Abstract
As a consequence of their increase in annual production and widespread distribution in the environment, nanoparticles potentially pose a significant public health risk. The sought-after catalytic activity granted by their physiochemical properties doubles as a hazard to physiological processes following exposure through inhalation, oral, transdermal, subcutaneous, and intravenous uptake. Upon uptake into the body, their size, morphology, surface charge, coating, and chemical composition augment the response of biological systems to the materials and enhance their toxicity. Identification of each property is necessary to predict the harm imposed by foreign nanomaterials in the body. Assay methods ranging from endotoxin and lactate dehydrogenase (LDH) signaling to apoptosis and oxidative stress detection supply valuable techniques for exposing biomarkers of nanoparticle-induced cellular damage. Spectroscopic investigation of epithelial barrier permeation and distribution within living cells reveals the proclivity of nanoparticles to penetrate the body’s natural defensive boundaries and deposit themselves in cytotoxic locations. Combination of the various characterization methodologies and assays is required for every new nanoparticulate system despite preexisting data for similar systems due to the lack of deterministic trends among investigated nanoparticles. The propensity of nanomaterials to denature proteins and oxidize substrates in their local environment generates significant concern for the applicability of several traditional in vitro assays, and the modification of susceptible approaches into novel methods suitable for the evaluation of nanoparticles comprises the focus of future work centered on nanoparticle toxicity analysis.
Keywords: Nanoparticle toxicity, In vitro assays, Mechanisms of toxicity, Nanoparticle-cell interaction, Nanomaterials, Biocompatibility, Nanoparticle characterization, Reactive oxygen species
1. Introduction
Due to their unique physiochemical properties, nanoparticles have received significant interest over the last several decades as therapeutic agents [1-3], catalysts [4, 5], and commercial material enhancers [6-8]. By combining their small size with individual chemistries, bulk material properties are altered or augmented to grant nanomaterials new features that are otherwise unattainable by their macroscale counterparts. In particular, the characteristic increase in surface area to volume ratio accompanying nanoparticles produces a substantial increase in catalytic activity. The size of nanomaterials also permits quantum effects to dominate the materials’ behavior and bestow fluorescent, magnetic, and electrical capacities that are foreign to their traditional chemistries [9, 10]. These properties have significantly improved the efficacy of everyday consumer products like food, paints, and household cleaners by supplying highly tuned amplification to the goods beyond their stand-alone limits [6, 7, 11, 12]. Albeit useful for improving commodities and the design of highly reactive agents for theranostics or catalysis, the surface reactivity of nanoparticles furthers their potential to interact with biological and environmental substances. In instances where magnified biological interaction is undesired or unforeseen, and even in cases emphasizing specific biological interplay, the materials can present unsought potency toward biosystems that can ultimately lead to adverse physiological consequences.
The frequent use of nanomaterials in everyday consumer products such as sunscreens, cosmetics, and pharmaceutical and food additives accelerates their widespread distribution in the environment [7, 8]. Once deposited therein, bystanders are at risk of exposure through inhalation, oral, or transdermal uptake. This risk can increase even further with intravenous or subcutaneous injection [13]. Inhalation puts the nasopharyngeal, tracheobronchial, and alveolar regions at risk for accumulation in the absence of alveolar–capillary barrier clearance. Oral ingestion of nanoparticles is typically shielded from toxicity by the physiochemical and cellular barriers inhabiting the gastrointestinal tract, but nanoparticles still exhibit size-dependent transition through the digestive system’s defenses into the bloodstream [13]. Upon entering the blood, nanoparticles follow the expected absorption, distribution, metabolism, and excretion (ADME) model of pharmacokinetics throughout the body. This model is, however, complicated by potential aggregation and surface absorption of blood plasma proteins which form a “corona” that greatly augments the biological response to these materials [14]. Nonetheless, target organs such as the spleen, kidney, and liver accumulate the nanomaterials for longer periods than traditional pharmaceutical agents with maximal half-lives extending into years. Without clearance, the particles can generate oxidative stress, inflammation, and subsequent cell death in and around the subjected organs and can form agglomerates within vessels that impede blood flow and rupture blockages [13]. As a result, methods for assessing the possible toxicity of newly developed nanoparticles are required to understand the particles’ threat to the health of both manufacturers and consumers before their final application.
As new nanomaterials are developed, rapid screening of their biological and health impacts must be performed to assess their potential risk and provide insights into proper handling and care. As part of this screening, in vitro nanoparticle toxicity assessment represents a de facto method of acute hazard identification for potentially dangerous nanoparticles. The methods are not a complete replacement for in vivo evaluation; whereas in vitro analysis records the acute damaging effects of nanoparticles in a specific cellular environment, in vivo animal models track biodistribution and bioaccumulation pathways in a manner that is currently unavailable to in vitro observation. Both methods are needed to fully investigate the toxicity and potential risk of a particular nanomaterial. Often, as a precursor for in vivo studies, in vitro assessment is employed to identify the minimum toxic dose from cell viability protocols. Complementary biomarker analysis is applied thereafter to detect signs of nonnecrotic cellular damage when cell viability is maintained. The absence of terminal markers combined with undisturbed cell viability offers possible categorization of the nanomaterial in question as inert, but the prognosis is moreover a herald for introduction into in vivo trials for confirmation. In vitro studies, moreover, screen for many of the known mechanisms of toxicity that can emerge in in vivo studies and, as a consequence, establish in vitro protocols as a necessary prerequisite. Readers interested in in vivo analysis of nanoparticles are recommended to read the review by Kumar et al. [15] and chapter by Clichici and Filip [16]. The methodology used for in vitro nanoparticle toxicity analysis is the focus of this book and will be discussed herein.
2. Mechanisms of Toxicity
Nanoparticle toxicity stems primarily from the particle’s physiochemical properties and individual architecture. Size, shape, surface charge, coating, and chemical constituency all play key roles in determining the particles’ uptake and biocompatibility. Established by the National Nanotechnology Initiative, nanoparticles are defined as having characteristic dimensions below 100 nm, but their size is generally recognized to include particles with dimensions of up to 1 μm [17]. In the lower strata of the size regime, particularly below 100 nm [18-20], nanoparticles can take advantage of the enhanced permeation and retention (EPR) effect attributed to highly vascularized tissues such as tumors which makes the size distribution desirable among the pharmacological community [21]. Additionally, as the size of nanoparticles decline, the energy barrier associated with uptake is also reduced, allowing for enhanced transdermal migration and cellular penetration. Decreasing the total surface area exposed to a phagocyte lowers the energy penalty required for energy-dependent endocytosis and raises the probability of phagocytic consumption. The relative increase in particle surface area linked to a reduction in volume also heightens the particle’s reactivity and enables further uptake through receptor-mediated endocytosis and nonphagocytic mechanisms.
The shape of nanoparticles plays a significant role in the progression of a material through the body in as much as the particles’ size. Morphologies can vary from homogeneous and heterogeneous solid spheres to hollow micellular rods depending on the material in question and the synthesis pathway employed [13, 17]. Spherical nanoparticles are reported to undergo phagocytosis and excretion at faster rates than their high aspect ratio counterparts. The phenomenon is assumed to be due to the minimization of contact area intersecting the cell membrane and the resulting added energy penalty associated with internalization [22]. When the long axis of a cylindrical particle is aligned perpendicular to the membrane surface, the enlarged and unsymmetrical deformation needed to envelope the incident particle is assumed to be energetically unfavorable in comparison to uniform spheres. In another conformation, if the face of the tube first imposes on the cell membrane, phagocytosis may commence without the opportunity for completion. The event can lead to frustrated phagocytosis with eventual cell rupture and localized inflammation as in the case of asbestos. In a countervailing study, however, Gratton et al. [23] identified high aspect ratio nanoparticles as having four-fold increased uptake for HeLa cells in comparison to low aspect ratio particles of similar size and chemistry. The explanation for the difference follows the same reasoning as presented for the rejection hypothesis, but the enlarged surface area affecting high aspect ratio particles is instead theorized to supply beneficial multivalent cationic interactions which promote nonspecific endocytosis. In either case, the morphology of the nanoparticles contributes heavily to their uptake mechanism and represents a major factor in their internalization and clearance potential. Additional shapes such as nonspherical, homogeneous and heterogeneous agglomerates, spherical and tubular micellular capsules, and dendritic structures individually conclude different preferential uptake mechanisms that complicate their ADME profile [13, 17]. Particles that evade phagocytic consumption and remain in circulation for longer periods of time are generally anticipated to possess increased systemic exposure and cytotoxic events, but vast generalizations are inappropriate for nanoparticulate systems due to incongruence among trends in their biocompatibility. Regardless of preexisting information from similar systems, new nanoparticle preparations necessitate in vitro study to verify the biocompatibility of slight morphological or chemical alterations.
