Abstract
Patients with breast cancer (BC) and lung cancer (LC) are prone to developing brain metastases, which are associated with devastating prognoses. Dormant tumor cells, a population of non-apoptotic quiescent cells and immunological escape mechanisms, including the Natural Killer Group 2 member D (NKG2D) receptor-ligand system, represent potential mechanisms of tumor recurrence. To date, the immunological characteristics of dormant tumor cells concerning the NKG2D system in cerebral malignancies are mostly unknown. In the present study, an extensive characterization of dormant and NKG2D ligand (NKG2DL)+ cells in cerebral metastases was performed. The expression profiles and localization patterns of various NKG2DL and several dormancy markers were analyzed in solid human brain metastases from patients with BC and LC using immunostaining and reverse transcription-quantitative polymerase chain reaction analyses. Statistical analysis was performed using Student's t-test and Bravais-Pearson correlation analysis. Not only 'peripheral', but also 'central' dormancy markers, which had been previously described in primary brain tumors, were identified in all cerebral metastases at detectable levels at protein and mRNA levels. Notably, the majority of NKG2DL+ cells were also positive for 'central' dormancy markers, but not 'peripheral' dormancy markers in both patient groups. This cell population may represent a promising future therapeutic target.
Keywords: cerebral metastases, breast cancer, lung cancer, dormancy, Natural Killer Group 2 member D
Introduction
Brain metastases are the most common intracerebral tumors. In the majority of cases, brain metastases have a devastating prognostic effect, as treatment options like chemotherapy and radiotherapy have only limited efficacy (1). Median survival is <1 year in most patients (2). The primary cancer types that frequently metastasize to the brain are lung and breast cancer, melanoma, renal and colorectal cancer (3). As the incidence is 2- to 10-fold higher compared with primary brain malignancies like glioblastoma (4), brain metastases have a major clinical relevance. The average age at diagnosis in western populations is increasing, and overall survival in a number of primary tumors improved due to new targeted treatment strategies and the increased availability of cranial magnetic resonance imaging. As a consequence, the incidence of cerebral metastases is also increasing (5). In a number of cases, brain metastases occur following successful treatment of the primary tumor including resection, chemotherapy and radiation therapy. This is mainly due to a small percentage of tumor cells, which hardly proliferate and are insensitive to chemotherapy, remaining in the patient following treatment (6,7). These dormant cells have the ability to be reactivated and therefore can be the source of recurrence. Dormancy is understood as a combination of three different mechanisms (8), including cellular dormancy (9-11) and tumor dormancy (12-15). Cellular dormancy is described as a state in which solitary disseminated cancer cells either circulate in the blood or settle at secondary sites in a quiescent state. Tumor dormancy is described when tumors, as an accumulation of cells, do not expand in size over a long period of time. Importantly, tumor dormancy crucially depends on the microenvironment and tumor stroma, which both induce tumor cell quiescence. Additionally, systemic factors, like the immune system, hormonal control or blockage and insufficiency of tumor angiogenesis may result in tumor dormancy (11,16-24). The majority of studies investigating dormant cells have focused on the bone marrow niche as a potential microenvironment.
Dormant cells are defined by their low proliferation rate, which is associated with a decreased expression of proliferation marker protein Ki-67, a protein that is exclusively expressed in the G1-, S-, G2- and M-phases of the cell cycle (25). Identification of dormant cells in vivo is challenging, but there are several markers that are known to be present on dormant cells. Induction of dormancy has been closely associated with the effect of fibroblast growth factor 2 (FGF-2) in breast cancer (26). Cells being stimulated with FGF-2 in the bone marrow niche turned into dormant cells, making FGF-2 one of the key regulators of dormancy (27). Other possible markers for dormant cells in breast cancer are thrombos-pondin-1 (25) and cyclin-dependent kinase inhibitor p27 (28). de Jong et al (29) indicated that, in invasive breast cancer, expression of platelet-derived growth factor β (PDGFβ) was positively correlated with the apoptotic index. Additionally, mice bearing microscopic dormant liposarcomas exhibited a significant increase in platelet-associated angiogenesis regulatory proteins including basic fibroblast growth factor (bFGF) and PDGFβ. These proteins have also been suggested to serve as potential biomarkers for dormant cells (30). PDGFβ serves, among other functions, an important role in the metastasis process (26). Hypoxia is considered to be an important inductor of dormancy, as upregulation of dormancy genes is closely associated with genes like glucose transporter, type 1 (GLUT1) and hypoxia-inducible factor 1-α (HIF1-α) (31). This has been described for disseminated tumor cells in the bone marrow for breast cancer, but additionally in lung cancer, where induction of dormancy is markedly associated with hypoxia (32). Under hypoxic conditions, which frequently occur on a cellular level in lung cancer, HIF1-α is upregulated and leads to a glutamate dehydrogenase-dependent increase in glutamine uptake, glutamate to α-ketoglutarate flux and generation of ATP, which serves an important role in survival and drug-resistance in lung cancer cells (33) and breast cancer (31). In summary, fibroblast growth factor 2 (FGF2), PDGFβ, and HIF1-α are dormancy markers that may be used to identify dormant cells outside the central nervous system. They are designated as 'peripheral' dormancy markers in the following text.
Almog et al (19) performed a genome wide transcriptional analysis of dormant breast cancer, glioblastoma, osteosarcoma and liposarcoma tumors derived from human cell lines. This led to, among the confirmation of known dormancy markers like thrombospondin-1, angiomotin and tropomyosin, the identification of novel dormancy specific biomarkers. Histone cluster 1 H2B family member K (H2BK), Ephrin receptor A5 (EphA5) and insulin-like growth factor-binding protein 5 (IGFBP5) were markedly upregulated in dormant cells derived from glioblastoma, which is a highly malignant primary brain tumor.
The Ephrin family of receptor tyrosine kinases and their ligands are involved in embryonic and adult neurogenesis (34,35). EphA5 itself is considered to be a membrane receptor, but is also identified at increased levels of dormant-tumor bearing mice. Levels decrease with increasing tumor stage in glioma.
Histone H2BK is a core component of the nucleosome. Whereas histone acetylation is well known to affect angiogenesis, the role of histone H2BK in tumor progression remains unclear (19).
The insulin-like growth factor (IGF) axis is known to be an important pathway in carcinogenesis (36,37). IGFPBs control the binding of IGF to its receptor and were demonstrated to serve a critical role in the conversion of dormant tumors to fast-growing angiogenic tumors (19).
Recently we were able to demonstrate that H2BK, IGFBP5 and EphA5 are also expressed in human glioblastoma cell lines in situ. Stimulation with the chemotherapeutic agent temozolomide, which is frequently used in clinical therapy of glioblastoma, leads to upregulation of IGFBP5 and EphA5 under certain conditions (38).
In summary, H2BK, IFGBP5 and EphA5 are markers of dormant cells in the central nervous system, and were termed 'central' dormancy markers in the present study.
The Natural Killer Group 2D (NKG2D) system is crucial for recognition of malignant cells. Induced by challenging environmental conditions, malignant cells in the brain express several stress inducible molecules like the Natural Killer Group 2D ligands (NKG2DL), which are usually recognized by the NKG2D receptor (NKG2DR) (39,40). NKG2DL are not expressed under healthy conditions, but are induced due to different types of cellular stress, for example viral infection, genotoxic stress, or malignant transformation (41). To date, 8 different NKG2DL are known: MHC class I chain-related protein A (MICA) and B (MICB) and 6 members of the UL16-binding protein family (ULBP1-6) (42). NKG2DR are located on effector cells of the immune system, including NK cells, γδ T cells, CD8+ T cells and a minor subset of immune regulatory CD4+ T cells (43). Binding of the ligand to the receptor results in killing of the NKG2DL-carrying cell. To avoid this, tumor cells can release NKG2DL by shedding them from the surface or releasing them in exosomes (44). In primary brain tumors, such as glioblastoma, the NKG2D system serves an important role in tumor immunosurveillance (45). In contrast to a number of other tumors, cells isolated from glioblastoma rarely express NKG2DL on their surface (40). Notably, glioma stem-like cells appear to carry certain subsets of NKG2DL on their surface, which they tend to upregulate when treated with temozolomide (45). As therapy itself might be part of the acquisition of dormancy (46), and chemotherapy has been suggested to be involved in the regulation of immune-associated factors like the NKG2D system, dormancy and immunosurveillance are closely connected. At present, the molecular mechanisms behind the survival of dormant cells in the neural niche and their invisibility to effector cells of the immune system are mainly unknown.
The aim of the present study was to investigate the NKG2DL expression pattern of dormant cells in cerebral metastases of breast and lung cancer patients in situ, and to determine whether there was a brain-specific 'central' dormancy phenotype, represented by markers like EphA5, H2BK and IGFBP5, which has been described primarily in glioblastoma, or a 'peripheral' dormancy phenotype, represented by markers like PDGFβ, FGF2 and HIF1-α in association with immunogenicity represented by the NKG2DL system. As dormant cells serve a crucial role in tumor recurrence and survival, deciphering the immunogenicity in cerebral metastases might assist to identify future therapeutic targets.
