Abstract
Printed electrolyte-gated transistors (EGTs) are an emerging biosensor platform that leverage the facile fabrication engendered by printed electronics with the low voltage operation enabled by ion gel dielectrics. The resulting label-free, nonoptical sensors have high gain and provide sensing operations that can be challenging for conventional chemical field effect transistor architectures. After providing an overview of EGT device fabrication and operation, we highlight opportunities for microfluidic enhancement of EGT sensor performance via multiplexing, sample preconcentration, and improved transport to the sensor surface.
I. INTRODUCTION
Surface-based adsorption, where an analyte of interest is pulled down from a solution by a specific capture agent, such as an antibody, is a standard biosensing method. These surface adsorption-based assays typically rely on an optical readout, such as the fluorescence-based measurements used in DNA microarrays1 or the changes in refractive index obtained by surface plasmon resonance (SPR).2 Electronic-based detection provides a potentially simpler readout of the binding event, obviating the need for the labeling steps in a fluorescence-based readout or the optical alignment required for sensitive methods such as SPR. Transistor-based methods, including field effect transistors (FETs) and electrolyte gated transistors (EGTs), provide one possible route toward electronic detection, where the binding event changes the threshold voltage required to turn the transistor ON.
This perspectives article focuses on recent advances in biosensing using electrolyte gated transistors and opportunities for improving the technology via microfluidics. In an EGT, the gate dielectric typical of a conventional FET is replaced with a liquid or gel electrolyte. There are two basic modes of EGT operation: electrochemical and electric double layer. In the electrochemical mode, application of a gate voltage drives electrochemical oxidation or reduction of the semiconductor channel with accompanying penetration of ions into the semiconductor. In the electric double layer mode, ions do not penetrate the semiconductor. Instead, the gate voltage drives the formation of a sheet of carriers (electrons or holes) in the semiconductor and an oppositely charged sheet of ions in the electrolyte immediately adjacent to the semiconductor. Both modes enhance the conductivity of the semiconductor channel by increasing the free carrier density. In most of our biosensing work, we have exploited EGTs based on the benchmark polymer semiconductor poly(3-hexylthiophene), or P3HT, which is permeable to ions. Thus, the P3HT EGTs operate in the electrochemical mode. Irrespective of the mode of operation, all EGTs have the property that they work at very low voltages because of the enormous capacitance of electrolytes; low voltage operation, in turn, makes the devices sensitive to label-free chemical binding to surfaces, as described below.
The EGT is an emerging platform, and much of the research to date has focused on proof-of-principle detection3–32 or fundamental studies of the device operation.17,28,33–41 However, the EGT platform and, in particular, the length scales accessible by printed electronics, make printed EGTs ideally suited for microfluidic integration. Microfluidics thus has the potential to dramatically improve EGT sensors via facile multiplexing, sample preconcentration, and improved transport to the surface.
This perspectives article is not intended to provide a comprehensive review of FET and EGT biosensors, and readers interested in a more thorough recent overview are referred elsewhere.42–45 Rather, this perspective aims to provide sufficient background on the sensing technology for the microfluidics community to identify and take advantage of opportunities in EGT sensing, with a particular emphasis on the floating-gate EGT platform developed in our labs.
II. CHEMICAL FIELD EFFECT TRANSISTORS
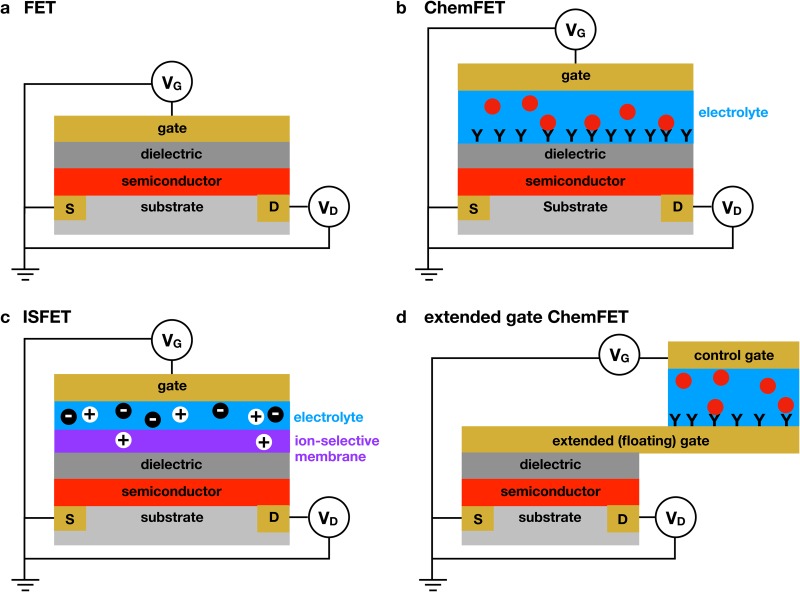
The principle of detecting a chemical in solution via transduction by a FET, known as a ChemFET, was introduced in the 1970s, with the earliest work pioneered by Bergveld46–48 and Janata.49,50 Figure 1 illustrates the basic principle behind different variants of a ChemFET. In the absence of any applied gate voltage, the FET is in the OFF state. As the gate voltage increases, the increasing electric field created within the semiconductor channel causes the transistor to turn ON at the threshold voltage, , producing a sharp increase in current between the source and the drain electrodes. The difference in current between the ON and OFF states is typically orders of magnitude. In the saturation regime, the drain current is given by51
| (1) |
where the width and length of the semiconductor channel are and , respectively, is the carrier mobility within the semiconductor, and is the specific capacitance of the gate dielectric. The latter quantity plays a key role in EGT-based sensing; we will return to the importance of in Sec. III A.
FIG. 1.
Different methods of FET-based detection. (a) Illustration of a standard field effect transistor (FET). The application of a gate voltage, , in excess of the threshold voltage, , causes the transistor to turn ON, leading to a measurable drain current, , between the source and drain electrodes under the application of the drain voltage, . (b) Illustration of a chemical field effect transistor (ChemFET).50 The aqueous electrolyte is contained between the dielectric and the gate electrode. Adsorption of a material to the dielectric surface, illustrated here as antibody/antigen binding, changes the properties of the dielectric, which then changes the threshold voltage required to turn the transistor ON. (c) Illustration of an ion-selective field effect transistor (ISFET).52 The ion selectivity can be provided by a selective membrane or an ion-selective oxide. Ions that penetrate the membrane polarize the dielectric. (d) Illustration of the extended-gate (or floating-gate) architecture.53 This approach decouples the transistor from the sensing medium, thereby greatly increasing the flexibility of the platform. The order of the layers for this extended gate example is chosen to provide easy comparison with the previous examples, not to reflect the actual fabrication of an extended gate device.
In a ChemFET, the solution containing the analyte is placed between the gate electrode [Fig. 1(b)] and the dielectric surface. Adsorption of the analyte to the dielectric produces a shift in the threshold voltage due to a change in the interfacial potential of the dielectric/solution surface. Measuring the difference in threshold voltage, , between the ChemFET in the presence and absence of the analyte thus provides a way to quantify chemisorption onto the FET surface. The particular example in Fig. 1(b) corresponds to the binding of an antibody to the dielectric surface, but one can use other capture agents, such as aptamers, or change the binding site to the semiconductor itself.54–59
In general, the ChemFET architecture in Fig. 1(b) poses significant materials science challenges for biological detection because (i) the capture agent must be bound directly to the thin dielectric and (ii) the dielectric is in contact with the aqueous medium. In particular, it is generally the case that the molecular grafting chemistry available for dielectric materials such as SiO is not as well-controlled or as convenient as the grafting chemistries applicable to noble metals such as silver, gold, or platinum. This limits the ability to attach capture molecules in an oriented fashion to the dielectric. Additionally, in order for the ChemFET to sense small threshold voltage shifts associated with chemical binding, the dielectric layer needs to be extremely thin (a few tens of nanometers), and this makes the whole device susceptible to leakage currents associated with ion migration in and out of pinholes in the thin dielectric. These considerations significantly limit the application of ChemFETs with the Fig. 1(b) architecture in applications where the analyte solution is a biofluid, such as blood, or situations where the reagents needed to extract the analyte from a complex medium also serve as etchants for the dielectric.
One approach to the material incompatibility problem is to use an ion-selective membrane between the FET and the aqueous medium [Fig. 1(c)], which is the basis of an ion-selective field effect transistor (ISFET).47 After initial demonstration by Bergveld,47,60 ISFETs have been used in a variety of applications,52 such as recent applications for pH sensing in coastal waters,61 blood glucose sensing,62,63 monitoring the metabolic activity of cell populations,64–66 and the detection of disease biomarkers in blood67 and sweat.68 The Ion Torrent DNA sequencer69 is arguably the most successful application of the ISFET principle for biological applications. This sequencer uses a tantalum oxide layer to provide proton sensitivity, along with a fluidic well above the oxide layer. The DNA templates are ligated to beads, and each well contains a single bead. This device uses a sequencing-by-synthesis protocol, where the ISFET detects the local change in pH due to the release of a proton when a base is added. The Ion Torrent sequencing approach leverages the ability to create a very dense array of identical sensors using CMOS manufacturing methods, with sensors on a single chip for massively parallel, nonoptical DNA sequencing.
A second approach to solve the material incompatibility problem is the extended gate53 concept illustrated in Fig. 1(d). Here, the gate electrode is extended away from the semiconductor, which allows one to create a separate aqueous environment. The extended gate architecture is also called a floating gate3 to highlight that the potential of the gate electrode in contact with the transistor is no longer directly imposed by the gate voltage. Rather, the gate voltage is applied at another electrode within the aqueous medium and capacitively coupled to the floating gate, which is then connected to the FET. Traditional FETs with extended gates have been utilized extensively for applications such as chemical sensing,53 pH sensing,70–72 and the detection of DNA,73,74 proteins,75–77 and small molecules.78,79
Both the ISFET and the extended gate ChemFET provide routes toward biosensing in challenging environments, but these methods also have their limitations. Clearly, while the ISFET architecture protects the transistor from the medium through the ion selective membrane, not all biosensing applications can be accomplished with an ISFET configuration. The most notable example is antibody/antigen binding, which underlies highly specific and highly sensitive immunogenic biosensing methods. The extended gate architecture is more flexible and can accommodate any capture agent that can be bound to the extended gate, which is often gold. Antibodies are often used as the capture agents in these devices,75,76 but aptamers77,79 have also been used for the detection of proteins and DNA with extended gate FETs.
