Abstract
Accumulating evidence suggests that the aberrant expression of long non-coding RNAs (lncRNAs) is involved in the initiation, development and metastasis of bladder cancer (BC). Although several differentially expressed lncRNAs have been identified via lncRNA expression profiling of BC tissues, their functions and the molecular mechanisms underlying these functions remain to be fully elucidated. In the present study, elevated levels of lncRNA breast cancer anti-estrogen receptor 4 (BCAR4) were identified in BC tissues compared with matched healthy tissues. Silencing of BCAR4 inhibited cell proliferation and induced apoptosis in BC cell lines 5637 and T24. Downregulation of BCAR4 led to the inactivation of Wnt signaling. Mechanistically, BCAR4 directly sponged microRNA (miR)-370-3p and elevated Wnt7a expression. Endogenous expression of Wnt7a reversed BCAR4 silencing-mediated cell growth arrest and induction of apoptosis in BC cells accompanied with a re-activation of Wnt signaling. Reverse transcription-quantitative PCR indicated that there was a strong association between BCAR4, miR-370-3p and Wnt7a expression in tumors from patients with BC compared with healthy control tissues. In conclusion, results of the present study suggest that lncRNA BCAR4 promoted proliferation and survival of BC cells via downregulation of miR-370-3p. Therefore, lncRNA BCAR4 may be a lncRNA of oncogenic potential in BC.
Keywords: lncRNA BCAR4, Wnt signaling, bladder cancer, miR-370-3p
Introduction
Bladder cancer (BC) is one of the most commonly diagnosed urological malignancies and originates from uncontrolled proliferation of abnormal bladder cells. Based on statistics from 2018, there are nearly 550,000 estimated newly diagnosed BC cases globally each year and BC causes approximately 200,000 deaths annually worldwide (1). Based on pathology, BC can be divided into two major categories, non-muscle invasive and muscle invasive (2). Low-grade non-muscle invasive BC is characterized by a weak invasive ability and is suitable for localized therapy (3). However, disease in patients with low-grade non-invasive BC progresses rapidly to become muscle invasive BC (4). Despite advances in therapeutic strategy the prognosis of patients with BC remains poor, particularly in those with muscle invasive BC (1). Therefore, unveiling the molecular mechanism of bladder cancer cells remains pivotal to facilitating the development of new therapeutic approach for patients with BC.
Long non-coding RNAs (lncRNAs) are defined as single-stranded transcripts of more than 200 nucleotides in length with no protein coding potential (5). Previously thought of as 'junk RNAs', previous findings have demonstrated that lncRNAs can regulate gene expression through a variety of mechanisms (6). One well-characterized mechanism of lncRNA action is that they can serve as competing endogenous RNAs, blocking miRNA binding sites in order to positively regulate gene expression (7). Several lncRNAs have been identified as oncogenes or tumor suppressors in BC. These lncRNAs act via regulation of the expression of key genes that control cell proliferation, migration, cell cycle progression and stemness (8). A high level of retrotransposon-derived lncRNA PEG10 (PEG10) expression was observed in BC tissues and experiments based on BC cell lines also suggested that PEG10 prevented miR-134 from inhibiting Wnt and JAK/STAT signaling pathways, leading to sustained cell proliferation and strong metastatic ability (9). A previous RNA sequencing-based profiling study of non-coding RNAs identified several lncRNAs that were differentially expressed between healthy and BC tissues (10). Among them, lncRNA maternally expressed 3 (MEG3) was indicated to act as a tumor suppressor or oncogene in BC cells (11,12). Also shown to be differentially expressed was lncRNA breast cancer anti-estrogen receptor 4 (BCAR4), a recognized oncogene in several cancer types (13,14). However, the role of BCAR4 in BC is unknown.
The Wnt signaling pathway is involved in BC cell proliferation, survival, metastasis and maintenance of stemness (15). Core components of the Wnt signaling pathway include the extracellular factor Wnt and the transmembrane receptor β-catenin (16). As a member of the Wnt family, upon activation, Wnt7a triggers signaling transduction and dephosphorylation of β-catenin, resulting in the translocation of β-catenin and the activation of target gene expression (15,17). In cancer cells, Wnt7a has been suggested to be a target gene of miRNAs such as miR-15b, miR-127 and miR-370-3p (15,18,19). It is unclear how miRNAs and Wnt7a are involved in BC.
The focus of the present study was to identify the role of BCAR4 in BC. RT-qPCR indicated an elevation of BCAR4 mRNA in BC tissues compared with matched healthy tissues. Downregulation of BCAR4 also inhibited cell proliferation and induced cell apoptosis in two BC cell lines. Mechanistically, the downregulation of BCAR4 led to inactivation of Wnt signaling via prevention of the binding of tumor suppressor miR-370-3p and subsequent elevation of Wnt7a expression. There was also a strong association between BCAR4, miR-370-3p and Wnt7a expression in tumors from patients with BC. Taken together, these data identified a BCAR4/miR-370-3p/Wnt7a axis in the promotion of the proliferation and survival of BC cells. Therefore, lncRNA BCAR4 is an lncRNA with oncogenic potential in BC.
Materials and methods
Collection of normal and tumor tissues
Tumor tissues and paired healthy tissues were collected from 40 patients diagnosed with BC at The First Hospital of Jilin University (Jilin, China) between January 2016 and May 2017 (patients were aged 40-75 years, with a median age of 61 years). The tumor tissues were histopathologically examined after surgical removal. None of the patients had received chemotherapy or radiotherapy prior to surgery. Written informed consent was obtained from all the participants. All the experiments were performed under the supervision of the Ethics Committee of The First Hospital of Jilin University. Tissues were stored at −80°C immediately after harvesting before RNA extraction.
