and they form the outgroup to the sulfurigaster clade, and D. niveifrons is most distantly related to all other flies investigated (fig. 4).
Fig. 4.
Phylogenetic relationships among species of the nasuta group. The autosome phylogeny (left) has the same species level topology as the X chromosome phylogeny (right) with the exception of Drosophila s. bilimbata, D. s. sulfurigaster, and D. pulaua. The colored lines correspond to all the strains that belong to the same species group.
Using molecular clock estimates, we dated several nodes that define major groups and distinct species. Our inferred date for the basal node suggests that this species group started to diverge ∼2 Ma. Assuming a neutral mutation rate of 3.46 ×10−9 per year (Keightley et al. 2009), we estimate that D. nasuta and D. albomicans diverged roughly 0.6 Ma (Ks = 0.030), and split from D. kepulauana ∼0.7 Ma (Ks = 0.034). The nasuta clade diverged from the sulfurigaster clades roughly 1 Ma (Ks = 0.047) and ∼1.8 Ma from D. niveifrons (Ks = 0.089). Thus, sequence divergence confirms that species within the nasuta group split only very recently, consistent with patterns of incomplete pre- and postzygotic isolation in this clade.
Heterogeneity in Patterns of Ancestry across the Genome
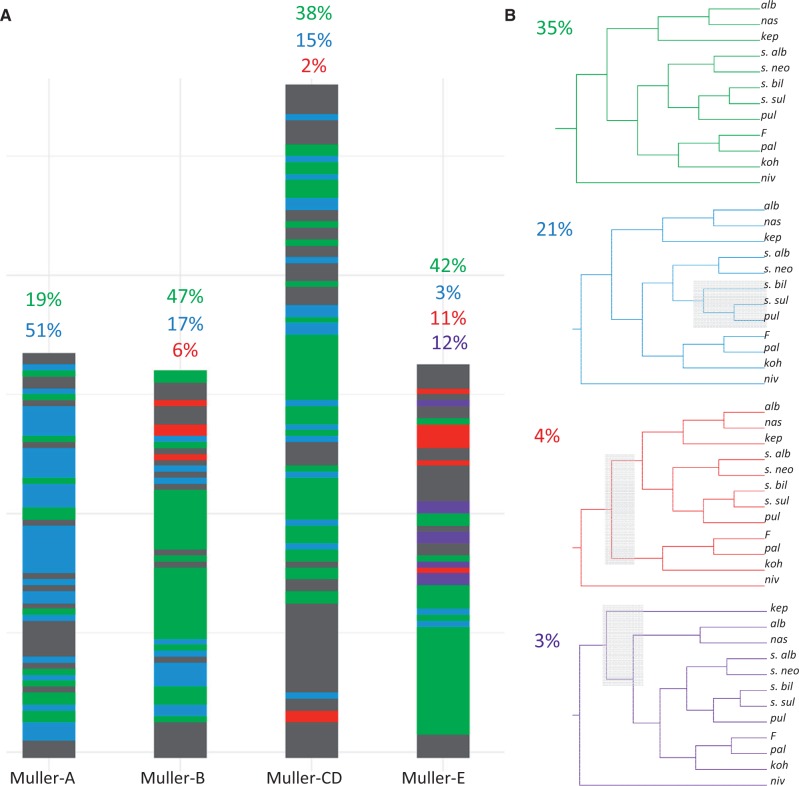
Although we generally find strong support for the inferred species tree, it conceals rampant phylogenetic complexity that is evident when examining the evolutionary history of more defined genomic regions. In particular, we analyzed the distribution of ancestry across the genome for the species (using a randomly selected individual from each group) by constructing trees in 500-kb (or 50-kb) sliding windows (fig. 5 and supplementary fig. S7, Supplementary Material online). Consistent with the inferred consensus trees, we find that the most prevalent topologies differ between the X and the autosomes. The most common topology is found in 35% of the windows (19% of the X windows, and 42% of the autosomal windows), and the second most common topology (21% of windows) dominates on the X chromosomes (51% of windows on X, vs. 12% of windows on autosomes). The third and fourth most common topologies are found in only 4% and 3% of windows, mostly on the autosomes. Conflicting signals in the distribution of ancestry across the genome may reflect incomplete lineage sorting and/or gene flow.
Fig. 5.
Local evolutionary history in the nasuta group varies across the genome. (A) Tree topology across the genome. For each 500-kb window, we color-code the topology recovered from that region (colors correspond to topologies in B). Note that while tree 1 (green) dominates on the autosomes, tree 2 (blue) dominates on the X. Coordinates are in terms of the Drosophila albomicans genome. Gray regions show alternative topologies. (B) Common topologies. The four most common trees are shown. The value in the top left corner is the percentage of all 500-kb windows that recover that topology.
