Abstract
Molecular biomarkers aim to stratify cancer patients into disease subtypes predictive of outcome, improving diagnostic precision beyond clinical descriptors such as tumour stage1. Transcriptomic intra-tumour heterogeneity (RNA-ITH) has been shown to confound existing expression-based biomarkers across multiple cancer types2–6. Here, we analyse multi-region whole-exome and RNA sequencing data for 156 tumour regions from 48 TRACERx patients to explore and control for RNA-ITH in non-small cell lung cancer (NSCLC). We find that chromosomal instability (CIN) is a major driver of RNA-ITH, and existing prognostic gene expression signatures are vulnerable to tumour sampling bias. To address this, we identify genes expressed homogeneously within individual tumours that encode expression modules of cancer cell proliferation and are often driven by DNA copy-number gains selected early in tumour evolution. Clonal transcriptomic biomarkers overcome tumour sampling bias, associate with survival independently of clinicopathological risk factors, and may provide a general strategy to refine biomarker design across cancer types.
Multiple attempts have been made to derive a prognostic gene expression signature for patients with lung adenocarcinoma (LUAD)7–16, the most common histological subtype of NSCLC. However, none has been successfully adopted in clinical practice due to poor reproducibility in independent patient cohorts or failure to provide molecular information beyond existing clinicopathological risk factors1,17.
Genomic intra-tumour heterogeneity (ITH) is prevalent across cancer types18. Previous multi-region sequencing studies have indicated that molecular biomarkers may be confounded by sampling bias arising from ITH2–6 (Fig. 1a). Therefore, addressing ITH as a confounding factor for biomarker design is an important challenge for precision oncology19–22 (Fig. 1b).
Fig. 1. Tumour sampling bias confounds lung cancer biomarkers.
a, Prognostic biomarkers classify tumour biopsies as high (red) or low risk (blue). The TRACERx trial samples multiple biopsies from each tumour (R1-4), however diagnosis is typically made using a single tumour biopsy (dashed triangle) in routine clinical practice. The hypothetical biomarker illustrated here exhibits discordant risk classification of tumour regions, thus the molecular read-out of the diagnostic biopsy (blue circle) is vulnerable to tumour sampling bias. b, Applied to a diagnostic biopsy (1) a prognostic biomarker stratifies lung cancer patients into more precise disease subtypes based on estimated survival risk (2), which may help inform therapeutic decision-making (3). For example correctly distinguishing high-risk patients (red), in need of adjuvant chemotherapy, from low-risk patients (blue) for whom surgery alone is curative. However, patients vulnerable to tumour sampling bias (gray) may be incorrectly stratified, resulting in assignment to a sub-optimal treatment and follow-up strategy. c, A published RNAseq prognostic signature for LUAD14 is evaluated in TRACERx (n = 28 LUAD patients, stage I-III). Each point represents a single tumour region and the vertical lines display the range for each patient. Points are coloured according to the risk classification of tumour regions within a patient: concordant low-risk (blue), concordant high-risk (red), or discordant (gray). d, Percentages of TRACERx patients (n = 28 LUAD patients, stage I-III) classified as concordant low-risk (blue), concordant high-risk (red), or discordant (gray) using two published RNAseq prognostic signatures for LUAD: Shukla et al14 (left), Li et al12 (right).
To explore the causes and consequences of RNA-ITH in NSCLC, we utilized three RNAseq-based expression datasets from patients with early-stage lung cancer (Extended Data 1a-b, Supplementary Table 1): (i) the TRACERx multi-region dataset23 to derive RNA intra- and inter-tumour heterogeneity scores (156 tumour regions from 48 TRACERx NSCLC patients [median 3 regions per tumour, range 2-7], stage I-III), (ii) The Cancer Genome Atlas (TCGA) NSCLC dataset24,25 to develop prognostic signatures (n = 959 NSCLC patients, stage I-III), and (iii) the Uppsala NSCLC dataset26 (n = 170 NSCLC patients, stage I-III) for validation purposes. All references to tumour stage are based on version 7 of the TNM groupings27. Four microarray-based expression datasets16,28–30 were also analysed as additional validation cohorts (Extended Data 1c).
Unsupervised hierarchical clustering on the top 500 most variant genes across all tumour samples in the TRACERx multi-region cohort (156 tumour regions, 48 NSCLC patients, stage I-III) revealed perfect clustering concordance between regions from the same tumour (Extended Data 2a), suggesting overall RNA inter-tumour heterogeneity exceeds intra-tumour heterogeneity. While this shows that each tumour has a uniquely identifiable expression profile, this gene set does not offer information on patient prognosis (log-rank P = 0.686, Extended Data 2b). To assess the impact of RNA-ITH on prognostic information, we investigated the effect of RNA-ITH and sampling bias on previously published prognostic gene expression signatures in LUAD using patients from the TRACERx cohort (89 tumour regions, 28 LUAD patients, stage I-III). First, we evaluated the performance of a recent RNAseq-based prognostic signature developed by Shukla et al14 (Fig. 1c). Using the RNAseq signature to classify tumour regions as either high-risk or low-risk, 43% of patients (12/28) exhibited discordant risk classification (Fig. 1d, left). Similarly, using an immune-related prognostic signature12, a discordance rate of 29% (8/28 patients) was observed (Fig. 1d, right). These data indicate that whether a patient is classified as low- or high-risk is frequently influenced by which tumour sample is analysed, thus potentially limiting the clinical utility of existing prognostic assays.
The majority of gene expression signatures in NSCLC have been derived using microarray expression profiling, not RNAseq. To assess the prevalence of sampling bias regardless of the original profiling platform, we used a previously described clustering method5 to evaluate sampling bias in the TRACERx cohort (89 tumour regions, 28 LUAD patients, stage I-III). Applied to 9 published prognostic signatures7–15, this analysis revealed the median discordance rate was 50% (15.5/28 LUAD tumours, range = 18-82%) at the level of individual patients, indicating that half of the tumours in this cohort could be at risk of misclassification due to sampling bias (Extended Data 2c-d). Although these analyses do not directly measure prognostic ability, taken together, the data suggest that existing signatures are commonly subject to sampling bias, which may contribute to the low validation rate of gene expression signatures in NSCLC.
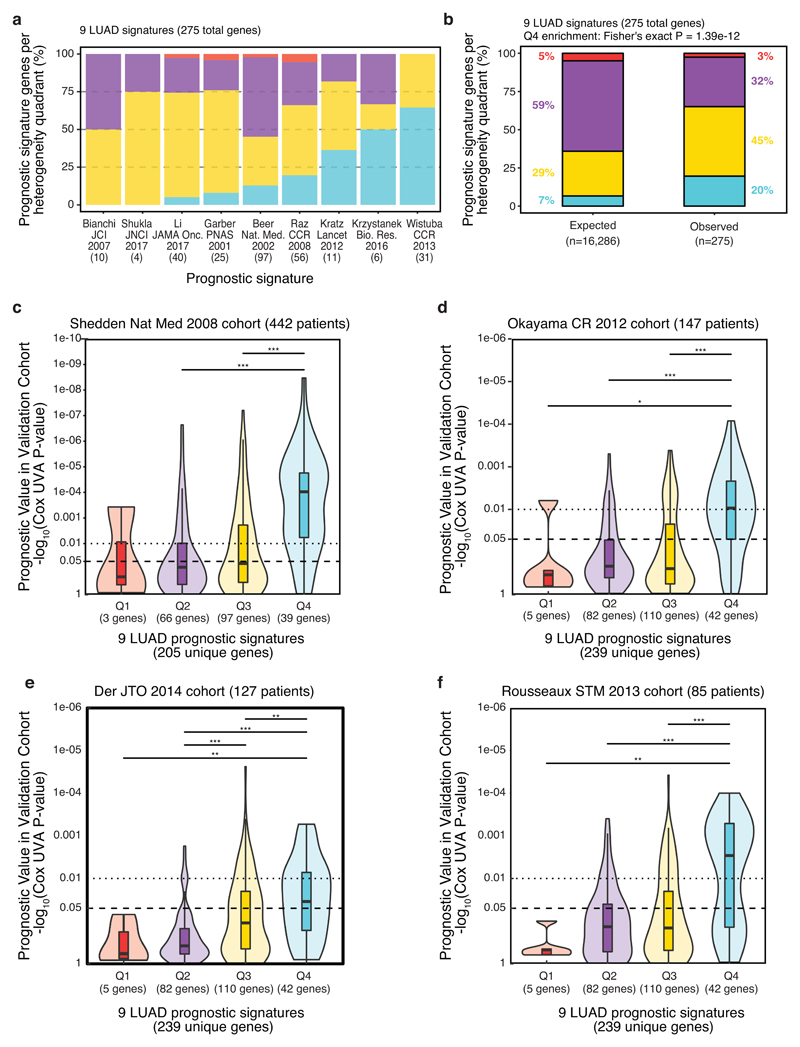
Biomarker design may be improved by limiting sampling bias (caused by intra-tumour heterogeneity) and maximising discriminatory power between tumours (inter-tumour heterogeneity), to identify prognostic RNA markers that offer superior reproducibility and clinical utility compared to existing prognostic signatures. To explore this hypothesis, we derived a per-gene metric for RNA intra- and inter-tumour heterogeneity and split both heterogeneity metrics by their mean (see Methods, Supplementary Table 2, Extended Data 3). This resulted in four RNA heterogeneity quadrants for LUAD (Fig. 2a): low inter- and high intra- (Q1 = 798 genes), low inter- and low intra- (Q2 = 9,642 genes), high inter- and high intra- (Q3 = 4,766 genes), high inter- and low intra- (Q4 = 1,080 genes). Genes in Q4 satisfy the desired criteria: exhibiting homogenous expression within tumours, restricting sampling bias, yet are highly variable between tumours, so may be informative for patient stratification. Determining clustering concordance scores for individual genes, we found Q4 genes best clustered TRACERx tumour regions by patient (Extended Data 4), indicating that these genes are the least vulnerable to sampling bias.