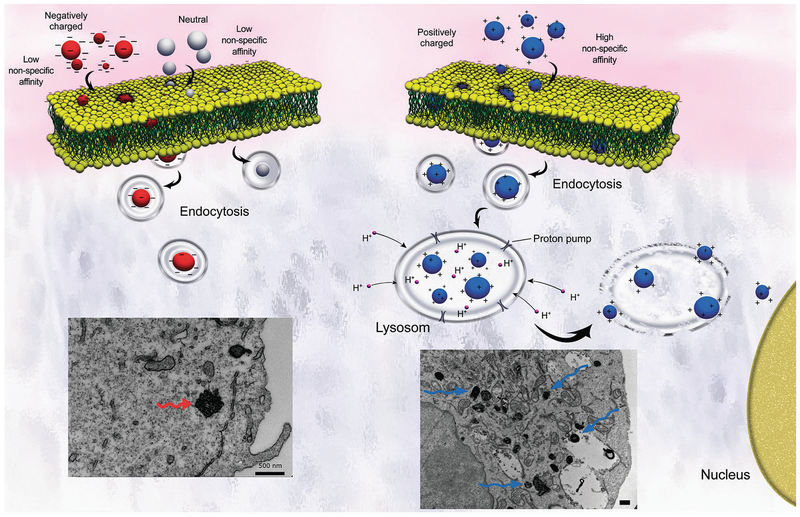
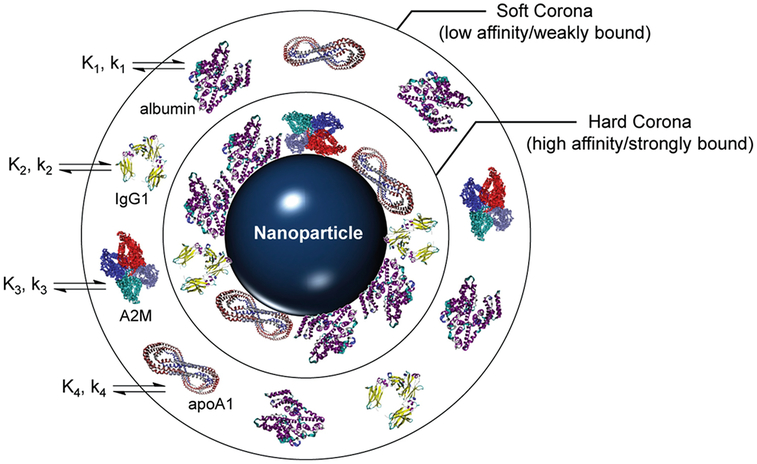
Surface charge likewise plays a deterministic role in cellular uptake that rivals the contributions of particle morphology. Both net cationic and anionic charge are correlated with increased toxicity while neutral surfaces are believed to have the greatest biocompatibility [13]. Zwitterionic particles, by contrast, are generally considered benign due to their self-regulated charge balance and have undergone extensive investigation as antimicrobial and antifouling agents [24]. Positively charged nanoparticles express affinity toward the negatively charged phospholipid heads populating the lipid bilayer and encourage endocytosis. Once internalized, the cationic surface charge acts as a proton sponge that disrupts normal lysosomal activity and initiates cell death as shown in Fig. 1 [13]. Negatively charged particles, by comparison, display greater potency in breaching the skin barrier via charge density and have the potential to signal coagulation cascades. Under sufficient doses, anionic nanoparticles can induce thrombosis and eventual embolism. In a similar manner, cationic particles assemble platelet aggregates known as “coronas” that disguise their unmasked chemistry and provide an alternative biological identity. Further complicating the functionality of a singular idealized “crown” covering the nanoparticles, protein adhesion to the particle surface operates by competitive affinity which changes the protein mixture surrounding the nanoparticle and modifies its new biological identity over time [14]. Illustrated in Fig. 2, a “hard” corona is initially formed by tightly bound protein aggregates and an exterior “soft” corona constantly exchanges proteins with the surrounding plasma [25]. The flux of adsorbed proteins forming the “soft” corona is regulated by a continual state of affinity competition termed the Vroman effect [14, 26]. The presence of a shielding corona can reduce toxicity by preventing the nonspecific cellular internalization of cationic particles and offsetting their membrane disruption and hemolytic capacities or increase toxicity by denaturing attached proteins and generating aberrations that elicit immunogenic and inflammatory responses [14]. Mimicking the myriad of proteins present in biological milieu for in vitro experimentation poses a significant challenge and comprises one of the major sources of disparity between in vitro and in vivo toxicity assessments.
Fig. 1.
Schematic displaying endocytosis of anionic (left) and cationic (right) nanoparticles and their resulting cytosolic formations. Cationic nanoparticles show stronger absorption capabilities than their anionic cousins, and, once endocytosed, they act as sponges that pump protons and signal cytotoxic cascades. The lower left transmission electron microscopy (TEM) images show aggregated anionic particles (bottom left) in HeLa cells compared to dispersed cationic particles with heightened uptake (bottom right). Reproduced from ref. 13 with permission from The Royal Society of Chemistry
Fig. 2.
Representative schematic of a fully developed protein corona surrounding a foreign nanoparticle. Proteins initially adhere to the surface strongly to form a hard corona followed by transient, competitive adsorption in an outer soft corona. Binding in the corona is kinetically (k) and thermodynamically (K) driven by serum proteins (with the common proteins albumin, immunoglobulin G1 (IgG1), alpha-2 macroglobulin (A2M), and apolipoprotein A-1 (apoA1) shown). Reproduced from [25]. Further permissions related to the material excerpted should be directed to the ACS. https:/pubs.acs.org/doi/10.1021/ar500190q
Coating nanoparticles with protein-resistant moieties or agglomeration inhibitors can mitigate the biomodification enveloping bare nanoparticles and counteract their unmodified toxicity. Uncoated nanoparticles often suffer from hydrophobic surfaces, and their preference to aggregate can form blockages that impeded circulation. Polyethylene glycol (PEG) is one of the most commonly employed surface coatings for therapeutic nanoparticles due to its ability to hide the particles from surveillance proteins [27]. The prevention of opsonization using a PEG steric barrier provides the nanoparticles with “stealth” properties that grant elongated circulation [28, 29]. As with shape regulating phagocytotic events, the lengthened biological half-life presents a double-edged sword: as the time in circulation extends, the opportunity for particles to affect both the desired target (e.g., a tumor) and unintended accumulation sites in susceptible tissues simultaneously increases [28]. Although heightened payload delivery to the target region significantly improves drug efficacy, the particles’ introduction to otherwise healthy areas can generate deleterious side effects that negate the therapy’s benefits. Moieties like chitosan and polylactic acid (PLA) have been combined with PEG to award both “stealth” and therapeutic properties concomitantly, and the modification of coatings to include environmentally responsive degradation cues can prompt even the relatively toxic nanoparticle cores to act as pseudotherapeutic agents. Protection against reactive oxygen species (ROS) generation by bare metallic nanoparticles using antioxidant-infused polymer coatings like poly(trolox ester) with controlled degradation offers a route for the combinatorial suppression of undesired oxidative damage and localized delivery of enzymatically hydrolyzed therapeutic agents while maintaining the core’s cytotoxic potential as a secondary mode of treatment [30]. Coatings can also harbor surface charges that selectively take advantage of cationic or anionic behavior without accruing their negative consequences. A cationic nanoparticle neutralized by a negatively charged shell can, for example, exploit extended circulation from a net neutral surface charge and the leaky vascular of tumors to specifically target tumor cells [31]. Shedding brought about by the acidic tumor microenvironment allows the nanocarrier to escape lysosomal destruction and reveal a reactive core. Alternatively, premature coating deterioration can turn an initially inert nanoparticle into a toxic agent by inciting immunogenic or inflammatory responses from its unshielded core prior to reaching its intended destination [13]. The untimely exposure provokes agglomeration, corona formation, and unabated chemical reactivity which potentially results in harmful physiological outcomes.
The chemical composition of nanoparticles governs their interaction with cells and milieu and shapes their capacity for oxidative stress production. Applying comparable size regimes, particles of varying fundamental compositions, especially in the case of metal oxide nanoparticles, display significantly different toxicity profiles due to alterations in their base constituencies [32]. Administered metallic nanoparticles are prone to dislodging toxic metal ions in the presence of fluctuating pH zones throughout the body, and the ions’ circulation into accumulation sites such as the liver and kidney present concerns for eventual genotoxic and cytotoxic effects [13]. Iron oxide and copper oxide nanoparticles pose as Fenton or Fenton-like catalysts for radical generation that contribute to lipid peroxidation and deoxyribonucleic acid (DNA) cleavage; gold, highly sought after as photothermal therapy and contrast agents, silver, traditionally used in antimicrobial prophylaxis, and zinc oxide, find use in a variety of applications ranging from filters to food additives, possess dose-dependent cytotoxicity; aluminum oxide and titanium dioxide, both employed in polymer and pharmaceutical industries, were initially considered inert but have since garnered attention as oxidative stress and inflammation promoters [32, 33]. Nanosized silica and carbon-based nanomaterials (e.g., carbon nanotubes, fullerenes, carbon black) constitute over 50% of airborne nanomaterials [34] and have extensive literature characterizing their metabolic resistance and size-dependent cytotoxicity [32, 35]. Polymeric nanoparticles propose additional considerations for their toxicity analysis, namely whether the nanoparticles are liable to degrade within the body and whether their metabolites are biocompatible. Although metallic nanoparticles hold similar concerns for metabolic degradation, therapeutic polymer-based nanoparticles often have the designed caveat of undergoing hydrolysis and breaking into their base monomers or analogs [17]. The feature allows on-demand release of medicinal agents into localized delivery sites to enhance the bioavailable fraction of therapeutics at the site of action while minimizing systemic circulation of drugs with deleterious side effects. Careful engineering is, however, required to ensure toxic degradants are prevented from entering the bloodstream and wreaking havoc downstream of the material’s degradation cue. The interplay of balancing the therapeutic and toxic potential of nanoparticles with their metabolites defines a foundational concern for minimizing ROS and inflammation production from therapeutic administration and environmental bioaccumulation, and their evaluation through in vitro testing marks a prerequisite study for any nanoparticle-based treatment.
3. Characterization and Toxicity Analysis
To properly evaluate the safety and impact of a nanomaterial, the fundamental physiochemical properties of the material and its interaction and impact upon living cellular systems must be understood. In this section, the most common methods to characterize nanomaterial properties and a set of in vitro toxicity screenings used to determine material safety are presented. The approaches presented herein do not represent the entirety of all available tools and methods but rather a sufficiently broad initial assessment for the evaluation of a new nanomaterial.
3.1. Size and Surface Charge Evaluation
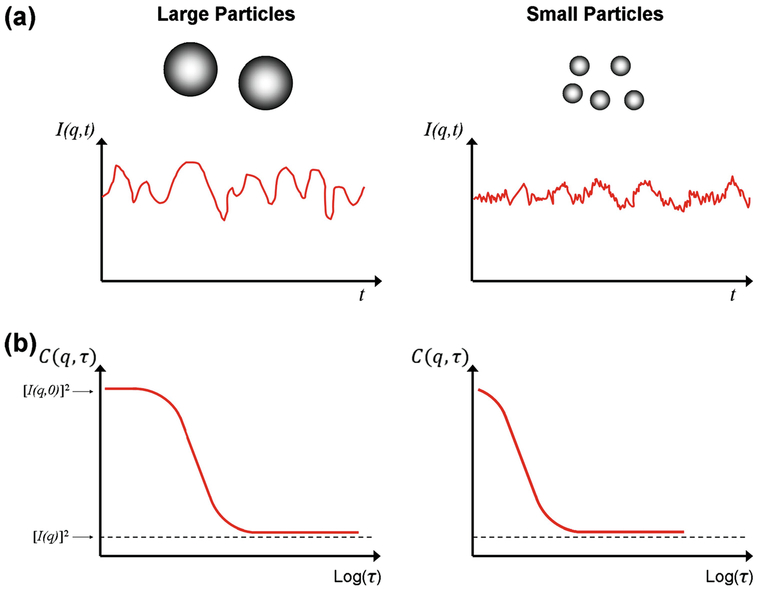
Numerous analytical techniques are available to characterize the toxicological aspects of nanoparticles, but two methods in particular are regularly used to grant critical quantitative information: dynamic light scattering (DLS) and zeta potential (ZP) analysis. With DLS, the random movements of dilute nanoparticles dispersed in solution caused by Brownian motion are detected by monitoring Rayleigh or Mie scattering generated from a monochromatic laser [36]. The intensity fluctuations recorded by the detector are translated into an autocorrelation function which tracks the intensity decay as a function of time. A representative image displaying the translation of scattering intensity into an autocorrelation function is displayed in Fig. 3. In the simplest case, the autocorrelation function is fit to an exponential decay whereby the translational diffusion coefficient is calculated in accordance with the wave vector relating the angle of excitation used by the DLS instrument. The diffusion coefficient is used in conjunction with the Stokes-Einstein equation to estimate the hydrodynamic radius of the particles assuming that the nanoparticles’ morphology is confined to a sphere [37]. Averaging the decay rate prior to evaluating the diffusion coefficient yields the ensemble translational diffusion coefficient which is applied to determine the commonly reported z-average nanoparticle diameter. Two primary analytical methods succinctly define the relationship between the autocorrelation function and the sample size distribution: the cumulant and CONTIN algorithms [36]. The cumulant algorithm fits the beginning of the autocorrelation function to a single exponential decay wherein the first cumulant term defines the z-average diameter and the second term indicates the polydispersity index (PDI). The PDI marks the homogeneity of a sample on a zero to one scale; a small PDI (≤0.1) is representative of a monodisperse sample while a large PDI (>0.4) designates a highly polydisperse sample [36]. The CONTIN algorithm employs a broader fitting timescale to isolate size distributions for multiple heterogeneous peaks in polydisperse samples. Modern DLS instruments apply both methods to provide the user with the z-average diameter, PDI, and heterogeneous peak distributions simultaneously. If the particles lack a spherical geometry and Mie scattering dominates, sample anisotropy will noticeably alter the calculated z-average diameter as a function of the irradiation angle used for the sample analysis. Post-measurement analysis can take advantage of equations derived for estimating the sample’s rotational diffusion coefficient together with its translational diffusion coefficient to approximate the aspect ratio of rod-like particles [38, 39].
