Abstract
A microarray analysis of an animal model with experimental sepsis induced by caecal ligation and puncture revealed that the level of microRNA-195 (miR-195) was upregulated. However, to the best of our knowledge, the role of miR-195 in sepsis remains unknown. The present study investigated the effect of miR-195 on apoptosis in sepsis and investigated the underlying mechanism. The level of miR-195 was measured in human intestinal epithelial cells following exposure to lipopolysaccharide (LPS). Cell viability and apoptosis were detected using Cell Counting kit-8 and flow cytometry assays. The expression levels of apoptosis-associated proteins were determined using western blot analysis. In addition, a dual-luciferase reporter assay was employed to verify the association between miR-195 and sirtuin 1 (SIRT1). Furthermore, the SIRT1 inhibitor EX527 was applied to further confirm the regulatory network of miR-195/SIRT1 in LPS-induced apoptosis. It was demonstrated that LPS significantly inhibited cell viability and promoted cell apoptosis in NCM460 cells in a dose-dependent manner. In addition, miR-195 was significantly upregulated following LPS treatment. The present results revealed that silencing miR-195 prevented apoptosis and alleviated cell injury in LPS-induced NCM460 cells. Further investigation demonstrated that miR-195 bound directly to and negatively regulated SIRT1. Inhibition of SIRT1 reversed the protective effects of miR-195-silencing on the apoptosis and viability of NCM460 cells. Furthermore, silencing miR-195 prevented endoplasmic reticulum (ER) stress-induced apoptosis via a downregulation of SIRT1 and its downstream effectors, including activating transcription factor 4, C/EBP homologous protein, glucose-regulated protein 78 and growth arrest and DNA-damage protein 34, as well as the phosphorylation of eukaryotic translation initiation factor 2A. In conclusion, the present study revealed a novel mechanism by which miR-195 regulates SIRT1-mediated downstream effectors in ER stress-induced apoptosis in sepsis.
Keywords: microRNA-195, apoptosis, sirtuin 1, endoplasmic reticulum
Introduction
Sepsis is a major cause of admission to the intensive care unit and mortality in the clinic (1), and it is usually characterized by dysregulation of the host response followed by infection, which can lead to organ failure and death (2). Currently, sepsis and its associated shock are major healthcare problems affecting millions of people worldwide each year. In Germany, the percentage of patients with severe sepsis was reported to rise from 27% in 2007 to >40% in 2013, and the in-hospital morality of sepsis rose from 2.7% to ~24% over the same period (3). Among these patients, at least one in four succumb to sepsis each year (4). Although there has been much focus on awareness campaigns and clinician-friendly care bundles, compliance with all six necessary steps within the first hour of admission remains unsatisfactory (5). Therefore, it is important to further investigate the exact molecular mechanism of sepsis. Interactions among the epithelium, local immune system and microbiome are essential for the maintenance of host health, and sepsis alters the intestinal environment to promote epithelial cell dysfunction (6). Apoptosis is considered to serve an important role in sepsis, which leads directly to metabolism dysfunction and organ failure (7,8), and results in severe immunosuppression (9,10). Therefore, it is essential to uncover the regulatory network of epithelial cell apoptosis in sepsis as it may provide an effective strategy for sepsis treatment.
MicroRNAs (miRNAs) are small non-coding RNAs that are ~22 nucleotides in length (11). A recent study has revealed that miRNAs serve critical roles in the progression of sepsis (12). Gao et al (13) reported that miR-146 attenuates cardiac dysfunction in polymicrobial sepsis by targeting interleukin-1 receptor-associated kinase 1 and TNF receptor-associated factor 6. Roderburg et al (14) demonstrated that circulating miR-150 serum levels predict the survival of patients with sepsis. Another study also confirmed that levels of circulating miR-133a serve as a biomarker to predict mortality in patients with severe sepsis (15). These findings suggest that miRNAs serve critical roles in the development of sepsis. miR-195 is regarded as a tumour suppressor in cancer development and progression (16). Almeida et al (17) reported that miR-195 regulates proliferation, osteogenesis and paracrine activity during angiogenesis in primary mesenchymal stromal/stem cells. In addition, Wu et al (18) revealed that the expression level of miR-195 is significantly increased in whole blood samples from a sepsis model compared with samples from a sham-operated group. However, to the best of our knowledge, the biological role of miR-195 in the development of sepsis remains largely unknown.
Sirtuin 1 (SIRT1) is a member of the mammalian sirtuin family, which is a conserved family of NAD+-dependent deacetylases and ADP-ribosyltransferases that serve critical roles in several cellular processes, including DNA repair, gene expression and metabolic regulation (19). A previous study revealed that SIRT1 restrains lung inflammasome activation in a sepsis animal model (20). Li et al (21) demonstrated that resveratrol relieves acute lung injury in a lipopolysaccharide (LPS)-induced sepsis animal model by activating SIRT1. Furthermore, enhancing the activation of SIRT1 markedly alters the transcription profiles in experimental sepsis models (22). Considering these findings, SIRT1 may serve a critical role in the process of sepsis; however, to the best of our knowledge, no studies on the role of miR-195 in the regulation of SIRT1 in sepsis have been conducted.
The present study investigated the function of miR-195 in an LPS-induced cell model of sepsis and the mechanism through which miR-195 modulates SIRT1 expression. The current study demonstrated that miR-195 promoted cell injury in sepsis via targeting SIRT1/eukaryotic translation initiation factor 2A (eIF2a) signalling, which suggests that miR-195 may be a potentially effective therapeutic target for patients with sepsis.
Materials and methods
Cell culture
NCM460 cells were obtained from American Type Culture Collection. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Sigma-Aldrich; Merck KGaA), 100 U/ml streptomycin (Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin (Thermo Fisher Scientific, Inc.) at 37°C with a 5% CO2 atmosphere in a humidified incubator. Sodium butyrate (purity >98.5) was purchased from Sigma-Aldrich; Merck KGaA. When cells reached >80% confluence, they were trypsinized with 0.25% trypsin-EDTA (Sigma-Aldrich; Merck KGaA).