However, the extended gate FET is not optimal from a fabrication perspective. Explicitly, the cost of CMOS fabrication is proportional to the area. This scaling is the reason, in part, why the Ion Torrent DNA sequencer69 is an exemplary use of FETs for chemical sensing; it is relatively easy to create a dense array of millions of identical sensors, and DNA sequencing benefits from such massively parallel operation. However, if one is making a dedicated biosensor for a single target,70–77,79 this advantage is lost. Rather, one needs to forgo substantial chip real estate to add the extended gate into an integrated device. Alternatively, one can connect a commercially available FET part to an extended gate electrode,70–73,80,81 design and fabricate a custom MOSFET for the sensor,74–77,79 or directly functionalize the leads of the FET device.82 The resulting devices are inexpensive but can lead to incommensurate sizing of the floating gate with respect to the FET channel. Essentially, one wants the capacitance of the sensing surface at the end of the extended gate electrode [ in Eqs. (2) and (3) appearing below] to be comparable to the capacitance of the gate/insulator/semiconductor stack in the FET. Such a situation ensures sensitivity to chemical binding events on the extended gate electrode. However, commercial FETs are tiny, and so one generally has a mismatch between the size of the extended gate, its sensing area, and the size of the commercial FET. This is not an insurmountable problem, but it is a consideration when one is considering high throughput, low cost manufacturing of relatively large, individual sensors.
III. PRINTED ELECTROLYTE-GATED TRANSISTORS
We have been pursuing an alternate strategy, based on printed electrolyte-gated transistors (EGTs), that is well suited for microfluidic sensing using a floating (extended) gate architecture. As the name implies, this device merges two concepts: printed electronics and electrolyte gating. We briefly describe each of these concepts below.
A. Electrolyte gating
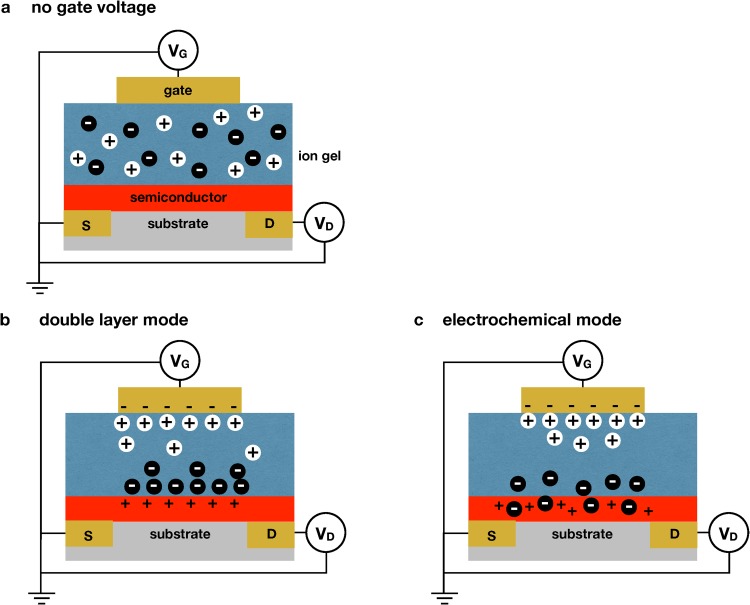
As its name implies, an EGT uses an electrolyte to gate the transistor, rather than a conventional dielectric. It is worthwhile to clearly define an EGT here, as there is some inconsistency in the literature (including in our own publications). The ISFET and extended gate FET in Fig. 1 can be considered “electrolyte-gated FETs;” the field effect transistor is still gated by a conventional dielectric, but there is a salt solution that is in series with the dielectric between the gate electrode and the semiconductor. We define an EGT (Fig. 2) to correspond to systems that do not have the dielectric layer such that the electrolyte is in direct contact with the semiconductor. The transistor is gated by either having ions from the electrolyte penetrate into the semiconductor or through the creation of an electric double-layer proximate to the semiconductor. Owing to the permeability of organic semiconductors, it is also possible to have mixtures of both current mechanisms within a single transistor.51
FIG. 2.
Mechanism of operation of an electrolyte gated transistor (EGT). (a) Schematic of an electrolyte gated transistor using a gel electrolyte as the dielectric in a stacked configuration. In the absence of an applied field, the ions are distributed uniformly within the gel. (b) If the ions cannot penetrate the semiconductor, application of an electric field to the gate electrode physically separates the ions within the gel to form double layers at the top and bottom of the gel. (c) If the ions can penetrate the semiconductor, application of an electric field to the gate electrode leads to the electrochemical doping of the semiconductor.
Our EGT sensors rely on the first approach by replacing the dielectric layer with an ion gel.83 An ion gel comprises an ionic liquid that is mechanically stabilized by a block polymer; our sensing experiments3–5,33,34 use 1-Ethyl-3-methylimidazolium bis(trifluoromethylsulfonyl)imide (EMIM/TFSI) as the ionic liquid and poly(styrene)--poly(ethyl acrylate)--poly(styrene) as the stabilizer.84 This system forms a gel because the poly(styrene) blocks of the copolymer are less miscible with the ionic liquid than the poly(ethyl acrylate) blocks, creating a network of nodes of poly(styrene) that are bridged by the poly(ethyl acrylate) chains within a matrix of ionic liquid. When an electric field is applied to the ion gel, the ions penetrate the semiconductor [Fig. 2(c)] to create a dense collection of holes. The 30 nm thickness of the semiconductor layer leads to very high specific capacitances of F/cm.51
Our EGTs have a lot of similarity to so-called organic electrochemical transistors (OECTs), which employ aqueous electrolytes instead of the ion gel and often use the conducting polymer PEDOT:PSS as the channel material.43,85 The mode of operation of PEDOT-based OECTs is electrochemical like our P3HT EGTs, the difference being that the PEDOT OECT is normally ON when the gate voltage is off. A positive gate voltage turns the device OFF. OECTs have been used in a variety of chemical sensing schemes which have recently been reviewed.43,44
In general, EGTs (and OECTs) have a high specific capacitance, owing to the charge separation illustrated in Figs. 2(b) and 2(c), and consequently operate at a lower voltage when compared to polarizing a conventional dielectric layer. In the context of Eq. (1) for the transistor drain current in the saturation regime, the resulting large value of implies that small changes in the threshold voltage created in a sensing environment are amplified into large changes in the drain current.51 This amplification is the origin of the high quasistatic gain achievable with EGTs.
One of the limitations of ion gels in EGTs is that the switching time (– Hz) in the best cases86 is considerably slower than conventional FETs. Slow switching is a limitation in computing applications. For example, typical modern central processing units (CPUs) have switching speeds in the GHz range, far faster than it is possible to polarize and depolarize an ion gel. Fortunately, such switching speeds are overkill for a biosensor. Rather, the rate-limiting step for biosensing is the mass transport to the sensor surface, which typically occurs on time scales of minutes. As a result, sweeping the gate voltage at circa 50 mV/s, which leads to good sensing performance, corresponds to sampling rates of circa 20 s. Such rates are sufficient to establish equilibrium binding to the surface.
B. Printed electronics
Additive manufacturing methods such as printing are particularly attractive for low cost production of high-volume, single-use sensors, especially because they can be implemented in continuous roll-to-roll formats common to the newsprint, publishing, and packaging industries.87–90 Widespread, commercially successful examples of printed sensors include glucose test strips and the home pregnancy test.91,92 Indeed, the increased availability of electronic inks based on conducting, semiconducting, and insulating materials makes printing of complete EGT-based sensors achievable. There are already examples in the literature of sheet-based aerosol-jet and ink jet printed FET- and EGT-based circuits that have more complexity than the sensor designs discussed here.51,93–95 However, a challenge is to translate these early results to roll-to-roll processes and such research is ongoing.
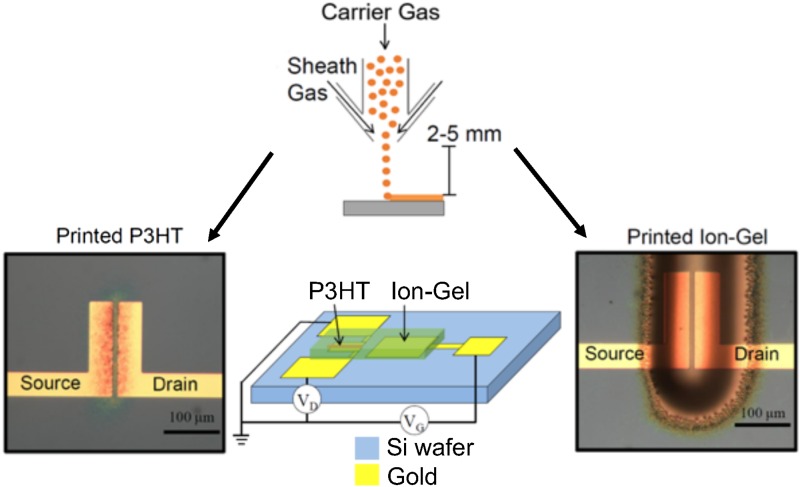
Currently, our printed EGT devices use a combination of conventional lithography and printed electronics3 illustrated in Fig. 3. In the first step, we lithographically pattern gold-on-chrome electrodes onto a silicon wafer in the desired locations. We then print the semiconductor, which in our case is the organic semiconductor poly(3-hexylthiophene), or P3HT. P3HT is dissolved in chloroform with terpineol added as a cosolvent. P3HT is then printed over the source and drain electrode using an aerosol-jet printer. The ion gel is printed subsequently over the channel and the gate electrode, thereby completing the EGT circuit. Following this, polstyrene is printed over the ion gel as an encapsulation layer. This prevents oxygen from diffusing through the ion gel and doping the semiconductor on the time scale of our experiments.
FIG. 3.
Printing of an EGT. The P3HT semiconductor and ion gel are aerosol-jet printed sequentially onto a silicon wafer. The aerosol-jet printing, illustrated schematically in the upper panel, uses a carrier flow containing the relevant ink (P3HT or ion gel) and a sheath flow that focuses the carrier jet onto the surface. The electrodes and contact pads are created prior to printing using the conventional photolithography.
In contrast to CMOS electronics, the length scales accessible by printed electronics are ideally suited for integration with microfluidics. The component size for printed electronics is set by the size of the spots of ink that can be deposited during printing; for aerosol-jet printing, this sets the feature sizes to be circa 10 m. Soft lithography in PDMS using SU-8 molds readily produces fluidics with these length scales.96 It is relatively easy to increase the feature size in printed electronics, which opens the possibility for creating molds from simpler methods (e.g., 3D printing97–100) rather than lithographically producing SU-8 molds.