Fig. 3.
Schematic scattering intensity as a function of time (a) and autocorrelation function variation with delay time (b) for large and small particle sizes, respectively. Large particles resist random force fluctuations and maintain smoother intensity plots; smaller particles move rapidly in solution and cause jitter in their intensities. Reproduced in part from [37]
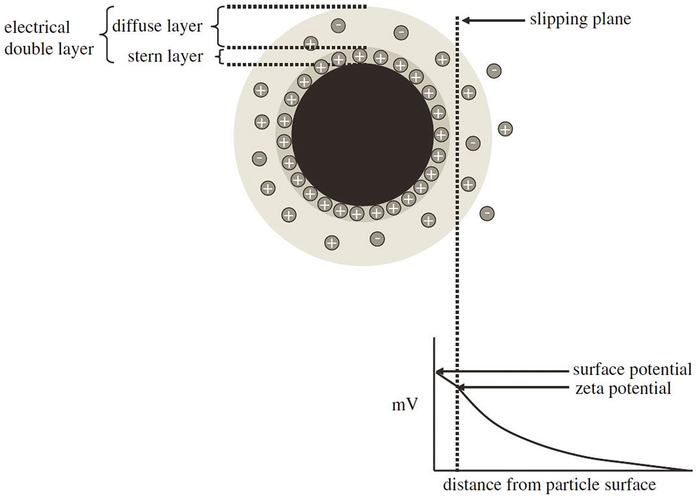
Zeta potential identifies the apparent surface charge of nanoparticles and is often a complementary capability of DLS systems. Particles dispersed in solution innately attract a closely packed layer of oppositely charged molecules to their surface known as the Stern layer, and the Stern layer is further surrounded by an ionically mixed diffuse layer extending outward toward a hypothetical boundary termed the slipping plane. Together, the Stern layer and slipping plane represent the electric double layer (EDL) enveloping a nanoparticle [40]. As shown in Fig. 4, the electric potential difference between the outermost slipping plane of the EDL and the potential of the dispersant comprises the zeta potential of a colloid. The measured ZP consequently does not signify the surface charge at the nanoparticle–Stern layer interface; the measured potential difference atop the EDL is substantially lower than the potential difference between the surface of the particle and the surrounding medium [41]. The decay of electrostatic force follows Debye’s law as an inverse exponential, and the true surface potential, called the Nernst potential, is only attainable through theoretical approximation [36]. The ZP is determined by correlating the speed of particles in transit to the device’s anode or cathode to the magnitude of an externally applied electric field. The velocity is assessed by monitoring the Doppler shift, which relates the frequency change between incident light shown onto electrically mobile particles and a reference laser to the velocity of the colloids in the medium [40]. The electrophoretic mobility of the nanoparticles is directly calculated from the experimentally determined particle velocity and electric field strength, and the mobility is thereafter related to the desired ZP with the Helmholtz–Smoluchowski equation for large particles (>100 nm) having relatively small EDLs with respect to their size or the Hückel equation for small particles (≤100 nm) with comparatively large EDLs [36]. The Helmholtz–Smoluchowski equation is viable for aqueous solutions with high salt concentrations (~10−2 M) while the Hückel equation requires minimal salt interference (<10−5 M) [36]. The dispersant pH plays a critical role in the stability of nanoparticle solutions by altering the ionic composition of the EDL and modifying the electrostatic repulsive forces between charged particles. As the solution pH approaches the isoelectric point of the nanoparticles, the electrostatic repulsion among particles of like charges diminishes and the van der Waals attractive forces between the floating bodies begins to dominate [42]. The magnitude of the measured ZP, therefore, does not provide a conclusive verification of colloid stability; ZP analyzes only particle surface charge without insight into interparticle attractive forces. Solutions with high ZP (±30 mV) are often conferred stability while low ZP solutions (≤10 mV) are considered unstable, but the scale of attractive van der Waals forces can (rarely) revoke traditional classifications [43]. The solution ionic strength also contributes to the nanoparticles’ stability by compressing the EDL with increasing ionic strength and lowering the measured ZP [41]. As mentioned earlier in this chapter, a particle’s calculated ZP and, by extension, surface charge play a significant role in deducing their possible toxicity, advancing integrated DLS-ZP systems as essential instruments for precursor biocompatibility evaluations.
Fig. 4.
Schematic representation of the EDL extending from the surface of an anionic nanoparticle into the hypothetical slipping plane. The zeta potential decreases exponentially from the surface outward toward the surrounding medium. Reproduced from [40]
Although DLS and ZP act as the primary characterization techniques for nanoparticles before their introduction to in vitro assays, numerous other techniques are available that offer comparable analyses or additional worthwhile information. Common practices include electron microscopy (e.g., transmission electron microscopy (TEM) and scanning electron microscopy (SEM)), laser diffraction, and atomic force microscopy (AFM) for sizing and geometry, Fourier-transform infrared spectroscopy (FTIR), X-ray diffraction (XRD), surface-enhanced Raman spectroscopy (SERS), and solid-state nuclear magnetic resonance spectroscopy (SSNMR) for composition analysis, and ultraviolet-visible spectroscopy (UV-Vis) and fluorometry for photonic properties [44]. Employing multiple methods in tandem with DLS and ZP analysis help identify some of the more critical parameters composing nanoparticles’ toxicity profiles, and their evaluation in concert with in vitro toxicity assessment can reveal the primary mechanisms generating toxicity that require resolution prior to their use.
3.2. Cellular Interaction Assays
An important screen for nanoparticle toxicity is the determination of a nanoparticle’s ability to transport across and interact with cellular barriers. Nanoparticle permeation across the cell membrane promoted by cationic surface charges and coatings enticing active transport [45] cultivate interaction between therapeutic or toxic agents and susceptible organelles in the cytoplasm. Assays surveying cellular infiltration desire quantitative analysis of particle positioning within cells and qualitative imaging of the material’s destination, but current instrumentation struggles to bridge the gap between the two analytical goals. In conventional procedures, cultivated cells are incubated with nanoparticles for 24 h followed by staining and microscopic analysis [46-48]. Fluorescent methods typically use anterior chemical modification [23, 45] or immunostaining [49] to couple tags such as fluorescein isothiocyanate (FITC), cyanines, and Alexa dyes with fluorescence-activated cell sorting (FACS), confocal laser scanning microscopy (CLSM), and imaging flow cytometry (IFC) for independent quantitative and qualitative identification of nanoparticle internalization following sample incubation. An excellent review by Ostrowski et al. [50] delivers an overview of traditional spectroscopic and microscopic imaging approaches for tissue and cellular internalization studies. Alternatively, methods using inductively coupled plasma mass spectroscopy (ICP-MS) [51], TEM, or transmission X-ray microscopy (TXM) in the case of metallic nanoparticles [52] eliminate the need for fluorophores by applying nonfluorometric instrumentation. None of the individual approaches offers a precise, high-resolution measure for internalization: CLSM and TXM reveal whether nanoparticles are internalized or externally adhered to the surface of cells, but the methods are only semiquantitative; FACS and ICP-MS provide quantitative results without visual resolution; and IFC offers a semiquantitative bridge by sacrificing the throughput of FACS and the resolution of CLSM [53]. Combination of two or more techniques is required to fully characterize the scale of nanoparticle intrusion while maintaining observations of their locality.
