Abstract
Feynman commented that “Everything that living things do can be understood in terms of the jiggling and wiggling of atoms”. Proteins can jiggle and wiggle large structural elements such as domains and subunits as part of their functional cycles. Single-molecule fluorescence resonance energy transfer (smFRET) is an excellent tool to study conformational dynamics and decipher coordinated large-scale motions within proteins. smFRET methods introduced in recent years are geared toward understanding the time scales and amplitudes of function-related motions. This review discusses the methodology for obtaining and analyzing smFRET temporal trajectories that provide direct dynamic information on transitions between conformational states. It also introduces correlation methods that are useful for characterizing intramolecular motions. This arsenal of techniques has been used to study multiple molecular systems, from membrane proteins through molecular chaperones, and we examine some of these studies here. Recent exciting methodological novelties permit revealing very fast, submillisecond dynamics, whose relevance to protein function is yet to be fully grasped.
Keywords: Protein conformational dynamics, Allostery, Molecular machines, Single-molecule FRET, Fluorescence correlation spectroscopy
Introduction
Living systems are physical entities marked by an out-of-equilibrium evolution of their microscopic degrees of freedom over time and characterized by the ability to harness energy from the environment to generate order out of disorder [1]. Proteins are the key functional molecules in the living system, governing nearly all cellular functions and biochemical tasks. These may include regulating ion passage across the cell membrane, chaperoning protein folding, transducing signals, carrying them into the cell nucleus, transcribing and translating DNA, catalyzing chemical reactions, and many other important cellular functions [2–5]. Remarkably, protein machines are tightly regulated and are coupled to cellular function in a spatial and temporal way. Studying the structural dynamics of protein machines is essential for deciphering how they function and in what way they are regulated. The knowledge acquired through such studies is relevant for engineering new proteins and is also important for drug discovery.
The amino acid sequence of a protein dictates its three-dimensional structure, which in turn controls its function [6,7]. Much energy has been invested in understanding protein structure and function since the first protein crystal structure was solved by Perutz [8]. Multiple methods now exist to obtain the structures of proteins at high resolution [9–11]. However, structural models provide only a static picture of a protein, from which function can be inferred only somewhat indirectly. Proteins are dynamic molecules, and they evolve in time at the atomic level, fluctuating between multiple conformational states [12,13]. The relative population of different states strongly depends on environmental conditions such as pH, salt concentration, and most importantly, interaction with other molecules (ligands) [14–17]. The molecular dynamics associated with these conformational states span multiple time scales and amplitudes. Local structural changes, such as bond vibrations and transitions between side chain rotamers, occur on very fast time scales (femtoseconds to nanoseconds). On the other hand, larger conformational changes, such as loop or helix motion, may take nanoseconds to microseconds. Even larger motions involving tertiary and quaternary structure elements (domains and subunits, respectively) may occur on much slower timescales, sometimes up to many seconds [13,18,19]. Interestingly, it was shown that fast local dynamics on the level of amino acid side chains may be connected and propagated to the level of larger and much slower conformational changes [20,21]. This demonstrates that local dynamics of the protein structure might be important for protein function [22]. However, the mechanism by which local motion couples to large-scale conformational dynamics is still not well understood.
Ligand binding can affect a protein’s conformational changes, which can in turn also affect the binding activity of other molecules at distinct sites on the protein. This phenomenon is referred to as allostery [23,24] and has been studied extensively [25,26]. On the basis of the original model proposed by Monod, Weiman and Changeux [23], it was suggested that allostery is mediated through the stabilization of one protein conformation over others, a phenomenon also known as “conformational selection” [19,26]. On the other hand, a model developed from the work of Koshland, Nemethy and Filmer [27], known as “induced fit”, asserts that the binding of an allosteric effector leads to a new, previously unpopulated conformation of the protein, with a modified binding activity at other sites [19,26]. While these conformation-based allostery models emphasized the thermodynamic states of the transforming proteins, Dryden and Cooper proposed that allostery can also involve changes in conformational dynamics [28,29]. There has been much interest in defining the potential role of dynamics in allosteric transitions and more generally, the dynamics of large-scale correlated motions of domains and subunits [30–33]. Interestingly, recent studies have proposed ways to control protein activity by altering dynamics using protein engineering methods [34].
Multiple experimental techniques have been applied to study protein structure and dynamics. High-resolution protein structures can be obtained using X-ray crystallography [10], nuclear magnetic resonance spectroscopy [9], and cryogenic electron microscopy [11]. Low-resolution techniques such as small angle X-ray scattering [35] can provide information on the overall shape of a protein and how it changes under various conditions. As for methods to probe dynamics, 2D infrared spectroscopy and time-resolved X-ray crystallography measure fast motions on the ps-ns timescales, reporting mainly on local structural rearrangements [36,37]. Nuclear magnetic resonance techniques can probe conformational changes on multiple time scales based on a range of different experiments that are sensitive to very short times or to longer times [38]. Recently, cryogenic electron microscopy techniques have been developed that can also report on conformational dynamics, either by using fast mixing methods [39] or by a combination with computer simulations [40]. To a large extent, these ensemble-based methodologies lack the ability to precisely characterize conformational heterogeneity, to observe in real time conformational changes between multiple protein conformers and to define the exact time constants characterizing these conformational changes.
Single-molecule experimental techniques can solve many of these problems and have therefore become critical to our understanding of molecular function, motion, and dynamics. Many studies of protein machines at the single-molecule level have been conducted using force spectroscopy tools [41], such as optical and magnetic tweezers, as well as atomic force microscopy, and have revealed multiple mechanistic details [42,43]. A second group of single-molecule methods, based on fluorescence, can be applied to study internal motions in proteins with less intervention than force spectroscopic techniques and can provide information on dynamics along multiple internal coordinates and on a broad range of time scales. In particular, single-molecule fluorescence resonance energy transfer (smFRET) spectroscopy, which is the topic of this review, can directly determine intramolecular distances in biological molecules and measure their modulation in time. The sensitivity of smFRET experiments stems from the strong dependence of the excitation energy transfer on the distance between fluorescent dyes [44,45].