Cell culture
Human BC cell lines 5637, T24 and SW780 and human immortalized ureter epithelial cell line SV-HUC-1 were purchased from the American Type Culture Collection (Manassas) and used within 6 months. These cell lines were cultured in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in an incubator with 5% CO2 at 37°C.
Silencing of BCAR4
BCAR4 siRNA (5′-GCU GCG AGG GUA GAC AUC UCU GUU U-3′) and control siRNA (5′-UAA GGC UAU GAA GAG AUA C-3′) were synthesized by and purchased from GenePharma Co. Ltd. The silencing of BCAR4 was achieved by the transfection of BCAR4 siRNA into 5637 and T24 cells using Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, 200 pM BCAR4 siRNA was incubated with 9 µl Lipofectamine RNAiMax in 500 µl Opti-MEM (Invitrogen; Thermo Fisher Scientific) for 5 mins. This mixture was then added into each well of 6-well plates seeded with cells. After 72 h, the cells were harvested for the subsequent experiments.
Cell viability assay
The viability of 5637 and T24 cells were determined using a cell counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) following the manufacturer's protocol. The cells were seeded at a density of 1×105 cells per well in each well of a 96-well plate. The following day, the cells were transfected with BRCA4 siRNA. CCK-8 solution was added after 0, 24, 48 or 72 h. CCK-8 solution (10 µl) was added into each well and the cells were incubated for a further 2 h. The medium containing the CCK-8 solution was then moved to another 96-well plate and the absorbance at 450 nM was measured by iMark Microplate Reader (Bio-Rad) to determine cell number.
RNA extraction and reverse transcription-quantitative PCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. The isolated RNA was quantified using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed to DNA using PrimeScript™ RT Master Mix (Takara Bio., Inc.) following the manufacturer's protocol. The qPCR was performed with TB Green® Premix Ex Taq™ II (Takara Bio., Inc.) on a CFX-96 RT-qPCR System (Bio-Rad Technologies, Inc.). The thermocycling conditions were as follows: Initial denaturation at 95°C for 30 sec followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. U6 and β-actin were used as internal controls for miRNA and lncRNA/mRNA respectively. The relative expression of genes was calculated with the 2−ΔΔCq method (20). The BCAR4 primers used were: Forward: 5′-ACA GCA GCT TGT TGC TCA TCT-3′; reverse: 5′-TTG CCT TGG GGA CAG TTC AC-3′. The Wnt7a primers used were: forward: 5′-CTC CGG ATC GGT GGC T-3′; reverse: 5′-CCC ATT TGT GAG CCT TCT CCT-3′. The β-actin primers used were: Forward: 5′-TCT GGC TGA GGC TGG TTG AC-3′; reverse: 5′-CTC CTT AAT GTC ACG CAC GAT-3′. The Stemloop primer used was: 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG ACC AGG TT-3′. The miR-370-3p primers used were: Forward: 5′-ACA CTC CAG CTG GGG CCT GCT GGG GTG GAA-3′; reverse: 5′-CTC AAC TGG TGT CGT GGA-3′. The U6 primers used were: Forward: 5′-CTC GCT TCG GCA GCA CA-3′; reverse: 5′-CTC GCT TCG GCA GCA CA-3′.
Flow cytometric analysis
The apoptotic cells were detected by flow cytometry with a Dead Cell Apoptosis kit with Annexin V Alexa Fluor™ 488 and Propidium Iodide (PI) kit (Invitrogen; Thermo Fisher Scientific) following manufacturer's protocol. Apoptotic cells were Annexin V positive with or without PI positive.
Protein extraction and western blotting
Antibodies against Wnt7a (rabbit polyclonal antibody, ab100792, 1:2,000), E-cadherin (mouse monoclonal antibody, ab1416, 1:2,000), Vimentin (mouse monoclonal antibody, ab8978, 1:1,000) and β-actin (mouse monoclonal antibody, ab8224, 1:5,000) were purchased from Abcam. Phospho-β-catenin (rabbit polyclonal antibody, no. 2009, 1:2,000) antibody was purchased from Cell Signaling Technology. Secondary antibodies against mouse (A3682, 1:50,000) and rabbit (A8275, 1:50,000) were obtained from Sigma-Aldrich. Proteins were extracted from cells using RIPA lysis buffer (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The protein concentration was determined with a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Lysate containing 20 µg proteins was loaded into each lane of an 8% SDS-PAGE gel (GenScript). After electrophoresis, proteins on the gel were transferred to a PVDF membrane. The membrane was blocked with 5% non-fat milk for 1 h at room temperature. The membrane was then incubated with primary antibodies overnight at 4°C. On the following day the membrane was incubated with secondary antibodies for 1 h at room temperature. The blots were developed using SuperSignal Western Blot Enhancer (Thermo Fisher Scientific, Inc.). The relative protein expression was determined with Image J software V 1.8.0 (National Institutes of Health) and β-actin served as the internal control.
Dual luciferase reporter assay
The BCAR4 sequences were amplified from the cDNA of T24 cells and ligated into a pGL3-basic luciferase reporter plasmid (Promega Corporation). Primer sequences used were: BCAR4-forward: 5′-CTC TAGA AGT TAG TGC TGG GAA ACAG-3′; reverse: 5′-CTC TAG ACA GAT TTT ATT TCTA TTTA-3′. Three site mutations were then introduced into the pGL3-BCAR4-wild type (WT) plasmid at the putative binding sites to construct pGL3-BCAR4-mutant (Mut). Then, 2 µg plasmids were transfected into cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). The luciferase activity was analyzed using the Dual-Glo Luciferase Reporter Assay kit (Promega Corporation) according to the manufacturer's protocol. The relative luciferase activity was determined 48 h after transfection. The Firefly luciferase was normalized to Renilla luciferase.