We have produced our devices on a silicon substrate, which easily bonds to the PDMS microfluidics. It is also possible to print devices on a wide range of substrates, including glass and flexible substrates.101–106 While glass is easily used with PDMS microfluidics, the polymeric substrates can pose integration challenges with the PDMS. A possible solution to the integration problem is to print both the electronics and the microfluidics, as done for a recently reported sensor.107
IV. OPERATION AND APPLICATIONS OF EGTS FOR BIOSENSING
A. Mechanism of detection in a floating gate EGT
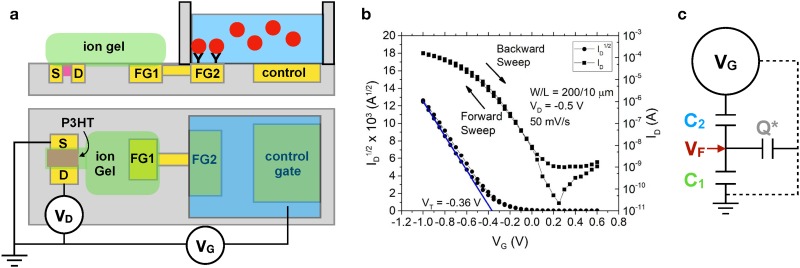
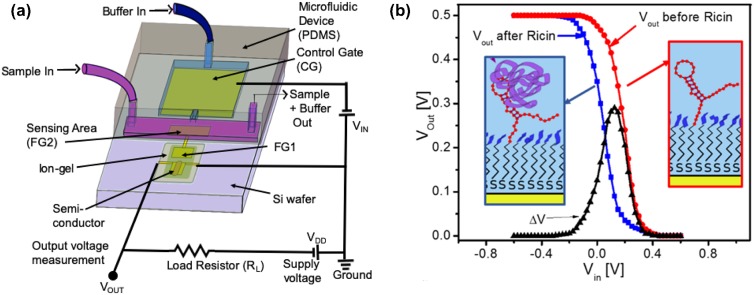
Figure 4(a) provides a schematic representation of an EGT biosensor using a floating gate architecture. The left side of the device is the EGT analogous to Fig. 2(a). The left arm (FG1) of the floating gate electrode communicates with the aqueous environment electronically by a via to the right arm (FG2) of the floating gate. The right arm of the floating gate is functionalized with the capture agent and communicates through the aqueous electrolyte to the control gate. This architecture is quite flexible with respect to the right arm of the floating gate, and we have demonstrated sensing functionality for DNA hybridization,3 protein binding to DNA aptamers,4,5 protein binding to antibodies,5 and self-assembled monolayers (SAMs)33,34 on the floating gate.
FIG. 4.
Model for EGT detection. (a) Schematic illustration of an EGT sensor using a static fluid reservoir. The P3HT is printed first to connect the source and drain electrodes, and the ion gel is printed second to connect the P3HT to the left arm of the floating gate (FG1). The floating gate extends into the aqueous environment, such that the right arm of the floating gate (FG2) can be functionalized for sensing. (b) Example of a transfer curve obtained by sweeping the control gate voltage, , at a rate of 50 mV/s and measuring the drain current, .34 The parameters and refer to the dimensions of the printed P3HT semiconductor. Reproduced with permission from Thomas et al., J. Phys. Chem. Lett. 9, 1335 (2018). Copyright 2018 American Chemical Society. (c) Model for the EGT sensor. The quantities and refer to the lumped capacitance of the EGT portion of the device and the aqueous portion of the device, respectively. Fitting the experimental data requires accounting for an additional parasitic capacitance via the stored charge . The model provides an approach to relate the applied gate voltage, , to the voltage at the floating gate.
In an experiment using the setup in Fig. 4(a), we sweep the control gate voltage, , and measure the drain current, , between the source and drain electrodes, producing a transfer curve such as the one in Fig. 4(b).34 As the gate voltage becomes more negative, the transistor turns ON and the drain current increases, which is the expected behavior for a p-type semiconductor such as P3HT. From Eq. (1) for a transistor in the saturation regime, the threshold voltage is obtained from a plot of vs by extrapolating the linear fit to zero current.
We have conducted a series of systematic experiments33,34 to determine how the voltage imposed on the control gate is related to the voltage at the floating gate, since is responsible for polarizing the ion gel and thus turning ON the EGT. The first set of experiments33 varied the size of the electrodes to investigate the role of the relative capacitances of each interface. These experiments revealed that the device operation is captured by the equivalent circuit model in Fig. 4(c). Applying voltage to the control gate sets up double layers at all of the interfaces within the device except the semiconductor/ion gel interface, where electrochemical doping occurs [e.g., Fig. 2(b)]. The double layers and electrochemical doping can be modeled by their capacitance. These interfaces can then be lumped into two overall capacitances, with representing the capacitance of interfaces on the ion gel side of the device in Fig. 4(a) and representing capacitances within the aqueous phase. A model with only these two capacitors predicts that
| (2) |
Unfortunately, this model fails to capture the experimental data as the ratio of the floating gate area to the P3HT area changes.33 Rather, we need to include an additional parasitic capacitance that accounts for a charge, , that can be stored elsewhere within the device. The exact nature of this parasitic capacitance remains a subject of investigation, but we have speculated33 that a fraction of the charge that would otherwise be stored on the floating gate is lost to the parasitic capacitor. As a result, that charge is no longer available to bias the transistor, shifting the response. The analysis of the complete circuit model in Fig. 4(c) furnishes33
| (3) |
where is the fraction of the charge on that is stored in the parasitic capacitor. Equation (3) fits the experimental data33 with , and a more detailed discussion of this effect appears in Ref. 33.
Equation (3) reveals that the sensor is capable of detecting the accumulation of the material near the floating gate surface FG2 since this material would affect and thus the relationship between the applied control gate voltage and the floating gate voltage that is turning the transistor ON. This effect is easily verified by creating self-assembled monolayers (SAMs) of alkane thiols of varying lengths at the gold FG2 surface33 since the capacitance of the SAM is an inverse to the thickness of the layer.108 In the context of biosensing, such changes could occur due to the capture of large neutral molecules.
Palazzo et al.,17 using an EGT with the gate electrode above the electrolyte [i.e., the EGT equivalent to the ChemFET configuration in Fig. 1(b)], proposed that there exists an additional capacitance within the device that was not considered directly in our previous work.33 Explicitly, the lumped capacitance in our model arises from the pair of capacitors due to the double layers formed at the control gate and the floating gate. Palazzo et al.17 proposed that, for biosensing, there exists a capacitor between the two in our model that accounts for the formation of a Donnan equilibrium created by the immobilized charges at the capture site. From an operational standpoint, the additional capacitance created by the Donnan layer does not affect Eq. (3) because it would appear within the lumped capacitance . However, the presence of a Donnan layer permits capacitive sensing outside the Debye layer, thereby overcoming a key limitation of field effect transistor detection.17
It is also worthwhile to understand how the sensor responds to the accumulation of charge, for example, during DNA hybridization or the capture of charged macromolecules. To decouple charge accumulation from changes in capacitance, subsequent experiments34 took advantage of a SAM of 11-mercaptoundecanoic acid (MUA) adsorbed onto the right arm of the floating gate. MUA is deprotonated by increasing pH, which leads to the accumulation of a charge close to the surface, without a change in capacitance.109 These experiments revealed that the model in Eq. (3) in the presence of charge accumulation becomes34
| (4) |
where accounts for potential changes on the right arm of the floating gate. While we have focused here on how arises due to charge accumulation at the sensor surface by deprotonation of a SAM,34 such potential changes could arise from the adsorption of a charged molecule such as DNA or from adsorption-induced surface dipoles.33,34 Equation (4) illustrates the advantage of working at low voltages, since we want the device to be sensitive to the term, which is typically a few hundred mV for SAMs and DNA.
Equation (4) represents our current working model for EGT detection, where the potential on the floating gate plays the role of the gate voltage in Eq. (1). Following Eq. (1), changes in capacitance lead to a change in the slope of vs , while changes in surface potential lead to a horizontal shift in the curve. We can thus discern the sensing mechanism directly from the inspection of the transfer curves obtained with and without a given analyte.
B. Device layout for biosensing
The device layout used in Fig. 4 is ideal for understanding the basis for EGT biosensing but is less useful for applications. First, the use of a quiescent fluid above the sensing pad leads to slow diffusional transport to the sensor surface. Second, comparing the threshold voltages for two different scenarios is suboptimal in terms of the signal gain.
Figure 5 provides a more robust biosensor layout for EGT detection.4 To improve detection time, the system uses flow over the sensing area of the floating gate. Since the floating gate needs to be capacitively coupled to the control gate, there is an additional flow from the control gate toward the floating gate, which keeps the control gate pristine. To improve detection sensitivity, the sensor is configured as an inverter by adding a resistor between the drain and the supply voltage. The output voltage, , is obtained between the drain and the load resistor. Figure 5(b) shows the advantage of operating the device as an inverter. As the input voltage to the control gate is swept to negative voltages, there is a rapid rise in the output signal over a very small change in , i.e., a high gain. Subtracting two inverter curves gives a distinct peak in the output voltage, which we interpret as the signal for sensing applications. Measuring curves or from an inverter provides equivalent information,111 but the inverter configuration is preferable due to the gain. We also find converting the inverter output to signal peaks, as in Fig. 5(b), to be an intuitive way to visualize the data when compared to analyzing differences between curves.
FIG. 5.
Improved EGT detection using flow and an inverter circuit. (a) Flow-based EGT detection configured as an inverter. The system has two connected microfluidic channels, one (purple) for sample flow over the right arm of the floating gate and a second (blue) to keep the control gate pristine. The addition of the load resistor between the drain and the supply voltage converts the circuit of Fig. 4(a) into an inverter. Instead of measuring the drain current to create a transfer curve, experiments using an inverter configuration measure the potential between the drain electrode of the EGT and the load resistor. (b) Example of inverter signals during the detection of ricin. The black signal is the difference between the measured before and after exposure to ricin. The insets illustrate that the sensor surface was functionalized with an aptamer for the B-subunit of ricin.110 Panel (a) adapted from and panel (b) reproduced with permission from White et al., ACS Sens. 1, 1213–1216 (2016). Copyright 2016 American Chemical Society.
C. Biosensing applications
Our first work using the EGT platform3 focused on the detection of DNA hybridization using the quiescent fluid device in Fig. 4(a). These experiments demonstrated the proof-of-principle for the platform; the hybridization equilibrium was well described by a Langmuir isotherm, with sensitivity to mismatches in the target strand. However, the sensitivity of the device was relatively low, especially when compared with fluorescence detection.
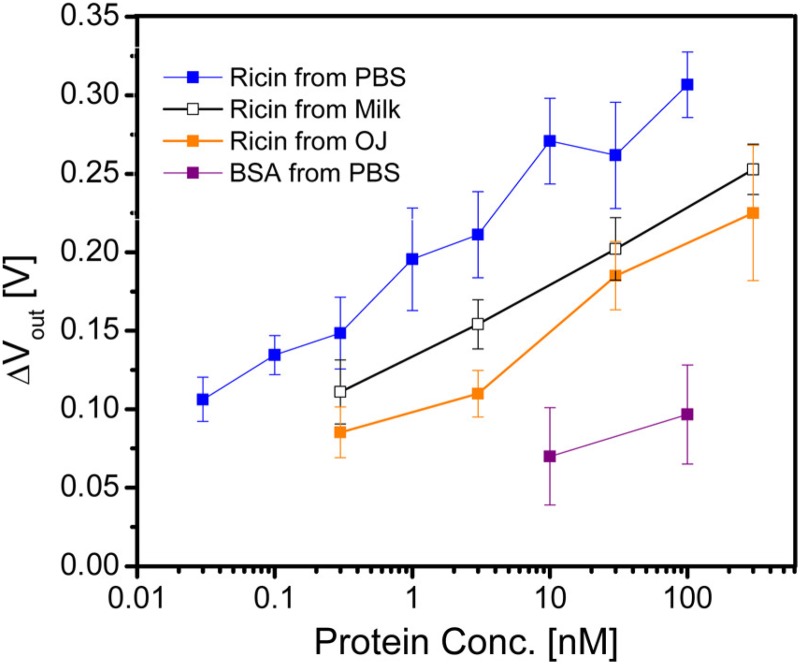
Our subsequent biosensing work4,5 thus took advantage of the flow-based inverter configuration in Fig. 5(a) for faster, more sensitive analysis. The first experiments4 demonstrated the ability to detect ricin subunit B using a DNA aptamer110 as the capture agent. Figure 6 shows that the EGT platform can successfully detect ricin at low concentrations, even from relatively complex food matrices such as milk and orange juice, without any sample cleanup or preconcentration. The approach to measure the inverter signal for these experiments is illustrated in Fig. 5(b), using the peak of as the signal. There is a slight reduction in the sensitivity of the device in complex media, which is consistent with previous experiments using this DNA aptamer110 and thus attributable to the binding agent rather than the sensor architecture.