In this review, we discuss how smFRET spectroscopy has contributed to studies of protein dynamics in recent years. We start by reviewing recent development in smFRET methodology for characterizing slow and fast conformational dynamics, and we then consider the biological implications of these dynamics through several important examples.
Recent developments in smFRET methodology for protein dynamics
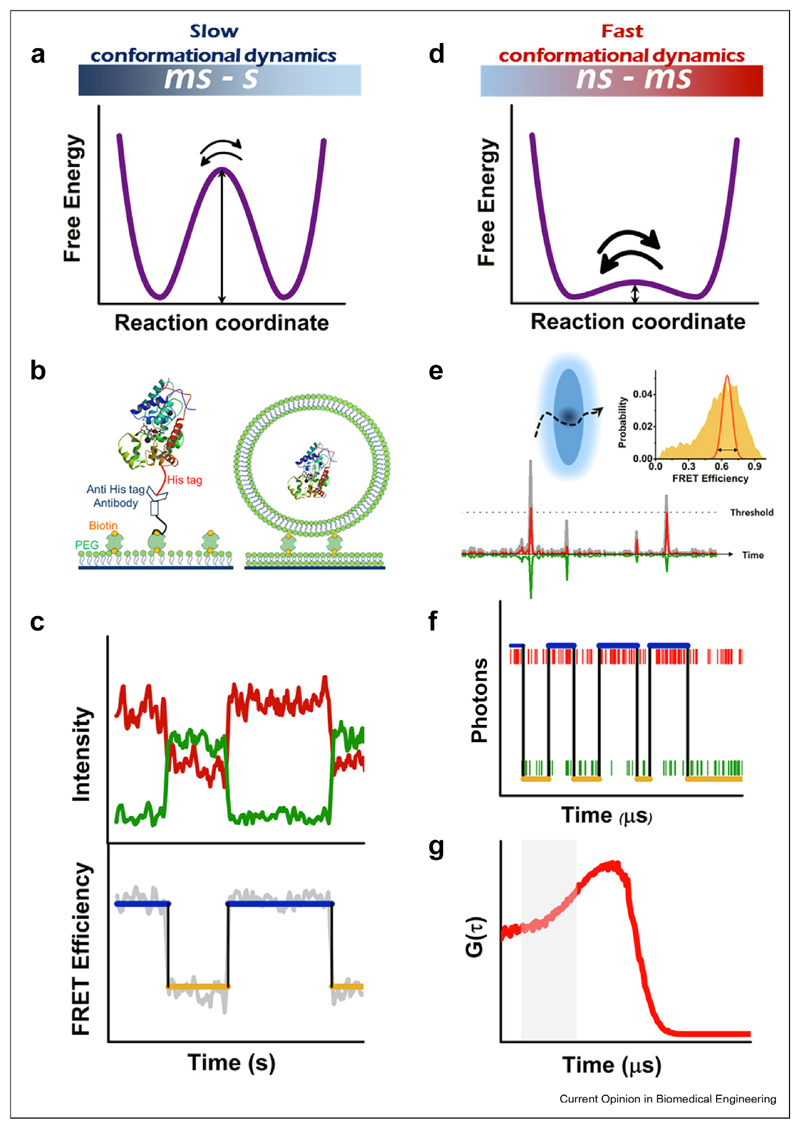
smFRET spectroscopy is a promising tool to reveal the basis of function-related conformational dynamics of proteins in action, and it can cover in principle the whole range of time scales from nanoseconds to seconds. It is possible to group the many different smFRET methods introduced over the years into two categories, roughly on the basis of the time scale of the dynamics probed: slow conformational dynamics (10 ms - s) are studied on molecules immobilized on a surface (Figure 1a–c), while fast conformational dynamics (ns - ms) are studied on molecules diffusing in solution (Figure 1d–g). In both cases, the molecules have to be labeled with two fluorescent tags, the donor and the acceptor. Recent years have also seen an increasing number of demonstrations of smFRET experiments with three [46–48] and even four dyes [49,50]. Such experiments are geared toward obtaining information about correlated motions, either within one molecule or involving several interacting molecules.
Figure 1. Probing proteins with smFRET spectroscopy: slow (a–c) vs fast (d–g) dynamics.
(a) Slow conformational dynamics involve a high free-energy barrier. (b) Two different methods to tether a single molecule to a surface, using direct tethering or vesicle encapsulation. (c) smFRET trajectory of an immobilized molecule. Top panel: donor (green) and acceptor (red) intensities; bottom panel: FRET efficiency (gray) and state assignments from a HMM analysis (blue and orange). (d) Fast conformational dynamics involve a low free-energy barrier. (e) Spectroscopy of freely diffusion molecule. Top: cartoon of a focal volume with a molecule passing through. Bottom: fluorescence bursts emanating from excited molecules. Inset; FRET efficiency histogram (orange) that is broader than shot noise (red), suggesting fast dynamics. (f) A photon-by-photon single-molecule trajectory, with donor and acceptor photons in green and red, respectively. Blue and orange lines are state assignments from an H2MM analysis. (g) Cross-correlation function of donor and acceptor fluorescence. The initial increase in the signal (shaded area) indicates conformational dynamics on the microsecond timescale. smFRET, single-molecule fluorescence resonance energy transfer; HMM, Hidden Markov Models
Many techniques have been devised to bind macromolecules to surfaces (Figure 1b), and each technique has its own advantage, as discussed in the literature [51,52]. Tethered molecules can be observed continuously for long times, limited only by photobleaching of fluorescent dyes. Such experiments are most often conducted using a total internal reflection microscope, and intensity trajectories of donor and acceptor dyes are collected on two parts of a sensitive camera [53]. Single-molecule temporal trajectories obtained in this manner can be used to generate a detailed dynamic model, which includes the number of relevant conformational states and the rates of exchange between them.