Materials and methods
Tissue samples
Cerebral metastases of 6 patients with breast cancer and 9 patients with lung cancer were surgically resected at the Department of Neurosurgery, University Medical Center Schleswig-Holstein. The present study was approved by the Ethics Committee of the University of Kiel (approval no. D536/15) and was in accordance with the Helsinki Declaration of 1964 and its later amendments. Informed consent was obtained from all individual patients included in the study. Patient information is provided in Tables I and II. Tissue was frozen in liquid nitrogen directly following resection. The median age of breast cancer patients was 61 years. All patients with breast cancer were female; brain metastases were located in the frontal lobe in 4 cases, and occipital and cerebellum in 1 case. A total of 55.6% of patients with lung cancer were female, and the median age was 57 years. The metastases were located in the frontal lobe in 1 cases, the temporal lobe in 2 cases, the cerebellum in 3 cases, the parietal lobe in 3 cases and the occipital lobe in 2 cases. Certain patients presented with >1 metastases or metastases on the border of two lobes.
Table I.
Patients with diagnosis of cerebral metastases of breast cancer.
| Sample number | Sex | Age, years | Area of localization | Histology | Receptor status |
|---|---|---|---|---|---|
| Bcm1 | Female | 64 | Left frontal lobe | Carcinoma metastasis | ER 20%; PR negative |
| Bcm2 | Female | 61 | Right fronto-parietal lobe | Carcinoma metastasis | ER >80%; PR negative |
| Bcm3 | Female | 65 | Right occipital lobe | Carcinoma metastasis | ER 10-50%; PR negative |
| Bcm4 | Female | 79 | Right cerebellar hemisphere | Carcinoma metastasis | ER positive; PR negative |
| Bcm5 | Female | 57 | Left frontal lobe | Carcinoma metastasis | ER negative; PR negative |
| Bcm6 | Female | 40 | Left frontal lobe | Carcinoma metastasis | ER <10%; PR <10% |
Bcm, breast cancer metastasis; ER, estrogen receptor; PR, progesterone receptor.
Table II.
Patients with diagnosis of cerebral metastases of lung cancer.
| Sample number | Gender | Age, years | Area of localization | Histology |
|---|---|---|---|---|
| Lcm1 | Female | 64 | left temporal lobe | Adenocarcinoma |
| Lcm2 | Male | 44 | Left cerebellar hemisphere | Adenocarcinoma |
| Lcm3 | Male | 51 | Right frontotemporal lobe | Adenocarcinoma |
| Lcm4 | Female | 54 | Right cerebellar hemisphere | Adenocarcinoma |
| Lcm5 | Female | 65 | Left occipital lobe | Adenocarcinoma |
| Lcm6 | Male | 72 | Right parietooccipital lobe, left occipital lobe | Adenocarcinoma |
| Lcm7 | Female | 49 | Left frontoparietal lobe, right frontal lobe | Adenocarcinoma |
| Lcm8 | Female | 46 | Right parietal lobe | Adenocarcinoma |
| Lcm9 | Male | 68 | Left cerebellar hemisphere | Adenocarcinoma |
Lcm, lung cancer metastasis.
A total of 4 of the 6 patients with breast cancer had received chemotherapy according to clinical guidelines prior to diagnosis of the cerebral metastasis. All patients with breast cancer were treated with mastectomy or segment resection of the primary tumor prior to surgical removal of the cerebral metastasis. Only 1 of the patients was diagnosed with brain metastases prior to the diagnosis of breast cancer. A total of 4 breast cancer metastases samples were positive for estrogen receptors. None were positive for progesterone receptors.
A total of 8 of 9 patients with lung cancer were diagnosed with cerebral metastases prior to diagnosis of the primary tumor. Histologically, adenocarcinoma was diagnosed in all patients with lung cancer. All diagnoses were verified by a pathologist.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was isolated using TRIzol® (Thermo Fisher Scientific, Inc.). The method used was first described by Chomczynski and Sacchi (47) following modifications made in accordance to the manufacturer's protocol. DNA was digested by RNase free DNase (Promega Corporation). cDNA synthesis and RT-qPCR were performed using TaqMan MasterMix and Primer Probes (Applied Biosystems; Thermo Fisher Scientific, Inc.): Human (h)GAPDH [hGAPDH; (assay ID: Hs99999905_m1)], hH2BK (assay ID: Hs00955067_g1), hIGFBP5 (assay ID: Hs00181213_m1), hEphA5 (assay ID: Hs00300724_m1), hPDGFβ (assay ID: Hs00966522_m1), hHIF1A (assay ID: Hs00153153_m1), hFGF2 (assay ID: Hs00266645_m1), hMICA (assay ID: Hs00792195_m1), hMICB (assay ID: Hs04187752_mH), hULBP1 (assay ID: Hs00360941_m1) and hULBP2 (assay ID: Hs00607609_mH) and hULBP3 (assay ID: Hs00225909_m1). Cycle of threshold values (Cq) were measured by a cycler (QuantStudio 5, Applied Biosystems by Thermo Fisher Scientific). Gene expression levels were normalized to the housekeeping gene GAPDH using the following formula: ΔCq=Cq (gene of interest)-Cq (GAPDH). As ΔCq values correspond to the exponential course of the RT-qPCR, ΔCq=3.33 corresponds to a 10-fold lower expression compared to GAPDH and ΔCq=6.67 corresponds to a 100-fold lower expression. The relative gene expression compared with GAPDH (2−ΔCq) was used for statistical analysis (48). Cq of n=41, which was the maximum cycle number, was defined as undetectable, as this would commonly represent the detection limit if 40 amplification cycles are run.
Immunohistochemistry
For in situ analysis of marker expression, 10 µm cryosections of 5 different patients with breast cancer and 4 different patients with lung cancer were stained by immunohistochemistry. Sections were fixated with an ice-cold acetone-methanol mixture (1:1) for 10 min. Blockade for autofluorescence was performed with sudan black (1% in 70% ethanol; Carl Roth GmbH + Co., KG). Subsequently, blockage of unspecific antibody binding was performed by adding 0.5% glycine/0.5% bovine serum albumin. The slides were then incubated with primary antibodies overnight at 4°C, followed by secondary antibodies at 37°C for 1 h. Nuclei were stained with DAPI (Sigma-Aldrich; Merck KGaA; 1:30,000), which was applied at room temperature for 30 min. All antibodies were diluted in TBS with 0.1% Tween (TBST). Sections were embedded with Immumount (Thermo Fisher Scientific, Inc.). Following each step, slides were washed with TBST. The primary antibodies used were rabbit anti-FGF2 (1:1,250; Abcam), mouse anti-H2BK (1:400, Biorbyt, Ltd), rabbit anti-IGFBP5 (1:400, Santa Cruz Biotechnology, Inc.), rabbit anti-EphA5 (1:400; Santa Cruz Biotechnology, Inc.), mouse anti-PDGFβ (1:20; Abcam), mouse anti-MICA (1:150; Santa Cruz Biotechnology, Inc.), mouse anti-MICB (1:500; R&D Systems, Inc.), rabbit anti-ULBP1 (1:200; ProteinTech Group, Inc.), rabbit anti-ULBP2 (1:500; Abcam) and mouse anti-ULBP3 (1:250; R&D Systems). When both primary antibodies were derived from the same species, unspecific binding was blocked by adding F(ab) fragments from the same species (donkey anti-mouse and anti-rabbit F(ab) fragments; 1:1,000; Jackson ImmunoResearch, Laboratories Inc.). Subsequently, secondary antibodies were added. Isotype controls were performed by applying mouse or rabbit IgG1 (both R&D Systems) in concentrations adapted to the other primary antibodies used (1:1,000; Fig. 1). As secondary antibodies, donkey anti-mouse or anti-rabbit IgGs labelled with Alexa Fluor 488 or Alexa Fluor 555 (1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.) were used (Fig. 1). For negative controls, the primary antibodies were omitted.
Figure 1.

Negative and isotype controls for all used antibodies. Negative controls for cryosections of cerebral breast cancer (left column) and lung cancer (right column) metastases. Mouse and rabbit Alexa Fluor 488 and Alexa Fluor 555 were used. Isotype controls for cryosections are presented in the lower panel. Scale bar=10 µm.
Microscopy
Images were obtained with a Zeiss Oberver. Z1 microscope at ×40 magnification (Carl Zeiss AG). A Plan-Apochromat 40x/1.4 Oil DIC (CN) VIS-IR M27 was used as a lens. The camera AxioCam MR MR5 (Zeiss) with a focus depth of 0.89 µm for 555 nm, 0.80 µm for 488 nm, and 0.72 µm for DAPI was used. For image processing, ZEN 2 (blue edition) software was used (Carl Zeiss AG). Images had a size of 1,388×1,040 pixels (223.82×167.70 µm) at an image bit depth of 12 bits.