FIG. 6.
EGT response4 in an inverter configuration to varying concentrations of ricin subunit B dissolved in phosphate buffered saline solution (PBS), milk, and orange juice, using a DNA aptamer110 for the capture agent. The control experiment uses the same device with bovine serum albumin (BSA) dissolved in PBS. The error bars correspond to the standard deviation between 3 and 5 devices. Reproduced with permission from White et al., ACS Sens. 1, 1213 (2016). Copyright 2016 American Chemical Society.
We have also demonstrated the detection of gluten in the EGT platform.5 This is a considerably more challenging sensing problem, due to the strongly reducing chemicals needed to extract gluten from food112 and the ill-defined chemical nature of gluten, which varies by source. In addition to detection using a DNA aptamer,113 we also used a commercially available antibody for wheat gluten114 and raised a custom antibody for barley gluten.5 These different capture agents were challenged with different concentrations of gluten extracted from barley, wheat, and corn, where the latter serves as the control experiment. In addition to achieving detection down to the “gluten-free” limit of 1 g/ml of gluten and negligible signal for the corn control, the three different sensors each yielded different responses depending on whether they were challenged with wheat or barley. This result opens up the potential for using an array of EGTs and capture agents to obtain chemical fingerprints for complicated analytes.5
Overall, we have demonstrated that EGTs can achieve competitive sensitivities in a label-free, nonoptical format from complex matrices without the need for any sample cleanup or preconcentration.4,5 The selectivity of the sensor arises from the dissociation constant, , for the capture agent, and we have generally been able to achieve a limit-of-detection of approximately 0.1, where has units of concentration. Since the sensing pad is a gold electrode, there are no impediments to using either DNA aptamers4,5 or antibodies5 as the capture agent. Moreover, it is relatively easy to passivate the surface with poly(ethylene glycol) to prevent nonspecific adsorption.4 There exist approaches to bind capture agents to and passivate other surfaces, but gold is particularly convenient to use because the chemistry for these reactions is very mature.
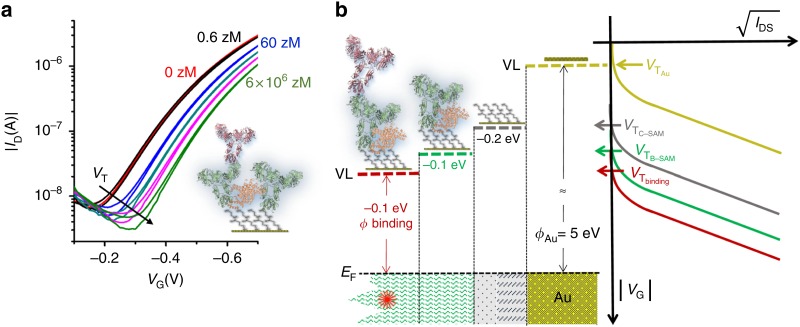
EGTs without an extended gate have also demonstrated competitive sensivities. Recent studies from the Torsi group have evidenced EGTs that can achieve a limit of detection down to a single protein via functionalization of the gate electrodes with chem-SAMs and bio-SAMs, using bovine serum albumin (BSA) to passivate the surface.28,31 Due to the limitations of the Debye length, de-ionized water was used as the gating electrolyte. The model proposed for the sensing mechanism of the single-molecule transistor (SiMoTs) assumes domains of SAMs that electrostatically cooperate through hydrogen bonding. Once the protein is bound to the bio-SAM, there is a change in the work function of the gate electrode that is propagated through the domain, which, in turn, changes the threshold voltage of the SiMoT, as shown in Fig. 7. Other gate-functionalized EGTs have also exploited SAM chemistry to selectively react with biomolecules, such as dopamine,13 while others have used different methods to immobilize antibodies37,38 aptamers,24 cells,27 and proteins.20
FIG. 7.
SiMoT sensing measurements. (a) Transfer curves with varied IgG concentrations of 6 zeptoMolar (zM, black curve), zM (blue curve), zM (dark curve), zM (magenta curve), and zM (light green curve) in PBS standard solutions. (b) Negative shifts in the work function with corresponding threshold voltage shifts at each functionalization step. EF is the gate electrochemical potential and VL is the vacuum level. Reproduced with permission from Macchia et al., Nat. Commun. 9, 3223 (2018). Copyright 2018, Author(s), licensed under Attribution 4.0 International (CC BY 4.0).
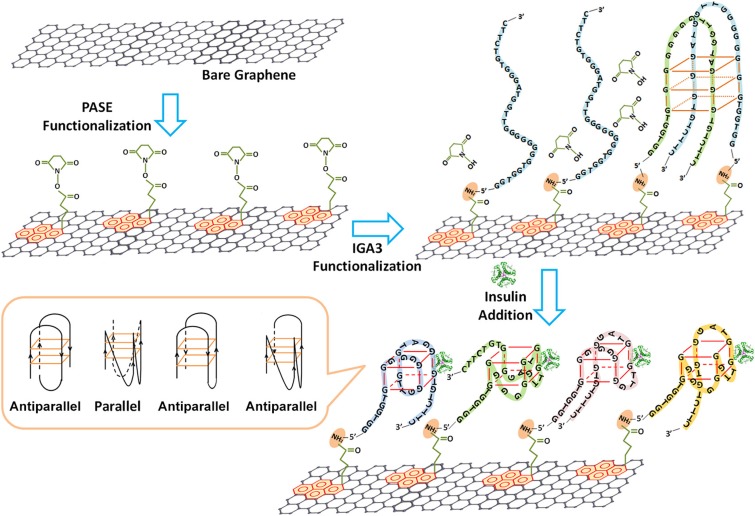
Similar to ChemFETs, the semiconductor of EGTs can be functionalized with capture agents at the semiconductor surface, and the chemistry can be tailored to expose a variety of bioreceptors.21,32 In addition to eliciting a change in capacitance upon binding, the semiconductor channel can also become doped. The response is thus a change in the drain current, or shift in the threshold voltage, depending on the type of doping. Hao et al. demonstrated this through functionalization of graphene with aptamers that specifically bind to insulin, causing deformation of the aptamer, as shown in Fig. 8.21 The pH of the electrolyte (7.4) was above the isoelectric point of insulin (5.8), so the insulin was weakly and negatively charged, causing the change in current and the Dirac voltage to be attributed to charge transfer from either the insulin or the deformed aptamer. In the case of organic electrochemical transistors, electrochemical doping of the ion permeable channel is responsible for the functionality of the sensor. Organic electrochemical transistors also elicit a change in the effective gating, or electrochemical doping, upon binding events at the functionalized channel surface and have been used to sense bacteria,11 metabolites,23 and viruses.25
FIG. 8.
Functionalization of graphene with 1-pyrenebutanoic acid succinimidyl ester (PASE), followed with IGA3 (aptamer) functionalization through a Schiff-base reaction. Upon binding to insulin, the aptamer complexes form both parallel and antiparallel G-quadruplexes, bringing the aptamer–insulin complex closer to the surface of graphene. Reproduced with permission from Hao et al., ACS Appl. Mater. Interfaces 9, 27504–27511 (2017). Copyright 2017 American Chemical Society.
The compatibility between the electrolyte and the semiconductor limits the versatility of this approach but can be overcome by the usage of an ion selective membrane16 or by passivating the surface with the bioreceptor.14,17 Functionalization of a phospholipid bilayer containing the capturing agents to the organic semiconductor surface causes capacitance changes at the semiconductor/electrolyte interface. The use of the phospholipid layer prevents ion diffusion from the electrolyte into the semiconductor, confirmed by low hysteresis in – curves along with low (nA) gate leakage currents. The sensing mechanism for the reported capture of negatively charged proteins, such as streptavidin and C-reactive protein, is the increase in drain current and shift in threshold voltage compared to EGTs with no exposure to the protein.28 This approach allows for both specific detection of biomolecules and detection beyond the Debye length in high ionic strength solutions ( nm) through the formation of Donnan equilibrium at the protein capture site.17
V. OPPORTUNITIES FOR MICROFLUIDICS IN EGT BIOSENSORS
Having provided an overview of EGT biosensing, we now explore a trio of possible approaches to improve EGT detection in the future via microfluidics. We consider both straightforward modifications, as well as some more speculative ideas related to control of the environment near the sensor surface.
A. Sensor multiplexing
Multiplexing of the sensors represents the most straightforward approach to improve sensor performance. There has been only minimal work toward multiplexing of EGT sensors, despite the relative simplicity of the process. For example, we have connected two sensors in the same inverter circuit4 to provide direct subtraction of the reference and test sensor during a measurement. We also fabricated multiple instances of the flow-based device in Fig. 5 on a single wafer for our multipixel approach to gluten detection.5 While useful steps forward, neither of these approaches takes advantage of the inherent advantages of microfluidics to create relatively complicated fluidic flow paths, especially in compact formats, that would enable improved device performance.
For a given target, it is desirable to make four measurements per sample, using all combinations of blank solution vs sample solution and functionalized sensor vs blank sensor. Such an experiment could be implemented using a multiplexed design. This quartet of experiments provides a complete set of control experiments. As indicated by the error bars in Fig. 6, there is also some variability in the signal obtained from a single EGT sensor. While the latter variability is sure to decrease as the EGT biosensor platforms mature, in particular, if they ultimately move to a commercial fabrication facility, it likely will remain desirable to add an additional layer of multiplexing to capture sensor-to-sensor variability as well. Even a 5-fold test would only involve 20 separate sensors on the chip, well below the massive levels of integration that have been achieved in microfluidic devices for almost 20 years now.115 Internal calibration can be achieved by subtracting the output of two EGTs to measure relative amounts.4
A key challenge in creating multiplexed devices is separately functionalizing different sensors on the device and alignment of the fluidics with the printed sensors. The former problem is readily solved by reversibly bonding a fluidic layer for flowing solution over the desired sample pads and then replacing that layer by the fluidic system for sample handling prior to use. Alternatively, if the pads are large enough, simply creating wells above each pad for functionalization is possible. The alignment of the microfluidics does not represent a significant challenge due to relatively large sizes of the control gate and floating gate required to optimize the device operation.33 We thus view multiplexing as the most natural microfluidic route for immediate improvements in EGT biosensors.
