Abstract
Shuangbai Tablets (SBT), a traditional herbal mixture, has shown substantial clinical efficacy. However, a systematic mechanism of its active ingredients and pharmacological mechanisms of action against proteinuria continues being lacking. A network pharmacology approach was effectual in discovering the relationship of multiple ingredients and targets of the herbal mixture. This study aimed to identify key targets, major active ingredients, and pathways of SBT against proteinuria by network pharmacology approach combined with thin layer chromatography (TLC). Human phenotype (HP) disease analysis, gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and molecular docking were used in this study. To this end, a total of 48 candidate targets of 118 active ingredients of SBT were identified. Network analysis showed PTGS2, ESR1, and NOS2 to be the three key targets, and beta-sitosterol, quercetin, and berberine were the three major active ingredients; among them one of the major active ingredients, quercetin, was discriminated by TLC. These results of the functional enrichment analysis indicated that the most relevant disease including these 48 candidate proteins is proteinuria, SBT treated proteinuria by sympathetically regulating multiple biological pathways, such as the HIF-1, RAS, AGE-RAGE, and VEGF signaling pathways. Additionally, molecular docking validation suggested that major active ingredients of SBT were capable of binding to HIF-1A and VEGFA of the main pathways. Consequently, key targets, major active ingredients, and pathways based on data analysis of SBT against proteinuria were systematically identified confirming its utility and providing a new drug against proteinuria.
1. Introduction
Proteinuria is an established marker of kidney damage and a risk factor for progression of chronic kidney disease (CKD) [1–3]. Albuminuria is classified as an indicator of advanced CKD [4] according to the Kidney Disease Improving Global Outcome (KDIGO) clinical practice guideline for glomerulonephritis [5]. Macroalbuminuria (>300 mg/24 h) is an extremely common feature in diabetic patients with advanced CKD, with a frequency range between 33% in stage 3B and nearly 100% in stage 5 [4, 6]. Proteinuria is associated with considerable mortality not only [7, 8] in the diabetic and nondiabetic population but also with cardiovascular events [9]. Therefore, controlling urinary albumin levels is important in treating proteinuria and its associated primary diseases [10]. Indeed, proteinuria pathogenesis is complex and multifactorial due to its relation with various diseases. However, some studies have found that proteinuria is associated with podocyte injury, endothelial cell injury, and an imbalance in the regulation of vascular endothelial growth factor (VEGF) [11–14].
Despite some advancements in the understanding of proteinuria pathogenesis, there is still a lack of therapies that work against proteinuria other than using the renin-angiotensin system (RAS) inhibitor as the gold standard [5], such as angiotensin-converting enzyme inhibitors (ACEI) or angiotensin receptor blockers (ARB). Unfortunately, RAS blocking slows down but does not preclude the progression of kidney disease [15] and increases the potential risk of hyperkalemia [16]. Immunosuppressive treatment with corticosteroids has shown some effect in remission of proteinuria although a certain percentage of patients do not respond to treatment and are labeled steroid resistant [17]. Glucocorticoid therapy can be resistant or dependent and can also cause many adverse reactions [17, 18]. Thus, it is crucial to search for more safe and effective drugs in treating proteinuria and its primary diseases based on their pharmacological mechanisms [15].
Recently, Traditional Chinese Medicine (TCM) has played an important role in the treatment of proteinuria via its multicomponent, multitarget, and multipathway approach [15, 19–23]. Shuangbai Tablet (SBT) is a classic TCM, which was approved by the Shanghai Food and Drug Administration. It has achieved satisfactory results for the treatment of mild to moderate proteinuria and its primary disease for more than twenty years in Ruijin Hospital, Shanghai Jiao Tong University [24]. SBT is composed of 15 traditional Chinese herbs: Imperatae Rhizoma (IRH), Hedysarum multijugum Maxim (HMM), Acanthopanax senticosus (ASE), Caragana sinica Rehd (CSR), Achyranthis Bidentatae Radix (ABR), Radix Paeoniae Rubra (RPR), Paeoniae Radix Alba (PRA), Fructus lipuidambaris (FRU), Lycopi Herba (LHE), Tetrapanacis Medulla (TME), Polygonum perfoliatum L (PPL), Sambucus williamsii Hance (SWH), Dipsaci Radix (DRA), Stephaniae Tetrandrae Radix (STR), and Chuanxiong Rhizoma (CRH). Furthermore, some single-flavor herbs or ingredients in SBT, such as Astragaloside IV of HMM, paeoniflorin of PRA, and CRH have also been revealed an exact therapeutic effect against proteinuria [25–27].
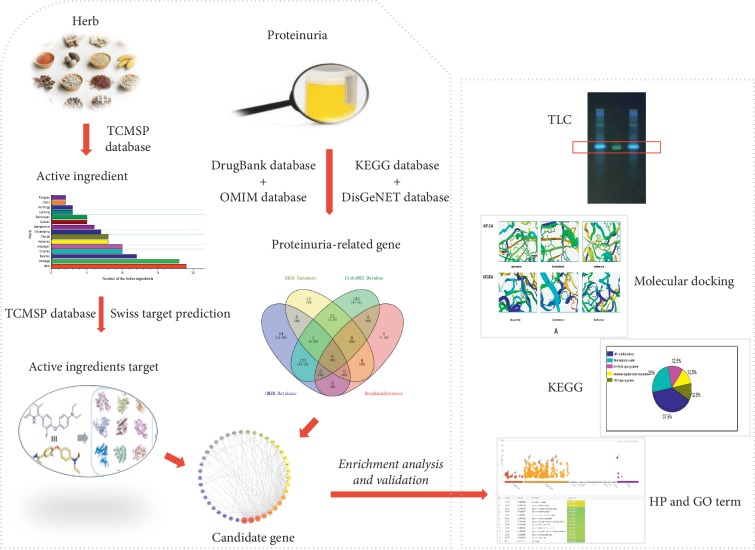
However, active components and potential pharmacological mechanisms of action of SBT against proteinuria have not been identified. In this study, human phenotype (HP) disease analysis, gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and molecular docking were applied to investigate the bioactive constituents and the underlying mechanisms of SBT for the treatment of proteinuria, combined with thin layer chromatography (TLC), to identify a single active ingredient. The flow diagram of this study is shown in Figure 1.
Figure 1.
Flow diagram of this study. Left: summary of the identification of representative ingredients of SBT and genes with efficacy against proteinuria. Right: summary of the determination and validation of the pharmacological mechanisms of SBT.
2. Materials and Method
SBT (MUST-1901001) was provided by Ruijin Hospital, Shanghai Jiaotong University, China. Quercetin (MUST-100081-200406, purity ≥97.3%) was purchased by China National Institute for Biological Products Standard. Reagent methanol (MUST-20180105), hydrochloric acid (MUST-20190104), ethyl acetate (MUST-20170609), formic acid (MUST-20140212), and toluene (MUST-20160106) supplied by Lingfeng Chemical Reagent Co., Ltd. (Shanghai, China). Ethanol (MUST-20190530) and aluminum trichloride (MUST-20100127) were supplied by Klings and Qiangshun Chemical Reagent Co., Ltd. (both in Shanghai, China), respectively. Distilled water was used, and all other reagents were of analytical reagent.
2.1. Active Ingredients Database Building
The active ingredients of each herb in SBT were obtained using TCMSP (http://lsp.nwsuaf.edu.cn/tcmsp.php) [28], according to bioavailability (OB) ≥30% [20] and drug-likeness (DL) ≥0.18 [29], as well as TCMGeneDIT (http://tcm.lifescience.ntu.edu.tw/) [30] and related studies [31–37].
2.2. Targets of Active Ingredients Database Building
The targets of active ingredients were searched in TCMSP and SwissTargetPrediction (http://www.swisstargetprediction.ch) databases with Homo sapiens [38, 39]. SwissTargetPrediction is a molecular similarity match tool based on simplified molecules which contain a larger collection of 376,342 compounds known to be experimentally active on an extended set of 3068 macromolecular targets [38].
2.3. Genes for Building a Proteinuria Database
Genes associated with proteinuria were searched on four existing human disease resources, including Online Mendelian Inheritance in Man Database (OMIM, http://www.omim.org/) [40], the Kyoto Encyclopedia of Genes and Genomes Pathway Database (KEGG, https://www.kegg.jp/) [41], DisGeNET (https://www.disgenet.org/) [42] database, and DrugBank database (https://www.drugbank.ca/) [43]. After deletion of duplications, Uniprot ID of each target of proteinuria was queried by the Uniport database (https://www.uniprot.org/) [44].
2.4. Candidate Target Identification and “Herbs-Active Ingredients-Candidate Targets” Comprehensive Network Construction
Candidate targets against proteinuria were identified by matching the targets of the active ingredients of SBT and the genes related to proteinuria. Next, the “herbs-active ingredients-candidate targets” comprehensive network was established using Cytoscape software (v3.7.1; https://www.nigms.nih.gov/). We calculated the number of overlapped active ingredients in 15 herbs, as well as the number of overlapped candidate targets in active ingredients, and the top three were considered as the major active ingredients and the key targets, respectively.