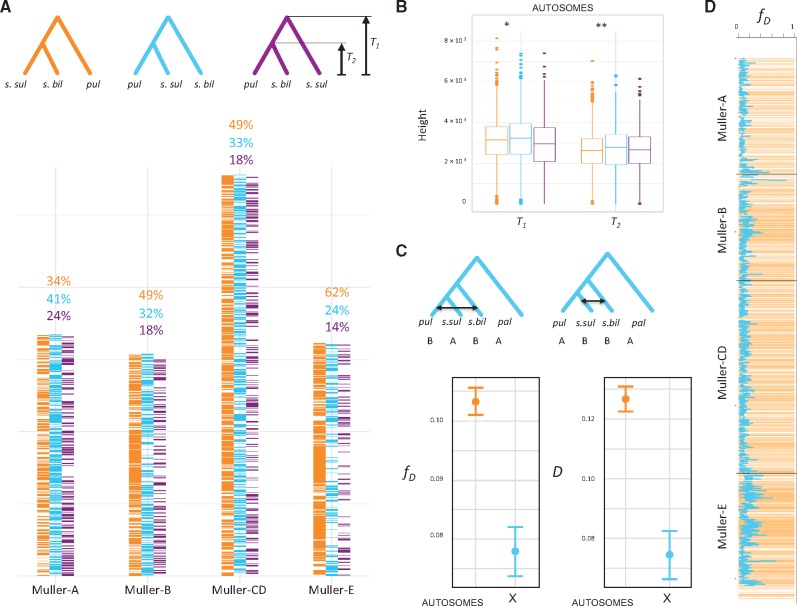
Increased introgression on the X chromosome between D. s. sulfurigaster and D. pulaua or autosomal introgression between D. s. sulfurigaster and D. s. bilimbata could account for the observed discrepancy of X-linked and autosomal topologies (Fontaine et al. 2015; fig. 6). The X chromosome often has a disproportionately large effects on hybrid sterility (the large X-effect; Masly and Presgraves 2007; Presgraves 2018), and autosomes may thus introgress more easily across species boundaries. Introgression will reduce sequence divergence between the species exchanging genes (Durand et al. 2011; Fontaine et al. 2015). Thus, gene trees constructed from nonintrogressed sequences should show deeper divergences than those constructed from introgressed sequences. To identify the correct species branching order, we inferred the length of autosomal topologies that support each of the possible groupings between D. s. sulfurigaster, D. s. bilimbata, and D. pulaua. If autosomal introgression resulted in conflicting phylogenetic signals, we would expect that topologies supporting the majority X chromosome grouping (i.e., (D. s. bilimbata, (D. s. sulfurigaster, D. pulaua))) to show higher divergence times than those supporting the majority autosomal topology (Fontaine et al. 2015). To estimate divergence times, we chose a random D. s. sulfurigaster, D. s. bilimbata, and D. pulaua strain and followed the procedure outlined in (Fontaine et al. 2015). In particular, there are three possible topologies and two divergence times for each tree (T1 and T2) for this trio (fig. 6A). We compared mean values of T1 and T2 between the three possible topologies, only focusing on trees derived from the autosomes (since many confounding factors differ between the X chromosome and autosomes; Fontaine et al. 2015). Indeed, the set of (autosomal) trees supporting the majority X chromosome topology (D. s. bilimbata, (D. s. sulfurigaster, D. pulaua)) had longer branches, as measured by both T1 and T2 (P < 0.05 and P < 10−4), than those supporting the majority autosomal tree (fig. 6B). This indicates that the species branching order inferred from the X chromosome is likely the correct topology, and that extensive autosomal introgression has resulted in a different majority phylogeny for the autosomes.
Fig. 6.
Autosomal introgression in the sulfurigaster clade. (A) Tree topology across the genome for Drosophila s. bilimbata, D. s. sulfurigaster, and D. pulaua in 50-kb windows. Topologies are color-coded, and tree heights T1 and T2 are indicated. (B) Tree height suggests that the majority X chromosome topology is the true phylogeny. T1 and T2 are shown for autosomal trees inferred from 50-kb windows. Trees with X majority relationship (s.bil, (s.sul, pul)) have significantly higher T1 and T2 than (s.bil, s.sul), pul) trees (P = 0.0037 and P = 2.55×10−11, Wilcoxon test), which is consistent with widespread introgression on the autosomes. (C) ABBA-BABA statistics (D and fD) to test for introgression between D. s. bilimbata, D. s. sulfurigaster and D. pulaua on the autosomes and the X chromosome (vertical bar shows the SE). Both test statistics are higher on the autosome compared with the X (D: P = 1.47×10−9; fD: P = 8.61×10−11; Wilcoxon test). (D) Genomic regions that show the autosome majority topology ((s.bil, s.sul), pul) show higher levels of introgression (as measured by fD; P < 2.2×10−16; Wilcoxon test). Shown is the autosomal tree topology across the genome (in yellow, as in panel A) across the genome and fD (blue line) in 50-kb windows.
