Abstract
Recent efforts in mapping spatial genome organization have revealed three evocative and conserved structural features of the inactive X in female mammals. First, the chromosomal conformation of the inactive X reveals a loss of topologically associated domains (TADs) present on the active X. Second, the macrosatellite DXZ4 emerges as a singular boundary that suppresses physical interactions between two large TAD-depleted “megadomains.” Third, DXZ4 reaches across several megabases to form “superloops” with two other X-linked tandem repeats, FIRRE and ICCE, which also loop to each other. Although all three structural features are conserved across rodents and primates, deletion of mouse and human orthologs of DXZ4 and FIRRE from the inactive X have revealed limited impact on X chromosome inactivation (XCI) and escape in vitro. In contrast, loss of Xist or SMCHD1 have been shown to impair TAD erasure and gene silencing on the inactive X. In this perspective, we summarize these results in the context of new research describing disruption of X-linked tandem repeats in vivo, and discuss their possible molecular roles through the lens of evolutionary conservation and clinical genetics. As a null hypothesis, we consider whether the conservation of some structural features on the inactive X may reflect selection for X-linked tandem repeats on account of necessary cis- and trans-regulatory roles they may play on the active X, rather than the inactive X. Additional hypotheses invoking a role for X-linked tandem repeats on X reactivation, for example in the germline or totipotency, remain to be assessed in multiple developmental models spanning mammalian evolution.
Keywords: tandem repeats, macrosatellite, X chromosome inactivation, chromosome conformation, chromatin loop extrusion, SMCHD1, XIST, intellectual disability
Introduction
Since its initial discovery over 70 years ago (Barr and Bertram, 1949), the singular nature of the inactive X (Xi) in the female mammalian nucleus has captured the imagination of cell biologists studying chromosome organization, localization, and chromatin condensation. These studies have revealed the Xi to form the condensed “Barr body”, which localizes to the repressed nuclear periphery and periodically attaches to the nucleolus. The human metaphase Xi reflects this peripheral and peri-nucleolar localization with, respectively, alternating bands of tri-methylated lysines 9 and 27 of histone 3 (H3K9me3 and H3K27me3) (Vallot et al., 2016). During interphase, these chromatin domains of the human Xi segregate into compartments facing the nuclear interior or -lamina (Chadwick and Willard, 2004), and yet are hypothesized to mutually reinforce repression across this bi-compartmental Xi (Pinter, 2016). Recent studies have identified SMCHD1 as the principal trans-acting factor that bridges both H3K9me3 (H3K9me2 for the mouse Xi) – and H3K27me3-rich compartments to mediate de novo CpG island methylation for long-term Xi silencing, as reviewed recently (Jansz et al., 2017).
Rapid technical advances have enabled zooming into the unique three-dimensional (3D) topology of the Xi by chromosome conformation capture. As discussed below, these experiments have: (a) revealed how/when the Xi adopts its unusual chromosome conformation, (b) attributed the erasure of active X (Xa) topology to the concerted action of SMCHD1 and X chromosome inactivation (XCI) master-regulator XIST/Xist, and (c) implicated two conserved X-linked tandem repeats in shaping the Xi. Here, we review how these structural features relate to each other, and integrate findings from current in vitro and in vivo perturbation experiments with recent epigenomics and clinical genetics studies.
The central thesis of this perspective examines which of these Xi structural features may have been conserved due to important functions in XCI or escape, and which may emerge as mere “by-products” of the conserved Xi remodeling during XCI. These early results suggest that the DXZ4/Dxz4 macrosatellite, while dispensable for XCI establishment, may have some limited impact on Xi choice. In contrast, accumulating evidence reveals that the FIRRE/Firre tandem repeat supplies critical cis- and trans-acting functions from its Xa allele. We therefore propose that FIRRE/Firre may have been conserved due to such XCI-independent roles. Alternatively, either or both of these conserved tandem repeats may participate in Xi biology in developmental contexts that have so far escaped analysis, for example in germline X reactivation or during zygotic genome activation.
Unique Structural Features of the Inactive X
Eukaryotic chromosomes are composed of topologically associated domains (TADs) that consist of concentrated 3D interactions and organize into euchromatic “A” and heterochromatic “B” compartments (Nora et al., 2013). TADs are often bounded by convergent CTCF sites at the base of chromatin loops. These distal interactions are thought to result from loop-extruding DNA complexes that terminate at specific sites when movement of ring-shaped cohesin is blocked by architectural DNA binding factors like CTCF and YY1 (Rao et al., 2014; Sanborn et al., 2015). Not all cohesin loops constitute TAD boundaries, as intra-TAD loops enable long-range contacts between promoters and their regulatory elements (e.g., enhancers) (Dixon et al., 2016; Gonzalez-Sandoval and Gasser, 2016). Conversely, boundaries between A/B-type TADs (A/B boundaries) are also defined by local transitions in replication timing, lamin-association and chromatin composition, possibly due to intrinsic liquid-liquid phase-separating properties of lamin-associated B-type heterochromatin (Di Pierro et al., 2017; Schwarzer et al., 2017; Falk et al., 2019; Mirny et al., 2019). Since not all TAD boundaries coincide with cohesin loops, such A/B boundaries remain stable even when cohesin is depleted (Rao et al., 2017; Schwarzer et al., 2017).
Cohesin Loop Erasure and TAD Attenuation
The first structural feature to distinguish the Xi from the Xa (Figure 1A) may therefore be separated into two mechanistically distinct observations: (1) near-complete loss of long-range cohesin loops outside of escapee genes (Splinter et al., 2011; Nora et al., 2012; Rao et al., 2014), and (2) attenuation or loss of most TADs across the human and mouse Xi, respectively (Nora et al., 2012; Rao et al., 2014; Deng et al., 2015). Both of these observations have been conclusively linked to Xist RNA in the mouse: initial loss of TADs during XCI depends on Xist-mediated gene silencing, which facilitates spreading of Xist RNA into active genes (Engreitz et al., 2013; Chen C.-K. et al., 2016; Giorgetti et al., 2016). Because mouse XCI is maintained via stable CpG methylation, loss of Xist after completed XCI does not undo gene silencing, but allows chromosome topology to recover across the Xi to mirror the Xa conformation (Csankovszki et al., 1999, 2001; Splinter et al., 2011; Minajigi et al., 2015).