2.5. Candidate Target Interacting Protein Network Construction
The interacting protein candidate targets against proteinuria were obtained from the String database (https://string-db.org/) [45–47] in which protein-protein interaction (PPI) information between two proteins or multiple proteins was given, and the combined score of each pair of interacting proteins was greater than or equal to 0.4 [47]. Next, the “candidate targets-proteins” network was built by Cytoscape 3.7.1 software [48–50]. Calculating the average degree of the nodes by using Network Analyzer plugin, all the nodes of the network represented central targets for enrichment analysis [50].
2.6. Enrichment Analysis Methods and Molecular Docking Simulation
2.6.1. Enrichment Analysis Method
We performed HP and online GO term enrichment analysis by placing all central targets into g:Profiler (https://biit.cs.ut.ee/gprofiler/) [51–53]. KEGG pathway enrichment analysis was performed by placing those targets into ClueGo plugin (ClueGo v2.5.4 + ClueGoPedia v1.5.4) [54].
HP enrichment analysis is a web that provides a standardized vocabulary of phenotypic abnormalities encountered in human disease which can reveal the correlation between central targets and proteinuria. GO term is able to classify functions into three aspects: molecular functions (molecular activities of gene products), cellular components (where gene products are active), and biological processes (pathways and larger processes made up of the activities of multiple gene products).
2.6.2. Molecular Docking
After identifying the pathways, in order to certify the reliability of the above enrichment analysis, we docked the factors in the pathways and the major active ingredients by SystemsDock (http://systemsdock.unit.oist.jp/) [55, 56], which classified the major active ingredients' activity by the criterion: a docking score greater than 5.52 or the score of the local ligands [55].
SystemsDock is a web server for network pharmacology-based prediction and analysis, which applies high-precision docking simulation and molecular pathway map to comprehensively characterize the ligand selectivity and to illustrate how a ligand acts on a complex molecular network. Structure files in the formats of 2D of the major active ingredients download from PubChem [57] (https://pubchem.ncbi.nlm.nih.gov/) and protein PDB ID of the factors in the pathways download from String and PDB database [58] (https://www.rcsb.org/).
2.7. Quercetin TLC Validation
2.7.1. Sample of SBT
30 tablets of SBT were dissolved in 50 ml (methanol-hydrochloric acid (25%) (4 : 1)) solution and heated under reflux for 1 hour. The residue was dissolved (20 ml distilled water) and extracted twice (20 ml of ethyl acetate). Next, ethyl acetate solution was washed (10 ml of distilled water), discarding the aqueous solution. Finally, the dissolved residue (2 ml of methanol) was prepared as SBT sample.
2.7.2. Standard Sample of Quercetin
Standard sample of quercetin (100081–200406, 2 mg/ml) was prepared by dissolving the solutes in methanol.
2.7.3. Thin Layer Chromatography
According to the thin layer chromatography (General Rule 0502, Part IV, Chinese Pharmacopoeia) test, the solutions of SBT samples (10 μl) and quercetin standard sample (2 μl) were spotted manually on the chromatographic plates. The mixture of toluene + ethyl acetate + formic acid (25 : 20 : 1) was used as the mobile phase. The plates were developed vertically at room temperature (20°C) to a distance of 10 cm and then dried for 20 h at room temperature (20°C). Finally, it was sprayed with 10% aluminum trichloride in ethanol and placed under ultraviolet light (356 nm) for examination.
3. Results
3.1. Active Ingredients of SBT
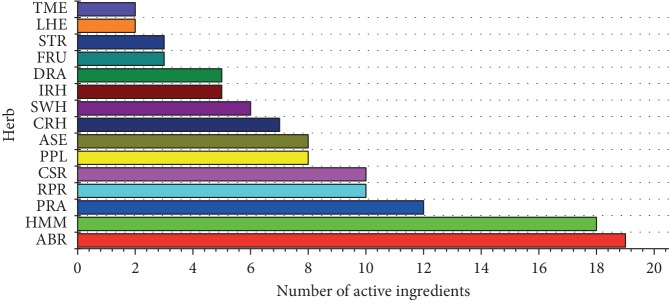
A total of 118 active ingredients (Supplementary ) were identified from 15 herbs in this study. The number of active ingredients in IRH, HMM, ASE, CSR, ABR, RPR, PRA, FRU, LHE, TME, PPL, SWH, DRA, STR, and CRH was 5, 18, 8, 10, 19, 10, 12, 3, 2, 2, 8, 6, 5, 3, and 7, respectively (shown in Figure 2). The top three herbs with higher active ingredients were ABR, HMM, and PRA (see Figure 2).
Figure 2.
The number of active ingredients in SBT.
3.2. The 8981 Targets of Active Ingredients
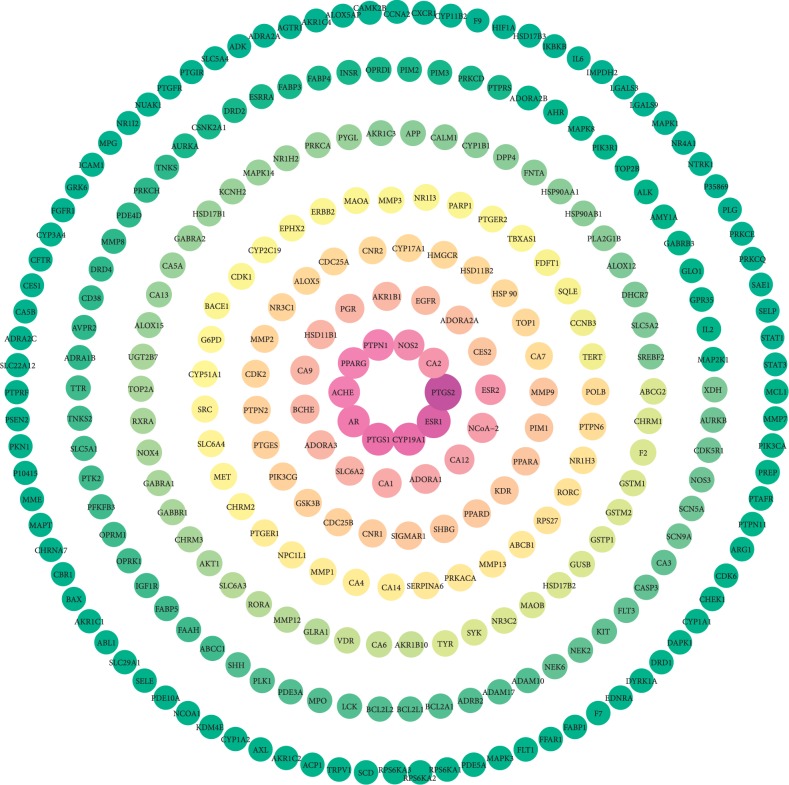
A total of 8981 human disease targets of the active ingredients in SBT (Supplementary ) were identified, among them 289 targets with the active ingredients frequency greater than 10 are shown in Figure 3. The top three active ingredients with higher number of targets are beta-sitosterol, quercetin, and berberine, and the top 10 overlapped targets are PTGS2, ERS1, CYP19A1, PTGS1, AR, ACHE, PPARG, PTPN1, NOS2, and CA2. The information of these targets is listed in Table 1.
Figure 3.
The targets with a frequency greater than 10; the color of the targets is shown in a gradient from red to green according to descending order of the frequency value with a yellow excess in the middle.
Table 1.
The information of the top 10 overlapped targets.
| Gene | Uniport ID | Description | Active ingredient category |
|---|---|---|---|
| PTGS2 | P35354 | Prostaglandin G/H synthase 2 | 106 kinds, kaempferol, quercetin, etc. |
| ESR1 | P03372 | Estrogen receptor | 83 kinds, beta-sitosterol, berberine, etc. |
| CYP19A1 | P11511 | Cytochrome P450 19A1 | 76 kinds, berberine, palmatine, etc. |
| PTGS1 | P23219 | Prostaglandin G/H synthase 1 | 75 kinds, palmatine, quercetin, etc. |
| AR | P10275 | Androgen receptor | 71 kinds, coniferin, kaempferol, etc. |
| ACHE | P22303 | Acetylcholinesterase | 67 kinds, beta-sitosterol, mairin, etc. |
| PPARG | P37231 | Peroxisome proliferator-activated receptor gamma | 66 kinds, coniferin, mairin, etc. |
| PTPN1 | P18031 | Protein-tyrosine phosphatase 1B | 66 kinds, daucosterol, hesperetin, etc. |
| NOS2 | P35228 | Nitric-oxide synthase, inducible | 62 kinds, sitosterol, coptisine, etc. |
| CA2 | P00918 | Carbonic anhydrase II | 59 kinds, albiflorin, kaempferol, etc. |
3.3. Genes Associated with Proteinuria
A total of 523 human disease genes associated with proteinuria (Supplementary ) were identified from four databases. The number of genes in the OMIM, KEGG, DisGeNET, and DrugBank databases was 182, 33, 303, and 5, respectively.