Overexpression of Wnt7a
The full length of the Wnt7a open reading frame was amplified from T24 cDNA and ligated into pcDNA3.1 (Shaanxi Yuanbang Biotech., Co., Ltd.). Primer sequences used were: Wnt7a-forward: 5′-CAA GCT TAT GAA CCG GAA AGC GCG GC-3′; reverse: 5′-GGA ATT CTC AGT TGC ACG TGT ACA TC-3′. For overexpression of Wnt7a, 2 µg pcDNA3.1-Wnt7a or pcDNA3.1 plasmids were transfected into cells with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were harvested 72 h later for subsequent experiments.
Bioinformatics analysis
The expression of BCAR4 in bladder tumors and normal tissues was analyzed on Starbase V3 (http://starbase.sysu.edu.cn/). The potential binding sites of miRNAs to BCAR4 were predicted in miRDB (http://mirdb.org/).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software, Inc.) and are presented as the mean ± SD. All the experiments were repeated three times. Correlations were analyzed using Pearson's correlation analysis. For comparisons, two-tailed Student's t-tests were performed where there were two groups and one-way ANOVA was applied to three groups followed by Newman Keuls post-hoc test. P<0.05 was considered to be statistically significant.
Results
High expression levels of BCAR4 in BC
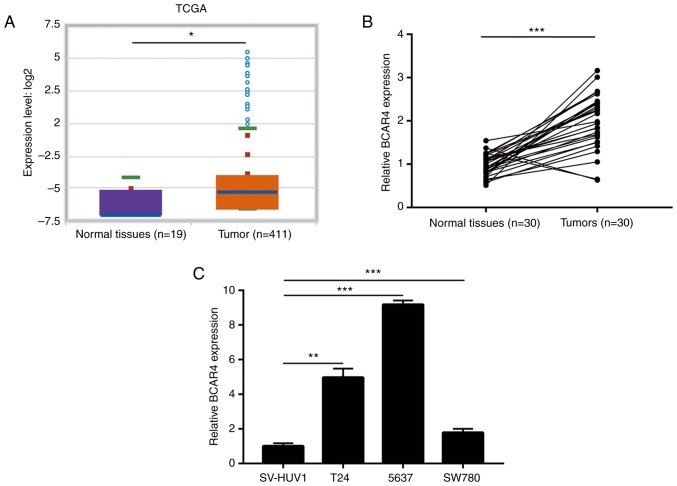
Previous lncRNA expression profiling identified that lncRNA-BCAR4 was one of several significantly upregulated lncRNAs in bladder tumors, when compared with normal tissues (10). To confirm this observation, StarBase V3 was used to compare the expression of BCAR4 in bladder tumors and normal tissues using data derived from The Cancer Genome Atlas. The result suggested that BCAR4 was overexpressed in 411 bladder tumors compared with 19 healthy tissues (Fig. 1A). For validation, 30 pairs of tumors and matched normal tissues from patients with BC were collected. RT-qPCR data showed that BCAR4 was increased in the majority of patients with BC (Fig. 1B). In addition, in a panel of BC cell lines (T24, 5637 and SW780), the expression of BCAR4 was significantly increased compared to the immortalized bladder cell line SV-HUV1 (Fig. 1C). These data indicated that BCAR4 was highly expressed in BC.
Figure 1.
lncRNA-BCAR4 is overexpressed in bladder cancer. (A) Expression of BCAR4 in 411 bladder tumors and 19 normal tissues was retrieved from the TCGA dataset. It was found that BCAR4 was overexpressed in bladder tumors. (B) RT-qPCR was used to detect BCAR4 expression in 30 pairs of bladder tumors and matched normal tissues. It was observed that BCAR4 was overexpressed in bladder tumors. (C) The expression of BCAR4 in normal bladder cell line (SV-HUV1) and bladder cancer cell lines (T24, 5637 and SW780) were detected with RT-qPCR. BCAR4 was overexpressed in bladder cancer cell lines. *P<0.05; **P<0.01; ***P<0.001.
Knockdown of BCAR4 inhibits cell proliferation and induces cell apoptosis in BC cells
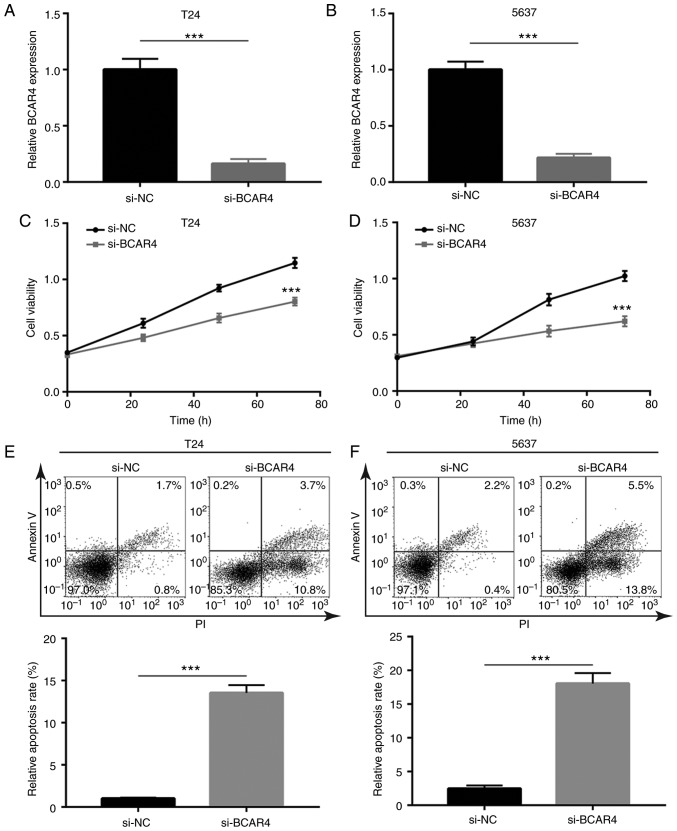
BCAR4 siRNA was transfected into BC cell lines to investigate the activity of BCAR4 in BC cells. Transfection of BCAR4 siRNA decreased BCAR4 expression in both T24 and 5637 cells (Fig. 2A and B). Knockdown of BCAR4 resulted in a reduction in cell viability in T24 cells (Fig. 2C). Similarly, the cell viability of 5637 cells was also greatly inhibited after BCAR4 knockdown (Fig. 2D). To clarify whether the reduction in cell viability caused by BCAR4 knockdown was associated with cell death, flow cytometric analysis was used to determine the cell apoptotic rate in T24 cells transfected with BCAR4 siRNA. A significant elevation of the percentage of cells in early [PI (propidium iodide)+/ AnnexinV-FITC−] and late (PI+/AnnexinV-FITC+) apoptosis was observed in BCAR4-silenced T24 cells (Fig. 2E). Similar to T24 cells, BCAR4 knockdown also induced cell apoptosis in 5637 cells (Fig. 2F). The data suggested that BCAR4 may promote BC cell proliferation and survival.