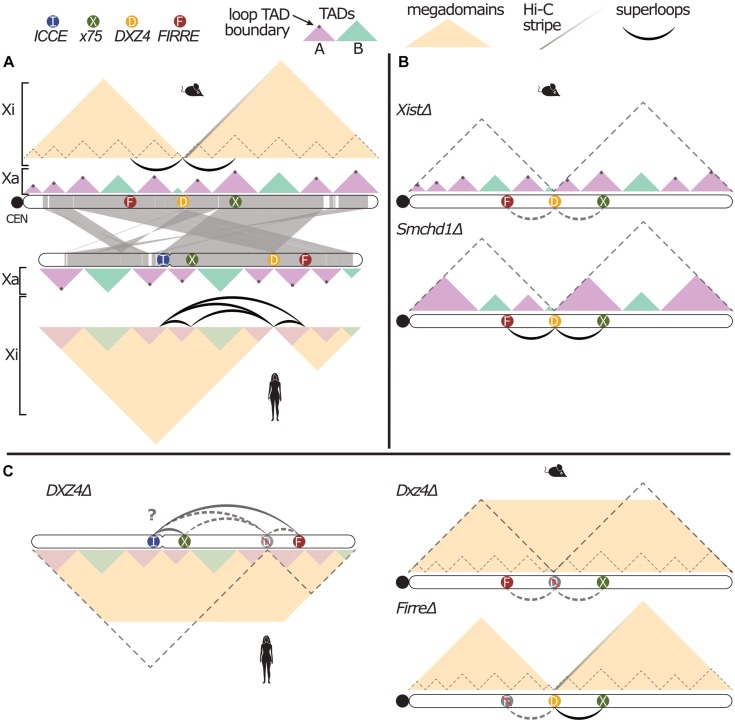
FIGURE 1.
Conserved structural features of the mammalian inactive X. (A) Illustration of human and mouse X chromosome 3D conformation and synteny (generated with http://bioinfo.konkuk.ac.kr/synteny_portal/). A-type TADs (lavender) have cohesin loops demarcating boundaries of euchromatic TADs (as shown by the dark purple dots of increased Hi-C interaction), while B-type (aqua) TADs are largely heterochromatic. TADs observed on the Xa are lost (mouse) or attenuated (human) on the Xi, leaving two large megadomains. The stripe of contacts emanating from Dxz4 on the mouse Xi represents the directionality of Dxz4 toward the telomeric end of mouse X chromosome, and long-range superloops indicate 3D proximity of tandem repeats on both mouse and human Xi. (B) Mouse Xi conformation upon loss of Xist (top) or Smchd1 (bottom). In XistΔ, there is a loss of megadomains (dashed gray lines), re-established strength of the TADs (as represented by the increased opacity of the TADs), and a loss of superloops (as represented by the dashed gray lines). In Smchd1Δ, there is a loss of megadomains, and a merging of lavender A-type TADs. (C) Xi conformation after DXZ4/Dxz4 or Firre deletions. Dashed gray lines represent loss of megadomains and superloops. Yellow trapezoid represents loss of contact isolation between megadomains. “D” or “F” are grayed and crossed out to indicate deletion of each superloop anchor. In both human (left) and mouse (right) Dxz4Δ, there is a loss of megadomain separation, Hi-C stripe and all superloops, with uncertain residual human superloops (“?”). In FirreΔ, there is only a loss of the Dxz4-Firre superloop.
How does Xist RNA ablate both cohesin loops and cohesin-independent A/B boundaries across the Xi? In a seminal study, Xist RNA was shown to interact directly with cohesin subunits and reduce cohesin across the Xi (Minajigi et al., 2015). While the mechanistic basis for this loss remains unclear, Xist RNA effectively ablates cohesin loops that separate cohesin-dependent TAD boundaries, thus merging TADs (Figure 1B). In contrast, remodeling of cohesin-independent (largely A/B) boundaries depends on Xist-mediated recruitment of SMCHD1 (Wang et al., 2018) via polycomb-repressive complex 1 (PRC1) (Jansz et al., 2018; Gdula et al., 2019; Wang et al., 2019). In the absence of SMCHD1, Xist RNA is trapped in merged A-type TADs with persistent A/B boundaries (Wang et al., 2018). This observation is consistent with a previously described two-step model for how Xist RNA first spreads across gene-rich A-type TADs before entering LINE1/lamin-rich B-type TADs (Simon et al., 2013). Together, these reports suggest that SMCHD1 may dissolve cohesin-independent A/B boundaries by merging phase-separated, lamin-rich B-type TADs with Xist/PRC1-enriched A-type TADs. Supporting evidence for such hypothesized modulation of chromosomal phase-separation by XIST/Xist was recently summarized (Cerase et al., 2019).
Megadomain Boundary: DXZ4/Dxz4
The second structural feature of the Xi emerges against the backdrop of this otherwise boundary-depleted chromosome conformation: the macrosatellite repeat DXZ4/Dxz4 forms the only remaining topological boundary, thus separating the Xi into two large megadomains on the mouse, rhesus and human Xi (Rao et al., 2014; Deng et al., 2015; Minajigi et al., 2015; Darrow et al., 2016). While the DXZ4/Dxz4 boundary is therefore conserved, gene content of the two megadomains is not evolutionarily fixed (Deng et al., 2015), as judged by mouse-human synteny maps (Figure 1A). Initially identified as a female-specific CpG-hypomethylated macrosatellite (Giacalone et al., 1992), a series of detailed studies by the Chadwick lab illuminated the enigmatic molecular configuration of this uniquely Xi-specific euchromatic region. On the Xa, transcription across both strands of its 3-kb long, CpG-rich repeat unit gives rise to small RNA transcripts that attract H3K9me3 heterochromatin and CpG-hypermethylation (Chadwick, 2008; Pohlers et al., 2014; Figueroa et al., 2015). In contrast, the CpG-hypomethylated Xi allele of DXZ4 is decorated by active H3K4me3 and H3K9 acetylation marks that form a privileged euchromatic hub inside the otherwise repressed Xi chromosome territory. In human, rhesus and mouse, this euchromatic Xi allele of DXZ4/Dxz4 binds CTCF and YY1 (McLaughlin and Chadwick, 2011; Horakova et al., 2012a, b; Moseley et al., 2012), and engages in Xi-specific long-range interactions. Notably, only the internal DXZ4/Dxz4 promoter element containing paired CTCF and YY1 sites is conserved across mammals, suggesting possible selection for Xi-specific and CTCF-mediated functions in XCI across mammalian evolution (Horakova et al., 2012b; Westervelt and Chadwick, 2018).