When the FRET efficiency states are well separated (Figure 1a), they can be extracted from the trajectories using some form of a statistical change-point analysis, which has the advantage that no kinetic model needs to be assumed a priori. Several new change-point techniques have been introduced in recent years, supplementing older methods [54–56]. Landes and coworkers [57] developed a change-point method that combines two calculation steps. The first is the detection of transition steps using Student’s t test, and the second is an iterative grouping of FRETsegments between transition points to determine the optimal number of states necessary to describe the data. Taylor et al. [58] introduced a sophisticated analysis tool on the basis of information theory and soft clustering to construct free energy landscapes in the form of disconnectivity graphs. They demonstrated the utility of their technique by analyzing experimental results on the dynamics of a glutamate receptor [58] and on the folding of a multidomain protein [59].
A different set of trajectory-analysis methods starts with a kinetic model, the parameters of which are inferred from the data using a statistical approach based on maximum-likelihood estimation using, for example, Hidden Markov Models (HMM) [60]. HMM has been demonstrated to be a powerful tool for analyzing smFRET trajectories [61] (Figure 1c). A limitation of HMM is the need to assume a priori the number of states involved in the dynamics. However, various methods for validating the correct number of states exist, and one can also use Bayesian methods that maximize the evidence rather than the likelihood, thereby optimizing also the number of states [62].
Many proteins are expected to show very fast conformational dynamics (Figure 1d) on a timescale of a few milliseconds or even microseconds, yet with a spatial amplitude that lends itself to FRET measurements. Such proteins can be studied in free diffusion in solution using a focused laser beam within a confocal microscope [63] (Figure 1e). The typical passage time of a molecule through the focal volume is ~1 ms, though it can be extended by defocusing the laser beam [64] or by connecting a protein molecule of interest to a freely diffusing large entity such as a lipid vesicle [65]. The burst of photons emitted by each molecule as it passes through the beam is collected using single-photon avalanche photodiodes, which allow recording donor and acceptor photon-arrival times with a picosecond time resolution [63]. The apparent FRET efficiency in each burst can then be calculated on the basis of the number of photons. Conformational dynamics on a time scale faster than the average burst length are identified by observing FRET efficiency histograms that are broader than expected from the shot noise [66,67] (Figure 1e, Inset).
The registration of photon-arrival times permits pushing the smFRETexperiment to the utmost time resolution, dictated only by the photon flux. Photon-by-photon analysis methods that take advantage of this capability have been introduced in recent years. Gopich and Szabo used a maximum-likelihood approach to optimize the parameters of a kinetic model [68]. They cleverly utilized the arrival times of photons as input for the analysis, so that variations in photon flux because of the position of the molecule in space could be ignored. Recently, this idea was adopted into an HMM-based algorithm, which facilitated likelihood maximization using analytical expressions for model parameters. This algorithm, H2MM [69] (Figure 1f), was shown to facilitate the extraction of kinetic data on a broad range of time scales, from microseconds to hundreds of milliseconds. Recent studies demonstrate the power and robustness of this algorithm to probe fast conformational dynamics [33,70], as will be discussed in the next section.
Fluorescence-correlation spectroscopy (FCS) can also be used to extract fast dynamics from smFRET data (Figure 1g). In particular, the donor and acceptor signals should be anticorrelated at short timescales if large-scale motions exist in the data [71,72]. Rate constants for transitions between conformational states are obtained by fitting correlation functions with an appropriate model [72]. Filtering of FCS curves based on different lifetime components allows removing scattering and other sources of noise and exposing the contributions of individual species [73,74]. An elegant method that does not require any prior knowledge about the fluorescence decay of different species was developed by Tahara et al. [75]. They generate a correlation map on the basis of the delay times of photon pairs and Laplace-transform this map to obtain a two-dimensional fluorescence–lifetime correlation map. By comparison of maps computed at different delay times one can infer the timescale of conversions between different fluorescent species, as demonstrated recently in studies of the conformational dynamics of cytochrome C [76] and of a photosynthetic protein [77]. High-order correlation functions were also shown recently to offer detailed kinetic information from microsecond smFRET data [78].
Implications of conformational dynamics for proteins function
The methods discussed in the previous section have been used extensively for operando observation of proteins, especially protein machines. Basic questions, such as the coupling between conformational changes and catalytic steps during functional cycles, have been addressed by smFRET experiments. In this section, we will discuss some notable examples from recent years, highlight how these studies were performed, and comment on how they contributed new insights to our understanding of functional dynamics in these proteins.
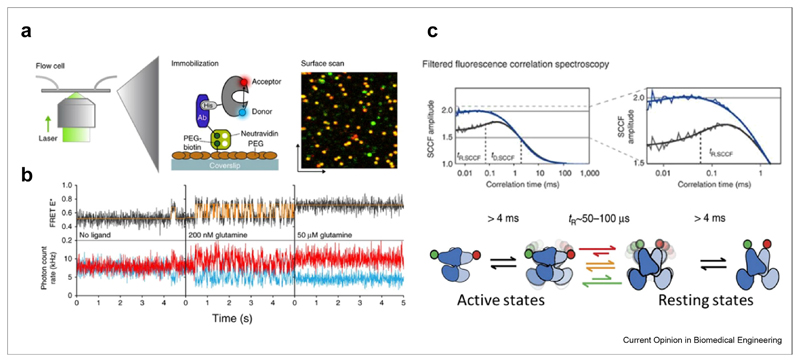
Membrane proteins were studied extensively with smFRET techniques. Interestingly, many of these studies tended to capture relatively slow conformational dynamics [79,80]. Several experiments focused on ATP-binding cassette (ABC) transporters, which are membrane protein machines that harness ATP energy to move molecules across the cell membrane [81–84]. ABC transporters are homodimers, each with a substrate-binding domain (SBD) or a separate substrate-binding protein (SBP), a transmembrane domain, and a cytoplasmic nucleotide-binding domain. Gouridis et al. [81] probed the dynamics of the SBDs of GlnPQ, an importer of asparagine, glutamine, and glutamate, both in free diffusion and when bound to a surface (Figure 2a). They found that 95% of the molecules are in an open conformation in the absence of substrates. The addition of the substrates shifts the population to the closed conformation, with transitions back to the open conformation characterized by time constants of hundreds of milliseconds (Figure 2b). Yang et al. [83] studied BtuCD, an importer of vitamin B12, both in a detergent solution and inserted into nanodiscs.