To identify genomic regions that have introgressed between species in the recent past, we used the Gmin statistics (Geneva et al. 2015). Gmin measures the ratio of the minimum pairwise sequence distance between species to the average pairwise distance between species, and is sensitive to genealogical configurations resulting from recent gene flow where the minimum pairwise divergence (and thus Gmin) is small relative to the mean pairwise distance (supplementary fig. S8, Supplementary Material online). A total of 11.9% of autosomal 50-kb windows (294 out of 2,464 windows) support significant introgression based on Gmin between D. s. sulfurigaster and D. s. bilimbata but only 0.1% of windows on the X (1 out of 669 windows). In contrast, we find a similar fraction of introgressed windows on the X and autosomes for the D. s. sulfurigaster and D. pulaua comparison: 7.7% of significant windows on autosomes and 5.4% on the X. Thus, patterns of introgression, as inferred by the Gmin statistic, indicate pervasive introgression at autosomes between D. s. sulfurigaster and D. s. bilimbata. Note, however, that most of the small autosomal Gmin values are caused by D. s. bilimbata strain 1821.03 (supplementary fig. S8, Supplementary Material online), which also show signatures of mixed ancestry in the structure analysis (supplementary fig. S6, Supplementary Material online). We also used the genealogy-based (ABBA-BABA) test, summarized by the D and fD statistic (Durand et al. 2011; Martin et al. 2015), to evaluate the distribution of shared derived variants between D. s. sulfurigaster and D. s. bilimbata on the X versus autosomes. Assuming a (((D. s. sulfurigaster, D. pulaua), D. s. bilimbata), D. pallidifrons) tree topology, we found significantly elevated values for both statistics on autosomes relative to the X chromosome (fig. 6C). This is indicative of a significant excess of shared derived sites between D. s. sulfurigaster and D. s. bilimbata on autosomes relative to the X, and provides complementary support for a history of increased levels of introgression on autosomes, potentially explaining the topological differences between the autosomal and X chromosome phylogeny. Indeed, we find that regions of the genome that support the alternative topology (D. pulaua, (D. s. sulfurigaster, D. s. bilimbata)) show elevated levels of introgression, as estimated by fD (fig. 6D, P < 10−4).
Discussion
Drosophila has long served as a prominent model in speciation research, from describing macroevolutionary patterns of diversification to identifying the molecular players involved in species incompatibilities (Dobzhansky 1937; Muller 1942; Orr 1993; Castillo and Barbash 2017). A large body of work to understand the genetic basis of reproductive isolation has focused on D. melanogaster and its sibling species (Presgraves et al. 2003; Brideau et al. 2006; Bayes and Malik 2009; Ferree and Barbash 2009; Phadnis and Orr 2009). These studies benefit from the amazing repertoire of genetic tools available in this model organism, and have allowed the dissection of hybrid incompatibilities at the molecular and cellular level. However, D. melanogaster and its siblings have split >5 Ma (Tamura et al. 2004), and have accumulated a large number of hybrid incompatibilities since their reproductive isolation (Presgraves 2003; Masly and Presgraves 2007). To identify the evolutionary forces and molecular pathways involved in the initial processes of species formation, it is necessary to investigate systems at the earliest stages of divergence (Phadnis and Orr 2009; Wong Miller et al. 2017). The nasuta radiation is therefore an ideal group to address questions on the genomics of speciation and adaptive radiations.
The nasuta species complex shows dramatic differences in patterns of pre- and postzygotic isolation, including divergence in courtship song and mating behavior, and male coloration (Spieth 1969; Wilson et al. 1969; Kitagawa et al. 1982). Yet many species pairs in this clade can form viable and often fertile hybrids, making it an ideal system to study the genetic basis of reproductive isolation. Our analyses establish phylogenetic relationships in this clade, and describe its evolutionary history, thereby providing a foundation for further detailed investigations of pre- and postzygotic barriers to gene flow. In addition, D. albomicans contains a recently formed sex chromosome, and genome-wide investigation of its young neo-X and neo-Y can yield important information about the initiation of sex chromosome divergence (Zhou and Bachtrog 2012; Zhou et al. 2012), and its contribution to the formation of species boundaries (Kitano et al. 2009; Bracewell et al. 2017).
We generated a chromosome-level high-quality genome assembly for D. albomicans and reference-based “pseudogenomes” for the other species in the nasuta species group, to resolve phylogenetic relationships in this clade, and describe global patterns of differentiation and gene flow. In addition to having all euchromatic chromosome arms contained within a single scaffold, our assembly also recovers large parts of repeat-rich regions. In particular, we assembled 4 Mb of the repeat-rich dot chromosomes, ∼1.25 Mb of the pericentromeric region on Muller B, and roughly 10 Mb of repeat-rich unmapped scaffolds (UH1-5, see fig. 2B) that presumably correspond to pericentromeric, heterochromatic regions. In total, our assembly contains ∼18 Mb of sequence that is composed mainly of repetitive DNA (defined as 50% or more bp repeat-masked in 10-kb windows). Many genome assemblies, and in particular those using short-read sequencing data, are highly fragmented, and repeat-rich regions are typically missing (Simpson and Pop 2015). Yet, several recent studies have suggested that repetitive DNA, or genes interacting with repeats and heterochromatin, play an important role in the evolution