Cell viability detection
Cell viability was determined using a Cell Counting kit-8 (CCK-8) assay. Briefly, NCM460 cells were seeded in a 96-well plate at a density of 3×103/well and maintained in DMEM for 24 h at 37°C. Subsequently, the cells were exposed to LPS (from Escherichia coli 0111:B4; Sigma-Aldrich; Merck KGaA) at concentrations of 0, 1, 5, 10, 25, 50 and 100 µg/ml. Following incubation for 24 h at 37°C, 10 µl/well CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to the medium and cultured for 3 h at 37°C. Subsequently, the plate was agitated for 5 min, and the absorbance of each well at 450 nm was determined using an automatic microplate reader (Multiskan MK3; Thermo Fisher Scientific, Inc.). Each sample was analysed in triplicate, and the mean absorbance was calculated as the final result.
Evaluation of apoptosis by flow cytometry
Firstly, 2×106 cells/well were seeded into a six-well plate for 24 h and then exposed to LPS at concentrations of 0, 1, 5 or 10 µg/ml for 24 h at 37°C. Next, the cells were washed with PBS three times and immediately fixed with 4% paraformaldehyde at 4°C for 15 min. Then, the cells were stained with an Annexin V-FITC/propidium iodide double staining apoptosis detection kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Subsequently, the apoptosis of cells was determined by an Accuri C6 flow cytometer (BD Biosciences) using FlowJo 7.2 software (FlowJo, LLC). Each experiment was performed in triplicate, and the mean value was calculated as the final result.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
After 24 h of transfection of miR-195 inhibitor, the cell medium was removed and total RNA was isolated using TRIzol® reagent (Takara Bio, Inc.) according to the manufacturer's protocol, and quantified using an SSP-3000 Nanodrop Spectrophotometer (Infinigen Biotechnology, Inc.). Subsequently, 1 µg RNA was reverse transcribed into complementary DNA using a Reverse Transcription kit (Takara Bio, Inc.), according to the manufacturer's protocol. Subsequently, the samples were amplified and analysed using a SYBR Premix Ex Taq™ kit (cat. no. RR820A; Takara Bio, Inc.) and an Applied Biosystems 7500 system (Thermo Fisher Scientific, Inc.). For mRNA, GAPDH was used as the internal control, and for miRNA, U6 was used as the internal control. The fold-change in gene expression was calculated using the 2−∆∆Cq method (23). The thermocycling conditions were as follows: 95°C for 3 min, followed by 35 cycles of 95°C for 30 sec, 59°C for 45 sec and 72°C for 40 sec. All PCR assays were replicated at least three times. The primers used in the RT-qPCR assays were as follows: miR-195 forward, 5′-GGC TAG CAG CAC AGA AAT-3′ and reverse, 5′-GTG CAG GGT CCG AGG T-3′; SIRT1 forward, 5′-CCA GAT CCT CAA GCC ATG T-3′ and reverse, 5′-TTG GAT TCC TGC AAC CTG-3′; U6 forward, 5′-CTC GCT TCG GCA GCA CAT ATA CT-3′ and reverse, 5′-ACG CTT CAC GAA TTT GCG TGT C-3′; and GAPDH forward, 5′-AAT GGG CAG CCG TTA GGA AA-3′ and reverse, 5′-TGA AGG GGT CAT TGA TGG CA-3′.
Plasmid construction and cell transfection
miR-195 mimic (5′-UAG CAG CAC AGA AAU GGC-3′), antagomir (5′-GCC AAU AUU UCU GUG CUG CUA-3′) and scrambled negative control (NC) miRNAs (NC mimic, 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and NC antagomir, 5′-CAG UAC UUU UGU GUA GUA CAA-3′) were purchased from GenePharma Co., Ltd. Cells were seeded into 96-well plates and cultured to 70-80% confluence. The cells were then transfected with miRNA mimic (100 nM) or antagomir (200 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 48 h after transfection, the cells were harvested.
Luciferase activity reporter assay
Bioinformatics analysis was performed to search for the potential targets of miR-195. TargetScan 5.0 software (http://www.targetscan.org/) validated SIRT1 as a putative target of miR-195. A luciferase vector including the 3′-untranslated region (3′-UTR) of human SIRT1 (WT luc-SIRT1), which contains the SIRT1-miR-195 response elements, was purchased from Addgene, Inc. A mutant containing two SIRT1-miR-195 response elements in the 3′-UTR of SIRT1 (mutant luc-SIRT1) was generated using site-directed gene mutagenesis. The mutant sequences were designed as follows: 5′-UAA UAU UUU GGA Cug cugUU-3′ (the five lowercase nucleotides were deleted) and 5′-TAA AGT ATT CCT CTG TAC GAT-3′ (the four italic nucleotides were substituted for TGCT). A reporter vector consisting of a luciferase gene followed by the miR-195 binding consensus sequence was purchased from Signosis, Inc. NCM460 cells (2×105 /well) were seeded in a 12-well plate for 24 h. Subsequently, 200 ng WT luc-SIRT1 or mutant luc-SIRT1 and miR-195-5p mimic, antagomir or scrambled oligonucleotide were co-transfected into NCM460 cells using Lipofectamine® 2000, according to the manufacturer's protocol. A pRL-CMV vector containing the CMV enhancer and early promoter elements, which express Renilla luciferase, served as the internal control. Luciferase activity was measured using a dual luciferase reporter assay system (Progema Corporation) 24 h after transfection. Renilla luciferase activity was used as the internal control.
