Abstract
Malaria pathogenesis is caused by the replication of Plasmodium parasites within the red blood cells (RBCs) of the vertebrate host. This selective pressure has favored the evolution of protective polymorphisms in erythrocyte proteins, a subset of which serve as cognate receptors for parasite invasion ligands. Recently, the generation of RBCs from immortalized hematopoietic stem cells (HSCs) has offered a more tractable system for genetic manipulation and long-term in vitro culture, enabling elucidation of the functional determinants of host susceptibility in vitro. Here we report the generation of an immortalized erythroid progenitor cell line (EJ cells) from as few as 100,000 peripheral blood mononuclear cells offering a robust method for the creation of customized model systems from small volumes of peripheral blood. EJ cell differentiation mirrored erythropoiesis of primary HSCs, yielding orthochromatic erythroblasts and enucleated RBCs after eight days (ejRBCs). ejRBCs supported invasion by both P. vivax and P. falciparum. To demonstrate the genetic tractability of this system, we used CRISPR/Cas9 to disrupt the Duffy Antigen/Receptor for Chemokines (DARC) gene, which encodes the canonical receptor of P. vivax in humans. Invasion of P. vivax into this DARC-knockout cell line was strongly inhibited providing direct genetic evidence that P. vivax requires DARC for RBC invasion. Further, genetic complementation of DARC restored P. vivax invasion. Taken together, the peripheral blood immortalization method presented here offers the capacity to generate biologically representative model systems for studies of blood-stage malaria invasion from the peripheral blood of donors harboring unique genetic backgrounds or rare polymorphisms.
Keywords: erythroid cell line, peripheral blood erythroid progenitor immortalization, Plasmodium invasion
Introduction
Malaria is responsible for more than 200 million cases and 400,000 deaths annually (World Malaria Report, 2017), ranking among the strongest selective pressures to confront the human species during recent human evolution (Carter & Mendis, 2002; Kwiatkowski, 2005). Plasmodium parasites are introduced into the blood stream via the bite of an Anopheles mosquito, migrate to the liver for one cycle of replication, and emerge to undergo repeated cycles of invasion, growth, and egress within the red blood cells (RBCs) of the vertebrate host (White et al., 2014). All malaria pathogenesis is attributable to complications of blood stage infection (Cowman et al., 2012), and proteins expressed upon the erythrocyte surface—which serve as receptors for parasite invasion ligands (Cowman & Crabb, 2006; Crosnier et al., 2011; Paul et al., 2015)—constitute considerable targets of parasite-induced selection (Carter & Mendis, 2002; Kwiatkowski, 2005; Leffler et al., 2017; Williams, 2006). The capacity to identify and elucidate these ligand-receptor pairings holds important implications for the development of blood-stage malaria vaccines and anti-malarial therapies that target these interactions (Grimberg et al., 2007; Ord et al., 2014; Reiling et al., 2012).
Efforts to identify the host determinants of malaria susceptibility have traditionally relied upon functional studies involving primary cells (reviewed in Bei & Duraisingh, 2012). Since the 1940s, epidemiological and population genetic analyses have associated naturally-occurring polymorphisms, such as sickle cell trait, glucose-6-phosphate dehydrogenase deficiency, thalassemia, and Duffy-negativity, with reduced susceptibility to P. falciparum and/or P. vivax (Allison, 1961; Haldane, 1949; Miller et al., 1976). Analysis of blood samples from donors harboring these RBC disorders have enabled researchers to elucidate the functional basis for these traits in vitro, and additional invasion pathways have been identified through biochemical manipulation of mature erythrocytes (Crosnier et al., 2011, 2013; Wanaguru et al., 2013; Williams, 2006). Finally, although the enucleated nature of the erythrocyte impedes genetic perturbation, targeted knockdown of erythrocyte membrane proteins has been achieved in nucleated CD34+ hematopoietic stem cells (HSCs) (Bei et al., 2010; Giarratana et al., 2005; Giarratana et al., 2011). Terminal differentiation of these HSCs in a high throughput genetic perturbation screen has facilitated the identification of novel P. falciparum receptors, such as CD44 and CD55 (Egan et al., 2015).
Although approaches based upon primary cells offer a degree of biological authenticity, their experimental utility is limited by several factors. First, the capacity to infer the invasion phenotype of a naturally-occurring RBC mutant requires experimental comparison to non-isogenic wildtype control RBCs. Such analyses could be confounded if the genetic background of mutant and wildtype blood is mismatched (e.g., other polymorphisms of relevance to parasite invasion) or if samples are handled differently (e.g., sample age, cryopreservation method) (Bei & Duraisingh, 2012). Additionally, because terminally differentiated RBCs are not capable of replication, analyses of natural polymorphisms require a continuous source of erythrocytes harboring genotypes of interest. While blood from donors with common genetic disorders may be more readily available, sources harboring rare polymorphisms (e.g., the CD55-null Inab phenotype) or unique genetic backgrounds are much more difficult to acquire. An additional suite of logistical issues complicates HSC-based approaches including inter-donor genetic variation, the short window of genetic tractability, inefficiencies in the generation of CRISPR-based knockouts, the long (~2.5-week) duration of terminal differentiation, and the need to repeat genetic perturbations for each assay.
In response to these challenges, Kanjee et al. (2017) developed a genetically tractable experimental system from a naturally-derived erythroleukemia cell line. This erythroid cancer cell line (deemed JK-1) can be expanded indefinitely as proerythroblasts and basophilic erythroblasts, can be experimentally induced to differentiate into polychromatic erythroblasts in 12 days, and supports invasion by P. falciparum (Kanjee et al., 2017) and P. vivax (Gruszczyk et al., 2018). The generation of CRISPR/Cas9-mediated genetic knockouts using this system has allowed for the identification of novel Plasmodium receptors (e.g., the P. vivax receptor, CD71) (Gruszczyk et al., 2018). Despite their considerable utility to studies of Plasmodium invasion, erythroleukemia cell lines are rare and exhibit considerable variation in cellular properties, limiting the capacity of researchers to investigate natural polymorphisms or multiple genetic backgrounds (Kanjee et al., 2017). Also, ~98% of terminally differentiated JK-1 cells arrest at the polychromatic erythroblast stage of erythropoiesis, raising questions as to whether cell lines that differentiate further along the erythroid lineage might more accurately reflect parasite invasion into more mature red blood cells.
Recently, several studies have demonstrated the capacity to immortalize novel erythroid progenitor cell lines via expression of the Human Papilloma Virus (HPV16) E6/E7 fusion protein in HSCs derived from induced pluripotent stem cells, umbilical cord blood, and bone marrow (Kurita et al., 2013, 2019; Trakarnsanga et al., 2017). By using a tet-inducible expression system, these immortalized lines can be induced to differentiate into orthochromatic erythroblasts and enucleated RBCs (Kurita et al., 2013, 2019; Trakarnsanga et al., 2017). Other groups have achieved similar results via the ectopic expression of genetic factors in embryonic stem cells (Hirose et al., 2013) and umbilical cord blood mononuclear cells (Huang et al., 2014). These immortalized cell lines have only been generated from a limited number of genetic backgrounds using material that can be difficult to source. Accordingly, replication of these approaches may be untenable for a variety of uncommon genotypes and globally-relevant genetic backgrounds of interest.