3.4. Candidate Target and “Herbs-Active Ingredients-Candidate Targets” Network
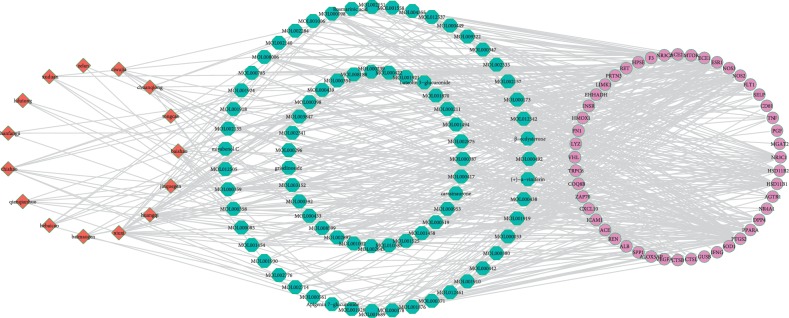
A total of 48 candidate targets (Supplementary ) were shown to have pharmacological effects against proteinuria. The comprehensive network of “herbs-active ingredients-candidate targets” had 141 nodes and 524 edges, as determined by the Cytoscape 3.7.1 software (see Figure 4). There were 16 active ingredients with the number of targets exceeding 8, of which, the top three active ingredients (beta-sitosterol (MOL000358), quercetin (MOL000098), and berberine (MOL001454)) were represented as the three major active ingredients. Information of those 16 active ingredients, including their inclusion in herbs and the number of targets they regulate, is listed in Table 2. As shown in Table 2, beta-sitosterol is contained in 9 herbs, such as ABR, HMM, and PRA; quercetin is contained in HMM and PPL; berberine is contained in ABR.
Figure 4.
The “herbs-active ingredients-candidate targets” network; the red diamond nodes represent the herbs, the blue hexagon nodes represent the active components, and the purple circle nodes represent the targets.
Table 2.
The information of the top 16 active ingredients with the herb number exceeding 8.
| Compound | Mol ID | Targets category | Herbs |
|---|---|---|---|
| MOL000358 | Beta-sitosterol | 15 kinds, NR3C1, HSD11B2, PPARA, etc. | ABR, IRH, PRA, RPR, STR, FRU, DRA, LHE, TME |
| MOL000098 | Quercetin | 11 kinds, DPP4, PPARA, PTGS2, etc. | HMM, PPL |
| MOL001454 | Berberine | 11 kinds, PTGS2, ALOX5AP, ICAM1, etc. | ABR |
| MOL008006 | Paryriogenin A | 11 kinds, NR3C1, ACE, NOS2, etc. | TME |
| MOL002897 | Epiberberine | 11 kinds, PTGS2, ESR1, ZAP70, etc. | ABR |
| MOL000296 | Hederagenin | 11 kinds, ALOX5AP, LYZ, NOS2, etc. | ASE, HMM, LHE |
| MOL000378 | 7-O-Methylisomucronulatol | 10 kinds, DPP4, GUSB, RET, etc. | IRH, HMM |
| MOL012542 | β-Ecdysterone | 10 kinds, AGTR1, NR3C2, TNF, etc. | ABR |
| MOL001494 | Mandenol | 9 kinds, PPARA, PTGS2, FLT1, etc. | CRH |
| MOL000519 | Coniferin | 8 kinds, AGTR1, ESR1, PTGS2, etc. | ASE, FRU, SWH |
| MOL000422 | Kaempferol | 8 kinds, ICAM1, HMOX1, DPP4, etc. | RPA, PPL, HMM, CSR, ABR |
| MOL000211 | Mairin | 8 kinds, ALOX5AP, CD81, NOS2, etc. | RPA, PPL, HMM |
| MOL002875 | Methyl oleate | 8 kinds, PPARA, PTGS2, HSD11B2, etc. | ASE |
| MOL000359 | Sitosterol | 8 kinds, ESR1, HSD11B1, NR3C2, etc. | SWH, RPA, RPR, CRH, FRU |
| MOL000449 | Stigmasterol | 8 kinds, PTGS2, ESR1, NOS2, etc. | IRH, RPR, ABR |
| MOL002157 | Wallichilide | 8 kinds, CTSL, ACE, CTSB, etc. | CRH |
There were 10 candidate targets with the number of active ingredients exceeding 14 (Table 3), of which the top 3 candidate targets (prostaglandin G/H synthase 2 (PTGS2), estrogen receptor (ESR1), and nitric oxide synthase (NOS2)) were represented as three key targets. As shown in Table 3, PTGS2, ESR1, and NOS2 are regulated by 74, 59, and 37 kinds of active ingredients, respectively, and all those three key targets can be regulated by three major active ingredients.
Table 3.
The information of the top 10 key targets with the active ingredient number exceeding 14.
| Gene | UniProt ID | Description | Active ingredients |
|---|---|---|---|
| PTGS2 | P35354 | Prostaglandin G/H synthase 2 | 74 kinds, beta-sitosterol, quercetin, berberine, etc. |
| ESR1 | P03372 | Estrogen receptor | 59 kinds, beta-sitosterol, quercetin, berberine, etc. |
| NOS2 | P35228 | Nitric oxide synthase, inducible | 37 kinds, beta-sitosterol, quercetin, berberine, etc. |
| NR3C1 | P04150 | Glucocorticoid receptor | 29 kinds, formononetin, coptisine, baicalein, etc. |
| HSD11B1 | P28845 | 11-Beta-hydroxysteroid dehydrogenase 1 | 28 kinds, stigmasterol, daucosterol, delta 7-stigmastenol, etc. |
| PPARA | Q07869 | Peroxisome proliferator-activated receptor alpha | 21 kinds, formononetin, coniferin, delta 7-stigmastenol, etc. |
| HSD11B2 | P80365 | 11-Beta-hydroxysteroid dehydrogenase 2 | 19 kinds, hederagenin, bidentatoside, mandenol, etc. |
| NR3C2 | P08235 | Mineralocorticoid receptor | 19 kinds, wallichilide, epiberberine, methyl oleate, etc. |
| DPP4 | P27487 | Dipeptidyl peptidase IV | 14 kinds, baicalein, quercetin, methyl oleate, etc. |
| NOS3 | P29474 | Nitric-oxide synthase, endothelial | 14 kinds, isorhamnetin, coptisine, syringin, etc. |
3.5. Candidate Targets-Proteins Network
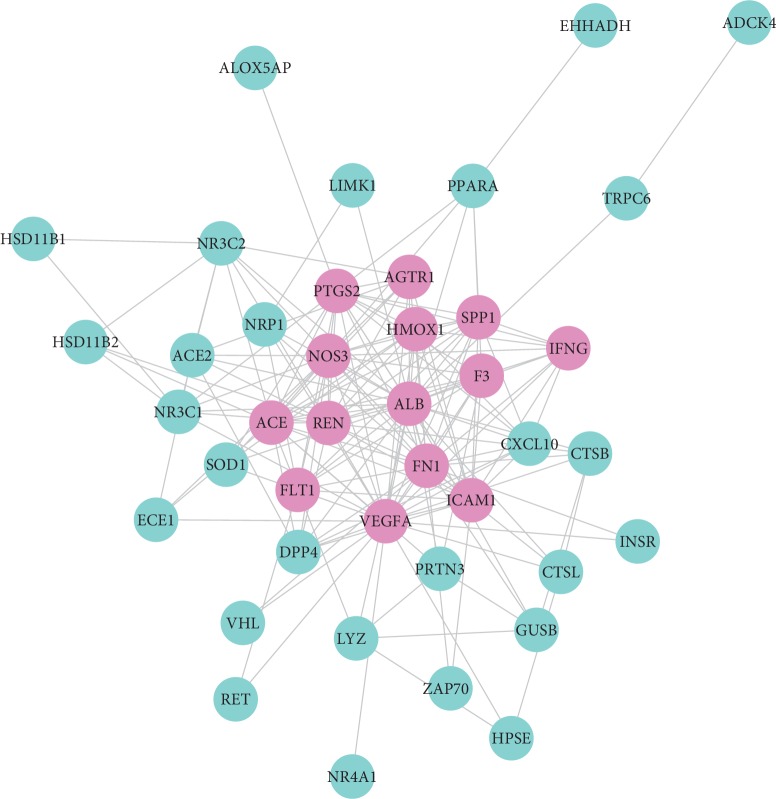
From the String database of “multiple proteins” interaction of targets, we gained 172 pairs of protein interactions of candidate targets (Supplementary ). Then, a network of “candidate targets-proteins” was constructed by Cytoscape 3.7.1 software (shown in Figure 5), and specific analytical parameters through NetworkAnalyzer plugin were in the supplemental material (Supplementary ). There were 41 nodes and 172 edges in this network, in which 41 nodes represented 41 central targets.
Figure 5.
The “Candidate Targets-Proteins” network; the red nodes represent candidate targets related proteins with degree greater than or equal to 10, and the blue nodes represent the degree of proteins less than 10.
3.6. Enrichment Analysis and Molecular Docking
3.6.1. Enrichment Results
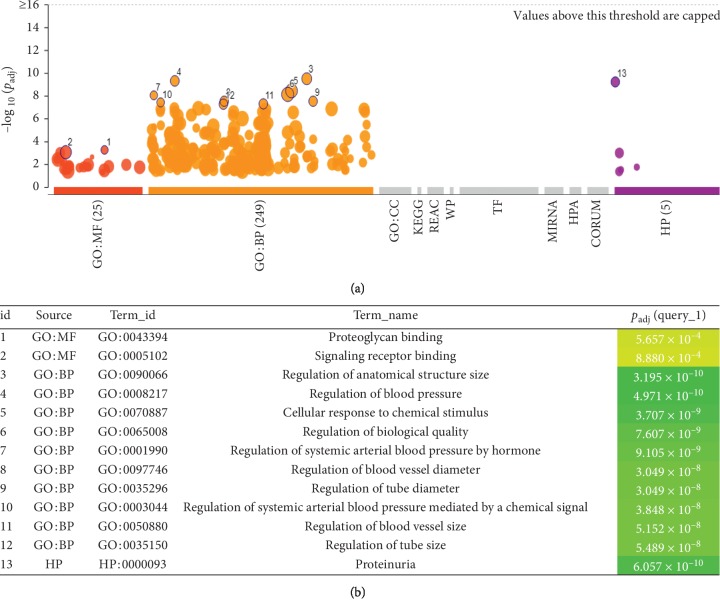
The HP enrichment analysis indicated that the most relevant human disease of the central targets was proteinuria (Figure 6, id 13).