B. Sample preconcentration
Our experiments4,5 revealed that our EGT sensor typically operates with a limit of detection of 0.1 for the capture agent. The limit of detection in the EGT is affected by the device-to-device variability in the signal, as indicated by the error bars in Fig. 6 and the noise in the control experiment. Both of these device-related issues can be ameliorated somewhat by improvements in both the fabrication process and the device design. But, it is also desirable to consider approaches to improve the limit of detection via microfluidics.
The standard approach to lower the limit of detection is sample preconcentration prior to analysis, such that the sample sent to the sensor is at a higher concentration than the original material. Sample preconcentration effectively acts as a multiplicative factor for the limit of detection.
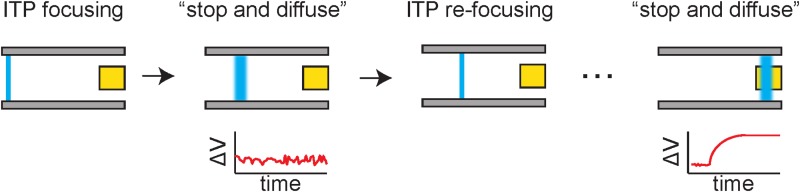
One particularly attractive method for sample preconcentration within a microfluidic device for charged molecules is isotachophoresis (ITP). Eid and Santiago116 have provided a thorough recent review of ITP, with a particular focus on how ITP can improve heterogeneous biomolecular reactions such as those arising from capture agents bound to the surface of a sensor. Briefly, ITP involves two electrolytes, a leading electrolyte with a high electrophoretic mobility and a trailing electrolyte with a low electrophoretic mobility. If the analyte of interest has an electrophoretic mobility that lies between the trailing and leading electrolytes, it will become concentrated at the interface between them. The thickness of the interface between the different electrolytes is set by molecular diffusion and is thus quite sharp. A particularly attractive feature of ITP is that it is a self-focusing method, since analytes in the leading electrolyte will fall behind (and vice versa for the trailing electrolyte), so there is no need to provide a sharp injection plug of the sample. Indeed, one of the limitations in ITP is that the sample plug may pass over the sensor so quickly that the time for reaction at the surface becomes a limiting factor. This effect can be attenuated by using a stop-and-diffuse approach,117 where the electric field is turned off when the sample plug reaches the sensor, thereby allowing additional time for reactions from the concentrated sample. Although million-fold increases in sample concentration are theoretically possible, typical enhancements in biological fluids are around 10 000-fold.116
Compared to a fluorescence-based assay, there are some minor challenges to implementing ITP in an EGT. It is unlikely one can simultaneously apply an electric field to drive the ITP while also modulating the gate voltage, as the ITP electric field would affect the potential at the gate electrode. Figure 9 illustrates a possible approach based on a pulsed ITP method. At the start of the process, ITP is used to focus the sample. Since we are using a label-free method, this band cannot be imaged in the manner used for fluorescence-based detection.117 If the sample has not reached the detector, then there should be no signal. The ITP can be turned on and off repeatedly while driving the analyte toward the sensor surface. We expect to see a sudden spike in the signal when the ITP band reaches the sensor surface. At this point, the pulsed ITP operation can be stopped and we can use a conventional EGT measurement by sweeping the gate voltage. Note that, since ITP is a focusing method, there is no loss in sensitivity by periodically releasing that electric potential and allowing the ions to diffuse; once the potential is reapplied, the bands will automatically sharpen again. As an effect, the proposed operating method is akin to running many “stop-and-diffuse” ITP protocols117 until the analyte band reaches the sensor surface. The key challenge is balancing the rate of switching between ITP and EGT sensing and the magnitude of the electric potential driving the electrophoresis of the analyte band to the sensor surface. The second challenge is identifying an appropriate leading and trailing electrolyte for new analytes of interest, although ITP recipes have been developed for a large number of useful biological moieties, in particular, for nucleic acids in complex media.118
FIG. 9.
Principle behind and ITP detection method using an EGT. The device could switch between steps of ITP (to focus and move the sample, illustrated by the blue color) and EGT measurement (to detect the analyte), thereby gaining the increase in detection sensitivity without the need to carefully control the sample preparation and transport. A signal is only measured when the sample is stopped over the floating gate (gold square).
C. Improved transport to the sensor surface
In addition to increasing the equilibrium binding to the surface, preconcentration also increases the kinetics of the reaction through an increase in the concentration gradient. Both of these factors are important for sensor performance; the equilibrium scenario sets the limit of detection, but also the kinetics of the binding reaction govern the speed of detection. Thus, it is also worthwhile to consider other microfluidic approaches to enhance detection speed in addition to detection sensitivity.
To a large extent, EGT sensors have worked with static reservoirs, such as the one in Fig. 4(a), or droplets.3,6–14,16–19,21–32,34–41 Squires et al.119 provide a basic understanding of how flow can enhance sensor operation by confining the diffusion-dominated region to a boundary layer near the sensor surface, building on classical chemical engineering analysis. There are considerable opportunities for improving the mass flux to the EGT sensor surface by using the results of scaling theory or insights gained by numerical solutions to the convection-diffusion-reaction equations.119 Naturally, any design optimization for the fluid flow will be constrained by the size scales imposed by the printed electronics and the operating mechanism for the EGT. In particular, the size of the floating gate for the EGT must be large compared to the semiconductor channel, which may pose challenges if that sensing pad needs to be confined within a thin microfluidic channel.
Another approach to enhance the reactions at the surface is to promote mixing within the microchannel near the sensor surface.119 One option is the herringbone mixer model in Fig. 10, which creates a chaotic flow pattern in the microchannel,120 but a myriad of other passive micromixers that have been developed (see Ref. 121 for a recent review). In this context, it is important to keep in mind the goal of the mixer. Much of the literature on microfluidic mixing focuses on the important problem of the homogenization of two inlet streams at different concentrations. This is not the challenge in sensing, since the initial inlet stream is already homogeneous. Rather, the goal is to break up the depletion layer near the sensor surface119 through convective transport from the bulk. In this respect, a herringbone mixer patterned onto the ceiling of the microchannel could be particularly effective at enhancing the sensor speed. Lamination-based mixing strategies based on spilt-and-recombining of streams, while effective for creating a homogeneous fluid, are not optimal for use in conjunction with the EGT sensor.
FIG. 10.
The flow in a staggered herringbone mixer can be used to break the boundary layers near the sensor surface. (a) Schematic illustration of a staggered herringbone mixer. The insets show the flow patterns generated by the herringbone. (b) Fluorescence microscopy images of the mixing between two fluid streams. Reproduced with permission from Stroock et al., Science 295, 647–651 (2002). Copyright 2002 AAAS.
Overall, we view enhancing transport to the sensor surface via fluid flow as the most promising avenue for improving EGT sensor performance via microfluidics. This opinion is based on the combination of (i) the absence of much engineering in this direction to date and (ii) the extensive literature in microfluidics in this area for other applications. Of particular interest is the work done to optimize depletion boundary layers in electrochemical reactors,122 which involve both the use of herringbone mixers and multiple inlets/outlets for replenishing depletion layers.
VI. CONCLUDING REMARKS
EGTs are an emerging platform for biosensing. This electronic-based method provides label-free, nonoptical detection of biomolecules from complex media4,5 without any sample cleanup or preconcentration. The key advantage of EGTs with respect to their FET counterparts is their easy manufacturing by printing, which is both advantageous for the prototyping work needed to develop the technology and, in the longer term, for commercial production. Indeed, if the promise of printing electronics using the same roll-to-roll techniques used, for example, in newspaper print are realized, this would lead to a dramatic reduction in cost and increase in throughput. Converting existing EGT sensors on silicon or glass onto flexible substrates could combine the portability of paper microfluidic devices123 with even more sophisticated electronics than existing electrochemical paper sensors,124 potentially enabling a wealth of new applications.
As an emerging sensing area, much of the work to date has focused on the electronics side of the EGT and proof-of-principle experimentation. The time is ripe to deploy the microfluidics toolbox to convert these prototype sensors into automated, high precision systems. We have identified here the low-hanging fruit that would come from multiplexing, sample preconcentration, and controlling transport to the surface. This is by no means an exhaustive list of possible ways in which microfluidics can improve EGT biosensors, and there is a wealth of knowledge in microfluidic sensors in other contexts that can be translated to the EGT platform. In general, printed electronics is a promising route for integration with microfluidics, since the electronic platforms tend to use planar layouts with length scales in the hundred micrometer range that are ideally suited for microfluidic integration.
We would be remiss if we concluded our discourse without a few comments on EGT detection in general, beyond those that are likely to be remedied by the microfluidic solutions discussed here. There are some improvements on the device side that are relatively straightforward. For example, our EGT biosensors could employ higher mobility semiconductors that can produce even higher gain.125 Similarly, the parasitic capacitance problem33 needs to be thoroughly understood and then remedied. This detective work will require careful, systematic experimentation, but it is likely a solvable problem using existing techniques. We also see two major outstanding challenges in EGT detection in general that do not have straightforward, generic solutions.
The first major challenge is EGT-based detection of small, neutral analytes. Owing to their small size relative to an antibody and the sparseness of the capture agents on the surface, such molecules are unlikely to create measurable changes in the capacitance in Eq. (4). One possible solution is the use of shape-changing DNA aptamers, following recent work with these molecules in conventional ChemFETs.29 In this approach, binding of the neutral molecules induces a change in the amount of DNA proximate to the surface and thus affects the term in Eq. (4), not only the capacitance . Shape-changing aptamers certainly can address the problem, but their use is limited because it is not obvious that such aptamers can be identified for all relevant targets. A generic solution to the small neutral molecule problem would have broad applicability, as there are a range of medically relevant targets (e.g., cortisol126,127) that fall within this class of analytes.
The second major challenge is the detection of small charged molecules. It is clear that charges located proximate to the surface will affect , as evidenced by our work with SAMs of MUA.34 However, an oft-cited problem with transistor-based detection is that charge detection is not operable outside the Debye layer, which is circa 1 nm in typical biological solutions. A number of possible solutions to this problem have appeared in the FET biosensor literature, including using antibody fragments128,129 or self-hybridizing DNA aptamers130 to reduce the capture agent size, employing high frequency gate voltages131 or quickly pulsing the gate voltage132–136 to avoid the formation of the static Debye layer on the measurement time, or the use of a porous poly(ethylene glycol) (PEG) layer at the sensor surface.81,137–139 The latter approach is especially attractive since PEG can also prevent nonspecific adsorption to the sensor surface. Initial reports for this strategy proposed that the PEG layer creates a region of lower dielectric constant and thus extends the Debye length. However, a recent analysis of the experimental data140 suggests that the key effect is the formation of a Donnan equilibrium for the antibody/antigen within the PEG layer, which leads to a different decay length and provides an interesting analogy with the capacitive sensing mechanism for EGTs proposed by Palazzo et al.17
While we have identified material-based and electronic-based solutions that might be translated from FETs to EGTs for both of these key challenges, it is worthwhile to contemplate whether microfluidics offers novel solutions that are orthogonal to approaches based on the electronics design or capture agent chemistry. Such flow-based solutions would not only improve EGT detection but could be translated broadly to all transistor-based detection schemes.