In this study, we generated an immortalized erythroid progenitor cell line from as few as 100,000 peripheral blood mononuclear cells (PBMCs), offering a robust method for the creation of customized model erythrocyte culture systems from small volumes of peripheral blood. This method yielded immortalized cell lines, which resembled basophilic erythroblasts in both morphology and cell surface markers. Differentiation of these cells mirrored erythropoiesis of primary HSCs, yielding orthochromatic erythroblasts and enucleated RBCs after eight days. These cells supported invasion by both P. vivax and P. falciparum. To demonstrate the genetic tractability of this system, we used a CRISPR/Cas9-mediated approach to disrupt the Duffy Antigen/Receptor for Chemokines (DARC)—the canonical RBC receptor of both P. vivax and P. knowlesi (Chitnis et al., 1996; Singh et al., 2006). Invasion of P. vivax into this DARC-knockout cell line was strongly inhibited. These results provide direct genetic evidence confirming DARC as a critical receptor of P. vivax and highlights the utility of this cell line as a tractable genetic model for studies of the host determinants of blood-stage malaria infection. This peripheral blood immortalization method offers the capacity to generate biologically representative model systems from small volumes of accessible starting material and will catalyze efforts to generate cell lines from a wider diversity of patients.
Methods
Generation of the EJ cell line
Anonymized discarded human blood samples used in this study were obtained under Boston Children’s Hospital IRB protocol # 04-02-017R. We centrifuged 50 mL of peripheral whole blood at 2000 x g for 5 minutes, collected the serum, and washed packed blood twice in RPMI media. Packed blood was then resuspended in 2 mM EDTA-PBS at 50% hematocrit. To enrich PBMCs, we gently layered 8 mL of this packed blood-EDTA-PBS suspension on 4 mL of Ficoll Paque (GE Healthcare) in 15 mL conical tubes (BD Falcon) and repeated this procedure for the remainder of the sample. Samples were centrifuged at 1200 x g for 25 minutes at room temperature using low acceleration/deceleration. Upon completion, we harvested the buffy coat and washed PBMCs twice in maintenance media (see below) without doxycycline. Cells were counted using a hemocytometer and light microscope, resuspended in T-flasks (BD Falcon) at a concentration of 5×105 cells/mL, and incubated at 37oC in a humidifier chamber with 5% (vol/vol) CO2. After 24 hours, PBMCs were washed in pIMDM media (see below) without octaplasma, divided into three experimental conditions (i.e., 1.0×105 PBMCs, 2.0×105 PBMCs, and 5.7×107 PBMCs) and transduced with the lentiviral vector CSIV-TRE-HPV16-E6/E7-UbC-KT (Kurita et al., 2013), which was provided by Dr. Kurita and Dr. Nakamura. Lentivirus was generated via transfection of HEK 293T cells (a kind gift of Robert Doms) using psPAX2 (Addgene plasmid # 12260) and VSV-G (Addgene plasmid # 12259) packaging plasmids (Doench et al., 2016). Lentivirus was then concentrated 27:1 via ultracentrifugation. Following transduction, cells were cultured in pIMDM media without doxycycline at a concentration of 5×105 cells/mL and incubated as above. After four days, 1 μg/mL doxycycline and 1 μM dexamethasone were added to the culture media, and cells were monitored for 65 days post-transduction. During this period, we changed culture media every 3–4 days and monitored cell concentration to ensure that cells did not exceed 1×106 cells/mL. Once cultures exhibited a stable rate of expansion and senescent cells had lysed, cells were cloned in 96-well plates via limiting dilution, yielding immortalized erythroid progenitor cell lines.
Maintenance and differentiation of EJ cells
During both maintenance and differentiation, EJ cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Biochrom) supplemented with 4 mM L-glutamine (Sigma-Aldrich), 330 μg/mL iron-saturated human holo-transferrin (BBI solutions), 10 μg/mL recombinant human insulin (Sigma-Aldrich), 2 IU/mL heparin sodium salt (Affymetrix), and 0.5% Pen/Strep (Life Technologies). Hereafter, we refer to this primary media as “pIMDM.” Maintenance media was comprised of pIMDM supplemented with 5% solvent/detergent virus-inactivated plasma (octaplasma; Octapharma), 3 IU/mL erythropoietin (EPO; Roche), 50 ng/mL stem cell factor (SCF; R&D systems), 1 μg/mL doxycycline (Sigma-Aldrich), and 1 μM dexamethasone (Sigma-Aldrich). We maintained EJ cells at a concentration of 5×104-7.5×105 cells/mL, changing media every 3–4 days. Every 2–3 weeks, or if cell concentration eclipsed 1×106 cells/mL, the culture was enriched for basophilic erythroblasts via a 40% (vol/vol) Percoll-PBS density gradient. To do so, cells were resuspended in 4 mL of pIMDM media supplemented with 0.1% bovine serum albumin (BSA), layered on 4 mL of 40% Percoll-PBS, and centrifuged at 1000 x g for 10 minutes with low acceleration/deceleration. Upon completion, we harvested and washed the interface three times in pIMDM before resuspending in culture media.
To induce differentiation, EJ cells were seeded at 2×105 cells/mL and cultured for 3 days in pIMDM supplemented with 5% octaplasma, 3 IU/mL EPO, 10 ng/mL SCF and 0.5 μg/mL doxycycline. On day 3, cells were counted and seeded at 5×105 cells/mL in pIMDM containing 5% octaplasma and 3 IU/mL EPO. Cells were maintained in this media for the remainder of differentiation. We changed media on day 5 and reseeded cells at 5×105 cells/mL. Cells were harvested on either day 8 or 9 of differentiation via 40% Percoll-PBS gradient. In this case, we retained the pellet, which contained terminally differentiated cells and discarded the interface. To monitor EJ cell differentiation, routine cytospins were prepared, fixed in methanol, and stained with May-Grünwald (Sigma-Aldrich) followed by Giemsa (Sigma-Aldrich) according to the manufacturer’s instructions as described previously (Egan et al., 2015). We used cytospins taken during differentiation to quantify the progression of EJ cells and primary HSCs along the erythroid lineage (basophilic erythroblast, polychromatic erythroblast, orthochromatic erythroblast, reticulocyte) using established morphological diagnostic criteria (Kaushansky et al., 2015; Rozenberg, 2011).
Flow cytometry
Cells were washed twice in 1x PBS and pelleted in a 96-well plate. For conjugated antibodies, cells were stained in 100 μl of flow buffer (0.1% BSA in 1x PBS) for 20 minutes at 4oC in the dark. Cells were washed three times in flow buffer and resuspended in 200 μl of flow buffer prior to reading. We used the following antibodies at the dilutions indicated by the manufacturer: anti-CD34 FITC (Miltenyi Biotec), anti-CD45 Violet Blue (Miltenyi Biotec), anti-CD117 APC (Clone REA996; Miltenyi Biotec), anti-CD49d PE-Violet 770 (Miltenyi Biotec), anti-CD36 Violet Blue (Miltenyi Biotec), anti-CD71 APC (Miltenyi Biotec), anti-CD235a FITC (Clone 2B7; Stem Cell Technologies), anti-basigin FITC (Invitrogen), anti-CD44 APC (Miltenyi Biotec), anti-CD55 APC (Miltenyi Biotec), and anti-glycophorin C FITC (BRIC10; Santa Cruz). To measure GPB expression, cells were trypsinized with 50 μl of 1 mg/mL trypsin (Sigma-Aldrich) in incomplete RPMI for 1 hour at 37oC to remove GPA. Trypsin was deactivated with complete RPMI (RPMI media with 0.5% albumax and 0.2% sodium bicarbonate) and cells were stained with anti-glycophorin A/B (1:8000; clone E3; Sigma-Aldrich) for 30 minutes at 4oC followed by Alexa Fluor 488 goat anti-mouse secondary antibody (1:1000; Invitrogen) for 30 minutes at 4oC. A non-trypsin control was included every time. To measure DARC, we incubated cells with an anti-DARC PE-conjugated primary antibody (mouse IgG2a; 1:10 dilution; R&D Systems) for 30 minutes at 4oC followed by IRDye® 680LT goat anti-mouse secondary antibody (1:50 dilution; LI-COR) for 30 minutes at 4oC. Prior to running cells on a flow cytometer, cells were resuspended in 200 μl staining buffer containing propidium iodide (PI, for the erythropoiesis markers) or in 200 μl of PI and Vybrant Dicycle Violet (1:10,000; Invitrogen, for the receptors) and incubated in the dark for 20 minutes at 37oC. Cells were run on a MACSQuant flow cytometer (Miltenyi Biotec) and data were analyzed using FlowJo (version 10.4). Cell volume was measured using the Moxi-GO II instrument (Orflo Technologies) and surface area for each cell type was calculated using the radius extrapolated from the volume measurements. Adjusted MFI values (MFI of receptor minus background MFI) for each receptor were divided by surface area to obtain a relative receptor expression between the different cell types (MFI/μm2).