Discovery of the Xi-specific megadomain boundary at DXZ4/Dxz4 (Rao et al., 2014; Deng et al., 2015) prompted several groups to test whether its deletion impacted mouse or human XCI and escape (Figure 1C). Darrow et al. (2016) demonstrated that human DXZ4 is required for boundary maintenance, and elegantly traced the 3D chromosome topology anchored at DXZ4, the XIST-proximal “X75” locus, and two distal tandem repeats, FIRRE and ICCE. Interestingly, in 2/3 RPE1 clones lacking DXZ4 on the Xi, a quarter of cells replaced the largest H3K27me3 domain on the Xi with H3K9me3 (∼15 Mb adjacent to DXZ4), with a concomitant delay in replication timing. However, Xi-DXZ4Δ cells maintained XCI and the bi-compartmental interphase organization with nuclear interior-and lamina facing Xi domains of H3K27me3 and H3K9me3, respectively (Darrow et al., 2016).
These results were mirrored on the mouse Xi by Giorgetti et al. (2016): the megadomain boundary was lost in all four Xi-Dxz4Δ clones, with negligible impact on XCI establishment or maintenance in neuronal progenitor cells (NPCs) differentiated from mESCs. A single Dxz4Δ NPC clone featured reduced expression and chromatin accessibility of cell-type specific (facultative) escapee genes, along with a collapse of their residual TADs on the Xi. However, constitutive escapees remained unaffected, indicating that Dxz4 is generally dispensable in mouse XCI and escape. Likewise, Froberg et al. (2018) found that Dxz4Δ abrogates the Xi megadomain boundary, but performed their experiments in the context of a Tsix mutation that rendered XCI non-random. This system enabled capturing the kinetics of megadomain boundary formation relative to Xist expression during XCI, while also assessing the impact of Xi-Dxz4Δ (-and FirreΔ) in differentiating mESC populations to exclude stochastic clonal phenomena. Indeed, the Dxz4-dependent megadomain boundary closely trails Xist-mediated gene silencing and TAD erasure, but chromatin accessibility and gene expression showed no significant changes in three independent Dxz4Δ clones, suggesting Dxz4 is not required for XCI establishment and escape. Finally, Bonora et al. (2018) generated a series of edited Dxz4 loci in immortalized Patski fibroblasts, separating its role from a proximal mouse-specific mini-satellite, DS-Tr. While Dxz4 was indeed responsible for boundary maintenance, these editing experiments also produced two clones that inverted Dxz4 and provided a critical mechanistic insight. As conserved CTCF sites in Dxz4 share a common polarity, inverted Dxz4 clones swapped the direction of 3D contacts anchored at Dxz4. Because CTCF sites block loop extrusion when paired in convergent fashion, the wildtype Dxz4 boundary must therefore arrest cohesin loops extruded from the telomeric side of the Xi. These orientation-dependent Hi-C stripes emanate from Dxz4 across ∼25–40 Mb of the Xi, indicating the remarkable persistence of cohesin rings traversing large swaths of the Xi (Figures 1A,C).
Superloop Formation: A Euchromatic Hub of DXZ4, FIRRE, and ICCE
How does cohesin move past other convergent CTCF sites to extrude such extraordinarily long loops on the Xi? Although most CTCF peaks seen on the Xa are maintained on the human and mouse Xi (Calabrese et al., 2012; Ding et al., 2014; Kung et al., 2015), they appear attenuated on the Xi, especially in specific cellular contexts (Bonora et al., 2018). While the mechanistic underpinnings of this observation remain unclear, the presence or silencing function of Xist RNA (Kung et al., 2015; Minajigi et al., 2015) may impact CTCF residence time on DNA, e.g., via its RNA-binding domain (Hansen et al., 2017, 2019; Saldaña-Meyer et al., 2019). Cohesin rings may thereby be favored to traverse most CTCF sites on the Xi, until they encounter an array of stable CTCF sites at Dxz4, which is depleted of Xist RNA (Simon et al., 2013). A corresponding (paired) cohesin ring may therefore arrest anywhere along the telomeric stripe that originates at Dxz4, reflecting cohesin “dispersal”, across the Xi (Figure 1). This interpretation, as first proposed in Bonora et al. (2018), may also explain how a seemingly cohesin-depleted Xi (Minajigi et al., 2015) avoids premature sister chromatid separation to remain mitotically stable. Notably, except in cancer (Carone and Lawrence, 2013; Xu et al., 2017), the Xi has not been reported to suffer general mitotic instability even in the absence of DXZ4/Dxz4, suggesting that even dispersed cohesin rings maintain cohesion (Darrow et al., 2016; Giorgetti et al., 2016; Bonora et al., 2018; Froberg et al., 2018).
In the context of such cohesin “dispersal,” it appears perhaps unsurprising that the cohesin rings remaining on the Xi may eventually anchor at tandem arrays of stable and Xist-depleted CTCF sites that resemble DXZ4/Dxz4. As first reported by Horakova et al. (2012b) and later by Rao et al. (2014), DXZ4 (at 115 Mb) is engaged in long-range, Xi-specific 3D contacts with two other X-linked repeats FIRRE (“X130”) and ICCE (“X56”), as well as the XIST-proximal “X75” locus (Figure 1A). Like the megadomain boundary, superloops between DXZ4/Dxz4 and FIRRE/Firre are conserved in human, rhesus and mouse (Darrow et al., 2016). Although many possible pair-wise human Xi superloops were first observed (Rao et al., 2014), Darrow et al. (2016) demonstrated requisite engagement of DXZ4 in most superloops by analysis of three-way proximity-ligated reads (COLA), and confirmed a co-localized DXZ4-FIRRE-ICCE hub at the single-cell level by FISH, as previously reported (Horakova et al., 2012b).
Like DXZ4/Dxz4, the FIRRE/Firre and ICCE tandem repeats reside inside Xa-transcribed genes (FIRRE/Firre and NBDY, respectively) and feature female-specific CpG hypo-methylation with paired CTCF-YY1 binding sites that are primarily occupied on the Xi (Ding et al., 2014; Hacisuleyman et al., 2014, 2016; Qu et al., 2015; Chen C. et al., 2016; Westervelt and Chadwick, 2018). ICCE is conserved across several mammals outside rodents, and likely derives from the ancestral DXZ4 macrosatellite (Westervelt and Chadwick, 2018). On the mouse Xi, both Dxz4 and Firre feature euchromatic H3K4me3 marks and are depleted of H3K27me3 and Xist RNA (Pinter et al., 2012; Simon et al., 2013), thereby sharing regulatory features of escapee genes despite residing inside genes that are subject to XCI (Berletch et al., 2010; Chen C. et al., 2016).