Fig. 2. RNA inter- and intra-tumour heterogeneity quadrants.
a, RNA heterogeneity quadrants calculated in TRACERx (n = 28 LUAD patients, stage I-III). RNA intra-tumour (y-axis) and inter-tumour heterogeneity (x-axis) are plotted on the axes as density curves. The plot is divided into quadrants by the mean intra-tumour (dashed horizontal line) and mean inter-tumour (dashed vertical line) heterogeneity scores. The quadrants are numbered and coloured (Q1=red, Q2=purple, Q3=yellow, Q4=blue) with the number of genes per quadrant indicated. b, Composition of published prognostic genes. Radar plot of the percentage of genes observed in 9 published LUAD prognostic signatures7–15 per quadrant (as indicated in Extended Data 5a-b), divided by the percentage of genes expected per quadrant (total no. genes, as indicated in Fig. 2a). c, Reproducible survival association of published prognostic genes. The ability of genes from published signatures7–15 to maintain prognostic value in an independent patient cohort (TCGA, n = 469 LUAD patients, stage I-III) was assessed as the gene-wise Cox univariate P-value (y-axis), stratified by RNA heterogeneity quadrant (x-axis). Boxplots represent the median, 25th and 75th percentiles and the vertical bars span the 5th to the 95th percentiles. Statistical significance was tested with a two-sided Wilcoxon signed-rank test. d, Cross-cohort prognostic significance of random signatures. 1000 random signatures were developed in the TCGA RNAseq cohort (n = 469 LUAD patients, stage I-III), derived using 20 genes randomly sampled from each RNA heterogeneity quadrant, then tested for prognostic ability across four microarray cohorts comprised of stage I-III LUAD patients (Shedden et al n = 442, Okayama et al n = 147, Der et al n = 127, Rousseaux et al n = 85).
Genes in Q4 comprise only 7% of all expressed genes (1,080/16,286, Fig. 2a), yet make up 20% of the genes collated from 9 published prognostic signatures7–15 (54/275, including 33 overlapping across multiple signatures, Extended Data 5a-b, Supplementary Table 3) - a three-fold enrichment (P = 1.39 x 10-12, Fig. 2b) suggesting that previous studies tend to select Q4 genes even in the absence of RNA-ITH information. We next evaluated the ability of genes from the 9 prognostic signatures7–15 (242 unique genes) to validate in an independent patient cohort (TCGA, n = 469 LUAD patients, stage I-III), finding those in Q4 reproducibly associate with survival significantly better than genes from other quadrants (Q2-vs-Q4 P = 6.5 x 10-8, Q3-vs-Q4 P = 4.0 x 10-4, Fig 2c; insufficient genes in Q1 for Q1-vs-Q4 comparison). Similar results were observed using microarray-based gene expression data from four cohorts (total n = 801 LUAD patients), despite the platform differences between microarray and RNAseq data (Extended Data 5c-f).
To further examine the ability of Q4 genes to reproducibly maintain prognostic information, we examined the cross-cohort performance of randomly generated signatures. Previous work has shown that a high proportion of random signatures significantly associate with survival upon assessment in independent validation datasets31,32. We derived 1,000 signatures in the TCGA RNAseq cohort (n = 469 LUAD patients, stage I-III), using 20 genes randomly drawn from each heterogeneity quadrant (defined in multi-region RNAseq data from the TRACERx LUAD cohort), then assessed their prognostic value across the four microarray-based cohorts (combined n = 801 LUAD patients, stage I-III). When based on Q4 genes, we observed a marked increase in the number of random signatures significantly associated with outcome across multiple cohorts (56% of Q4 signatures significant across 4 cohorts, versus 0%, 0.7% and 7.3% for Q1, Q2 and Q3 genes respectively, Fig. 2d). These results provide evidence that Q4 is highly enriched for genes with a reproducible survival association relative to the other RNA heterogeneity quadrants.
To assess the relevance of our findings for biomarker design, we replicated a range of methods previously taken to derive prognostic signatures in LUAD10,14,33,34 (Extended Data 6a, Supplementary Table 4), using the TCGA RNAseq cohort (n = 469 LUAD patients, stage I-III) for signature development. Conventional biomarker design involves the selection of survival-associated genes, and the fitting of a prognostic model using a machine learning algorithm (such as stepwise regression14, tree classification33, random forest regression34, or elastic-net regression10) to generate a gene expression signature. In parallel, we implemented a clonal version of each signature using the same methodology but including only Q4 genes in the prognostic model (Extended Data 6a, Supplementary Table 4). The survival association of each signature was evaluated in the Uppsala RNAseq dataset (n = 103 LUAD patients, stage I-III) as an independent patient cohort for validation. Only the clonal version of the signature based on elastic-net regression was significant in the validation dataset (Fig. 3a), highlighting the limited reproducibility of conventional prognostic signature design.
Fig. 3. Clonal gene selection improves prognostic accuracy over conventional biomarker design and beyond clinicopathological risk factors.
a, Prognostic value of conventional versus clonal biomarker design. Prognostic signature development requires gene selection, followed by the application of a machine learning algorithm (see Extended Data 6a). Several conventional methods are replicated from published studies10,14,33,34 (orange), and a clonal version of each signature is generated in parallel (blue). In addition, a de novo strategy, prioritizing the selection of clonally expressed genes (see Extended Data 6b-d and Methods for details), is used to derive the Outcome Risk Associated Clonal Lung Expression (ORACLE) biomarker. All signatures are developed in the TCGA RNAseq dataset (n=469 LUAD patients, stage I-III). Prognostic accuracy of the resulting signatures was assessed in the Uppsala RNAseq dataset (n=103 LUAD patients, stage I-III) as an independent cohort of patients.
b, Prognostic value of ORACLE assessed in a meta-analysis across five validation cohorts of LUAD patients. Univariate Cox analysis was performed in one RNAseq dataset (Uppsala) and four microarray datasets (Shedden et al, Okayama et al, Der et al, Rousseaux et al). Hazard ratios with a 95% confidence interval are shown for each cohort and are plotted on a natural log scale. The diamond indicates the hazard ratio for the meta-analysis of five validation cohorts. c, Prognostic value over known risk factors. Multivariate Cox analysis was performed in the Uppsala RNAseq dataset (n=103 LUAD patients, stage I-III), incorporating ORACLE risk score, tumour stage, therapy status, patient age, WHO performance status, smoking status, patient gender and Ki67 staining percentage. Hazard ratios with a 95% confidence interval are shown for each variable and are plotted on a natural log scale. d, Prognostic value in stage I patients. The ability of substaging criteria (left) versus ORACLE (right) to split patients into prognostically informative groups is tested in stage I patients. Kaplan-Meier plots with log-rank P-values calculated in the Uppsala RNAseq dataset (n=60 stage I LUAD patients). All statistical tests were two-sided.
The approach based on elastic-net regression restricted signature design to a gene list of published prognostic genes10 (Extended Data 6a). However, we observed above that a high proportion of Q4 genes have a reproducible survival association (Fig. 2d), suggesting the potential for novel biomarker discovery. We thus designed a de novo strategy, inputting a list of Q4 genes (ranked by clustering concordance in the TRACERx cohort, Extended Data 4) to the elastic-net algorithm. This generated a 23-gene prognostic signature (Supplementary Table 5) we termed the Outcome Risk Associated Clonal Lung Expression (ORACLE) biomarker (Extended Data 6b-d and Methods for details). Only 11% of TRACERx LUAD patients (3/28) exhibited discordant classification using ORACLE (Extended Data 6e), which compares favourably with the discordance rates for existing prognostic signatures (43% and 29% for two RNAseq-based signatures12,14, Fig. 1d). Moreover, the ORACLE risk score significantly associated with mortality in the Uppsala validation cohort (HR = 3.16 [1.4-7.0], Cox UVA P-value = 0.00474, Fig. 3a). 3-year overall survival was 80% [68-94%] in the ORACLE low-risk group and 57% [46-71%] in the ORACLE high-risk group (Extended Data 7a).
To investigate concordance across multiple cohorts, we applied ORACLE to the four microarray datasets. We expected ORACLE’s performance to be poorer, considering that we were only able to match 19/23 genes to the microarray probe sets and we applied signature weights trained on RNAseq data. However, ORACLE significantly associated with survival in 3/4 microarray datasets (univariate Cox regression: Okayama et al cohort P = 0.002, HR = 5.4; Rousseaux et al cohort P = 0.003, HR = 2.9; Shedden et al cohort P = 2.3 x 10-8, HR = 3.6; Der et al cohort P = 0.3, HR = 1.6), and in a meta-analysis considering all validation cohorts (combined n = 904 LUAD patients, stage I-III) ORACLE was significantly associated with outcome (overall HR = 3.57 [2.94-3.54], P < 0.0001, Fig. 3b). These data indicate that survival associations resistant to differences in expression profiling technology can be obtained by controlling for RNA-ITH in biomarker design.
In the Uppsala RNAseq validation cohort (n = 103 LUAD patients, stage I-III) ORACLE was significantly associated with overall survival in multivariate analysis adjusting for TNM stage, adjuvant treatment status, age, WHO performance status, smoking history, gender and Ki67 staining percentage (adjusted HR = 2.64 [1.15-6.05], Cox MVA P = 0.0216, Fig. 3c). This analysis suggests that ORACLE could provide prognostic information independently of known clinicopathological risk factors.
Stage I LUAD patients with tumour diameters <4cm are not routinely offered adjuvant chemotherapy due to lack of treatment benefit35,36. Biomarker based stratification in this patient population could identify a higher risk subgroup that might benefit from adjuvant therapy1,17. Therefore, we specifically explored this subgroup (Uppsala cohort, n = 60 stage I LUAD patients). Dividing this cohort by sub-stage parameters (n = 42 IA patients, n = 18 IB patients; Fig. 3d) was not prognostically informative, likely due to the small sample size. However, ORACLE separated stage I patients into high-risk (n = 32 LUAD patients, stage I) and low-risk (n = 28 stage I LUAD patients) groups with significantly different survival times (log-rank P = 0.02, Fig. 3d). This result was replicated using the updated version 8 TNM criteria37 (Extended Data 7b-c). While these data must be considered hypothesis generating due to cohort size restrictions, our findings suggest that ORACLE is associated with mortality in stage I LUAD patients.