CRISPR/Cas9-mediated perturbation of the DARC gene
CRISPR/Cas9-mediated genetic perturbations of the EJ cell line were generated via adaptation of previously published protocols (Sanjana et al., 2014; Shalem et al., 2015). We first expressed the lentiCas9-Blast plasmid (Addgene plasmid #52962) (Sanjana et al., 2014) in EJ cells via lentiviral transduction. Lentivirus was produced in HEK 293T using psPAX2 (Addgene plasmid # 12260) and VSV-G (Addgene plasmid # 12259) packaging plasmids (Doench et al., 2016). One million EJ cells were transduced with 1 mL of lentiCas9-Blast lentivirus via spinfection for 2 hours at 1000 x g in the presence of 4 μg/mL polybrene. The transduced cells were then selected on 10 μg/mL blasticidin for 2–4 weeks. Although the stable EJ-Cas9 line was not cloned, Cas9 cutting efficiency was confirmed using the pXPR_011-sgEGFP lentivirus (Addgene plasmid #59702). We next used the lentiGuide-Puro plasmid (Sanjana et al., 2014) to express a single-guide RNA (sgRNA; 5’-TGGGGCTCACTGTGGGAATT-3’) targeting the second exon of the DARC gene (lentiGuide-Puro was a gift from Feng Zhang (Addgene plasmid # 52963)). Lentivirus generation and transduction were performed as described above. After one week of selection on 2 μg/mL puromycin, cells expressing the transgene were cloned via limiting dilution, and clones were screened for indels in DARC via Sanger sequencing and Tracking of Indels by Decomposition (TIDE) (Brinkman et al., 2014) using the DARCseqgRNA3_Fwd (5’-TGCTGGATGACT-CTGCACTG-3’) and DARCseqgRNA3_Rev (5’-GTTCAGCAGCAGGTCCAG-AG-3’) primer set. The chromatograms of clones that exhibited evidence of bi-allelic indels via TIDE analysis were manually inspected to identify the location of indels within the sequence. Sequences were then translated to identify the effect of these genetic perturbations on the sequence of the protein. Finally, we compared DARC expression in wildtype and DARC knockout lines via flow cytometry and confirmed the phenotype via Plasmodium invasion assays (see below) involving the DARC-dependent parasite P. vivax.
To validate these results, we complemented the ΔDARC cell line by cloning the Fyb allele into the pLVX-FADD-DD vector containing a hygromycin resistance cassette (pLVX-DARCFyb-Hygro; a kind gift from Joan Massague; Addgene plasmid #58263) (Valiente et al., 2014). We included synonymous mutations in the sgRNA binding site of these DARC sequences to prevent the constitutively-expressed Cas9 from cutting these transgenes. We also substituted the CMV promoter for a minimal beta globin promoter to increase expression of the transgene in erythroid cells. The beta globin promoter was amplified from human genomic DNA using the primers 5’-GGACCATCGATGGTACGGCTGTCATCACTTAGACC-3’ and 5’-GGACCCTCGAGGGTGTCTGTTTGAGGTTGCTAGTG-3’ (restriction sites in primers are underlined) and ClaI/XhoI cloned into pLVX-DARCFyb-Hygro. We then expressed the transgenic DARC allele in the ΔDARC cell line via lentiviral transduction. Cells were selected on 200 μg/mL Hygromycin B (Sigma-Aldrich) for two weeks, and expression was confirmed via flow cytometry.
Invasion assays
Plasmodium falciparum cultures were maintained in human O-positive erythrocytes (Interstate Blood bank) at 2% hematocrit in complete RPMI media with 0.5% albumax and 0.2% sodium bicarbonate at 37oC with 5% CO2 and 1% O2 mixed gas. Plasmodium vivax isolates were obtained from Brazil, and were maintained and enriched as previously described (Rangel et al., 2018). Plasmodium falciparum lines were synchronized one cycle before the invasion assay in a 4-hour window using the magnetic-activated cells sorting (MACS) purification (Miltenyi Biotec) followed by treatment with heparin. The synchronized schizonts were purified again using the MACS method prior to setting up the invasion assay.
Prior to performing invasion assays, ejRBCs were passed through a 40% Percoll-PBS gradient to remove dead cells and undifferentiated erythroblasts as described above. The pellet was retained and washed in complete RPMI media. In addition, reticulocytes were isolated from peripheral blood using CD71 microbeads (Miltenyi Biotec) according to manufacturer’s instructions.
Invasion assays were performed in the respective parasite maintenance media in 2–3 technical replicates at 0.1–0.25% hematocrit and 0.5–2% starting parasitemia. Invasion assays were set up in half-area 96-well plates (Corning Inc.). We took cytospins immediately after initiation of the invasion assay, as well as 16–24 hours post-invasion. Parasitemia was quantified using a reticle and light microscopy and presented either as PEMR (parasitized erythrocyte multiplication rate calculated by dividing parasitemia post invasion by starting schizontemia) or invasion efficiency (PEMRs were normalized to the corresponding control as indicated). All invasion assays were performed three times independently.
Statistical analyses
Principal components analysis (PCA) of erythropoiesis markers CD34, CD45, CD36, CD49d, CD71, and GPA was performed in R (version 3.4.3) using the ‘pcaMethods’ package of Bioconductor (https://www.bioconductor.org/packages/release/bioc/html/pcaMethods.html). MFI input data for PCA were derived from flow cytometry of EJ cells and primary HSCs at multiple points during differentiation (EJ cells: day 0, day 3, day 5, day 8, and day 9; primary HSCs: day 7, day 10, day 12, and day 18). All other statistical tests were also performed in R (version 3.4.3).
Results
Generation of an immortalized erythroid cell line from small volumes of peripheral blood
Previous efforts to generate erythroid progenitor cell lines have relied upon the immortalization of HSCs derived from induced pluripotent stem cells, embryonic stem cells, umbilical cord blood, or bone marrow (Hirose et al., 2013; Huang et al., 2014; Kurita et al., 2013, 2019; Trakarnsanga et al., 2017). Because these materials may be difficult to source from a wide variety of patients harboring particular genotypes of interest, we sought to adapt this method to immortalize peripheral blood-derived HSCs. To do so, we used a Ficoll Paque density gradient to isolate PBMCs from 50 mL of anonymized and discarded human peripheral blood from a hemochromatosis patient (Fig. 1A). There are no known changes in eyrthropoiesis and the number of circulating erythroid progenitors in hemochromatosis patients (Crownover & Covey, 2013). Moreover, after culturing cells for 24 hours in maintenance media without doxycycline, 5.73×107 PBMCs were harvested. The concentration of PBMCs harvested from this patient is comparable to that which would be expected in healthy donors (Corkum et al., 2015). PBMCs were split into three experimental conditions, comprising a total of 1.0×105 cells, 2.0×105 cells, and 5.7×107 cells, respectively. For each condition, we expressed the HPV16-E6/E7 gene in PBMCs under a tet-inducible promoter via lentiviral transduction and cultured cells in erythroid media with doxycycline, changing media every 3–4 days. After 15–46 days in culture, senescent cells had lysed and immortalized cells (deemed the “EJ cell line”) expanded in all three experimental conditions (Fig. 1B). Cells were then cloned via limiting dilution, yielding isogenic immortalized erythroid progenitor cell lines. Although we generated multiple clonal lines, in this study we characterized the EJ-E3 clone—derived from the 1.0×105 PBMC condition (corresponding to ~100 microliters of peripheral blood). To validate expression of the HPV16-E6/E7 protein in the EJ-E3 clone, we measured expression of humanized kusabira orange 1 (hKO1)—which is linked to HPV16-E6/E7 in the CSIV-TRE-HPV16-E6/E7-UbC-KT construct by a Thosea asigna virus 2A peptide sequence under the control of a human ubiquitin C promoter—via flow cytometry. As expected, hKO1 (and by extension HPV16-E6/E7) expression declined as EJ cells differentiated (Fig. S1).