What is the function of this conserved euchromatic hub and does it depend on superloops? To address the latter question, Froberg et al. (2018) deleted Firre on the mouse Xi to remove the Firre-Dxz4 superloop without disrupting the Dxz4 anchor directly, demonstrating that the superloop is dispensable for Dxz4 boundary function (Figure 1C). Moreover, neither Dxz4Δ, FirreΔ or double Xi-knockout cells reveal a consistent impact on XCI or escape. Likewise, female Xi-FirreΔ embryonic fibroblasts lose the Firre-Dxz4 superloop, but maintain the syntenic mouse x75-Dxz4 superloop (Barutcu et al., 2018). These data indicate that each Xi-linked superloop generally depends only on its own pair of anchors and the presence of the DXZ4/Dxz4 hub. Yet, the role of this conserved euchromatic hub in Xi biology remains to be addressed, as do mutual dependencies of DXZ4/Dxz4, FIRRE/Firre and ICCE on CTCF binding and euchromatin maintenance.
Dissecting Tandem Repeat DNA and RNA Functions In Vivo
While the results cited above focus on the Xi-linked roles of DXZ4/Dxz4 and FIRRE/Firre in XCI in vitro, two new reports from the Rinn lab address their possible in vivo functions in development and XCI (Andergassen et al., 2019; Lewandowski et al., 2019). These studies touch on the critical question of whether there may be crucial XCI-specific functions of DXZ4/Dxz4 or FIRRE/Firre that have been missed to-date, due to inaccessibility of certain developmental contexts with current mouse and human cell-based systems. However, discussion of these results necessitates drawing a distinction between cis-specific functions of these tandem repeats, and trans-acting roles of their RNA products.
While the DXZ4/Dxz4 macrosatellite is thought to merely produce cis-acting short RNAs to maintain its heterochromatin on the Xa (Pohlers et al., 2014; Figueroa et al., 2015), the active FIRRE/Firre locus on the Xa also gives rise to multiple species of nuclear non-coding (nc)RNAs, which regulate autosomal genes, likely at the post-transcriptional level (Hacisuleyman et al., 2014, 2016; Bergmann et al., 2015; Izuogu et al., 2018). Firre RNA primarily regulates autosomal genes in the hematopoietic system, including in common lymphoid progenitors (Andergassen et al., 2019; Lewandowski et al., 2019). This observation may prove relevant to sex differences in autoimmune disorders (Syrett and Anguera, 2019), as the FIRRE locus was also recently identified as differentially methylated in CD4+ memory T cells of twins discordant for multiple sclerosis (Souren et al., 2019). While global trans-acting FIRRE/Firre RNA functions are well beyond the scope of this perspective, it appears that many autosomal targets function in RNA splicing, processing and transport, likely due to FIRRE/Firre RNAs physical association with HNRNPU (Hacisuleyman et al., 2014; Bergmann et al., 2015). This member of the large heterogeneous nuclear ribonucleoprotein family functions in mRNA splicing and processing, and has recently discovered roles in general genome architecture (Geuens et al., 2016; Zhang et al., 2019). Such roles may relate to the unusually stable trans-chromosomal hub anchored at Firre on the mouse Xa (Hacisuleyman et al., 2014). In addition, HNRNPU has fascinating but highly context-dependent roles in XCI (Hasegawa et al., 2010; Kolpa et al., 2016), as reviewed previously (Hasegawa and Nakagawa, 2011; Cerase et al., 2015; Pinter, 2016; Creamer and Lawrence, 2017).
Complicating the distinction from the CTCF-bound Firre locus on the Xi, predominantly Xa-transcribed Firre RNA was recently shown to play a role in tethering the Xi to the nucleolus via CTCF for maintenance of H3K27me3 (Yang et al., 2015). A new report from the Disteche Lab confirms that Firre cDNA expression rescues such H3K27me3 maintenance defects in trans (Fang et al., 2019). Interestingly, H3K27me3 dependency on Firre RNA appears to be confined to Patski and primary embryonic fibroblasts of the same interspecific cross (Mus spretus × Mus musculus), whereas H3K27me3-enrichment of the Xi in differentiating Xa-FirreΔ mESCs and primary tissues of Xa-FirreΔ mice appears unaffected in a pure M. musculus background (Yang et al., 2015; Fang et al., 2019). These differences between intra- and inter-specific F1 hybrids may be reconciled by global trans-acting changes encoded by the M. spretus genome. Such evolutionary divergence of trans-regulation (Signor and Nuzhdin, 2018) may therefore reveal a function for Firre RNA acting on the Xi. For example, compared with other mouse clades, the M. spretus genome appears to feature an overall reduction of stable CTCF binding (Kentepozidou et al., 2019), which may underpin the pronounced sensitivity to both CTCF and Firre RNA levels for nucleolar tethering of the Xi (Yang et al., 2015; Fang et al., 2019). Although the mechanism of this phenomenon remains to be explored, indeed CTCF binds Firre RNA (Graindorge et al., 2019), and CTCF peaks on the Xi are diminished in FirreΔ Patski cells (Fang et al., 2019). Another plausible explanation to reconcile differences between Firre RNA knockdowns and knockout mice/mESCs, could be that acute loss of Firre RNA affects H3K27me3 maintenance only on the Xi of differentiated cells, but may be compensated for if lost prior to initiation of XCI (Fang et al., 2019).
Yet, two new reports by the Rinn lab assess, but find no evidence for sex ratio distortion, or any in vivo defect in random or imprinted XCI establishment or maintenance in FirreΔ (Lewandowski et al., 2019), as well as single and double Dxz4Δ and FirreΔ mice (Andergassen et al., 2019). However, the latter study reports increased skewing of random XCI toward the Dxz4Δ Xi in these single- and double-knockout Dxz4Δ mice that merits follow-up. In sum, neither Dxz4 nor Firre appear to be generally required for XCI establishment, maintenance or escape. Even double Xi knockouts of these tandem repeats, abolishing both megadomain boundary and superloops, appear to have little to no impact on XCI in mESCs (Froberg et al., 2018) and mice (Andergassen et al., 2019).
Firre Duplication as a Potential X-Linked Intellectual Disability (XLID) Candidate?
If these X-linked tandem repeats are largely dispensable on the Xi, why were their sequence elements, chromatin composition and topology conserved across mammalian evolution? The FIRRE/Firre locus in particular illustrates how tandem repeats and macrosatellites compound special challenges intrinsic to molecular dissection of ncRNA loci (DNA vs. RNA/cis- vs. trans) (Bassett et al., 2014). Human (clinical) genetics studies can help link FIRRE to novel roles outside of XCI. These results have revealed increased CTCF binding and chromatin accessibility across the FIRRE tandem repeat in females (Ding et al., 2014; Qu et al., 2015), as well as differential CpG methylation in multiple sclerosis (Souren et al., 2019). In closing, we want to raise the question whether conservation of CTCF-mediated superloops on the Xi may be merely a consequence of evolutionary selection for critical regulatory functions originating from the Xa?