Figure 2.
Functional effects of BCAR4 knockdown in bladder cancer cells. (A) RT-qPCR showed that transfection of BCAR4 siRNA decreased BCAR4 expression in T24 cells. (B) RT-qPCR showed that transfection of BCAR4 siRNA decreased BCAR4 expression in 5637 cells. Silencing of BCAR4 decreased cell viability of (C) T24 and (D) 5637 cells. Flow cytometric analysis suggested that BCAR4 silencing elevated apoptotic cell percentage in (E) T24 cells and (F) 5637 cells. ***P<0.001.
Silencing of BCAR4 inactivates Wnt/β-catenin signaling in BC cells
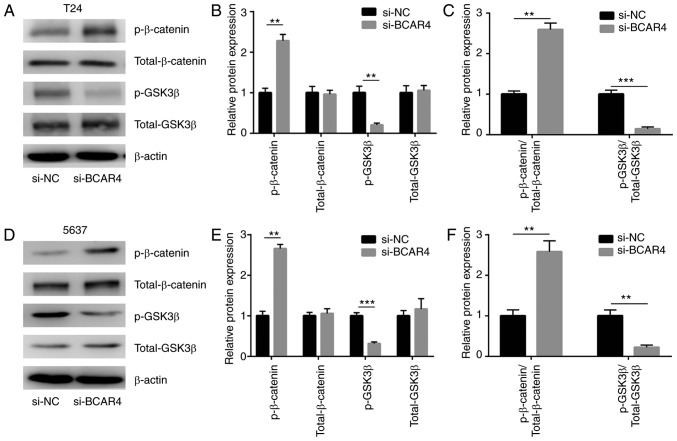
Wnt/β-catenin signaling is a well-characterized oncogenic pathway in BC. Sustained activation of Wnt/β-catenin signaling is critical for the survival of BC cells. To determine whether Wnt/β-catenin signaling was involved in BCAR4-mediated BC cell survival, the expression levels of the inactive form of β-catenin [phosphorylated (p)-β-catenin] and the active form of GSK3β (p-GSK3β) were assessed in cells transfected with BCAR4 siRNA. Western blot analysis revealed that p-β-catenin levels were increased and p-GSK3β levels were decreased in T24 cells after BCAR4 silencing (Fig. 3A and B). The ratio of p-β-catenin to total β-catenin was increased, while the ratio of p-GSK3β to GSK3β was decreased, suggesting the shutdown of Wnt/β-catenin signaling (Fig. 3C). As in T24 cells, the level of p-β-catenin was increased and the p-GSK3β level was decreased in 5637 cells after BCAR4 silencing (Fig. 3D and E). Wnt/β-catenin signaling was inactivated after BCAR4 silencing, as the ratio of p-β-catenin to total β-catenin was increased and the ratio of p-GSK3β to GSK3β was decreased (Fig. 3F). BCAR4 positively regulated Wnt7a expression via the repression of miR-370-3p in BC cells.
Figure 3.
Activity of Wnt/β-catenin signaling is mediated by BCAR4 in bladder cancer cells. (A) Western blot analysis of p-β-catenin, total-β-catenin, p-GSK3β and total-GSK3β expression in T24 cells transfected with BCAR4 siRNA. (B) Quantitative analysis of protein expression p-β-catenin, total-β-catenin, p-GSK3β and total-GSK3β expression in (A) showed that p-β-catenin was increased and p-GSK3β was decreased. (C) Quantitative analysis of protein expression in (A) showed that p-β-catenin/total-β-catenin was increased and p-GSK3β/total-GSK3β was decreased. (D) Western blot analysis of p-β-catenin, total-β-catenin, p-GSK3β and total-GSK3β expression in 5637 cells transfected with BCAR4 siRNA. (E) Quantitative analysis of protein expression in (D) showed that p-β-catenin was increased and p-GSK3β was decreased. (F) Quantitative analysis of protein expression in (D) showed that p-β-catenin/ total-β-catenin was increased and p-GSK3β/total-GSK3β was decreased. **P<0.01; ***P<0.001.