Figure 1:
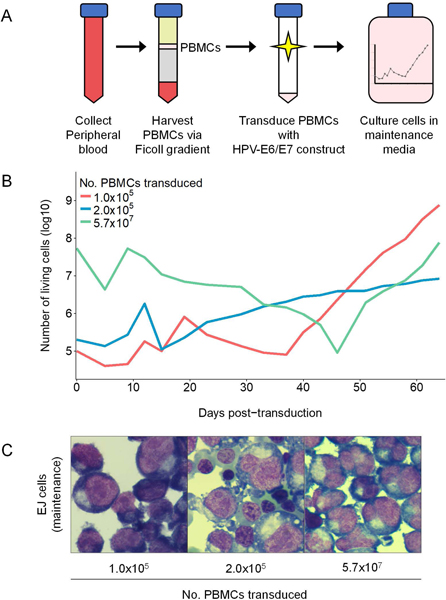
Generation of an immortalized erythroid progenitor cell line from peripheral blood. (A) Immortalization approach. PBMCs were harvested from peripheral blood via density gradient and immortalized through expression of the tet-inducible HPV16-E6/E7 construct. Immortalized cells were cultured with doxycycline and cloned via limiting dilution. (B) Immortalized PBMC population dynamics. Cells derived from the 1.0×105 PBMC (red), 2.0×105 PBMC (blue), and 5.7×107 PBMC (green) transduction conditions are shown. (C) Cytospin images of cell populations after 60 days in maintenance culture. Bulk populations exhibited a heterogeneous combination of basophilic, polychromatic, and orthochromatic erythroblasts.
During expansion, we maintained EJ cells at a concentration of 5×104-7.5×105 cells/mL. At this concentration, EJ cells exhibited a doubling rate of ~14 hours, and cells exhibited morphological characteristics of the early basophilic erythroblast stage of erythropoiesis (Fig. 1C). However, if the concentration of cells in culture eclipsed 1×106 cells/mL, we observed an increase in the developmental heterogeneity of the cell line due to an elevated proportion of polychromatic cells. In these circumstances, we performed a 40% (vol/vol) Percoll-PBS gradient and retained the interface to eliminate these spontaneously differentiating cells.
Differentiation of the EJ cell line to produce orthochromatic and enucleated ejRBCs in eight days
Erythropoiesis is the process by which nucleated erythroid progenitor cells divide and differentiate into enucleated reticulocytes. During the terminal phase of erythropoiesis, each successive division of the proerythroblast yields a morphologically distinguishable erythroid stage: (1) basophilic erythroblast, (2) polychromatic erythroblast, (3) orthochromatic erythroblast, and (4) enucleated reticulocyte (Chen et al., 2009; Dzierzak & Philipsen, 2013; Hu et al., 2013). Each of these stages is characterized by a progressive decrease in cell size, an increase in chromatin condensation and hemoglobinization, and dynamic changes in proteins expressed upon the erythrocyte surface.
By expressing the HPV16-E6/E7 transgene under a tet-inducible promoter, the EJ cell line can be induced to differentiate into orthochromatic erythroblasts and reticulocytes (deemed “ejRBCs”) via the removal of doxycycline from the culture media. The kinetics of cell growth during differentiation exhibited three general phases: (1) expansion from day 0–3 (mean: 6.5-fold expansion; range: 5.5–8.7, n=5 experiments), (2) plateau from day 3–5 (mean: 1.3-fold; range: 0.8–1.8; n=5 experiments), and (3) senescence from day 5–8 (mean: 0.7-fold; range: 0.5–1.1; n=5 experiments), yielding a mean net expansion of 6.7-fold (range: 1.6–14.1; n=5 experiments) during this procedure (Fig. 2A). Although we continued differentiation beyond day 8 in a subset of experiments, we observed a decrease in cell counts of ~50% between days 8 and 9. Given this tradeoff between terminal differentiation and cell senescence, we deemed the optimal point of cell harvest to be eight days after inducing erythropoiesis, and all experiments involving ejRBCs presented hereafter correspond to cells harvested at this time point.
Figure 2:
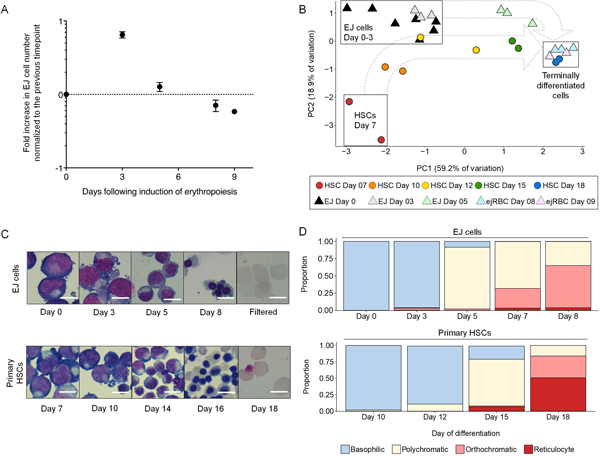
Differentiation of the ejRBC cell line mirrors erythropoiesis of primary HSCs. (A) Dynamics of EJ cells during the differentiation protocol. Data points reflect fold increase in cell number relative to the previous time point (log10 scale). Average and standard error of mean (SEM) from five experiments are presented. (B) Principal components analysis (PCA) of erythropoiesis marker median fluorescent intensity (MFI) data (adjusted for the background of unstained controls). PCA reflects expression of CD34, CD45, CD36, CD49d, CD71, and GPA, derived from two primary HSC differentiation experiments (circles) and 3–7 EJ cell differentiation experiments (triangles). Principal components 1 and 2 (PC1 and PC2, respectively) are plotted. Dotted arrows outline trajectory of erythropoiesis. PCA variable loadings are presented in Fig. S4. Representative flow cytometry plots of all erythropoiesis markers are presented in Fig. S3. (C) Cell morphology during erythropoiesis. Immortalized EJ cells (top panel) and primary HSCs (bottom panel) are represented at multiple time points during erythropoiesis. Scale bars are 10 μm in length. (D) Representation of erythropoiesis stages during terminal differentiation. Relative frequencies of basophilic (blue), polychromatic (pale yellow), orthochromatic (pink), and enucleated (red) stages are represented at multiple time points during differentiation. EJ cells are represented in the top panel, and primary HSCs are displayed in the bottom panel.
Throughout differentiation, the morphology of EJ cells recapitulated the stages of erythropoiesis as modeled by primary CD34+ HSCs (Fig. 2C). Basophilic erythroblasts comprised over 96% of the culture after three days of differentiation, which resembled primary HSCs after 10–12 days of differentiation (Fig. 2D). At day 5 of differentiation, 89.6% of EJ cells resembled polychromatic erythroblasts, which was similar to primary HSCs after 15 days of differentiation (Fig. 2D). After 8 days of differentiation, 63.3% of EJ cells were orthochromatic erythroblasts by morphology, resembling the age of terminally differentiated primary HSCs (Fig. 2D). However, in comparison to primary cells, the EJ cell line exhibits a substantially lower rate of enucleation (Fig. 2D). On day 8, ejRBCs exhibited an average rate of enucleation of 3.1% (range: 2.2–3.8%; n=5 experiments), which increased to 5% after nine days in culture (Fig. S2). Filtration of these terminally differentiated cells yielded a pure population of enucleated reticulocyte-like cells (Fig. 2C).