At present, there are two identified developmental roles that can serve as a basis for positive selection: (1) an established trans-acting function for FIRRE RNA in common lymphoid progenitors (Lewandowski et al., 2019), and (2) a hypothesized role in brain development, for which we refer readers to clinical reports of patients with copy-number gains in Xq26 (Schroer et al., 2012; Abe et al., 2014; Ha et al., 2019; Herriges et al., 2019). The latest of these reports summarizes sex-biased ID associated with duplications in this genomic region (Herriges et al., 2019), the shortest of which overlaps only IGSF1, olfactory receptor gene OR1H, and FIRRE (Abe et al., 2014).
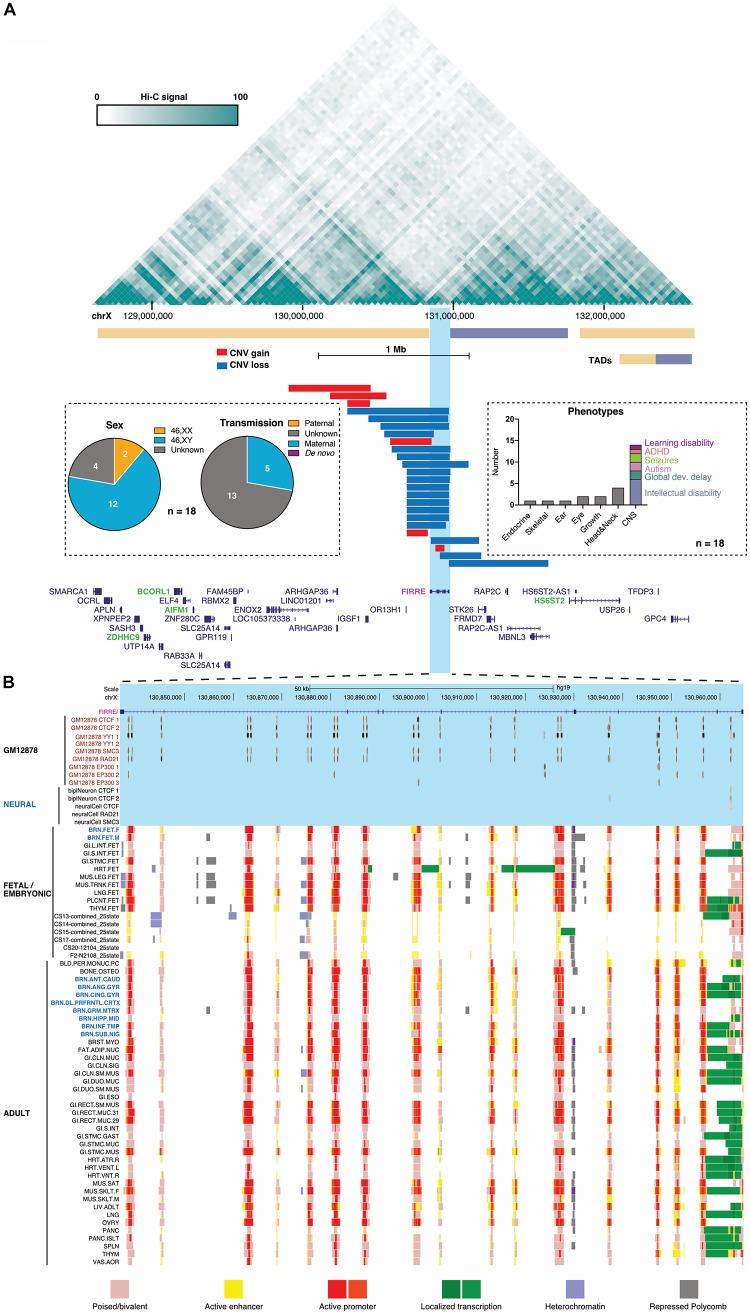
Because additional cases may help to better resolve this region, we queried the DECIPHER database (Firth et al., 2009) and collated all short (<1 Mb) overlapping CNVs (Figure 2A). Tabulating phenotypes, transmission and sex only in patients who lack any other CNVs, these entries illustrate that: (1) almost all CNVs are gains that overlap FIRRE, (2) the associated phenotypes primarily involve the nervous system, and (3) most (12/14) patients are males who inherited the FIRRE-overlapping gain from weakly -or non-manifesting maternal carriers, where reported (Abe et al., 2014; Herriges et al., 2019). In contrast, among >5000 control individuals without pediatric disease in gnomAD-SV (Collins et al., 2019), 8/12 FIRRE duplications were present in heterozygous females, leaving one homozygous female and three males. While these low counts preclude strong conclusions at this time, and FIRRE duplications on the Xa are clearly not incompatible with neurotypical development, hemizygous FIRRE duplications appear to be over-represented in DECIPHER relative to this gnomAD-SV control cohort. Yet, certain biases in DECIPHER entries or an indirect contribution of FIRRE’s repeats to structural variation occurring in this region cannot be ruled out at this time.
FIGURE 2.
Conformation, copy-number variation (CNV) and chromatin signatures of the human FIRRE locus. (A) Prefrontal cortex Hi-C contact and TAD structure map from Schmitt et al. (2016) (top, generated with http://promoter.bx.psu.edu/hi-c/) of a 4-Mb region harboring FIRRE-overlapping CNVs in DECIPHER entries (gains blue, deletions red), in which no additional structural variation was detected. Names of genes located in the 4-Mb FIRRE-harboring region (bottom), with genes previously implicated in intellectual disability highlighted in green (OMIM #): ZDHHC9 (300646), BCORL1 (300688), AIFM1 (310490), HS6ST2 (300545). Insets tabulate the sex, transmission, and grouped clinical representations observed in DECIPHER patients carrying these FIRRE-overlapping gains. (B) Chromatin states and CTCF, YY1, cohesin, and P300 binding sites of the 130-kb human FIRRE gene. Poised/bivalent (Salmon), active enhancer (Yellow), heterochromatin (Pale Turquoise) or repressed polycomb (Silver) chromatin states form an array across FIRRE with localized transcription (Green/Lime Green) and active promoter activity (Red/Orange red). Data from across a variety of adult [Roadmap Epigenome (Yen and Kellis, 2015)] and developing human craniofacial (Wilderman et al., 2018) tissues.
