Abstract
Development of resistance to endocrine therapy, such as tamoxifen, remains a tricky clinical problem during the treatment of breast cancer. Accumulating evidence suggested that dysregulation of long noncoding (lnc_RNAs contributes to the development of tamoxifen resistance. In the current study, via screening, cytoskeleton regulator RNA (CYTOR) was identified as the most significantly elevated lncRNA in the established tamoxifen resistant MCF7 cell lines (MCF7/TAM1 and MCF7/TAM2) compared with the parental MCF7 cells (MCF7-P). The CCK-8 assay indicated that silencing of CYTOR increased the sensitivity of MCF7/TAM1 and MCF7/TAM2 to tamoxifen treatment. Using bioinformatic analysis, it was predicted that microRNA (miR)-125a-5p might bind to CYTOR and the expression of miR-125a-5p was negatively correlated with CYTOR in the tumor tissues of breast cancer. In addition, RT-qPCR and dual luciferase assays validated that CYTOR directly repressed miR-125a-5p expression in breast cancer cells. Through regulation of miR-125a-5p, CYTOR elevated serum response factor (SRF) expression and activated Hippo and mitogen associated protein kinase signaling pathways to promote breast cancer cell survival upon tamoxifen treatment. In the collected tumor tissues of breast cancer in the present study, high expression of CYTOR was detected in tissues from patients with no response to tamoxifen compared with those from patients who were not treated with tamoxifen. A positive correlation between CYTOR and SRF mRNA expression was observed in tissues collected from patients with breast cancer. In conclusion, the results of the present study demonstrated a pivotal role of CYTOR in mediating tamoxifen resistance in breast cancer.
Keywords: cytoskeleton regulator RNA, tamoxifen resistance, miR-125a-5p, serum response factor, TAZ
Introduction
Breast cancer is the most commonly diagnosed cancer type and the leading cause of cancer-related mortality in women worldwide (1). Overexpression of estrogen receptor α (ERα), is a feature of ERα+ breast cancer, a major subtype of breast cancer (2). ER is a hormonal transcription factor that is activated by binding to estrogen, thus promoting cell cycle progression (3). For patients with ERα+ breast cancer, endocrine therapy is a conventional therapeutic approach (4). Tamoxifen, a frequently used agent for hormone treatment, inhibits ER transcription program by competing with estrogen for binding to ER protein (5). Although tamoxifen greatly improves the prognosis of patients with breast cancer, de novo and acquired tamoxifen resistance are frequently observed during clinical treatment, which lead to the recurrence or metastasis of breast cancer, and eventually resulting in patient deaths (6). Aberrant ER transcriptional activity and activation of pro-survival signaling pathways are proposed to mediate tamoxifen resistance (7). Several key drivers are identified via experimental studies (8,9). Understanding the mechanisms underlying tamoxifen resistance is still urgent to fulfil clinical needs.
Long non-coding RNAs (lncRNAs) are non-coding transcripts that are usually more than 200 nucleotides in length (10). Accumulating evidences suggest that lncRNAs are pivotal for regulating gene expression via directly binding to mRNA, non-coding RNA and protein (11). With RNA sequencing, numerous differentially expressed lncRNAs have been discovered between tumor tissues and normal tissues (12). Several lncRNAs have proved to be key regulators during cancer initiation and development (13,14). Overexpression of lncRNA- cytoskeleton regulator RNA (CYTOR) has been detected in several cancer types (15,16), which has been experimentally identified as a driver of cell proliferation, migration and invasion (17).
In the current study, overexpression of CYTOR was identified to contribute to the development of tamoxifen resistance in breast cancer cells. In the established tamoxifen resistant sublines (MCF7/TAM1 and MCF7/TAM2), CYTOR was significantly increased and silencing of CYTOR re-sensitized tamoxifen resistant breast cancer cells to tamoxifen. It was further demonstrated that CYTOR functioned as a competitive endogenous (ce)RNA to sponge microRNA (miR)-125a-5p, leading to the upregulation of serum response factor (SRF) and activation of Hippo and mitogen associated protein kinase (MAPK)/extracellular signal activated kinase (ERK) signaling. Moreover, high expression of CYTOR was detected in tissues from patients who exhibited no response to tamoxifen compared with those from patients who were not treated with tamoxifen. The data demonstrated a pivotal role of CYTOR in mediating tamoxifen resistance in breast cancer.
Materials and methods
Collection of tumor and normal tissues
A total of 40 pairs of tumor and normal tissues were collected from patients (28 cases were not treated with tamoxifen and 12 cases were resistant to tamoxifen treatment, aged from 26-72 years old) with ER+ breast cancer who underwent surgery at Cancer Hospital of China Medical University during September 2015 to April 2018. Written consents were provided by all the participants before starting the study. Patients who received chemotherapy treatment prior to surgery were excluded. All experiments were performed under the supervision of the Ethic Committee of Cancer Hospital of China Medical University. The tissues were immediately snap-frozen in liquid nitrogen before subjection to the following experiments.
Cell culture and establishment of tamoxifen resistant breast cancer cell lines
The human breast cancer cell lines MCF7 was purchased from American Type Culture Collection. Cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (HyClone; Thermo Fisher Scientific, Inc.) in a humidified incubator with 5% CO2. MCF7 cells were exposed to tamoxifen (1 µM; Selleck Chemicals) for over 12 months and two colonies were picked and cultured as MCF7/TAM1 and MCF7/TAM2 cells. MCF7/TAM1 and MCF7/TAM2 cells were then maintained in RMPI 1640 supplemented with 1 µM tamoxifen. E2 (1 mM in ethanol) was purchased from Selleck Chemicals. E2 was diluted to 10 nM in Phenol Red-free RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) to investigate the dependency of cells on the activity of ER.
RNA extraction and reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from MCF7 cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following manufacturer's protocol. After that, RNA was reverse transcribed to first-stranded cDNA with PrimeScript™ First Strand cDNA Synthesis kit (Takara Bio, Inc.) at 30°C for 10 min. The RT-qPCR was conducted with SYBR Premix Ex Taq (Takara Bio, Inc.) on a CFX96 System (Bio-Rad Laboratories, Inc.). The thermocycling condition was: Predenaturation at 95°C for 30 sec; denature at 95°C for 15 sec, annealing and elongation at 55°C for 30 sec, repeated for 35 cycles. U6 and GAPDH were used as internal controls for miRNA and mRNA/lncRNA, respectively. The relative expression of genes was calculated using 2−ΔΔCq method (18). The primer sequences were listed in Table I.
Table I.