Next, we used a panel of antibodies targeting stage-diagnostic markers CD34, CD45, thrombospondin receptor (CD36), α−4 integrin (CD49d), transferrin receptor (CD71), and glycophorin A (GPA, CD235a) to characterize the dynamic changes in the expression of cell surface proteins in EJ cells during erythropoiesis. The proerythroblast—the first committed erythroblast stage—is characterized by weak CD45 expression and the initiation of GPA expression. Proerythroblasts are also positive for CD71, α−4 integrin, and CD36 (Dzierzak & Philipsen, 2013; Fajtova et al., 2013; Hu et al., 2013; Van Lochem et al., 2004; Wood, 2004). During erythropoiesis, the proerythroblast divides and differentiates, in the process losing CD45 expression, followed successively by CD36, CD49d, and CD71, while GPA expression increases (Dzierzak & Philipsen, 2013; Fajtova et al., 2013; Hu et al., 2013; Van Lochem et al., 2004; Wood, 2004). This analysis revealed that undifferentiated EJ cells are characterized by a CD34-CD235a+CD45-CD36+CD49d+CD71+ expression profile (Fig. S3). The morphology and expression profile of the EJ cell line in maintenance culture is consistent with early basophilic erythroblasts derived from primary HSCs. Principal component analysis (PCA) of surface protein expression data revealed that these undifferentiated EJ cells cluster with primary HSCs after 10–12 days of differentiation (Fig. 2B; Fig. S4). After five days of differentiation, the expression profile of EJ cells resembled day 15 HSCs, consistent with their polychromatic morphology (Fig. 2B). By day 8, cells were primarily poly- and orthochromatic erythroblasts, which exhibited a CD49lowCD71lowGPAhigh expression profile that strongly resembled day 18 HSCs (Fig. 2B; Fig. S3). Taken together, these results demonstrate that the eight-day ejRBC differentiation protocol adequately replicates erythropoiesis of primary HSCs.
ejRBCs support efficient invasion of P. falciparum and P. vivax
Efforts to elucidate the host determinants of malaria susceptibility have traditionally relied upon either primary cells or erythroleukemia cell lines as the substrate for parasite invasion assays. While the former grants a degree of biological authenticity, the latter offers a more tractable system for genetic manipulation and long-term in vitro culture. To evaluate whether ejRBCs constitute a representative model system for studies of the host determinants of malaria invasion, we next sought to compare the expression of previously identified Plasmodium receptors among ejRBCs, CD71-enriched reticulocytes, and fully mature normocytes. To do so, we used a panel of antibodies targeting receptors of both P. falciparum (GPA, Glycophorin B, Glycophorin C, Basigin, CD44, and CD55) and P. vivax (DARC and CD71). When adjusting for cell size, expression of these Plasmodium receptors in ejRBCs was comparable to reticulocytes (Fig. 3A). Notably, CD71 (an essential P. vivax receptor (Gruszczyk et al., 2018)) and basigin (a strain-transcendent P. falciparum receptor (Crosnier et al., 2011)) were expressed more highly in ejRBCs than in reticulocytes (CD71: ~14.5-fold higher; BSG: ~7.5-fold higher, Figure 3A). Given that CD71 and Basigin expression is downregulated with erythroid maturation (Hu et al., 2013), this observation is consistent with the comparatively younger stage of differentiated ejRBCs (primarily orthochromatic erythroblasts and early reticulocytes). Moreover, the persistence of elevated Basigin levels could also be explained by the high metabolic requirements of immortalized erythroid cell lines since Basigin interacts with monocarboxylate transporters to transport glycolysis byproducts such as lactate, outside of the cell (Kirk et al., 2000). Additionally, DARC expression was lower in ejRBCs than in primary cells. Taken together, these results demonstrate that ejRBCs express the full complement of known Plasmodium receptors.
Figure 3:
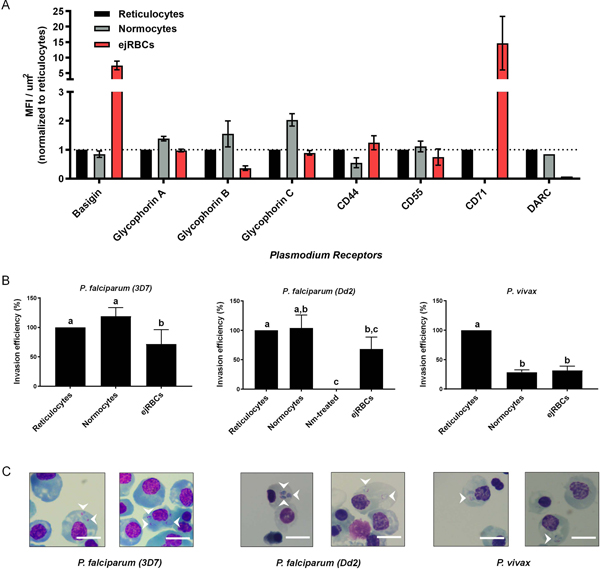
Terminally differentiated ejRBCs constitute a representative model system for studies of malaria invasion. (A) Flow cytometry-based comparison of Plasmodium receptor expression among ejRBCs (red), reticulocytes (black), and normocytes (gray) relative to surface area of respective cell. Adjusted median fluorescence intensity (MFI) (MFI of receptor minus background MFI) for each receptor were divided by surface area to obtain a relative receptor expression between the different cell types (MFI/μm2). The MFI/μm2 values were normalized to reticulocytes. The mean and SEM of data derived from three experiments are presented. (B) Comparison of invasion efficiency of P. falciparum (strains 3D7 and Dd2) and P. vivax into ejRBCs, reticulocytes, and normocytes. In vitro invasion assays were setup at 0.1–0.25% hematocrit and 0.5–2.0% initial parasitemia. Parasitized erythrocyte multiplication rate (PEMR) was estimated by dividing percent ring parasitemia at 16–24 hours by percent initial schizontemia. PEMR of normocytes and ejRBCs were normalized to PEMR of reticulocytes. Data represents mean and SEM from three independent experiments (9–15 technical replicates in total). P-values were estimated using a nested ANOVA and Tukey’s HSD adjusted for multiple comparisons. Letters correspond to significantly different pairwise comparisons after Tukey adjustment (p<0.05). (C) Representative images of P. falciparum (strains 3D7 and Dd2) and P. vivax invasion into ejRBCs. Ring-stage parasites are highlighted with white arrows. Scale bars are 10 μm in length.
Although ejRBCs exhibit low rates of enucleation (Fig. S2), previous studies have demonstrated that nucleated RBCs, particularly orthochromatic erythroblasts, support Plasmodium invasion (Bei et al., 2010; Egan et al., 2015; Gruszczyk et al., 2018; Kanjee et al., 2017; Tamez et al., 2009). To elucidate the utility of this system for functional studies of malaria invasion in vitro, we compared invasion of P. falciparum and P. vivax into ejRBCs, reticulocytes, and normocytes. Plasmodium falciparum lines 3D7 and Dd2 invaded ejRBCs with an average parasitized erythrocyte multiplication rate (PEMR) of 1.55 and 1.21, respectively, which was lower than PEMR into CD71-enriched retics (2.98 and 2.51, respectively) and normocytes (2.93 and 2.17, respectively). P. vivax PEMR into ejRBCs was also lower than that into CD71-enriched reticulocytes (0.13 vs. 0.44) but similar to normocytes (0.12).