lncRNAs regulate gene expression via sponging miRNAs in cancer cells
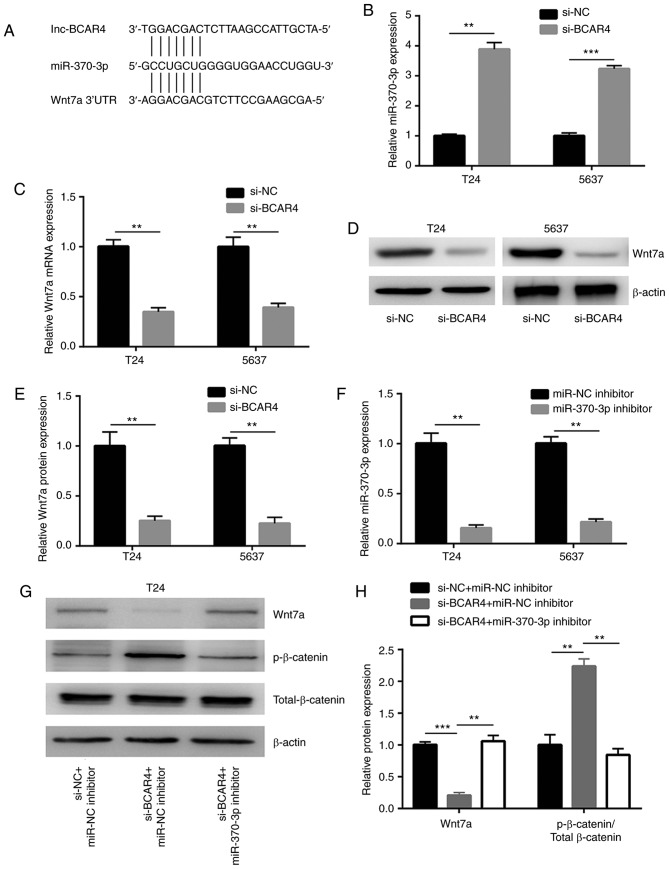
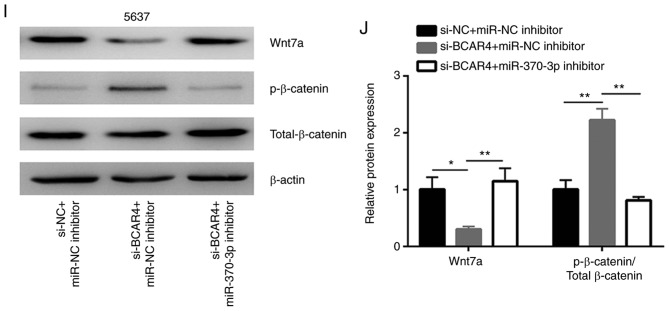
The potential binding sites of miRNAs to BCAR4 were predicted in miRDB. There were 22 miRNAs predicted to have binding sites on BCAR4. Through searching the literature, miR-370-3p was identified as a tumor suppressor in BC via targeting Wnt7a, a positive regulator of Wnt/β-catenin signaling (Fig. 4A). Silencing of BCAR4 led to a significant upregulation of the miR-370-3p level in T24 and 5637 cells (Fig. 4B). In addition, the downregulation of BCAR4 decreased the Wnt7a mRNA level in these cells (Fig. 4C). A reduction in Wnt7a protein expression was also found in T24 and 5637 cells after BCAR4 silencing (Fig. 4D and E). To explore whether BCAR4 regulated Wnt7a and downstream targets via control of miR-370-3p expression, miR-370-3p inhibitor was transfected into T24 and 5637 cells to downregulate miR-370-3p expression (Fig. 4F). As hypothesized, the downregulation of miR-370-3p reversed the reduction in Wnt7a protein expression observed in T24 cells treated with BCAR4 siRNA (Fig. 4G and H). In addition, upregulation of p-β-catenin/Total β-catenin induced by BCAR4 silencing was also reversed following miR-370-3p inhibition (Fig. 4G and H). As observed in T24 cells, the downregulation of miR-370-3p also reversed reduction of the Wnt7a protein expression and upregulation of p-β-catenin/Total β-catenin in 5637 cells treated with BCAR4 siRNA (Fig. 4I and J).
Figure 4.
BCAR4 regulates Wnt/β-catenin signaling through miR-370-3p. (A) Putative binding sites for miR-370-3p on BCAR4 sequence and 3′UTR of Wnt7a mRNA. (B) Silencing of BCAR4 increased miR-370-3p levels in T24 and 5637 cells. (C) Silencing of BCAR4 decreased Wnt7a mRNA expression in T24 and 5637 cells. (D) Western blot analysis of Wnt7a protein expression in T24 and 5637 cells transfected with BCAR4 siRNA. (E) Quantitative analysis of Wnt7a protein expression in (D) showed that BCAR4 silencing reduced Wnt7a protein expression. (F) Transfection of miR-370-3p inhibitor decreased miR-370-3p in T24 and 5637 cells. (G) Silencing of BCAR4 decreased Wnt7a and increased p-β-catenin protein expression in T24 cells, which was reversed after miR-370-3p inhibition. (H) Quantitative analysis of Wnt7a and p-β-catenin/Total β-catenin protein expression in (G). (I) Silencing of BCAR4 decreased Wnt7a and increased p-β-catenin protein expression in 5637 cells, which was reversed after miR-370-3p inhibition. (J) Quantitative analysis of Wnt7a and p-β-catenin/Total β-catenin protein expression in (I). *P<0.05; **P<0.01; ***P<0.001.