Sequences of primers.
| Primer | Sequence |
|---|---|
| PgR-Forward | 5′-ACCCGCCCTATCTCAACTACC-3′ |
| PgR-Reverse | 5′-AGGACACCATAATGACAGCCT-3′ |
| pS2-Forward | 5′-CCCCGTGAAAGACAGAATTGT-3′ |
| pS2-Reverse | 5′-GGTGTCGTCGAAACAGCAG-3′ |
| ATB-Forward | 5′-TCTGGCTGAGGCTGGTTGAC-3′ |
| ATB-Reverse | 5′-ATCTCTGGGTGCTGGTGAAGG-3′ |
| BCAR4-Forward | 5′-ATACAATGGCGTAATCATAGC-3′ |
| BCAR4-Reverse | 5′-AGACATTCAGAGCAAGACA-3′ |
| CCAT2-Forward | 5′-CCAGGCAATAACTGTGCAACTC-3′ |
| CCAT2-Reverse | 5′-ACTTACGTAGGGCATGCCAAA-3′ |
| CYTOR-Forward | 5′-ACCGAAAATCACGACTCAGCCC-3′ |
| CYTOR-Reverse | 5′-AATGGGAAACCGACCAGACCAG-3′ |
| GAS5-Forward | 5′-CTTCTGGGCTCAAGTGATCCT-3′ |
| GAS5-Reverse | 5′-TTGTGCCATGAGACTCCATCAG-3′ |
| H19-Forward | 5′-TGCTGCACTTTACAACCACTG-3′ |
| H19-Reverse | 5′-ATGGTGTCTTTGATGTTGGGC-3′ |
| HOTAIR-Forward | 5′-CAGTGGGGAACTCTGACTCG-3′ |
| HOTAIR-Reverse | 5′-GTGCCTGGTGCTCTCTTACC-3′ |
| MALAT1-Forward | 5′-GACGGAGGTTGAGATGAAGC-3′ |
| MALAT1-Reverse | 5′-ATTCGGGGCTCTGTAGTCCT-3′ |
| NEAT1-Forward | 5′-TTCTTAGCCTGATGAAATAACTTGG-3′ |
| NEAT1-Reverse | 5′-AGCGTTTAGCACAACACAATGAC-3′ |
| PVT1-Forward | 5′-AGCACTCTGGACGGACTTGAGA-3′ |
| PVT1-Reverse | 5′-CCACTAGCAGCAACAGGAGAAG-3′ |
| TUG1-Forward | 5′-AGGTAGAACCTCTATGCATTTTGTG-3′ |
| TUG1-Reverse | 5′-ACTCTTGCTTCACTACTTCATCCAG-3′ |
| UCA1-Forward | 5′-CTCTCCATTGGGTTCACCATTC-3′ |
| UCA1-Reverse | 5′-GCGGCAGGTCTTAAGAGATGAG-3′ |
| SRF-Forward | 5′-CGAGATGGAGATCGGTATGGT-3′ |
| SRF-Reverse | 5′-GGGTCTTCTTACCCGGCTTG-3′ |
| GAPDH-Forward | 5′-GGAGCGAGATCCCTCCAAAAT-3′ |
| GAPDH-Reverse | 5′-GGCTGTTGTCATACTTCTCATGG-3′ |
| Stem-loop | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCACAG-3′ |
| miR-125a-5p-Forward | 5′-TCGGCAGGTCCCTGAGACCCTT-3′ |
| miR-125a-5p-Reverse | 5′-CTCAACTGGTGTCGTGGA-3′ |
| U6-Forward | 5′-CGCTTCGGCAGCACATATAC-3′ |
| U6-Reverse | 5′-CAGGGGCCATGCTAATCTT-3′ |
Downregulation of CYTOR in breast cancer cells
CYTOR siRNA and control siRNA were synthesized and bought from Guangzhou RiboBio, Co., Ltd. For silencing of CYTOR, cells were transfected with CYTOR siRNA at a concentration of 20 nM with Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The cells were maintained for 48 h before subjection to the following experiments. The sequences were: Control siRNA: 5′-UUC UCC GAA CGU GUC ACG UTT-3′; CYTOR siRNA1: 5′-CAG UCU CUA UGU GUC UUA ATT-3′; CYTOR siRNA2: 5′-CAC ACU UGA UCG AAU AUG ATT-3′.
Western blotting
TAZ (cat. no. 83669; 1:1,000) ERK1/2 (cat. no. 4695; 1:2,000) and phosphorylated (p)-ERK1/2 (cat. no. 4370; 1:2,000) antibodies were products of Cell Signaling Technology, Inc. SRF (cat. no. ab53147; 1:2,000) and GAPDH (cat. no. ab8245; 1:5,000) primary antibodies were bought from Abcam. Horseradish peroxidase-conjugated secondary antibodies against rabbit (cat. no. SA00001-2; 1:10,000) and mouse (cat. no. SA00001-1; 1:10,000) were products of Proteintech Group, Inc. RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) was used to prepare protein lysis buffer. The protein concentration was determined with the BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.; Sigma-Aldrich; Merck KGaA). The western blotting was performed in a standard procedure. A total of 20 µg protein was loaded on the 8% SDS-PAGE gel and separated with electrophoresis. The PVDF membrane was incubated with primary antibody and secondary antibody for 1 h at room temperature sequentially. The blots were developed with ECL Western Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.; Sigma-Aldrich; Merck KGaA). Densitometry analysis was performed using ImageJ software (version 1.51J8, National Institute of Science).
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) was used to analyze cell proliferation ability. Briefly, 5,000 cells were plated in each well of 96-well plates. At 24 h after transfection, 10 µl CCK-8 solution was added into the well and sustained for another 2 h. After that, the medium was transferred to another 96-well plate and the absorbance at 450 nM was detected by a microplate reader to reflect cell proliferation ability.
Overexpression and downregulation of miR-125a-5p in breast cancer cells
miR-125a-5p mimic (5′-UCC CUG AGA CCC UUU AAC CUG UGA-3′), miR-NC mimic (5′-AUU GGA ACG AUA CAG AGA AGA UU-3′), miR-125a-5p inhibitor (5′-UCA CAG GUU AAA GGG UCU CAG GGA-3′) and miR-NC inhibitor (5′-CAG UAC UUU UGU GUA GUA CAA-3′) were synthesized and purchased from Guangzhou RiboBio Co., Ltd. 50 nM miR-125a-5p mimic, miR-NC mimic, miR-125a-5p inhibitor and miR-NC inhibitor were transfected into cells using Lipofectamine RNAiMax following manufacturers' protocol and the cells were harvested for the following experiments after 48 h.
Dual luciferase reporter assay
The full length of SRF 3′untranslated region (UTR) and fragment from CYTOR that contained the predicted miR-125a-5p binding site were amplified from MCF7 cDNA and ligated into pGL3 plasmid (Promega Corporation) respectively. Two site mutations were introduced into pGL3-SRF 3′UTR-wild-type (WT) and pGL3-CYTOR-WT plasmid to establish pGL3-SRF 3′UTR-mutant (Mut) and pGL3-CYTOR-Mut using Quick Site-Mutation kit (Agilent Technologies, Inc.). miR-125a-5p mimic (Guangzhou RiboBio Co., Ltd.) or miR-NC (Guangzhou RiboBio Co., Ltd.) in combination with pGL3-SRF 3′UTR-WT or pGL3-SRF 3′UTR-Mut or pGL3-CYTOR-WT or pGL3-CYTOR-Mut were transfected into cells and maintained for 48 h. The firefly luciferase and Renilla luciferase activity was detected with the Dual Luciferase Reporter Assay System (Promega Corporation) following manufacturer's protocol, and the relative luciferase activity was calculated via normalizing firefly luciferase activity to Renilla luciferase activity.
SRF overexpression in breast cancer cells
The SRF open reading frame was amplified from MCF7 cDNA and inserted into pcDNA3.1 plasmid (YouBio). For overexpression of SRF, 2 µg pcDNA3.1-SRF was transfected to 1×106 cells with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) following manufacture's protocol. A total of 48 h later, the cells were harvested and subjected to western blotting to detect overexpression of SRF.