Western blot analysis
Following treatment, the medium was removed and the cells were lysed using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) at 4°C for 15 min. The cell lysates were then harvested and centrifuged for 10 min at 11,000 × g at 4°C. The supernatants were collected and quantified using the BCA method. Subsequently, 30 µg total protein from each sample was separated by 10% SDS-PAGE and transferred to PVDF membranes. Next, the PVDF membranes were blocked with 5% non-fat milk dissolved in TBS and 20% Tween-20 (TBST) at room temperature for 1 h. After rinsing with TBST three times for 5 min each time, the membranes were incubated at 4°C overnight with specific antibodies against the following: (1:100; cat. no. sc-74465, Santa Cruz Biotechnology, Inc.), Bcl-2 (1:1,000; cat. no. 15071, Cell Signaling Technology, Inc.), Bax (1:1,000; cat. no. 14796, Cell Signaling Technology, Inc.), phosphorylated-eIF2a (1:1,000; cat. no. 3398, Cell Signaling Technology, Inc.), eIF2a (1:1,000; cat. no. 5324, Cell Signaling Technology, Inc.), activating transcription factor 4 (ATF4; 1:1,000; cat. no. 11815, Cell Signaling Technology, Inc.), C/EBP homologous protein (CHOP; 1:1,000; cat. no. 2895, Cell Signaling Technology, Inc.) GRP78 (1:1,000; cat. no. ab28615, Abcam), growth arrest and DNA damage-inducible protein (GADD34; cat. no. ab9869, 1:1,000; Abcam), activating transcription factor 6 (ATF6; 1:1,000; cat. no. ab37149, Abcam), IRE1 (1:1,000; cat. no. ab37073, Abcam) and GAPDH (1:5,000; cat. no. 5174, Cell Signaling Technology, Inc.). After rinsing with TBST three times for 5 min each time, the PVDF membranes were incubated with horseradish peroxidase-conjugated goat-anti-rabbit secondary antibody (1:2,000; cat. no. 32460; Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for 1 h. Subsequently, the membranes were rinsed with TBST three times (10 min each time) and visualized using an ECL plus kit (GE Healthcare). Data were analyzed by ImageJ software (version 1.6; National Institutes of Health).
Statistical analyses
In the current study, each experiment was repeated at least three times with consistent results. All data are presented as the mean ± standard deviation, and were analyzed using GraphPad Prism (version 7.0; GraphPad Software, Inc.). One-way ANOVA followed by the Newman-Keuls test was conducted for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
LPS treatment upregulates the expression of miR-195 in NCM460 cells
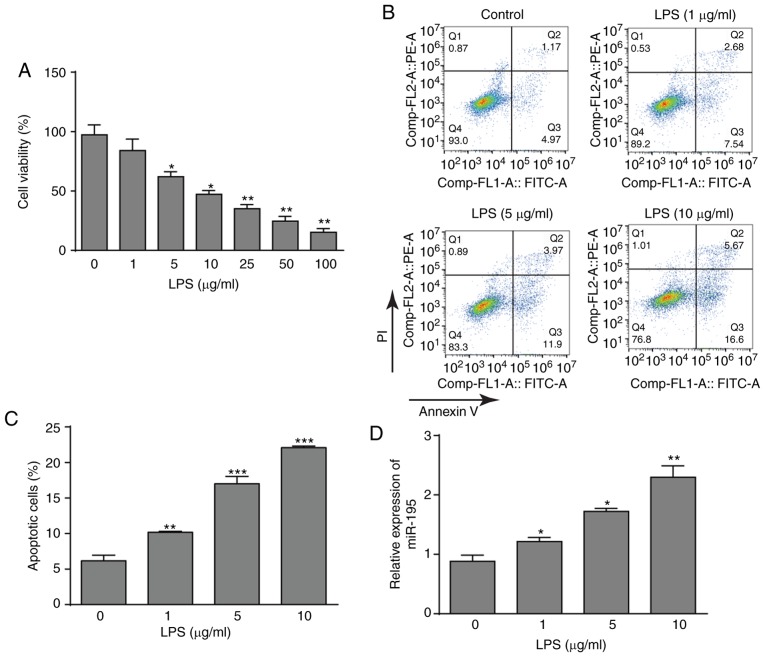
To investigate the effects of LPS on NCM460 cells, cell viability after treatment with LPS was detected using a CCK-8 assay. LPS significantly decreased the viability of NCM460 cells in a dose-dependent manner (Fig. 1A). According to the data, three LPS concentrations (1, 5 and 10 µg/ml) were selected for further investigation. Subsequently, the flow cytometry assay also demonstrated that LPS could promote the apoptosis of NCM460 cells with increasing LPS concentrations (Fig. 1B and C). In addition, the expression pattern of miR-195 was determined. As presented in Fig. 1D, LPS treatment significantly induced the upregulation of miR-195 in NCM460 cells. Thus, these data indicated that LPS treatment inhibited NCM460 cells and increased miR-195 expression, which may indicate that miR-195 serves an important role in LPS-induced apoptosis of NCM460 cells.
Figure 1.
Effects of LPS on cell viability, apoptosis and miR-195 expression. (A) Cultured NCM460 cells were incubated with LPS (0, 1, 5, 10, 25, 50 or 100 µg/ml) for 24 h, and cell viability was then examined by CCK-8 assay. (B) Apoptosis of NCM460 cells following exposure to LPS (0, 1, 5 or 10 µg/ml) was detected by flow cytometry. (C) Quantification of the LPS-induced apoptosis rate of NCM460 cells. (D) miR-195 expression level in NCM460 cells following exposure to LPS (0, 1, 5 or 10 µg/ml) was examined by reverse transcription-quantitative PCR and compared with that in the control group. The miRNA expression level is expressed as a ratio of U6. *P<0.05, **P<0.01 and ***P<0.001 vs. control (0 µg/ml). LPS, lipopolysaccharide; miR-195, microRNA-195; PI, propidium iodide.