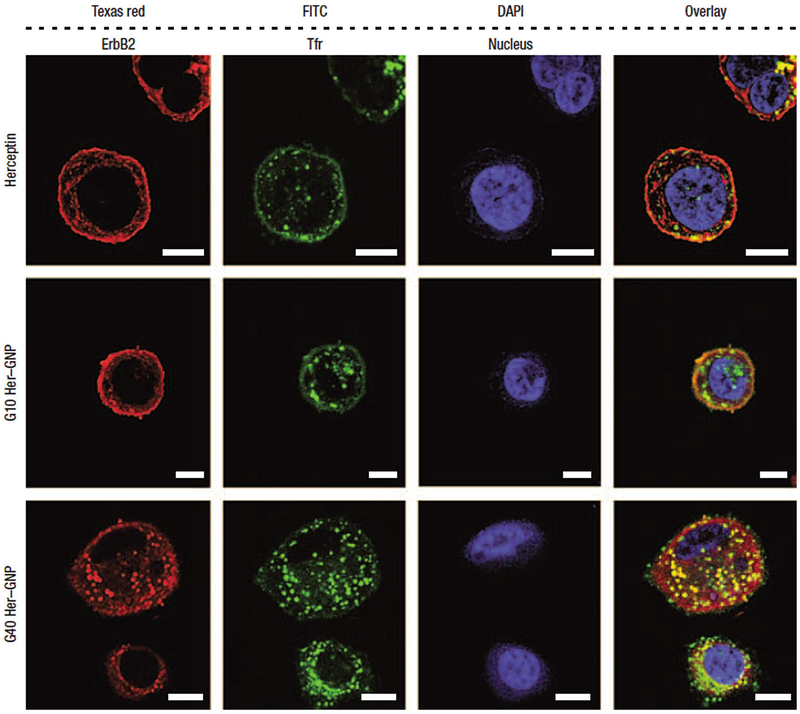
Alongside cellular internalization assays evaluating a nanomaterial’s proclivity to bypass the cell membrane, quantifying the material’s preference to localize in the cytosol or in the nucleus provides further understanding of the nanoparticle’s potential routes for therapy and toxicity. After passage into the cytosol, large molecule and particle migration into the nucleus is primarily regulated through active transport mechanisms involving the cytoskeletal motor proteins kinesin and dynein along cellular microtubules [54]. Kinesin-mediated transport exports agents from the centrosome and nucleus while dynein shuttles molecules in the opposite direction, ultimately facing the nuclear pore complex as a gateway into the nucleoplasm. Imaging and quantifying particles that pass the size restrictions of the nuclear envelope (≤40 nm) [54] are typically performed with electron microscopy and fluorometry to visualize nanoparticle populations in the nucleoplasm and surrounding cytosol. Images of Herceptin-loaded gold nanoparticles depositing in both locations depending upon the size of the nanocarrier are presented in Fig. 5. In the case of gold nanoparticles (which have undergone extensive exploration as radio-sensitizers), positioning within or outside the nucleus is deduced from TEM while uptake concentrations are determined with ICP-MS [46, 55]. Fluorescent tags for polymeric nanoparticles allow observation of their localization patterns using confocal fluorescence microscopy, epifluorescence microscopy, and CLSM [47, 48]. Beyond transient analysis of internalization behavior, real-time evaluation of nanoparticle transport into cells is available with fluorescence correlation spectroscopy (FCS), raster image correlation spectroscopy (RICS), and time-resolved confocal microscopy [56]. Each method offers insight into the pervasiveness of nanoparticles within the cellular interior and possible explanation for the cytotoxic or biocompatible nature of specific particles.
Fig. 5.
Herceptin transcytosis into human breast cancer SK-BR-3 cells treated for 3 h promoted by Herceptin-functionalized gold nanoparticles (Her-GNP). Stains from left to right: anti-ErbB2 (Texas red), anti-transferrin receptor (Tfr) antibodies (FITC), and nuclear counterstain (DAPI). Vehicles from top to bottom: free Herceptin, 10 nm Her-GNP, and 40 nm Her-GNP. Each stain was overlaid in the fourth panel. The Her-GNP vehicles display enhanced cytosolic uptake of Herceptin, and the smaller 10 nm Her-GNP carriers demonstrate greater nuclear internalization than their 40 nm counterparts. Scale bars are 10 μM. Reproduced from [20]
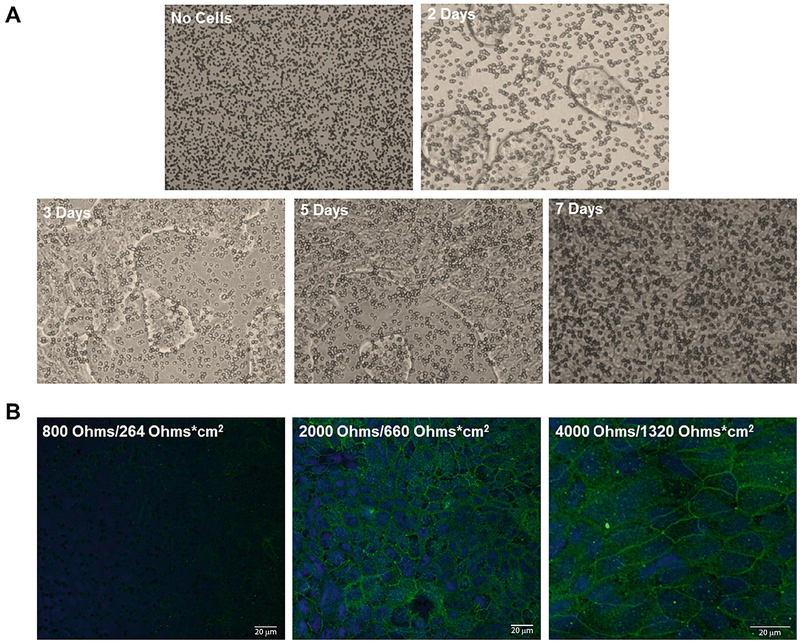
As a complement to the cellular internalization and distribution schemes offered by microscopic techniques, transcytosis assays identify the proficiency of particles to permeate through epithelial barriers like the lungs, gastrointestinal tract, and blood-brain barrier (BBB). Epithelial cells generally congregate into tight junctions once plated, and measurement of the transepithelial electrical resistance (TEER) across the junction denotes the integrity of the barrier as shown in Fig. 6. If nanoparticles permeate across the barrier and tear the junctions or if construction of the junction is otherwise impaired, the acquired TEER value is typically observed to decrease from the range of 500–2000 Ω/cm2 to less than 500 Ω/cm2 [57]. Isolating the apical and basolateral compartment concentrations of nanoparticles as a function of time following barrier exposure effectively relates the permeability of the epithelial membrane with respect to the tested nanoparticles [58], and incorporation of the particle translocation tendency with TEER results can decide whether the particles are constructed to innately bypass the junction or whether the transmission occurs as a result of nanoparticle-induced membrane rupture. Basolateral sampling with one of the aforementioned fluorescent stains and fluorometry methods [59] allows direct quantification of the permeated particle count and the opportunity for inline characterization. Altogether, data from TEER and migrated nanoparticle populations compose satisfactory reports for the particle’s transcytosis performance, and the assay’s coupling with other assays describing cellular cytotoxicity details the exposure hazard for both therapeutic and environmental nanoparticles.
Fig. 6.
Human lung epithelial Calu-3 cell confluence over a single week’s culture in 10% FBS imaged using an inverted microscope with 10× magnification (a). Monolayer formation hides Transwell insert pores throughout the culture duration as seen from the initial image at time zero (top left) to the completion of monolayer formation at the conclusion of a week (bottom right). Confocal microscopy images of ZO-1 (green) and DAPI (blue) stained junctions and nuclei, respectively, display monolayer formation as a function of increased TEER resistance (b). Reproduced from [57]. https:/creativecommons.org/licenses/by-nc-nd/4.0/
3.3. Viability Assays
Lactate dehydrogenase (LDH) is an enzyme produced by living cells to regulate pyruvate and lactate levels through nicotinamide adenine dinucleotide (NAD) oxidation. When cells undergo hemolysis or necrosis, LDH is released into the surrounding extracellular environment while maintaining enzymatic activity. Conversion of pyruvate to lactate follows NADH + Pyruvate ↔ NAD+ + Lactate in the presence of LDH where NADH and NAD+ represent the reduced and oxidized forms of NAD, respectively, and complementary reaction of NADH with tetrazolium salts permits spectroscopic monitoring of LDH enzymatic activity using known initial concentrations of lactate and NAD. The time-dependent change in spectroscopic absorbance of the reduced tetrazolium compound measured by an enzyme-linked immunosorbent assay (ELISA) reader or UV-Vis spectrometer is directly proportional to the sample LDH concentration [60] and, by inference, the extent of cellular trauma. Tetrazolium salts were originally investigated as nonradioactive replacements for the traditional radioactive chromium (51Cr) assay technique [61] and as methods to enhance the sensitivity of detecting NADH absorbance at 340 nm. Common tetrazolium salts include iodonitrotetrazolium (INT), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) which rival the throughput of the 51Cr assay and exhibit a threefold sensitivity increase over ultraviolet NADH assays [62, 63]. Numerous salt-based commercial kits like the Pierce™ LDH Cytotoxicity Assay Kit and CytoTox 96® Non-Radioactive Cytotoxicity Assay are available, and nonsalt fluorometric kits utilizing the reduction of resazurin (nonfluorescent) to resorufin (fluorescent) by NADH offer alternatives to colorimetric assays. Additionally, the luminogenic glycylphenylalanyl-aminofluorocoumarin (GF-AFC) and firefly luciferase ATP assays provide nontoxic and highly sensitive options for assessing cell viability [64]. Regardless of the assay employed, nanoparticles can interfere with viability assay results through several mechanisms: static adsorption of LDH and related proteins to the nanoparticle surface can denature a fixed amount of enzyme prior to surface saturation, dynamic adsorption can continuously inactive LDH throughout the duration of the assay, and ROS generation can prematurely oxidize NADH to skew absorbance values [65]. Careful sample preparation is necessary to minimize perturbations from reactive particles that can severely distort outcomes from in vitro LDH analysis.
3.4. Apoptosis Assays
Acceleration of apoptosis due to nanoparticle exposure can jeopardize cellular regeneration and hasten aging in mammalian hosts. Markers of programmed cell death include two major trackable factors for in vitro analysis: phosphatidylserine (PS) migration to the outer leaflet of the cell membrane and caspase activation into initiator and effector enzymes. Prior to apoptosis initiation, flippases hold PS exclusively on the cytosolic face of the asymmetric cell membrane; during the execution phase of apoptosis, scramblases transport PS from the inner leaflet to the exoplasmic face [66]. The loss of phospholipid asymmetry accompanying PS exchange maintains membrane integrity while signaling for macrophage consumption and platelet aggregation. To prevent rampant blood coagulation occurring from the natural cell cycle, calcium-mediated annexin V binds to exposed PS as a shield from coagulation cascades. Attaching hapten moieties (e.g., FITC) to annexin V permits apoptotic cell labeling, and secondary stains that identify cellular necrosis such as membrane impermeable propidium iodide (PI) combined with fluorescence microscopy or flow cytometry constitute the annexin V-affinity assay, which is capable of quantifying healthy (FITC and PI negative), apoptotic (FITC positive and PI negative), and dead (FITC and PI positive) cells simultaneously [67]. The annexin Vassay monitors a seemingly ubiquitous apoptotic event in PS translocation resulting from a number of causal factors [66], establishing the method as a reliable measure for apoptosis signaling. Caspase activation is, however, a precedent to PS transport and a complementary event for verification of premature apoptosis onset. Caspases represent a family of 14 cysteine proteases initially lodged as inactive zymogens (procaspases) within the cellular cytoplasm [68]. Following a host of stimuli-induced signaling pathways, procaspases are oligomerized from encoded prodomains containing large and small subunits through intrinsic or extrinsic pathways. Active caspases offer several assay analysis approaches, including: cleavable substrates with fluorogen and chromogen labels, immunoblotting, immunofluorescence, and affinity assays with attached reporter moieties. Readers are encouraged to refer to the informative review by Kaufmann et al. [69] for detailed explanations and methodologies on each procedure. None of the caspase assays offer definitive evidence for individual quantitative caspase activity, and multiple assays are required for full characterization. The multimodal capability of caspases to cleave their unpreferred substrates hinders isolation with labels; immunoblots are not quantitative and cannot discern non-cleavage-based enzymatic activity; immunofluorescence necessitates specific conformational antibodies with the potential for cross-reactivity; and affinity labels often use expensive reagents that are susceptible to nucleophilic competition [69]. Nonetheless, the caspase assays provide a useful technique for identifying cellular apoptosis signals preceding PS translocation.