Figure 2. Membrane protein conformational dynamics measured with smFRET.
(a) Single-molecule dynamics of a surface-tethered SBD of the ABC importer GlnPQ probed by confocal scanning microscopy. (b) Single molecule trajectories at different ligand concentrations. Figure 2 A-B reprinted from Ref. [81] with permission from Nature publishing group. (c) Dynamics of the G-protein-coupled receptor mGluR, reprinted from Ref. [85] with permission. Top: filtered fluorescence cross-correlation curves, indicating microsecond conformational dynamics in the wild type (grey) but not in the constitutively active mutant (blue). Bottom: kinetic model of mGluR dynamics. SBD, substrate-binding domain; smFRET, single-molecule fluorescence resonance energy transfer; ABC, ATP-binding cassette.
Using multiple FRET pairs, they demonstrated the tight coupling between the binding of a nucleotide or a substrate-binding protein and the conformational changes that occur on each domain of the machine (transmembrane domain and nucleotide-binding domain). They also described transient conformational changes that apparently allow the substrate to pass through the transporter.
Much faster, submillisecond conformational dynamics of a membrane protein were discovered in a study of the metabotropic glutamate receptor (mGluR), a G-protein-coupled receptor essential for synaptic activity [85]. FRET between the two parts of the ligand binding domain homodimer of mGluR was used to trace conformational transitions between resting and active states (Figure 2c). Using filtered FCS spectroscopy, it was found that the protein continuously shuttles between the two states on a time scale of <100 μs. Ligand binding was found to tune the ratio between the two states rather than stabilizing a static active state.
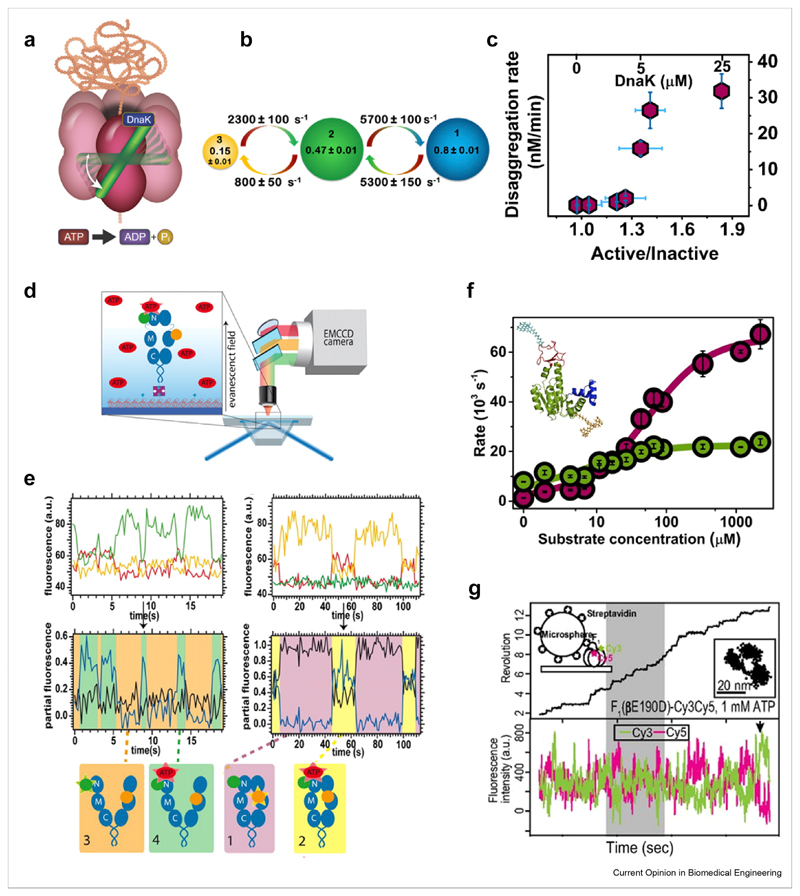
Ultrafast conformational dynamics were also found by Mazal et al. [70] in their studies of the AAA+ disaggregation chaperone ClpB [86] (Figure 3a). Studying ClpB’s middle domain (M domain), which serves as a regulatory switch, they used a photon-by-photon H2MM analysis on FRET trajectories of diffusing molecules to reveal transitions between the inactive and active states of the M domain on a time scale of ~150 μs (Figure 3b). This timescale is much faster than the activity of the machine, suggesting that the ratio of the two conformations serves to tune disaggregation activity. Indeed, factors that change this ratio, such as the concentration of the co-chaperone DnaK or nucleotides, were found to also modify the rate of disaggregation (Figure 3c). The mechanism of tunable allosteric switching reported by Olofsson et al. [85] and by Mazal et al. [70] likely involves a low-energy barrier between the active and inactive states (Figure 1d) and may be a general way to modulate machine activity, to be corroborated in future studies.
Figure 3. Dynamics of soluble protein machines measured with smFRET.
