ABSTRACT
The selective clearance of cellular components by macroautophagy (hereafter autophagy) is critical for maintaining cellular homeostasis. In this punctum, we summarize and discuss our recent findings regarding a novel type of selective autophagy that targets centriolar satellites (CS) for degradation, a process we termed doryphagy from the Greek word “doryphoros”, standing for “satellite”. CS are microtubule-associated protein complexes that regulate centrosome composition. We show that CS degradation is mediated through a direct interaction between GABARAPs and an LC3-interacting region (LIR) motif in the CS protein PCM1. Autophagy-deficient systems accumulate large abnormal CS and consequently display centrosome reorganization and abnormal mitoses. Our findings provide a mechanistic link between autophagy deficiency and centrosome abnormalities and exemplify how mammalian Atg8-family proteins (mATG8s) can regulate substrate specificity.
KEYWORDS: Centriolar satellites, centrosome, doryphagy, mitosis, PCM1, selective autophagy
Whereas macroautophagy (hereafter autophagy) was long considered a largely nonspecific process, it is now well demonstrated that a wide range of substrates are specifically recruited to the developing autophagosome/phagophore for engulfment and degradation. Substrate specificity can be achieved through specialized autophagy receptors that simultaneously bind ubiquitin tags on the substrate and mATG8 proteins. Alternatively, substrate-resident proteins may directly bind mATG8 proteins through LIR motifs.
The centriolar satellites (CS) are large protein complexes associated with microtubules surrounding the centrosome that appear as cloud-like structures by fluorescent immunodetection. The CS are primarily composed of centrosome proteins, and they regulate centrosome composition through dynein-mediated trafficking along the microtubules. The centrosome is the major microtubule-organizing center of animal cells and also organizes the mitotic spindle that mediates the equal distribution of DNA during cell division. CS assembly depends on the PCM1 protein that provides a scaffold for their formation. As the CS have only begun to attract attention in recent years, many aspects of their biological function and regulation remain to be discovered.
We began our study after observing abnormal mitoses following genetic or pharmacological inhibition of autophagy [1]. To specify the nature of the abnormalities, we performed time-lapse imaging and detailed microscopy analyses. We found that the centrosomes were intact prior to cell division but then pulled apart in mitosis during chromosome alignment, a phenotype termed centrosome fragmentation. Consequently, autophagy-deficient cells display mitotic delay, unbalanced chromosome segregation, formation of micronuclei and post-mitotic cell death.
During mitosis, the centrosomes are subjected to a high level of stress from motor protein-generated traction forces, that align the chromosomes on the metaphase plate. We therefore reasoned that mitotic centrosome fragmentation could reflect an inherent instability in the centrosomes, making them unable to withstand such forces. A detailed analysis of centrosomes in autophagy-deficient vs. autophagy-proficient cells revealed that the centrosome composition varies between the two conditions. In particular, specific centrosome proteins accumulate in the CS, suggesting CS dysfunction. Indeed, a CS analysis showed that autophagy-deficient cells accumulate large dysmorphic CS, leading us to hypothesize that autophagy may mediate their degradation.
To identify putative CS substrates, we conducted mass spectrometry analyses using 3 mATG8 variants: MAP1LC3/LC3B, GABARAP and GABARAPL2 as bait. By this approach, we identified 2 strong CS hits, PCM1 and CEP131, that interact with GABARAPs but not LC3B, with a clear preference for GABARAPL2. Interestingly, PCM1 and CEP131 are precipitated as a complex, with PCM1 being the interaction point. As additional CS components show a similar mATG8 specificity, we concluded that the entire CS complex interacts with GABARAPs and found that the PCM1-GABARAP interaction is direct and occurs through a C-terminal LIR motif in PCM1. As the determinants of LIR binding to distinct mATG8s are not well-understood, we used modeling and molecular dynamics simulations to shed light on this topic. This approach allowed us to conclude that PCM1 specificity toward GABARAP depends on residues inside and upstream the core PCM1 LIR, as well as on specific polar and negatively-charged GABARAP residues (Y25 and E8, respectively).
Next, we determined that the CS are indeed autophagy substrates. CS proteins accumulate following inhibition of basal autophagy and decrease upon starvation. In addition, CS proteins clearly colocalize with both autophagosomes and lysosomes.
To link the PCM1-GABARAP interaction to CS degradation, we took advantage of a PCM1 LIR mutant, with strongly reduced binding affinity for GABARAPs. Unlike wild type PCM1, the PCM1 LIR mutant does not distribute to autophagosomes or lysosomes, confirming that the PCM1-GABARAP interaction is required for PCM1 degradation. Furthermore, depletion of GABARAPL2, but not LC3B or GABARAP, results in PCM1 accumulation and abnormal mitoses, in agreement with the highest binding affinity being observed for GABARAPL2. Thus, CS accumulation and mitotic phenotypes display the same mATG8 selectivity as the PCM1-mATG8 interaction.
