ABSTRACT
Cellular homeostasis requires selective autophagic degradation of damaged or defective organelles, including the endoplasmic reticulum (ER). Previous studies have shown that specific ER transmembrane receptors recruit LC3 on autophagic membranes by using LC3-interacting domains. In this study, we showed that the N-degron pathway mediates ubiquitin (Ub)-dependent reticulophagy. During this 2-step process, the ER transmembrane E3 ligase TRIM13 undergoes auto-ubiquitination via lysine 63 (K63) linkage chains and acts as a ligand for the autophagic receptor SQSTM1/p62 (sequestosome 1). In parallel, ER-residing molecular chaperones, such as HSPA5/GRP78/BiP, are relocated to the cytosol and conjugated with the amino acid L-arginine (Arg) at the N-termini by ATE1 (arginyltransferase 1). The resulting N-terminal Arg (Nt-Arg) binds the ZZ domain of SQSTM1, inducing oligomerization of SQSTM1-TRIM13 complexes and facilitating recruitment of LC3 on phagophores to the sites of reticulophagy. We developed small molecule ligands to the SQSTM1 ZZ domain and demonstrate that these chemical mimics of Nt-Arg facilitate reticulophagy and autophagic protein quality control of misfolded aggregates in the ER.
KEYWORDS: Alpha1-antitrypsin deficiency, ER homeostasis, ER protein quality control, ER stress response, ER-phagy, N-degron pathway, N-terminal arginylation, SQSTM1/p62, TRIM13, ubiquitination
Macroautophagy mediates degradation of unwanted or cytotoxic substrates such as misfolded protein aggregates and subcellular organelles. The targeting of protein cargoes requires specific receptors such as SQSTM1 and NBR1 that bind Ub chains on cargoes via their Ub-associated (UBA) domains and LC3 on autophagic membranes via an LC3-interacting regions (LIR). The targeted cargoes are sequestered within autophagosomes, which in turn fuse with lysosomes for cargo degradation. In addition to proteins, macroautophagy is responsible for degradation of subcellular organelles as well as invading pathogens such as bacteria and viruses. Mechanistically, autophagic degradation of organelles are divided into 2 types. In Ub-dependent pathways, as exemplified by the PINK1-PRKN/Parkin pathway in mitophagy, ubiquitination of transmembrane ligands generates a molecular beacon that attracts autophagic receptors, which bring phagophores to the sites of organellophagy. In contrast, Ub-independent pathways involve specific transmembrane ligands that act as receptors whose LIR domains directly recruit autophagic membranes. Recent studies identified LIR-containing transmembrane receptors of reticulophagy such as RETREG1/FAM134B, SEC62, CCPG1, and TEX264 (Figure 1). However, it has remained unknown whether and how degradation signals such as Ub regulate reticulophagy.
Figure 1.
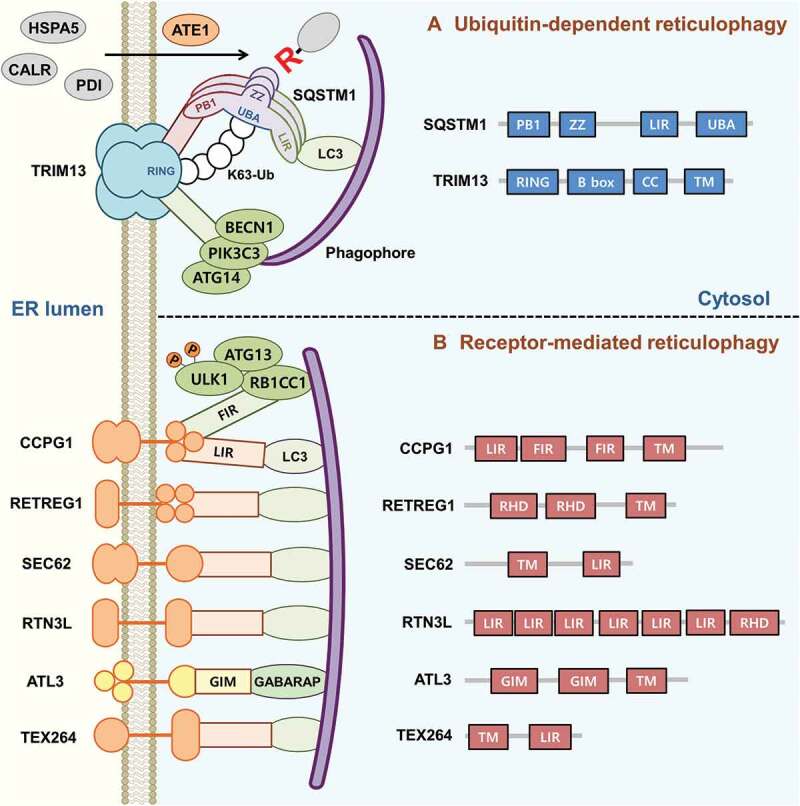