Figure 6.
The results of the enrichment result; red points: id 1 and id 2 are the top 2 of GO molecular function; orange points: id 3 to id 12, the top 10 of GO biological process and purple points: id 13, HP results of the candidate targets.
The results of the GO term-molecules function showed that all the central targets were associated with proteoglycan binding and signal receptor binding (Figure 6, id 1 and id 2). The GO term-biological process showed that these targets were highly related to systemic arterial blood pressure by RAS system and so forth (Figure 6, id 3 to id 12).
The KEGG pathway enrichment results showed that these central targets were mainly concentrated in the hypoxia-inducible factor-1 (HIF-1) signaling pathway (37.5%), renin-angiotensin system (RAS) signaling pathway (25%), advanced glycation end products-receptor of advanced glycation end products (AGE-RAGE) signaling pathway (12.5%), aldosterone-regulated sodium reabsorption (12.5%), and vascular endothelial growth factor (VEGF) signaling pathway (12.5%) (see Figure 7).
Figure 7.
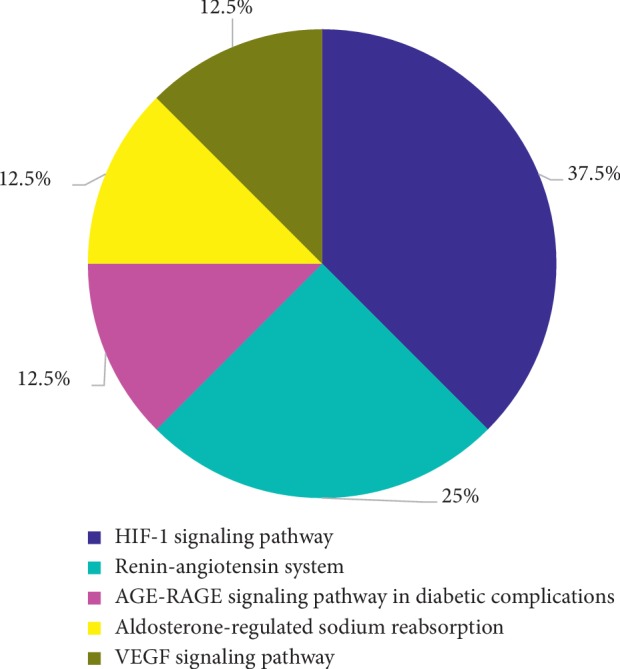
KEGG pathway enrichment results of the candidate targets. Blue, light blue, pink, yellow, and brown represent the HIF-1 signaling pathway, the renin-angiotensin system, the AGE-RAGE signaling pathway, aldosterone-mediated sodium reabsorption, and VEGF signaling, respectively.
3.6.2. Molecular Docking
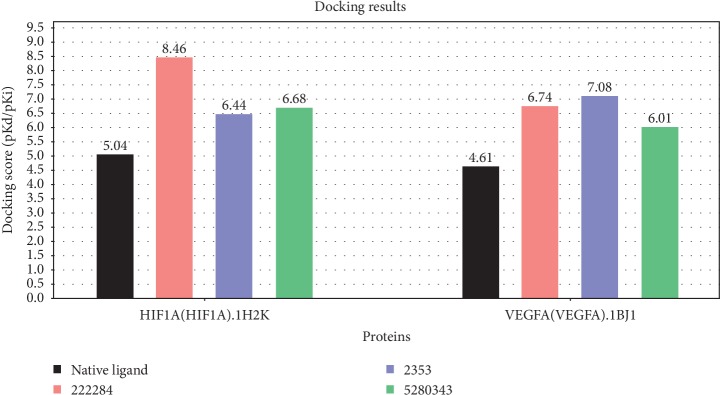
Molecular docking of bate-sitosterol, quercetin, and berberine with HIF-1A and VEGFA, respectively, all docking scores were not only greater than local ligand scores, but also greater than 5.52 (shown in Figure 8). The results showed that three major active ingredients could be well binding with those two factors, respectively. The 3D binding mode of 3 major active ingredients in the active site of HIF-1A and VEGFA, respectively, is represented in Figure 9(a), while schematic 2D representation is shown in Figure 9(b).
Figure 8.
Results of the 3 major active compounds docking with 2 factors in SystemsDock. Black, pink, blue, and green column charts represent the score of native ligands, beta-sitosterol (222284), berberine (2353), and quercetin (5280343), respectively.
Figure 9.
Results of the 3D and 2D docking models of 3 major active compounds docking with 2 factors in SystemsDock. (a) 3D Docking model, quercetin, β-sitosterol, and berberine are shown as stick with blue, and HIF-1A and VEGFA are represented as strips. (b) 2D docking model, 3 major active compounds are shown as stick with purple, and residues of 2 factors are represented as pink curves.
3.7. TLC Validation Results
In the TLC chromatogram, there are fluorescent spots in the chromatogram of the SBT sample corresponding with quercetin standard sample both in position and color (shown in Figure 10).
Figure 10.
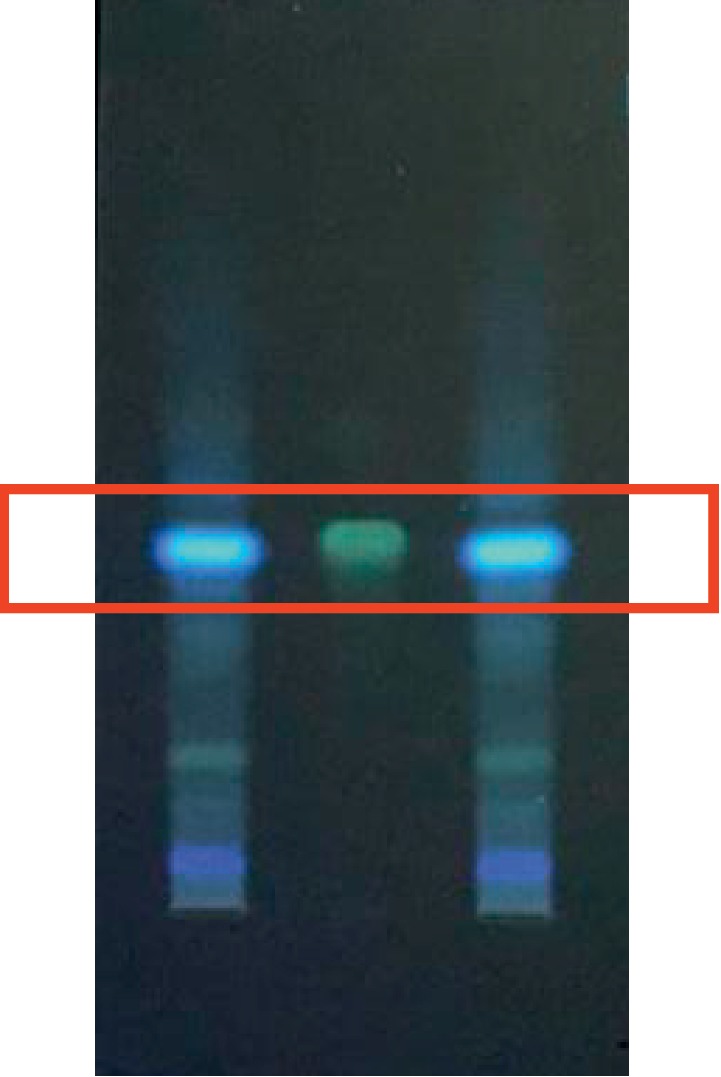
Results of TLC validation of quercetin in SBT. Middle line is the fluorescent point of the quercetin standard sample, and the SBT sample is on both sides.
4. Discussion
Network pharmacology is a mechanism research method for TCM, based on the characteristics of the comprehensive regulation of “multi-ingredient, multitarget, and multipathway” [59–62]. In this study, synergistic pharmacological mechanisms of SBT against proteinuria were identified through 3 major active ingredients (beta-sitosterol, quercetin, and berberine), 48 candidate targets including 3 key targets and 3 pathways including HIF-1 signaling pathway, VEGF signaling pathway, RAS signaling pathway, and AGE-RAGE signaling pathway. In particular, those candidate targets were highly correlated with proteinuria in our results of HP enrichment, and those 3 major active ingredients were capable of binding to HIF-1A and VEGFA protein. Together with one of the major active ingredients, quercetin in SBT was discriminated by TLC, and all the results confirmed the reliability of our prediction.
4.1. Beta-Sitosterol, Quercetin, and Berberine as the Major Active Ingredients of SBT
In our study, beta-sitosterol contained in 9 herbs (HMM and PRA), quercetin contained in HMM and PPL, and berberine contained in ABR acted on 15, 11, and 11 candidate targets against proteinuria, respectively. In addition, 92.1% of 118 active ingredients acted on at least two candidate targets (Figure 4). More importantly, the three major active ingredients had tighter combinations than the local ligand with HIF-1A and VEGFA in our docking results, which demonstrates that SBT worked against proteinuria through a multi-ingredient synergistic way.