Statistical analysis
Data are presented as the mean ± standard deviation (SD). For testing significance of increase or decrease of expression, Student's t-test was used. P<0.05 were considered to indicate a statistically significant difference. A Pearson correlation analysis was performed to analyze the associations between marker expression levels. Microsoft Excel 2016 (Microsoft Corporation) and SPSS Statistics (version 24.0; IBM Corp) were used for all statistical analyses.
Results
Expression and cellular localization of NKG2DL and dormancy markers in cerebral metastases of breast and lung cancer in situ
The expression of NKG2DL and the different 'central' and 'peripheral' dormancy markers mentioned above were examined at mRNA and protein levels by RT-qPCR and double immunohistochemistry, respectively. Samples were obtained directly from metastases tissues of patients with breast and lung cancer, who underwent surgery at the Department of Neurosurgery, University Hospital Schleswig-Holstein, Campus Kiel.
mRNA expression of NGK2DL and different dormancy markers
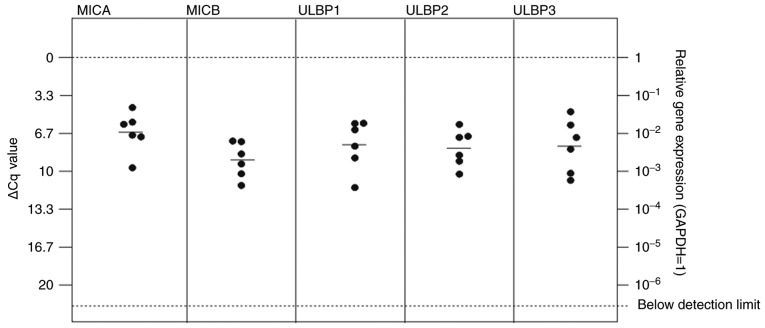
All NKG2L were identified to be expressed at detectable mRNA levels in breast and lung cancer metastases, with ULBP1 exhibiting the lowest expression level in the patients with lung cancer [Figs. 2 and 3; median ΔCq values in breast cancer tissues: MICA=6.57 (SD, 1.78); MICB=9.00 (SD, 1.56); ULBP1=7.66 (SD, 2.20); ULBP2=7.96 (SD, 1.62); ULBP3=7.79 (SD, 2.36); median ΔCq values in lung cancer tissues: MICA=6.90 (SD, 1.28); MICB=5.83 (SD, 1.98); ULBP1=9.84 (SD, 3.12); ULBP2=9.76 (SD, 1.37); and ULBP3=3.49 (SD, 1.51)].
Figure 2.
In vitro expression of Natural Killer Group 2 member D ligands MICA, MICB, ULBP1, 2 and 3 in cerebral metastases in patients with breast cancer. All ligands are expressed at a stable level without any significant differences. MICA, MHC class I chain-related protein A; MICB, MHC class I chain-related protein B; ULBP, UL16-binding protein family.
Figure 3.
In vitro expression of NKG2DL MICA, MICB, ULBP1, 2 and 3 in cerebral metastases of lung cancer. All NKG2DL are expressed on a stable level. Expression of ULBP1 was lowest compared with the other NKG2DL. MICA and MICB exhibited the highest expression levels, with significantly increased expression of MICA compared with ULBP2 and ULBP3. *P≤0.05. NKG2DL, Natural Killer Group 2 member D ligands; MICA, MHC class I chain-related protein A; MICB, MHC class I chain-related protein B; ULBP, UL16-binding protein family.
In the breast cancer metastases tissues, no significant differences between mRNA expression of the NKG2DL MICA, MICB, ULBP1, 2 and 3 were observed (Fig. 2). In lung cancer, MICA and MICB were highly expressed, with a significantly increased expression level of MICA compared with ULPB2 and 3 (Fig. 3).
The majority of the other dormancy markers were also expressed at detectable mRNA levels in breast and lung cancer metastases [Figs. 4 and 5; median ΔCq values in breast cancer tissues: H2BK=4.77 (SD, 2.56); IGFBP5=2.88 (2.50); EphA5=12.04 (SD, 2.36); PDGFβ=3.98 (SD, 2.85); FGF2=9.08 (SD, 2.71); HIF1-α=2.20 (SD, 1.85); median ΔCq values in lung cancer: H2BK=3.50 (SD, 1.51); IGFBP5=3.50 (SD, 2.66); EphA5=9.60 (SD, 1.42); PDGFβ=4.58 (SD, 1.34); FGF2=7.96 (SD, 1.40); HIF1-α=1.00 (1.06)]. EphA5 was expressed at the lowest levels in both cancer types. In breast cancer metastases, the 'peripheral' dormancy markers PDGFβ and HIF1-α were expressed at significantly increased levels compared with the 'central' dormancy marker EphA5 (Fig. 4). Similar results were obtained in lung cancer metastases tissues, where HIF1-α and PDGFβ also exhibited significantly increased expression levels compared with EphA5 (Fig. 5). In lung cancer metastases tissues, HIF1-α also was expressed at a significantly increased level compared with the 'central' dormancy marker H2BK. Among the 'central' dormancy markers, H2BK and IGFBP5 exhibited the highest expression levels, with significantly increased H2BK expression levels compared with the 'peripheral' dormancy marker FGF2 (Fig. 5).
Figure 4.
In vitro expression of central (H2BK, IGFBP5, EphA5) and peripheral (FGF2, PDGFβ, HIF1-α) dormancy markers in the cerebral metastasis tissues from patients with breast cancer. mRNA levels were analyzed by reverse transcription-quantitative polymerase chain reaction. Comparable with measurements in the patients with lung cancer metastases, all dormancy markers were expressed at a stable level. The 'peripheral' dormancy markers HIF1-α and PDGFβ exhibited significantly increased expression levels compared with the 'central' dormancy marker EphA5. The expression level of EphA5 was decreased compared with H2BK and IGFBP5, but this was not statistically significant. *P≤0.05. H2BK, Histone cluster 1 H2B family member K; IGFBP5, insulin-like growth factor-binding protein 5; EphA5, Ephrin receptor A5; FGF2, fibroblast growth factor 2; PDGFβ, platelet-derived growth factor β; HIF1-α, hypoxia-inducible factor 1-α.
Figure 5.
In vitro expression of 'central' (H2BK, IGFBP5, EphA5) and 'peripheral' (FGF2, PDGFβ, HIF1-α) dormancy markers in the cerebral metastasis tissues from patients with lung cancer. All dormancy markers were expressed at a stable level. The 'peripheral' dormancy marker HIF1-α was expressed at a significantly increased level compared with H2BK and EphA5, which are both 'central' dormancy markers. The 'peripheral' dormancy marker PDGFβ was expressed at a significantly increased level compared with the 'central' dormancy marker EphA5. Among the 'central' dormancy makers, H2BK and IGFBP5 were expressed at the highest levels. H2BK exhibited a significantly higher expression level compared with FGF2, which was expressed at relatively low levels. Overall, the expression of EphA5, one of the 'central' dormancy markers, was the lowest and was not detectable in 4 of the probes. *P≤0.05 and **P≤0.01. H2BK, Histone cluster 1 H2B family member K; IGFBP5, insulin-like growth factor-binding protein 5; EphA5, Ephrin receptor A5; FGF2, fibroblast growth factor 2; PDGFβ, platelet-derived growth factor β; HIF1-α, hypoxia-inducible factor 1-α.
Bravais-Pearson correlation analysis
A Bravais-Pearson correlation analysis of the RT-qPCR data, including all NKG2DL and dormancy markers, was performed.
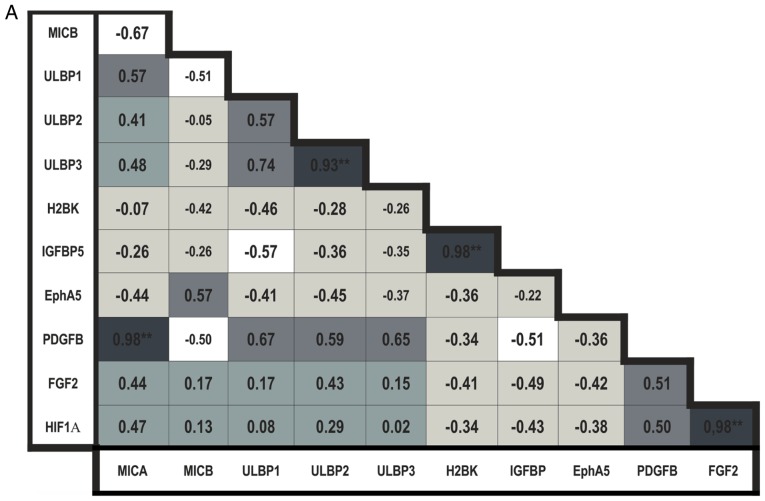
In the breast cancer metastases tissues, the expression of MICB was negatively correlated with expression of all other NKG2DL. The expression levels of MICA, ULBP1, 2 and 3 were positively correlated with each other; the correlation between ULBP2 and 3 was observed to be significant (P<0.01; Fig. 6A).