Silencing miR-195 inhibits the apoptosis of NCM460 cells
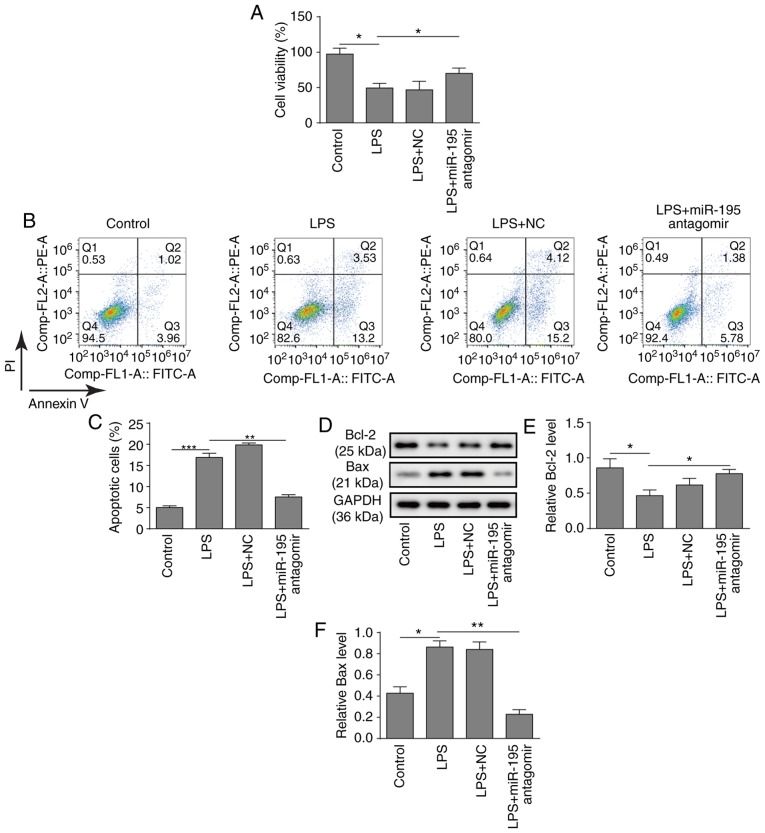
To further investigate the potential role of miR-195, a miR-195 antagomir was applied to silence the expression of miR-195 in NCM460 cells. Subsequently, the biological behaviours of NCM460 cells were investigated. First, a CCK-8 assay demonstrated that silencing miR-195 significantly relieved the inhibition of cell viability induced by LPS in NCM460 cells (Fig. 2A). Consistently, silencing miR-195 also significantly decreased the apoptosis of NCM460 cells induced by LPS (Fig. 2B and C). Furthermore, a western blot analysis of apoptosis-related proteins revealed that LPS treatment significantly inhibited the expression of Bcl-2 and promoted the expression of Bax, while silencing miR-195 significantly increased the expression of Bcl-2 and suppressed the expression of Bax (Fig. 2D-F). In summary, these findings suggest that silencing miR-195 could attenuate the apoptosis of NCM460 cells induced by LPS.
Figure 2.
Role of miR-195 in LPS-induced apoptosis of NCM460 cells (A) Cultured NCM460 cells were transfected with miR-195 antagomir or a scrambled oligonucleotide as a control for 24 h and then incubated with LPS (5 µg/ml) for 24 h. Cell viability was determined by CCK-8 assay kit. (B) Flow cytometry was used to detect the apoptosis of LPS-treated NCM460 cells following transfection with miR-195 antagomir. (C) Quantification of LPS-induced apoptosis of NCM460 cells following transfection with miR-195 antagomir. (D) Bcl-2 and Bax protein levels in LPS-treated NCM460 cells following transfection with miR-195 antagomir were measured by western blot analysis. Quantification of (E) Bcl-2 and (F) Bax protein levels. *P<0.05, **P<0.01 and ***P<0.001. LPS, lipopolysaccharide; miR-195, microRNA-195; PI, propidium iodide; NC, negative control.
miR-195 directly regulates the expression of SIRT1
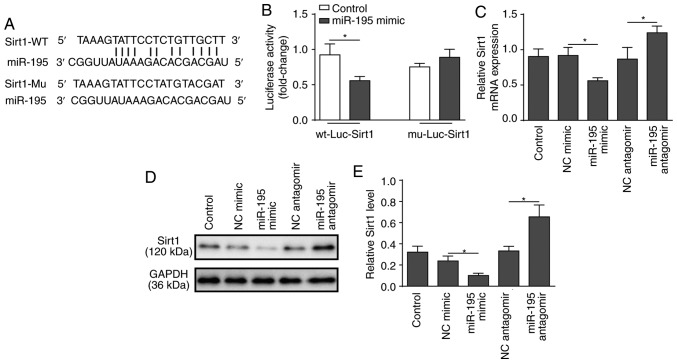
SIRT1 was identified as a predicted target of miR-195 using bioinformatics analysis (Fig. 3A). To verify the association between miR-195 and SIRT1, a dual-luciferase reporter assay was performed. miR-195 mimic and luciferase reporter plasmids with cloned miR-195 binding sites of WT SIRT1-3′UTR or MUT SIRT1-3′UTR were co-transfected. The results revealed that miR-195 mimic significantly reduced the lucif-erase activity of the WT SIRT1-3′UTR, but had no effect on the MUT SIRT1-3′UTR (Fig. 3B). Following that, loss or gain-of-function of miR-195 was employed using miR-195 mimic and antagomir (Fig. S1). RT-qPCR and western blot analyses demonstrated that silencing miR-195 significantly increased the expression of SIRT1, while overexpression of miR-195 significantly suppressed the expression of SIRT1 (Fig. 3C-E). These findings support the hypothesis that SIRT1 is a direct target of miR-195.
Figure 3.