Both parasite species invaded primary reticulocytes more efficiently (P. falciparum strain 3D7, nested ANOVA: F2,27=7.60, p=0.0024; P. falciparum strain Dd2, nested ANOVA: F3,23=8.67, p=0.0005; P. vivax, nested ANOVA: F2,10=21.83, p<0.0001; Fig. 3B). Nevertheless, these results demonstrate that terminally differentiated ejRBCs support invasion of both sialic acid-dependent (Dd2, 68.1% invasion efficiency relative to reticulocytes) and sialic acid-independent (3D7, 71.8% invasion efficiency relative to reticulocytes) P. falciparum laboratory strains, as well as clinical P. vivax isolates (31.5% invasion efficiency relative to reticulocytes; Fig. 3B, Fig. 3C). Expression of the full complement of known Plasmodium receptors and support of Plasmodium invasion indicate that ejRBCs constitute an adequate in vitro model system for the investigation of research questions related to P. falciparum and P. vivax invasion. We also assessed whether ejRBCs supported P. falciparum growth. After 24 hours, parasites developed to late rings in ejRBCs, resembling growth in normocyte culture. However, we also observed an apparent reduction in the percentage of parasites (~30–50%) that transitioned to the trophozoite and schizont stages in ejRBCs (Fig. S5).
CRISPR/Cas9-mediated disruption of DARC limits P. vivax invasion into ejRBCs
Although the ancient origin of human P. vivax lineages has been traced to African great ape reservoirs (i.e., chimpanzees and gorillas) (Liu et al., 2014), this parasite has been largely excluded from human populations in sub-Saharan Africa by a widespread polymorphism in the promoter of the DARC gene, which abrogates expression on erythrocytes (Hamblin & Di Rienzo, 2000; Howes et al., 2011). Researchers have used naturally occurring DARCNULL erythrocytes to confirm the functional role of the DARC receptor during P. vivax invasion (Barnwell et al., 1989; Grimberg et al., 2007). However, a genetically tractable system is necessary to elucidate synergistic effects of other P. vivax receptors (e.g., CD71 (Gruszczyk et al., 2018)), which may help to explain recent observations of DARCNULL P. vivax invasion (Cavasini et al., 2007; Ménard et al., 2010; Mendes et al., 2011; Ryan et al., 2006). Thus, we next sought to evaluate whether the ejRBC model system is conducive to genetic perturbation.
As a proof-of-concept, we generated a CRISPR/Cas9-mediated disruption of the DARC gene in EJ cells. To do so, we first expressed the lentiCas9-Blast plasmid (Sanjana et al., 2014) in EJ cells via lentiviral transduction, yielding stable Cas9 expression after 2–4 weeks of selection on blasticidin. Cas9 activity was confirmed using an EGFP reporter assay (Fig. S6). We next used the lentiGuide-Puro plasmid (Sanjana et al., 2014) to express a single-guide RNA (sgRNA) targeting the second exon of the DARC gene. After one week of selection on puromycin, cells expressing the transgene were cloned via limiting dilution, and clones were screened for indels in DARC. In one clone, Sanger sequencing and Tracking of Indels by Decomposition (TIDE) (Brinkman et al., 2014) revealed deletions of 13- and 20-bps, yielding a premature stop codon and truncated protein (Fig. 4A-B). Due to the low expression of DARC in ejRBCs we were not able to detect a difference in expression between the control Cas9 line and the ΔDARC line by flow cytometry as their signal was very low and overlapped with the isotype controls (Fig. 4C). Therefore, we performed a western which confirmed the very low expression of DARC in the control Cas9 ejRBCs and its absence in the ΔDARC ejRBCs (Fig. S7).
Figure 4:
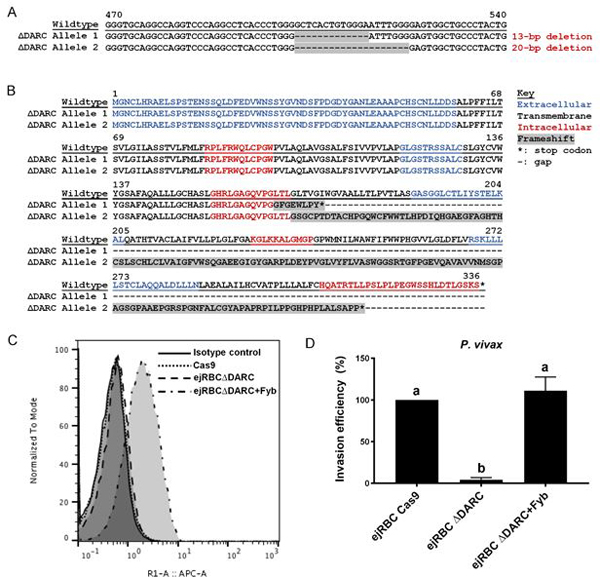
CRISPR/Cas9-mediated disruption of the DARC gene in ejRBCs restricts invasion of P. vivax. (A) Partial sequence of DARC gene spanning experimentally-induced deletions. Wildtype sequence is presented, along with each allele of the ΔDARC line. Cas9-mediated deletions are highlighted in gray. (B) Protein sequence of DARC gene in wildtype and ΔDARC EJ cells. Extracellular (blue), transmembrane (black), and intracellular (red) domains are represented. Cas9-mediated perturbations in the ΔDARC line (highlighted in gray) resulted in two frameshift mutation resulting in premature stop codons and truncated proteins. (C) Flow cytometry-based comparison of DARC protein expression in Cas9, ΔDARC, and DARC-complemented ejRBCs. (D) Invasion into ΔDARC ejRBCs was strongly inhibited relative to wildtype, while complementation of the DARC gene via overexpression restored susceptibility of the cell line to wildtype levels. Invasion efficiency of each parasite relative to the wildtype is presented. Data represents mean and SEM from three independent experiments (six technical replicates in total). P-values were estimated using a nested ANOVA and Tukey’s HSD adjusted for multiple comparisons. Letters correspond to significantly different pairwise comparisons after Tukey adjustment (p<0.05).
Previous work has demonstrated that DARC constitutes a critical RBC receptor of P. vivax (Chitnis et al., 1996; Cowman et al., 2012; Singh et al., 2006). We confirmed the dependence of P. vivax on the DARC receptor by comparing invasion into wildtype and ΔDARC ejRBCs. Relative to control ejRBCs, our analyses demonstrated that ΔDARC cells are largely refractory to P. vivax invasion (P. vivax nested ANOVA: F2,8=28.1, p<0.0001, Fig. 4D). We found that ejRBCs express almost undetectable levels of surface DARC (Fig. 4C, Fig. S7), which is nevertheless required for P. vivax invasion. Critically, complementation of the DARC receptor in the ΔDARC cell line via expression under a minimal beta-globin promoter increased expression of this protein (Fig. 4C, Fig. S7). Despite the increase in protein levels, invasion into ejRBCs complemented with DARC was restored to the same levels as those of control ejRBCs (Fig. 4D). These results indicate that the specific disruption of the DARC gene underlies the differential susceptibility to P. vivax invasion, highlighting the dependence of this parasite on the DARC receptor.
Discussion
This study presents a robust method for the generation of immortalized erythroid progenitor cell lines from as little as 100 microliters of peripheral blood. The wide accessibility of these inputs stands to catalyze studies of the functional determinants of malaria susceptibility in a greater diversity of genetic backgrounds than had previously been available. As a proof of concept for the viability of this assay, we have immortalized erythroid progenitor cells from peripheral blood (EJ cells), which differentiate similarly to primary HSCs, are genetically tractable, and allow for efficient invasion of both P. falciparum and P. vivax.