Bioinformatic analysis
The list of oncogenic and tumor suppressive lncRNA for breast cancer was obtained from LncRNADisease database (http://www.cuilab.cn/lncrnadisease). StarBase v3.0 (http://starbase.sysu.edu.cn/) was used to predict potential binding miRNAs for lncRNA. The potential target gene of miRNA was predicted by TargetScan software, version 7.2 (http://www.targetscan.org/vert_72/).
Statistical analysis
All experiments were repeated at least three times. Data were analyzed by GraphPad Prism 6.0 (Graphpad Software, Inc.) and presented as the mean ± standard deviation. Difference between two groups was compared with a Student's t test. Three groups were analyzed with one-way analysis of variance, followed by Newman-Keuls analysis. Pearson correlation analysis was applied for the analysis of the correlation between SRF and CYTOR. P<0.05 was considered to indicate a statistically significant difference.
Results
Tamoxifen resistant breast cancer cells exhibit overactivation of the MAPK/ERK pathway and elevation of TAZ
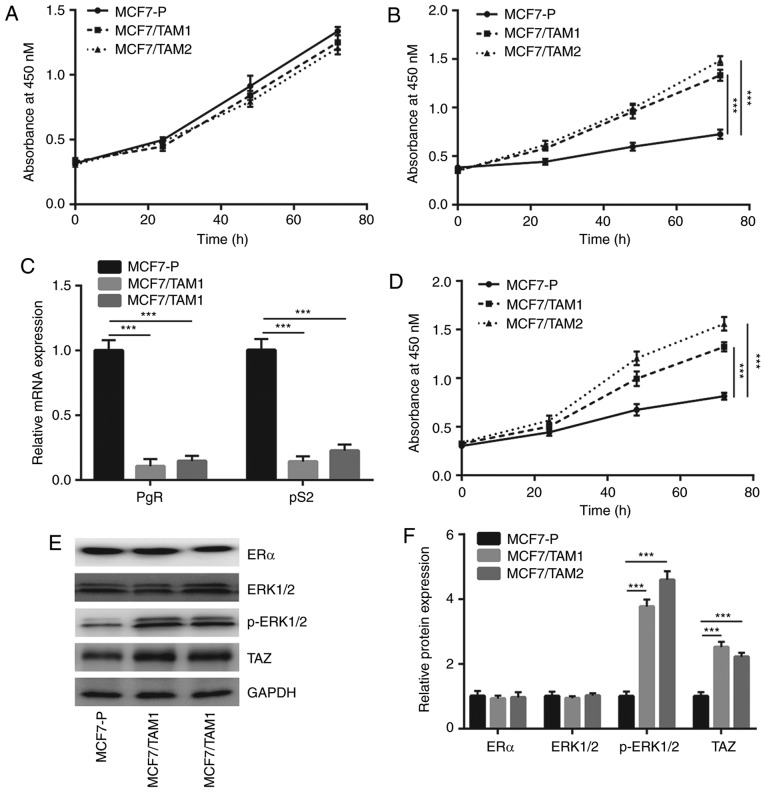
To study the mechanism of tamoxifen resistance, tamoxifen resistant MCF7 sublines (MCF7/TAM1 and MCF7/TAM2) were constructed by continuously exposed MCF7 cells to tamoxifen. In complete medium, the proliferation of MCF7/TAM1 and MCF7/TAM2 cells was similar to MCF7-P cells (Fig. 1A). However, in phenol red-free medium (without E2), the growth rate of MCF7-P was significantly decreased compared with MCF7/TAM1 and MCF7/TAM2 (P<0.001; Fig. 1B), suggesting the proliferation of tamoxifen resistant sublines no longer relied on activity of ERα. RT-qPCR results showed that the mRNA expression of PgR and pS2, two well-documented target genes of ERα, were significantly decreased in MCF7/TAM1 and MCF7/TAM2 (P<0.001; Fig. 1C). Additionally, in the presence of tamoxifen, the growth of MCF7-P cells was significantly inhibited compared to tamoxifen resistant sublines (P<0.001; Fig. 1D). the expression of ERα and effectors of several key pathways (MAPK/ERK and Hippo pathways) were next detected in MCF7-P and tamoxifen resistant sublines. Western blotting showed that ERα and ERK1/2 expression were not changed whereas p-ERK1/2 and TAZ levels were significantly elevated in both MCF7/TAM1 and MCF7/TAM2 cells (P<0.001; Fig. 1E and F). The data suggested that altered activity of MAPK/ERK and Hippo pathways might contribute to development of tamoxifen resistance.
Figure 1.
Characterization of tamoxifen resistant MCF7 cells. (A) The cell proliferation assay indicated that the growth rate of MCF7-P was similar to that of MCF7/TAM1 and MCF7/TAM2. (B) Different from MCF7-P, the growth of MCF7/TAM1 and MCF7/TAM2 did not rely on the addition of E2. (C) The mRNA levels of ER target genes (PgR and pS2) were decreased in MCF7/TAM1 and MCF7/TAM2 cells compared with parental cells. (D) MCF7/TAM1 and MCF7/TAM2 were relatively insensitive towards tamoxifen treatment compared with MCF7-P cells. (E) Western blotting showed that the protein expression of TAZ and p-ERK1/2 was increased in MCF7/TAM1 and MCF7/TAM2 compared with MCF7-P, whereas the expression of ER and ERK1/2 was not altered. (F) Quantification analysis of TAZ, p-ERK1/2, ER and ERK1/2 expression in E was presented. ***P<0.001. MCF7-P, parental MCF7 cells; p-ERK1/2, phosphorylated-extracellular signal regulated kinase 1/2; MCF7/TAM1, tamoxifen-resistant MCF7 subline1; MCF7/TAM2, tamoxifen-resistant MCF7 subline2; ER, estrogen receptor.