3.5. Oxidative Stress/Inflammation Assays
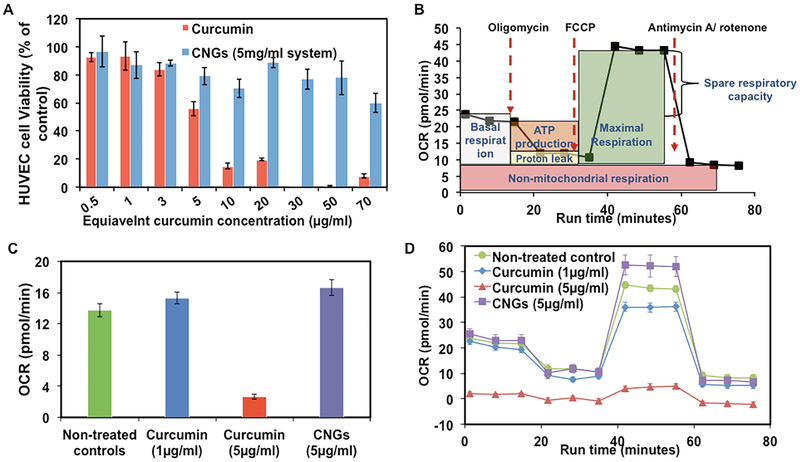
The enhanced reactivity of nanoparticles imposed by their high surface area to volume ratio boosts cellular oxidative stress and promotes intracellular damage. Detection of oxidation events through direct measurement of ROS, indicators of oxidative damage such as protein carbonyl content and genotoxicity, and inflammatory markers reveals the in vitro potential of nanoparticles to produce detrimental cellular oxidants. Generation of ROS within the cytoplasm beyond natural levels is quantifiable via introduction of ROS sensitive dyes such as the nonionic, nonpolar, membrane-permeable fluorophore 2′,7′-dichlorofluorescein diacetate (DCFH-DA) as described by Keston and Brandt [70]. Upon cellular internalization of nonfluorescent DCFH-DA, the fluorophore is enzymatically hydrolyzed by cytosolic esterases into its nonfluorescent polar analog dichlorofluorescein (DCFH), whereby the analog becomes trapped within the cytosol. Hydroxyl radicals and comparable cellular ROS oxidize DCFH into highly fluorescent dichlorofluorescein (DCF) which is monitored using fluorescence microscopy or flow cytometry [71]. Albeit relatively easy to implement and quantitative, the DCFH-DA method for evaluating ROS content is susceptible to inaccuracy due to nonspecific enzymatic oxidation and photooxidation [71, 72]. Catalase offers possible inhibition for enzymatic oxidation, but the results from DCF assays must, nevertheless, be interpreted with caution to avoid overestimating nanoparticles’ ROS generation capability. In a comparable method for analyzing mitochondrial respiration, commercial Seahorse Extracellular Flux (XF) Analyzers measure the oxygen consumption rate (OCR) and energy production potential of cells subjected to chemical inhibitors as markers of mitochondrial oxidative stress. Checking the OCR following incubation with antioxidants and subsequent oxidizers tracks oxidation-induced mitochondrial damage inhibition capacity, and the offset in OCR decline observed by the introduction of a therapeutic agent can signify the efficacy of a treatment [73]. Post-analysis treatment with cell viability kits can verify ROS mitigation and the inherent safety of proposed treatments as in the case of curcumin versus curcumin nanogels displayed in Fig. 7.
Fig. 7.
Cytotoxicity of curcumin and curcumin nanogels (CNGs) for human umbilical vein endothelial cells (HUVECs) following 24 h of treatment at specified concentrations. (a) Cell viability results using calcein AM red-orange live cell tracer assay. (b) Seahorse XF96 calculated OCR for untreated cells undergoing a typical mitochondrial stress assay under the conditions shown. (c) Endpoint basal OCR rates for select samples from a. (d) Mitochondrial stress assay profiles for curcumin and CNG treatments following inhibitor additions as described in b. Reproduced from [73]
The carbonyl content of proteins increases in response to oxidation from ROS and provides a general indicator of oxidative damage. Several approaches are available for measuring protein carbonyl levels, but two are particularly notable: tritiated borohydride and 2,4-dinitrophenylhydrazine (DNPH). Borohydride reduces protein carbonyls to alcohols with stable tritium labels detectable by spectrophotometric absorbance at 340 nm [74]. As with other carbonyl reagents, extraneous nucleic acids necessitate removal prior to labeling; streptomycin treatment is typically recommended as a pretreatment to precipitate nucleic acids [75]. Employing DNPH as the carbonyl reagent offers a nonradiochemical labeling method, but larger quantities of sampled protein are required for successful analysis. Reaction of the protein carbonyl group with DNPH generates a stable 2,4-dinitrophenyl (DNP) hydrazone product that is detected from its maximum absorbance between 360 and 390 nm and translated to a carbonyl content value using a molar absorption coefficient of 22,000 M−1 cm−1 [74]. Both assays produce identical values for the evaluated carbonyl content assuming protein-bound chromophores with absorbances in the range of the applied reagent are subtracted via a blank. The sensitivity and specificity of the DNPH assay is further improved while eliminating the expenditure of excess sample by applying high-performance liquid chromatography (HPLC) or Western blotting with a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) system [76]. Immunoassays in the form of ELISA and slot blotting can likewise be applied as highly sensitive techniques for analyzing carbonyl content, but the approaches are less common than the standard DNPH and borohydride methods [75].
Genotoxicity assays survey DNA and chromosomal damage and gene mutations occurring as a consequence of toxin-induced oxidative stress. The comet, micronucleus, Ames, and chromosome aberration assays are comprehensively reviewed by Golbamaki et al. [77] for metal oxide and silica nanomaterials and will be related for their general use herein. The single-cell gel electrophoresis, or comet, assay is a routine method for quantifying DNA breaks using fluorescence microscopy. Under alkaline electrophoresis conditions, supercoiled loops of DNA straighten into tails resembling comets, and the comet’s head-to-tail distance reveals the number of DNA breaks in the sample. The Olive tail moment (OTM), first described by Olive et al. [78], relates the amount of damage in a standard format by reporting the comet tail’s length multiplied by its DNA encapsulation percentage. Micronucleus (MN) assays regularly utilize cytochalasin B under the direction of the Organisation for Economic Co-operation and Development (OECD) Test Guideline no. 487 to test chromosomal breakage by preventing daughter cell separation following mitosis [79]. Visualization of binucleate cells using fluorescent stains like acridine orange permits quantification of micronucleus frequency among treated mitotic cell populations [80]. The Ames test uses Salmonella (S. typhimurium) and Escherichia coli (E. coli) bacterial strains to test for amino acid production among genetically defunct bacteria as prescribed by OECD Test Guideline no. 471 [81]. In the test, mutated bacteria that are incapable of producing an essential amino acid are incubated with the tested substance and the growth of revertant bacteria that regain the ability to synthesize the amino acid is observed. Implementation is rapid and facile but often considered inappropriate for nanoparticles due to the materials’ meager uptake by bacterial cells [77]. The chromosome aberration test monitors chromosomal repair process malfunctions by halting mammalian cell cycles using a metaphase-arresting substance as per OECD Test Guideline no. 473 and registering metaphase chromosome aberrations microscopically [82]. The test is limited to clastogen identification and is, therefore, considered inferior to MN assays that also allow for aneugen detection [83].
Lipid peroxidation presents another quantifiable effect of ROS overproduction through evaluation of two major secondary peroxidation products in malondialdehyde (MDA) and 4-hydroxyl-2-nonenal (4-HNE). The more mutagenic of the products, MDA, arises from the decomposition of large polyunsaturated fatty acids (PUFAs) and the metabolism of arachidonic acid (AA) during thromboxane A2 synthesis [84]. Reaction of the by-product with thiobarbituric acid (TBA) at pH 3.5 forms a MDA-TBA adduct that is detected fluorescently at 553 nm with an excitation of 515 nm or spectrophotometrically at 532 nm as part of the thiobarbituric acid reactive substances (TBARS) assay [85]. The assay has received intense scrutiny for its lack of specificity; TBA reacts with a number of substances other than MDA ranging from oxidized lipids to urea, altogether producing drastic overestimations of ROS lipid peroxidation. Efforts to raise the specificity of the assay include using HPLC to isolate the desired MDA-TBA adduct and eliminate background signals from conflicting reactive species [86]. The more cytotoxic lipid peroxidation product, 4-HNE, occurs as a by-product of AA decomposition and enzymatic and nonenzymatic PUFA peroxidation. The compound’s high reactivity toward primary amines to form Schiff bases and thiol or amino compounds to make Michael adducts [87] enables detection using HNE-protein adduct ELISA assays [88]. In samples containing both MDA and 4-HNE, the TBA assay approach described for MDA analysis normally assembles fluorescent adducts for both peroxidation markers due to the assay’s nonspecific nature, but substitution of TBA with 1-methyl-2-phenylindole in a hydrochloric acid reaction medium has been shown to promote MDA adduct yields over 4-HNE [89]. Despite its misgivings, the TBARS assay remains the predominant method for determining toxin lipid peroxidation and a viable approach for nanoparticles’ peroxidation proclivity.
Inflammatory response markers include numerous families of cytokines activated by the immune system to combat hazardous invaders like reactive nanoparticles. One family in particular, interleukins, hosts 43 cytokines expressed by proinflammatory and antiinflammatory macrophages that promote T-helper type 1 (Th1) and T-helper type 2 (Th2) cell responses, respectively [90]. Activity by Th1 and Th2 cells delivers tumor necrosis factor alpha (TNF-α) and interleukin-10 (IL-10) into the extracellular milieu whereby the concentrations are readily measured using ELISA [90] to gauge inflammatory responses occurring from treating macrophages with nanoparticles. As a precursor to interleukin upregulation, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activity controls the cytokine production pathway and presents a foundational inflammation test using electrophoretic mobility shift assays (EMSAs). Binding of NF-κB to DNA response elements forms a bulky complex which, when exposed to an electrophoretic gel, inhibits migration and provides a gap between complexes that is frequently visualized with 32P radiolabeling [91, 92]. Confirmation of NF-κB activity offers a qualitative or, at best, semiquantitative signal of proinflammation. As a quantitative alternative, firefly-derived luciferase assays, which use expression vectors cloned upstream of the Photinus pyralis luciferase gene, encode enzymes capable of measuring the genetic expression of inflammatory response markers such as the nuclear factor for interleukin-6 (NF-IL6) [93]. Following conjugation of clones containing the regulatory region of the inflammatory marker with the interleukin gene-of-interest during cell incubation, the produced luciferase reporter enzymes transiently oxidize injected luciferin into oxyluciferin which is quantified using a luminometer. Adding coenzyme A to the post-incubation luciferin spike greatly enhances the assay’s sensitivity by augmenting and sustaining the emitted luminescence [94]. Applying quantitative luciferase assays in combination with inflammatory response and ROS assays contributes vital information for characterizing the oxidative and inflammatory potential of any nanoparticle species.