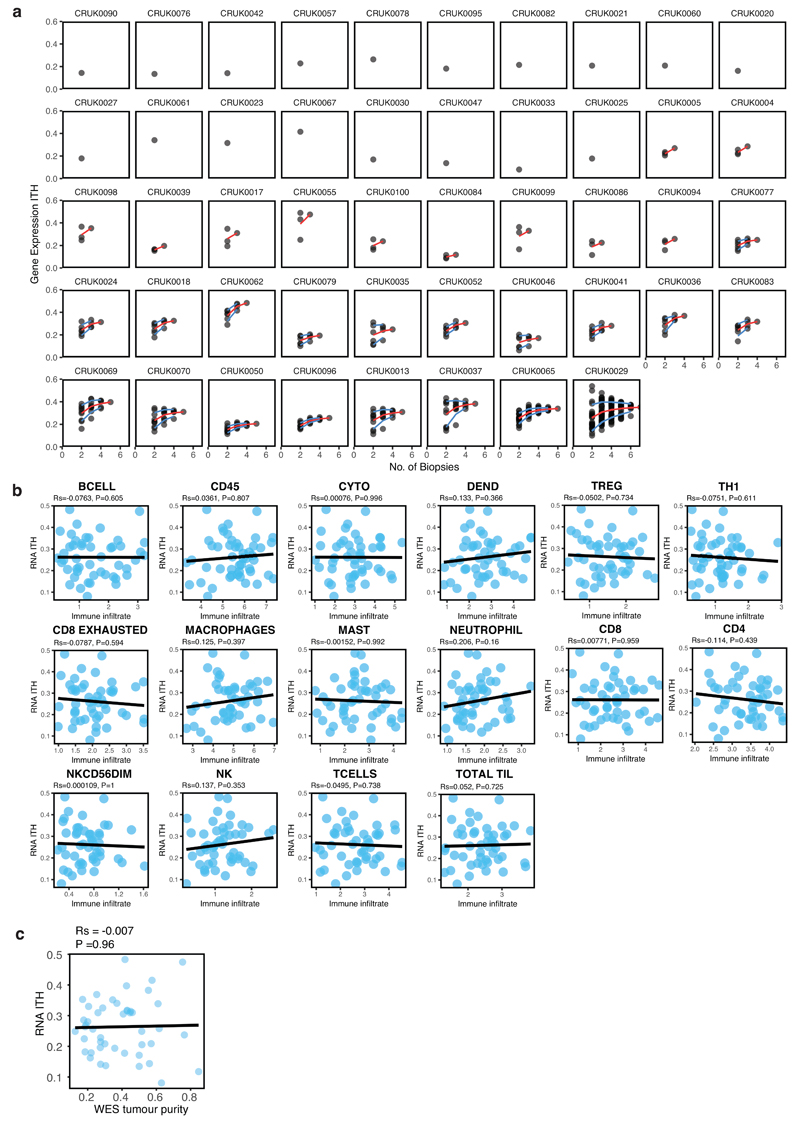
Exploring the biological underpinnings of the ORACLE signature, we observed that ORACLE risk scores increased with primary tumour stage (Uppsala, n=103 LUAD patients, stage I-III) and were significantly higher in metastatic samples (MET500 dataset38, n=8 patients with RNAseq data from primary LUAD tumours) (Extended Data 7d). ORACLE expression also positively correlated with Ki67 staining in TRACERx (89 regions from 28 LUAD patients, stage I-III), a histological marker of cancer cell proliferation (Rs = 0.44, P = 0.0000205, Extended Data 7e). To clarify whether the signature was predominantly expressed in cancer cells, we explored the relationship between ORACLE risk score and metrics of immune infiltration in the TRACERx cohort (89 tumour regions from 28 LUAD patients, stage I-III). There was a significant negative correlation between ORACLE risk scores and most (11/16) immune cell-subsets defined using an RNAseq-based metric of immune infiltration39 (Extended Data 8a) and a non-significant, but trending negative, correlation with a WES-based measure of tumour purity40 (Extended Data 8b). In gene expression “clusters”, previously defined for stromal cells using single-cell RNAseq from lung tumours41, the expression levels of most (20/23) ORACLE genes was negligible compared to known microenvironmental cell-type marker genes (Extended Data 8c). We examined whether there was a relationship in the TRACERx cohort (89 tumour regions from 28 LUAD patients, stage I-III) between the expression of individual ORACLE genes and the tumour copy-number state at the corresponding gene locus, revealing a positive correlation for most (21/23) ORACLE genes (Extended Data 8d). Taken together, these data suggest that ORACLE is derived principally from genes expressed in cancer cells and may serve as a molecular read-out for tumour aggressiveness and metastatic potential.
Next, we investigated whether the design of clonal biomarkers may hold prognostic relevance across other cancer types. To ensure our heterogeneity quadrant approach was not biased to LUAD specific genes, we leveraged the full multi-region RNAseq dataset from TRACERx (n = 48 NSCLC patients), incorporating data from multi-region LUSC tumours and other NSCLC histologies, to calculate NSCLC RNA heterogeneity scores. Using pan-cancer prognostic scores from PRECOG42, a meta-dataset summarizing 166 microarray datasets covering 39 distinct malignant histologies, the proportion of genes that were found to give a pan-cancer significant prognostic value was assessed within each quadrant. Consistent with our analysis in LUAD, genes within Q4 exhibited significantly higher pan-cancer prognostic ability compared to all other quadrants (Q1-vs-Q4 P = 8.9 x 10-27, Q2-vs-Q4 P = 9.3 x 10-08, Q3-vs-Q4 P = 1.9 x 10-18; Fig. 4a). Moreover, we found that Q4 genes were significantly enriched for prognostic genes in 49% (19/39) of cancer types (Fig. 4b, bottom-right panel, indicated in red) and only significantly depleted in head and neck cancer (3% of cancer types, 1/39; Fig. 4b, bottom-right panel, indicated in blue). Conversely, Q1 (high within, low between tumour variability) was significantly depleted in 56% (22/39) of cancer types, but enriched in 0% (0/39). Both Q2 (low within, low between variation) and Q3 (high within, high between variation) showed similar numbers of depleted and enriched cancer types (Fig 4b, Supplementary Table 6). Concordant with this analysis, we found that Q4 exhibited enrichment for prognostic genes across cancer types (Supplementary Table 6) using prognostic scores calculated from 19 distinct malignant histologies, as part of the Human Pathology Atlas study43.
Fig. 4. Pan-cancer prognostic relevance and the genomic underpinning of RNA heterogeneity quadrants.
a, Survival association of RNA heterogeneity quadrants across cancer types. Gene-wise pan-cancer survival associations are evaluated by NSCLC RNA heterogeneity quadrants. Z-scores were sourced from the PRECOG database42 (n = 17,808 tumours from 39 malignant histologies). A |z| score > 1.96 is equivalent to a two-sided P < 0.05. Boxplots represent the median, 25th and 75th percentiles and the vertical bars span the 5th to the 95th percentiles. Statistical significance was tested with a two-sided t-test. b, Survival association of RNA heterogeneity quadrants for individual cancer types. Each point corresponds to 1 out of 33 cancer types sourced from the PRECOG database42 (n = 17,808 tumours from 39 malignant histologies). The number of prognostically significant genes (|z| score > 1.96, equivalent to a P value < 0.05) per NSCLC RNA heterogeneity quadrant is indicated for each cancer type as non-significant (gray), significantly enriched (red, odds ratio > 1) or significantly depleted (blue, odd ratio < 1). Odds ratios are plotted on a natural log scale. Statistical significance was tested with a two-sided Fisher’s exact test. No corrections were made for multiple comparisons. c, Gene expression ITH correlated with copy-number ITH. The scatter plot shows the Spearman correlation between patient-wise RNA-ITH scores and patient-wise SCNA-ITH scores calculated in the TRACERx cohort (n = 28 LUAD patients, stage I-III).d, Association between subclonal chromosomal copy-number changes and gene expression. This analysis was performed using 118,943 paired SCNA and RNA values in TRACERx (143 regions from 44 NSCLC tumours; sample selection criteria described in Methods). Boxplots represent the median, 25th and 75th percentiles and the vertical bars span the 5th to the 95th percentiles. Statistical significance was tested with a two-sided paired t-test. e, Enrichment or depletion of specific copy number states by heterogeneity quadrant. All genes were assigned a copy number state across all samples (clonal/subclonal gain or loss, or no change). Genes were then tested for enrichment or depletion of a specific category by RNA heterogeneity quadrant. Odds ratios are plotted on a natural log scale. Statistical significance was tested with a two-sided Fisher’s exact test. f, Pathway analysis on Q4 genes using Reactome, showing the top 5 pathways most significantly enriched in Q4 genes (low intra- and high inter-tumour heterogeneity).
To explore the mechanisms underpinning RNA-ITH, we calculated tumour-level scores for RNA-ITH (as illustrated in Extended Data 3a) and evaluated how these scores relate to immune infiltration44 and genetic ITH metrics23. To determine the dependence of RNA-ITH on multi-region sequencing, we assessed the effect of varying the number of samples per tumour on the RNA-ITH estimate for each patient. RNA-ITH scores saturate with increasing sample number, plateauing at around four samples for most tumours (Extended Data 9a). Examining the relationship between RNA-ITH and tumour cellular composition in TRACERx NSCLC patients (n = 48 NSCLC patients, stage I-III), RNA-ITH did not correlate with any of the immune cell subsets defined using an RNAseq-based metric of immune infiltration39 (Extended Data 9b), and also did not associate with a WES-based measure of tumour purity40 (Extended Data 9c). By contrast, a significant correlation between the median RNA-ITH score per tumour and somatic copy-number alterations (SCNA) ITH per tumour23 was observed for TRACERx NSCLC tumours (Rs = 0.48, P = 0.0162, Fig. 4c), indicating that SCNA-ITH may contribute to transcriptomic heterogeneity. Consistent with this, we found that subclonal copy-number gains or losses associated with a corresponding change in expression (P < 2.2 x 10-16, Fig. 4d). These data indicate that RNA-ITH may reflect on-going chromosomal instability and the selection of heterogeneous DNA copy number events.