Overexpression of CYTOR is essential for tamoxifen resistance of breast cancer cells
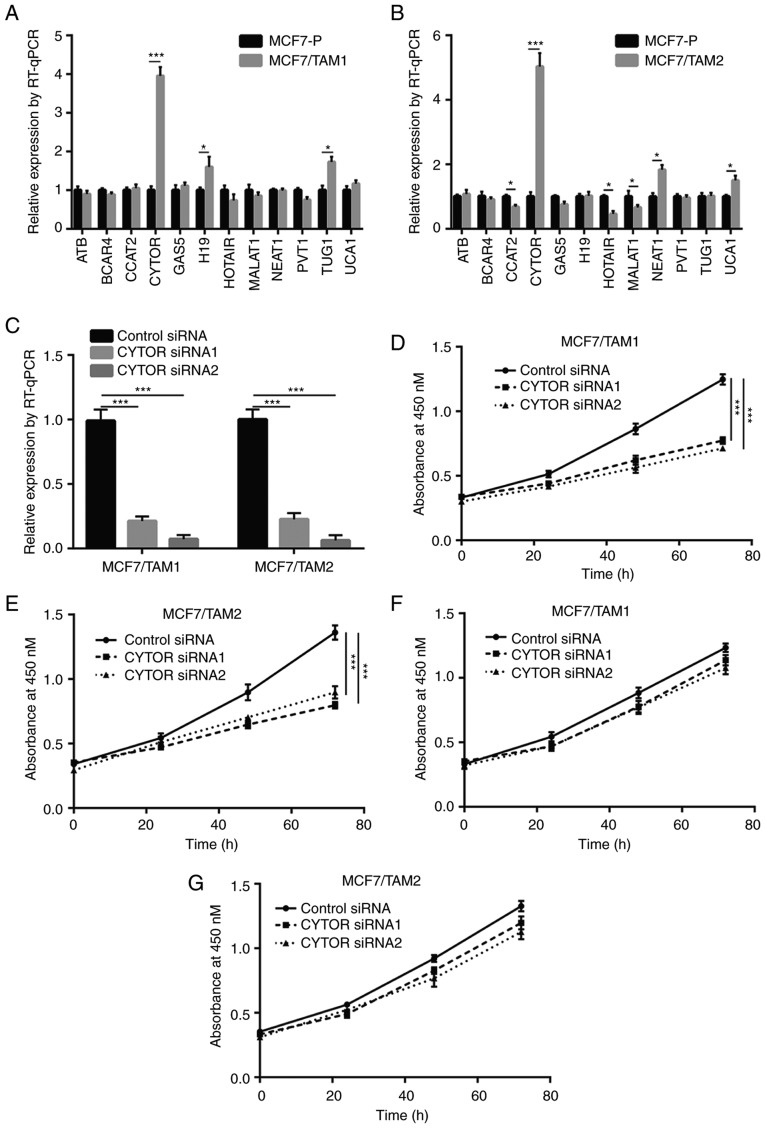
To explore whether lncRNA was involved in tamoxifen resistance, RT-qPCR was used to detect expression of several lncRNAs which have been identified as oncogenes or tumor suppressors in breast cancer that were listed in LncRNADisease database. Among the 12 lncRNAs, CYTOR expression was the most significantly elevated lncRNA with a 4-fold increase in MCF7/TAM1 and MCF7/TAM2 compared with MCF7-P (P<0.001; Fig. 2A and B). Transfection of CYTOR siRNA1 or siRNA2 significantly decreased CYTOR expression in MCF7/TAM1 and MCF7/TAM2 cells (P<0.001; Fig. 2C). Downregulation of CYTOR inhibited MCF7/TAM1 cell proliferation in the presence of tamoxifen (Fig. 2D). Similarly, CYTOR silencing also sensitized MCF7/TAM2 to tamoxifen (Fig. 2E). More importantly, silencing of CYTOR did not inhibit cell proliferation of MCF7/TAM1 and MCF7/TAM2 without tamoxifen treatment (Fig. 2F and G), indicating that CYTOR regulated tamoxifen sensitivity instead of cell proliferation ability in breast cancer cells.
Figure 2.
lncRNA CYTOR promoted tamoxifen resistance in breast cancer cells. (A) Screen of breast cancer-associated lncRNA (from LncRNADisease database) showed that several lncRNAs were differentially expressed between MCF7-P and MCF7/TAM1. (B) Screen of breast cancer-associated lncRNA (from LncRNADisease database) showed that several lncRNAs were differentially expressed between MCF7-P and MCF7/TAM2. (C) CYTOR siRNA decreased CYTOR expression in MCF7/TAM1 cells and MCF7/TAM2 cells. (D) Silencing of CYTOR re-sensitized MCF7/TAM1 cells to tamoxifen treatment. (E) Silencing of CYTOR re-sensitized MCF7/TAM2 cells to tamoxifen treatment. (F) Silencing of CYTOR showed no effect on the proliferation of MCF7/TAM1 without treatment of tamoxifen. (G) Silencing of CYTOR showed no effect on the proliferation of MCF7/TAM2 without treatment of tamoxifen. *P<0.05, ***P<0.001. CYTOR, cytoskeleton regulator RNA; lnc, long noncoding; si, small interfering; RT-q, reverse transcription; MCF7/TAM1, tamoxifen-resistant MCF7 subline1; MCF7/TAM2, tamoxifen-resistant MCF7 subline2.
CYTOR sponges miR-125a-5p in breast cancer cells
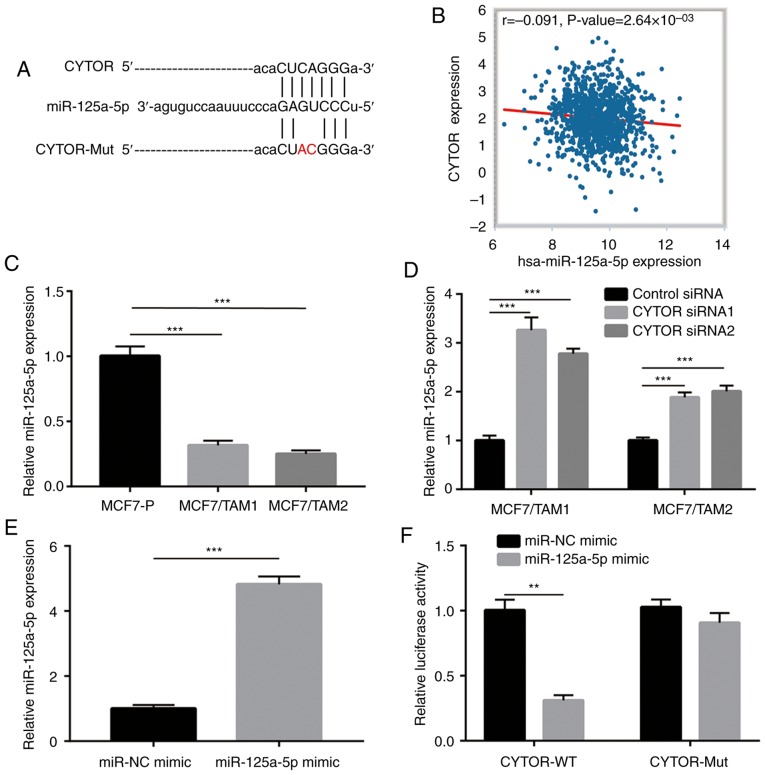
Using StarBase software, several miRNAs were predicted to harbor binding sites for CYTOR, such as miR-125a-5p (Fig. 3A). It was observed that the expression of miR-125a-5p was negatively correlated with CYTOR levels in dataset which contained the expression data of 1085 invasive breast cancer tissues (Fig. 3B). In addition, miR-125a-5p expression was significantly decreased in both MCF7/TAM1 and MCF7/TAM2 cells compared with MCF7-P cells (P<0.001; Fig. 3C). Furthermore, silencing of CYTOR increased miR-125a-5p expression in MCF7/TAM1 and MCF7/TAM2 cells (Fig. 3D). More importantly, in the dual luciferase reporter assay, overexpression of miR-125a-5p decreased relative luciferase activity in MCF7/TAM1 cells transfected with CYTOR-WT (Fig. 3E and F), suggesting that CYTOR directly interacted with miR-125a-5p in breast cancer cells.
Figure 3.
CYTOR directly regulates miR-125a-5p in breast cancer cells. (A) Bioinformatic analysis showed that miR-125a-5p harbored a putative binding site for CYTOR. (B) Bioinformatic analysis of The Cancer Genome Atlas data suggested that expression of CYTOR was negatively correlated with miR-125-5p in breast cancer. (C) miR-125a-5p expression was decreased in MCF7/TAM1 and MCF7/TAM2 compared with parental cells. (D) Silencing of CYTOR increased miR-125a-5p expression in MCF7/TAM1 and MCF7/TAM2 cells. (E) Transfection of miR-125a-5p mimic increased miR-125a-5p levels in MCF7/TAM1 cells. (F) Dual luciferase reporter assay indicated that miR-125a-5p could directly bind to CYTOR in MCF7/TAM1 cells. **P<0.01 and ***P<0.001. CYTOR, cytoskeleton regulator RNA; miR, microRNA; Mut, mutant; NC, negative control.