If confirmed as a possible XLID risk locus, one interpretation of these data may suggest that FIRRE duplications, when present on the Xa, may tend to impact neurodevelopment: either by changing FIRRE RNA expression or cis-regulation of nearby genes by FIRRE’s array of CTCF sites. Importantly, FIRRE’s repetitive DNA elements were shown to confer enhancer activity in vitro (Hacisuleyman et al., 2016), contact neighboring genes in the human cortex (Schmitt et al., 2016) (Figure 2A), and attract a poised or active enhancer chromatin signature (Figure 2B) across a variety of adult (Yen and Kellis, 2015) and developing human tissues (Wilderman et al., 2018). Interestingly, the embryonic forebrain of FirreΔ mice shows significantly reduced expression of the neighboring Hs6st2 gene (Lewandowski et al., 2019), the human ortholog of which was recently identified as a cause of X-linked ID in highly dosage-sensitive fashion (Paganini et al., 2019). Duplication-associated “re-wiring” of FIRRE-anchored promoter contacts may also increase or decrease expression of other neighboring genes, three of which are implicated in ID phenotypes in OMIM. All four of these XLID loci contact FIRRE in the developing human cortex (Figure 2). The compilation of these data are merely meant to caution against ruling out a potentially important and conserved cis-regulatory role for the FIRRE locus at this time (Lewandowski et al., 2019).
Whether the limited impact of DXZ4/Dxz4 and Firre deletions on XCI in vivo (Andergassen et al., 2019) suggests an XCI-independent basis for conservation of these tandemly repeated CTCF/YY1 arrays, remains an open question for now. Alternative XCI-specific hypotheses may include cis-acting functions for X-linked tandem repeats in biological contexts that were not specifically explored by in vitro or in vivo experiments discussed here. In view of common chromatin features that DXZ4, FIRRE, and ICCE share with the DUX4-encoding D4Z4 macrosatellite (Chadwick, 2009), we remain particularly curious about potential contributions of X-linked tandem repeats to human X reactivation phenomena in totipotency (Iturbide and Torres-Padilla, 2017), pluripotency (Geens and Chuva De Sousa Lopes, 2017), and primordial germ cell development (Payer, 2016).
Data Availability Statement
All datasets generated and analyzed for this study are cited in the article/supplementary files.
Author Contributions
PB and YK contributed equally and generated the figures. All authors wrote and edited the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by NIH grant R35GM123926 to SP.
References
- Abe Y., Kikuchi A., Kobayashi S., Wakusawa K., Tanaka S., Inui T., et al. (2014). Xq26.1-26.2 gain identified on array comparative genomic hybridization in bilateral periventricular nodular heterotopia with overlying polymicrogyria. Dev. Med. Child Neurol. 56 1221–1224. 10.1111/dmcn.12553 [DOI] [PubMed] [Google Scholar]
- Andergassen D., Smith Z. D., Lewandowski J. P., Gerhardinger C., Meissner A., Rinn J. L. (2019). In vivo Firre and Dxz4 deletion elucidates roles for autosomal gene regulation. eLife 8:e47214. 10.7554/eLife.47214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M. L., Bertram E. G. (1949). A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163:676. 10.1038/163676a0 [DOI] [PubMed] [Google Scholar]
- Barutcu A. R., Maass P. G., Lewandowski J. P., Weiner C. L., Rinn J. L. (2018). A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 9:1444. 10.1038/s41467-018-03614-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Akhtar A., Barlow D. P., Bird A. P., Brockdorff N., Duboule D., et al. (2014). Considerations when investigating lncRNA function in vivo. eLife 3:e03058. 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J. H., Li J., Eckersley-Maslin M. A., Rigo F., Freier S. M., Spector D. L. (2015). Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 25 1336–1346. 10.1101/gr.189027.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch J. B., Yang F., Disteche C. M. (2010). Escape from X inactivation in mice and humans. Genome Biol. 11:213. 10.1186/gb-2010-11-6-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora G., Deng X., Fang H., Ramani V., Qiu R., Berletch J. B., et al. (2018). Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome. Nat. Commun. 9:1445. 10.1038/s41467-018-03694-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese J. M., Sun W., Song L., Mugford J. W., Williams L., Yee D., et al. (2012). Site-specific silencing of regulatory elements as a mechanism of x inactivation. Cell 151 951–963. 10.1016/j.cell.2012.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone D. M., Lawrence J. B. (2013). Heterochromatin instability in cancer: from the Barr body to satellites and the nuclear periphery. Semin. Cancer Biol. 23 99–108. 10.1016/J.SEMCANCER.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerase A., Armaos A., Neumayer C., Avner P., Guttman M., Tartaglia G. G. (2019). Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 26 331–334. 10.1038/s41594-019-0223-0 [DOI] [PubMed] [Google Scholar]
- Cerase A., Pintacuda G., Tattermusch A., Avner P. (2015). Xist localization and function: new insights from multiple levels. Genome Biol. 16:166. 10.1186/s13059-015-0733-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B. P. (2008). DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 18 1259–1269. 10.1101/gr.075713.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B. P. (2009). Macrosatellite epigenetics: the two faces of DXZ4 and D4Z4. Chromosoma 118 675–681. 10.1007/s00412-009-0233-5 [DOI] [PubMed] [Google Scholar]
- Chadwick B. P., Willard H. F. (2004). Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. U. S. A. 101 17450–17455. 10.1073/pnas.0408021101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-K., Blanco M., Jackson C., Aznauryan E., Ollikainen N., Surka C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354 468–472. 10.1126/science.aae0047 [DOI] [PubMed] [Google Scholar]
- Chen C., Shi W., Balaton B. P., Matthews A. M., Li Y., Arenillas D. J., et al. (2016). YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci. Rep. 6:37324. 10.1038/srep37324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. L., Brand H., Karczewski K. J., Zhao X., Alföldi J., Khera A. V., et al. (2019). gnomAD-SV An open resource of structural variation for medical and population genetics The Genome Aggregation Database (gnomAD) Production Team 7, The gnomAD Consortium. bioRxiv [Preprint] 10.1101/578674 [DOI] [Google Scholar]
- Creamer K. M., Lawrence J. B. (2017). XIST RNA: a window into the broader role of RNA in nuclear chromosome architecture. Philos. Trans. R. Soc. B Biol. Sci. 372:20160360. 10.1098/rstb.2016.0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G., Nagy A., Jaenisch R. (2001). Synergism of Xist Rna, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153 773–784. 10.1083/jcb.153.4.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G., Panning B., Bates B., Pehrson J. R., Jaenisch R. (1999). Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 22 323–324. 10.1038/11887 [DOI] [PubMed] [Google Scholar]