To define the genetic basis for the stable expression of Q4 genes, we assessed the relative enrichment of clonal or subclonal copy-number changes in the genes present within each RNA heterogeneity quadrant (Fig. 4e) using the TRACERx cohort (n = 48 NSCLC patients, stage I-III). We observed a highly significant enrichment of Q4 genes subject to clonal copy number gain events (OR = 1.64, P=1.18 x 10-5), and Q3 genes to a lesser extent (OR = 1.26, P = 1.1 x 10-4), while we observed a depletion of Q2 genes (OR = 0.74, P = 6.86 x 10-8). When we investigated individual SCNA events, we found most were sample-specific and not shared across the cohort. Indeed, the mean percentage of samples showing clonal SCNA for any given Q4 gene was only 12% (data not shown). These data suggest that homogeneous expression across a tumour is likely derived from sample-specific clonal DNA copy number gains, selected early in tumour evolution.
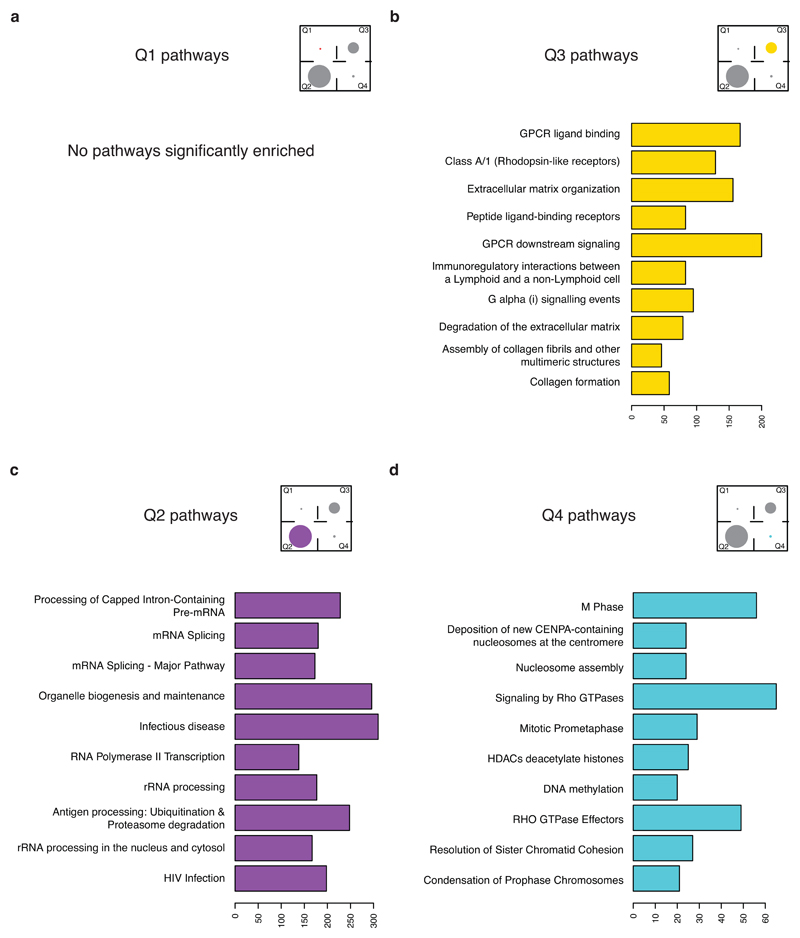
Finally, we investigated whether Q4 genes may also be linked to specific biological features of tumour aggressiveness that might explain their discriminatory prognostic properties. In a Reactome pathway analysis (Extended Data 10, Supplementary Table 7), Q1 (low inter- and high intra-) showed no significant enrichment, the top pathways enriched in Q2 (low inter- and low intra-) showed involvement in RNA splicing processing, and top pathways enriched in Q3 (high inter- and high intra-) showed involvement in GPCR ligand binding and extracellular matrix organization. Notably, Q4 genes (high inter- and low intra-) were significantly enriched for pathways involved in cell proliferation, including mitosis, nucleosome assembly and epigenetic regulation (Fig. 4f).
Overall, these data suggest Q4 genes - which retain uniform expression levels within individual tumours (low intra-tumour heterogeneity) despite ongoing CIN, yet vary greatly between tumours (high inter-tumour heterogeneity) - tend to correlate with clinical outcome and encode cell proliferation modules. Such genes, a subset of which are incorporated into ORACLE, may be optimal candidates for the development of biomarker assays.
Tumour evolution, fostered by ITH, has been shown to bias the application of molecular biomarkers to diagnostic tumour samples2–6, and is an unaddressed confounding factor for biomarker design19–22. Here, we leveraged multi-region RNAseq data from the TRACERx lung study23 to minimize the confounding effects of RNA-ITH in biomarker design. We defined a core set of clonally expressed genes in lung cancer that reproducibly maintain prognostic value in multiple patient cohorts, and are subject to clonal chromosomal gains in genes encoding cell proliferation. In addition, we find RNA-ITH is associated with spatially separated subclonal chromosomal aberrations. Overall, these data suggest the early evolutionary selection for high-risk DNA copy number changes driving proliferation may contribute to poor clinical outcome, with ongoing CIN giving rise to RNA-ITH as a confounding factor for biomarker discovery.
Previous recommendations have suggested that multi-region sequencing may be more informative for prognostication, either by pooling multiple samples per tumour to take the average molecular read-out20, or to identify the “lethal” subclone22 with maximal immune-evasive45 or metastatic46–48 potential. As this is impractical for routine clinical use, we suggest ORACLE as a pragmatic solution that may be applicable to single-region tumour samples if validated in further cohorts.
A subset of stage I LUAD patients have disease relapse following surgery leading to death, yet stage I patients with tumour diameters <4cm do not exhibit improved survival outcomes with adjuvant therapy35,36. Conceivably, this may reflect a limitation of the TNM staging system in accurately assigning risk to stage I patients that could be overcome by biomarker-driven risk stratification1,17. Here ORACLE enabled division of a small cohort of stage I patients (Uppsala cohort, n = 60 LUAD patients) into two groups with differing overall survival risk. A prospective clinical trial would be required to ascertain whether offering adjuvant therapy to the ORACLE high-risk group could improve post-surgical outcomes through the reduction of cancer associated death.
Future analyses may further hone the RNA-ITH metric described here, possibly through explicitly modelling expression levels as a function of spatial coordinates49. The design of clonal biomarkers may be extended, incorporating domain knowledge to focus on a single expression module (such as cell cycle genes15 or immune pathways12) or ensuring the equal representation of multiple expression modules (using manual50 or “blind”51 dimensionality reduction methods). Lastly, we note that existing expression-based predictive biomarkers for checkpoint blockade immunotherapy52,53 exhibit substantial sampling bias in the TRACERx cohort44, possibly indicating that the clonal expression approach developed here could help refine the prediction of patient responses to specific therapies, including those manipulating the immune microenvironment.
Methods
NSCLC Datasets
TRACERx WES and RNAseq
Tumour samples and clinical data were collected from 100 NSCLC patients enrolled in the TRACERx lung cancer study and subjected to complete surgical resection with curative intent23. The TRACERx study (Clinicaltrials.gov no: NCT01888601) is sponsored by University College London (UCL/12/0279) and has been approved by an independent Research Ethics Committee (13/LO/1546). Multi-region sampling was performed to obtain DNA and RNA sequentially from the same tissue. Whole exome sequencing was performed on DNA samples, as described by Jamal-Hanjani et al23. RNA was extracted from the TRACERx 100 cohort using a modification of the AllPrep kit (Qiagen), as previously described23, and RNA integrity was assessed by TapeStation (Agilent Technologies). Of the cohort of 100 TRACERx tumours, RNA samples of sufficient quality (RNA integrity score ≥5) were obtained for 174 regions from 68 tumours and were sent to the Oxford Genomics Centre for whole-RNA (RiboZero depleted) paired-end sequencing. Of these, at least two regions were available from 48 tumours, yielding the TRACERx RNA M-seq cohort (Extended Data 1a). Alignment was performed using the STAR package54 version 2.5.2b to map reads to the human genome (GRCh37/hg19). Transcript expression was quantified using the RSEM package55 version 1.3.0 to generate count and Transcript Per Million (TPM) expression values. An expression filter was applied, keeping genes with an expression value of at least 1 TPM in at least 20% (30/156) of tumour samples in the TRACERx multi-region RNAseq dataset. In total, 16,286 genes were filtered out of the 25,343 unique genes outputted by RSEM. Lastly, a variance stabilizing transformation was applied to counts from filtered genes using the DESeq2 package56 version 1.14.1, assuming a negative binomial distribution for count values, to output homoscedastic and library size normalized count values.
TCGA RNAseq
Pre-processed RNAseq and clinical data were downloaded for 959 NSCLC patients (469 LUAD + 490 LUSC) enrolled in The Cancer Genome Atlas (TCGA) research network lung trials24,25 using the TCGA2STAT package57 version 1.2. An expression filter was applied, keeping genes with at least 0.5 counts per million in at least 2 tumour samples, before normalized count values were obtained for filtered genes using a variance stabilizing transformation from the DESeq2 package56 version 1.14.1.