3.6. Endotoxin Assays
A common challenge for nanoparticles arises from their ability to absorb ambient contaminants to their surface, which can result in an enhanced inflammatory response over what would be expected if the absorbed chemicals were simply free in solution. This is most commonly seen in the case of endotoxins that cause acute inflammatory responses in humans. Endotoxins, or lipopolysaccharides (LPS), are environmentally prevalent pyrogens found in the outer cell wall of gram-negative bacteria that elicit inflammatory cytokines following activation of the coagulation signaling cascade in mammalian systems. The particular cascade sequence discovered for the analogous Atlantic horseshoe crab (Limulus polyphemus) by Bang [95] capitalizes upon the crabs’ lethal intravascular coagulation when exposed to Vibrio endotoxin, and the reaction was developed into a routine series of in vitro assay procedures identified under the umbrella of the Limulus amebocyte lysate (LAL) assay using the blood cells (amebocytes) of the Limulus polyphemus or the homologous Japanese horseshoe crab (Tachypleus tridentatus). Three main assays stem from the LAL approach: the gel clot assay, the coagulogen-based (turbidity) assay, and the chromogenic assay [96]. The gel clot method employs endotoxin-activated enzymatic coagulogen cleavage by combining a portion of LAL solution with the endotoxin sample solution and checking for clotting following sufficient incubation. The technique is somewhat subjective due to confirmation arising from a simple positive or negative tube inversion to visualize clot formation, and quantitation requires inference from serial dilutions. Coagulogen-based assays vary in their quantitation by examining either changes in turbidity, coagulogen reduction during clotting, or peptide fragmentation [96]. Chromogenic approaches substitute chromogens for coagulogen that release chromophores when cleaved. Traditionally, para-nitroaniline is attached to an amino acid sequence resembling the clotting enzyme cleavage site in LAL and colors the solution upon liberation. Absorbance at 405 nm indicates the available concentration of clotting enzyme, which, in turn, reveals the concentration of endotoxin [96, 97]. Chromogenic LAL assays are included in commercial kits such as the Endosafe®-PTS (Portable Test System) for routine endotoxin analysis. In an alternative approach to using the blood of horseshoe crabs for assays, recombinant Factor C (rFC), the priming agent for coagulation, has developed into a standard reagent for endotoxin detection as a method for improving the sensitivity of LAL assays and sparing horseshoe crabs from endangerment [98]. Isolation and implementation of endotoxin-sensitive rFC eliminates possible false-positive reads from β-glucan contamination in LAL assays by eliminating glucan-reactive factor G that acts as a secondary clotting cascade pathway [99]. Additional methodologies have explored using immunoassays and rabbit pyrogen as alternatives to the popular LAL and rFC assays, but insensitivity and difficulty quantifying biological activity have curtailed their use in comparison to bioassays. Notably, evidence has demonstrated that nanoparticles can perturb LAL assay approaches and disrupt their accuracy [97], which has generated interest in identifying alternative routes for endotoxin detection to accommodate nanoparticles. Advances include using toll-like receptor 4 (TLR4) as a reporter protein, and the technology has made its way into commercial kits like the HEK-Blue™ detection system [97].
From the assays reviewed in the previous sections, the toxins and biomarkers corresponding to each assay are summarized in Table 1.
Table 1.
Summary of toxins or injuries surveyed by each in vitro assay, their corresponding biomarkers, and select references applying the assays
| Toxin/injury | Assays | Biomarkers | References |
|---|---|---|---|
| Barrier degradation | Cellular internalization, cytosol/nuclear localization, TEER | Epithelial barrier damage, pervasive nanoparticle cellular internalization | [23, 45-49, 51-53, 55-57, 59] |
| Hemolysis or necrosis | INT, MTT, resazurin, NADH, GF-AFC, firefly luciferase ATP | Extracellular LDH | [60, 62-65] |
| Apoptosis | Annexin V, fluorogenic and chromogenic caspase, caspase immunoblotting, caspase immunofluorescence, caspase affinity | PS transport across to the outer leaflet of the cell membrane, procaspase oligomerization | [67, 69] |
| Hydroxyl radicals (ROS) | DCF, Seahorse XF Analyzers Tritiated borohydride, DNPH Comet, micronucleus, Ames, chromosome aberration |
Heightened intracellular ROS Heightened protein carbonyl content DNA/chromosome damage, gene mutations |
[70-73] [74, 76] [78-83] |
| Lipid peroxidation | TBARS, HNE-protein ELISA | MDA, 4-HNE | [85, 86, 88, 89] |
| Inflammation | ELISA, EMSA luciferase | Interleukin and nuclear factor release | [90-94] |
| Endotoxin/LPS | Gel clot LAL, coagulogen-based LAL, chromogenic LAL, rFC, rabbit pyrogen, TLR4 | Release of inflammatory cytokines IL-1β, TNF-α, and IL-6 following CD14 binding and MD2/TLR4 association | [96, 97, 99] |
4. Conclusions/Outlook
Despite similarities in size, nanoparticles threaten multimodal forms of toxicity to unfortunate hosts through numerous mechanisms. Nanomaterial toxicity occurs due to the physicochemical properties of the material determining its proclivity to interact with the various proteins and cells comprising the biological milieu. Catalyzing ROS generation forms one of the most potent contributors to nanoparticles’ toxicological components and sparks cascades of oxidative stress and inflammatory signals that ultimately lead to necrosis, expedited apoptosis, or carcinogenesis. The roots of the disastrous cascades are uncovered by observing the interplay between the particles’ size and morphology and their chemical composition and apparent surface charge. Evaluation of each property is required to fully understand the toxicity profile of the queried material, and, even with exhaustive characterization, causes for toxicity can be shrouded by the complexity of biological systems. No individual gold standard exists for foreshadowing the toxicity of a unique nanomaterial; rather, combinations of several techniques are necessary to adequately describe the material’s toxicological profile. Albeit ubiquitous, sizing from DLS does not provide an insurmountable threshold to bar cellular infiltration and epithelial barrier penetration, and ZP relates only the particle’s apparent surface charge. Deduction of cellular uptake, protein corona formation, catalysis potential, and protein denaturation proficiency from routine nanoparticle analysis is, at best, inferential and necessitates in vitro study for verification.
Akin to the limitations plaguing characterization of physiochemical properties, in vitro assays also lack a one-size-fits-all method for exposing evidence regarding the toxicity or biocompatibility of tested nanoparticles. The myriad of cell types and assays impose a formidable array of tests for identifying individual markers of toxicity, but none of the studies provides an ideal pass-or-fail qualification. Multiple cell types should ideally be screened to replicate the heterogeneity of in vivo conditions, and singular signs of apparent inertness do not exclude alternative toxicological mechanisms from remaining in play. Combinations of multiple assays are, therefore, needed to sufficiently elucidate physiological responses to the nanoparticle system in question. Issues still persist for several of the established in vitro assays, namely: a method for the parallel high-resolution imaging and precise quantification of nanoparticle internalization beyond the sacrifices present for IFC has yet to be drawn into a standalone instrument; protein-based catalysis assays frequently assume a risk of cross-reactivity with secondary analytes that can mar data from the assays; and the enhanced reactivity of nanoparticles can preemptively oxidize analytical reagents or denature proteins that constitute the basis of numerous in vitro approaches. Resolution to the analytical and application-based problems involved with in vitro assays, particularly those associated with nanoparticulate systems, embodies the focus of future endeavors into nanoparticle toxicity analysis.