of species boundaries. For example, several of the known “speciation genes” in Drosophila associate with satellite DNA and repeats. HMR and LHR interact with heterochromatin at centromeres and telomeres, and are needed for transposable element repression (Brideau et al. 2006); ZHR is a protein that localizes to a chromosome-specific satellite (Ferree and Barbash 2009) and OdsH encodes a heterochromatin-associated protein that binds to the repeat-rich Y chromosome (Bayes and Malik 2009). Additionally, transposable elements have been found to be misexpressed in hybrids between closely related species, including Drosophila (Lopez-Maestre et al. 2017), fish (Dion-Côté et al. 2014), mammals (O’Neill et al. 1998), or plants (Wu et al. 2015). Finally, the rapid evolution of centromeric satellite DNA and the centromere-specific histone protein CENP-A has led to the proposal that these two components evolve under genetic conflict, and may result in hybrid incompatibilities (Henikoff et al. 2001; Brown and O’Neill 2010). Homologous chromosomes may compete for inclusion in the oocyte, and centromere DNA may act as a selfish element and exploit asymmetric female meiosis to promote transmission to the egg. Coevolution of CENP-A may restore meiotic parity, but could result in segregation problems in hybrids (Henikoff et al. 2001; Brown and O’Neill 2010; Rosin and Mellone 2017). Work in monkeyflowers provides empirical support for the centromere drive hypothesis (Fishman and Saunders 2008). Interspecific monkeyflower hybrids exhibit strong transmission advantage of one parental allele via female meiosis, and divergence of centromere-associated repeats is thought to be responsible for this drive (Fishman and Saunders 2008). Centromere drive has also been detected in mice. Here, selfish centromeres exploit asymmetry of the meiotic spindle and preferentially orient toward the egg pole, thereby achieving preferential transmission into the next generation (Akera et al. 2017; Iwata-Otsubo et al. 2017). A candidate meiotic driver in a centromere-linked region that shows a moderate increase in transmission frequency has also been found in Drosophila using a quantitative sequencing approach (Wei et al. 2017). Together, these studies provide empirical support that repetitive DNA can play an important role in the evolution of reproductive isolation. High-quality genomes will be necessary to study the impact of heterochromatin and repetitive DNA on the evolution of species boundaries.
Previous studies have obtained conflicting results on the phylogenetic relationships among members of the nasuta species group (summarized in Yu et al. 1999). These phylogenies were based on both phenotypic data, such as hybrid sterility (Kitagawa et al. 1982), courtship song (Shao et al. 1997), male frons coloration (Yu et al. 1999), or genetic markers, such as isozymes (Kitagawa et al. 1982), mitochondrial loci (Yu et al. 1999), or a handful of nuclear genes (Bachtrog 2006). Our phylogenomic approach reveals that while phylogenetic relationships vary dramatically across the genome, we find overall strong support for the inferred species trees. Our analysis, using both population genetic and phylogenetic inferences, reveals consistent species groupings. Drosophila albomicans, D. nasuta, and D. kepulauana form one cluster. These species all show similar male frons coloration (fig. 4), and produce viable (though partially sterile) offspring. Another cluster consists of D. pulaua, D. s. sulfurigaster, D. s. bilimbata, D. s. albostrigata, and D. s. neonasuta, and most crosses between these species result in viable hybrids (Kitagawa et al. 1982). Drosophila s. albostrigata and D. s. neonasuta have been described as different subspecies (Yu et al. 1999) but are genetically indistinguishable in our analysis. Previous studies have typically placed D. pulaua as the sister group to the D. sulfurigaster semi-species, but our genomic analysis clearly places D. s albostrigata and D. s. neonasuta as the sister species to D. s. sulfurigaster, D. s. bilimbata, and D. pulaua. These taxa also show differences in their frons colorations: D. s albostrigata and D. s. neonasuta have thicker frons markings than D. s. bilimbata and D. s. sulfurigaster, and D. pulaua males have very faintly marked frons (fig. 4). Drosophila pallidifrons, Taxon F and D. kohkoa form a distinct cluster, and are the sister to the sulfurigaster species group, and D. niveifrons forms the outgroup to this radiation.
Interestingly, however, signals involved in prezygotic isolation (i.e., courtship song, mating behavior and male frons coloration) do not always follow the species phylogeny. For example, frons marking on male forehead seems to have evolved convergently in different groups (see figs. 1 and 4). The silvery markings on the frons were either present in an ancestor of the nasuta species complex, and modified or lost in some species, or gained independently in different clades. Drosophila pallidifrons, which is most closely related to D. kohkoa, completely lacks silvery markings on its forehead, while D. kohkoa males have a continuous silver patch on their frons similar to D. albomicans/D. nasuta. Interestingly, D. pallidifrons is also the only species in this group in which the male never faces the female in his courtship (Spieth 1969), which may suggest that the frons marking and courtship display coevolved. Drosophila pulaua, on the other hand, is very closely related to D. s. bilimbata and D. s. sulfurigaster, yet its frons are extremely faintly marked, and male courtship song is also drastically different in this species relative to all the D. sulfurigaster flies (Nalley MJ and Bachtrog D, unpublished data). Introgression between lineages, or independent sorting of ancestral variation may be responsible for convergent evolution of signals involved in prezygotic isolation.