Uppsala II RNAseq
Pre-processed Uppsala RNAseq and clinical data were downloaded for 170 NSCLC patients (103 LUAD + 67 LUSC) enrolled in the Uppsala NSCLC II cohort26 from the Gene Expression Omnibus (GSE81089). ENSEMBL gene IDs were converted to HGNC IDs using the biomaRt package58 version 2.30.0 and maximum values were selected for multi-mapping probes. Genes, identified as lowly expressed in the TRACERx RNAseq dataset were filtered, then a variance stabilizing transform was applied using the DESeq2 package56 version 1.14.1 to output normalized count values. Additional clinical information (therapy status, patient age, WHO performance status, smoking status, patient gender and Ki67 staining) was provided in private communication with the authors.
Microarray cohorts
Microarray data (.RMA files) and clinical data were downloaded from the Gene Expression Omnibus for four patient cohorts: 442 LUAD patients enrolled by Shedden et al16 (GSE68465); 85 LUAD patients enrolled by Rousseaux et al29 (GSE30219); 147 LUAD patients enrolled by Okayama et al28 (GSE31210); 127 LUAD patients enrolled Der et al30 (GSE50081). Affy IDs were mapped to HGNC IDs, and the “best” probe was selected using the Jetset package59 version 3.4.0.
MET500
Gene expression data was downloaded via dbGaP (accession number phs000673.v2.p1.) for metastatic samples from patients in the MET500 cohort38 with LUAD primary tumours and RNAseq data available (n=8). Alignment was performed using STAR package54 version 2.5.2 to map reads to the human genome (Ensembl GRCh38-release-89). Transcript expression was quantified using the RSEM package55 version 1.3.0 to generate count expression values. Normalized count values were obtained using a variance stabilizing transformation from the DESeq2 package56 version 1.14.1.
Pan-cancer Datasets
PRECOG
Pan-cancer gene-wise prognostic values were downloaded from the PRECOG resource (http://precog.stanford.edu). Gentles et al42 had applied univariate Cox regression to microarray data from ~18,000 tumours across 39 cancer types, quantifying gene-wise survival associations as Z-scores (a |z| score > 1.96 is equivalent to a two-sided P < 0.05).
Human Pathology Atlas
As part of the Human Protein Atlas effort (www.proteinatlas.org/pathology), Uhlen et al43 had calculated gene-wise survival associations as log-rank P values for RNAseq datasets from 17 different cancer types. The pan-cancer gene-wise prognostic values were downloaded as supplementary information from the Uhlen et al43.
LUAD Prognostic Signatures
Literature search
A Pubmed search was performed to identify articles describing prognostic gene expression signatures for LUAD Each article was manually reviewed: if the list of genes comprising the prognostic signature was fully specified, then the signature was included in subsequent analysis. This process yielded two RNAseq signatures12,14, six microarray signatures7–9,13,15, and one qPCR signature10 (Extended Data 5a). Several of the gene names from microarray signatures were updated, to ensure compatibility with RNAseq data (see Supplementary Table 3).
Sampling bias of RNAseq signatures
Two RNAseq LUAD prognostic signatures12,14 were assessed for tumour sampling bias in the TRACERx cohort (n = 28 LUAD patients, stage I-III), by calculating a risk score for each tumour region, then classifying each patient as “concordant low”, “concordant high” or “discordant” survival risk. For the signature described by Shukla et al14, regression coefficients were re-derived from supplementary data provided in the original publication, then applied to TRACERx TPM data, using the risk score cut-off specified in the original publication to stratify tumour regions as “high” or “low” risk. For the signature described by Li et al12, the method to calculate risk scores from “immune-related gene pairs” was applied to TRACERx TPM data, using the median risk score in the TRACERx cohort as the cut-off to stratify tumour regions as “high” or “low” risk.
Sampling bias of RNAseq and non-RNAseq signatures
The sampling bias of nine LUAD prognostic signatures7–15 was assessed in the cohort of TRACERx LUAD patients using a metric invariant of gene expression profiling platform. Hierarchical clustering was performed for each prognostic signature using the Ward method on the Manhattan metric, as in the method described by Gyanchandani et al5. For a given number of clusters, clustering concordance was quantified as the percentage of TRACERx patients with all tumour regions in the same cluster. This analysis was run iteratively from 2 to 28 clusters; 28 is the total number of TRACERx LUAD patients, hence clustering concordance of 100% at 28 clusters is the theoretical upper limit using this metric.
RNA heterogeneity scores
Intra-tumour RNA heterogeneity scores
Gene-wise and patient-wise RNA-ITH scores were calculated using multi-region RNAseq data (normalized count values) from TRACERx tumours. For a given tumour, the standard deviation of expression values for a particular gene across tumour regions was calculated yielding a gene-specific, patient-specific measure of RNA-ITH (σg,p). This was repeated for all genes, then all tumours, generating a matrix of σg,p values (see Extended Data 3a). Gene-wise RNA-ITH values are summarised as the average (median) value per gene across all tumours in the cohort (σg). Conversely, patient-wise RNA-ITH values are summarised as the average (median) value per tumour across all expressed genes (σp). Consideration of median absolute deviation (MAD), and coefficient of variation (CV), as alternative metrics for the quantification of gene-wise RNA-ITH, found these to show good agreement with scores based on the standard deviation (see Extended Data 3b).
Inter-tumour RNA heterogeneity scores
For the TRACERx data-set, an inter-tumour heterogeneity measure is derived for each gene by randomly sampling one region per patient and taking the standard deviation across the resulting single-biopsy cohort, then repeating this process 10 times to take the average score across iterations (see Extended Data 3c). We applied the same method to the TCGA NSCLC data-set, a true single-biopsy cohort, finding good agreement with scores calculated within the TRACERx cohort (PMCC=0.94, P<0.001, Extended Data 3d), which indicates calculation of inter-tumour heterogeneity scores is reproducible.
RNA heterogeneity quadrants
Splitting intra-tumour RNA heterogeneity and inter-tumour RNA heterogeneity by their respective average (mean) value generates RNA heterogeneity quadrants.
Pathway analysis
Pathway enrichment analysis was performed on genes in LUAD Q1-Q4 quadrants using the ReactomePA package60 version 1.24.0. Significance was evaluated based on Bonferroni-adjusted P-value < 0.01.
Random signature analysis
Probe sets were matched to gene symbols using the Jetset package59 version 3.4.0, and all four microarray cohorts were subsetted to the genes present in all four cohorts (7720 genes in total, Q1 = 327, Q2 = 4912, Q3 = 1983, Q4 = 498). From each quadrant, 20 genes were randomly picked, trained in the TCGA RNAseq cohort (n = 469 LUAD patients, stage I-III), and then tested for the ability to reproducibly associate with survival across all 4 microarray cohorts (combined n = 801 LUAD patients, stage I-III). Patients were stratified based on the median value of the first principle component, as in the method described by Venet et al31. This approach was repeated 1000 times.
Copy number analysis
Tumour purity
As previously described for the TRACERx cohort23, tumour purity was quantified by an exome-sequencing based metric generated using ASCAT40.
SCNA calling
As previously described for the TRACERx cohort23, segmented allele-specific copy number states were defined based on the WES data. To determine genome-wide copy number gain and loss, copy number data for each sample was divided by the sample mean ploidy, and log2-transformed. Gain and loss were defined as log2(2.5/2) and log2(1.5/2), respectively. Gene-level clonal copy number gain or loss was defined as all regions from an individual tumour showing either gain or loss, in the same direction. Gene-level subclonal copy number gain or loss was defined as at least one region but not all regions from an individual tumour showing copy number gain or loss. Only subclonally gained or lost copy number segments were used to analyse the effect of copy number alterations on gene expression. To ensure proper copy-number variation, only samples with an absolute copy number difference of 0.5 on a log2 scale were included.
Linking subclonal SCNA to gene expression changes
We identified genes with a heterogeneous copy number state between regions of an individual tumour. We then examined the paired RNAseq data for evidence of an expression difference between copy number aberrant and non-aberrant tumour regions for the corresponding transcripts by subtracting the log2 expression value of non-aberrant genes from the aberrant genes. Statistical significance was tested with a two-sided paired t-test. SCNA-ITH scores were determined as previously described for the TRACERx cohort23.
Enrichment of SCNA per heterogeneity quadrant
To determine enrichment of genes with clonal gain across all four heterogeneity quadrants, we first classify all genes within an individual tumour as either “clonal gain” or “not clonal gain”, based on whether or not the specific gene demonstrate copy number gain across all tumour regions. For each gene, we determine the percentage of samples that demonstrate clonal gain, then determine if the top 25% of genes most commonly subjected to clonal copy number gain are enriched for genes in each heterogeneity quadrant relative to the 25% of genes least commonly subjected to clonal copy number gain. Statistical significance was tested with a two-sided Fisher’s exact test.
Prognostic signature construction
Stepwise regression
A previously published prognostic signature construction pipeline, described by Shukla et al14, was replicated. In the training cohort (TCGA, n = 469 LUAD patients, stage I-III) univariate Cox regression analysis was performed, then a primary prognostic filter was applied (univariate Cox analysis P < 0.00025) identifying 108 genes to take forward for signature construction. Next, a secondary prognostic filter (univariate Cox analysis FDR < 0.02) identified 15 genes that were taken as input for forward conditional stepwise (AIC) regression, yielding a 6-gene prognostic signature (“Signature A”); alternatively, stepwise (BIC) regression yielded a 3-gene signature (“Signature B”). In parallel, a secondary “clonal expression” filter (selecting Q4 genes using heterogeneity scores calculated in TRACERx, n = 28 LUAD patients, stage I-III) also identified 15 genes, yielding a 7-gene signature by stepwise (AIC) regression (“Signature A-clonal”); stepwise (BIC) regression generated a 6-gene signature (“Signature B-clonal”). In the validation cohort (Uppsala, n = 103 LUAD patients, stage I-III), a linear combination of gene expression values, weighted by stepwise regression coefficients, was used to calculate a risk score for each patient. Patients were classified as “high” or “low” risk using the median risk score as a cut-off value. Stepwise regression was performed using the MASS package (https://CRAN.R-project.org/package=MASS) version 7.3.48 to select a prognostic signature by the Akaike Information Criterion (“Signature A” and “Signature A-clonal”) or by the Bayesian Information Criterion (“Signature B” and “Signature B-clonal”).