SRF is a target gene of miR-125a-5p in breast cancer cells
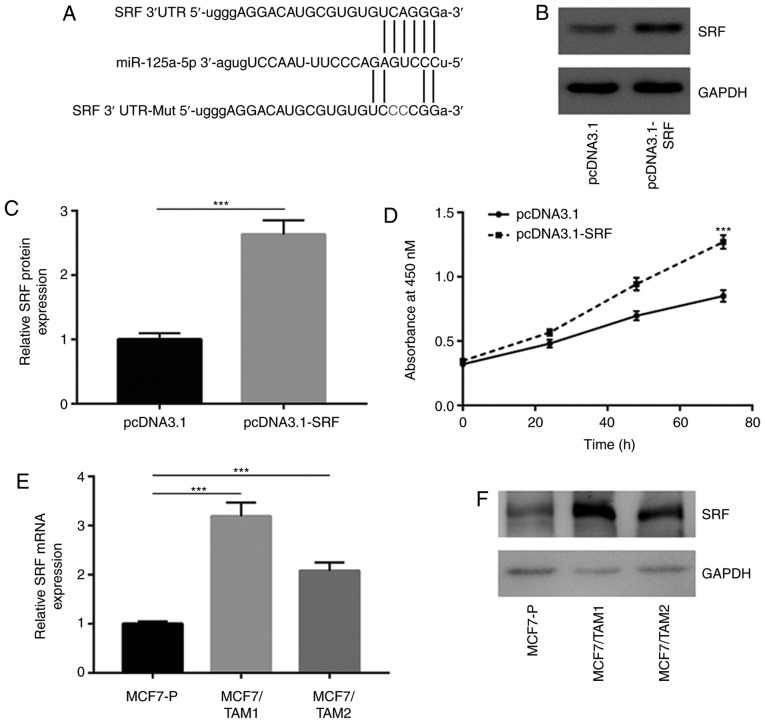
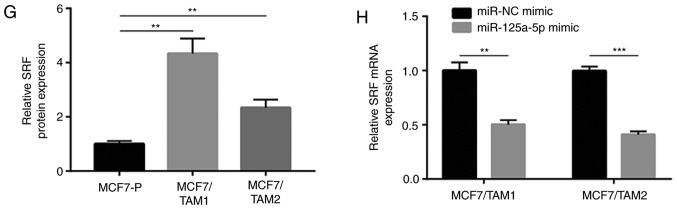
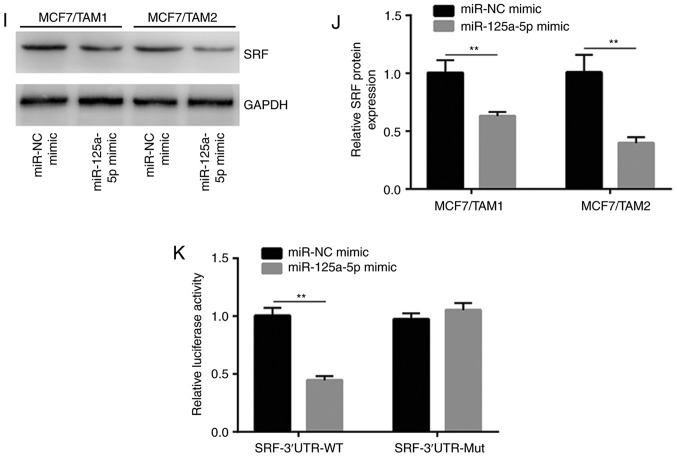
The TargetScan software was used to predict potential target genes of miR-125a-5p. Among them, it was noticed that the sequence of SRF 3′UTR, a regulator of Hippo and MAPK/ERK pathways, could complementarily bind to miR-125a-5p (Fig. 4A). Recombinant SRF was transfected to overexpress SRF in MCF7 cells (Fig. 4B and C). Interestingly, it was found that SRF overexpression decreased tamoxifen sensitivity in MCF7 cells (Fig. 4D). RT-qPCR showed that SRF mRNA levels were increased in MCF7/TAM1 and MCF7/TAM2 (Fig. 4E). Western blotting results indicated that SRF protein levels were increased in MCF7/TAM1 and MCF7/TAM2 (Fig. 4F and G). Overexpression of miR-125a-5p decreased SRF mRNA expression in MCF7/TAM1 and MCF7/TAM2 (Fig. 4H). Meanwhile, upregulation of miR-125a-5p also reduced SRF protein levels in the two tamoxifen resistant sublines (Fig. 4I and J). The dual luciferase reporter assay showed that transfection of miR-125a-5p mimic repressed relative luciferase activity in MCF7/TAM1 cells transfected with SRF 3′UTR-WT (Fig. 4K).
Figure 4.
SRF is upregulated and acts as a target gene of miR-125a-5p in tamoxifen resistant MCF7 cells. (A) There was a putative binding site for miR-125a-5p on the 3′UTR of SRF. (B) Transfection of recombinant SRF increased SRF protein expression in MCF7 cells. (C) Quantification analysis of SRF protein expression in B was presented. (D) Overexpression of SRF decreased sensitivity of MCF7 cells towards tamoxifen treatment. (E) RT-qPCR showed that SRF was overexpressed in MCF7/TAM1 and MCF7/TAM2 compared with parental cells. (F) Western blotting showed that SRF protein levels were increased in MCF7/TAM1 and MCF7/TAM2. (G) Quantification analysis of SRF protein expression in F was presented. (H) RT-qPCR suggested that SRF mRNA levels were decreased towards miR-125a-5p overexpression in MCF7/TAM1 and MCF7/TAM2 cells. **P<0.01 and ***P<0.001. SRF is upregulated and acts as a target gene of miR-125a-5p in tamoxifen resistant MCF7 cells. (I) Western blotting showed that SRF protein levels were decreased towards miR-125a-5p overexpression in MCF7/TAM1 and MCF7/TAM2 cells. (J) Quantification analysis of SRF protein expression in I was presented. (K) Dual luciferase reporter assay showed that SRF was a target gene of miR-125a-5p in MCF7/TAM1 cells. **P<0.01. SRF, serum response factor; RT-q, reverse transcription-quantitative; UTR, untranslated region; MCF7/TAM1, tamoxifen-resistant MCF7 subline1; MCF7/TAM2, tamoxifen-resistant MCF7 subline2; NC, negative control; miR, microRNA; WTT, wild-type; Mut, mutant.