Throughout erythropoiesis, EJ cells mirrored primary HSCs with respect to both morphology and the expression of stage-diagnostic cell surface proteins (Fig. 2B-D). While several erythroid lines produced by other groups exhibit terminal differentiation defects and very little enucleation (Hirose et al., 2013; Kurita et al., 2013), ejRBCs enucleate at a rate of ~3–5% (Fig. S2). Although this enucleation rate is eclipsed by erythroid lines generated by Huang et al. (2014) (~30%), Trakarnsanga et al. (2017) (“up to 30%”), and Kurita et al. (2019) (18%), these cell lines were immortalized at an earlier stage of erythropoiesis and, thus, require up to 16 and 18 days for terminal differentiation, respectively. By contrast, terminal differentiation of ejRBCs is accomplished in eight days of culture, reducing the effort and resources necessary to derive orthochromatic erythroblasts. Thus, this shortened differentiation time offers a practical advantage over other cell lines in applications for which enucleation is not critical (e.g., studies of malaria invasion; forward genetic screens), and facilitate synchronization through erythroid differentiation. For applications that prioritize enucleated cells, the EJ cell culture can be expanded, differentiated, and filtered to produce a pure population of enucleated cells as appropriate. Nevertheless, efforts to further optimize the low and variable enucleation rate of the EJ cell line are underway using stromal cell-lines, which have been previously shown to improve enucleation (Giarratana et al., 2005; Vinjamur & Bauer, 2018). Moreover, a recent study demonstrated that enucleation rates of CD34+ HSC lines derived from the same donor may range from 0–25% when differentiated for 10 days, suggesting that complex mechanisms regulate the enucleation process in vitro (Kurita et al., 2019). We are currently working to identify small molecules and genetic or epigenetic modifications that can elevate enucleation in the EJ cell line.
Given that malaria parasites can successfully invade nucleated RBCs, including orthochromatic erythroblasts, the EJ line constitutes a useful model system for studies of malaria invasion. However, it should be noted that ejRBCs appear to be less permissive to invasion than primary cells (P. falciparum: ~60% efficiency in ejRBCs relative normocytes; P. vivax: ~30% invasion efficiency in ejRBCs relative to reticulocytes). Terminally differentiated ejRBCs express all known Plasmodium spp. receptors at comparable levels to reticulocytes (Fig. 3A), with the exception of DARC, which is expressed at considerably lower levels in ejRBCs (Fig. 3A, Fig. S7). Nevertheless, CRISPR/Cas9-mediated disruption of the DARC gene in this cell line strongly inhibited P. vivax invasion (Fig. 4D). These findings highlight the dependence of P. vivax invasion on the DARC receptor, supporting previous studies (Barnwell et al., 1989; Grimberg et al., 2007) and suggest that minimal levels of DARC expression are sufficient and necessary for P. vivax invasion. Interestingly, although DARC protein expression in the complementation line was increased by roughly 10–20 fold compared to the Cas9 line (Fig. S7), P. vivax invasion was only restored to wildtype levels. This apparent saturation of invasion at low levels of DARC expression suggests that P. vivax is relatively insensitive to natural variation in the levels of DARC expression.
Malaria pathogenesis ranks among the strongest selective pressures to confront the human species during the last 50,000 years (Carter & Mendis, 2002; Hedrick, 2012; Williams, 2006), and proteins expressed upon the RBC membrane have been particular targets of parasite-induced selection. The capacity to generate immortalized erythroid progenitor cell lines from small volumes of peripheral blood will allow for the development of customized model systems from a wider diversity of patients. The EJ cell line produced via this method expands indefinitely, differentiates rapidly, is representative of primary HSCs, supports Plasmodium invasion, and is conducive to genetic manipulation. Application of this peripheral blood immortalization method to human populations living in malaria endemic regions will grant researchers access to a renewable source of erythroid cells harboring natural polymorphisms of interest. Further, the genetic tractability of these cell lines will enable researchers to isolate the genetic determinants of malaria susceptibility via the experimental introduction of candidate polymorphisms in alternative isogenic backgrounds. Thus, by offering to expand the diversity of cell lines available to researchers, this study stands to catalyze follow-up research into the host determinants of malaria susceptibility.
Supplementary Material
Acknowledgements
Funding for this study was provided by a Broad Institute Broadnext10 Research Grant and National Institutes of Health research grants R01HL139337 and R01AI140751. The authors are also thankful for support provided by a National Science Foundation Graduate Research Fellowship (EJS), a Swiss National Science Foundation Postdoctoral Fellowship (CG), and a senior researcher scholarship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (MUF).
References
- Allison AC (1961). Genetic factors in resistance to malaria. Annals of the New York Academy of Sciences, 91(1), 710–729. [DOI] [PubMed] [Google Scholar]
- Barnwell JW, Nichols ME, & Rubinstein P (1989). In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. Journal of Experimental Medicine, 169(5), 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei AK, Brugnara C, & Duraisingh MT (2010). In vitro genetic analysis of an erythrocyte determinant of malaria infection. The Journal of Infectious Diseases, 202(11), 1722–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei AK, & Duraisingh MT (2012). Functional analysis of erythrocyte determinants of Plasmodium infection. International Journal for Parasitology, 42(6), 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, & van Steensel B (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research, 42(22), e168–e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, & Mendis KN (2002). Evolutionary and Historical Aspects of the Burden of Malaria. Clinical Microbiology Reviews, 15(4), 564–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasini CE, Mattos L. C. de, Couto ÁAD, Bonini-Domingos CR, Valencia SH, Neiras W. C. de S., … Machado RLD (2007). Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Transactions of the Royal Society of Tropical Medicine and Hygiene, 101(10), 1042–1044. [DOI] [PubMed] [Google Scholar]
- Chen K, Liu J, Heck S, Chasis J. a, An X, & Mohandas N (2009). Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proceedings of the National Academy of Sciences of the United States of America, 106(41), 17413–17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, & Miller LH (1996). The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. Journal of Experimental Medicine, 184(4), 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corkum CP, Ings DP, Burgess C, Karwowska S, Kroll W, & Michalak TI (2015). Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPTTM) and standard density gradient. BMC Immunology, 16(1), 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Berry D, & Baum J (2012). The cell biology of disease: The cellular and molecular basis for malaria parasite invasion of the human red blood cell. The Journal of Cell Biology, 198(6), 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman Alan F, & Crabb BS (2006). Invasion of red blood cells by malaria parasites. Cell, 124(4), 755–766. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, … Wright GJ (2011). Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature, 480(7378), 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Wanaguru M, McDade B, Osier FH, Marsh K, Rayner JC, & Wright GJ (2013). A library of functional recombinant cell-surface and secreted P. falciparum merozoite proteins. Molecular & Cellular Proteomics, 12(12), 3976–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crownover BK, & Covey CJ (2013). Hereditary hemochromatosis. American Family Physician, 87(3), 183–190. [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, … Orchard R (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature Biotechnology, 34(2), 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E, & Philipsen S (2013). Erythropoiesis: Development and Differentiation In Weatherall D, Schechter AN, & Nathan DG (Eds.), Additional Perspectives on Hemoglobin and Its Diseases (pp. 1–16). Cold Springs Harbor Perspectives in Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ES, Jiang RHY, Moechtar MA, Barteneva NS, Weekes MP, Nobre LV, … Wiegand RC (2015). A forward genetic screen identifies erythrocyte CD55 as essential for Plasmodium falciparum invasion. Science, 348(6235), 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajtova M, Kovarikova A, Svec P, Kankuri E, & Sedlak J (2013). Immunophenotypic profile of nucleated erythroid progenitors during maturation in regenerating bone marrow. Leukemia & Lymphoma, 54(11), 2523–2530. [DOI] [PubMed] [Google Scholar]