Figure 6.
Bravais-Pearson correlation analysis. (A) Correlation analysis in the cerebral metastasis tissues from patients with breast cancer. (B) Correlation analysis in the cerebral metastasis tissues from patients with lung cancer. Colors indicate a positive (dark grey) or negative (light grey and white) correlation as follows: Very dark grey (ρ>0,8), mid-dark grey (0.8>ρ>0.5) and dark gray (0.5>ρ>0) colors indicate a positive correlation coefficient, and white (ρ<−0,8), mid-light grey (−0.8>ρ<−0,5) and light grey (−0.5>ρ<0) colors indicate a negative correlation coefficient. *P≤0.05 and **P≤0.01.
When analyzing the 'central' dormancy markers in the breast cancer metastases tissues, the expression levels of H2BK and IGFBP5 were identified to be positively correlated (P<0.01; Fig. 6A).
The expression levels of the 'peripheral' dormancy markers in breast cancer metastases tissues were positively correlated with each other, but this result was not statistically significant (Fig. 6A). There was a significantly positive correlation between the expression of FGF2 and HIF1-α (P<0.01; Fig. 6A).
The expression of 'central' dormancy markers was negatively correlated with expression of 'peripheral' dormancy markers in breast cancer.
The expression levels of all NKG2DL were positively correlated with the expression of nearly all 'peripheral' dormancy markers, with exclusion of the association between MICB and PDGFβ; the correlation between MICA and PDGFβ was observed to be statistically significant (P<0.01; Fig. 6A) in breast cancer. The expression levels of all NKG2DL were negatively correlated with the expression of 'central' dormancy markers, with exception of MICB, whose expression exhibited a slightly positive correlation with expression of EphA5 (Fig. 6A).
When examining the cerebral metastases tissues from the patients with lung cancer, the results of the correlation analysis were more complex. Similar to the data from the metastases tissues from the patients with breast cancer, the expression of MICB was negatively correlated with expression of all other NKG2DL. The expression of MICA, ULBP1 and 3 were positively correlated. A significant correlation in lung cancer was observed in the expression of ULBP1 and 3, which were positively correlated (P<0.05; Fig. 6B).
When examining the associations between the expression levels of the dormancy markers, mostly positive correlations were observed between the expression levels of all 'peripheral' dormancy markers with all other 'central' dormancy markers in lung cancer metastases. Only the expression of EphA5 was negatively correlated with the expression of the other 'peripheral' dormancy markers FGF2 and HIF1-α, but this was not significantly different (Fig. 6B).
The correlation analysis concerning NKG2DL and dormancy marker expression in lung cancer metastases was heterogeneous. The MICA and ULBP3 expression levels were negatively correlated with the expression of 'central' and 'peripheral' dormancy markers, with the exception of FGF2, which exhibited a moderate, non-significant positive correlation. Expression of MICB was significantly positively correlated with the expression of the 'central' dormancy marker IGFBP5 and the 'peripheral' marker HIF1-α. The expression levels of ULBP1 and 2 were significantly positively correlated with the expression of the 'central' dormancy marker EphA5. There was no significant correlation between MICB and EphA5 observed.
Double immunostaining
To specify the in situ distribution and localization of dormancy markers, cryosections of cerebral metastases from patients with breast and lung cancer were stained. The NKG2D ligands MICA, MICB, ULBP1 and ULBP2 were stained in combination with the 'central' dormancy markers EphA5, H2BK and IGFBP5 and the 'peripheral' dormancy markers PDGFβ, FGF2 and HIF1-α. The staining results are summarized in Fig. 7A and B. The numbers of stained tumors is also presented in Fig. 7A and B. A high degree of consistency concerning the staining results between the different patients and between different areas of the tumors was observed. Example images of the staining results are presented in Figs. 8-11. In order to provide a respresentative impression, only one image per combination of markers and per tumor entity has been presented. In cases where this one image does not represent the results obtained in all areas the tumors, a second image was added in a white box from a second area of a second tumor. If the image in the white box was taken from another tumor of the same entity, it was marked with an asterix.
Figure 7.
Staining patterns in the breast and lung cancer patients. (A) Overview of staining pattern in breast cancer metastases. (B) Overview of staining pattern in lung cancer metastases. The numbers in the box represent the number of different tumors that were examined. Light grey color represents no co-staining, normal grey represents partial co-staining and dark grey represents complete co-staining. MICA, MHC class I chain-related protein A; MICB, MHC class I chain-related protein B; ULBP, UL16-binding protein family; H2BK, Histone cluster 1 H2B family member K; IGFBP5, insulin-like growth factor-binding protein 5; EphA5, Ephrin receptor A5; FGF2, fibroblast growth factor 2; PDGFβ, platelet-derived growth factor β; HIF1-α, hypoxia-inducible factor 1-α.
Figure 8.


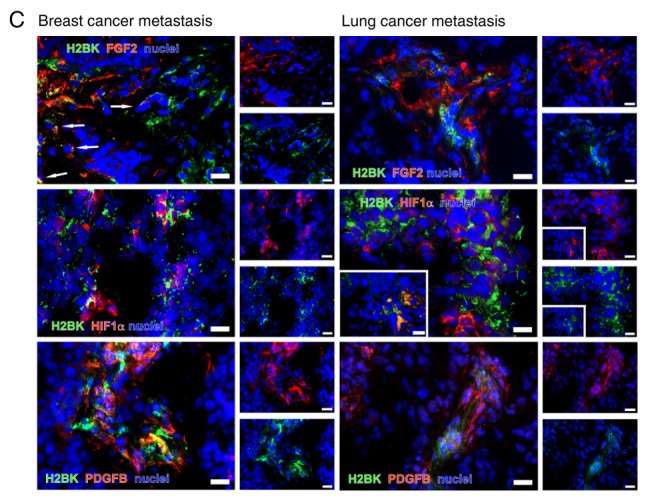
Double immunostaining of dormancy markers. Selected images from different cerebral metastases of lung and breast cancer. (A) Double immunos-taining for 'central' dormancy markers EphA5, H2BK and IGFBP5 in representative cryosections from cerebral metastases of breast (left column) and lung cancer (right column). (B) Double immunostaining for 'peripheral' dormancy markers PDGFβ, FGF2 and HIF1-α in exemplary cryosections from cerebral metastases of breast (left column) and lung cancer (right column). (C) Double immunostaining for 'central' with 'peripheral' dormancy markers in exemplary cryosections from cerebral metastases of breast (left column) and lung cancer (right column). Scale bar=20 µm. The images in the white boxes show another representative image in the same tumor. H2BK, Histone cluster 1 H2B family member K; IGFBP5, insulin-like growth factor-binding protein 5; EphA5, Ephrin receptor A5; FGF2, fibroblast growth factor 2; PDGFβ, platelet-derived growth factor β; HIF1-α, hypoxia-inducible factor 1-α.
Figure 11.
Double immunostaining for dormancy markers with Ki-67. Representative results are presented for staining of HIF1-α with Ki-67. Neither of the breast (left) and lung (right) cancer metastases tissues exhibited of HIF1-α and Ki-67 co-staining. Scale bar=20 µm. HIF1-α, hypoxia-inducible factor 1-α; Ki-67, proliferation marker protein Ki-67.
Double immunostaining for dormancy markers among each other
All dormancy markers were detectable using double immunostaining (Fig. 8A-C). The majority of the marker-positive cells were identified as single cells or in small clusters.
When staining for 'central' dormancy markers exclusively, a broad co-staining of IGFBP5 with H2BK and IGFBP5 with EphA5 was observed in breast cancer metastases. In the lung cancer tissues, a broad co-staining of all 'central' dormancy markers was identified. This was the case in different areas of one tumor, but also when comparing tumors from different patients. The distribution pattern was similar in breast and lung cancer metastases tissues (Fig. 8A).
When considering the 'peripheral' dormancy markers, several partial co-staining patterns were observed in breast cancer: HIF1-α with FGF2; HIF1-α with PDGFβ; and PDGFβ with FGF2 (Fig. 8B), and a partial or complete co-staining in lung cancer metastases (Fig. 8B).
When staining for 'peripheral' and 'central' dormancy markers in breast cancer, a rare but partial co-staining of most markers was observed. Representative images for H2BK co-staining with 'peripheral' dormancy markers are presented in Fig. 8C.
In the lung cancer metastases tissues, the majority of the 'central' and 'peripheral' dormancy markers were partially co-stained with each other in a small number of samples. Representative sections are presented in Fig. 8C.
Clearly observable co-staining between the majority of the 'central' dormancy markers and a lack of co-staining between the 'central' and 'peripheral' dormancy markers in both tumor types suggested that there may be a circumscribed population of dormant cells in each tumor type, which is defined by the expression of 'central' dormancy markers. As only a partial co-staining between the 'peripheral' dormancy markers was observed in both tumor types, these appear mark non-overlapping populations of dormant cells.