Intriguingly, we observed a large amount of phylogenetic discordance between trees generated from the autosomes and X chromosome for D. s. sulfurigaster, D. s. bilimbata, and D. pulaua. The autosomes, which make up the majority of the genome, largely supported the grouping of D. s. bilimbata and D. s. sulfurigaster being sister species, while on the X chromosome, D. pulaua and D. s. sulfurigaster are more often placed as sister species. Our analysis suggests that the most common topology on the X reflects the true species branching order, and introgression on the autosomes has contributed to the incongruent topologies between the X chromosome and autosomes in this species clade. Lower rates of introgression on the X are expected, since X chromosomes from different species generally have disproportionately large effects on hybrid sterility (the large X-effect; Masly and Presgraves 2007; Presgraves 2018). The large X-effect results from the hemizygous expression of recessive X-linked hybrid sterility factors in XY hybrids and the higher density of hybrid sterility factors on the X relative to the autosomes. Thus, strong selection against hybrid sterility factors would disproportionately eliminate incompatible X-linked variation in species hybrids. Indeed, reduced introgression on the X chromosomes has been reported in multiple systems. For example, hybridizing subspecies of rabbits show elevated levels of differentiation on the X compared with autosomes (Carneiro et al. 2014). Likewise, the X chromosomes of house mouse subspecies are more highly differentiated than the autosomes (Phifer-Rixey et al. 2014). Interspecific gene flow has also been found to be lower on X chromosomes in various Drosophila clades (Turissini and Matute 2017; Meiklejohn et al. 2018). Thus, our data support the notion that X chromosomes are less permeable to cross species boundaries. Extensive autosomal introgression between D. s. bilimbata and D. s. sulfurigaster paradoxically has the effect that most of the trees derived from autosomes do not recover the correct species branching order. This resembles patterns of genomic differentiation between mosquito species (Fontaine et al. 2015). Mosquito species also show discordant X-linked and autosomal phylogenies, with the X chromosome reflecting the species branching order while pervasive autosomal introgression groups nonsister species together (Fontaine et al. 2015).
Materials and Methods
Fly Strains
We investigated a total of 67 nasuta group fly strains, and one D. immigrans strain as an outgroup. Supplementary table S1, Supplementary Material online, gives an overview of the species and strains used, and their geographic location. We chose the inbred D. albomicans 15112-1751.03 strain to generate a high-quality genome assembly using PacBio sequencing and Hi-C scaffolding.
PacBio DNA Extraction and Genome Sequencing
We used a mix of 15112-1751.03 females and extracted high-molecular weight DNA using a QIAGEN Gentra Puregene Tissue Kit (Cat #158667). DNA was sequenced on the PacBio RS II platform. In total, this produced 11.6-Gb spanning 531,638 filtered subreads with a mean read length of 12,992 bp.
Chromatin-Conformation Capture
Hi-C libraries were created from sexed female third instar larvae of D. albomicans, adapted from (Stadler et al. 2017). Single larvae were first homogenized, washed, and fixed with final concentration of 1% formaldehyde for 30 min. Fixed chromatin was then digested overnight with HpyCH4IV at 37 °C. The resulting sticky ends were then filled in and marked with biotin-14-dCTP, and dilute blunt end ligation was performed for 4 h at room temperature. Cross-links were then reversed by incubation at 65 °C with Proteinase K. DNA was purified through phenol/chloroform extraction and sheared using a Covaris instrument S220. Biotinylated fragments were enriched using streptavidin beads and subsequent washes. Library preparation (end repair, A-tailing, adapter ligation, library amplification) was performed off the DNA on the streptavidin beads. The final amplified library was purified using Ampure XP beads.
Whole-Genome Resequencing of nasuta Group Flies
We extracted DNA from all flies from supplementary table S1, Supplementary Material online, using either Illumina TruSeq or Nextera libraries. Illumina TruSeq Nano libraries were prepared from 100 ng genomic DNA according to Illumina’s protocol for 350-bp inserts. Libraries were pooled and sequenced on a HiSeq 4000 with 100-bp paired-end reads. Nextera libraries were prepared from genomic DNA, following Illumina’s protocol with the following modification: reaction volumes were scaled to 10 ng input DNA. Two-sided Ampure XP size selections removed fragments <200 bp and minimized fragments >800 bp. Libraries were pooled and sequenced on a HiSeq 4000 with 100-bp paired-end reads or 150-bp single-end reads.
Genome Assembly and Annotation
The genome assembly was generated as described in (Michael et al. 2018). Briefly, long reads were assembled into contigs using Minimap and Miniasm (Li 2016). This draft assembly was polished three times with RACON (Vaser et al. 2017) and once with Pilon (Walker et al. 2014). Juicer (Durand et al. 2016) and 3D DNA (Dudchenko et al. 2017) were used to process Hi-C reads and reorder contigs from the draft assembly based on levels of short range interactions. Blocks of ordered contigs which showed short-range interactions were stitched together into chromosome level scaffolds. Juicer’s bash script was modified to run on our cluster and job scheduling system. 3D DNA was used with the following options: “-m haploid -t 10000 -s 0 -c 3.” We looked at synteny between our scaffolded assembly and a previously published D. albomicans genome assembly (Zhou et al. 2012) using MUMmer3 (Kurtz et al. 2004). Scaffolds from our assembly were assigned to Muller elements based on synteny. To confirm that the sex chromosome, Muller A, was correctly assembled, we mapped 20× male and female D. albomicans reads with BWA (Li and Durbin 2009) using default options and obtained coverage data for 10-kb windows using bedtools genomecov (Quinlan and Hall 2010) and an in-house Python script. Female coverage was also compared with male/female coverage to identify uncollapsed heterozygosities in our assembly (i.e., regions where both haplotypes were assembled independently). Uncollapsed haplotypes can be identified based on reduced genomic coverage (by half; Mahajan et al. 2018), and were removed from our assembly, and resulting gaps in our scaffolds were stitched over. The final genome assembly was annotated using Maker (Campbell et al. 2014). RNA-seq data from the following tissues (male and female head, third instar larvae, carcass; and ovary, spermatheca, accessory glands, and testis) was mapped to the D. albomicans genome assembly with HiSat2 version 2.1.0 (Kim et al. 2015) using default parameters and the -dta option. A transcriptome assembly was then generated with the alignments using StringTie version 1.3.3b (Pertea et al. 2015) with default parameters. Finally, fasta sequences of the transcripts were extracted and used as the input for Maker.