(a–c) The disaggregation machine ClpB, reprinted from Ref. [70] with permission. (a) Schematic of ClpB and its dynamic M domain (green), which toggles between three states on the microsecond timescale (b). (c) DnaK binding tunes the ratio between the two major states of the M domain and in turn disaggregation activity. (d–e) The chaperone Hsp90, reprinted from Ref. [47] with permission. (d) ATP-binding related dynamics of Hsp90 are studied using three-color smFRET. (e) Top: single molecule trajectories in three channels upon donor excitation. Middle: FRET efficiency trajectories in different states of Hsp90, as depicted in the bottom cartoons. (f) Adenylate kinase, reprinted from Ref. [33] with permission. Rates of domain closing and opening (cherry and green, respectively) as a function of substrate concentration. Inset: structure with attached labels. (g) F1-ATPase, Reprinted from Ref. [91] with permission. Simultaneous measurement of rotational motion of the ɣ-shaft using a bead (top panel, black line) and of FRET between αβ dimers (bottom panel, donor and acceptor in green and red). smFRET, single-molecule fluorescence resonance energy transfer.
Another molecular chaperone that has been studied extensively by smFRET is Hsp90, a homodimeric protein that acts at the last stages of folding of a diverse set of client proteins and consumes ATP as fuel [87]. Hsp90 dimers adopt a V-shaped open conformation and undergo a large conformational change during their active cycle to form a closed conformation. Employing three-color smFRET, with two dyes on the chaperone and a third on ATP, Hugel and coworkers demonstrated that ATP binding couples only weakly to the large conformational changes of Hsp90 [47] (Figure 3d–e). They also found negative cooperativity between the two ATP binding sites. Extending their studies to four colors, they added a labeled co-chaperone and concluded from data analysis that the presence of this co-chaperone and of nucleotides leads to forward and backward transition probabilities that are not equal, indicating directionality in the machine cycle [50]. In a recent study on freely diffusing Hsp90, Hellenkamp et al. analyzed distance distribution widths to obtain a qualitative indication for fast (submillisecond) conformational dynamics of the dimer, the extent of which varied between different states of the machine [88]. This observation awaits a more direct investigation of the time scales of fast conformational changes.
A direct observation of the modulation of conformational dynamics by substrate binding was achieved in a recent study of the domain closure of the ubiquitous enzyme adenylate kinase (AK) [33]. AK is key to the maintenance of ATP levels in cells, catalyzing the reaction ATP+AMP ⇌ 2ADP. It has been thought that domain closure is rate limiting for the enzymatic reaction of AK [89]. Aviram et al. [33] used smFRET spectroscopy of freely diffusing AK molecules, combined with a photon-by-photon H2MM analysis, to obtain a detailed picture of the time scales of domain closure (Figure 3f). It was found that ATP binding increases domain closing and opening rates, making the corresponding reaction times as short as 15 and 45 μs, respectively (Figure 3f). Domain closure is thus two orders of magnitude faster than the catalytic rate of AK. It was suggested that the ultrafast dynamics serve to facilitate proper orientation of the two substrates for the catalytic step.
Finally, a recent interesting trend, which is likely to grow, is the combination of smFRET spectroscopy with an additional single-molecule method that provides complementary information. For example, Comstock et al. [90] combined smFRETwith force microscopy to reveal the correlation of conformational changes and motion along DNA of a repair helicase, UvrD. Sugawa et al. [91] studied the coupling between the conformational changes in the β subunits of the rotary machine F1-ATPase and the rotation of its ɣ-shaft. To this end, they combined smFRET with bead rotation measurements (Figure 3g). Their results shed new light on the rotational mechanism of F1-ATPase and could be nicely connected to the available crystal structures of the protein.
Conclusion and future directions
smFRET spectroscopy has become a valuable method for the study of protein conformational dynamics. In general, studies on the single-molecule level are unique in their ability to go beyond ensemble averages not only in terms of structure but also in terms of motion. A question that is often very difficult to directly answer by most ensemble methods, i.e. “what is the dynamic connectivity of conformations in a complex protein reaction”, is resolved readily through single-molecule measurements. smFRET spectroscopy is particularly well poised to answer this and similar questions regarding structural dynamics related to function. Further, it has the advantage, stated already in preceding sections, that it can cover a very broad range of time scales, from seconds down to microseconds. In this review, we discussed new developments in smFRET methodology, as well as new applications that shed light on the function of specific protein machines. We hope that we were able to convey the spirit of excitement in this field and convince the reader of the huge potential still awaiting its students.
We believe that a particularly important future direction of smFRET spectroscopy will be its application to study very fast conformational transitions and related allosteric pathways. It is now possible to use smFRET experiments to obtain information on the microsecond time scale and to shed light on ultrafast conformational transitions to which other techniques have been blind. We have mentioned several examples for this capability, and we note briefly that ultrafast smFRET methodologies have also been applied to study protein folding [76], particularly in relation to the measurement of the transition path time, i.e. the time it takes molecules to pass over the folding free-energy barrier [92–94].
We close by hypothesizing that large-scale microsecond motions in proteins might be much more ubiquitous than is currently assumed. It is likely that relatively low free-energy barriers connect many conformations of proteins and that the high free-energy barriers required for slow dynamics are provided by chemical steps such as ATP hydrolysis. If our hypothesis is correct, then the future will no doubt unearth multiple additional examples for domain and subunit motions on the microsecond time scale. We also expect that further methodology developments will make the measurement of such motions simpler and more robust, both on the level of the experiment and of data analysis.
Acknowledgements
GH is funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 742637). The PhD research of HM is supported by Planning and Budgeting Committee of the Council for Higher Education of Israel.