Determination of SIRT1 as a direct target of miR-195. (A) A segment of the SIRT1 3′UTR was inserted downstream of the luciferase coding sequence. Sequence alignment of miR-195 and the 3′UTR of SIRT1 shows complementarity at the 5′ end of miR-195, where the crucial seed region is located. Sequence alignment of miR-195 and the mutated 3′UTR of SIRT1 showed no complementarity at the 5′ end of miR-195. Sequence alignment of miR-195 and the 3′UTR of SIRT1 or the mutated 3′UTR of SIRT1. The four nucleotides TGCT were substituted for ACGA. (B) NCM460 cells were co-transfected with miR-195 mimic or a scrambled oligonucleotide as a control and a plasmid containing the wild-type 3′UTR segment of SIRT1 or the mutated 3′UTR segment of SIRT1. A luciferase activity assay was performed in NCM460 cells to confirm the interaction between miR-195 and SIRT1. (C) NCM460 cells were transfected with miR-195 antagomir, mimic or a scrambled oligonucleotide as a control for 48 h. mRNA expression levels of SIRT1 were determined by reverse transcription-quantitative PCR in NCM460 cells. (D) Protein expression levels of SIRT1 were examined by western blot analysis in NCM460 cells. (E) Quantification of SIRT1 protein levels in NCM460 cells. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05. SIRT1, sirtuin 1; miR-195, microRNA-195; 3′UTR, 3′-untranslated region; WT, wild-type; Mu, mutant; NC, negative control; Luc, luciferase.
Inhibition of SIRT1 reverses the effects of miR-195 antagomir on LPS-induced NCM460 cells
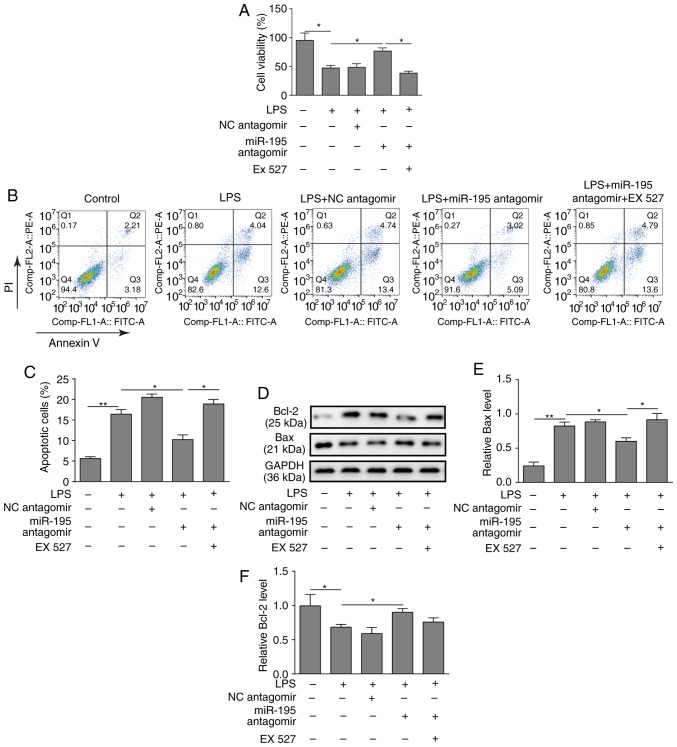
To investigate the function of the miR-195/SIRT1 regulatory network in LPS-induced apoptosis of intestinal epithelial cells, NCM460 cells were pre-treated with the SIRT1 inhibitor EX527. As presented in Fig. 4A, the results of the CCK-8 assay demonstrated that silencing miR-195 increased the viability of NCM460 cells treated with LPS, while the SIRT1 inhibitor EX527 inhibited this effect (Fig. 4A). Similarly, a flow cytometry assay revealed that the SIRT1 inhibitor EX527 reversed the protective effects of miR-195-silencing on NCM460 cell survival (Fig. 4B and C). Furthermore, western blotting demonstrated that silencing miR-195 could promote the expression of Bcl-2 but reduce the expression of Bax, while application of EX527 significantly reversed the effects on Bax level (Fig. 4D-F).
Figure 4.
Effects of the SIRT1 inhibitor EX527 on NCM460 cell apoptosis. NCM460 cells were transfected with miR-195 antagomir or a scrambled oligo-nucleotide as a control before exposure to LPS. A total of 24 h after transfection, the cells were incubated with the SIRT1 inhibitor EX527 for 24 h. (A) Cell viability was measured using a CCK-8 assay kit. The SIRT1 inhibitor EX527 decreased the viability of NCM460 cells. (B) Apoptosis of NCM460 cells was determined by flow cytometry. (C) Quantification of NCM460 cell apoptosis. (D) Bax and Bcl-2 protein levels were measured by western blot analysis. (E) Bax and (F) Bcl-2 protein levels in NCM460 cells were determined and quantified. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01. LPS, lipopolysaccharide; NC, negative control; PI, propidium iodide; SIRT1, sirtuin 1; miR-195, microRNA-195.