SNP Calling and Filtering
Repeat libraries for D. albomicans 15112-1751.03 were generated using RepeatModeler version 1.0.5 (Smith and Hubley 2008–2015) and REPdenovo (Chu et al. 2016) using default parameters. RepeatModeler was run with default parameters. REPdenovo was run with the following parameters: “MIN_REPEAT_FREQ 3, RANGE_ASM_FREQ_DEC 2, RANGE_ASM_FREQ_GAP 0.8, K_MIN 30, K_MAX 50, K_INC 10, K_DFT 30, READ_LENGTH 100, READ_DEPTH 185.099490, THREADS 20, GENOME_LENGTH 172728670, ASM_NODE_LENGTH_OFFSET -1, MIN_CONTIG_LENGTH 100, IS_DUPLICATE_REPEATS 0.85, COV_DIFF_CUTOFF 0.5, MIN_SUPPORT_PAIRS 20, MIN_FULLY_MAP_RATIO 0.2, TR_SIMILARITY 0.85, and RM_CTN_CUTOFF 0.9.” The D. albomicans genome was then repeat masked with RepeatMasker version 3.3.0 (Smith et al. 2013–2015) using default parameters. Reads from each fly strain were mapped separately to the D. albomicans genome. Read alignment files of strains from the same species were combined. We then call SNPs and indels for each strain using GATK’s haplotype caller (DePristo et al. 2011). SNPs were filtered out with the following cutoffs (Gilks et al. 2016): “QD < 2.0,” “MQ < 58.0,” “FS > 60.0,” “SOR > 3.0,” “MQRankSum < −7.0,” and “ReadPosRankSum < −5.0”—SNPs that fail to meet these thresholds are subsequently masked. These SNPs were used to perform phylogenetic analyses. However, they were pruned using PLINK1.9 (Chang et al. 2015) to minimize the effects of LD in our clustering analyses and demographic inference using the following option: “–indep-pairwise 5 kb 50 0.1.”
Phylogenetic Reconstruction and Analysis
To create a phylogeny, we generated pseudogenomes for each strain by replacing sites on the D. albomicans genome assembly with their called SNPs. Sites that are heterozygous and where there is <20× coverage were masked, and the reference D. albomicans genome was excluded from this analysis, due to reference genome biases. The pseudogenomes were split into 50-kb bins, and a maximum likelihood (ML) phylogeny was created for each bin using RAxML 8.2.11 (Stamatakis 2014), and a consensus tree was created with ASTRAL-III (Zhang et al. 2018). We used FigTree (https://github.com/rambaut/figtree/) to visualize the phylogeny. To test for heterogeneity in evolutionary history across the genome, we randomly selected one representative strain for each species, and calculated topologies in 50-kb or 500-kb windows, as described earlier. To calculate tree heights in the sulfurigaster subgroup, we followed an approach outlined in (Fontaine et al. 2015). We randomly selected (four times) one representative strain for D. s. sulfurigaster, D. pulaua, D. s. bilimbata, and D. pallidifrons and generated phylogenies using nonoverlapping 50-kb windows along the autosomes with RAxML using the same parameters as mentioned earlier. With the topology, ((a, b), c), we calculated the more shallow divergence time (T2) using the equation, , and the more deep divergence time (T1) using the equation, , where is the distance between strains a and b in branch lengths. We used the phytools R package (Revell 2012) to infer the topologies and obtain terminal branch length for each phylogeny.
Divergence Time Estimates
We used the set of coding sequences (CDS) from the genome annotation to derive Ks (the number of synonymous substitutions per site) values between species. To obtain the coding sequences from non-D. albomicans species, we used the corresponding sites from the pseudogenome used to create a phylogeny. Ks values were calculated using KaKs_Calculator (Zhang et al. 2006). We used a neutral mutation rate estimate of 3.46×10−9 per base per generation, which was experimentally determined from D. melanogaster (Keightley et al. 2009). The species studied here have a generation time that is slightly longer than D. melanogaster and we therefore used an intermediate estimate of the number of generations per year for Drosophilids (7 generations; Cutter 2008) to convert the mutation rate to time-based units (2.42×10−8 mutations per base per year).