Double immunostaining of NKG2DL
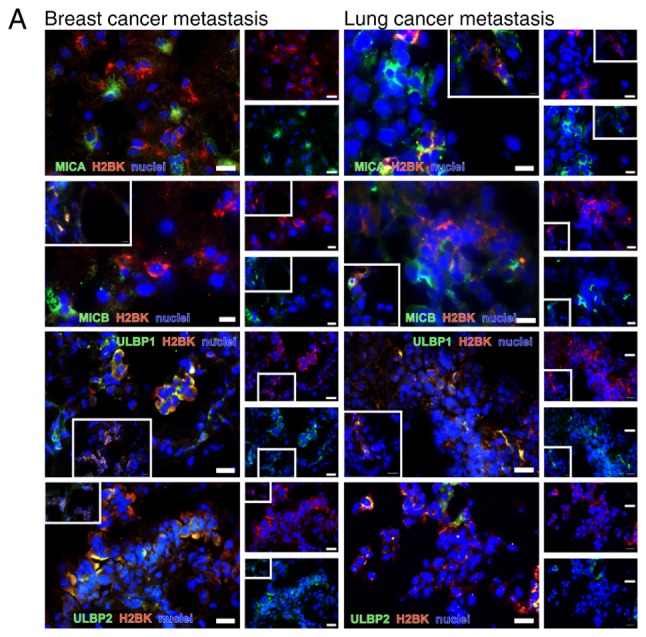
The majority of the NKG2DL+ cells were identified in small groups or as single cells scattered over the tumor. Notably, most tumors exhibited a similar staining pattern, irrespective of the tumor type. MICA and MICB were identified to be co-stained. MICA was very rarely co-stained with ULBP1 and 2 in the breast cancer tissues. In the lung cancer samples, MICA and ULBP2 were observed as co-stained. ULBP1 was co-stained with ULBP2 in both tumor types. MICB was not co-stained with ULBP1 and 2 in breast cancer and partially co-stained with ULBP1 in lung cancer (Fig. 9). As observed previously in glioma stem-like cells (45), only a subset of NKG2DL was identified to be expressed simultaneously in each cell.
Figure 9.

Double immunostaining of NKG2DL. Double immunostaining of the NKG2DL MICA, MICB, ULBP1 and ULBP2 in the cerebral metastasis tissues from patients with breast (first column) and lung cancer (second column). Both tumor types exhibited a similar staining pattern. Scale bar=20 µm. The image in the white box shows in the first row shows a rare costaining between ULBP1 and MICA in a breast cancer metastasis. NKG2DL, Natural Killer Group 2 member D ligands; MICA, MHC class I chain-related protein A; MICB, MHC class I chain-related protein B; ULBP, UL16-binding protein family.
Double immunostaining of dormancy markers with NKG2DL
Staining pattern for 'central' and 'peripheral' dormancy markers and NKG2DL was similar in breast and lung cancer metastases tissues.
In breast cancer metastases, co-staining for MICA, MICB and ULBP1 with 'central' dormancy markers H2BK and IGFBP5 was observed in a small number of samples. EphA5 was frequently co-stained with MICB, ULBP1 and 2. ULBP2 was identified to be co-stained with 'central', but not 'peripheral' dormancy markers in breast cancer tissues. Representative images are presented in Fig. 10A.
Figure 10.

Double immunostaining of dormancy markers with NKG2DL. (A) Double immunostaining of 'central' dormancy markers with H2BK as a representative marker in combination with NKG2DL MICA, MICB, ULBP1 and ULBP2 in cryosections from cerebral metastases tissues from patients with breast (left column) and lung cancer (right column). Staining analyses for EphA5 and IGFBP5 with all NKG2D ligands were also performed. (B) Double immunostaining of 'peripheral' dormancy markers, here with PDGFβ as an example, in combination with the NKG2DL MICA, MICB, ULBP1 and 2 in cryosections from cerebral metastases of breast (left column) and lung cancer (right column). *Representative staining of different tumor tissues compared with those in the larger picture. Scale bar=20 µm. The images in the white boxes show representative areas of the markers in the same tumor. If the image in the white box was taken from another tumor, it was marked with an asterix. NKG2DL, Natural Killer Group 2 member D ligands; MICA, MHC class I chain-related protein A; MICB, MHC class I chain-related protein B; ULBP, UL16-binding protein family; H2BK, Histone cluster 1 H2B family member K; PDGFβ, platelet-derived growth factor β.
NKG2DL+ cells therefore appear to be hallmarked by a spectrum of 'central' dormancy markers in breast cancer metastases tumors.
In the lung cancer metastases tumors, co-staining for MICB and ULBP1 with IGFBP5 was observed in a small number of samples; representative images are presented in Fig. 10A. MICA and MICB were co-stained with H2BK and EphA5 in some, but not all cells. ULBP1 was co-stained with H2BK and EphA5 in examined all cells. Unlike in breast cancer metastases, ULBP2 was co-stained with H2BK, IGFBP5 and EphA5 (Fig. 10A).
When analyzing the 'peripheral' dormancy markers, similar staining patterns were observed when comparing breast and lung cancer metastases tissues. Representative images are presented in Fig. 10B. MICA was completely co-stained with PDGFβ and HIF1-α. MICB was completely co-stained with FGF2, but only partly with PDGF and HIF1-α. ULBP1 and 2 were rarely co-stained with PDGFβ in both tumor types, completely co-stained with HIF1-α in breast cancer, and partially co-stained with HIF1-α in lung cancer. ULBP1 and 2 were not co-stained with FGF2 (Fig. 10).
Although a positive NKG2DL and HIF1-α co-staining result was considered to be representative of 'peripheral' dormancy markers, the majority of the NKG2DL+ cells were identified to express 'central' dormancy markers in lung cancer metastases.
Double immunostaining of dormancy markers with Ki-67
As previously described, Ki-67 is a protein primarily expressed during the G1-, S-, G2- and M-phases of the cell cycle (25). Therefore, is not expected to be expressed in dormant cells. In the present study, HIF1-α was co-stained with Ki-67 (Fig. 11). Ki-67 was observed to be primarily identified in the nucleus. As indicated in Fig. 11, HIF1-α and Ki-67 were not observed to be co-stained.
Discussion
Prognosis of patients with cerebral metastases is poor (49,50). Mechanisms of tumor escape in the periphery involve the NKG2D system and therapy resistance of dormant cells. NKG2DL+ cells are usually detected by NKG2DR+ cells and then killed by cytotoxic T-cells and NK cells. In cells that proliferate normally, this mechanism is avoided by the shedding of NKG2DL and its subsequent secretion in exosomes. Dormancy, as a universal mechanism of chemo-resistance, crucially depends on the microenvironment that surrounds the disseminated tumor cells. The bone marrow and peri-vascular niches in particular have been closely investigated as important sites for the development of dormant tumor cells (46,51). To the best of our knowledge, the role of the NKG2D system in dormant cells in the brain has not been investigated fully.
EphA5, H2BK and IGFBP5 serve a role in glioblastoma and cerebral metastases
RT-qPCR analysis and IHC on cerebral metastases tissues from patients with breast and lung cancer indicated that the 'central' dormancy markers EphA5, H2BK and IGFBP5 were expressed at a stable level in both tumor entities.
Immunohistochemistry analysis revealed that these 'central' dormancy markers were co-stained with each other, but only occasionally with 'peripheral' dormancy markers. 'Central' dormancy markers therefore appeared to be expressed by a special cell population in cerebral metastases, which may also be identified in other types of tumors of the brain. EphA5, H2BK and IGFBP5 as dormancy markers have been identified to be relevant in glioblastoma (19,38). None of these 'central' dormancy markers have been demonstrated to serve a role in the dormancy processes occurring outside of the central nervous system. Nevertheless, IGFBP5 has been identified in metastases of lung cancer and low levels of IGFBP5 were observed to be associated with improved survival in non-small cell lung cancer (52). In breast cancer, IGFBP5 serves a role in peripheral tumor progression and is associated with tumor cell differentiation, apoptosis and regulation of cellular growth (53,54). EphA5 has been suggested to be involved in anticancer processes in breast cancer, and a deletion of EphA5 leads to a significantly increased susceptibility towards chemotherapeutics (55). EphA5 has been demonstrated to be expressed in ~70% of all lung cancer cells and is markedly associated with radio resistance of lung cancer cells; the expression of EphA5 is markedly correlated with an increased mortality rate (56). To the best of our knowledge, H2BK expression has not been identified in the development of peripheral cancer.
Notably, the microenvironment of the neural niche appears to lead to the development of a central type of dormancy in brain metastases, which is different from the type of dormancy observed in cells in the periphery. This confirms the importance of the microenvironment on the development of dormancy and may explain the varying susceptibility of tumor cell populations in different anatomical sites to chemo- and radiotherapy.