Previous studies have demonstrated that beta-sitosterol has been widely used in the treatment of proteinuria resulting from diabetes and kidney disease [63–65]. Berberine has also been reported to treat proteinuria and kidney damage caused by diabetes and hypertension [66–68], which provided evidence for our results.
It is worth mentioning that quercetin in SBT was detected in our TLC results, and studies confirmed it was able to treat proteinuria as well as improve renal function through regulating HIF-1A and VEGFA factors in rat models [69–71], which is in line with our docking results. Interestingly, other active compounds such as Astragaloside IV (contained in HMM) and paeoniflorin (contained in PRA) with a concentration of 3.20–64.00 μg·mL−1 were detected in SBT in our previously quality standard study [72]. Studies have experimentally proved that Astragaloside IV was able to reduce proteinuria in diabetic nephropathy mice [73] and paeoniflorin could improve proteinuria in chronic renal failure rats [25]. All of the above supports our result that SBT worked against proteinuria in a multi-ingredient way.
4.2. PTGS2, ESR1, and NOS2 as the Key Targets of SBT against Proteinuria
In our study, PTGS2, ESR1, and NOS2 were overlapped by 74, 59, and 37 active ingredients, respectively. Moreover, 68.75 % of 48 candidate targets of SBT can be overlapped by at least two active ingredients in SBT (Figure 4), which demonstrated the active ingredient in SBT worked against proteinuria through a multitarget synergistic way.
It is reported that PTGS2 (also known as COX-2) was involved in podocyte injury and various renal pathological processes such as renal interstitial fibrosis, showing a potential therapeutic target against lupus nephritis-induced nephropathy [74, 75]. Coincidentally, several studies have proved that PTGS2 was inhibited by beta-sitosterol, quercetin, and berberine inhibiting the expression of PTGS2 in mouse colitis, the small intestine mucosa of rats, and human endometrial cells, respectively [76–78]. ESR1 is a potential therapy target for proteinuria caused by IgA nephropathy through the proliferation of glomerular mesangial cells [79]. Related research showed that quercetin regulated the activity of ESR1 in HepG2 (hepatocytes) and Caco-2 (intestinal) [80].
NOS2 could produce NO which simultaneously stimulates the increased expression of COX-2 and transforming growth factor-β1, causing damage in renal function, vascular and structural [81]. Related scholars have confirmed that NOS2 can be inhibited by beta-sitosterol, quercetin, and berberine in insulin rats, mouse hepatocyte, and mouse 264.7 macrophages, respectively [82–85]. Other candidate targets can also play a curative role in SBT against proteinuria, such as HSD11B1 [86] and PPAPA [87].
All these studies supported our results that SBT worked against proteinuria in a multitarget synergistic way. Whether three active ingredients act against proteinuria through regulating PTGS2, ESR1, and NOS2, it needs further study.
4.3. HIF-1, RAS, VEGF, and AGE-RAGE Signaling Pathway Playing an Important Role in SBT against Proteinuria
The HIF-1 signaling pathway and RAS signaling pathway were identified as mechanisms of SBT against proteinuria from ClueGO results. Studies have reported that high expression of HIF-1A in renal tubular epithelial cells can aggravate glomerular hypertrophy and extracellular matrix accumulation, increasing urinary protein excretion [88]. More importantly, HIF-1 signaling mediated pathways, such as the mitogen-activated protein kinase (MAPK) signaling pathway, and the phosphatidylinositol-3-kinase protein kinase B (PI3K-Akt) signaling pathway [89] both directly damaged renal function through contributing to renal fibrosis or proteinuria-induced renal epithelial-mesenchymal transition [90–92]. Fortunately, berberine was reported to activate HIF-1A to adapt to the stressful conditions and protected tubular epithelial cells from apoptosis in diabetic nephropathy rats [93]. However, it needs further study of whether berberine worked against proteinuria through activating HIF-1A.
In addition, many studies [94–97] have confirmed that the RAS system is closely related to proteinuria caused by various diseases, such as diabetic nephropathy and CKD. CKD is associated with hypoxia which can activate HIF-1 [98] and induce profibrogenic changes in proximal tubular epithelial cells and interstitial fibroblasts, leading to further aggravation of proteinuria [99]. Additionally, it has been demonstrated that proteinuria can activate the local RAS system in the kidneys, which can then cause vasoconstriction and subsequent hypoxia [100]. To sum up, this cross talk between proteinuria and hypoxia can cause a vicious circle of kidney destruction. Therefore, it can be speculated that the inhibition of the RAS system and HIF-1 activation may be the potential treatment for proteinuria. Fortunately, it has been reported that quercetin inhibits the RAS system and inhibits the expression of growth-transforming factor-β which promotes glomerular sclerosis and renal tubular interstitial fibrosis in progressive nephropathy [101]. However, there is still a lack of studies on whether quercetin works against proteinuria through inhibiting the RAS system.
Inhibition of RAS can also regulate the high glucose-induced VEGF signaling pathway [102]. VEGF-A, the most specific and prominent angiogenic factor of VEGF family, has been reported to be involved in the process of angiogenesis, migration, and proliferation of endothelial cells [8, 88, 103, 104]. In addition, autocrine secretion of VEGF was caused by the interactions of AGEs and their receptor RAGE, which were new risk factors in the pathogenesis of end-stage renal disease [105, 106]. In this background, beta-sitosterol was reported to inhibit the expression of VEGF in kidney cancer rats, protecting functions in renal tissues [107]. Quercetin can also inhibit RAGE and reduce cardiovascular damage in diabetic nephropathy [108]. Therefore, it can be speculated that inhibition of VEGF and AGE-RAGE signaling pathways may be potential treatments for proteinuria.
5. Conclusion
In conclusion, the “multi-ingredient, multitarget, and multipathway” of SBT against proteinuria were effectively elucidated by the network pharmacology approach. Beta-sitosterol is one of the major active ingredients used to treat proteinuria caused by IgA nephropathy, by targeting ERS1 through its anti-immune effects. Berberine reduces proteinuria and kidney damage caused by diabetes and hypertension through two key targets, PTGS2 and NOS2, which improve podocyte injury. Quercetin, contained in SBT through TLC verification, reduces proteinuria by regulating HIF-1 and VEGF in nephrons. Additionally, blocking RAS, a gold standard for treating proteinuria caused by CKD, can prevent HIF-1 and prevent VEGF from being activated. Blocking AGE-RAGE to prevent the autocrine signaling of VEGF can treat proteinuria caused by diabetic nephropathy. Thus, we predicted that SBT plays a therapeutic role in proteinuria caused by IgA nephropathy, CKD, and diabetic nephropathy and prevents complications such as cardiovascular diseases. All these contribute to the utility of SBT and may provide a new drug against proteinuria. However, more experimental studies are still needed to verify these findings.
Acknowledgments
This project was supported by the construction of important weak disciplines in Shanghai of Shanghai Health and Wellness Committee (2016ZB0304-01) and Pharmaceutical and Clinical Research of Double White Tablets, Shanghai Natural Science Foundation (08ZR141400).
Data Availability
The data of our research can be acquired from the Supplementary Materials uploaded with this article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Xiaoqin Ma and Meixiang Yu contributed equally to this manuscript. Xiaoqin Ma, Meixiang Yu, and Wanhua Yang designed, wrote, and revised the manuscript. Chenxia Hao revised the manuscript and provided technical or material support.
Supplementary Materials
Supplementary Table 1: 118 active ingredients in Shuangbai Tablets. Supplementary Table 2: 8981 targets of the active ingredients in Shuangbai Tablets. Supplementary Table 3: 110 523 genes associated with proteinuria. Supplementary Table 4: candidate targets with therapeutic proteinuria. Supplementary Table 5: 172 pairs of protein interactions of candidate targets. Supplementary Table 6: specific analytical parameters of 41 central targets.