Population Genetic Analysis
We used Ohana (Cheng et al. 2017) with default options to quantify population structure, and calculate admixture proportions between species in the two major clades found in the phylogenetic analysis: the albomicans subclade consisting of D. albomicans, D. nasuta, and D. kepulauana as well as the sulfurigaster subclade consisting of D. s. albostrigata, D. s. neonasuta, D. s. bilimbata, D. s. sulfurigaster, and D. pulaua. FlashPCA (Abraham and Inouye 2014) was used to perform PCA with all strains. To test for introgression in the sulfurigaster subgroup, we calculated the Gmin and ABBA-BABA statistics (Durand et al. 2011; Geneva et al. 2015; Martin et al. 2015). Aligned reads from D. s. bilimbata, D. s. sulfurigaster, and D. pulaua were processed in 50-kb windows with the POPBAM package (Garrigan 2013), and Gmin was calculated using POPBAMTools (https://github.com/geneva/POPBAMTools). We also calculated the D and fD statistic (Green et al. 2010; Martin et al. 2015), to test for introgression between (((D. s. bilimbata, D. s. sulfurigaster), D. pulaua), D. pallidifrons). The genome was split into 50-kb windows and a Wilcoxon test was used to determine if the median values are statistically different between X-linked and autosomal windows. We calculated values of average pairwise diversity π along the genome using nonoverlapping 50-kb windows. Mean and median values of the entire genome for species with more than one sequenced individual are reported. Software to calculate both the D and fD statistic as well as π was obtained from (https://github.com/simonhmartin/genomics_general).
Supplementary Material
Acknowledgments
We thank Masayoshi Watada for providing nasuta flies and Carolus Chan for help generating data. This research was supported by NIH grants (R01GM076007, R01GM101255, and R01GM093182) to D.B. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant.
References
- Abraham G, Inouye M.. 2014. Fast principal component analysis of large-scale genome-wide data. PLoS One 9(4):e93766.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T, Chmátal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA.. 2017. Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358(6363):668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2006. The speciation history of the Drosophila nasuta complex. Genet Res. 88(1):13–26. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Charlesworth B.. 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416(6878):323–326. [DOI] [PubMed] [Google Scholar]
- Bayes JJ, Malik HS.. 2009. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326(5959):1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracewell RR, Bentz BJ, Sullivan BT, Good JM.. 2017. Rapid neo-sex chromosome evolution and incipient speciation in a major forest pest. Nat Commun. 8(1):1593.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513(7518):375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA.. 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314(5803):1292–1295. [DOI] [PubMed] [Google Scholar]
- Brown JD, O’Neill RJ.. 2010. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genom Hum Genet. 11(1):291–316. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M.. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 48:4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Albert FW, Afonso S, Pereira RJ, Burbano H, Campos R, Melo-Ferreira J, Blanco-Aguiar JA, Villafuerte R, Nachman MW, et al. 2014. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 10(8):e1003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM, Barbash DA.. 2017. Moving speciation genetics forward: modern techniques build on foundational studies in Drosophila. Genetics 207(3):825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JY, Mailund T, Nielsen R.. 2017. Fast admixture analysis and population tree estimation for SNP and NGS data. Bioinformatics 33(14):2148–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Nielsen R, Wu Y.. 2016. REPdenovo: inferring de novo repeat motifs from short sequence reads. PLoS One 11(3):e0150719.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 25(4):778–786. [DOI] [PubMed] [Google Scholar]
- Dannemann M, Racimo F.. 2018. Something old, something borrowed: admixture and adaptation in human evolution. Curr Opin Genet Dev. 53:1–8. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion-Côté A-M, Renaut S, Normandeau E, Bernatchez L.. 2014. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol Biol Evol. 31(5):1188–1199. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. 1937. Genetics and the origin of species. New York: Columbia University Press. [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28(8):2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL.. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3(1):95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA.. 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7(10):e1000234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Saunders A.. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322(5907):1559–1562. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217):1258524–1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Leonard CJ, Young RE, Schaeffer SW, Phadnis N.. 2018. Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLoS Genet. 14(7):e1007526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D. 2013. POPBAM: tools for evolutionary analysis of short read sequence alignments. Evol Bioinformatics Online. 9:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneva AJ, Muirhead CA, Kingan SB, Garrigan D.. 2015. A new method to scan genomes for introgression in a secondary contact model. PLoS One 10(4):e0118621.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks WP, Pennell TM, Flis I, Webster MT, Morrow EH.. 2016. Whole genome resequencing of a laboratory-adapted Drosophila melanogaster population sample. F1000Res. 5:2644.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JM, Ritchie MG.. 1998. Evolution of courtship song and reproductive isolation in the Drosophila willistoni species complex: do sexual signals diverge the most quickly? Evolution 52(5):1493–1500. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz M-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE.. 2017. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 27(15):2365–2373.e2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter ML.. 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19(7):1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa O, Wakahama KI, Fuyama Y, Shimada Y, Takanashi E, Hatsumi M, Uwabo M, Mita Y.. 1982. Genetic study of Drosophila nasuta subgroup, with notes on distribution and morphology. Jpn J Genet. 57(2):113–141. [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461(7267):1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL.. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin C-J, Wang C, Zamani N, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518(7539):371–375. [DOI] [PubMed] [Google Scholar]
- Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32(14):2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maestre H, Carnelossi EAG, Lacroix V, Burlet N, Mugat B, Chambeyron S, Carareto CMA, Vieira C.. 2017. Identification of misexpressed genetic elements in hybrids between Drosophila-related species. Sci Rep. 7(1):40618.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Wei K-C, Nalley MJ, Gibilisco L, Bachtrog D.. 2018. De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biol. 16(7):e2006348.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD.. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Jiggins CD.. 2017. Interpreting the genomic landscape of introgression. Curr Opin Genet Dev. 47:69–74. [DOI] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC.. 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5(9):e243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Gordon KE, Rzatkiewicz T, Kingan SB, Geneva AJ, Vedanayagam JP, Muirhead CA, Garrigan D, Stern DL, et al. 2018. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. Elife 7:610.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Jupe F, Bemm F, Motley ST, Sandoval JP, Lanz C, Loudet O, Weigel D, Ecker JR.. 2018. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat Commun. 9(1):541.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T.. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. 1942. Isolation mechanism, evolution and temperature. Biol Symp. 6:71–125. [Google Scholar]
- Nagaraja Nagaraju J, Ranganath HA.. 2004. Molecular phylogeny of the nasuta subgroup of Drosophila based on 12S rRNA, 16S rRNA and CoI mitochondrial genes, RAPD and ISSR polymorphisms. Genes Genet Syst. 79:293–299. [DOI] [PubMed] [Google Scholar]
- O’Neill RJ, O’Neill MJ, Graves JA.. 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–72. [DOI] [PubMed] [Google Scholar]
- Orr HA. 1993. Haldane’s rule has multiple genetic causes. Nature 361(6412):532–533. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Orr HA.. 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323(5912):376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, Bomhoff M, Nachman MW.. 2014. Genome-wide patterns of differentiation among house mouse subspecies. Genetics 198(1):283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163(3):955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2018. Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol Ecol. 27(19):3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA.. 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423(6941):715–719. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Rosin LF, Mellone BG.. 2017. Centromeres drive a hard bargain. Trends Genet. 33(2):101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Li D, Zhang X, Yu H, Li X, Zhu D, Zhou Y, Geng Z.. 1997. Study on the recognition and evolutionary genetics of the courtship song of species in Drosophila nasuta species subgroup. Yi Chuan Xue Bao. 24(4):311–321. [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Pop M.. 2015. The theory and practice of genome sequence assembly. Annu Rev Genomics Hum Genet. 16:153–172. [DOI] [PubMed] [Google Scholar]
- Smith A, Hubley R.. 2008. –2015. RepeatModeler Open-1.0. Available from: httpwww.repeatmasker.org.
- Smith A, Hubley R, Green P.. 2013. –2015. RepeatMasker Open-4.0. Available from: httpwww.repeatmasker.org.
- Spieth HT. 1969. Courtship and mating behavior of the Drosophila nasuta subgroup of species. Univ Texas Publ. 6918:255–270. [Google Scholar]
- Stadler MR, Haines JE, Eisen MB.. 2017. Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogaster embryo. Elife 6:637662.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S.. 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 21(1):36–44. [DOI] [PubMed] [Google Scholar]
- Turissini DA, Matute DR.. 2017. Fine scale mapping of genomic introgressions within the Drosophila yakuba clade. PLoS Genet. 13(9):e1006971.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M.. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27(5):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K-C, Reddy HM, Rathnam C, Lee J, Lin D, Ji S, Mason JM, Clark AG, Barbash DA.. 2017. A pooled sequencing approach identifies a candidate meiotic driver in Drosophila. Genetics 206(1):451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson FD, Wheeler MR, Harget M, Kambysellis M.. 1969. Cytogenetic relations in the Drosophila nasuta subgroup of the immigrans group of species. Univ Texas Publ. 6918:207–253. [Google Scholar]
- Wong Miller KM, Bracewell RR, Eisen MB, Bachtrog D.. 2017. Patterns of genome-wide diversity and population structure in the Drosophila athabasca species complex. Mol Biol Evol. 34(8):1912–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sun Y, Shen K, Sun S, Wang J, Jiang T, Cao S, Josiah SM, Pang J, Lin X, et al. 2015. Immediate genetic and epigenetic changes in F1 hybrids parented by species with divergent genomes in the rice genus (Oryza). PLoS One 10(7):e0132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wang W, Fang S, Zhang YP, Lin FJ, Geng ZC.. 1999. Phylogeny and evolution of the Drosophila nasuta subgroup based on mitochondrial ND4 and ND4L gene sequences. Mol Phylogenet Evol. 13(3):556–565. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S.. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(Suppl 6):153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao X-Q, Wang J, Wong G-S, Yu J.. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4(4):259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D.. 2012. Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr Biol. 22(6):522–525. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D.. 2015. Ancestral chromatin configuration constrains chromatin evolution on differentiating sex chromosomes in Drosophila. PLoS Genet. 11(6):e1005331.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhu H-M, Huang Q-F, Zhao L, Zhang G-J, Roy SW, Vicoso B, Xuan Z-L, Ruan J, Zhang Y, et al. 2012. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics 13(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.