Dormancy marker expression of cerebral metastases is tumor type-specific, but homogeneous when comparing patients
All examined markers, in particular H2BK and IGFBP5, were expressed at detectable levels in RT-qPCR analysis.
In the breast cancer tissues, a negative correlation between the expression levels of the 'central' and peripheral' dormancy markers was observed. Therefore, 'central' and 'peripheral' dormancy marker expression was used to distinguish between the two populations of dormant cells. In the lung cancer tumor tissues, a positive correlation between 'central' and 'peripheral' dormancy markers was observed, with exception of EphA5, suggesting that the population of dormant cells cannot be divided into two separate populations based on these markers.
Breast and lung cancer metastases differ concerning the dormancy marker expression. This heterogeneity may, in part, explain the differences observed in chemo-resistance patterns (49), which is frequently observed in numerous patients with brain metastases, particularly in breast cancer (57).
Despite the clear tumor type-specific patterns observed, the staining results were highly homogeneous among different patients with the same tumor type. In contrast to glioblastoma, which exhibited a very high level of inter- and intratumoral heterogeneity, metastases clearly follow a distinct evolutionary path, which depends on their parent tumor type (58). The present study demonstrated that the microenvironment of the brain affected the dormancy marker and NKG2DL expression levels in a site-specific manner. This is in concordance with the data from Kienast et al (59), who demonstrated that the unique microenvironmental architecture of the brain exerts a selective drive on metastasized cells and leads to an induction of vascular dormancy in the perivascular neural niche. Therefore, it is not clear whether the distinct features of dormant metastatic cells in the brain are induced by the surrounding cerebral microenvironment, or whether they are selected from a minority of cells already expressing 'central' dormancy markers.
NKG2DL are differentially expressed in dormant cells
Similar to observations in glioblastoma and glioma stem-like cells (43,45), the expression levels of NKG2DL in dormant cells of cerebral metastases appear to differ on a cellular level. The present study demonstrated that, in breast cancer tissues, the populations of 'central' dormant cells contain a subset of NKG2DL+ cells, which differs from the 'peripheral' dormant cells. In the breast cancer tissues, 'central' dormancy marker-expressing cells also expressed MICB and 'peripheral' dormancy marker-expressing cells also expressed MICA, ULBP1 and 2, as demonstrated by the RT-qPCR data. In the lung cancer cases, MICB appeared to be expressed in cells that also expressed 'central' dormancy markers, primarily IGFBP5, as demonstrated by the RT-qPCR data. Notably, MICB expression was correlated with HIF1-α expression in lung cancer metastases. This may be due to the overall high expression level of HIF1-α in the breast cancer metastases tissues, which was hypothesized to have occurred due to the hypoxic cerebral microenvironment. HIF1-α is a known key factor in the hypoxic response in cancer (60). Concerning the other NKG2DL, the expression profile patterns of NKG2DL in the dormant cells was not as able to distinguish between central and peripheral dormancy as in breast cancer.
Malladi et al (61) indicated that the depletion of NK cells in athymic mice resulted in permissive outgrowth of dormant tumors cells of breast and lung cancer and an increased number of bone metastases. They also demonstrated that dormant cells that survived NK cell surveillance exhibited a downregulation of several UL16 binding proteins (ULBPs). As the present study identified a preserved expression of NKG2DL in cells expressing 'central' dormancy markers, it was hypothesized that these cells may be susceptible to therapeutic approaches targeting 'central' dormancy markers. In addition, the mechanisms leading to downregulation of ULBPs in dormant cells in the periphery may not occur in the central nervous system.
Analysis of the expression of NKG2DL provides potential new therapeutic approaches. As a mechanism of tumor immune escape, tumor cells shed NKG2DL from their surface using a disintegrin metalloproteinases (ADAMs) (62). There are several known inhibitors of ADAMs (63). These may be effective in targeting a subgroup of dormant cells in the central nervous system.
The heterogeneity of NKG2DL expression in dormant cells may be an important immunological escape mechanism and may explain the heterogenous treatment responses and survival curves of patients with cerebral metastases (49). Additionally, heterogeneity in NKG2DL expression has been described in other tumor entities including lymphoma (64), melanoma (65), leukemia (66), colorectal cancer (67), ovarian carcinoma (68) and glioblastoma (45). The data from the present study demonstrates that this pattern of heterogeneity was also present in cerebral metastases tissues from patients with breast and lung cancer.
NKG2DL and dormancy marker RNA expression levels differ from surface expression
In the breast cancer cases, the mRNA expression levels of 'peripheral' dormancy markers were increased compared with the expression levels of the 'central' dormancy markers. The increased mRNA levels of the 'peripheral' dormancy markers were associated with a high expression level of NKG2DL. At a protein level, NKG2DL expression occurred primarily in cells carrying 'central' dormancy markers. In the lung cancer metastases tissues, the absolute mRNA expression levels of the 'peripheral' dormancy markers were also increased compared with the expression levels of the 'central' dormancy markers; however, a positive correlation between mRNA expression levels was observed. At the protein level, NKG2DL+ cells were identified to express mostly 'central' dormancy markers.
Localization in-situ and protein expression levels, which were examined using IHC, and mRNA expression, which was examined using RT-qPCR, are not directly comparable.
Wolpert et al demonstrated that NKG2DL mRNA and surface expression levels may differ, which is most likely due to posttranslational and posttranscriptional modification (69). Other mechanisms, including the induction of surface loss of MICB via the secretory pathway by human cytomegalovirus, may also serve a role (70,71) in this process.
Study limitations
As a very limited number of patients were included in the present study, the results are only preliminary. However, the staining results were highly consistent between the different patients. Nevertheless, due to the reproducibility of the results, they may be considered as representative. Due to the limited availability of frozen tissue and corresponding DNA, immunocytochemistry was only performed in 5 of the 6 patients breast cancer and in 4 of the 9 patients with lung cancer. A quantitative analysis of protein expression would be valuable, but would not be meaningful in the low number of patients analyzed in the present study. The majority of the patients with breast cancer metastases in the present cohort received chemotherapy prior to the development and consequent surgical resection of brain metastases. This was not the case in the patients with lung cancer; in these patients, the brain metastases were the first detected manifestation of the tumor in 8 of the 9 patients with lung cancer who were included into the study. Chemotherapy is known to induce dormancy, and therefore may have changed the expression patterns of the markers observed in the patients with breast cancer. Additionally, the cohort of lung cancer patients analyzed in the present study only represented a subgroup of patients, as the majority of cases of pulmonary adenocarcinoma are diagnosed prior to manifestation of brain metastases. It therefore cannot be excluded that the cohort of the present study is susceptible to systematic bias. An additional important limitation of the present study is that no healthy brain tissues were available to compare the dormancy markers and NKG2DL expression profiles. This is due to the fact that living cerebral cells and cryoprotected tissue from healthy donors are difficult to obtain. Using tissue from patients who underwent surgery for conditions such as epilepsy or brain trauma would incur a significant risk of bias, as trauma, ischemia and other pathologic stimuli are known to alter NKG2DL expression (41). Using tissue from cadavers is also not an acceptable alternative, as the hypoxia occurring during death may also alter NKG2DL expression (72).
In conclusion, breast and lung cancer brain metastases were demonstrated to express 'peripheral' dormancy markers at increased levels compared with 'central' dormancy markers at an mRNA level, which was predicted, due to the peripheral origin of the tumors. Notably, EphA5, H2BK and IGFBP5, which have been demonstrated to serve roles in glioblastoma, were observed to be important factors in cerebral metastases. NKG2DL are expressed at high levels, with tumor type-specific differences observed between different members of the family. NKG2DL were identified to be predominantly co-localized with 'central', not 'peripheral', dormancy markers in both tumor types examined. This cell population, which was demonstrated to be positive for NKG2DL and dormancy markers, may represent a highly attractive target for therapy in the future.
Acknowledgments
The authors would like to thank the medical-technical assistants Mrs Fereshteh Ebrahim and Mrs Brigitte Rehmke (both from the Department of Neurosurgery, University Hospital of Schleswig-Holstein, Campus Kiel, Germany) for their expert technical support.
Funding
The present study was supported by a sponsorship from the University Hospital of Schleswig-Holstein, Campus Kiel (Forschungsförderung 2016, grant no. F356971; Juniorförderung 2018, grant no. F358916) which was awarded to CF. Additionally, the study was funded by the German Research Foundation as part as of the Research Training Group 'Materials4Brain' (grant no. RTG2154; P8).