References
- 1.Pesola G. R., Argos M., Chen Y., et al. Dipstick proteinuria as a predictor of all-cause and cardiovascular disease mortality in Bangladesh: a prospective cohort study. Preventive Medicine. 2015;78:72–77. doi: 10.1016/j.ypmed.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster A. C., Nagler E. V., Morton R. L., Masson P. Chronic kidney disease. The Lancet. 2017;389(10075):1238–1252. doi: 10.1016/s0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 3.Ying T., Clayton P., Naresh C., Chadban S. Predictive value of spot versus 24-hour measures of proteinuria for death, end-stage kidney disease or chronic kidney disease progression. BMC Nephrology. 2018;19:p. 55. doi: 10.1186/s12882-018-0853-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P.-M., Wada T., Chiang C.-K. Prognostic value of proteinuria and glomerular filtration rate on Taiwanese patients with diabetes mellitus and advanced chronic kidney disease: a single center experience. Clinical and Experimental Nephrology. 2017;21(2):307–315. doi: 10.1007/s10157-016-1290-8. [DOI] [PubMed] [Google Scholar]
- 5.MBalk E., Raman G., Dana C. M., Deo A., Amy E., Haynes KDIGO clinical practice guideline for glomerulonephritis. Kidney International Supplements. 2012;142:137–194. [Google Scholar]
- 6.Wada T., Haneda M., Furuichi K., et al. Clinical impact of albuminuria and glomerular filtration rate on renal and cardiovascular events, and all-cause mortality in Japanese patients with type 2 diabetes. Clinical and Experimental Nephrology. 2014;18(4):613–620. doi: 10.1007/s10157-013-0879-4. [DOI] [PubMed] [Google Scholar]
- 7.Wen C. P., Cheng T. Y. D., Tsai M. K., et al. All-cause mortality attributable to chronic kidney disease: a prospective cohort study based on 462 293 adults in Taiwan. The Lancet. 2008;371(9631):2173–2182. doi: 10.1016/s0140-6736(08)60952-6. [DOI] [PubMed] [Google Scholar]
- 8.Shujun Chen H. C., Qi L., Ma Q. Effect of simvastatin on the expression of nephrin, podocin, and vascular endothelial growth factor (VEGF) in podocytes of diabetic rat. Nefrologia. 2015;35:131–138. [PMC free article] [PubMed] [Google Scholar]
- 9.Röthlisberger S., Pedroza-Diaz J. Urine protein biomarkers for detection of cardiovascular disease and their use for the clinic. Expert Review of Proteomics. 2017;14(12):1091–1103. doi: 10.1080/14789450.2017.1394188. [DOI] [PubMed] [Google Scholar]
- 10.Hiremath S., Fergusson D. A., Fergusson N., Bennett A., Knoll G. A. Renin-angiotensin system blockade and long-term clinical outcomes in kidney transplant recipients: a meta-analysis of randomized controlled trials. American Journal of Kidney Diseases. 2017;69(1):78–86. doi: 10.1053/j.ajkd.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Liu M., Liang K., Zhen J., et al. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nature Communications. 2017;8:p. 413. doi: 10.1038/s41467-017-00498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cellesi F., Li M., Rastaldi M. P. Podocyte injury and repair mechanisms. Current Opinion in Nephrology and Hypertension. 2015;24(3):239–244. doi: 10.1097/mnh.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 13.Timothy H. E. M., Sutton A., Campos B., et al. Injury of the renal microvascular endothelium alters barrier function after ischemia. American Journal of Physiology-Renal Physiology. 2003;285 doi: 10.1152/ajprenal.00042.2003. [DOI] [PubMed] [Google Scholar]
- 14.Eremina V., Sood M., Haigh J., et al. Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. Journal of Clinical Investigation. 2003;111(5):707–716. doi: 10.1172/jci17423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y. M., Chiang W. C., Lin S. L., Tsai T. J. Therapeutic efficacy of pentoxifylline on proteinuria and renal progression: an update. Journal of Biomedical Science. 2017;24:p. 84. doi: 10.1186/s12929-017-0390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu F.-Y., Lin F.-J., Ou H.-T., Huang S.-H., Wang C.-C. Renoprotective effect of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers in diabetic patients with proteinuria. Kidney and Blood Pressure Research. 2017;42(2):358–368. doi: 10.1159/000477946. [DOI] [PubMed] [Google Scholar]
- 17.Dogra S., Kaskel F. Steroid-resistant nephrotic syndrome: a persistent challenge for pediatric nephrology. Pediatric Nephrology. 2017;32(6):965–974. doi: 10.1007/s00467-016-3459-5. [DOI] [PubMed] [Google Scholar]
- 18.Funder J. W. Minireview: aldosterone and mineralocorticoid receptors: past, present, and future. Endocrinology. 2010;151(11):5098–5102. doi: 10.1210/en.2010-0465. [DOI] [PubMed] [Google Scholar]
- 19.Gao X., Shang J., Liu H., Yu B. A meta-analysis of the clinical efficacy of TCM decoctions made from formulas in the liuwei dihuang wan categorized formulas in treating diabetic nephropathy proteinuria, evidence-based complementary and alternative medicine. eCAM. 2018;2018 doi: 10.1155/2018/2427301.2427301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong X.-G., An Z.-M., Guo Y., Zhou J.-L., Qin T. Effect of triptolide on expression of oxidative carbonyl protein in renal cortex of rats with diabetic nephropathy. Journal of Huazhong University of Science and Technology. 2017;37(1):25–29. doi: 10.1007/s11596-017-1689-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X. M., Zhang Y., He X. H., et al. Chinese herbal medicine Shenzhuo Formula treatment in patients with macroalbuminuria secondary to diabetic kidney disease: study protocol for a randomized controlled trial. Trials. 2018;19:p. 200. doi: 10.1186/s13063-018-2573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Deng Y., Ni Z., et al. Efficacy and safety of traditional Chinese medicine (Shenqi particle) for patients with idiopathic membranous nephropathy: a multicenter randomized controlled clinical trial. American Journal of Kidney Diseases. 2013;62(6):1068–1076. doi: 10.1053/j.ajkd.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y., Cai G., Sun X., Chen X. Treatment of chronic kidney disease using a traditional Chinese medicine, flos abelmoschus manihot (linnaeus) medicus (malvaceae) Clinical and Experimental Pharmacology and Physiology. 2016;43(2):145–148. doi: 10.1111/1440-1681.12528. [DOI] [PubMed] [Google Scholar]
- 24.Yang W. H., Yuan Y. F., Wang G. Y., Chen X. N. Overview of western medicine intervention in proteinuria. Chinese Pharmacist. 2008;76:p. 11. [Google Scholar]
- 25.La L., Wang L., Qin F., et al. Zhen-wu-tang ameliorates adenine-induced chronic renal failure in rats: regulation of the canonical Wnt4/beta-catenin signaling in the kidneys. Journal of Ethnopharmacology. 2018;219:81–90. doi: 10.1016/j.jep.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Yang W.-J., Li Y.-R., Gao H., et al. Protective effect of the ethanol extract from Ligusticum chuanxiong rhizome against streptozotocin-induced diabetic nephropathy in mice. Journal of Ethnopharmacology. 2018;227:166–175. doi: 10.1016/j.jep.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 27.Yu Z. K., Yang B., Zhang Y., Li L. S., Zhao J. N., Hao W. Modified Huangqi Chifeng decoction inhibits excessive autophagy to protect against Doxorubicin-induced nephrotic syndrome in rats via the PI3K/mTOR signaling pathway. Experimental and Therapeutic Medicine. 2018;16:2490–2498. doi: 10.3892/etm.2018.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai F.-F., Bian Y.-Q., Wu R., et al. Yinchenhao decoction suppresses rat liver fibrosis involved in an apoptosis regulation mechanism based on network pharmacology and transcriptomic analysis. Biomedicine & Pharmacotherapy. 2019;114:p. 108863. doi: 10.1016/j.biopha.2019.108863. [DOI] [PubMed] [Google Scholar]
- 30.Fang Y. C., Huang H. C., Chen H. H., Juan H. F. TCMGeneDIT: a database for associated traditional Chinese medicine, gene and disease information using text mining. BMC Complementary and Alternative Medicine. 2008;8:p. 58. doi: 10.1186/1472-6882-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan D., Zhou X., Zhao C., Chen H., Zhao Y., Gong X. Anti-inflammatory, antiviral and quantitative study of quercetin-3-O-β-D-glucuronide in Polygonum perfoliatum L. Fitoterapia. 2011;82(6):805–810. doi: 10.1016/j.fitote.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Shao Hua M. Y. B. W., Ji Kai L. The chemical constituents from toricellia angulata. Acta Botanica Yunnanica. 2000;22:214–218. [Google Scholar]
- 33.Zhang Q. G., Wei F., Liu Q., et al. The flavonoid from Polygonum perfoliatum L. inhibits herpes simplex virus 1 infection. Acta Virologica. 2014;58(4):368–373. doi: 10.4149/av_2014_04_368. [DOI] [PubMed] [Google Scholar]
- 34.Xu L., Huang G., Guo X., Zhou Q., He S. Total flavonoids, extracted from Polygonum knotweed L, exert beneficial hepatoprotection against liver injury. Journal of Cellular Biochemistry. 2019;120 doi: 10.1002/jcb.28535. [DOI] [PubMed] [Google Scholar]
- 35.Cho H., Park J.-H., Ahn E.-K., Oh J. S. Kobophenol A isolated from roots of Caragana sinica (Buc’hoz) rehder exhibits anti-inflammatory activity by regulating NF-κB nuclear translocation in J774A.1 cells. Toxicology Reports. 2018;5:647–653. doi: 10.1016/j.toxrep.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S. R., Kwak J. H., Kim H. J., Pyo S. Neuroprotective effects of kobophenol A against the withdrawal of tropic support, nitrosative stress, and mitochondrial damage in SH-SY5Y neuroblastoma cells. Bioorganic & Medicinal Chemistry Letters. 2007;17(7):1879–1882. doi: 10.1016/j.bmcl.2007.01.078. [DOI] [PubMed] [Google Scholar]