References
- 1.Salata OV (2004) Applications of nanoparticles in biology and medicine. J Nanobiotechnol 2:3–3. 10.1186/1477-3155-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eifler AC, Thaxton CS (2011) Nanoparticle Therapeutics: FDA Approval, Clinical Trials, Regulatory Pathways, and Case Study In: Hurst SJ (ed) Biomedical Nanotechnology: Methods and Protocols. Humana, Totowa, NJ, pp 325–338. 10.1007/978-1-61779-052-2_21 [DOI] [PubMed] [Google Scholar]
- 3.Priyadarsini S, Mukherjee S, Mishra M (2018) Nanoparticles used in dentistry: a review. J Oral Biol Craniofac Res 8(1):58–67. 10.1016/j.jobcr.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copéret C, Héroguel F (2017) Recent advances in surface organometallic chemistry In: Cornils B, Herrmann WA, Beller M, Paciello R (eds) Applied homogeneous catalysis with organometallic compounds. Wiley-VCH Verlag GmbH & Co, Weinheim, pp 1069–1084. 10.1002/9783527651733.ch15 [DOI] [Google Scholar]
- 5.Yan N, Xiao C, Kou Y (2010) Transition metal nanoparticle catalysis in green solvents. Coord Chem Rev 254(9):1179–1218. 10.1016/j.ccr.2010.02.015 [DOI] [Google Scholar]
- 6.Weiss J, Takhistov P, McClements DJ (2006) Functional materials in food nanotechnology. J Food Sci 71(9):R107–R116. 10.1111/j.1750-3841.2006.00195.x [DOI] [Google Scholar]
- 7.Contado C (2015) Nanomaterials in consumer products: a challenging analytical problem. Front Chem 3:48 10.3389/fchem.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raj S, Jose S, Sumod US, Sabitha M (2012) Nanotechnology in cosmetics: opportunities and challenges. J Pharm Bioallied Sci 4 (3) :186–193. 10.4103/0975-7406.99016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boxberg F, Tulkki J (2004) Quantum dots: phenomenology, photonic and electronic properties, modeling and technology the handbook of nanotechnology. Nanometer structures: theory, modeling, and simulation. SPIE Press, Bellingham: 10.1117/3.537698.ch4 [DOI] [Google Scholar]
- 10.Akbarzadeh A, Samiei M, Davaran S (2012) Magnetic nanoparticles: preparation, physical properties, and applications in biomedicine. Nanoscale Res Lett 7(1):144–144. 10.1186/1556-276X-7-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smulders S, Luyts K, Brabants G, Landuyt KV, Kirschhock C, Smolders E, Golanski L, Vanoirbeek J, Hoet PHM (2014) Toxicity of nanoparticles embedded in paints compared with pristine nanoparticles in mice. Toxicol Sci 141(1):132–140. 10.1093/toxsci/kfu112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rai M, Yadav A, Gade A (2009) Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv 27(1):76–83. 10.1016/j.biotechadv.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 13.Sharifi S, Behzadi S, Laurent S, Forrest ML, Stroeve P, Mahmoudi M (2012) Toxicity of nanomaterials. Chem Soc Rev 41 (6):2323–2343. 10.1039/c1cs15188f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbo C, Molinaro R, Parodi A, Furman NET, Salvatore F, Tasciotti E (2016) The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 11(1):81–100. 10.2217/nnm.15.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar V, Sharma N, Maitra SS (2017) In vitro and in vivo toxicity assessment of nanoparticles. Int Nano Lett 7(4):243–256. 10.1007/s40089-017-0221-3 [DOI] [Google Scholar]
- 16.Clichici S, Filip A (2015) In vivo assessment of nanomaterials toxicity. Nanomaters toxicity and risk assessment. 10.5772/60707 [DOI] [Google Scholar]
- 17.Vasilakes AL, Dziubla TD, Wattamwar PP (2013) Polymeric nanoparticles In: Bader RA (ed) Engineering polymer systems for improved drug delivery. John Wiley & Sons, Inc., pp 117–161. 10.1002/9781118747896.ch5 [DOI] [Google Scholar]
- 18.Cabral H, Matsumoto Y, Mizuno K, Chen Q, Murakami M, Kimura M, Terada Y, Kano MR, Miyazono K, Uesaka M, Nishiyama N, Kataoka K (2011) Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat Nanotechnol 6:815 10.1038/nnano.2011.166 https://www.nature.com/articles/nna-no.2011.166#supplementary-information [DOI] [PubMed] [Google Scholar]
- 19.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WCW (2009) Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett 9(5):1909–1915. 10.1021/nl900031y [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Kim BYS, Rutka JT, Chan WCW (2008) Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol 3:145 10.1038/nnano.2008.30 https://www.nature.com/articles/nnano.2008.30#supplementary-information [DOI] [PubMed] [Google Scholar]
- 21.Jain RK, Stylianopoulos T (2010) Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol 7:653 10.1038/nrclinonc.2010.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldorera-Moore M, Guimard N, Shi L, Roy K (2010) Designer nanoparticles: incorporating size, shape and triggered release into nanoscale drug carriers. Expert Opin Drug Deliv 7 (4) :479–495. 10.1517/17425240903579971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM (2008) The effect of particle design on cellular internalization pathways. Proc Natl Acad Sci 105 (33):11613–11618. 10.1073/pnas.0801763105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo S, Jiang Y, Gupta A, Jiang Z, Landis RF, Hou S, Liang XJ, Rotello VM (2016) Fully zwitterionic nanoparticle antimicrobial agents through tuning of core size and ligand structure. ACS Nano 10(9):8732–8737. 10.1021/acsnano.6b04207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischer CC, Payne CK (2014) Nanoparticle–cell interactions: molecular structure of the protein corona and cellular outcomes. Acc Chem Res 47(8):2651–2659. 10.1021/ar500190q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vroman L, Adams AL (1969) Identification of rapid changes at plasma–solid interfaces. J Biomed Mater Res 3(1):43–67. 10.1002/jbm.820030106 [DOI] [PubMed] [Google Scholar]
- 27.Owens DE, Peppas NA (2006) Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm 307 (1):93–102. 10.1016/j.ijpharm.2005.10.010 [DOI] [PubMed] [Google Scholar]
- 28.Li S-D, Huang L (2010) Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. J Control Release 145 (3) :178–181. 10.1016/j.jconrel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry JL, Reuter KG, Kai MP, Herlihy KP, Jones SW, Luft JC, Napier M, Bear JE, DeSimone JM (2012) PEGylated PRINT nanoparticles: the impact of peg density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano Lett 12 (10):5304–5310. 10.1021/nl302638g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cochran DB, Wattamwar PP, Wydra R, Hilt JZ, Anderson KW, Eitel RE, Dziubla TD (2013) Suppressing iron oxide nanoparticle toxicity by vascular targeted antioxidant polymer nanoparticles. Biomaterials 34 (37):9615–9622. 10.1016/j.biomaterials.2013.08.025 [DOI] [PubMed] [Google Scholar]
- 31.Fan M, Zeng Y, Ruan H, Zhang Z, Gong T, Sun X (2017) Ternary nanoparticles with a sheddable shell efficiently deliver microRNA-34a against CD44-positive melanoma. Mol Pharm 14(9):3152–3163. 10.1021/acs.molpharmaceut.7b00377 [DOI] [PubMed] [Google Scholar]
- 32.Bahadar H, Maqbool F, Niaz K, Abdollahi M (2016) Toxicity of nanoparticles and an overview of current experimental models. Iran Biomed J 20(1):1–11. 10.7508/ibj.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han SG, Newsome B, Hennig B (2013) Titanium dioxide nanoparticles increase inflammatory responses in vascular endothelial cells. Toxicology 306:1–8. 10.1016/j.tox.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donaldson K, Stone V (2003) Current hypotheses on the mechanisms of toxicity of ultrafine particles. Ann Ist Super Sanita 39 (3):405–410 [PubMed] [Google Scholar]
- 35.Napierska D, Thomassen LC, Lison D, Martens JA, Hoet PH (2010) The nanosilica hazard: another variable entity. Part Fibre Toxicol 7(1):39 10.1186/1743-8977-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharjee S (2016) DLS and zeta potential – what they are and what they are not? J Control Release 235:337–351. 10.1016/j.jconrel.2016.06.017 [DOI] [PubMed] [Google Scholar]
- 37.Lim J, Yeap SP, Che HX, Low SC (2013) Characterization of magnetic nanoparticle by dynamic light scattering. Nanoscale Res Lett 8 (1):381–381. 10.1186/1556-276X-8-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pecora R (1968) Spectrum of light scattered from optically anisotropic macromolecules. J Chem Phys 49(3):1036–1043. 10.1063/1.1670189 [DOI] [Google Scholar]
- 39.Rodríguez-Fernández J, Pérez-Juste J, Liz--Marzán LM, Lang PR (2007) Dynamic light scattering of short Au rods with low aspect ratios. J Phys Chem C 111(13):5020–5025. 10.1021/jp067049x [DOI] [Google Scholar]
- 40.Kaszuba M, Corbett J, Watson FM, Jones A (2010) High-concentration zeta potential measurements using light-scattering techniques. Philos Transact A Math Phys Eng Sci 368(1927):4439–4451. 10.1098/rsta.2010.0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown MA, Goel A, Abbas Z (2016) Effect of electrolyte concentration on the stern layer thickness at a charged interface. Angew Chem Int Ed 55(11):3790–3794. 10.1002/anie.201512025 [DOI] [PubMed] [Google Scholar]
- 42.Pfeiffer C, Rehbock C, Hühn D, Carrillo-Carrion C, de Aberasturi DJ, Merk V, Barcikowski S, Parak WJ (2014) Interaction of colloidal nanoparticles with their local environment: the (ionic) nanoenvironment around nanoparticles is different from bulk and determines the physico-chemical properties of the nanoparticles. J R Soc Interface 11 (96):20130931 10.1098/rsif.2013.0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clogston JD, Patri AK (2011) Zeta potential measurement In: McNeil SE (ed) Characterization of nanoparticles intended for drug delivery. Humana Press, Totowa, NJ, pp 63–70. 10.1007/978-1-60327-198-1_6 [DOI] [Google Scholar]
- 44.Lin P-C, Lin S, Wang PC, Sridhar R (2014) Techniques for physicochemical characterization of nanomaterials. Biotechnol Adv 32 (4) :711–726. 10.1016/j.biotechadv.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muro S, Garnacho C, Champion JA, Leferovich J, Gajewski C, Schuchman EH, Mitragotri S, Muzykantov VR (2008) Control of endothelial targeting and intracellular delivery of therapeutic enzymes by modulating the size and shape of ICAM-1-targeted carriers. Mol Ther 16(8):1450–1458. 10.1038/mt.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lechtman E, Pignol JP (2017) Interplay between the gold nanoparticle sub-cellular localization, size, and the photon energy for radiosensitization. Sci Rep 7(1):13268 10.1038/s41598-017-13736-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh L, Parboosing R, Kruger HG, Maguire GEM, Govender T (2016) Intracellular localization of gold nanoparticles with targeted delivery in MT-4 lymphocytes. Adv Nat Sci Nanosci Nanotechnol 7(4). 10.1088/2043-6262/7/4/045013 [DOI] [Google Scholar]
- 48.Tammam SN, Azzazy HM, Breitinger HG, Lamprecht A (2015) Chitosan nanoparticles for nuclear targeting: the effect of nanoparticle size and nuclear localization sequence density. Mol Pharm 12(12):4277–4289. 10.1021/acs.molpharmaceut.5b00478 [DOI] [PubMed] [Google Scholar]