ACKNOWLEDGMENTS
This work was supported by the Michael H. Baker Family Foundation. Demetra Z. Adrahtas was supported by a Biotechnology Training (Grant No. NIH T32GM008347).
Contributor Information
Kevin D. Dorfman, Email: .
C. Daniel Frisbie, Email: .
REFERENCES
- 1.Schena M., Shalon D., Davis R. W., and Brown P. O., Science 270, 467 (1995). 10.1126/science.270.5235.467 [DOI] [PubMed] [Google Scholar]
- 2.Anker J. N., Hall W. P., Lyandres O., Shah N. C., Zhao J., and van Duyne R. P., Nat. Mater. 7, 442 (2008). 10.1038/nmat2162 [DOI] [PubMed] [Google Scholar]
- 3.White S. P., Dorfman K. D., and Frisbie C. D., Anal. Chem. 87, 1861 (2015). 10.1021/ac503914x [DOI] [PubMed] [Google Scholar]
- 4.White S. P., Sreevatsan S., Frisbie C. D., and Dorfman K. D., ACS Sens. 1, 1213 (2016). 10.1021/acssensors.6b00481 [DOI] [Google Scholar]
- 5.White S. P., Frisbie C. D., and Dorfman K. D., ACS Sens. 3, 395 (2018). 10.1021/acssensors.7b00810 [DOI] [PubMed] [Google Scholar]
- 6.Zhu Z., Mabeck J. T., Zhu C., Cady N. C., Batt C. A., and Malliaras G. G., Chem. Commun. 2004, 1556 10.1039/b403327M [DOI] [PubMed] [Google Scholar]
- 7.Bernards D. A., Macaya D. J., Nikolou M., DeFranco J. A., Takamatsu S., and Malliaras G. G., J. Mater. Chem. 18, 116 (2008). 10.1039/B713122D [DOI] [Google Scholar]
- 8.Shim N. Y., Bernards D. A., Macaya D. J., DeFranco J. A., Nikolou M., Owens R. M., and Malliaras G. G., Sensors 9, 9896 (2009). 10.3390/s91209896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohno Y., Maehashi K., Yamashiro Y., and Matsumoto K., Nano Lett. 9, 3318 (2009). 10.1021/nl901596m [DOI] [PubMed] [Google Scholar]
- 10.Buth F., Donner A., Sachsenhauser M., Stutzmann M., and Garrido J. A., Adv. Mater. 24, 4511 (2012). 10.1002/adma.201201841 [DOI] [PubMed] [Google Scholar]
- 11.He R. X., Zhang M., Tan F., Leung P. H. M., Zhao X., Chan H. L. W., Yang M., and Yan F., J. Mater. Chem. 22, 22072 (2012). 10.1039/c2jm33667g [DOI] [Google Scholar]
- 12.Suspène C., Piro B., Reisberg S., Pham M., Toss H., Berggren M., Yassar A., and Horowitz G., J. Mater. Chem. B 1, 2090 (2013). 10.1039/c3tb00525a [DOI] [PubMed] [Google Scholar]
- 13.Casalini S., Leonardi F., Cramer T., and Biscarini F., Org. Electron. 14, 156 (2013). 10.1016/j.orgel.2012.10.027 [DOI] [Google Scholar]
- 14.Magliulo M., Mallardi A., Mulla M. Y., Cotrone S., Pistillo B. R., Favia P., Vikholm-Lundin I., Palazzo G., and Torsi L., Adv. Mater. 25, 2090 (2013). 10.1002/adma.201203587 [DOI] [PubMed] [Google Scholar]
- 15.Schmoltner K., Kofler J., Klug A., and List-Kratochvil E. J. W., Adv. Mater. 25, 6895 (2013). 10.1002/adma.201303281 [DOI] [PubMed] [Google Scholar]
- 16.Bandiello E., Sessolo M., and Bolink H. J., J. Mater. Chem. C 2, 10277 (2014). 10.1039/C4TC02075H [DOI] [Google Scholar]
- 17.Palazzo G., de Tullio D., Magliulo M., Mallardi A., Intranuovo F., Mulla M. Y., Favia P., Vikholm-Lundin I., and Torsi L., Adv. Mater. 27, 911 (2015). 10.1002/adma.201403541 [DOI] [PubMed] [Google Scholar]
- 18.Bihar E., Deng Y., Miyake T., Saadaoui M., Malliaras G. G., and Rolandi M., Sci. Rep. 6, 27582 (2016). 10.1038/srep27582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magliulo M., De Tullio D., Vikholm-Lundin I., Albers W. M., Munter T., Manoli K., Palazzo G., and Torsi L., Anal. Bioanal. Chem. 408, 3943 (2016). 10.1007/s00216-016-9502-3 [DOI] [PubMed] [Google Scholar]
- 20.Pappa A. M., Curto V. F., Braendlein M., Strakosas X., Donahue M. J., Fiocchi M., Malliaras G. G., and Owens R. M., Adv. Healthc. Mater. 5, 2295 (2016). 10.1002/adhm.201600494 [DOI] [PubMed] [Google Scholar]
- 21.Hao Z., Zhu Y., Wang X., Rotti P. G., DiMarco C., Tyler S. R., Zhao X., Engelhardt J. F., Hone J., and Lin Q., ACS Appl. Mater. Interfaces 9, 27504 (2017). 10.1021/acsami.7b07684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minamiki T., Hashima Y., Sasaki Y., and Minami T., Chem. Commun. 54, 6907 (2018). 10.1039/C8CC02462F [DOI] [PubMed] [Google Scholar]
- 23.Pappa A. M., Ohayon D., Giovannitti A., Maria I. P., Savva A., Uguz I., Rivnay J., McCulloch I., Owens R. M., and Inal S., Sci. Adv. 4, eaat0911 (2018). 10.1126/sciadv.aat0911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saraf N., Woods R. E., Peppler M., and Seal S., Biosens. Bioelectron. 117, 40 (2018). 10.1016/j.bios.2018.05.031 [DOI] [PubMed] [Google Scholar]
- 25.Hai W., Goda T., Takeuchi H., Yamaoka S., Horiguchi Y., Matsumoto A., and Miyahara Y., Sens. Actuators B 260, 635 (2018). 10.1016/j.snb.2018.01.081 [DOI] [Google Scholar]
- 26.Seshadri P., Manoli K., Schneiderhan-Marra N., Anthes U., Wierzchowiec P., Bonrad K., Di Franco C., and Torsi L., Biosens. Bioelectron. 104, 113 (2018). 10.1016/j.bios.2017.12.041 [DOI] [PubMed] [Google Scholar]
- 27.Chen L., Fu Y., Wang N., Yang A., Li Y., Wu J., Ju H., and Yan F., ACS Appl. Mater. Interfaces 10, 18470 (2018). 10.1021/acsami.8b01987 [DOI] [PubMed] [Google Scholar]
- 28.Macchia E., Manoli K., Holzer B., Di Franco C., Ghittorelli M., Torricelli F., Alberga D., Mangiatordi G. F., Palazzo G., Scamarcio G., and Torsi L., Nat. Commun. 9, 3223 (2018). 10.1038/s41467-018-05235-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatsuka N., Yang K. A., Abendroth J. M., Cheung K. M., Xu X., Yang H., Zhao C., Zhu B., Rim Y. S., Yang Y., Weiss P. S., Stojanović M. N., and Andrews A. M., Science 362, 319 (2018). 10.1126/science.aao6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos R., Borme J., Guerreiro J. R., Machado G., Cerqueira M. F., Pertovykj D. Y., and Alpuim P., ACS Sens. 4, 286 (2019). 10.1021/acssensors.8b00344 [DOI] [PubMed] [Google Scholar]
- 31.Macchia E., Manoli K., Holzer B., Di Franco C., Picca R. A., Cioffi N., Scamarcio G., Palazzo G., and Torsi L., Anal. Bioanal. Chem. 411, 4899 (2019). 10.1007/s00216-019-01778-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J., Chen X., Wan Q., Xiao M., Zhong D., Sun W., Zhang G., and Zhang Z., Nano Lett. 19, 1437 (2019). 10.1021/acs.nanolett.8b03818 [DOI] [PubMed] [Google Scholar]
- 33.White S. P., Dorfman K. D., and Frisbie C. D., J. Phys. Chem. C 120, 108 (2016). 10.1021/acs.jpcc.5b10694 [DOI] [Google Scholar]
- 34.Thomas M. S., White S. P., Dorfman K. D., and Frisbie C. D., J. Phys. Chem. Lett. 9, 1335 (2018). 10.1021/acs.jpclett.8b00285 [DOI] [PubMed] [Google Scholar]
- 35.Lin P., Yan F., and Chan H. L. W., ACS Appl. Mater. Interfaces 2, 1637 (2010). 10.1021/am100154e [DOI] [PubMed] [Google Scholar]
- 36.Toss H., Suspène B., Piro C., Yassar A., Crispin X., Kergoat L., Pham M., and Berggren M., Org. Electron. 15, 2420 (2014). 10.1016/j.orgel.2014.06.017 [DOI] [Google Scholar]
- 37.Casalini A., Dumitru A. C., Leonardi F., Bortolotti C. A., Herruzo E. T., Campana A., de Oliveira R. F., Cramer T., Garcia R., and Biscarni F., ACS Nano 9, 5051 (2015). 10.1021/acsnano.5b00136 [DOI] [PubMed] [Google Scholar]
- 38.Berto M., Casalini S., Di Lauro M., Marasso S. L., Cocuzza M., Perrone D., Pinti M., Cossarizza A., Pirri C. F., Simon D. T., Berggren M., Zerbetto F., Bortolotti C. A., and Biscarini F., Anal. Chem. 88, 12330 (2016). 10.1021/acs.analchem.6b03522 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q., Leonardi F., Casalini S., Temiño I., and Mas-Torrent M., Sci. Rep. 6, 39623 (2016). 10.1038/srep39623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuda S., Ono T., Kanai Y., Ikuta T., Shimatani M., Ogawa S., Maehashi K., Inoue K., and Matsumoto K., ACS Sens. 3, 200 (2018). 10.1021/acssensors.7b00851 [DOI] [PubMed] [Google Scholar]
- 41.Sensi M., Berto M., Candini A., Liscio A., Cossarizza A., Beni V., Biscarini F., and Bortolotti C. A., ACS Omega 4, 5374 (2019). 10.1021/acsomega.8b03319 [DOI] [Google Scholar]
- 42.Kaisti M., Biosens. Bioelectron. 98, 437 (2018). 10.1016/j.bios.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 43.Rivnay J., Inal S., Salleo A., Owens R. M., Berggren M., and Malliaras G. G., Nat. Rev. Mater. 3, 17086 (2018). 10.1038/natrevmats.2017.86 [DOI] [Google Scholar]