- Darrow E. M., Huntley M. H., Dudchenko O., Stamenova E. K., Durand N. C., Sun Z., et al. (2016). Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. U.S.A. 113 E4504–E4512. 10.1073/pnas.1609643113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Ma W., Ramani V., Hill A., Yang F., Ay F., et al. (2015). Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16:152. 10.1186/s13059-015-0728-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro M., Cheng R. R., Lieberman Aiden E., Wolynes P. G., Onuchic J. N. (2017). De novo prediction of human chromosome structures: epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. U.S.A. 114 12126–12131. 10.1073/pnas.1714980114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Ni Y., Timmer S. W., Lee B.-K., Battenhouse A., Louzada S., et al. (2014). Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 10:e1004798. 10.1371/journal.pgen.1004798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Gorkin D. U., Ren B., Alipour E., Marko J. F., Austenaa L. M., et al. (2016). Chromatin domains: the unit of chromosome organization. Mol. Cell 62 668–680. 10.1016/j.molcel.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J. M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341:1237973. 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M., Feodorova Y., Naumova N., Imakaev M., Lajoie B. R., Leonhardt H., et al. (2019). Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570 395–399. 10.1038/s41586-019-1275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Bonora G., Lewandowski J. P., Thakur J., Filippova G. N., Henikoff S., et al. (2019). Trans- and cis-acting effects of the lncRNA Firre on epigenetic and structural features of the inactive X chromosome. bioRxiv [Preprint] 10.1101/687236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa D. M., Darrow E. M., Chadwick B. P. (2015). Two novel DXZ4-associated long noncoding RNAs show developmental changes in expression coincident with heterochromatin formation at the human (Homo sapiens) macrosatellite repeat. Chromosom. Res. 23 733–752. 10.1007/s10577-015-9479-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth H. V., Richards S. M., Bevan A. P., Clayton S., Corpas M., Rajan D., et al. (2009). DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84 524–533. 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froberg J. E. J. E., Pinter S. F. S. F., Kriz A. J. A. J., Jégu T., Lee J. T. J. T. (2018). Megadomains and superloops form dynamically but are dispensable for X-chromosome inactivation and gene escape. Nat. Commun. 9:5004. 10.1038/s41467-018-07446-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula M. R., Nesterova T. B., Pintacuda G., Godwin J., Zhan Y., Ozadam H., et al. (2019). The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 10:30. 10.1038/s41467-018-07907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geens M., Chuva De Sousa Lopes S. M. (2017). X chromosome inactivation in human pluripotent stem cells as a model for human development: back to the drawing board? Hum. Reprod. Update 23 520–532. 10.1093/humupd/dmx015 [DOI] [PubMed] [Google Scholar]
- Geuens T., Bouhy D., Timmerman V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135 851–867. 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacalone J., Friedes J., Francke U. (1992). A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1 137–143. 10.1038/ng0592-137 [DOI] [PubMed] [Google Scholar]
- Giorgetti L., Lajoie B. R., Carter A. C., Attia M., Zhan Y., Xu J., et al. (2016). Structural organization of the inactive X chromosome in the mouse. Nature 535 575–579. 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Gasser S. M. (2016). On TADs and LADs: spatial control over gene expression. Trends Genet. 32 485–495. 10.1016/j.tig.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Graindorge A., Pinheiro I., Nawrocka A., Mallory A. C., Tsvetkov P., Gil N., et al. (2019). In-cell identification and measurement of RNA-protein interactions. Nat. Commun. 10:5317. 10.1038/s41467-019-13235-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. K., Mardy A. H., Beleford D., Spanier A., Wayman B. V., Penon-Portmann M., et al. (2019). X-linked duplication copy number variation in a familial overgrowth condition. Am. J. Med. Genet. Part C Semin. Med. Genet. 181 644–649. 10.1002/ajmg.c.31756 [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E., Goff L. A., Trapnell C., Williams A., Henao-Mejia J., Sun L., et al. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21 198–206. 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E., Shukla C. J., Weiner C. L., Rinn J. L. (2016). Function and evolution of local repeats in the Firre locus. Nat. Commun. 7:11021. 10.1038/ncomms11021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. S., Hsieh T.-H. S., Cattoglio C., Pustova I., Saldaña-Meyer R., Reinberg D., et al. (2019). Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol. Cell. 76:395-411.e13. 10.1016/j.molcel.2019.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. S., Pustova I., Cattoglio C., Tjian R., Darzacq X. (2017). CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6:e25776. 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y., Brockdorff N., Kawano S., Tsutui K. K. K., Tsutui K. K. K., Nakagawa S., et al. (2010). The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19 469–476. 10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y., Nakagawa S. (2011). Revisiting the function of nuclear scaffold/matrix binding proteins in X chromosome inactivation. RNA Biol. 8 735–739. 10.4161/rna.8.5.16367 [DOI] [PubMed] [Google Scholar]
- Herriges J. C., Arch E. M., Burgio P. A., Baldwin E. E., LaGrave D., Lamb A. N., et al. (2019). Delineating the clinical spectrum associated with Xq25q26.2 duplications: report of 2 families and review of the literature. J. Child Neurol. 34 86–93. 10.1177/0883073818811454 [DOI] [PubMed] [Google Scholar]
- Horakova A. H., Calabrese J. M., McLaughlin C. R., Tremblay D. C., Magnuson T., Chadwick B. P. (2012a). The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 13:R70. 10.1186/gb-2012-13-8-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horakova A. H., Moseley S. C., McLaughlin C. R., Tremblay D. C., Chadwick B. P. (2012b). The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum. Mol. Genet. 21 4367–4377. 10.1093/hmg/dds270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbide A., Torres-Padilla M.-E. (2017). Starting embryonic transcription for the first time. Nat. Genet. 49 820–821. 10.1038/ng.3880 [DOI] [PubMed] [Google Scholar]
- Izuogu O. G., Alhasan A. A., Mellough C., Collin J., Gallon R., Hyslop J., et al. (2018). Analysis of human ES cell differentiation establishes that the dominant isoforms of the lncRNAs RMST and FIRRE are circular. BMC Genomics 19:276. 10.1186/s12864-018-4660-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansz N., Chen K., Murphy J. M., Blewitt M. E. (2017). The epigenetic regulator SMCHD1 in development and disease. Trends Genet. 33 233–243. 10.1016/j.tig.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Jansz N., Nesterova T., Keniry A., Iminitoff M., Hickey P. F., Pintacuda G., et al. (2018). Smchd1 targeting to the inactive X Is dependent on the Xist-HnrnpK-PRC1 pathway. Cell Rep. 25:1912-1923.e9. 10.1016/j.celrep.2018.10.044 [DOI] [PubMed] [Google Scholar]