- 49.Hewitt RE, Vis B, Pele LC, Faria N, Powell JJ (2017) Imaging flow cytometry assays for quantifying pigment grade titanium dioxide particle internalization and interactions with immune cells in whole blood. Cytometry A 91(10):1009–1020. 10.1002/cyto.a.23245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrowski A, Nordmeyer D, Boreham A, Holzhausen C, Mundhenk L, Graf C, Meinke MC, Vogt A, Hadam S, Lademann J, Ruhl E, Alexiev U, Gruber AD (2015) Overview about the localization of nanoparticles in tissue and cellular context by different imaging techniques. Beilstein J Nanotechnol 6:263–280. 10.3762/bjnano.6.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao D, Oh ZG, Chen Z (2016) Characterization of silver nanoparticles internalized by Arabidopsis plants using single particle ICP-MS analysis. Front Plant Sci 7:32 10.3389/fpls.2016.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H-H, Chien C-C, Petibois C, Wang C-L, Chu YS, Lai S-F, Hua T-E, Chen Y-Y, Cai X, Kempson IM, Hwu Y, Margaritondo G (2011) Quantitative analysis of nanoparticle internalization in mammalian cells by high resolution X-ray microscopy. J Nanobiotechnol 9(1):14 10.1186/1477-3155-9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gottstein C, Wu G, Wong BJ, Zasadzinski JA (2013) Precise quantification of nanoparticle internalization. ACS Nano 7(6):4933–4945. 10.1021/nn400243d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen O, Granek R (2014) Nucleus-targeted drug delivery: theoretical optimization of nanoparticles decoration for enhanced intracellular active transport. Nano Lett 14 (5) :2515–2521. 10.1021/nl500248q [DOI] [PubMed] [Google Scholar]
- 55.McNamara AL, Kam WW, Scales N, McMahon SJ, Bennett JW, Byrne HL, Schuemann J, Paganetti H, Banati R, Kuncic Z (2016) Dose enhancement effects to the nucleus and mitochondria from gold nanoparticles in the cytosol. Phys Med Biol 61(16):5993–6010. 10.1088/0031-9155/61/16/5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemmerich PH, von Mikecz AH (2013) Defining the subcellular interface of nanoparticles by live-cell imaging. PLoS One 8(4): e62018. 10.1371/journal.pone.0062018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Derk R, Davidson DC, Manke A, Stueckle TA, Rojanasakul Y, Wang L (2015) Potential in vitro model for testing the effect of exposure to nanoparticles on the lung alveolar epithelial barrier. Sens Biosensing Res 3:38–45. 10.1016/j.sbsr.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ (2015) TEER measurement techniques for in vitro barrier model systems. J Lab Autom 20(2):107–126. 10.1177/2211068214561025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bannunah AM, Vllasaliu D, Lord J, Stolnik S (2014) Mechanisms of nanoparticle internalization and transport across an intestinal epithetlial cell model: effect of size and surface charge. Mol Pharm 11(12):4363–4373. 10.1021/mp500439c [DOI] [PubMed] [Google Scholar]
- 60.Ponsoda X, Jover R, Castell JV, Gómez-Lechón MJ (1991) Measurement of intracellular LDH activity in 96-well cultures: a rapid and automated assay for cytotoxicity studies. J Tissue Cult Methods 13(1):21–24. 10.1007/bf02388199 [DOI] [Google Scholar]
- 61.Brunner KT, Mauel J, Cerottini JC, Chapuis B (1968) Quantitative assay of the lytic action of immune lymphoid cells of (51)Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 14 (2):181–196 [PMC free article] [PubMed] [Google Scholar]
- 62.Babson AL, Babson SR (1973) Kinetic colorimetric measurement of serum lactate dehydrogenase activity. Clin Chem 19(7):766. [PubMed] [Google Scholar]
- 63.Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65(1):55–63. 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- 64.Riss T, Moravec R, Niles A, Duellman S, Benink H, Worzella T, Minor L (2013, Updated 1 Jul 2016) Cell viability assays. [Internet] [Google Scholar]
- 65.Han X, Gelein R, Corson N, Wade-Mercer P, Jiang J, Biswas P, Finkelstein JN, Elder A, Oberdorster G (2011) Validation of an LDH assay for assessing nanoparticle toxicity. Toxicology 287(1–3):99–104. 10.1016/j.tox.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Engeland M, Nieland LJW, Ramaekers FCS, Schutte B, Reutelingsperger CPM (1998) Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry 31(1):1–9. [DOI] [PubMed] [Google Scholar]
- 67.Koopman G, Reutelingsperger C, Kuijten G, Keehnen R, Pals S, van Oers M (1994) Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84(5):1415–1420 [PubMed] [Google Scholar]
- 68.Fan T-J, Han L-H, Cong R-S, Liang J (2005) Caspase family proteases and apoptosis. Acta Biochim Biophys Sin 37(11):719–727. 10.1111/j.1745-7270.2005.00108.x [DOI] [PubMed] [Google Scholar]
- 69.Kaufmann SH, Lee SH, Meng XW, Loegering DA, Kottke TJ, Henzing AJ, Ruchaud S, Samejima K, Earnshaw WC (2008) Apoptosis-associated caspase activation assays. Methods 44(3):262–272. 10.1016/j.ymeth.2007.11.005 [DOI] [PubMed] [Google Scholar]
- 70.Keston AS, Brandt R (1965) The fluorometric analysis of ultramicro quantities of hydrogen peroxide. Anal Biochem 11(1):1–5. 10.1016/0003-2697(65)90034-5 [DOI] [PubMed] [Google Scholar]
- 71.Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds mC, Thomas M (1983) Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J Immunol 130(4):1910–1917 [PubMed] [Google Scholar]
- 72.Wang H, Joseph JA (1999) Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader11Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee by the United States Department of Agriculture and does not imply its approval to the exclusion of other products that may be suitable. Free Radic Biol Med 27(5):612–616. 10.1016/S0891-5849(99)00107-0 [DOI] [PubMed] [Google Scholar]
- 73.Gupta P, Jordan CT, Mitov MI, Butterfield DA, Hilt JZ, Dziubla TD (2016) Controlled curcumin release via conjugation into PBAE nanogels enhances mitochondrial protection against oxidative stress. Int J Pharm 511 (2):1012–1021. 10.1016/j.ijpharm.2016.07.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz A-G, Ahn B-W, Shaltiel S, Stadtman ER (1990) Determination of carbonyl content in oxidatively modified proteins In: Abelson JN (ed) Methods in enzymology, vol 186 Academic, Cambridge, MA, pp 464–478. 10.1016/0076-6879(90)86141-H [DOI] [PubMed] [Google Scholar]
- 75.Dalle-Donne I, Rossi R, Giustarini D, Milzani A, Colombo R (2003) Protein carbonyl groups as biomarkers of oxidative stress. Clin Chim Acta 329(1–2):23–38. 10.1016/s0009-8981(03)00003-2 [DOI] [PubMed] [Google Scholar]
- 76.Levine RL, Williams JA, Stadtman EP, Shacter E (1994) Carbonyl assays for determination of oxidatively modified proteins In: Abelson JN (ed) Methods in enzymology, vol 233 Academic, Cambridge, MA, pp 346–357. 10.1016/S0076-6879(94)33040-9 [DOI] [PubMed] [Google Scholar]
- 77.Golbamaki N, Rasulev B, Cassano A, Marchese Robinson RL, Benfenati E, Leszczynski J, Cronin MT (2015) Genotoxicity of metal oxide nanomaterials: review of recent data and discussion of possible mechanisms. Nanoscale 7 (6) :2154–2198. 10.1039/c4nr06670g [DOI] [PubMed] [Google Scholar]
- 78.Olive PL, Banath JP, Durand RE (2012) Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. 1990. Radiat Res 178 (2):AV35–AV42 [DOI] [PubMed] [Google Scholar]
- 79.OECD. Test no. 487: in vitro mammalian cell micronucleus test. OECD, Paris [Google Scholar]
- 80.Ellard S, Parry EM (1993) A modified protocol for the cytochalasin B in vitro micronucleus assay using whole human blood or separated lymphocyte cultures. Mutagenesis 8 (4):317–320. 10.1093/mutage/8.4.317 [DOI] [PubMed] [Google Scholar]
- 81.OECD. Test no. 471: bacterial reverse mutation test. OECD, Paris [Google Scholar]
- 82.OECD. Test no. 473: in vitro mammalian chromosome aberration test. OECD, Paris [Google Scholar]
- 83.Doak SH, Manshian B, Jenkins GJ, Singh N (2012) In vitro genotoxicity testing strategy for nanomaterials and the adaptation of current OECD guidelines. Mutat Res 745 (1–2):104–111. 10.1016/j.mrgentox.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ayala A, Munoz MF, Arguelles S (2014) Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxidative Med Cell Longev 2014:360438 10.1155/2014/360438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohkawa H, Ohishi N, Yagi K (1979) Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem 95 (2):351–358. 10.1016/0003-2697(79)90738-3 [DOI] [PubMed] [Google Scholar]
- 86.Moselhy HF, Reid RG, Yousef S, Boyle SP (2013) A specific, accurate, and sensitive measure of total plasma malondialdehyde by HPLC. J Lipid Res 54(3):852–858. 10.1194/jlr.D032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esterbauer H, Schaur RJ, Zollner H (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11(1):81–128. 10.1016/0891-5849(91)90192-6 [DOI] [PubMed] [Google Scholar]
- 88.Weber D, Milkovic L, Bennett SJ, Griffiths HR, Zarkovic N, Grune T (2013) Measurement of HNE-protein adducts in human plasma and serum by ELISA-Comparison of two primary antibodies. Redox Biol 1:226–233. 10.1016/j.redox.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gérard-Monnier D, Erdelmeier I, Régnard K, Moze-Henry N, Yadan J-C, Chaudière J (1998) Reactions of 1-methyl-2-phenylindole with malondialdehyde and 4-hydroxyalkenals. Analytical applications to a colorimetric assay of lipid peroxidation. Chem Res Toxicol 11 (10):1176–1183. 10.1021/tx9701790 [DOI] [PubMed] [Google Scholar]
- 90.Bartosh TJ, Ylostalo JH (2014) Macrophage inflammatory assay. Bio Protoc 4(14):e1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fujita T, Nolan GP, Ghosh S, Baltimore D (1992) Independent modes of transcriptional activation by the p50 and p65 subunits of NF-kappa B. Genes Dev 6(5):775–787. 10.1101/gad.6.5.775 [DOI] [PubMed] [Google Scholar]
- 92.L-f C, Fischle W, Verdin E, Greene WC (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293 (5535):1653–1657. 10.1126/science.1062374 [DOI] [PubMed] [Google Scholar]
- 93.Kinoshita S, Akira S, Kishimoto T (1992) A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc Natl Acad Sci 89(4):1473–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smale ST (2010) Luciferase assay. Cold Spring Harb Protoc 2010(5):pdb prot5421. 10.1101/pdb.prot5421 [DOI] [PubMed] [Google Scholar]
- 95.Bang FB (1956) A bacterial disease of Limulus polyphemus. Bull Johns Hopkins Hosp 98 (5) :325–351 [PubMed] [Google Scholar]
- 96.Hurley JC (1995) Endotoxemia: methods of detection and clinical correlates. Clin Microbiol Rev 8(2):268–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smulders S, Kaiser JP, Zuin S, Van Landuyt KL, Golanski L, Vanoirbeek J, Wick P, Hoet PH (2012) Contamination of nanoparticles by endotoxin: evaluation of different test methods. Part Fibre Toxicol 9:41 10.1186/1743-8977-9-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ding JL, Ho B (2010) Endotoxin detection – from limulus amebocyte lysate to recombinant factor C In: Wang X, Quinn PJ (eds) Endotoxins: structure, function and recognition. Springer, Dordrecht, pp 187–208. 10.1007/978-90-481-9078-2_9 [DOI] [PubMed] [Google Scholar]
- 99.Alwis KU, Milton DK (2006) Recombinant factor C assay for measuring endotoxin in house dust: comparison with LAL, and (1 → 3)-β-D-glucans. Am J Ind Med 49 (4):296–300. 10.1002/ajim.20264 [DOI] [PubMed] [Google Scholar]