Footnotes
Conflict of interest statement
Nothing declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* * of outstanding interest
- 1.Schrödinger E. What is life? The physical aspect of the living cell. Cambridge University Press; 1944. [Google Scholar]
- 2.Nelson DL, Cox MM, Lehninger AL. Lehninger principles of biochemistry. 2017 [Google Scholar]
- 3.Mavroidis C, Dubey A, Yarmush ML. Molecular machines. Annu Rev Biomed Eng. 2004;6:363–395. doi: 10.1146/annurev.bioeng.6.040803.140143. [DOI] [PubMed] [Google Scholar]
- 4.Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 5.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 6.Gruebele M, Dave K, Sukenik S. Globular protein folding in vitro and in vivo. Annu Rev Biophys. 2016;45:233–251. doi: 10.1146/annurev-biophys-062215-011236. [DOI] [PubMed] [Google Scholar]
- 7.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 8.Fersht AR. From the first protein structures to our current knowledge of protein folding: delights and scepticisms. Nat Rev Mol Cell Biol. 2008;9:650–654. doi: 10.1038/nrm2446. [DOI] [PubMed] [Google Scholar]
- 9.Kanelis V, Forman-Kay JD, Kay LE. Multidimensional NMR methods for protein structure determination. IUBMB Life. 2001;52:291–302. doi: 10.1080/152165401317291147. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y. A glimpse of structural biology through X-ray crystallography. Cell. 2014;159:995–1014. doi: 10.1016/j.cell.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y, Grigorieff N, Penczek PA, Walz T. A primer to single-particle cryo-electron microscopy. Cell. 2015;161:438–449. doi: 10.1016/j.cell.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 13.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 14.O’Brien EP, Brooks BR, Thirumalai D. Effects of pH on proteins: predictions for ensemble and single-molecule pulling experiments. J Am Chem Soc. 2012;134:979–987. doi: 10.1021/ja206557y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maity H, Muttathukattil AN, Reddy G. Salt effects on protein folding thermodynamics. J Phys Chem Lett. 2018;9:5063–5070. doi: 10.1021/acs.jpclett.8b02220. [DOI] [PubMed] [Google Scholar]
- 16.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 17.Lorimer GH, Horovitz A, McLeish T. Allostery and molecular machines. Philos Trans R Soc Lond B Biol Sci. 2018;373 doi: 10.1098/rstb.2017.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei G, Xi W, Nussinov R, Ma B. Protein ensembles: how does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem Rev. 2016;116:6516–6551. doi: 10.1021/acs.chemrev.5b00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Zhou HX. Protein allostery and conformational dynamics. Chem Rev. 2016;116:6503–6515. doi: 10.1021/acs.chemrev.5b00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchli B, Waldauer SA, Walser R, Donten ML, Pfister R, Blochliger N, Steiner S, Caflisch A, Zerbe O, Hamm P. Kinetic response of a photoperturbed allosteric protein. Proc Natl Acad Sci U S A. 2013;110:11725–11730. doi: 10.1073/pnas.1306323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan D, Mathies RA. Chromophore structure in lumirhodopsin and metarhodopsin I by time-resolved resonance Raman microchip spectroscopy. Biochemistry. 2001;40:7929–7936. doi: 10.1021/bi010670x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Hage K, Brickel S, Hermelin S, Gaulier G, Schmidt C, Bonacina L, van Keulen SC, Bhattacharyya S, Chergui M, Hamm P, et al. Implications of short time scale dynamics on long time processes. Struct Dyn. 2017;4 doi: 10.1063/1.4996448. 061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 24.Monod J, Jacob F. General conclusions: teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harbor Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 25.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Changeux JP. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 27.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 28.Ahuja LG, Taylor SS, Kornev AP. Tuning the "violin" of protein kinases: the role of dynamics-based allostery. IUBMB Life. 2019;71:685–696. doi: 10.1002/iub.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper A, Dryden DTF. Allostery without conformational change. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 30.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 32.Boehr DD, Schnell JR, McElheny D, Bae SH, Duggan BM, Benkovic SJ, Dyson HJ, Wright PE. A distal mutation perturbs dynamic amino acid networks in dihydrofolate reductase. Biochemistry. 2013;52:4605–4619. doi: 10.1021/bi400563c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aviram HY, Pirchi M, Mazal H, Barak Y, Riven I, Haran G. Direct observation of ultrafast large-scale dynamics of an enzyme under turnover conditions. Proc Natl Acad Sci U S A. 2018;115:3243–3248. doi: 10.1073/pnas.1720448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boehr DD, D’Amico RN, O’Rourke KF. Engineered control of enzyme structural dynamics and function. Protein Sci. 2018;27:825–838. doi: 10.1002/pro.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh A, Ostrander JS, Zanni MT. Watching proteins wiggle: mapping structures with two-dimensional infrared spectroscopy. Chem Rev. 2017;117:10726–10759. doi: 10.1021/acs.chemrev.6b00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srajer V, Schmidt M. Watching proteins function with time-resolved X-ray crystallography. J Phys D Appl Phys. 2017;50 doi: 10.1088/1361-6463/aa7d32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaledhonkar S, Fu Z, Caban K, Li W, Chen B, Sun M, Gonzalez RL, Jr, Frank J. Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature. 2019;570:400–404. doi: 10.1038/s41586-019-1249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonomi M, Pellarin R, Vendruscolo M. Simultaneous determination of protein structure and dynamics using cryo-electron microscopy. Biophys J. 2018;114:1604–1613. doi: 10.1016/j.bpj.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veigel C, Schmidt CF. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat Rev Mol Cell Biol. 2011;12:163–176. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- 43.Smith DE. Single-molecule studies of viral DNA packaging. Curr Opin Virol. 2011;1:134–141. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann Phys. 1948;437:55–75. [Google Scholar]