CYTOR regulates SRF via sponging miR-125a-5p
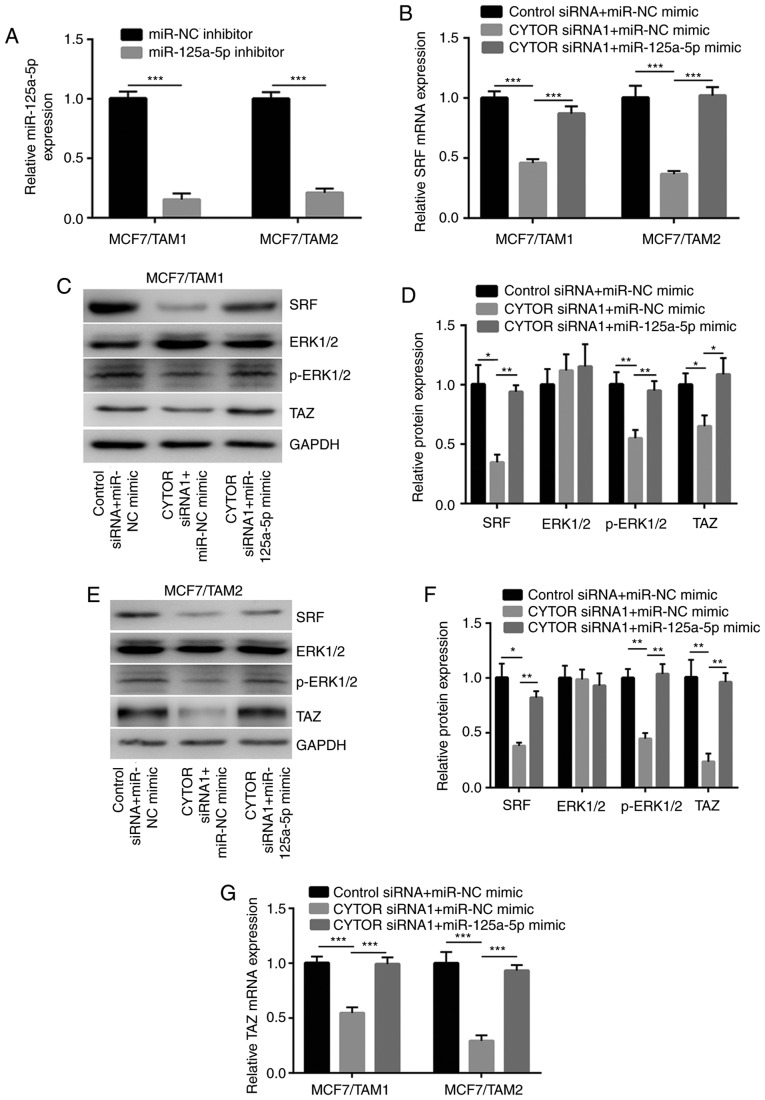
To investigate whether miR-125-5p was pivotal for regulation of SRF by CYTOR, miR-125a-5p was downregulated by transfection of an miR-125a-5p inhibitor (Fig. 5A). RT-qPCR showed that CYTOR silencing decreased SRF expression which was reversed after miR-125a-5p inhibition in MCF7/TAM1 and MCF7/TAM2 cells (Fig. 5B). Moreover, silencing of CYTOR1 decreased TAZ and p-ERK1/2 protein expression which was reversed towards transfection of miR-125a-5p inhibitor in MCF7/TAM1 cells (Fig. 5C and D). Similar results were also observed in MCF7/TAM2 cells (Fig. 5E and F). In addition, RT-qPCR suggested that TAZ mRNA expression was decreased upon CYTOR silencing which was reversed by miR-125a-5p overexpression (Fig. 5G).
Figure 5.
CYTOR regulates SRF expression in a miR-125a-5p dependent manner. (A) Transfection of miR-125a-5p inhibitor decreased miR-125a-5p levels in MCF7/TAM1 and MCF7/TAM2 cells. (B) Silencing of CYTOR decreased SRF mRNA levels in MCF7/TAM1 and MCF7/TAM2 cells, which was reversed upon miR-125a-5p inhibition. (C) Silencing of CYTOR decreased SRF, TAZ and p-ERK1/2 protein levels in MCF7/TAM1 cells, which was reversed upon miR-125a-5p inhibition. (D) Quantification of SRF, p-ERK1/2, ERK1/2 and TAZ expression in C was exhibited. (E) Silencing of CYTOR decreased SRF and p-ERK1/2 protein levels in MCF7/TAM2 cells, which was reversed upon miR-125a-5p inhibition. (F) Quantification of SRF, p-ERK1/2, ERK1/2 and TAZ expression in E was exhibited. (G) Silencing of CYTOR decreased TAZ mRNA levels in MCF7/TAM1 and MCF7/TAM2 cells, which was reversed upon miR-125a-5p inhibition. *P<0.05; **P<0.01 and ***P<0.001. SRF, serum response factor; CYTOR, cytoskeleton regulator RNA; miR, microRNA; p-ERK1/2, phosphorylated-extracellular signal regulated kinase; NC, negative control.
CYTOR regulates tamoxifen sensitivity via SRF in breast cancer cells
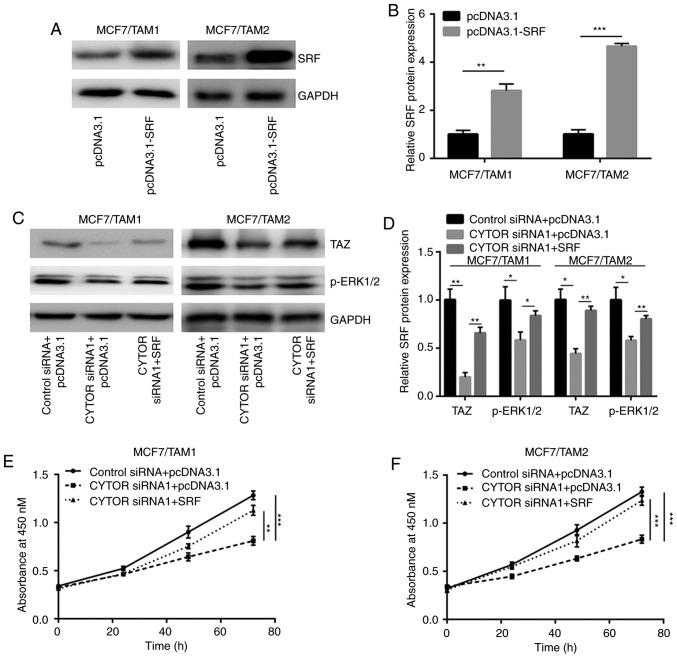
A recombinant SRF plasmid was constructed to study the role of CYTOR and SRF during the development of tamoxifen resistance. Transfection of recombinant SRF significantly elevated SRF protein expression in MCF7/TAM1 and MCF7/TAM2 cells (P<0.01; Fig. 6A and B). Silencing of CYTOR decreased TAZ and p-ERK1/2 which was reversed after SRF overexpression in MCF7/TAM1 and MCF7/TAM2 cells (Fig. 6C and D). In the cell proliferation assay, downregulation of CYTOR sensitized MCF7/TAM1 and MCF7/TAM2 cells to tamoxifen which was attenuated by SRF overexpression (Fig. 6E and F).
Figure 6.
CYTOR regulates tamoxifen sensitivity via targeting SRF. (A) Transfection of recombinant SRF increased SRF protein expression in MCF7/TAM1 and MCF7/TAM2 cells. (B) Quantification analysis of SRF protein expression in A was exhibited. (C) In MCF7/TAM1 and MCF7/TAM2 cells, silencing of CYTOR decreased TAZ and p-ERK1/2 expression, which was reversed upon transfection of recombinant SRF. (D) Quantification of protein expression in C was exhibited. (E) Overexpression of SRF attenuated CYTOR siRNA induced elevation of tamoxifen sensitivity in MCF7/TAM1 cells. (F) Overexpression of SRF attenuated CYTOR siRNA induced elevation of tamoxifen sensitivity in MCF7/TAM2 cells. *P<0.05; **P<0.01 and ***P<0.001. SRF, serum response factor; CYTOR, cytoskeleton regulator RNA; si, small interfering; MCF7/TAM1, tamoxifen-resistant MCF7 subline1; MCF7/TAM2, tamoxifen-resistant MCF7 subline2; p-ERK 1/2, phosphorylated-extracellular protein kinase 1/2.