Two distinct mechanisms of reticulophagy. (A) Ub-dependent reticulophagy is mediated by the N-degron pathway, which involves the SQSTM1-Nt-Arg-TRIM13 circuit. On the ER membrane, the receptor TRIM13, which undergoes auto-ubiquitination via K63-linked Ub chains, recruits SQSTM1. In parallel, ER-residing proteins are translocated to the cytosol and Nt-arginylated by ATE1. The Nt-Arg of these proteins binds the SQSTM1-ZZ domain and thereby induces the oligomerization of SQSTM1 in complex with TRIM13. Oligomeric SQSTM1-TRIM13 complexes mark the sites of reticulophagy and recruit LC3 on phagophores through Nt-Arg-induced interaction of SQSTM1 with LC3. Oligomerized TRIM13 also serves as a platform of morphogen complexes composed of PIK3C3/VPS34, BECN1 and ATG14, which induce ER membrane curvature for efficient dissociation and subsequent reticulophagy. (B) Receptor-mediated reticulophagy is mediated by ER transmembrane receptors carrying LIR or GIM domains that can directly recruit LC3 or GABARAP on phagophores. These receptors include CCPG1, RETREG1/FAM134B, SEC62, RTN3 long isoform (RTN3L), TEX264 and ATL3.
The N-degron pathway modulates the half-lives of substrates based on the identity of their N-terminal destabilizing residues, called N-degrons. N-degrons include the Nt-Arg, which is generated through conjugation of L-Arg to Nt-Asp, Nt-Glu, and oxidized Nt-Cys by ATE1. N-recognins, such as UBR box-carrying E3 ligases, bind the Nt-Arg and induce the ubiquitination and proteasomal degradation of the Nt-arginylated substrate. We have recently identified SQSTM1 as an N-recognin whose ZZ domain binds Nt-Arg to induce cargo degradation by lysosomal hydrolases via autophagy. Upon binding to Nt-Arg, SQSTM1 undergoes conformational changes, facilitating PB1 domain-dependent self-polymerization and its interaction with LC3 on phagophores. This dual mechanism modulates autophagic protein quality control for cytosolic misfolded proteins and aggregates.
In this study, we show that the N-degron pathway regulates reticulophagy [1] (Figure 1). In this process, the E3 ligase TRIM13 acts as an ER transmembrane receptor that auto-ubiquitinates each other via a K63 linkage. Ubiquitinated TRIM13 binds cytosolic SQSTM1 whose ZZ domain, in turn, binds the Nt-Arg of arginylated proteins. Upon binding to the Nt-Arg, SQSTM1 undergoes PB1 domain-dependent self-oligomerization in complex with TRIM13 and interacts with LC3 on autophagic membranes, which marks the site of reticulophagy on both the smooth and the rough ER. Our results suggest that the N-degron pathway is a major mechanism underlying reticulophagy in normal as well as stressed conditions. Where are the Nt-Arg from? Our earlier studies show that under cellular stresses, a subpopulation of ER-residing molecular chaperones, such as HSPA5, CALR (calreticulin), and PDI (protein disulfide isomerase), are increasingly Nt-arginylated by R-transferases, leading to their cytosolic accumulation. Under these stress conditions, a large number of Nt-Arg-exposing proteins in the cytosol, which would otherwise be degraded via the Ub-proteasome system (UPS), are also metabolically stabilized and serve as active ligands to the SQSTM1 ZZ domain, modulating SQSTM1 activity and omegasome biogenesis. Perhaps, the Nt-Arg residues of arginylated proteins may represent a unique proteome that modulates autophagic proteolysis as well as organellophagy at the crossroad between the UPS (as a degron) and autophagy (as an autophagy-modulating ligand). The physiological role of the Nt-Arg in organellophagy and other macroautophagic processes remains to be further investigated.