Tree classification
A previously published prognostic signature construction pipeline, described by Chen et al33, was replicated. In the training cohort (TCGA, n = 469 LUAD patients, stage I-III), genes reported to be associated with invasive activity61 (656 could be recovered with HUGO gene symbols out of 672 genes reported in the original study) were selected as a primary prognostic filter. Next, a secondary prognostic filter was used (univariate Cox analysis P < 0.00005), as in the original study33, selecting the 8 genes with highest prognostic significance in the training cohort. These were taken as input for tree classification, yielding a 8-gene prognostic signature (“Signature C”). In parallel, a secondary “clonal expression” filter (selecting Q4 genes using heterogeneity scores calculated in TRACERx, n = 28 LUAD patients, stage I-III) also identified 9 genes, yielding a 9-gene signature by tree classification (“Signature C-clonal”). In the validation cohort (Uppsala, n = 103 LUAD patients, stage I-III), patients were classified as “high” or “low” risk using predictions from the tree models. Tree classification was performed using the rpart package (https://CRAN.R-project.org/package=rpart) version 4.1.13.
Random forest regression
A previously published prognostic signature construction pipeline, described by Reka et al34, was replicated. First, 97 genes associated with an EMT secretory phenotype, listed by Reka et al34, were selected in the training cohort (TCGA, n = 469 LUAD patients, stage I-III) as a primary prognostic filter. A random forest model was fitted using all 97 genes, then the resulting variable importance scores were used as a secondary prognostic filter to select and re-fit a model using the top 10 most informative genes, yielding a 10-gene prognostic signature (“Signature D”). In parallel, a secondary “clonal expression” filter (selecting Q4 genes using heterogeneity scores calculated in TRACERx, n = 28 LUAD patients, stage I-III) identified 9 genes, yielding a 9-gene signature by random forest regression regression (“Signature D-clonal”). The random forest models were then used to calculate a risk score for each patient in the validation cohort (Uppsala, n = 103 LUAD patients, stage I-III). Patients were classified as “high” or “low” risk using the median risk score as a cut-off value. Random forest regression is performed using the randomForestSRC package62 version 2.5.1.
Elastic-net (lasso) regression
A previously published prognostic signature construction pipeline, described by Kratz et al10, was replicated. First, a list of genes (249 genes) was collated from previously published LUAD prognostic signatures7–15; these genes were selected in the training cohort (TCGA, n = 469 LUAD patients, stage I-III) as a primary prognostic filter. Next, a short-list of cancer-related genes - previously identified by manual review10,13 - was used as a secondary prognostic filter, identifying 56 genes for input to lasso regression, which then yielded a 24-gene prognostic signature (“Signature E”). In parallel, a secondary “clonal expression” filter (selecting Q4 genes using heterogeneity scores calculated in TRACERx, n = 28 LUAD patients, stage I-III) identified 44 genes, yielding a 14-gene signature by lasso regression (“Signature E-clonal”). In the validation cohort (Uppsala, n = 103 LUAD patients, stage I-III), a linear combination of gene expression values, weighted by lasso regression coefficients, was used to calculate a risk score for each patient. Patients were classified as “high” or “low” risk using the median risk score as a cut-off value. Elastic-net regression is performed using the glmnet package63 version 2.0.13 applying the lasso penalty (alpha=1).
ORACLE signature
An analysis pipeline was developed to select genes that are both prognostic and clonally expressed on the basis of four criteria (Extended Data 6b). First, starting with all expressed genes (19,024 genes) in the TCGA cohort (n = 469 LUAD patients, stage I-III), genes with a below median expression were removed (9,512/19,024 genes). This expression threshold is commonly used as a pre-processing step in cancer expression studies64, as lowly expressed genes are more vulnerable to noise due to the detection limits of expression profiling technologies. Second, significantly prognostic genes (2,023/9,512) were identified as genes correlated with overall survival (Cox univariate P-value < 0.05, not corrected for multiple hypothesis testing), a standard method for identifying significantly prognostic genes14,33. Third, to restrict RNA-ITH, Q4 genes were selected (176/2,023 genes) using heterogeneity scores calculated in TRACERx (n = 28 LUAD patients, stage I-III). Fourth, we scored the “clustering concordance” of each gene (Extended Data 4a) and picked the highest scoring genes (90/176 genes) as an additional step to select clonally expressed genes. The optimal number of genes to take forward at this step was determined using ten-fold cross-validation in the training cohort (Extended Data 6c). Using the genes selected by these four criteria, a prognostic signature was generated through elastic-net (lasso) regression against overall survival. This machine learning algorithm performs further gene selection, and also calculates a model coefficient for each gene, yielding an Outcome Risk Associated Clonal Lung Expression signature (23/90 genes, Supplementary Table 5). As is standard for prognostic signatures with an underlying linear model, a molecular risk score was calculated for each patient as a linear combination of gene expression values, weighted by the model coefficients fitted in the training cohort. To dichotomise the continuous risk score variable, and classify patients as “high” or “low” molecular risk, a threshold was determined in the training cohort as the average (median) risk score value amongst significant (log-rank P < 0.01) cut-off values (Extended Data 6d). To the best of our knowledge, only 30% of the list of genes comprising ORACLE (7/23 genes, Supplementary Table 5) has previously been used in LUAD prognostic signatures (ASPM, FURIN, PLK1, PNP, PRKCA, PTTG1, TPBG). For the meta-analysis univariate Cox regression was performed on ORACLE risk scores determined in the Uppsala RNAseq cohort and in the 4 microarray cohorts (Fig. 3b). In the microarray cohorts, 19/23 genes were available for analysis (ASPM, CDCA4, FURIN, GOLGA8A, ITGA6, JAG1, LRP12, MAFF, MRPS17, PLK1, PNP, PPP1R13L, PRKCA, PYGB, SCPEP1, SLC46A3, SNX7, TPBG, XBP1). Weights applied to ORACLE genes were not modified across cohorts. Meta-analysis was performed using the rmeta package (https://CRAN.R-project.org/package=rmeta) version 3.0.
Stromal expression
Immune infiltration scores
Bulk RNAseq data from TCGA and the TRACERx cohort was used to calculate infiltration scores for 16 immune subsets using the method described by Danaher et al39.
scRNAseq data
Lambrechts et al performed single-cell RNAseq on 52,698 cells sourced from 5 NSCLC patients, then defined 7 clusters of stromal cell genes and provided a per-cluster expression measure for every gene. Gene-wise relative expression levels were downloaded as supplementary information from Lambrechts et al41.
Immunohistochemistry data
Ki67 staining
As previously described for the TRACERx cohort23, staining was performed for Ki67.
Statistical analysis
All statistical tests were performed in R version 3.3.1. No statistical methods were used to predetermine sample size. Tests involving correlations were done using ‘cor.test’ with either the Spearman’s method or Pearson’s method as specified. Tests involving comparisons of distributions were done using ‘wilcox.test’ or ‘t.test’ as stated. Hazard ratios and P values were calculated using the survival package (https://CRAN.R-project.org/package=survival) version 2.41.3, through univariate or multivariate Cox regression analyses as stated. Kaplan-Meier plots were generated using the survminer package (https://CRAN.R-project.org/package=survminer) version 0.4.2. All statistical tests are two-sided, unless otherwise stated, and the number of data points included are plotted and/or annotated in the corresponding figure.
Extended Data
Extended Data 1. Patient cohorts included in the study.
a, CONSORT diagram for patient recruitment (left) and composition by tumour stage (right) of the TRACERx cohort. b, Patient composition of two RNAseq datasets: The Cancer Genome Atlas cohort (left), and the Uppsala cohort (right). c, Patient composition of four microarray datasets: Der et al, GSE50081 (top left); Okayama et al, GSE31210 (top right); Rousseaux et al, GSE30219 (bottom left); Shedden et al, GSE68465 (bottom right). Tumour stage (x-axis) and therapy status (colour) is indicated for all patient composition bar charts. LUAD = lung adenocarcinoma, LUSC = lung squamous cell carcinoma.
Extended Data 2. Analysis of the most variably expressed genes in TRACERx.
a, The dendrogram and coloured heatmap (top) shows the hierarchical clustering of tumour regions (columns) in the TRACERx multi-region RNAseq cohort (156 tumour regions, 48 NSCLC patients, stage I-III) according to the top 500 variably expressed genes (rows). The sparse heatmap (bottom) shows tumour regions (coloured by histology) per patient (rows). b, Kaplan-Meier survival analysis of the largest two patient clusters from the dendrogram in (a). Statistical significance was tested with a two-sided log-rank test. c, The hierarchical clustering approach taken to quantify discordance rates for published signatures is illustrated for a non-RNAseq signature, Kratz et al10, in TRACERx (n = 28 LUAD patients, stage I-III). As previously described by Gyanchandani et al5, this clustering approach provides a metric that is invariant of gene expression profiling platform. For a given number of clusters, clustering concordance was quantified as the percentage of TRACERx patients with all tumour regions in the same cluster. This analysis was run iteratively from 2 to 28 clusters; 28 is the total number of TRACERx LUAD patients, hence clustering concordance of 100% at 28 clusters is the theoretical upper limit using this metric. The dendrogram and coloured heatmap (top) shows the clustering of tumour regions (columns) according to the expression pattern of genes comprising the prognostic signature (rows). The grayscale heatmap (bottom left) shows tumour regions per patient (rows). For a range of clusters (2, 3, 14, 28), the coloured bars (middle left) show the assignment of tumour regions to clusters, the grayscale bars (bottom right) show which patients have their tumour regions discordantly assigned (gray) across clusters, and the pie charts (middle right) show the percentage of discordantly classified patients. d, Discordance rates for 9 published LUAD prognostic signatures7–15 plotted as the percentage of patients with tumour regions clustering together against the number of clusters. Vertical dashed lines mark a range of clusters (2, 3, 14, 28) as highlighted in (c).