In sum, our findings identify a novel selective autophagy pathway of CS, a process we term doryphagy from the Greek word “doryphoros”, standing for “satellite” (Figure 1). While our results led us to focus on cell division, doryphagy may potentially affect all centrosome-related functions, such as cellular migration, ciliogenesis, cell polarity and cell adhesion, affecting cell fate and cell survival in addition to proliferation. Interestingly, Sharon Tooze’s lab has identified PCM1 as a key factor in modulating, the other way around, autophagosome formation, implying the existence of a dual activity of this molecule. Indeed, the presence of regulatory feedback pathways in controlling autophagy is a quite common scenario, with, for example, the upstream autophagy regulators MTOR/mTOR, AMPK, AKT and ULK1 being reciprocally and finely modulated by post-translational modifications.
Figure 1.
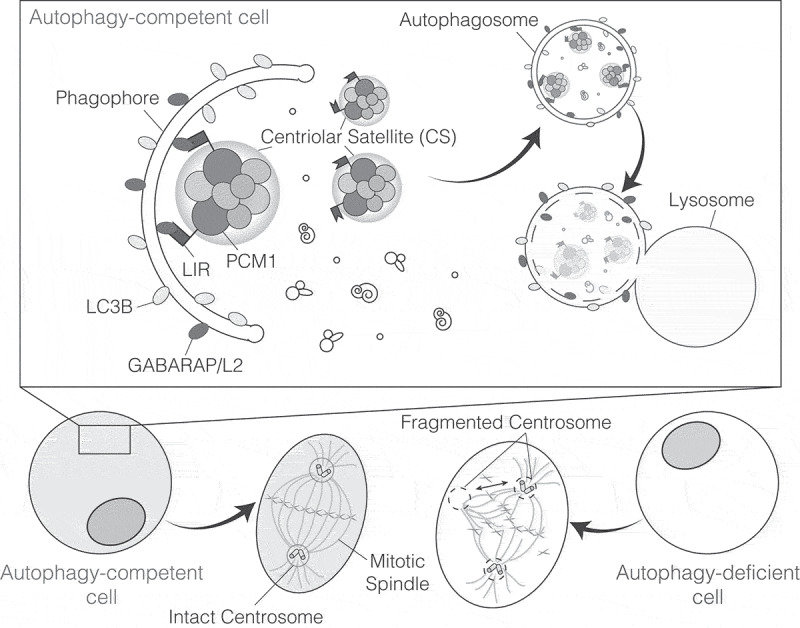
Centriolar satellites are substrates of selective autophagy. The centriolar satellite (CS) protein PCM1 directly binds GABARAP/L2 through a LIR motif. This interaction mediates the selective engulfment of the CSs targeted for degradation into phagphores. Notably, autophagy inhibition results in loss of pericentriolar material integrity, which has an impact on centrosome stability and causes aberrant mitoses.
In recent years, an increasing number of selective autophagy pathways have been discovered, in which mATG8 proteins directly interact with substrate-resident proteins or even individual long-living proteins. Examples include FASN/FAS (fatty acid synthase), CBL/c-Cbl for SRC/Src kinase degradation, RETREG1/FAM134B for endoplasmic reticulum disposal and BNIP3L/Nix, BNIP3, FUNDC1, and Atg32 for mitophagy. In the case of doryphagy, we have identified PCM1 as a CS-specific autophagy receptor. While we show that CS degradation is mediated through a direct LIR-dependent interaction, a number of questions remain unanswered. Is the interaction and thus the degradation regulated? Does it occur without the requirement of post-translational modification on the key regulators GABARAPL2 or PCM1? Are the CS targeted for degradation continuously as a baseline control system or are abnormal CS somehow flagged for removal? Also, can CS degradation be (specifically) induced in response to stimuli (other than starvation) and if so, what is the functional significance? Interestingly, the CS have previously been reported to disperse in response to stress stimuli. Doryphagy may therefore be part of a larger stress network regulating the CS. How this interplay is regulated and how it affects the centrosome and possibly cell cycle progression is an interesting topic for further studies.
Another interesting aspect is the selectivity toward GABARAPs and in particular GABARAPL2. With the identification of new selective degradation pathways, it will be interesting to link mATG8 proteins to individual cellular functions and, possibly, to map mATG8 expression patterns to specific cell types, tissues, developmental stages or pathological conditions. The continuous improvement of techniques for identification of functional LIRs and their mATG8 specificity may aid this process.
Finally, the relevance of autophagy in controlling centrosome homeostasis by acting on CS activity may highlight additional roles for this process in a number of pathological conditions. Due to the key function of centrosomes in cell division, polarity and migration, the relationship between doryphagy and cancer biology is particularly relevant, and this novel selective autophagy process may turn out in the near future to be of the highest importance in biomedicine.
Funding Statement
This work was supported by the Danish Cancer Society (KBVU R72-A4408, R146-A9364), the Novo Nordisk Foundation (7559, 22544), the Lundbeckfonden (R233-2016-3360), the LEO Foundation (LF17024), and “Associazione Italiana per la Ricerca sul Cancro” (AIRC IG-23543) to Francesco Cecconi. Valentina Cianfanelli was supported by the Lundbeckfonden (R209–20153505) and the Danish Cancer Society (KBVU R146-A9471).
Disclosure statement
No potential conflict of interest was reported by the authors.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Reference
- [1].Holdgaard SG, Cianfanelli V, Pupo E, et al. Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites. Nat Commun. 2019;10. DOI: 10.1038/s41467-019-12094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