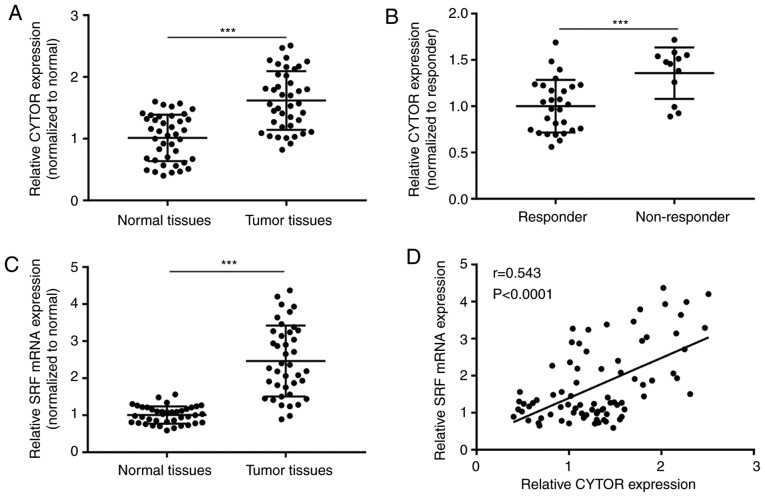
CYTOR expression is positively correlated with SRF expression in breast tumors
To validate whether the CYTOR/miR-125a-5p/SRF axis was relevant to clinical situations, 40 pairs of normal and tumor tissues were collected from patients with ER+ breast cancer. It was observed that CYTOR was significantly overexpressed in tumors compared with normal tissues (P<0.001; Fig. 7A). More importantly, CYTOR expression was significantly increased in tumors from tamoxifen non-responders compared with those from tamoxifen untreated patients (P<0.001; Fig. 7B). SRF mRNA levels were also significantly increased in tumors compared with normal tissues (P<0.001; Fig. 7C). The Pearson correlation analysis suggested that the CYTOR expression was positively correlated with SRF expression in tissues collected from patients (Fig. 7D).
Figure 7.
CYTOR expression is increased in tamoxifen non-responder of patients with breast cancer. (A) RT-qPCR indicated that CYTOR expression was increased in tumor tissues compared with normal tissues from patients with breast cancer. (B) The CYTOR expression was increased in tumor tissues from tamoxifen resistant patients compared with those from tamoxifen untreated patients. (C) RT-qPCR showed that SRF levels were increased in tumor tissues compared with normal tissues from patients with breast cancer. (D) The Pearson correlation analysis showed a significant positive correlation between CYTOR expression and SRF mRNA levels. ***P<0.001. SRF, serum response factor; RT-q, reverse transcription-quantitative; CYTOR, cytoskeleton regulator RNA.
Discussion
Endocrine therapy is the most commonly used approach for the treatment of ERα positive breast cancer, however, its efficacy is greatly limited by de novo and acquired tamoxifen resistance in patients with breast cancer (19). For the past two decades, several molecules have been identified as mediators of tamoxifen resistance or predictors of response to tamoxifen treatment (20,21). Most recently, lncRNA has attracted the attention of researchers and some lncRNAs were proved to be regulators of tamoxifen sensitivity in breast cancer (22). For example, Xu et al (23) found that lncRNA-UCA1 is overexpressed in tamoxifen resistant breast cancer cells compared to tamoxifen sensitive breast cancer cells and tamoxifen resistant breast cancer cells can deliver UCA1 to tamoxifen sensitive breast cancer via secretion of exosome. In the two established tamoxifen resistant sublines, the present study observed elevation of UCA1 in MCF7/TAM2 but not in MCF7/TAM1 cells, suggested that UCA1 might promote tamoxifen resistance in a subset of breast cancer. It was found that CYTOR was elevated in both two tamoxifen resistant sublines. Silencing of CYTOR sensitized MCF7/TAM1 and MCF7/TAM2 cells to tamoxifen treatment. The present study to the best of our knowledge for the first time showed that CYTOR drove tamoxifen resistance in breast cancer.
lncRNAs can function as ceRNA to bind specific miRNAs, thereby regulating miRNA-mediated gene silencing (24). Previously, CYTOR was discovered as a ceRNA for miR-155 and miR-195 to regulate gene expression (25,26). In breast cancer, CYTOR was initially identified as an oncogene via a genome-wide transcriptional survey to explore the lncRNA landscape across 995 breast tumors (27). CYTOR is involved in several key pathways such as epidermal growth factor receptor and MAPK/ERK pathways and its high expression predicts poor prognosis in patients with breast cancer (27). CYTOR is overexpressed in triple negative breast cancer (TNBC) and promotes cell proliferation via induction of PTEN ubiquitination and degradation (28). The overexpression of CYTOR is associated with transcriptional inhibition by YY1 in TNBC cells (28). In the present study, increased CYTOR expression accompanied with elevated activity of the MAPK/ERK pathway was observed in tamoxifen resistant sublines. Silencing of CYTOR decreased p-ERK1/2 expression in tamoxifen resistant sublines, indicating that CYTOR might contribute to tamoxifen resistance via activation of the MAPK/ERK pathway.
miR-125a-5p is a tumor suppressor in breast cancer. miR-125a-5p directly repressed BAP1, TSTA3, PTPN18 in breast cancer cells, leading to cell proliferation inhibition, induction of cell apoptosis and imatinib resistance (29-31). lncRNAs such as UCA1 and ANRIL have been known to sponge miR-125a-5p in cancer cells (32,33). In the present study, it was found that miR-125a-5p also shared sequence with CYTOR. Decreased expression of miR-125a-5p was observed in tamoxifen resistant sublines and silencing of CYTOR increased expression level of miR-125a-5p. The direct binding between CYTOR and miR-125a-5p was further validated in a dual luciferase reporter assay. The data indicated CYTOR as a new ceRNA for miR-125a-5p in breast cancer cells.
SRF is a transcription factor that regulates expression of hundreds of genes (34). The activity of SRF is controlled by serum concentration and cytoskeleton assembly (35), which is also governed by CYTOR. High expression of SRF contributes to stemness of breast cancer stem cells (36). In breast cancer cells, TAZ and YAP, two effectors of Hippo signaling, are both regulated by SRF. SRF recruits YAP to the promoters of genes to enhance YAP activity and directly activate TAZ transcription via binding to its promoter (36,37). As Hippo pathway is known to regulate tamoxifen resistance via elevation of cancer cell stemness, the overexpression of TAZ might contribute to tamoxifen resistance in the studied system in the current study. The present study revealed that the upregulation of TAZ was induced by aberrant expression of the CYTOR/miR-125a-5p/SRF axis. It was recently reported that DLG5 inhibited TAZ expression to sensitize breast cancer cells to tamoxifen (38). The results further extended the understandings on how TAZ expression was increased during development of tamoxifen resistance in breast cancer cells.
In conclusion, the current study revealed a CYTOR/miR-125a-5p/SRF axis in mediating tamoxifen resistance in breast cancer cells. Mechanistically, the CYTOR/miR-125a-5p/SRF axis activated MAPK/ERK and Hippo pathways during development of tamoxifen resistance. The data might provide valuable information for the development of alternative approaches to diagnose tamoxifen resistant breast cancer.