- 45.Lerner E, Cordes T, Ingargiola A, Alhadid Y, Chung S, Michalet X, Weiss S. Toward dynamic structural biology: two decades of single-molecule Forster resonance energy transfer. Science. 2018;359 doi: 10.1126/science.aan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratzke C, Berkemeier F, Hugel T. Heat shock protein 90’s mechanochemical cycle is dominated by thermal fluctuations. Proc Natl Acad Sci U S A. 2012;109:161–166. doi: 10.1073/pnas.1107930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo J, Louis JM, Gopich IV, Chung HS. Three-color single-molecule FRET and fluorescence lifetime analysis of fast protein folding. J Phys Chem B. 2018;122:11702–11720. doi: 10.1021/acs.jpcb.8b07768. [** Three dyes label a single protein molecule in this sophisticated work in order to study folding and demonstrate that unique information can be extracted by measuring three FRET channels.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Single-molecule four-color FRET. Angew Chem Int Ed Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratzke C, Hellenkamp B, Hugel T. Four-colour FRET reveals directionality in the Hsp90 multicomponent machinery. Nat Commun. 2014;5 doi: 10.1038/ncomms5192. 4192. [** Four FRET channels are measured in this work, with two dyes on Hsp90, on an ATP molecule and one on a cochaperone, to demonstrate the out-of-equilibrium operation of the machine.] [DOI] [PubMed] [Google Scholar]
- 51.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi UB, Weninger KR, Bowen ME. Immobilization of proteins for single-molecule fluorescence resonance energy transfer measurements of conformation and dynamics. In: Uversky VN, Dunker AK, editors. Intrinsically disordered protein analysis: volume 2, methods and experimental tools. New York: Springer; 2012. pp. 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voith von Voithenberg L, Lamb DC. Single pair forster resonance energy transfer: a versatile tool to investigate protein conformational dynamics. Bioessays. 2018;40 doi: 10.1002/bies.201700078. [DOI] [PubMed] [Google Scholar]
- 54.Watkins LP, Yang H. Detection of intensity change points in time-resolved single-molecule measurements. J Phys Chem B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 55.Kalafut B, Visscher K. An objective, model-independent method for detection of non-uniform steps in noisy signals. Comput Phys Commun. 2008;179:716–723. [Google Scholar]
- 56.Ensign DL, Pande VS. Bayesian detection of intensity changes in single molecule and molecular dynamics trajectories. J Phys Chem B. 2010;114:280–292. doi: 10.1021/jp906786b. [DOI] [PubMed] [Google Scholar]
- 57.Shuang B, Cooper D, Taylor JN, Kisley L, Chen J, Wang W, Li CB, Komatsuzaki T, Landes CF. Fast step transition and state identification (STaSI) for discrete single-molecule data analysis. J Phys Chem Lett. 2014;5:3157–3161. doi: 10.1021/jz501435p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor JN, Li CB, Cooper DR, Landes CF, Komatsuzaki T. Error-based extraction of states and energy landscapes from experimental single-molecule time-series. Sci Rep. 2015;5 doi: 10.1038/srep09174. 9174. [** A novel method based on rate-distortion theory is introduced in order to extract the underlying state sequences from experimental single-molecule time series.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor JN, Pirchi M, Haran G, Komatsuzaki T. Deciphering hierarchical features in the energy landscape of adenylate kinase folding/unfolding. J Chem Phys. 2018;148 doi: 10.1063/1.5016487. 123325. [DOI] [PubMed] [Google Scholar]
- 60.Talaga DS. COCIS: Markov processes in single molecule fluorescence. Curr Opin Colloid Interface Sci. 2007;12:285–296. doi: 10.1016/j.cocis.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aviram HY, Pirchi M, Barak Y, Riven I, Haran G. Two states or not two states: single-molecule folding studies of protein L. J Chem Phys. 2018;148 doi: 10.1063/1.4997584. 123303. [DOI] [PubMed] [Google Scholar]
- 62.Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr, Wiggins CH. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holmstrom ED, Holla A, Zheng W, Nettels D, Best RB, Schuler B. Accurate transfer efficiencies, distance distributions, and ensembles of unfolded and intrinsically disordered proteins from single-molecule FRET. Methods Enzymol. 2018;611:287–325. doi: 10.1016/bs.mie.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalinin S, Felekyan S, Valeri A, Seidel CA. Characterizing multiple molecular States in single-molecule multiparameter fluorescence detection by probability distribution analysis. J Phys Chem B. 2008;112:8361–8374. doi: 10.1021/jp711942q. [DOI] [PubMed] [Google Scholar]
- 65.Kim JY, Kim C, Lee NK. Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nat Commun. 2015;6 doi: 10.1038/ncomms7992. 6992. [** The authors extend smFRET studies of freely diffusing molecules to longer times by tethering them to vesicles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gopich IV, Szabo A. Single-molecule FRET with diffusion and conformational dynamics. J Phys Chem B. 2007;111:12925–12932. doi: 10.1021/jp075255e. [DOI] [PubMed] [Google Scholar]
- 67.Torella JP, Holden SJ, Santoso Y, Hohlbein J, Kapanidis AN. Identifying molecular dynamics in single-molecule FRET experiments with burst variance analysis. Biophys J. 2011;100:1568–1577. doi: 10.1016/j.bpj.2011.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gopich IV, Szabo A. Decoding the pattern of photon colors in single-molecule FRET. J Phys Chem B. 2009;113:10965–10973. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pirchi M, Tsukanov R, Khamis R, Tomov TE, Berger Y, Khara DC, Volkov H, Haran G, Nir E. Photon-by-Photon hidden Markov model analysis for microsecond single-molecule FRET kinetics. J Phys Chem B. 2016;120:13065–13075. doi: 10.1021/acs.jpcb.6b10726. [** This paper introduces the H2MM method that extends standard HMM single-molecule analysis to photon-by-photon data and paves the way to studies of microsecond protein dynamics.] [DOI] [PubMed] [Google Scholar]