miR-195 promotes apoptosis of NCM460 cells via targeting SIRT1/eIF2a
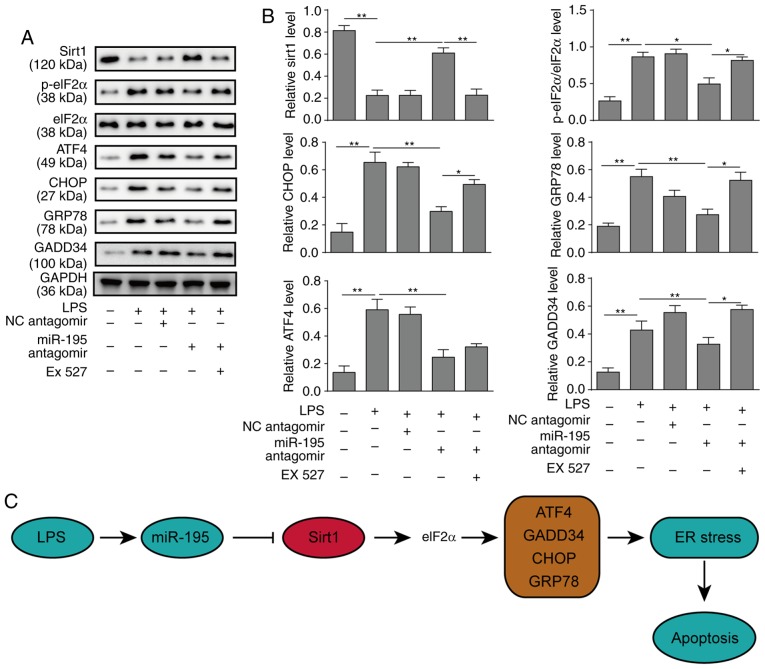
A previous study indicated that SIRT1 serves a protective role in myocardial cells by regulating eIF2a and its downstream effectors (24). Therefore, the signalling pathway of SIRT1/eIF2a was investigated in the endoplasmic reticulum (ER) stress response of NCM460 cells in the present study. Western blot analysis revealed that silencing miR-195 could inhibit apoptosis derived from ER stress via upregulating the expression of SIRT1 and downregulating the expression of ATF4, CHOP, GRP78 and GADD34 and reducing the phosphorylation of eIF2a. However, the SIRT1 inhibitor EX527 significantly increased the expression of CHOP, GRP78 and GADD34, as well as the phosphorylation of eIF2a, which reversed the protective effects of miR-195-silencing (Fig. 5A and B). In addition, the other two pathways (ATF6 and IRE1) of ER stress were further investigated, and the results were similar to the PERK pathway markers (Fig. S2A and B). Taken together, it can be concluded that silencing miR-195 serves a positive role in inhibiting NCM460 cell apoptosis via regulating SIRT1/eIF2a. A schematic figure presenting the association of miR-195, SIRT1 and ER stress is presented in Fig. 5C.
Figure 5.
miR-195 promotes apoptosis of NCM460 cells via targeting SIRT1/eIF2a. NCM460 cells were transfected with miR-195 antagomir or a scrambled oligonucleotide as a control. A total of 24 h after transfection, the cells were incubated with LPS. To further investigate the underlying mechanisms of miR-195, the cells were incubated with the SIRT1 inhibitor EX527 after transfection. (A) The expression levels of markers of the SIRT1/eIF2a signalling pathway were measured by western blot analysis. (B) Quantification of markers in the SIRT1/eIF2a signalling pathway. (C) A schematic figure demonstrating the roles of miR-195, Sirt and ER stress response. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01. LPS, lipopolysaccharide; NC, negative control; SIRT1, sirtuin 1; miR-195, microRNA-195; p-, phosphorylated; eIF2a, eukaryotic translation initiation factor 2A; ATF4, activating transcription factor 4; CHOP, C/EBP homologous protein; GADD34, growth arrest and DNA damage-inducible protein; ER, endoplasmic reticulum.
Discussion
In the present study, miR-195 was identified to be significantly upregulated in an LPS-induced cell sepsis model. Furthermore, transfection with a miR-195 antagomir could significantly decrease apoptosis but increase the viability of intestinal epithelial NCM460 cells. Further analyses revealed that miR-195 could bind to the 3′UTR of SIRT1 and regulate the expression of SIRT1. In addition, silencing miR-195 protected the viability of NCM460 cells treated with LPS, but the SIRT1 inhibitor EX527 reversed this effect by silencing miR-195. These findings indicate that miR-195 regulates the apoptosis of intestinal epithelial cells in LPS-induced sepsis via SIRT1.
Sepsis is a major public health concern and is usually characterized by intravascular or extravascular microbial infection, systemic inflammation and microcirculatory dysfunction, which results in tissue damage, organ failure or even death (25). Notably, dysregulated apoptosis and increased neutrophil function lead to immune and organ dysfunction in sepsis and its associated multiple organ failure (26). LPS, a membrane component of gram-negative bacteria, is typically used to induce sepsis in cell and animal models, in which pro-inflammatory cytokines in the serum are increased with septic clinic manifestations (27). A previous review has documented that miRNAs, such as miR-195, serve critical roles in the process of sepsis (28). Wu et al (18) revealed that miR-195 is significantly upregulated in a caecal ligation and puncture-induced experimental sepsis model. Another study revealed that serum miR-195 serves as a biomarker for Chinese patients with sepsis. The present study also detected upregulation of miR-195 in an LPS-induced sepsis cell model, which was in line with other work. Furthermore, Zheng et al (29) demonstrated that inhibition of miR-195 suppresses apoptosis and multiple organ injury in a sepsis mouse model. The present study demonstrated that silencing miR-195 could protect NCM460 cells from LPS-induced apoptosis. This suggests that miR-195 may serve a critical role in the pathogenesis of sepsis.
SIRT1 is a NAD+-dependent histone deacetylase that serves critical roles in various biological processes, including the ER stress response and cell survival. During sepsis adaptation, SIRT1 switches monocyte energy sources from glycolysis to fatty acid oxidation (30). Gao et al (20) revealed that SIRT1 could restrain inflammasome activation in the lungs of a murine sepsis model. Li et al (21) reported that resveratrol attenuates acute lung injury via activating SIRT1 in LPS-induced sepsis. Vachharajani et al (31) revealed that inhibition of SIRT1 improves immunity and outcomes of the hypo-inflammatory phenotype of sepsis. Thus, accumulating evidence supports the protective role of SIRT1 in sepsis. In addition, miR-195 has been reported to regulate the initiation and development of tumours by targeting multiple genes. For example, Zhou et al (32) demonstrated that miR-195 inhibits cervical cancer migration and invasion via binding Smad3. miR-195 has also been identified to suppress colon cancer proliferation by targeting Wnt family member 3A (33). Furthermore, Zhu et al (34) have reported that miR-195 promotes palmitate-induced apoptosis in cardiomyocytes by reducing the expression of SIRT1. Considering these findings, we speculated whether there was such a similar connection between miR-195 and SIRT1 in sepsis. The present study verified that SIRT1 is a target of miR-195 in NCM460 cells, and inhibition of miR-195 could significantly upregulate the expression of SIRT1. To further confirm the underlying mechanism, the SIRT1 inhibitor EX527 was used to investigate the biofunctional changes in LPS-treated NCM460 cells. The results demonstrated that silencing miR-195 could reduce LPS-induced apoptosis in NCM460 cells, while pre-treatment with EX527 could promote apoptosis. These findings demonstrated that SIRT1 may be a critical downstream target of miR-195-mediated apoptosis in intestinal epithelial cells.