- 37.Meng Q., Niu Y., Niu X., Roubin R. H., Hanrahan J. R. Ethnobotany, phytochemistry and pharmacology of the genus Caragana used in traditional Chinese medicine. Journal of Ethnopharmacology. 2009;124(3):350–368. doi: 10.1016/j.jep.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 38.Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Research. 2019;47 doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong M., Cai Z., Song L., Liu Y., Wang Q., Feng X. Gynostemma pentaphyllum attenuates the progression of nonalcoholic fatty liver disease in mice: a biomedical investigation integrated with in silico assay. Evidence-Based Complementary and Alternative Medicine. 2018;2018 doi: 10.1155/2018/8384631.8384631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amberger J. S., Hamosh A. Searching online mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Current Protocols in Bioinformatics. 2017;58 doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piñero J., Bravo À., Queralt-Rosinach N., et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wishart D. S., Feunang Y. D., Marcu A., et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Research. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Research. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franceschini A., Szklarczyk D., Frankild S., et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Mering C., Jensen L. J., Kuhn M., et al. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Research. 2007;35(D1):D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H.-Y., Chen J.-Q., Li J.-Y., et al. Deep learning and random forest approach for finding the optimal traditional chinese medicine formula for treatment of alzheimer’s disease. Journal of Chemical Information and Modeling. 2019;59(4):1605–1623. doi: 10.1021/acs.jcim.9b00041. [DOI] [PubMed] [Google Scholar]
- 48.Brysbaert G., Lorgouilloux K., Vranken W., Lensink M. F. RINspector: a cytoscape app for centrality analyses and dynamine flexibility prediction. Bioinformatics. 2017;34 doi: 10.1093/bioinformatics/btx586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y. W., Wang Y., Guo Z. F., Du K. C., Meng D. L. Systems pharmacological approach to investigate the mechanism of ohwia caudata for application to alzheimer’s disease. Molecules. 2019;24 doi: 10.3390/molecules24081499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anitha P., Anbarasu A., Ramaiah S. Gene network analysis reveals the association of important functional partners involved in antibiotic resistance: A report on an important pathogenic bacterium Staphylococcus aureus. Gene. 2016;575(2):253–263. doi: 10.1016/j.gene.2015.08.068. [DOI] [PubMed] [Google Scholar]
- 51.Reimand J., Arak T., Adler P., et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Research. 2016;44(W1):W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raudvere U., Kolberg L., Kuzmin I., et al. Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Research. 2019;47 doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimand J., Isserlin R., Voisin V., et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and enrichmentmap. Nature Protocols. 2019;14(2):482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J., Ma X., Liu C., et al. Exploring the mechanism of danshen against myelofibrosis by network pharmacology and molecular docking, evidence-based complementary and alternative medicine. eCAM. 2018;2018 doi: 10.1155/2018/8363295.8363295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsin K.-Y., Matsuoka Y., Asai Y., et al. systemsDock: a web server for network pharmacology-based prediction and analysis. Nucleic Acids Research. 2016;44(W1):W507–W513. doi: 10.1093/nar/gkw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong Q., Ma Y., Yu J., Chen X. Predicted molecular targets and pathways for germacrone, curdione, and furanodiene in the treatment of breast cancer using a bioinformatics approach. Scientific Reports. 2017;7:p. 15543. doi: 10.1038/s41598-017-15812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Bryant S. H., Cheng T., et al. Pubchem bioassay: 2017 update. Nucleic Acids Research. 2017;45(D1):D955–D963. doi: 10.1093/nar/gkw1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burley S. K., Berman H. M., Kleywegt G. J., Markley J. L., Nakamura H., Velankar S. Protein data bank (PDB): the single global macromolecular structure archive. Methods in Molecular Biology. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W., Yuan G., Pan Y., Wang C., Chen H. Network pharmacology studies on the bioactive compounds and action mechanisms of natural products for the treatment of diabetes mellitus: a review. Frontiers in Pharmacology. 2017;8:p. 74. doi: 10.3389/fphar.2017.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hopkins A. L. Network pharmacology. Nature Biotechnology. 2007;25(10):1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 61.Yuan H., Ma Q., Cui H., et al. How can synergism of traditional medicines benefit from network pharmacology? Molecules. 2017;22 doi: 10.3390/molecules22071135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong M., Li S., Wang N., Tan H. Y., Cheung F., Feng Y. Gynostemma pentaphyllum attenuates the progression of nonalcoholic fatty liver disease in mice: a biomedical investigation integrated with in silico assay effect of radix salviae miltiorrhizae and network pharmacology-based prediction of the active compounds and molecular targets. International Journal of Molecular Sciences. 2017;18 doi: 10.3390/ijms18030620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahmoud F., Al-Ozairi E., Haines D., et al. Effect of diabetea tea consumption on inflammatory cytokines and metabolic biomarkers in type 2 diabetes patients. Journal of Ethnopharmacology. 2016;194:1069–1077. doi: 10.1016/j.jep.2016.10.073. [DOI] [PubMed] [Google Scholar]
- 64.Gutierrez Nava Z. J., Jimenez-Aparicio A. R., Herrera-Ruiz M. L., Jimenez-Ferrer E. Immunomodulatory effect of agave tequilana evaluated on an autoimmunity like-SLE model induced in Balb/c mice with pristane. Molecules. 2017;22 doi: 10.3390/molecules22060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mashnafi S., Plat J., Mensink R. P., Baumgartner S. Non-cholesterol sterol concentrations as biomarkers for cholesterol absorption and synthesis in different metabolic disorders: a systematic review. Nutrients. 2019;11 doi: 10.3390/nu11010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang K., Feng X., Chai L., Cao S., Qiu F. The metabolism of berberine and its contribution to the pharmacological effects. Drug Metabolism Reviews. 2017;49(2):139–157. doi: 10.1080/03602532.2017.1306544. [DOI] [PubMed] [Google Scholar]
- 67.Wang B., Xu X., He X., Wang Z., Yang M. Berberine improved aldo-induced podocyte injury via inhibiting oxidative stress and endoplasmic reticulum stress pathways both In Vivo and In Vitro. Cellular Physiology and Biochemistry. 2016;39(1):217–228. doi: 10.1159/000445618. [DOI] [PubMed] [Google Scholar]
- 68.Guo Z., Sun H., Zhang H., Zhang Y. Anti-hypertensive and renoprotective effects of berberine in spontaneously hypertensive rats. Clinical and Experimental Hypertension. 2015;37:332–339. doi: 10.3109/10641963.2014.972560. [DOI] [PubMed] [Google Scholar]
- 69.Yang H., Song Y., Liang Y.-N., Li R. Quercetin treatment improves renal function and protects the kidney in a rat model of adenine-induced chronic kidney disease. Medical Science Monitor. 2018;24:4760–4766. doi: 10.12659/msm.909259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomes I. B. S., Porto M. L., Santos M. C. L. F. S., et al. The protective effects of oral low-dose quercetin on diabetic nephropathy in hypercholesterolemic mice. Frontiers in Physiology. 2015;6:p. 247. doi: 10.3389/fphys.2015.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dos Santos M., Poletti P. T., Favero G., et al. Protective effects of quercetin treatment in a pristane-induced mouse model of lupus nephritis. Autoimmunity. 2018;51(2):69–80. doi: 10.1080/08916934.2018.1442828. [DOI] [PubMed] [Google Scholar]
- 72.Wan-hua Y., Juan L., Bing C. Study on quality standard of shuangbai tablets. China Pharmacy. 2010;21:2549–2551. [Google Scholar]
- 73.Sun H., Wang W., Han P., et al. Astragaloside IV ameliorates renal injury in db/db mice. Scientific Reports. 2016;6(1):p. 32545. doi: 10.1038/srep32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agrawal S., Guess A. J., Chanley M. A., Smoyer W. E. Albumin-induced podocyte injury and protection are associated with regulation of COX-2. Kidney International. 2014;86(6):1150–1160. doi: 10.1038/ki.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin J., Zhao L., Zou W., Shen W., Zhang H., He Q. Activation of cyclooxygenase-2 by ATF4 during endoplasmic reticulum stress regulates kidney podocyte autophagy induced by lupus nephritis. Cellular Physiology and Biochemistry. 2018;48(2):753–764. doi: 10.1159/000491904. [DOI] [PubMed] [Google Scholar]
- 76.Tóth Š., Jonecová Z., Čurgali K., et al. Quercetin attenuates the ischemia reperfusion induced COX-2 and MPO expression in the small intestine mucosa. Biomedicine & Pharmacotherapy. 2017;95:346–354. doi: 10.1016/j.biopha.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y., Zhang S. Berberine suppresses growth and metastasis of endometrial cancer cells via miR-101/COX-2. Biomedicine & Pharmacotherapy. 2018;103:1287–1293. doi: 10.1016/j.biopha.2018.04.161. [DOI] [PubMed] [Google Scholar]