- 44.Bai L. M., Elosegui C. G., Li W. Q., Yu P., Fei J. J., and Mao L. Q., Front. Chem. 7, 313 (2019). 10.3389/fchem.2019.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Shi W., Song J., Jang H. J., Dailey J., Yu J., and Katz H. E., Chem. Rev. 119, 3 (2019). 10.1021/acs.chemrev.8b00016 [DOI] [PubMed] [Google Scholar]
- 46.Bergveld P., IEEE Trans. Biomed. Eng. BME-17, 70 (1970). 10.1109/TBME.1970.4502688 [DOI] [PubMed] [Google Scholar]
- 47.Bergveld P., IEEE Trans. Biomed. Eng. 19, 342 (1972). 10.1109/TBME.1972.324137 [DOI] [PubMed] [Google Scholar]
- 48.Toumazou C., Georgiou P., and Bergveld P., Electron. Lett. 47, S7 (2011). 10.1049/el.2011.3231 [DOI] [Google Scholar]
- 49.Caras S. and Janata J., Anal. Chem. 52, 1935 (1980). 10.1021/ac50062a035 [DOI] [Google Scholar]
- 50.Janata J., Electroanalysis 16, 1831 (2004). 10.1002/elan.200403070 [DOI] [Google Scholar]
- 51.Kim S. H., Hong K., Xie W., Lee K. H., Zhang S., Lodge T. P., and Frisbie C. D., Adv. Mater. 25, 1822 (2013). 10.1002/adma.201202790 [DOI] [PubMed] [Google Scholar]
- 52.Bergveld P., Sens. Actuators B 88, 1 (2003). 10.1016/S0925-4005(02)00301-5 [DOI] [Google Scholar]
- 53.van der Spiegel J., Lauks I., Chan P., and Babic D., Sens. Actuators 4, 291 (1983). 10.1016/0250-6874(83)85035-5 [DOI] [Google Scholar]
- 54.Mao S., Lu G., Yu K., Bo Z., and Chen J., Adv. Mater. 22, 3521 (2010). 10.1002/adma.201000520 [DOI] [PubMed] [Google Scholar]
- 55.Kim J. M., Jha S. K., Chand R., Lee D. H., and Kim Y. S., Biosens. Bioelectron. 26, 2264 (2011). 10.1016/j.bios.2010.09.047 [DOI] [PubMed] [Google Scholar]
- 56.Hagen J., Lyon W., Chushak Y., Tomczak M., Naik R., Stone M., and Kelley-Loughnane N., ACS Chem. Neurosci. 4, 444 (2013). 10.1021/cn300159e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammock M. L., Knopfmacher O., Naab B. D., Tok J. B. H., and Bao Z., ACS Nano 7, 3970 (2013). 10.1021/nn305903q [DOI] [PubMed] [Google Scholar]
- 58.Park H., Han G., Lee S. W., Lee H., Jeong S. H., Naqi M., AlMutairi A., Kim Y. J., Lee J., Kim W., Kim S., Yoon Y., and Yoo G., ACS Appl. Mater. Interfaces 9, 43490 (2017). 10.1021/acsami.7b14479 [DOI] [PubMed] [Google Scholar]
- 59.Lee H. W., Kang D., Cho J. H., Lee S., Jun D., and Park J., ACS Appl. Mater. Interfaces 10, 17369 (2018). 10.1021/acsami.8b03432 [DOI] [PubMed] [Google Scholar]
- 60.Bergveld P., Sens. Actuators 1, 17 (1981). 10.1016/0250-6874(81)80004-2 [DOI] [Google Scholar]
- 61.McLaughlin K., Dickson A., Weisberg S. B., Coale K., Elrod V., Hunter C., Johnson K. S., Kram S., Kudela R., Martz T., Negrey K., Passow U., Shaughnessy F., Smith J. E., Tadesse D., Washburn L., and Weis K. R., Reg. Stud. Mar. Sci. 12, 11 (2017). 10.1016/j.rsma.2017.02.008 [DOI] [Google Scholar]
- 62.Kiumra J., Ito N., Kuriyama T., Kikuchi M., Arai T., Negishi N., and Tomita Y., J. Electrochem. Soc. 136, 1744 (1989). 10.1149/1.2097003 [DOI] [Google Scholar]
- 63.Luo X. L., Xu J. J., Zhao W., and Chen H. Y., Sens. Actuators B 97, 249 (2004). 10.1016/j.snb.2003.08.024 [DOI] [Google Scholar]
- 64.Sakata T., Sugimoto H., and Saito A., Anal. Chem. 90, 12731 (2018). 10.1021/acs.analchem.8b03070 [DOI] [PubMed] [Google Scholar]
- 65.Martinoia S., Rosso N., Grattarola M., Lorenzelli L., Margesin B., and Zen M., Biosens. Bioelectron. 16, 1043 (2001). 10.1016/S0956-5663(01)00202-0 [DOI] [PubMed] [Google Scholar]
- 66.Bettaieb F., Ponsonnet L., Lejeune P., Ouada H. B., Martelet C., Bakhrouf A., Jaffrézic-Renault N., and Othmane A., Bioelectrochem. 71, 118 (2007). 10.1016/j.bioelechem.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 67.Kutova O., Dusheiko M., Klyui N. I., and Skryshevsky V. A., Microelectron. Eng. 215, 110993 (2019). 10.1016/j.mee.2019.110993 [DOI] [Google Scholar]
- 68.Nakata S., Arie T., Akita S., and Takei K., ACS Sens. 2, 443 (2017). 10.1021/acssensors.7b00047 [DOI] [PubMed] [Google Scholar]
- 69.Rothberg J. M., Hinz W., Rearick T. M., Schultz J., Mileski W., Davey M., Leamon J. H., Johnson K., Milgrew M. J., Edwards M., Hoon J., Simons J. F., Marran D., Myers J. W., Davidson J. F., Branting A., Nobile J. R., Puc B. P., Light D., Clark T. A., Humber M., Branciforte J. T., Stoner I. B., Cawley S. E., Lyons M., Fu Y., Homer N., Sedova M., Miao X., Reed B., Sabina J., Feierstein E., Schorn M., Alanjary M., Dimalanta E., Dressman D., Kasinskas R., Sokolsky T., Fidanza J. A., Namsaraev E., McKernan K. J., Williams A., Roth G. T., and Bustillo J., Nature 475, 348 (2011). 10.1038/nature10242 [DOI] [PubMed] [Google Scholar]
- 70.Al-Hardan N. H., Abdul Hamid M. A., Ahmed N. M., Jalar A., Shamsudin R., Othman N. K., Kar Keng L., Chiu W., and Al-Rawi H. N., Sensors 16, 839 (2016). 10.3390/s16060839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin L. T., Chou J. C., Chung W. Y., Sun T. P., and Hsiung S. K., Sens. Actuators B 71, 106 (2000). 10.1016/S0925-4005(00)00613-4 [DOI] [Google Scholar]
- 72.Batista P. D. and Mulato M., Appl. Phys. Lett. 87, 143508 (2005). 10.1063/1.2084319 [DOI] [Google Scholar]
- 73.Chen C. P., Ganguly A., Lu C. Y., Chen T. Y., Kuo C. C., Chen R. S., Tu W. H., Fishcer W. B., Chen K. H., and Chen L. C., Anal. Chem. 83, 1938 (2011). 10.1021/ac102489y [DOI] [PubMed] [Google Scholar]
- 74.Sakata T., Matsumoto S., Nakajima Y., and Miyahara Y., Jpn. J. Appl. Phys. 44, 2860 (2005). 10.1143/JJAP.44.2860 [DOI] [Google Scholar]
- 75.Kamahori M., Ishige Y., and Shimoda M., Biosens. Bioelectron. 22, 3080 (2007). 10.1016/j.bios.2007.01.011 [DOI] [PubMed] [Google Scholar]
- 76.Xue Q., Bian C., Tong J., Sun J., Zhang H., and Xia S., Sens. Actuators A 169, 282 (2011). 10.1016/j.sna.2011.01.017 [DOI] [Google Scholar]
- 77.Goda T. and Miyahara Y., Biosens. Bioelectron. 45, 89 (2013). 10.1016/j.bios.2013.01.053 [DOI] [PubMed] [Google Scholar]
- 78.Guan W., Duan X., and Reed M. A., Biosens. Bioelectron. 51, 225 (2014). 10.1016/j.bios.2013.07.061 [DOI] [PubMed] [Google Scholar]
- 79.Aliakbarinodehi N., Jolly P., Bhalla N., Miodek A., de Micheli G., Estrela P., and Carrara S., Sci. Rep. 7, 44409 (2017). 10.1038/srep44409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tarasov A., Gray D. W., Tsai M.-Y., Sheilds N., Montrose A., Creedon N., Lovera P., O’Riordan A., Mooney M. H., and Vogel E. M., Biosens. Bioelectron. 79, 669 (2016). 10.1016/j.bios.2015.12.086 [DOI] [PubMed] [Google Scholar]
- 81.Gutiérrez-Sanz Ó., Andoy N. M., Filipiak M. S., Haustein N., and Tarasov A., ACS Sens. 2, 1278 (2017). 10.1021/acssensors.7b00187 [DOI] [PubMed] [Google Scholar]
- 82.Zafar S., Lu M., and Jagtiani A., Sci. Rep. 7, 41430 (2017). 10.1038/srep41430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho J. H., Lee J., Xia Y., Kim B., He Y., Renn M. J., Lodge T. P., and Frisbie C. D., Nat. Mater. 7, 900 (2008). 10.1038/nmat2291 [DOI] [PubMed] [Google Scholar]
- 84.Tang B., White S. P., Frisbie C. D., and Lodge T. P., Macromolecules 48, 4942 (2015). 10.1021/acs.macromol.5b00882 [DOI] [Google Scholar]
- 85.Khodagholy D., Rivnay J., Sessolo M., Gurfinkel M., Leleux P., Jimison L. H., Stavrinidou E., Herve T., Sanaur S., Owens R. M., and Malliaras G. G., Nat. Commun. 4, 2133 (2013). 10.1038/ncomms3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bidoky F. Z., Tang B., Ma R., Jochem K. S., Hyun W. J., Song D., Koester S. J., Lodge T. P., and Frisbie C. D., Adv. Funct. Mater. 29, 1902028 (2019). 10.1002/adfm.201902028 [DOI] [Google Scholar]
- 87.Abbel R., Galagan Y., and Groen P., Adv. Eng. Mater. 20, 1701190 (2018). 10.1002/adem.201701190 [DOI] [Google Scholar]
- 88.Arias A. C., MacKenzie J. D., McChulloch I., Rivnay J., and Salleo A., Chem. Rev. 110, 3 (2010). 10.1021/cr900150b [DOI] [PubMed] [Google Scholar]
- 89.Tobjork D. and Osterbacka R., Adv. Mater. 23, 1935 (2011). 10.1002/adma.201004692 [DOI] [PubMed] [Google Scholar]
- 90.Sondergaard R. R., Hosel M., and Krebs F. C., J. Polym. Sci. B Polym. Phys. 51, 16 (2013). 10.1002/polb.23192 [DOI] [Google Scholar]
- 91.Taleat Z., Khoshroo A., and Mazlou-Ardakani M., Microchim. Acta 181, 865 (2014). 10.1007/s00604-014-1181-1 [DOI] [Google Scholar]