- Giarratana MC, Kobari L, Lapillonne H, Chalmers D, Kiger L, Cynober T, … Douay L (2005). Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nature Biotechnology, 23(1), 69. [DOI] [PubMed] [Google Scholar]
- Giarratana M, Rouard H, Dumont a, Kiger L, Safeukui I, Le Pennec PY, … Douay L (2011). Proof of principle for transfusion of in vitro generated red blood cells. Blood, 118(19), 5071–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, Sattabongkot J, … King CL (2007). Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Medicine, 4(12), 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszczyk J, Kanjee U, Chan L-J, Menant S, Malleret B, Lim NTY, … Pearson RD (2018). Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science, 359(6371), 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS (1949). The rate of mutation of human genes. Hereditas, 35(S1), 267–273. [Google Scholar]
- Hamblin MT, & Di Rienzo a. (2000). Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. American Journal of Human Genetics, 66(5), 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW (2012). Resistance to malaria in humans: the impact of strong, recent selection. Malaria Journal, 11(1), 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S, Takayama N, Nakamura S, Nagasawa K, Ochi K, Hirata S, … Eto K (2013). Immortalization of Erythroblasts by c-MYC and BCL-XL Enables Large-Scale Erythrocyte Production from Human Pluripotent Stem Cells. Stem Cell Reports, 1(6), 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, … Hay SI (2011). The global distribution of the Duffy blood group. Nature Communications, 2, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, … An X (2013). Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood, 121(16), 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Shah S, Wang J, Ye Z, Dowey SN, Tsang KM, … Cheng L (2014). Extensive ex vivo expansion of functional human erythroid precursors established from umbilical cord blood cells by defined factors. Molecular Therapy, 22(2), 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjee U, Grüring C, Chaand M, Lin K-M, Egan E, Manzo J, … Weekes MP (2017). CRISPR/Cas9 knockouts reveal genetic interaction between strain-transcendent erythrocyte determinants of Plasmodium falciparum invasion. Proceedings of the National Academy of Sciences, 114(44), E9356–E9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky K, Lichtman MA, Prchal JT, Levi MM, Press OW, Burns LJ, & Caligiuri M (2015). Williams Hematology (9th Ed.). New York, NY: McGraw-Hill. [Google Scholar]
- Kirk P, Wilson MC, Heddle C, Brown MH, Barclay AN, & Halestrap AP (2000). CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. The EMBO Journal, 19(15), 3896–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita R, Funato K, Abe T, Watanabe Y, Shiba M, Tadokoro K, … Satake M (2019). Establishment and characterization of immortalized erythroid progenitor cell lines derived from a common cell source. Experimental Hematology, 69, 11–16. [DOI] [PubMed] [Google Scholar]
- Kurita R, Suda N, Sudo K, Miharada K, Hiroyama T, Miyoshi H, … Nakamura Y (2013). Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PloS One, 8(3), e59890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP (2005). How malaria has affected the human genome and what human genetics can teach us about malaria. American Journal of Human Genetics, 77(2), 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Band G, Busby GBJ, Kivinen K, Le QS, Clarke GM, … Kwiatkowski DP (2017). Resistance to malaria through structural variation of red blood cell invasion receptors. Science, 356(6343). Retrieved from http://science.sciencemag.org/content/356/6343/eaam6393.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke J. a, … Sharp PM (2014). African origin of the malaria parasite Plasmodium vivax. Nature Communications, 5, 3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, … Zimmerman P. a. (2010). Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proceedings of the National Academy of Sciences of the United States of America, 107(13), 5967–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, … Arez AP (2011). Duffy negative antigen is no longer a barrier to Plasmodium vivax - molecular evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Neglected Tropical Diseases, 5(6), 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Clyde DF, & McGinniss MH (1976). The Resistance Factor to Plasmodium vivax in Blacks. New England Journal of Medicine, 295(6), 302–304. [DOI] [PubMed] [Google Scholar]
- Ord RL, Caldeira JC, Rodriguez M, Noe A, Chackerian B, Peabody DS, … Lobo CA (2014). A malaria vaccine candidate based on an epitope of the Plasmodium falciparum RH5 protein. Malaria Journal, 13(1), 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Egan ES, & Duraisingh MT (2015). Host–parasite interactions that guide red blood cell invasion by malaria parasites. Current Opinion in Hematology, 22(3), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel GW, Clark MA, Kanjee U, Lim C, Shaw-Saliba K, Menezes MJ, … Rathod PK (2018). Enhanced ex vivo Plasmodium vivax intraerythrocytic enrichment and maturation for rapid and sensitive parasite growth assays. Antimicrobial Agents and Chemotherapy, 62(4), e02519–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling L, Richards JS, Fowkes FJI, Wilson DW, Chokejindachai W, Barry AE, … Donelson J (2012). The Plasmodium falciparum erythrocyte invasion ligand Pfrh4 as a target of functional and protective human antibodies against malaria. PLoS One, 7(9), e45253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg G (2011). Microscopic Haematology: A Practical Guide for the Laboratory (3rd Ed.). Chatswood, Australia: Elsevier Health Sciences. [Google Scholar]
- Ryan JR, Stoute JA, Amon J, Dunton RF, Mtalib R, Koros J, … Rosenberg R (2006). Evidence for transmission of Plasmodium vivax among a Duffy antigen negative population in western Kenya. The American Journal of Tropical Medicine and Hygiene, 75(4), 575–581. [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, & Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods, 11(8), 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, & Zhang F (2015). High-throughput functional genomics using CRISPR–Cas9. Nature Reviews Genetics, 16(5), 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hora R, Belrhali H, Chitnis CE, & Sharma A (2006). Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature, 439(7077), 741–744. [DOI] [PubMed] [Google Scholar]
- Tamez PA, Liu H, Fernandez-Pol S, Haldar K, & Wickrema A (2009). Stage-specific susceptibility of human erythroblasts to Plasmodium falciparum malaria infection. Blood, 114(17), 3652–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakarnsanga K, Griffiths RE, Wilson MC, Blair A, Satchwell TJ, Meinders M, … Frayne J (2017). An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nature Communications, 8(May 2016), 14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH-F, Lee DJ, … Brogi E (2014). Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell, 156(5), 1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lochem EG, Van der Velden VHJ, Wind HK, Te Marvelde JG, Westerdaal NAC, & Van Dongen JJM (2004). Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: Reference patterns for age-related changes and disease-induced shifts. Cytometry Part B : Clinical Cytometry, 60(1), 1–13. [DOI] [PubMed] [Google Scholar]
- Vinjamur DS, & Bauer DE (2018). Growing and Genetically Manipulating Human Umbilical Cord Blood-Derived Erythroid Progenitor (HUDEP) Cell Lines BT - Erythropoiesis: Methods and Protocols. In Lloyd JA (Ed.) (pp. 275–284). New York, NY: Springer New York. [DOI] [PubMed] [Google Scholar]
- Wanaguru M, Crosnier C, Johnson S, Rayner JC, & Wright GJ (2013). Biochemical analysis of the Plasmodium falciparum erythrocyte-binding antigen-175 (EBA175)-glycophorin-A interaction: Implications for vaccine design. Journal of Biological Chemistry, 288(45), 32106–32117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu O. a, & Dondorp AM (2014). Malaria. Lancet, 383(9918), 723–735. [DOI] [PubMed] [Google Scholar]
- Williams TN (2006). Human red blood cell polymorphisms and malaria. Current Opinion in Microbiology, 9(4), 388–394. [DOI] [PubMed] [Google Scholar]
- Wood B (2004). Multicolor immunophenotyping: human immune system hematopoiesis In Methods in cell biology (Vol. 75, pp. 559–576). Elsevier. [DOI] [PubMed] [Google Scholar]
- World Malaria Report. (2015). World Health Organization. https://doi.org/ISBN9789241564403
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