- 70.Mazal H, Iljina M, Barak Y, Elad N, Rosenzweig R, Goloubinoff P, Riven I, Haran G. Tunable microsecond dynamics of an allosteric switch regulate the activity of a AAA+ disaggregation machine. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09474-6. 1438. [** A photon-by-photon study of the regulatory coiled-coil domain of the disaggregation machine ClpB reveals microsecond dynamics that affect the much slower functional activity of the protein.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres T, Levitus M. Measuring conformational dynamics: a new FCS-FRET approach. J Phys Chem B. 2007;111:7392–7400. doi: 10.1021/jp070659s. [DOI] [PubMed] [Google Scholar]
- 72.Felekyan S, Sanabria H, Kalinin S, Kuhnemuth R, Seidel CA. Analyzing Forster resonance energy transfer with fluctuation algorithms. Methods Enzymol. 2013;519:39–85. doi: 10.1016/B978-0-12-405539-1.00002-6. [DOI] [PubMed] [Google Scholar]
- 73.Kapusta P, Wahl M, Benda A, Hof M, Enderlein J. Fluorescence lifetime correlation spectroscopy. J Fluoresc. 2007;17:43–48. doi: 10.1007/s10895-006-0145-1. [DOI] [PubMed] [Google Scholar]
- 74.Felekyan S, Kalinin S, Sanabria H, Valeri A, Seidel CA. Filtered FCS: species auto- and cross-correlation functions highlight binding and dynamics in biomolecules. ChemPhysChem. 2012;13:1036–1053. doi: 10.1002/cphc.201100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishii K, Tahara T. Two-dimensional fluorescence lifetime correlation spectroscopy. 1. Principle. J Phys Chem B. 2013;117:11414–11422. doi: 10.1021/jp406861u. [** An impressive correlation analysis technique that employs data on photon arrival times to observes protein states and their sequence of interconversion on very short time scales.] [DOI] [PubMed] [Google Scholar]
- 76.Otosu T, Ishii K, Tahara T. Microsecond protein dynamics observed at the single-molecule level. Nat Commun. 2015;6 doi: 10.1038/ncomms8685. 7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kondo T, Gordon JB, Pinnola A, Dall’Osto L, Bassi R, Schlau-Cohen GS. Microsecond and millisecond dynamics in the photosynthetic protein LHCSR1 observed by single-molecule correlation spectroscopy. Proc Natl Acad Sci U S A. 2019;116:11247–11252. doi: 10.1073/pnas.1821207116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Phelps C, Israels B, Marsh MC, von Hippel PH, Marcus AH. Using multiorder time-correlation functions (TCFs) to elucidate biomolecular reaction pathways from microsecond single-molecule fluorescence experiments. J Phys Chem B. 2016;120:13003–13016. doi: 10.1021/acs.jpcb.6b08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dyla M, Terry DS, Kjaergaard M, Sorensen TL, Lauwring Andersen J, Andersen JP, Rohde Knudsen C, Altman RB, Nissen P, Blanchard SC. Dynamics of P-type ATPase transport revealed by single-molecule FRET. Nature. 2017;551:346–351. doi: 10.1038/nature24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gouridis G, Schuurman-Wolters GK, Ploetz E, Husada F, Vietrov R, de Boer M, Cordes T, Poolman B. Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat Struct Mol Biol. 2015;22:57–64. doi: 10.1038/nsmb.2929. [DOI] [PubMed] [Google Scholar]
- 82.Goudsmits JMH, Slotboom DJ, van Oijen AM. Single-molecule visualization of conformational changes and substrate transport in the vitamin B12 ABC importer BtuCD-F. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01815-7. 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang M, Livnat Levanon N, Acar B, Aykac Fas B, Masrati G, Rose J, Ben-Tal N, Haliloglu T, Zhao Y, Lewinson O. Single-molecule probing of the conformational homogeneity of the ABC transporter BtuCD. Nat Chem Biol. 2018;14:715–722. doi: 10.1038/s41589-018-0088-2. [DOI] [PubMed] [Google Scholar]
- 84.de Boer M, Gouridis G, Vietrov R, Begg SL, Schuurman-Wolters GK, Husada F, Eleftheriadis N, Poolman B, McDevitt CA, Cordes T. Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. Elife. 2019;8 doi: 10.7554/eLife.44652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olofsson L, Felekyan S, Doumazane E, Scholler P, Fabre L, Zwier JM, Rondard P, Seidel CA, Pin JP, Margeat E. Fine tuning of sub-millisecond conformational dynamics controls metabotropic glutamate receptors agonist efficacy. Nat Commun. 2014;5 doi: 10.1038/ncomms6206. 5206. [** Analysis of cross-correlation FRET functions reveals microsecond dynamics in a G-protein coupled receptor.] [DOI] [PubMed] [Google Scholar]
- 86.Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol. 2013;14:617–629. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- 87.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 88.Hellenkamp B, Wortmann P, Kandzia F, Zacharias M, Hugel T. Multidomain structure and correlated dynamics determined by self-consistent FRET networks. Nat Methods. 2017;14:174–180. doi: 10.1038/nmeth.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanson JA, Duderstadt K, Watkins LP, Bhattacharyya S, Brokaw J, Chu JW, Yang H. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc Natl Acad Sci U S A. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Comstock MJ, Whitley KD, Jia HF, Sokoloski J, Lohman TM, Ha T, Chemla YR. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348:352–354. doi: 10.1126/science.aaa0130. [** Simulataneous smFRET and force spectroscopy to study protein conformational changes and movement along the DNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sugawa M, Okazaki K, Kobayashi M, Matsui T, Hummer G, Masaike T, Nishizaka T. F1-ATPase conformational cycle from simultaneous single-molecule FRET and rotation measurements. Proc Natl Acad Sci U S A. 2016;113:E2916–E2924. doi: 10.1073/pnas.1524720113. [** A bold simultaneous smFRET and bead rotation study to reveal novel information on the functional cycle of F1-ATPas.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chung HS, Piana-Agostinetti S, Shaw DE, Eaton WA. Structural origin of slow diffusion in protein folding. Science. 2015;349:1504–1510. doi: 10.1126/science.aab1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sturzenegger F, Zosel F, Holmstrom ED, Buholzer KJ, Makarov DE, Nettels D, Schuler B. Transition path times of coupled folding and binding reveal the formation of an encounter complex. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim JY, Meng FJ, Yoo J, Chung HS. Diffusion-limited association of disordered protein by non-native electrostatic interactions. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06866-y. [DOI] [PMC free article] [PubMed] [Google Scholar]