BCAR4 represses miR-370-3p activity in BC cells
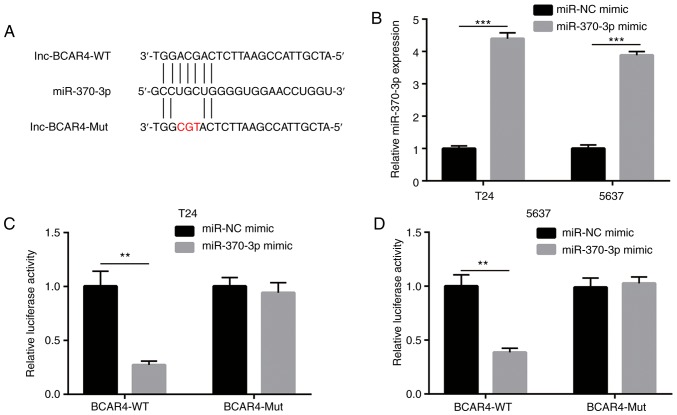
To determine whether BCAR4 directly regulates miR-370-3p expression, luciferase plasmids containing BCAR4-WT BCAR4-Mut were constructed with a mutation at the putative binding sites of miR-370-3p (Fig. 5A). miR-370-3p mimic was transfected into T24 and 5637 cells to overexpress miR-370-3p (Fig. 5B). Overexpression of miR-370-3p reduced relative luciferase activity of T24 cells with BCAR4-WT, but not BCAR4-Mut (Fig. 5C). Similarly, miR-370-3p mimic also reduced relative luciferase activity of 5637 cells with BCAR4-WT, but not BCAR4-Mut (Fig. 5D). Collectively, the data demonstrated that BCAR4 directly repressed miR-370-3p expression via the prevention of miR-370-3p binding in BC cells.
Figure 5.
BCAR4 sponges miR-370-3p in bladder cancer cells. (A) Sequence alignment of wild-type BCAR4 (BCAR4-WT) and mutant BCAR4 (BCAR4-Mut) to miR-370-3p. (B) Transfection of miR-370-3p increased miR-370-3p expression in T24 and 5637 cells. (C) The dual luciferase reporter assay showed that overexpression of miR-370-3p repressed relative luciferase activity of T24 cells transfected with BCAR4-WT. (D) The dual luciferase reporter assay showed that overexpression of miR-370-3p repressed relative luciferase activity of 5637 cells transfected with BCAR4-WT. **P<0.01; ***P<0.001.
BCAR4 controls cell proliferation and survival via sponging miR-370-3p in BC cells
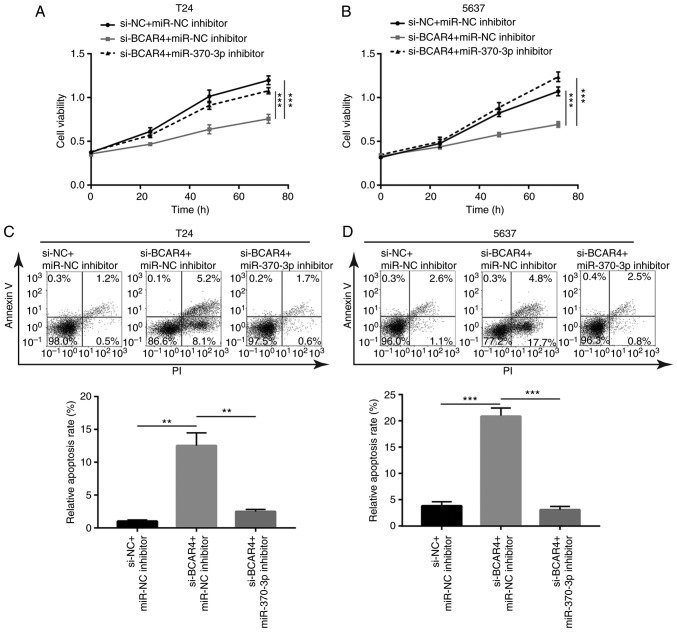
To examine the role of miR-370-3p in the function of BCAR4 in BC, T24 cells were transfected with BCAR4 siRNA with or without miR-370-3p inhibitor. The inhibitory effect of BCAR4 silencing on cell viability was reversed after miR-370-3p inhibition in T24 cells (Fig. 6A). Additionally, the cell viability reduction led by BCAR4 silencing was reversed following miR-370-3p inhibition in 5637 cells (Fig. 6B). Additionally, the cell apoptosis induced by BCAR4 silencing was reversed after miR-370-3p inhibition in T24 cells (Fig. 6C), which was also observed in 5637 cells (Fig. 6D).
Figure 6.
BCAR4 is dependent on the regulation of miR-370-3p to control cell proliferation and survival. (A) Silencing of BCAR4 reduced cell viability and inhibition of miR-370-3p reversed cell viability reduction in T24 cells. (B) Silencing of BCAR4 reduced cell viability and inhibition of miR-370-3p reversed cell viability reduction in 5637 cells. (C) Silencing of BCAR4 induced cell apoptosis and inhibition of miR-370-3p reversed cell apoptosis in T24 cells. (D) Silencing of BCAR4 induced cell apoptosis and inhibition of miR-370-3p reversed cell apoptosis in 5637 cells. **P<0.01; ***P<0.001.
BCAR4 expression is associated with miR-370-3p and Wnt7a expression in bladder tumors
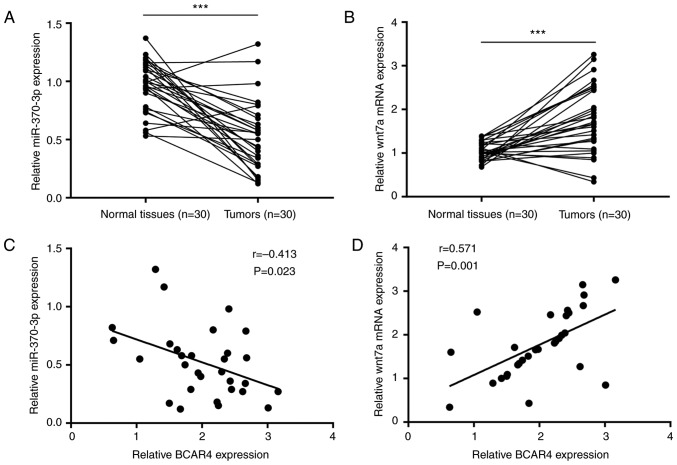
The clinical association between BCAR4, miR-370-3p and Wnt7a was then examined. RT-qPCR results showed that miR-370-3p was reduced in bladder tumors compared with matched healthy tissues (Fig. 7A). By contrast, Wnt7a mRNA expression was significantly increased in bladder tumors (Fig. 7B). Pearson's correlation analysis suggested that BCAR4 expression negatively correlated with miR-370-3p expression (Fig. 7C) and positively correlated with Wnt7a mRNA expression (Fig. 7D).