Although the underlying mechanisms by which silencing miR-195 affects SIRT1 and reduces apoptosis in sepsis remain to be investigated, other studies may provide some suggestions. Liu et al (35) identified that downregulating Smad3/ATF4 is essential for the SIRT1 suppression of ER stress-induced apoptosis. Furthermore, Koga et al (36) reported that ER stress promotes hepatocellular injury via increasing the expression of SIRT1 in the PI3K/Akt-GSK3β signalling pathway. Prola et al (37) demonstrated that SIRT1 attenuates ER stress-induced cell apoptosis in cardiomyocytes via eIF2a deacetylation. These findings suggest that SIRT1 may serve critical roles in ER stress-induced apoptosis. In the present study, the activation of the ER stress response was also revealed. The results demonstrated that silencing miR-195 could significantly increase the expression level of SIRT1, while decreasing the expression levels of ATF4, CHOP, GRO78 and GADD34 and inhibiting the phosphorylation of eIF2a. By contrast, inhibiting SIRT1 increased the expression levels of ATF4, CHOP, GRO78 and GADD34 and the phosphorylation of eIF2a. The present results confirmed that miR-195 could promote cell apoptosis by downregulating the SIRT1/eIF2a signalling pathway in the ER.
In summary, the present study revealed that upregulation of miR-195 in a sepsis cell model could serve a pivotal role in promoting intestinal epithelial cell apoptosis and result in sepsis aggravation. Furthermore, molecular mechanism experiments indicated that miR-195 directly targets SIRT1 and promotes apoptosis in intestinal epithelial cells via the SIRT1/eIF2a signalling pathway.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China. (grant no. 81400031).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JGL is responsible for the integrity of the entire study, designed the study, analyzed the data, performed statistical analysis, and edited and reviewed the manuscript. YT designed the study, performed the literature research and experimental studies, acquired the data, performed data and statistical analyses, and prepared the manuscript. SY performed the experimental studies and statistical analysis, and acquired the data. SYD performed the experimental studies. LZ performed the experimental studies and data analysis, and prepared the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kaukonen KM, Bailey M, Pilcher D, Cooper DJ, Bellomo R. Systemic inflammatory response syndrome criteria in defining severe sepsis. New Engl J Med. 2015;372:1629–1638. doi: 10.1056/NEJMoa1415236. [DOI] [PubMed] [Google Scholar]
- 2.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleischmann C, Thomas-Rueddel DO, Hartmann M, Hartog CS, Welte T, Heublein S, Dennler U, Reinhart K. Hospital incidence and mortality rates of sepsis. Dtsch Arztebl Int. 2016;113:159–166. doi: 10.3238/arztebl.2016.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]
- 5.Hunter JG, Pritchett C, Pandya D, Cripps A, Langford R. Sim-sepsis: Improving sepsis treatment in the emergency department? BMJ Simul Technol Enhanc Learn. 2018;2018:bmjstel. doi: 10.1136/bmjstel-2018-000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fay KT, Ford ML, Coopersmith CM. The intestinal micro-environment in sepsis. Biochim Biophysica Acta Mol Basis Dis. 2017;1863:2574–2583. doi: 10.1016/j.bbadis.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoseph BP, Klingensmith NJ, Liang Z, Breed ER, Burd EM, Mittal R, Dominguez JA, Petrie B, Ford ML, Coopersmith CM. Mechanisms of intestinal barrier dysfunction in sepsis. Shock. 2016;46:52–59. doi: 10.1097/SHK.0000000000000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gill SE, Rohan M, Mehta S. Role of pulmonary microvascular endothelial cell apoptosis in murine sepsis-induced lung injury in vivo. Respir Res. 2015;16:1–13. doi: 10.1186/s12931-015-0266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luan Y, Yao Y, Xiao X, Sheng Z. Insights into the apoptotic death of immune cells in sepsis. J Interferon Cytokine Res. 2015;35:17–22. doi: 10.1089/jir.2014.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delano MJ, Ward PA. Sepsis-induced immune dysfunction: Can immune therapies reduce mortality? J Clin Invest. 2016;126:23–21. doi: 10.1172/JCI82224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang W, Bartel DP. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol Cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giza DE, Fuentesmattei E, Bullock MD, Tudor S, Goblirsch MJ, Fabbri M, Lupu F, Yeung SJ, Vasilescu C, Calin GA. Cellular and viral microRNAs in sepsis: Mechanisms of action and clinical applications. Cell Death Differ. 2016;23:1906–1918. doi: 10.1038/cdd.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao M, Wang X, Zhang X, Ha T, Ma H, Liu L, Kalbfleisch JH, Gao X, Kao RL, Williams DL. Attenuation of cardiac dysfunction in polymicrobial sepsis by MicroRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J Immunol. 2015;195:672–682. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roderburg C, Luedde M, Vargas Cardenas DV, Vucur M, Scholten D, Frey N, Koch A, Trautwein C, Tacke F, Luedde T. Circulating MicroRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One. 2013;8:e54612. doi: 10.1371/journal.pone.0054612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tacke F, Roderburg C, Benz F, Cardenas DV, Luedde M, Hippe HJ, Frey N, Vucur M, Gautheron J, Koch A, et al. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients. Crit Care Med. 2014;42:1096–1104. doi: 10.1097/CCM.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y, Liu Y, Li S, Liang Z, Xu X, et al. MicroRNA-195-5p a new regulator of Fra-1, suppresses the migration and invasion of prostate cancer cells. J Transl Med. 2015;13:289. doi: 10.1186/s12967-015-0650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almeida MI, Silva AM, Vasconcelos DM, Almeida CR, Caires H, Pinto MT, Calin GA, Santos SG, Barbosa MA. MiR-195 in human primary mesenchymal stromal/stem cells regulates proliferation, osteogenesis and paracrine effect on angiogenesis. Oncotarget. 2016;7:7–22. doi: 10.18632/oncotarget.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu SC, Yang JC, Rau CS, Chen YC, Lu TH, Lin MW, Tzeng SL, Wu YC, Wu CJ, Hsieh CH. Profiling circulating MicroRNA expression in experimental sepsis using cecal ligation and puncture. PLoS One. 2013;8:e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, et al. SIRT1 is required for AMPK activation and the beneficial effects of resve-ratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao R, Ma Z, Hu Y, Chen J, Shetty S, Fu J. Sirt1 restrains lung inflammasome activation in a murine model of sepsis. Am J Physiol Lung Cell Mol Physiol. 2015;308:L847–L853. doi: 10.1152/ajplung.00274.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Zhang J, Feng J, Li Q, Wu L, Ye Q, Sun J, Lin Y, Zhang M, Huang R, et al. Resveratrol reduces acute lung injury in a LPS-induced sepsis mouse model via activation of Sirt1. Mol Med Rep. 2013;7:1889–1895. doi: 10.3892/mmr.2013.1444. [DOI] [PubMed] [Google Scholar]
- 22.Opal S, Ellis JL, Suri V, Freudenberg JM, Vlasuk GP, Li Y, Chahin AB, Palardy JE, Parejo N, Yamamoto M, et al. Sirt1 activation markedly alters transcription profiles and improves outcome in experimental sepsis. Shock. 2015;28:559–567. doi: 10.1097/SHK.0000000000000528. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Prola A, Pires Da Silva J, Guilbert A, Lecru L, Piquereau J, Ribeiro M, Mateo P, Gressette M, Fortin D, Boursier C, et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2α deacetylation. Cell Death Differ. 2017;24:343–356. doi: 10.1038/cdd.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djoumerska-Alexieva I, Pashova S, Vassilev T, Pashov A. The protective effect of modified intravenous immunoglobulin in LPS sepsis model is associated with an increased IRA B cells response. Autoimmun Rev. 2013;12:653–656. doi: 10.1016/j.autrev.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Paunel-Görgülü A, Flohé S, Scholz M, Windolf J, Lögters T. Increased serum soluble Fas after major trauma is associated with delayed neutrophil apoptosis and development of sepsis. Critical Care. 2011;15:R20. doi: 10.1186/cc9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai B, Deitch EA, Ulloa L. Novel insights for systemic inflammation in sepsis and hemorrhage. Med Inflamm. 2010;2010:642462. doi: 10.1155/2010/642462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingsley SMK, Bhat BV. Role of microRNAs in sepsis. Inflamm Res. 2017;66:1–17. doi: 10.1007/s00011-017-1031-9. [DOI] [PubMed] [Google Scholar]
- 29.Zheng D, Yu Y, Li M, Wang G, Chen R, Fan GC, Martin C, Xiong S, Peng T. Inhibition of MicroRNA 195 prevents apoptosis and multiple-organ injury in mouse models of sepsis. J Infect. 2016;213:1661–1670. doi: 10.1093/infdis/jiv760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu TF, Vachharajani V, Millet P, Bharadwaj MS, Molina AJ, Mccall CE. Sequential actions of SIRT1-RELB-SIRT3 coordinate nuclear-mitochondrial communication during immunometabolic adaptation to acute inflammation and sepsis. J Biol Chem. 2015;290:396–408. doi: 10.1074/jbc.M114.566349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vachharajani VT, Liu T, Brown CM, Wang X, Buechler NL, Wells JD, Yoza BK, Mccall CE. SIRT1 inhibition during the hypoinflammatory phenotype of sepsis enhances immunity and improves outcome. J Leuk Biol. 2014;96:785–796. doi: 10.1189/jlb.3MA0114-034RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Q, Han LR, Zhou YX, Li Y. MiR-195 suppresses cervical cancer migration and invasion through targeting smad3. Int J Gynecol Cancer. 2016;26:817–824. doi: 10.1097/IGC.0000000000000686. [DOI] [PubMed] [Google Scholar]
- 33.Li B, Wang S, Wang S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol Genet Genomics. 2018;293:1245–1253. doi: 10.1007/s00438-018-1457-y. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 2011;92:75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z, Gu H, Lu G, Xu Y, Fei F, Saeed M, Chao S. Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget. 2016;8:9267–9279. doi: 10.18632/oncotarget.14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koga T, Suico MA, Shimasaki S, Watanabe E, Kai Y, Koyama K, Omachi K, Morino-Koga S, Sato T, Shuto T, et al. Endoplasmic reticulum (ER) stress induces sirtuin 1 (SIRT1) expression via the PI3K-Akt-GSK3β signaling pathway and promotes hepatocellular injury. J Biol Chem. 2015;290:30366–30374. doi: 10.1074/jbc.M115.664169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prola A, Silva JP, Guilbert A, Lecru L, Piquereau J, Ribeiro M, Mateo P, Gressette M, Fortin D, Boursier C, et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2α deacetylation. Cell Death. 2016;24:343–356. doi: 10.1038/cdd.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.