Although ER-associated degradation (ERAD) efficiently removes soluble misfolded ER clients through ubiquitination and proteasomal degradation, it remains unclear how misfolded aggregates are removed from the ER lumen for degradation. Indeed, a number of human genetic disorders are caused by ERAD-resistant protein aggregates, such as mutant SERPINA1/α1-antitrypsin/A1AT in α1-antitrypsin deficiency and mutant CFTR (CF transmembrane conductance regulator) in cystic fibrosis. In this study, we developed synthetic ligands to the SQSTM1 ZZ domain and showed that these compounds accelerate reticulophagy and, moreover, alleviate ER stress and promote ER proteostasis by clearing ER-resident misfolded aggregates. Our results suggest that the N-degron pathway mediates ER protein quality control via autophagy, functioning as a second defense line against aggregation-prone misfolded proteins that escape from ERAD. As ER stress is implicated in not only the pathogenesis of various diseases including cancer, metabolic disorders and proteinopathies but also the death of cells tasked with extreme protein synthesis such as with antibody production, SQSTM1 may be a potential drug target for a myriad of therapeutic applications.
One outstanding question concerns the role of Nt-Arg in inducing oligomerization of SQSTM1 in complex with TRIM13. Although SQSTM1 binds K63-ubiquitinated TRIM13 independent of Nt-Arg, their monomeric complex appears to be insufficient to trigger reticulophagy, instead requiring self-polymerization driven by Nt-Arg as an N-degron. Given that TRIM13 recruits the BECN1-PIK3C3/VPS34 complex that signals ER membrane curvature and phagophore nucleation from the omegasome, N-degron-dependent reticulophagy is likely to occur in the proximity of maturing phagophores. If so, oligomerization of organellular receptors may be a common signal to initiate autophagic removal of subcellular structures.Another question to be addressed is the relative role of Ub-dependent and Ub-independent mechanisms for reticulophagy. Given our finding that inactivation of any part of the SQSTM1-Nt-Arg-TRIM13 circuit significantly blocks basal and stress-induced reticulophagy, the N-degron pathway plays a dominating role in reticulophagy. However, we do not exclude the possibility that the two systems cooperatively work for reticulophagy. Finally, given the roles of the N-degron pathway in aggrephagy and reticulophagy, it remains to be seen whether the N-degron pathway mediates other types of organellophagy, such as mitophagy, pexophagy, and lipophagy.
Funding Statement
This work was supported by National Research Council of Science and Technology (NST) [grant number CAP-16-03-KRIBB], Ministry of Science, ICT, and Future Planning (MSIP) [grant number NRF-2016R1A2B3011389], and the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program;Korea Research Institute of Bioscience and Biotechnology [Research Initiative Program]; Ministry of Science, ICT, and Future Planning [NRF-2016R1A2B3011389]; National Research Council of Science and Technology (KR) [CAP-16-03-KRIBB].
Acknowledgments
This work was supported by the R&D Convergence Program (CAP-16-03-KRIBB) of the National Research Council of Science & Technology (NST) of Korea, the KRIBB Research Initiative Program (to B.Y.K.), the Basic Science Research Programs of the NRF funded by the Ministry of Science, ICT, and Future Planning (MSIP) (NRF-2016R1A2B3011389 to Y.T.K.), Seoul National University (SNU) Nobel Laureates Invitation Program, and the Brain Korea 21 PLUS Program (to SNU).
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- [1].Ji CH, Kim HY, Heo AJ, et al. The N-degron pathway mediates ER-phagy. Mol Cell. 2019. September 05;75(5):1058–1072. PubMed PMID: 31375263. [DOI] [PubMed] [Google Scholar]