- 92.Mak W. C., Beni V., and Turner A. P. F., TRAC Trends Anal. Chem. 79, 297 (2016). 10.1016/j.trac.2015.10.017 [DOI] [Google Scholar]
- 93.Nayak L., Mohanty S., Nayak S. K., and Ramadoss A., J. Mater. Chem. C 7, 8771 (2019). 10.1039/C9TC01630A [DOI] [Google Scholar]
- 94.Arias A. C., Daniel J., Krusor B., Ready S., Sholin V., and Street R., J. Soc. Inf. Display 15, 485 (2007). 10.1889/1.2759554 [DOI] [Google Scholar]
- 95.Ha M. J., Zhang W., Braga D., Renn M. J., Kim C. H., and Frisbie C. D., ACS Appl. Mater. Interfaces 5, 13198 (2013). 10.1021/am404204q [DOI] [PubMed] [Google Scholar]
- 96.Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., Anal. Chem. 70, 4974 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 97.Comina G., Suska A., and Filppini D., Lab Chip 14, 424 (2014). 10.1039/C3LC50956G [DOI] [PubMed] [Google Scholar]
- 98.Kamei K., Mashimo Y., Koyama Y., Fockenberg C., Nakashima M., Nakajima M., Li J., and Chen Y., Biomed. Microdev. 17, 36 (2015). 10.1007/s10544-015-9928-y [DOI] [PubMed] [Google Scholar]
- 99.Hwang Y., Paydar O. H., and Candler R. N., Sens. Actuators A 226, 137 (2015). 10.1016/j.sna.2015.02.028 [DOI] [Google Scholar]
- 100.Chan H. N., Chen Y., Shu Y., Chen Y., Tian Q., and Wu H., Microfluid. Nanofluid. 19, 9 (2015). 10.1007/s10404-014-1542-4 [DOI] [Google Scholar]
- 101.Ahn B. Y., Duoss E. B., Motala M. J., Guo X., Park S. I., Xiong Y., Yoon J., Nuzzo R. G., Rogers J. A., and Lewis J. A., Science 323, 1590 (2009). 10.1126/science.1168375 [DOI] [PubMed] [Google Scholar]
- 102.Kim S. H., Hong K., Lee K. H., and Frisbie C. D., ACS Appl. Mater. Interfaces 5, 6580 (2013). 10.1021/am401200y [DOI] [PubMed] [Google Scholar]
- 103.Gandhiraman R. P., Jayan V., Han J.-W., Chen B., Koehne J. E., and Meyyappan M., ACS Appl. Mater. Interfaces 6, 20860 (2014). 10.1021/am505325y [DOI] [PubMed] [Google Scholar]
- 104.Smith M., Choi Y. S., Boughey C., and Kar-Narayan S., Flex. Print. Electron. 2, 015004 (2017). 10.1088/2058-8585/aa5af9 [DOI] [Google Scholar]
- 105.Kahn S., Nguyen T. P., Lubej M., Thiery L., Vairac P., and Briand D., Microelectron. Eng. 194, 71 (2018). 10.1016/j.mee.2018.03.013 [DOI] [Google Scholar]
- 106.Rahaman M. T., Cheng C. Y., Karagoz B., Renn M., Schrandt M., Gellman A., and Panat R., ACS Appl. Nano Mater. 2, 3280 (2019). 10.1021/acsanm.9b00628 [DOI] [Google Scholar]
- 107.Di Novo N. G., Cantù E., Tonello S., Sardini E., and Serpelloni M., Sensors 19, 1842 (2019). 10.3390/s19081842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rampi M. A., Schueller O. J. A., and Whitesides G. M., Appl. Phys. Lett. 72, 1781 (1998). 10.1063/1.121183 [DOI] [Google Scholar]
- 109.Kakiuchi T., Iida M., Imabayashi S., and Niki K., Langmuir 16, 5397 (2000). 10.1021/la991358f [DOI] [Google Scholar]
- 110.Lamont E. A., He L., Warriner K., Labuza T. P., and Sreevatsan S., Analyst 136, 3884 (2011). 10.1039/c1an15352h [DOI] [PubMed] [Google Scholar]
- 111.Thomas M. S., Dorfman K. D., and Frisbie C. D., Flex. Print. Electron. 4, 044001 (2019). 10.1088/2058-8585/ab4dcf [DOI] [Google Scholar]
- 112.García E., Llorente M., Hernando A., Kieffer R., Wieser H., and Méndez E., Eur. J. Gastroenterol. Hepatol. 17, 529 (2005). 10.1097/00042737-200505000-00010 [DOI] [PubMed] [Google Scholar]
- 113.Amaya González S., de Los Santos Álvarez N., Miranda Ordieres A. J., and Lobo Castañón M. J., Anal. Chem. 86, 2733 (2014). 10.1021/ac404151n [DOI] [PubMed] [Google Scholar]
- 114.Hochegger R., Mayer W., and Prochaska M., Foods 4, 654 (2015). 10.3390/foods4040654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thorsen T., Merkl S. J., and Quake S. R., Science 298, 580 (2002). 10.1126/science.1076996 [DOI] [PubMed] [Google Scholar]
- 116.Eid C. and Santiago J. G., Lab Chip 18, 11 (2018). 10.1039/C7LC00852J [DOI] [PubMed] [Google Scholar]
- 117.Paratore F., Kalman T. Z., Rosenfeld T., Kaigala G. V., and Bercovici M., Anal. Chem. 89, 7373 (2017). 10.1021/acs.analchem.7b00725 [DOI] [PubMed] [Google Scholar]
- 118.Rogacs A., Marshall L. A., and Santiago J. G., J. Chromatogr. A 1335, 105 (2014). 10.1016/j.chroma.2013.12.027 [DOI] [PubMed] [Google Scholar]
- 119.Squires T. M., Messinger R. J., and Manalis S. R., Nat. Biotechnol. 26, 417 (2008). 10.1038/nbt1388 [DOI] [PubMed] [Google Scholar]
- 120.Stroock A. D., Dertinger S. K. W., Ajdari A., Mezic I., Stone H. A., and Whitesides G. M., Science 295, 647 (2002). 10.1126/science.1066238 [DOI] [PubMed] [Google Scholar]
- 121.Lee C. Y., Wang W. T., Liu C. C., and Fu L. M., Chem. Eng. J. 288, 146 (2016). 10.1016/j.cej.2015.10.122 [DOI] [Google Scholar]
- 122.Yoon S. K., Fichtl G. W., and Kenis P. J. A., Lab Chip 6, 1516 (2006). 10.1039/b609289f [DOI] [PubMed] [Google Scholar]
- 123.Liana D. D., Raguse B., Gooding J. J., and Chow E., Sensors 12, 11505 (2012). 10.3390/s120911505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dungchai W., Chailapakul O., and Henry C. S., Anal. Chem. 81, 5821 (2009). 10.1021/ac9007573 [DOI] [PubMed] [Google Scholar]
- 125.Hong K., Kim S. H., Lee K. H., and Frisbie C. D., Adv. Mater. 24, 3413 (2013). 10.1002/adma.201300211 [DOI] [PubMed] [Google Scholar]
- 126.Kaushik A., Vasudev A., Arya S. K., Pasha S. K., and Bhansali S., Biosens. Bioelectron. 53, 499 (2014). 10.1016/j.bios.2013.09.060 [DOI] [PubMed] [Google Scholar]
- 127.Parlak O., Keene S. T., Marais A., Curto V. F., and Salleo A., Sci. Adv. 4, eaar2904 (2018). 10.1126/sciadv.aar2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Elnathan R., Kwiat M., Pevzner A., Engel Y., Burstein L., Khatchtourints A., Lichtenstein A., Kantaev R., and Patolsky F., Nano Lett. 12, 5245 (2012). 10.1021/nl302434w [DOI] [PubMed] [Google Scholar]
- 129.Cheng S., Hotani K., Hideshima S., Kuroiwa S., Nakanishi T., Hashimoto M., Mori Y., and Osaka T., Materials 7, 2490 (2014). 10.3390/ma7042490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maehashi K., Katsura T., Kerman K., Takamura Y., Matsumoto K., and Tamiya E., Anal. Chem. 79, 782 (2007). 10.1021/ac060830g [DOI] [PubMed] [Google Scholar]
- 131.Kulkarni G. S. and Zhong Z., Nano Lett. 12, 719 (2012). 10.1021/nl203666a [DOI] [PubMed] [Google Scholar]
- 132.Chu C. H., Sarangadharan I., Regmi A., Chen Y. W., Hsu C. P., Chang W. H., Lee G. Y., Chyi J. I., Chen C. C., Shiesh S. C., Lee G. B., and Wang Y. L., Sci. Rep. 7, 5256 (2017). 10.1038/s41598-017-05426-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang J., Carey P., Ren F., Wang Y. L., Good M. L., Jang S., Mastro M. A., and Pearton S. J., Appl. Phys. Lett. 111, 202104 (2017). 10.1063/1.5011151 [DOI] [Google Scholar]
- 134.Sarangadharan I., Regmi A., Chen Y. W., Hsu C. P., Chen P. C., Chang W. H., Lee G. Y., Chyi J. I., Shiesh S. C., Lee G. B., and Wang Y. L., Biosens. Bioelectron. 100, 282 (2018). 10.1016/j.bios.2017.09.018 [DOI] [PubMed] [Google Scholar]
- 135.Chen Y. W., Tai T. Y., Hsu C. P., Sarangadharan I., Pulikkathodi A. K., Wang H. L., Sukesan R., Lee G. Y., Chyi J. I., Chen C. C., Lee G. B., and Wang Y. L., Sens. Actuators B 271, 110 (2018). 10.1016/j.snb.2018.05.119 [DOI] [Google Scholar]
- 136.Tai T. Y., Sinha A., Sarangadharan I., Pulikkathodi A. K., Wang S. L., Lee G. Y., Chyi J. I., Shiesh S. C., Lee G. B., and Wang Y. L., Anal. Chem. 91, 5953 (2019). 10.1021/acs.analchem.9b00353 [DOI] [PubMed] [Google Scholar]
- 137.Gao N., Zhou W., Jiang X., Hong G., Fu T. M., and Lieber C. M., Nano Lett. 15, 2143 (2015). 10.1021/acs.nanolett.5b00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gao N., Gao T., Yang X., Dai X., Zhou W., Zhang A., and Lieber C. M., Proc. Natl. Acad. Sci. U.S.A. 113, 14633 (2016). 10.1073/pnas.1625010114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ma S., Li X., Lee Y. K., and Zhang A., Biosens. Bioelectron. 117, 276 (2018). 10.1016/j.bios.2018.05.061 [DOI] [PubMed] [Google Scholar]
- 140.Haustein N., Gutiérrez-Sanz Ó., and Tarasov A., ACS Sens. 4, 874 (2019). 10.1021/acssensors.8b01515 [DOI] [PubMed] [Google Scholar]