Availability of data and materials
The datasets analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
CF performed the histological examination of the cryosections, performed the statistical analysis, analyzed and interpreted patient data, generated the figures and wrote the manuscript. VM performed the reverse transcription-quantitative polymerase chain reaction, immunohistochemical staining and quantitative analysis of immunohistochemical staining. VA assisted with the immunohistochemical staining procedure. MS and JF supervised the project and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the University of Kiel (approval no. D536/15) and was in accordance with the Helsinki Declaration of 1964 and its later amendments. Informed consent was obtained from all individual patients included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Doron H, Pukrop T, Erez N. A Blazing Landscape: Neuroinflammation shapes brain metastasis. Cancer Res. 2019;79:423–436. doi: 10.1158/0008-5472.CAN-18-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cagney DN, Martin AM, Catalano PJ, Redig AJ, Lin NU, Lee EQ, Wen PY, Dunn IF, Bi WL, Weiss SE, et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. Neuro Oncol. 2017;19:1511–1521. doi: 10.1093/neuonc/nox077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berghoff AS, Schur S, Füreder LM, Gatterbauer B, Dieckmann K, Widhalm G, Hainfellner J, Zielinski CC, Birner P, Bartsch R, Preusser M. Descriptive statistical analysis of a real life cohort of 2419 patients with brain metastases of solid cancers. ESMO Open. 2016;1:e000024. doi: 10.1136/esmoopen-2015-000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekici K, Temelli O, Dikilitas M, Halil Dursun I, Bozdag Kaplan N, Kekilli E. Survival and prognostic factors in patients with brain metastasis: Single center experience. J BUON. 2016;21:958–963. [PubMed] [Google Scholar]
- 5.Takei H, Rouah E, Ishida Y. Brain metastasis: Clinical characteristics, pathological findings and molecular subtyping for therapeutic implications. Brain Tumor Pathol. 2016;33:1–12. doi: 10.1007/s10014-015-0235-3. [DOI] [PubMed] [Google Scholar]
- 6.Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC. Multistep nature of metastatic inefficiency: Dormancy of solitary cells after successful extrava-sation and limited survival of early micrometastases. Am J Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gužvić M, Klein CA. Cancer dormancy: Time to explore its clinical relevance. Breast Cancer Res. 2013;15:321. doi: 10.1186/bcr3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naumov GN, Bender E, Zurakowski D, Kang SY, Sampson D, Flynn E, Watnick RS, Straume O, Akslen LA, Folkman J, Almog N. A model of human tumor dormancy: An angiogenic switch from the nonangiogenic phenotype. J Natl Cancer Inst. 2006;98:316–325. doi: 10.1093/jnci/djj068. [DOI] [PubMed] [Google Scholar]
- 9.Townson JL, Chambers AF. Dormancy of solitary metastatic cells. Cell Cycle. 2006;5:1744–1750. doi: 10.4161/cc.5.16.2864. [DOI] [PubMed] [Google Scholar]
- 10.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 11.Barkan D, El Touny LH, Michalowski AM, Smith JA, Chu I, Davis AS, Webster JD, Hoover S, Simpson RM, Gauldie J, Green JE. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res. 2010;70:5706–5716. doi: 10.1158/0008-5472.CAN-09-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wikman H, Vessella R, Pantel K. Cancer micrometastasis and tumour dormancy. APMIS. 2008;116:754–770. doi: 10.1111/j.1600-0463.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 13.Holmgren L, O'Reilly MS, Folkman J. Dormancy of micro-metastases: Balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 14.Achilles EG, Fernandez A, Allred EN, Kisker O, Udagawa T, Beecken WD, Flynn E, Folkman J. Heterogeneity of angiogenic activity in a human liposarcoma: A proposed mechanism for 'no take' of human tumors in mice. J Natl Cancer Inst. 2001;93:1075–1081. doi: 10.1093/jnci/93.14.1075. [DOI] [PubMed] [Google Scholar]
- 15.Udagawa T, Fernandez A, Achilles EG, Folkman J, D'Amato RJ. Persistence of microscopic human cancers in mice: Alterations in the angiogenic balance accompanies loss of tumor dormancy. FASEB J. 2002;16:1361–1370. doi: 10.1096/fj.01-0813com. [DOI] [PubMed] [Google Scholar]
- 16.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimbrone MA, Jr, Leapman SB, Cotran RS, Folkman J. Tumor dormancy in vivo by prevention of neovascularization. J Exp Med. 1972;136:261–276. doi: 10.1084/jem.136.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339:58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- 19.Almog N, Ma L, Raychowdhury R, Schwager C, Erber R, Short S, Hlatky L, Vajkoczy P, Huber PE, Folkman J, Abdollahi A. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Res. 2009;69:836–844. doi: 10.1158/0008-5472.CAN-08-2590. [DOI] [PubMed] [Google Scholar]
- 20.Quesnel B. Dormant tumor cells as a therapeutic target? Cancer Lett. 2008;267:10–17. doi: 10.1016/j.canlet.2008.02.055. [DOI] [PubMed] [Google Scholar]
- 21.Moserle L, Amadori A, Indraccolo S. The angiogenic switch: Implications in the regulation of tumor dormancy. Curr Mol Med. 2009;9:935–941. doi: 10.2174/156652409789712800. [DOI] [PubMed] [Google Scholar]
- 22.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 23.Kenny PA, Bissell MJ. Tumor reversion: Correction of malignant behavior by microenvironmental cues. Int J Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkan D, Green JE, Chambers AF. Extracellular matrix: A gatekeeper in the transition from dormancy to metastatic growth. Eur J Cancer. 2010;46:1181–1188. doi: 10.1016/j.ejca.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomis RR, Gawrzak S. Tumor cell dormancy. Mol Oncol. 2017;11:62–78. doi: 10.1016/j.molonc.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker ND, Patel J, Munoz JL, Hu M, Guiro K, Sinha G, Rameshwar P. The bone marrow niche in support of breast cancer dormancy. Cancer Lett. 2016;380:263–271. doi: 10.1016/j.canlet.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 27.Tivari S, Korah R, Lindy M, Wieder R. An in vitro dormancy model of estrogen-sensitive breast cancer in the bone marrow: A tool for molecular mechanism studies and hypothesis generation. J Vis Exp. 2015:e52672. doi: 10.3791/52672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel P, Chen EI. Cancer stem cells, tumor dormancy, and metastasis. Front Endocrinol (Lausanne) 2012;3:125. doi: 10.3389/fendo.2012.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jong JS, van Diest PJ, van der Valk P, Baak JP. Expression of growth factors, growth factor receptors and apoptosis related proteins in invasive breast cancer: Relation to apoptotic rate. Breast Cancer Res Treat. 2001;66:201–208. doi: 10.1023/A:1010650305100. [DOI] [PubMed] [Google Scholar]
- 30.Cervi D, Yip TT, Bhattacharya N, Podust VN, Peterson J, Abou-Slaybi A, Naumov GN, Bender E, Almog N, Italiano JE, Jr, et al. Platelet-associated PF-4 as a biomarker of early tumor growth. Blood. 2008;111:1201–1207. doi: 10.1182/blood-2007-04-084798. [DOI] [PubMed] [Google Scholar]
- 31.Fluegen G, Avivar-Valderas A, Wang Y, Padgen MR, Williams JK, Nobre AR, Calvo V, Cheung JF, Bravo-Cordero JJ, Entenberg D, et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat Cell Biol. 2017;19:120–132. doi: 10.1038/ncb3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endo H, Okami J, Okuyama H, Nishizawa Y, Imamura F, Inoue M. The induction of MIG6 under hypoxic conditions is critical for dormancy in primary cultured lung cancer cells with activating EGFR mutations. Oncogene. 2017;36:2824–2834. doi: 10.1038/onc.2016.431. [DOI] [PubMed] [Google Scholar]
- 33.Jiang ZF, Wang M, Xu JL, Ning YJ. Hypoxia promotes mitochondrial glutamine metabolism through HIF1α-GDH pathway in human lung cancer cells. Biochem Biophys Res Commun. 2017;483:32–38. doi: 10.1016/j.bbrc.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Abdollahi A, Hahnfeldt P, Maercker C, Gröne HJ, Debus J, Ansorge W, Folkman J, Hlatky L, Huber PE. Endostatin's antiangiogenic signaling network. Mol Cell. 2004;13:649–663. doi: 10.1016/S1097-2765(04)00102-9. [DOI] [PubMed] [Google Scholar]
- 35.Yancopoulos GD, Klagsbrun M, Folkman J. Vasculogenesis, angiogenesis, and growth factors: Ephrins enter the fray at the border. Cell. 1998;93:661–664. doi: 10.1016/S0092-8674(00)81426-9. [DOI] [PubMed] [Google Scholar]