- Kentepozidou E., Aitken S. J., Feig C., Stefflova K., Ibarra-Soria X., Odom D. T., et al. (2019). Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. bioRxiv[Preprint] 10.1101/668855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpa H. J., Fackelmayer F. O., Lawrence J. B. (2016). SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell 39 9–10. 10.1016/j.devcel.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J. T., Kesner B., An J. Y., Ahn J. Y., Cifuentes-rojas C., Colognori D., et al. (2015). Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57 361–375. 10.1016/j.molcel.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski J. P., Lee J. C., Hwang T., Sunwoo H., Goldstein J. M., Groff A. F., et al. (2019). The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat. Commun. 10:5137. 10.1038/s41467-019-12970-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin C. R., Chadwick B. P. (2011). Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 12:R37. 10.1186/gb-2011-12-4-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minajigi A., Froberg J. E., Wei C., Sunwoo H., Kesner B., Colognori D., et al. (2015). A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349:6245. 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny L. A., Imakaev M., Abdennur N. (2019). Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 58 142–152. 10.1016/J.CEB.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley S. C., Rizkallah R., Tremblay D. C., Anderson B. R., Hurt M. M., Chadwick B. P. (2012). YY1 associates with the macrosatellite DXZ4 on the inactive X chromosome and binds with CTCF to a hypomethylated form in some male carcinomas. Nucleic Acids Res. 40 1596–1608. 10.1093/nar/gkr964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E. P., Dekker J., Heard E. (2013). Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? BioEssays 35 818–828. 10.1002/bies.201300040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., Servant N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganini L., Hadi L. A., Chetta M., Rovina D., Fontana L., Colapietro P., et al. (2019). A HS6ST2 gene variant associated with X-linked intellectual disability and severe myopia in two male twins. Clin. Genet. 95 368–374. 10.1111/cge.13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B. (2016). Developmental regulation of X-chromosome inactivation. Semin. Cell Dev. Biol. 56 88–99. 10.1016/j.semcdb.2016.04.014 [DOI] [PubMed] [Google Scholar]
- Pinter S. F. (2016). A tale of two cities: how Xist and its partners localize to and silence the bicompartmental X. Semin. Cell Dev. Biol. 56 19–34. 10.1016/j.semcdb.2016.03.023 [DOI] [PubMed] [Google Scholar]
- Pinter S. F. S. F., Sadreyev R. I. R. I., Yildirim E., Jeon Y., Ohsumi T. K. T. K., Borowsky M., et al. (2012). Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 22 1864–1876. 10.1101/gr.133751.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlers M., Calabrese J. M., Magnuson T. (2014). Small RNA expression from the human macrosatellite DXZ4. G3 Genes Genomes Genet. 4 1981–1989. 10.1534/G3.114.012260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K., Zaba L. C., Giresi P. G., Li R., Longmire M., Kim Y. H., et al. (2015). Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 1 51–61. 10.1016/j.cels.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. P., Huang S.-C., Glenn St Hilaire B., Engreitz J. M., Perez E. M., Kieffer-Kwon K.-R., et al. (2017). Cohesin loss eliminates all loop domains. Cell 171:305-320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. P., Huntley M. H. H., Durand N. C. C., Stamenova E. K. K., Bochkov I. D., Robinson J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña-Meyer R., Rodriguez-Hernaez J., Escobar T., Nishana M., Jácome-López K., Nora E. P., et al. (2019). RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell. 76:412-422.e5. 10.1016/j.molcel.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn A. L., Rao S. S. P., Huang S.-C., Durand N. C., Huntley M. H., Jewett A. I., et al. (2015). Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112:201518552. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A. D., Hu M., Jung I., Xu Z., Qiu Y., Tan C. L., et al. (2016). A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17 2042–2059. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer R. J., Beaudet A. L., Shinawi M., Sahoo T., Patel A., Sun Q., et al. (2012). Duplication of OCRL and adjacent genes associated with autism but not Lowe syndrome. Am. J. Med. Genet. A 158A 2602–2605. 10.1002/ajmg.a.35566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., et al. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor S. A., Nuzhdin S. V. (2018). The Evolution of Gene Expression in cis and trans. Trends Genet. 34 532–544. 10.1016/J.TIG.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. D. M. D., Pinter S. F. S. F., Fang R., Sarma K., Rutenberg-Schoenberg M., Bowman S. K. S. K., et al. (2013). High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504 465–469. 10.1038/nature12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souren N. Y., Gerdes L. A., Lutsik P., Gasparoni G., Beltrán E., Salhab A., et al. (2019). DNA methylation signatures of monozygotic twins clinically discordant for multiple sclerosis. Nat. Commun. 10:2094. 10.1038/s41467-019-09984-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., de Wit E., Nora E. P., Klous P., van de Werken H. J. G., Zhu Y., et al. (2011). The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25 1371–1383. 10.1101/gad.633311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C. M., Anguera M. C. (2019). When the balance is broken: X-linked gene dosage from two X chromosomes and female-biased autoimmunity. J. Leukoc. Biol. 106 919–932. 10.1002/JLB.6RI0319-094R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot C., Ouimette J.-F., Rougeulle C. (2016). Establishment of X chromosome inactivation and epigenomic features of the inactive X depend on cellular contexts. BioEssays 38 869–880. 10.1002/bies.201600121 [DOI] [PubMed] [Google Scholar]
- Wang C.-Y., Colognori D., Sunwoo H., Wang D., Lee J. T. (2019). PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 10:2950. 10.1038/s41467-019-10755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-Y., Jégu T., Chu H.-P., Oh H. J., Lee J. T. (2018). SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174:406-421.e25. 10.1016/J.CELL.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westervelt N., Chadwick B. P. (2018). Characterization of the ICCE repeat in mammals reveals an evolutionary relationship with the DXZ4 macrosatellite through conserved CTCF binding motifs. Genome Biol. Evol. 10 2190–2204. 10.1093/gbe/evy176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman A., VanOudenhove J., Kron J., Noonan J. P., Cotney J. (2018). High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 23 1581–1597. 10.1016/j.celrep.2018.03.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Peng X., Chen Y., Zhang Y., Ma Q., Liang L., et al. (2017). Free-living human cells reconfigure their chromosomes in the evolution back to uni-cellularity. eLife 6:e28070. 10.7554/eLife.28070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Deng X., Ma W., Berletch J. B., Rabaia N., Wei G., et al. (2015). The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16 1–17. 10.1186/s13059-015-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen A., Kellis M. (2015). Systematic chromatin state comparison of epigenomes associated with diverse properties including sex and tissue type. Nat. Commun. 6:7973. 10.1038/ncomms8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Song D., Zhu B., Wang X. (2019). The role of nuclear matrix protein HNRNPU in maintaining the architecture of 3D genome. Semin. Cell Dev. Biol. 90 161–167. 10.1016/j.semcdb.2018.07.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated and analyzed for this study are cited in the article/supplementary files.