Figure 7.
BCAR4 expression negatively correlates with miR-370-3p expression and positively correlates with Wnt7a mRNA expression in bladder tumors. (A) RT-qPCR detection of miR-370-3p expression in 30 pairs of bladder tumors and normal tissues. It was observed that miR-370-3p was decreased in bladder tumors. (B) RT-qPCR detection of Wnt7a expression in 30 pairs of bladder tumors and normal tissues. It was observed that Wnt7a was decreased in bladder tumors. (C) BCAR4 expression negatively correlated with miR-370-3p expression in 30 bladder tumors. (D) BCAR4 expression positively correlated with Wnt7a expression in 30 bladder tumors. ***P<0.001.
Discussion
lncRNAs have been demonstrated to act as oncogenes or tumor suppressors in BC and have been indicated to control cell proliferation, migration, cell cycle progression and stemness (8). lncRNA BCAR4 has been identified to be an oncogene in numerous types of cancer. BCAR4 was shown to induce proliferation, invasion and metastasis of non-small cell lung cancer cells by regulating epithelial-mesenchymal transition (13). Additionally, elevation of BCAR4 was demonstrated to induce the proliferation and migration of cervical cancer cells (14). BCAR4 was found to induce cisplatin resistance and predicts poor prognosis in patients with gastric cancer via activating Wnt/β-catenin signaling pathway (21). BCAR4 also promoted the progression of colon cancer through activation of the Wnt/β-catenin signaling pathway as well as castration resistant prostate cancer by activation of GLI2 signaling (22,23). Nevertheless, its function in BC is unknown.
The present study identified an elevation of BCAR4 in BC compared with matched healthy tissues. Thus, the data also supported elevation of BCAR4 in various types of cancer as observed in non-small cell lung cancer, cervical cancer and gastric cancer (13,14,21). In two BC cell lines, our data showed that downregulation of BCAR4 inhibited cell proliferation, which was also reported in several cancer types such as glioma and non-small cell lung cancer (24,25). In addition, downregulation of BCAR4 induced cell apoptosis in BC cells. The involvement of BCAR4 in cell apoptosis was previously identified in glioma and non-small cell lung cancer (13,25). These findings suggested the pivotal role of BCAR4 in the progression of BC.
There is a regulatory relationship between lncRNAs and miRNAs (26). lncRNAs regulate gene methylation, transcription/translation and other processes by conjugation with mRNAs and miRNAs (27). miRNAs are small non-coding RNAs (20-22 nucleotides in length), which target the 3′-UTR of target mRNAs by sequence complementarity (28). miRNAs function as oncogenes and tumor suppressors in carcinogenesis (29). miRNAs are also involved in the apoptosis, proliferation, and metastasis of cancer cells (30). BCAR4 was reported to sponge miR-665 to upregulate expression of STAT3 in colorectal cancer cells (31). Thus, whether miRNAs/ mRNAs could be regulated by BCAR4 in BC has become a topic of interest.
BCAR4 regulates the activation of the Wnt/β-catenin signaling pathway in multiple cancer types; for instance, BCAR4 promotes the progression of colon cancer through activation of the Wnt/β-catenin signaling pathway (22). BCAR4 is elevated in osteosarcoma and promotes the development of osteosarcoma through the Wnt/β-catenin signaling pathway (32). In addition, elevation of BCAR4 in gastric cancer induced the expression of tumor stem cell-related biomarkers through the Wnt signaling pathway (21). However, although those studies only mentioned that BCAR4 controls the activity of the Wnt/β-catenin signaling pathway in cancer cells, the molecular mechanism by which BCAR4 regulated Wnt/β-catenin signaling pathway was not previously reported. As a member of the Wnt family, Wnt7a, which regulates the dephosphorylation and translocation of β-catenin, is targeted by miR-370-3p to promote BC cell proliferation, survival, metastasis and maintenance of stemness (15). Furthermore, an increasing number of studies have shown that miR-370 is involved in the progression of BC. miR-370 is expressed at reduced levels in BC clinical specimens when compared with controls and miR-370 activates p21 expression by targeting the p21 promoter, thereby suppressing the proliferation and metastasis of human BC cells (33-35). miR-370 also inhibits cell growth of BC by targeting DNA replication complex GINS protein SLD5 (36). By contrast, it is unclear whether BCAR4 regulates miR-370-3p and Wnt7a expression in BC. Bioinformatic analysis revealed miR-370-3p as a potential target miRNA of BCAR4, which was further validated by RT-qPCR and dual luciferase reporter assay. Downregulation of miR-370-3p attenuated BCAR4 siRNA-mediated cell proliferation inhibition and cell apoptosis in BC cells. These data indicated that a high expression of miR-370-3p was due to overexpression of BCAR4 in BC. Moreover, via sponging miR-370-3p, BCAR4 upregulated Wnt7a and activated Wnt signaling. Thus, our results indicated that BCAR4 activated Wnt signaling by directly sponging miR-370-3p and elevated Wnt7a expression in BC cells. Furthermore, there was a strong association between BCAR4, miR-370-3p and Wnt7a expression in tumors from patients with BC, indicating a BCAR4/ miR-370-3p/Wnt7a axis in BC tumors.