- 36.Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, Church D, Hassan AB. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol. 2005;205:145–153. doi: 10.1002/path.1712. [DOI] [PubMed] [Google Scholar]
- 37.Lassalle P, Molet S, Janin A, Heyden JV, Tavernier J, Fiers W, Devos R, Tonnel AB. ESM-1 is a novel human endothelial cell-specific molecule expressed in lung and regulated by cytokines. J Biol Chem. 1996;271:20458–20464. doi: 10.1074/jbc.271.34.20458. [DOI] [PubMed] [Google Scholar]
- 38.Adamski V, Hempelmann A, Flüh C, Lucius R, Synowitz M, Hattermann K, Held-Feindt J. Dormant glioblastoma cells acquire stem cell characteristics and are differentially affected by Temozolomide and AT101 treatment. Oncotarget. 2017;8:108064–108078. doi: 10.18632/oncotarget.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung TY, Choi YD, Kim YH, Lee JJ, Kim HS, Kim JS, Kim SK, Jung S, Cho D. Immunological characterization of glioblastoma cells for immunotherapy. Anticancer Res. 2013;33:2525–2533. [PubMed] [Google Scholar]
- 40.Chitadze G, Flüh C, Quabius ES, Freitag-Wolf S, Peters C, Lettau M, Bhat J, Wesch D, Oberg HH, Luecke S, et al. In-depth immunophenotyping of patients with glioblastoma multiforme: Impact of steroid treatment. Oncoimmunology. 2017;6:e1358839. doi: 10.1080/2162402X.2017.1358839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene. 2008;27:5944–5958. doi: 10.1038/onc.2008.272. [DOI] [PubMed] [Google Scholar]
- 42.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chitadze G, Lettau M, Luecke S, Wang T, Janssen O, Fürst D, Mytilineos J, Wesch D, Oberg HH, Held-Feindt J, Kabelitz D. NKG2D- and T-cell receptor-dependent lysis of malignant glioma cell lines by human γδ T cells: Modulation by temozolomide and A disintegrin and metalloproteases 10 and 17 inhibitors. Oncoimmunology. 2015;5:e1093276. doi: 10.1080/2162402X.2015.1093276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chitadze G, Bhat J, Lettau M, Janssen O, Kabelitz D. Generation of soluble NKG2D ligands: Proteolytic cleavage, exosome secretion and functional implications. Scand J Immunol. 2013;78:120–129. doi: 10.1111/sji.12072. [DOI] [PubMed] [Google Scholar]
- 45.Flüh C, Chitadze G, Adamski V, Hattermann K, Synowitz M, Kabelitz D, Held-Feindt J. NKG2D ligands in glioma stem-like cells: Expression in situ and in vitro. Histochem Cell Biol. 2018;149:219–233. doi: 10.1007/s00418-018-1633-5. [DOI] [PubMed] [Google Scholar]
- 46.Werner-Klein M, Klein CA. Therapy resistance beyond cellular dormancy. Nat Cell Biol. 2019;21:117–119. doi: 10.1038/s41556-019-0276-7. [DOI] [PubMed] [Google Scholar]
- 47.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–498. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak L, Peters S, Arvold ND, Harsh GR, Steeg PS, Chang SD. Brain metastases. Nat Rev Dis Primers. 2019;5:5. doi: 10.1038/s41572-018-0055-y. [DOI] [PubMed] [Google Scholar]
- 50.Hall WA, Djalilian HR, Nussbaum ES, Cho KH. Long-term survival with metastatic cancer to the brain. Med Oncol. 2000;17:279–286. doi: 10.1007/BF02782192. [DOI] [PubMed] [Google Scholar]
- 51.Carlson P, Dasgupta A, Grzelak CA, Kim J, Barrett A, Coleman IM, Shor RE, Goddard ET, Dai J, Schweitzer EM, et al. Targeting the perivascular niche sensitizes disseminated tumour cells to chemotherapy. Nat Cell Biol. 2019;21:238–250. doi: 10.1038/s41556-018-0267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shersher DD, Vercillo MS, Fhied C, Basu S, Rouhi O, Mahon B, Coon JS, Warren WH, Faber LP, Hong E, et al. Biomarkers of the insulin-like growth factor pathway predict progression and outcome in lung cancer. Ann Thorac Surg. 2011;92:1805–1811. doi: 10.1016/j.athoracsur.2011.06.058. discussion 1811. [DOI] [PubMed] [Google Scholar]
- 53.Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): A critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sureshbabu A, Okajima H, Yamanaka D, Tonner E, Shastri S, Maycock J, Szymanowska M, Shand J, Takahashi S, Beattie J, et al. IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J Cell Sci. 2012;125:1693–1705. doi: 10.1242/jcs.092882. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Chu J, Feng W, Yang M, Zhang Y, Zhang Y, Qin Y, Xu J, Li J, Vasilatos SN, et al. EPHA5 mediates trastuzumab resistance in HER2-positive breast cancers through regulating cancer stem cell-like properties. FASEB J. 2019;33:4851–4865. doi: 10.1096/fj.201701561RRRR. [DOI] [PubMed] [Google Scholar]
- 56.Staquicini FI, Qian MD, Salameh A, Dobroff AS, Edwards JK, Cimino DF, Moeller BJ, Kelly P, Nunez MI, Tang X, et al. Receptor tyrosine kinase EphA5 is a functional molecular target in human lung cancer. J Biol Chem. 2015;290:7345–7359. doi: 10.1074/jbc.M114.630525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sperduto PW, Kased N, Roberge D, Chao ST, Shanley R, Luo X, Sneed PK, Suh J, Weil RJ, Jensen AW, et al. The effect of tumor subtype on the time from primary diagnosis to development of brain metastases and survival in patients with breast cancer. J Neurooncol. 2013;112:467–472. doi: 10.1007/s11060-013-1083-9. [DOI] [PubMed] [Google Scholar]
- 58.Perus LJM, Walsh LA. Microenvironmental heterogeneity in brain malignancies. Front Immunol. 2019;10:2294. doi: 10.3389/fimmu.2019.02294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 60.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, de Stanchina E, Massagué J. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;165:45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chitadze G, Lettau M, Bhat J, Wesch D, Steinle A, Fürst D, Mytilineos J, Kalthoff H, Janssen O, Oberg HH, Kabelitz D. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: Heterogeneous involvement of the 'a disintegrin and metalloproteases' 10-17. Int J Cancer. 2013;133:1557–1566. doi: 10.1002/ijc.28174. [DOI] [PubMed] [Google Scholar]
- 63.Malemud CJ. Inhibition of MMPs and ADAM/ADAMTS. Biochem Pharmacol. 2019;165:33–40. doi: 10.1016/j.bcp.2019.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pende D, Rivera P, Marcenaro S, Chang CC, Biassoni R, Conte R, Kubin M, Cosman D, Ferrone S, Moretta L, Moretta A. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: Analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–6186. [PubMed] [Google Scholar]
- 65.Vetter CS, Groh V, thor Straten P, Spies T, Bröcker EB, Becker JC. Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol. 2002;118:600–605. doi: 10.1046/j.1523-1747.2002.01700.x. [DOI] [PubMed] [Google Scholar]
- 66.Salih HR, Antropius H, Gieseke F, Lutz SZ, Kanz L, Rammensee HG, Steinle A. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102:1389–1396. doi: 10.1182/blood-2003-01-0019. [DOI] [PubMed] [Google Scholar]
- 67.Watson NF, Spendlove I, Madjd Z, McGilvray R, Green AR, Ellis IO, Scholefield JH, Durrant LG. Expression of the stress-related MHC class I chain-related protein MICA is an indicator of good prognosis in colorectal cancer patients. Int J Cancer. 2006;118:1445–1452. doi: 10.1002/ijc.21510. [DOI] [PubMed] [Google Scholar]
- 68.Castriconi R, Dondero A, Negri F, Bellora F, Nozza P, Carnemolla B, Raso A, Moretta L, Moretta A, Bottino C. Both CD133+ and CD133- medulloblastoma cell lines express ligands for triggering NK receptors and are susceptible to NK-mediated cytotoxicity. Eur J Immunol. 2007;37:3190–3196. doi: 10.1002/eji.200737546. [DOI] [PubMed] [Google Scholar]
- 69.Wolpert F, Tritschler I, Steinle A, Weller M, Eisele G. A disintegrin and metalloproteinases 10 and 17 modulate the immunogenicity of glioblastoma-initiating cells. Neuro Oncol. 2014;16:382–391. doi: 10.1093/neuonc/not232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu J, Chalupny NJ, Manley TJ, Riddell SR, Cosman D, Spies T. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glyco-protein. J Immunol. 2003;170:4196–4200. doi: 10.4049/jimmunol.170.8.4196. [DOI] [PubMed] [Google Scholar]
- 71.Spreu J, Stehle T, Steinle A. Human cytomegalovirus-encoded UL16 discriminates MIC molecules by their alpha2 domains. J Immunol. 2006;177:3143–3149. doi: 10.4049/jimmunol.177.5.3143. [DOI] [PubMed] [Google Scholar]
- 72.Zhang ZX, Wang S, Huang X, Min WP, Sun H, Liu W, Garcia B, Jevnikar AM. NK cells induce apoptosis in tubular epithelial cells and contribute to renal ischemia-reperfusion injury. J Immunol. 2008;181:7489–7498. doi: 10.4049/jimmunol.181.11.7489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the present study are available from the corresponding author upon reasonable request.