- 78.Das N., Bhattacharya A., Kumar Mandal S., et al. Ichnocarpus frutescens (L.) R. Br. root derived phyto-steroids defends inflammation and algesia by pulling down the pro-inflammatory and nociceptive pain mediators: An in-vitro and in-vivo appraisal. Steroids. 2018;139:18–27. doi: 10.1016/j.steroids.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Mirfazeli E. S., Marashi S.-A., Kalantari S. In silico prediction of specific pathways that regulate mesangial cell proliferation in IgA nephropathy. Medical Hypotheses. 2016;97:38–45. doi: 10.1016/j.mehy.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 80.Haas M. J., Onstead-Haas L. M., Szafran-Swietlik A., et al. Induction of hepatic apolipoprotein A-I gene expression by the isoflavones quercetin and isoquercetrin. Life Sciences. 2014;110(1):8–14. doi: 10.1016/j.lfs.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 81.Bautista-Garcia P., Sanchez-Lozada L. G., Cristobal-Garcia M., et al. Chronic inhibition of NOS-2 ameliorates renal injury, as well as COX-2 and TGF- 1 overexpression in 5/6 nephrectomized rats. Nephrology Dialysis Transplantation. 2006;21(11):3074–3081. doi: 10.1093/ndt/gfl444. [DOI] [PubMed] [Google Scholar]
- 82.Cristina Cerquetti M., Hovsepian E., Sarnacki S. H., Goren N. B. Salmonella enterica serovar enteritidis dam mutant induces low NOS-2 and COX-2 expression in macrophages via attenuation of MAPK and NF-κB pathways. Microbes and Infection. 2008;10(14-15):1431–1439. doi: 10.1016/j.micinf.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 83.Thirugnanasampandan R., Ramya G., Bhuvaneswari G., Aravindh S., Vaishnavi S., Gogulramnath M. Preliminary phytochemical analysis and evaluation of antioxidant, cytotoxic and inhibition of lipopolysaccaride—induced NOS (iNOS) expression in BALB/c mice liver by Ziziphus oenoplia mill. fruit. Journal of Complementary and Integrative Medicine. 2017;14(2) doi: 10.1515/jcim-2016-0009. [DOI] [PubMed] [Google Scholar]
- 84.Kim K.-W., Ha K.-T., Park C.-S., et al. Polygonum cuspidatum, compared with baicalin and berberine, inhibits inducible nitric oxide synthase and cyclooxygenase-2 gene expressions in RAW 264.7 macrophages. Vascular Pharmacology. 2007;47(2-3):99–107. doi: 10.1016/j.vph.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 85.Radika M. K., Viswanathan P., Anuradha C. V. Nitric oxide mediates the insulin sensitizing effects of β-sitosterol in high fat diet-fed rats. Nitric Oxide. 2013;32:43–53. doi: 10.1016/j.niox.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 86.Christakoudi S., Runglall M., Mobillo P., et al. Steroid regulation: An overlooked aspect of tolerance and chronic rejection in kidney transplantation. Molecular and Cellular Endocrinology. 2018;473:205–216. doi: 10.1016/j.mce.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 87.Donegan D., Bale L. K., Conover C. A. PAPP-A in normal human mesangial cells: effect of inflammation and factors related to diabetic nephropathy. Journal of Endocrinology. 2016;231(1):71–80. doi: 10.1530/joe-16-0205. [DOI] [PubMed] [Google Scholar]
- 88.Suyama M., Miyazaki Y., Matsusaka T., et al. Forced expression of vascular endothelial growth factor-A in podocytes decreases mesangial cell numbers and attenuates endothelial cell differentiation in the mouse glomerulus. Clinical and Experimental Nephrology. 2018;22(2):266–274. doi: 10.1007/s10157-017-1450-5. [DOI] [PubMed] [Google Scholar]
- 89.Liu H., Lu Z., Tian C., et al. Mechanism of Shenbing decoction in the treatment of proteinuria in chronic kidney disease: a network pharmacology-based study. Nan Fang Yi Ke Da Xue Xue Bao. 2019;39:227–234. doi: 10.12122/j.issn.1673-4254.2019.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhattacharjee N., Barma S., Konwar N., Dewanjee S., Manna P. Mechanistic insight of diabetic nephropathy and its pharmacotherapeutic targets: An update. European Journal of Pharmacology. 2016;791:8–24. doi: 10.1016/j.ejphar.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 91.Yoshida S., Nagase M., Shibata S., Fujita T. Podocyte injury induced by albumin overload in vivo and in vitro: involvement of TGF-beta and p38 MAPK. Nephron Experimental Nephrology. 2008;108(3):e57–e68. doi: 10.1159/000124236. [DOI] [PubMed] [Google Scholar]
- 92.Liang Y., Jing Z., Deng H., et al. Soluble epoxide hydrolase inhibition ameliorates proteinuria-induced epithelial-mesenchymal transition by regulating the PI3K-Akt-GSK-3β signaling pathway. Biochemical and Biophysical Research Communications. 2015;463(1-2):70–75. doi: 10.1016/j.bbrc.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 93.Xiuli Zhang T. G., Yang B., Chi Z. Protective effect of berberine on high glucose and hypoxia-induced apoptosis via the modulation of HIF-1α in renal tubular epithelial cells. American Journal of Translational Research. 2019;11:669–682. [PMC free article] [PubMed] [Google Scholar]
- 94.Viazzi F., Bonino B., Cappadona F., Pontremoli R. Renin-angiotensin-aldosterone system blockade in chronic kidney disease: current strategies and a look ahead. Internal and Emergency Medicine. 2016;11(5):627–635. doi: 10.1007/s11739-016-1435-5. [DOI] [PubMed] [Google Scholar]
- 95.Wang M., Zhang X., Song X., et al. Nodular glomerulosclerosis and renin angiotensin system in Chinese patients with type 2 diabetes. Molecular and Cellular Endocrinology. 2016;427:92–100. doi: 10.1016/j.mce.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 96.Xu R., Sun S., Huo Y., et al. Effects of ACEIs versus ARBs on proteinuria or albuminuria in primary hypertension. Medicine. 2015;94(39):p. e1560. doi: 10.1097/md.0000000000001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heleniak Z., Kuźmiuk-Glembin I., Adrych D., et al. Management of renin-angiotensin-aldosterone system blockade in kidney transplant recipients. Transplantation Proceedings. 2018;50(6):1842–1846. doi: 10.1016/j.transproceed.2018.03.027. [DOI] [PubMed] [Google Scholar]
- 98.Bogdanovski D. A., DiFazio L. T., Bogdanovski A. K., et al. Hypoxia-inducible-factor-1 in trauma and critical care. Journal of Critical Care. 2017;42:207–212. doi: 10.1016/j.jcrc.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 99.Norman J. T., Fine L. G. Intrarenal oxygenation in chronic renal failure. Clinical and Experimental Pharmacology and Physiology. 2006;33(10):989–996. doi: 10.1111/j.1440-1681.2006.04476.x. [DOI] [PubMed] [Google Scholar]
- 100.Katavetin P., Inagi R., Miyata T., et al. Albumin suppresses vascular endothelial growth factor via alteration of hypoxia-inducible factor/hypoxia-responsive element pathway. Biochemical and Biophysical Research Communications. 2008;367(2):305–310. doi: 10.1016/j.bbrc.2007.12.086. [DOI] [PubMed] [Google Scholar]
- 101.Li M., Jiang Y., Jing W., Sun B., Miao C., Ren L. Quercetin provides greater cardioprotective effect than its glycoside derivative rutin on isoproterenol-induced cardiac fibrosis in the rat. Canadian Journal of Physiology and Pharmacology. 2013;91(11):951–959. doi: 10.1139/cjpp-2012-0432. [DOI] [PubMed] [Google Scholar]
- 102.Ho C., Hsu Y.-C., Tseng C.-C., Wang F.-S., Lin C. L., Wang J. Y. Simvastatin alleviates diabetes-induced VEGF-mediated nephropathy via the modulation of Ras signaling pathway. Renal Failure. 2008;30(5):557–565. doi: 10.1080/08860220802064457. [DOI] [PubMed] [Google Scholar]
- 103.Costache M. I., Ioana M., Iordache S. VEGF expression in pancreatic cancer and other malignancies: a review of the literature. Romanian Journal of Internal Medicine. 2015;10 doi: 10.1515/rjim-2015-0027. [DOI] [PubMed] [Google Scholar]
- 104.Tanabe K., Maeshima Y., Sato Y., Wada J. Antiangiogenic therapy for diabetic nephropathy. BioMed Research International. 2017;2017:12. doi: 10.1155/2017/5724069.5724069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamagishi S.-I., Matsui T., Nakamura K., et al. Olmesartan blocks advanced glycation end products (AGEs)-induced angiogenesis in vitro by suppressing receptor for AGEs (RAGE) expression. Microvascular Research. 2008;75(1):130–134. doi: 10.1016/j.mvr.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 106.Prasad K., Dhar I., Zhou Q., Elmoselhi H., Shoker M., Shoker A. AGEs/sRAGE, a novel risk factor in the pathogenesis of end-stage renal disease. Molecular and Cellular Biochemistry. 2016;423(1-2):105–114. doi: 10.1007/s11010-016-2829-4. [DOI] [PubMed] [Google Scholar]
- 107.Sharmila R., Sindhu G. Modulation of angiogenesis, proliferative response and apoptosis by β-sitosterol in rat model of renal carcinogenesis. Indian Journal of Clinical Biochemistry. 2017;32(2):142–152. doi: 10.1007/s12291-016-0583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamagishi S.-I., Ishibashi Y., Isami F., Abe Y., Sakaguchi T., Higashimoto Y. Phytochemicals against advanced glycation end products (AGEs) and the receptor system. Current Pharmaceutical Design. 2017;23:1135–1141. doi: 10.2174/1381612822666161021155502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: 118 active ingredients in Shuangbai Tablets. Supplementary Table 2: 8981 targets of the active ingredients in Shuangbai Tablets. Supplementary Table 3: 110 523 genes associated with proteinuria. Supplementary Table 4: candidate targets with therapeutic proteinuria. Supplementary Table 5: 172 pairs of protein interactions of candidate targets. Supplementary Table 6: specific analytical parameters of 41 central targets.
Data Availability Statement
The data of our research can be acquired from the Supplementary Materials uploaded with this article.