Extended Data 3. Intra- and inter-tumour RNA heterogeneity scores.
a, Gene-wise and patient-wise RNA-ITH scores were calculated using multi-region RNAseq data (normalized count values) from TRACERx tumours (n=28 LUAD patients, 89 tumour regions, stage I-III). For a given tumour, the standard deviation of expression values for a particular gene across tumour regions was calculated yielding a gene-specific, patient-specific measure of RNA-ITH (σg,p). This was repeated for all genes, then all tumours, generating a matrix of σg,p values. Gene-wise RNA-ITH values are summarised as the average (median) value per gene across all tumours in the cohort (σg). Conversely, patient-wise RNA-ITH values are summarised as the average (median) value per tumour across all expressed genes (σp). Dashed lines indicate mean values. b, The scatter plots show the Spearman correlation between the chosen metric of intra-tumour expression variability (standard deviation) and alternative metrics, median absolute deviation (left) or coefficient of variation (right), as calculated in the TRACERx cohort (n=28 LUAD patients, 89 tumour regions, stage I-III). c, Diagram illustrating the calculation of gene-wise inter-tumour RNA heterogeneity scores through the random sampling of tumour regions from the TRACERx cohort (n=28 LUAD patients, 89 tumour regions, stage I-III; see Methods). d, The scatter plot shows the Spearman correlation between inter-tumour RNA heterogeneity scores calculated in TRACERx (n=28 LUAD patients, 89 tumour regions, stage I-III), randomly sampled to yield a sham single-biopsy cohort, and TCGA (n = 469 LUAD patients, stage I-III), a true single-biopsy cohort.
Extended Data 4. Clustering concordance and published prognostic signatures.
a, Clustering concordance scores calculated in TRACERx (n=28 LUAD patients, 89 tumour regions, stage I-III) using the same method taken to estimate the sampling bias of microarray signatures as described by Gyanchandani et al5 (see Extended Data 2c-d). For each gene, a curve is calculated for the number of patients with all regions in the same cluster against the number of clusters (2-28 clusters). Curves for five genes (minimum = CKMT2, lower quartile = CYSLTR2, median = MCM2, upper quartile = MFSD1, maximum = HOXC11) are shown (top), in addition to summarised clustering concordance scores for all genes (bottom). b, Gene-wise clustering concordance scores stratified by RNA heterogeneity quadrant, both calculated in TRACERx (n=28 LUAD patients, 89 tumour regions, stage I-III). Boxplots represent the median, 25th and 75th percentiles and the vertical bars span the 5th to the 95th percentiles. Statistical significance was tested with a two-sided Wilcoxon signed rank sum test. “*” indicates a P-value < 0.05, “**” indicates a P-value < 0.01, “***” indicates a P-value < 0.001.
Extended Data 5. Analysis of published prognostic signatures for LUAD by RNA heterogeneity quadrant.
a, The composition of published prognostic signatures by RNA heterogeneity quadrant, plotted in order of increasing percentage of Q4 genes (low intra- and high inter-tumour heterogeneity). b, Percentage of genes expected (total no. genes, as indicated in Fig. 2a) versus observed (in 9 published LUAD prognostic signatures7–15) per RNA heterogeneity quadrant. Statistical significance was tested with a two-sided Fisher’s exact test. The ability of published prognostic genes for LUAD (the combined gene list from nine published signatures, 242 unique genes) to maintain prognostic value across patient cohorts is assessed (using Cox univariate survival analysis) in four microarray datasets: Shedden et al, GSE68465 (c); Okayama et al, GSE31210 (d); Der et al, GSE50081 (e); Rousseaux et al, GSE30219 (f). Boxplots represent the median, 25th and 75th percentiles and the vertical bars span the 5th to the 95th percentiles. Statistical significance was tested with a two-sided Wilcoxon signed rank sum test. “*” indicates a P-value < 0.05, “**” indicates a P-value < 0.01, “***” indicates a P-value < 0.001.
Extended Data 6. Prognostic signature design.
a, Biomarkers are designed using state-of-the-art signature construction methods, replicated from Shukla et al14 (signature A and B), Chen et al33 (signature C), Reka et al34 (Signature D) and Kratz et al10 (signature E). In parallel, the “prognostic significance” filters (present in each signature construction method) were substituted with “clonal expression” filters, generating corresponding clonal signatures (signatures A-clonal, B-clonal, C-clonal, D-clonal, and E-clonal). Published signature construction methods are indicated in orange, novel methods integrating clonal biomarker design are indicated in blue. All signatures are developed in TCGA LUAD patients (n=469, stage I-III) as the training dataset. b, Flow diagram illustrating the gene selection steps for ORACLE. Criteria to identify prognostic and clonally expressed genes, and the number of genes selected at each step are indicated. c, Optimization of the number of genes to select at the clustering concordance step through 10-fold cross-validation in the training cohort (TCGA, n=469 LUAD patients, stage I-III). The optimal number of genes, with the lowest cross-validation error, is shown by the vertical red line. d, The cut-off to dichotomize the ORACLE risk-score into ‘high’ and ‘low’ risk groups is optimized in the training cohort (TCGA, n=469 LUAD patients, stage I-III). The horizontal blue line indicates a log-rank P-value = 0.01 and the optimal cut-off is shown by the vertical red line. Statistical significance was tested with a two-sided log-rank test. e, Tumour sampling bias of the ORACLE signature assessed using multi-region RNAseq data from TRACERx (n=28 LUAD patients, 89 tumour regions, stage I-III). Each point represents a single tumour region, vertical lines display the range for each patient, and patients are ordered by predicted survival risk score. Points are coloured according to the risk classification of tumour regions within a patient: concordant low-risk (blue), concordant high-risk (red), or discordant (gray).
Extended Data 7. Risk stratification using ORACLE.
a, Kaplan-Meier plot of ORACLE in the RNAseq-based validation cohort (Uppsala, n=103 LUAD patients, stage I-III). Statistical significance was tested with a two-sided log-rank test. The ability of substaging criteria (b) and ORACLE (c) to split patients into prognostically informative groups is tested in stage I patients using the updated TNM version 8 criteria37, shown as Kaplan-Meier plots for the Uppsala RNAseq dataset (n=53 LUAD patients, stage I, TNMv8). Statistical significance was tested with a two-sided log-rank test. d, The distribution of ORACLE risk scores by disease stage, shown for the Uppsala cohort (n=103 LUAD patients, stage I-III) and the MET500 cohort38 (n=8 metastatic samples from patients with LUAD primary tumours). Boxplots represent the median, 25th and 75th percentiles and the vertical bars span the 5th to the 95th percentiles. Statistical significance was tested with a Wilcoxon signed rank sum test. No corrections were made for multiple comparisons. e, The scatter plot shows the Spearman correlation between Ki67 staining % and ORACLE risk-scores in the TRACERx cohort (n=28 LUAD patients, 89 tumour regions, stage I-III).
Extended Data 8. ORACLE as a cancer cell expression signature.
a, Spearman correlations between the infiltration of immune cell subsets, calculated from RNAseq data using the method described by Danaher et al39, and ORACLE risk-scores in the TCGA dataset (n=469 patients, stage I-III). b, The scatter plot shows the Spearman correlation between ORACLE risk score and tumour purity assessed from whole-exome sequencing data using ASCAT, as described by Van Loo et al40, in TRACERx (n=28 LUAD patients, 84 tumour regions, stage I-III). c, Lambrechts et al41 performed single-cell RNAseq on 52,698 cells sourced from 5 NSCLC patients, then defined 7 clusters of stromal cell genes and provided a per-cluster expression measure for every gene. The relative expression levels (y-axis) for each stromal cluster (coloured by cell-type, see figure legend) is plotted for all 23 genes comprising the ORACLE signature (bottom 3 rows). To aid interpretation, a marker gene for each of the 7 stromal cell clusters is also plotted (top row) for comparison: alveolar (AGER), B cell (MS4A1), epithelial (EPCAM), fibroblast (COL6A2), myeloid (CD68), T cell (CD3D), and vascular (FLT1) cell-types. d, Pearson correlations between the expression of individual ORACLE genes and copy-number state at the corresponding gene locus in the TRACERx cohort (n=28 LUAD patients, 89 tumour regions, stage I-III). Significant correlations (P<0.05) are marked in red, non-significant correlations are marked in blue.
Extended Data 9. Patient-level estimates of RNA-ITH and association with tumour cellular composition.
a, RNA-ITH scores calculated from each tumour by sampling one to N biopsies (where N is the total number of biopsies yielded by that tumour) in TRACERx (n=48 NSCLC patients, 156 tumour regions, stage I-III). For each patient the RNA-ITH score (y-axis) is plotted for all possible subgroups of tumour regions against the number of biopsies (x-axis). The mean (red line) and standard deviation (blue lines) are shown for each tumour. b, The scatter plots show the Spearman correlation between patient-level RNA-ITH scores and RNAseq-based immune infiltration measures, calculated from RNAseq data using the method described by Danaher et al39 in TRACERx (n=48 NSCLC patients, 156 tumour regions, stage I-III). c, The scatter plot shows the Spearman correlation between patient-level RNA-ITH scores and tumour purity assessed from whole-exome sequencing data using ASCAT, as described by Van Loo et al40, in TRACERx (n=48 NSCLC patients, 156 tumour regions, stage I-III).
Extended Data 10. Pathway analysis by RNA heterogeneity quadrant.
The top 10 Reactome pathways for each RNA heterogeneity quadrant are plotted: low inter- and high intra- (Q1, a), low inter- and low intra- (Q2, b), high inter- and high intra- (Q3, c), high inter- and low intra- (Q4, d).