In summary, the results of the present study indicate that the BCAR4/miR-370-3p/Wnt7a axis has a role in promoting the proliferation and survival of BC cells. Therefore, lncRNA BCAR4 was a lncRNA with oncogenic potential in BC.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of materials and data
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CC conceived and designed the study. CC, RZ, JW and EJ performed the experiments. JZ, NL and RZ analyzed the data, and wrote the manuscript, CC revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
This current study was approved by the Ethics Committee of The First Hospital of Jilin University. Written informed consent was obtained from all the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Babjuk M, Bohle A, Burger M, Capoun O, Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 3.Anastasiadis A, de Reijke TM. Best practice in the treatment of nonmuscle invasive bladder cancer. Ther Adv Urol. 2012;4:13–32. doi: 10.1177/1756287211431976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah JB, McConkey DJ, Dinney CP. New strategies in muscle-invasive bladder cancer: On the road to personalized medicine. Clin Cancer Res. 2011;17:2608–2612. doi: 10.1158/1078-0432.CCR-10-2770. [DOI] [PubMed] [Google Scholar]
- 5.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Adelman K, Egan E. Non-coding RNA: More uses for genomic junk. Nature. 2017;543:183–185. doi: 10.1038/543183a. [DOI] [PubMed] [Google Scholar]
- 7.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Jiang F, Qi W, Wang Y, Wang W, Fan L. lncRNA PEG10 promotes cell survival, invasion and migration by sponging miR-134 in human bladder cancer. Biomed Pharmacother. 2019;114:108814. doi: 10.1016/j.biopha.2019.108814. [DOI] [PubMed] [Google Scholar]
- 10.Seitz AK, Christensen LL, Christensen E, Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM, Palmfeldt J, Thomson M, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7:395. doi: 10.1038/s41598-017-00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng SQ, Zhang XY, Fan HT, Sun QJ, Zhang M. Up-regulation of LncRNA MEG3 inhibits cell migration and invasion and enhances cisplatin chemosensitivity in bladder cancer cells. Neoplasma. 2018;65:925–932. doi: 10.4149/neo_2018_180125N55. [DOI] [PubMed] [Google Scholar]
- 12.Pan J, Li X, Wu W, Xue M, Hou H, Zhai W, Chen W. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382:64–76. doi: 10.1016/j.canlet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Gao WJ, Liu NS. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21:2075–2086. [PubMed] [Google Scholar]
- 14.Zou R, Chen X, Jin X, Li S, Ou R, Xue J, Yan X, Chen L, Hu Y, Zhu H. Up-regulated BCAR4 contributes to proliferation and migration of cervical cancer cells. Surg Oncol. 2018;27:306–313. doi: 10.1016/j.suronc.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Huang X, Zhu H, Gao Z, Li J, Zhuang J, Dong Y, Shen B, Li M, Zhou H, Guo H, et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J Biol Chem. 2018;293:6693–6706. doi: 10.1074/jbc.RA118.001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 17.Teng Y, Wang X, Wang Y, Ma D. Wnt/beta-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem Biophys Res Commun. 2010;392:373–379. doi: 10.1016/j.bbrc.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 18.MacLean JA, II, King ML, Okuda H, Hayashi K. WNT7A Regulation by miR-15b in ovarian cancer. PLoS One. 2016;11:e0156109. doi: 10.1371/journal.pone.0156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Wang X, Jiang X. miR-127 suppresses gastric cancer cell migration and invasion via targeting Wnt7a. Oncol Lett. 2019;17:3219–3226. doi: 10.3892/ol.2019.9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Chunyan Q, Zhou Y, He Q, Ma Y, Ga Y, Wang X. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064–4070. [PubMed] [Google Scholar]
- 22.Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M. LncRNA BCAR4 promotes colon cancer progression via activating Wnt/β-catenin signaling. Oncotarget. 2017;8:92815–92826. doi: 10.18632/oncotarget.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Z, Wu Y, Li Y, Ren J, Wang L. BCAR4 activates GLI2 signaling in prostate cancer to contribute to castration resistance. Aging (Albany NY) 2018;10:3702–3712. doi: 10.18632/aging.101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei L, Yi Z, Guo K, Long X. Long noncoding RNA BCAR4 promotes glioma cell proliferation via EGFR/PI3K/AKT signaling pathway. J Cell Physiol. 2019;234:23608–23617. doi: 10.1002/jcp.28929. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Yan L, Sun K, Sun X, Zhang X, Cai K, Song T. lncRNA BCAR4 increases viability, invasion, and migration of non-small cell lung cancer cells by targeting glioma-associated oncogene 2 (GLI2) Oncol Res. 2019;27:359–369. doi: 10.3727/096504018X15220594629967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: Orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8:12472–12483. doi: 10.18632/oncotarget.13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15:1171–1180. doi: 10.1016/j.autrev.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Lawrie CH. MicroRNAs and haematology: Small molecules, big function. Br J Haematol. 2007;137:503–512. doi: 10.1111/j.1365-2141.2007.06611.x. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Ouyang S, Zhou X, Chen Z, Wang M, Zheng X, Xie M. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019;19:72. doi: 10.1186/s12935-019-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: Biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419. doi: 10.1159/000479205. [DOI] [PubMed] [Google Scholar]
- 33.Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu H, Ye Z, Li LC. Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS Lett. 2014;588:4654–4664. doi: 10.1016/j.febslet.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Ge Q, Chen Z, Hu J, Li F, Ye Z. Promoter-associated endogenous and exogenous small RNAs suppress human bladder cancer cell metastasis by activating p21 (CIP1/WAF1) expression. Tumour Biol. 2016;37:6589–6598. doi: 10.1007/s13277-015-4571-z. [DOI] [PubMed] [Google Scholar]
- 36.Yamane K, Naito H, Wakabayashi T, Yoshida H, Muramatsu F, Iba T, Kidoya H, Takakura N. Regulation of SLD5 gene expression by miR-370 during acute growth of cancer cells. Sci Rep. 2016;6:30941. doi: 10.1038/srep30941. [DOI] [PMC free article] [PubMed] [Google Scholar]