Acknowledgments
Not applicable.
Funding
The present study was supported by National key research and development program, China (grant. no. 2016YFC1303000).
Availability of data and materials
They are available at special request.
Authors' contributions
HP, YL, ML and HY participated in the design and performance of the experiments; ML and HY contributed to the collection of samples and clinical data; HP supervised and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in the present study involving human participants were approved by the Ethic Committee of Cancer Hospital of China Medical University. Written consents were provided by all the participants before starting the study.
Patient consent for publication
Written informed consent for the publication of any associated data and accompanying images was provided by all patients prior to surgery.
Competing interests
There was not any type of conflict of interests in current study.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Li J, Wang H, Wang Y, He Q, Xia X, Hu ZY, Ouyang Q. Clinical and genetic risk factors for Fulvestrant treatment in post-menopause ER-positive advanced breast cancer patients. J Transl Med. 2019;17:27. doi: 10.1186/s12967-018-1734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doisneau-Sixou SF, Sergio CM, Carroll JS, Hui R, Musgrove EA, Sutherland RL. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- 4.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer-An overview and update. Mol Cell Endocrinol. 2015;418(Pt 3):220–234. doi: 10.1016/j.mce.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/S0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 6.Roop RP, Ma CX. Endocrine resistance in breast cancer: Molecular pathways and rational development of targeted therapies. Future Oncol. 2012;8:273–292. doi: 10.2217/fon.12.8. [DOI] [PubMed] [Google Scholar]
- 7.Giuliano M, Schifp R, Osborne CK, Trivedi MV. Biological mechanisms and clinical implications of endocrine resistance in breast cancer. Breast. 2011;20(Suppl 3):S42–S49. doi: 10.1016/S0960-9776(11)70293-4. [DOI] [PubMed] [Google Scholar]
- 8.Piggott L, Silva A, Robinson T, Santiago-Gómez A, Simões BM, Becker M, Fichtner I, Andera L, Young P, Morris C, et al. Acquired resistance of ER-Positive breast cancer to endocrine treatment confers an adaptive sensitivity to TRAIL through posttranslational downregulation of c-FLIP. Clin Cancer Res. 2018;24:2452–2463. doi: 10.1158/1078-0432.CCR-17-1381. [DOI] [PubMed] [Google Scholar]
- 9.Anurag M, Punturi N, Hoog J, Bainbridge MN, Ellis MJ, Haricharan S. Comprehensive profiling of DNA repair defects in breast cancer identifies a novel class of endocrine therapy resistance drivers. Clin Cancer Res. 2018;24:4887–4899. doi: 10.1158/1078-0432.CCR-17-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 14.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 15.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Q, Wang ZQ, Wang SH, Li C, Zhu ZG, Quan ZW, Zhang WJ. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am J Transl Res. 2016;8:4068–4081. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Yu H, Sun W, Kong J, Zhang L, Tang J, Wang J, Xu E, Lai M, Zhang H. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol Cancer. 2018;17:110. doi: 10.1186/s12943-018-0860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Fan W, Chang J, Fu P. Endocrine therapy resistance in breast cancer: Current status, possible mechanisms and overcoming strategies. Future Med Chem. 2015;7:1511–1519. doi: 10.4155/fmc.15.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foxo3a re-expression overcomes tamoxifen resistance in breast cancer. J Biol Regul Homeost Agents. 2018;32(4 Suppl 1):S43. [PubMed] [Google Scholar]
- 21.Gao A, Sun T, Ma G, Cao J, Hu Q, Chen L, Wang Y, Wang Q, Sun J, Wu R, et al. LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERα pathway. Nat Commun. 2018;9:4180. doi: 10.1038/s41467-018-06309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi YF, Lu H, Wang HB. Downregulated lncRNA ADAMTS9-AS2 in breast cancer enhances tamoxifen resistance by activating microRNA-130a-5p. Eur Rev Med Pharmacol Sci. 2019;23:1563–1573. doi: 10.26355/eurrev_201902_17115. [DOI] [PubMed] [Google Scholar]
- 23.Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:4362–4368. [PubMed] [Google Scholar]
- 24.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Li W. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci Rep. 2018;38:BSR20181599. doi: 10.1042/BSR20181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Y, Wang J, Chen Q, Wu Q, Deng W, Zhou H, Shen D. Long non-coding RNA cytoskeleton regulator RNA (CYTOR) modulates pathological cardiac hypertrophy through miR-155-mediated IKKi signaling. Biochim Biophys Acta Mol Basis Dis. 2019;1865:1421–1427. doi: 10.1016/j.bbadis.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Van Grembergen O, Bizet M, de Bony EJ, Calonne E, Putmans P, Brohée S, Olsen C, Guo M, Bontempi G, Sotiriou C, et al. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016;2:e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, Zhong J, Yu P, Zhao Q, Huang T. YY1-regulated LINC00152 promotes triple negative breast cancer progression by affecting on stability of PTEN protein. Biochem Biophys Res Commun. 2019;509:448–454. doi: 10.1016/j.bbrc.2018.12.074. [DOI] [PubMed] [Google Scholar]
- 29.Yan L, Yu MC, Gao GL, Liang HW, Zhou XY, Zhu ZT, Zhang CY, Wang YB, Chen X. MiR-125a-5p functions as a tumour suppressor in breast cancer by downregulating BAP1. J Cell Biochem. 2018;119:8773–8783. doi: 10.1002/jcb.27124. [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Liu X, Zhang Q, Mao X, Feng L, Su P, Chen H, Guo Y, Jin F. Oncogenic potential of TSTA3 in breast cancer and its regulation by the tumor suppressors miR-125a-5p and miR-125b. Tumour Biol. 2016;37:4963–4972. doi: 10.1007/s13277-015-4178-4. [DOI] [PubMed] [Google Scholar]
- 31.Huang WK, Akçakaya P, Gangaev A, Lee L, Zeljic K, Hajeri P, Berglund E, Ghaderi M, Åhlén J, Bränström R, et al. miR-125a-5p regulation increases phosphorylation of FAK that contributes to imatinib resistance in gastrointestinal stromal tumors. Exp Cell Res. 2018;371:287–296. doi: 10.1016/j.yexcr.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 32.Hu X, Jiang H, Jiang X. Downregulation of lncRNA ANRIL inhibits proliferation, induces apoptosis, and enhances radiosensitivity in nasopharyngeal carcinoma cells through regulating miR-125a. Cancer Biol Ther. 2017;18:331–338. doi: 10.1080/15384047.2017.1310348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu Y, Xu X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J Cell Biochem. 2018;119:6296–6308. doi: 10.1002/jcb.26899. [DOI] [PubMed] [Google Scholar]
- 34.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 35.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/S0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 36.Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S, Kang K, Ahn J, Lee D, Kim MY, et al. A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun. 2015;6:10186. doi: 10.1038/ncomms10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu CY, Chan SW, Guo F, Toloczko A, Cui L, Hong W. MRTF/SRF dependent transcriptional regulation of TAZ in breast cancer cells. Oncotarget. 2016;7:13706–13716. doi: 10.18632/oncotarget.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Li J, Li P, Jiang Y, Chen H, Wang R, Cao F, Liu P. DLG5 suppresses breast cancer stem cell-like characteristics to restore tamoxifen sensitivity by inhibiting TAZ expression. J Cell Mol Med. 2019;23:512–521. doi: 10.1111/jcmm.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
They are available at special request.