Supplementary Material
Acknowledgements
D.B. is a recipient of a Jean Shanks Foundation MBPhD studentship and also receives funding from the MBPhD programme at University College London, and the NIHR BRC at University College London Hospitals. N.J.B. is a fellow of the Lundbeck Foundation and acknowledges funding from the Aarhus University Research Foundation and the Danish Cancer Society. J.M., B.D. and J.F. are supported by the Hungarian Science Foundation (OTKA-K129065). I.C. is supported by NVKP_16–1–2016-0004. Z.S. is supported by NAP2-2017-1.2.1-NKP-0002 and the Breast Cancer Research Foundation (BCRF-18-159). N.M. is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (Grant Number 211179/Z/18/Z), and also receives funding from CRUK, Rosetrees, and the NIHR BRC at University College London Hospitals. C.S. is Royal Society Napier Research Professor. This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001169, FC001202), the UK Medical Research Council (FC001169, FC001202), and the Wellcome Trust (FC001169, FC001202). C.S. is funded by Cancer Research UK (TRACERx and CRUK Cancer Immunotherapy Catalyst Network), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees Trust, Butterfield and Stoneygate Trusts, NovoNordisk Foundation (ID16584), the Prostate Cancer Foundation, the Breast Cancer Research Foundation (BCRF). The research leading to these results has received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) Consolidator Grant (FP7-THESEUS-617844), European Commission ITN (FP7-PloidyNet 607722), ERC Advanced Grant (PROTEUS) has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 835297), Chromavision – this project has received funding from the European’s Union Horizon 2020 research and innovation programme (grant agreement No. 665233). Support was also provided to C.S. by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre, and the Cancer Research UK University College London Experimental Cancer Medicine Centre.
Footnotes
Data availability.
Sequence data used during the study Sequence data used during the study are available through the Cancer Research UK & University College London Cancer Trials Centre (ctc.tracerx@ucl.ac.uk) for noncommercial research purposes, and access will be granted upon review of a project proposal that will be evaluated by a TRACERx data access committee and entering into an appropriate data access agreement subject to any applicable ethical approvals.
Code availability.
Code is available at: https://github.com/dhruvabiswas/tracerx-oracle.
Author Contributions
D.B. and N.J.B. conceived the project, designed experiments, performed bioinformatics analyses and wrote the manuscript. R.R., E.L.L., K.P., S.B., M.K., T.B.K.W. and G.A.W. performed data processing and bioinformatics analyses. C.T.H., Y.W., D.A.M., M.S., C.A. and M.A.B. gave advice on clinical interpretation. D.D., L.L.F., M.G., B.D., J.F., H.B. and J.M. performed sample collection, curated clinical data and helped with interpretation. K.L., I.C., Z.S. and J.H. helped to direct avenues of bioinformatics analysis. S.V. performed sample preparation and RNA extraction. M.J.-H. designed TRACERx study protocols and helped to analyse patient clinical characteristics. J.Botling, A.M.C., P.M. and J.Bartek. provided access to additional RNAseq datasets and gave feedback on the manuscript. A.H. provided statistical advice. N.M. and C.S. conceived the project, designed experiments and helped to write the manuscript. N.J.B., N.M. and C.S. supervised the study. All authors reviewed and approved the manuscript.
Competing Interests
C.S. receives grant support from Pfizer, AstraZeneca, BMS, Roche and Ventana. C.S. has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, BMS, Celgene, AstraZeneca, Illumina, Genentech, Roche-Ventana, GRAIL, Medicxi, the Sarah Cannon Research Institute and is an Advisor for Dynamo Therapeutics. C.S. is a shareholder of Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options in and is co-founder of Achilles Therapeutics. R.R. has stock options in and has consulted for Achilles Therapeutics. C.A. has received speaking honorarium or expenses from Novartis, Roche, AstraZeneca and BMS. M.A.B. has consulted for Achilles Therapeutics. G.A.W. is a shareholder of Achilles Therapeutics. M.J.-H. has consulted for and is an advisor for Achilles Therapeutics. D.B., N.J.B., N.M., and C.S. are co-inventors on a UK patent application (1901439.8) filed by Cancer Research Technology relating to methods of predicting survival rates for cancer patients.
References
- 1.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer. 2016;16:525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee W-C, et al. Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer. Mod Pathol. 2018:1. doi: 10.1038/s41379-018-0029-3. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gulati S, et al. Systematic Evaluation of the Prognostic Impact and Intratumour Heterogeneity of Clear Cell Renal Cell Carcinoma Biomarkers. Eur Urol. 2014;66:936–948. doi: 10.1016/j.eururo.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gyanchandani R, et al. Intratumor Heterogeneity Affects Gene Expression Profile Test Prognostic Risk Stratification in Early Breast Cancer. Clin Cancer Res. 2016;22:5362–5369. doi: 10.1158/1078-0432.CCR-15-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulati S, Turajlic S, Larkin J, Bates PA, Swanton C. Relapse models for clear cell renal carcinoma. Lancet Oncol. 2015;16:e376–e378. doi: 10.1016/S1470-2045(15)00090-X. [DOI] [PubMed] [Google Scholar]
- 7.Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi F, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117:3436–3444. doi: 10.1172/JCI32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber ME, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kratz JR, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. The Lancet. 2012;379:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzystanek M, Moldvay J, Szüts D, Szallasi Z, Eklund AC. A robust prognostic gene expression signature for early stage lung adenocarcinoma. Biomark Res. 2016;4:4. doi: 10.1186/s40364-016-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Cui Y, Diehn M, Li R. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non–Small Cell Lung Cancer. JAMA Oncol. 2017 doi: 10.1001/jamaoncol.2017.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raz DJ, et al. A Multigene Assay Is Prognostic of Survival in Patients with Early-Stage Lung Adenocarcinoma. Clin Cancer Res. 2008;14:5565–5570. doi: 10.1158/1078-0432.CCR-08-0544. [DOI] [PubMed] [Google Scholar]
- 14.Shukla S, et al. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. JNCI J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wistuba II, et al. Validation of a Proliferation-Based Expression Signature as Prognostic Marker in Early Stage Lung Adenocarcinoma. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shedden K, et al. Gene expression–based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian J, Simon R. Gene Expression–Based Prognostic Signatures in Lung Cancer: Ready for Clinical Use? JNCI J Natl Cancer Inst. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 19.Boutros PC. The path to routine use of genomic biomarkers in the cancer clinic. Genome Res. 2015;25:1508–1513. doi: 10.1101/gr.191114.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackhall FH, et al. Stability and Heterogeneity of Expression Profiles in Lung Cancer Specimens Harvested Following Surgical Resection. Neoplasia N Y N. 2004;6:761–767. doi: 10.1593/neo.04301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachtiary B, et al. Gene Expression Profiling in Cervical Cancer: An Exploration of Intratumor Heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 22.Barranco SC, et al. Intratumor Variability in Prognostic Indicators May Be the Case of Conflicting Estimates of Patient Survival and Response to Therapy. Cancer Res. 1994;54:5351–5356. [PubMed] [Google Scholar]
- 23.Jamal-Hanjani M, et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djureinovic D, et al. Profiling cancer testis antigens in non–small-cell lung cancer. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstraw P, et al. The IASLC Lung Cancer Staging Project: Proposals for the Revision of the TNM Stage Groupings in the Forthcoming (Seventh) Edition of the TNM Classification of Malignant Tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 28.Okayama H, et al. Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-Negative Lung Adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 29.Rousseaux S, et al. Ectopic Activation of Germline and Placental Genes Identifies Aggressive Metastasis-Prone Lung Cancers. Sci Transl Med. 2013;5:186ra66–186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Der SD, et al. Validation of a Histology-Independent Prognostic Gene Signature for Early-Stage, Non–Small-Cell Lung Cancer Including Stage IA Patients. J Thorac Oncol. 2014;9:59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 31.Venet D, Dumont JE, Detours V. Most Random Gene Expression Signatures Are Significantly Associated with Breast Cancer Outcome. PLOS Comput Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang H, et al. Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies. Ann Oncol. 2017;28:733–740. doi: 10.1093/annonc/mdw683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H-Y, et al. A Five-Gene Signature and Clinical Outcome in Non–Small-Cell Lung Cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 34.Reka AK, et al. Epithelial–mesenchymal transition-associated secretory phenotype predicts survival in lung cancer patients. Carcinogenesis. 2014;35:1292–1300. doi: 10.1093/carcin/bgu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauss GM, et al. Adjuvant Paclitaxel Plus Carboplatin Compared With Observation in Stage IB Non–Small-Cell Lung Cancer: CALGB 9633 With the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pignon J-P, et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 37.Goldstraw P, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017 doi: 10.1038/nature23306. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danaher P, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loo PV, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018:1. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 42.Gentles AJ, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhlen M, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. eaan2507. [DOI] [PubMed] [Google Scholar]
- 44.Rosenthal R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019:1. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mlecnik B, et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. JNCI J Natl Cancer Inst. 2018;110:97–108. doi: 10.1093/jnci/djx123. [DOI] [PubMed] [Google Scholar]
- 46.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim T-M, et al. Subclonal Genomic Architectures of Primary and Metastatic Colorectal Cancer Based on Intratumoral Genetic Heterogeneity. Clin Cancer Res. 2015;21:4461–4472. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed] [Google Scholar]
- 49.Svensson V, Teichmann SA, Stegle O. SpatialDE: identification of spatially variable genes. Nat Methods. 2018 doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang H, et al. A 12-Gene Set Predicts Survival Benefits from Adjuvant Chemotherapy in Non–Small Cell Lung Cancer Patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleary B, Cong L, Cheung A, Lander ES, Regev A. Efficient Generation of Transcriptomic Profiles by Random Composite Measurements. Cell. 2017 doi: 10.1016/j.cell.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018:1. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hugo W, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan Y-W, Allen GI, Liu Z. TCGA2STAT: simple TCGA data access for integrated statistical analysis in R. Bioinforma Oxf Engl. 2016;32:952–954. doi: 10.1093/bioinformatics/btv677. [DOI] [PubMed] [Google Scholar]
- 58.Durinck S, Spellman PT, Birney E, Huber W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G, He Q-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol BioSyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 61.Chen JJW, et al. Global Analysis of Gene Expression in Invasion by a Lung Cancer Model. Cancer Res. 2001;61:5223–5230. [PubMed] [Google Scholar]
- 62.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 63.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell JD, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48 doi: 10.1038/ng.3564. ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.