Abstract
Haloferax volcanii is an obligate halophilic archaeon with its origin in the Dead Sea. Simple laboratory culture conditions and a wide range of genetic tools have made it a model organism for studying haloarchaeal cell biology. Halophilic enzymes of potential interest to biotechnology have opened up the application of this organism in biocatalysis, bioremediation, nanobiotechnology, bioplastics and the biofuel industry. Functionally active halophilic proteins can be easily expressed in a halophilic environment, and an extensive genetic toolkit with options for regulated protein overexpression has allowed the purification of biotechnologically important enzymes from different halophiles in H. volcanii. However, corrosion mediated damage caused to stainless-steel bioreactors by high salt concentrations and a tendency to form biofilms when cultured in high volume are some of the challenges of applying H. volcanii in biotechnology. The ability to employ expressed active proteins in immobilized cells within a porous biocompatible matrix offers new avenues for exploiting H. volcanii in biotechnology. This review critically evaluates the various application potentials, challenges and toolkits available for using this extreme halophilic organism in biotechnology.
Keywords: Haloferax volcanii, Biotechnology, Protein expression, Immobilization, Model organism
Introduction
Halophiles are extremophilic microbes that are widespread in all three domains of life—Eukarya, Bacteria and in particular, Archaea (Kamekura 1998; Oren 2013). They are found in hypersaline environments around the world where the salt concentrations is higher than sea water (around 3.5% dissolved salt), in various geographical niches such as salt lakes, deep sea, salt pans, saline soils, salt mines or salt marshes (Oren 2015). Based on their salt preference, halophiles can be split into three different categories: mild halophiles growing optimally at 1–3% (0.2–0.5 M) NaCl, moderate halophiles with optimum growth at 3–15% (0.5–2.5 M) and extreme halophiles with optimal growth at 15–20% (2.5–5.2 M) NaCl (Amoozegar et al. 2017; De Lourdes et al. 2013). Below 0.2 M NaCl (< 1%), microbes are classified as non-halophiles and a key difference is that the growth of mesophilic organisms is impaired at higher salinities, where archaeal species dominate. All halophilic archaea belong to the Euryarchaeota kingdom (Oren et al. 2009).
Halophiles have been important in natural salt production since they facilitate salt crystallization through the absorption of solar radiation using their carotenoid pigments, which confers distinct pink-reddish hues to the saltern crystallizer ponds they inhabit. More recently, halophiles have been used in biotechnology as producers of antimicrobials, bioplastics, biofuel, extremozymes, extracellular polysaccharides, retinal proteins, coloured pigments and compatible solutes (Amoozegar et al. 2019; Atanasova et al. 2013; Koller 2019; Patel and Saraf 2015; Torregrosa-Crespo et al. 2017). This has led to their applications in astrobiology, agriculture, food and nutrition industry, biosensor development and photochemical industry, bioremediation, textile and tanning industry, aquaculture, nanotechnology, sustainable chemical production and medical application (Le Borgne et al. 2008; Lee 2013; Margesin and Schinner 2001; Rodrigo-Baños et al. 2015). Several reviews on the biotechnologically valuable products generated by halophiles and their potential applications have been recently published (Antunes et al. 2017; DasSarma et al. 2010a; Mohammadipanah et al. 2015; Oren 2010; Yin et al. 2015).
Halophiles are under-represented in biotechnology compared with other extremophiles such as thermophiles or alkaliphiles, which have been used extensively for valuable product generation. This is surprising considering the biotechnological potential of halophiles and their great diversity across all three domains of life. While some halophiles are mechanically fragile, prone to lysis in low salt environment, and their culturing can lead to possible damage of the stainless equipment (due to the required molar concentrations of salt), some examples of their successful application in industrial biotechnology are represented by the application of purple membrane protein bacteriorhodopsin (from Halobacterium salinarum) in photochemical application, ectoine, an enzyme immobilizer used in the cosmetic industry (from Halomonas elongata) and β-carotene production (from green algae Dunaliella salina) (Margesin and Schinner 2001; Oren 2010).
H. volcanii is an extremely halophilic archaeon belonging to the family Haloferacaceae. Easy growth conditions, a short generation time and facile genetics have made it a model organism for haloarchaeal biology (Allers and Mevarech 2005; Leigh et al. 2011). It encodes numerous biotechnologically attractive halophilic enzymes with application potentials in sustainable fine chemical synthesis, bioprocessing, bioremediation, biofuel and bioplastic production (Amoozegar et al. 2019; Koller 2019; Lobasso et al. 2015; Timpson et al. 2013; Uthandi et al. 2009). Developments of tools such as bioreactor-scale overexpression of functionally active proteins and immobilization of cells within a porous matrix have opened up new possibilities in exploiting H. volcanii for biotechnological applications (Haque et al. 2019; Strillinger et al. 2016). This review summarizes the biotechnologically relevant features of this organism, the challenges involved and the tools available for realizing its true biotechnological potential.
Genetics and biochemistry of H. volcanii
H. volcanii was first isolated from the sediment of the Dead Sea (Mullakhanbhai and Larsen 1975). The organism originally described as Halobacterium volcanii was named after the microbiologist Benjamin Elazari-Volcani who reported the presence of indigenous microbial life in the salt rich Dead Sea (Elazari-Volcani 1943). It grows optimally at 45 °C with a generation time of 2 h (Robinson et al. 2005). It is simple to culture in the laboratory since it grows aerobically in complex, minimal and synthetic media (Mevarech and Werczberger 1985). Compared with other extreme halophiles, H. volcanii tolerates a wide range of salt concentrations (1.8–3.5 M NaCl) (Allers 2010). It is comparatively resistant to contamination since few other organisms can survive the molar concentration of salt present in the growth media. It can grow up to a temperature of 50 °C (Dyall-Smith 2009). It lacks a rigid cell wall and instead presents a single layer of glycoprotein known as surface layer or S-layer (Rodrigues-Oliveira et al. 2017). Protein subunits in S-layers are held together by divalent cation such as Mg2+ (Cohen et al. 1991). Hence, the S-layer can be completely removed by treatment of H. volcanii with chelating agents such as EDTA.
The H. volcanii genome consists of a main chromosome (2.848 Mb) and three smaller mini-chromosomes (85.1 kb pHV1, 438 kb pHV3 and 636 kb pHV4) along with a 6.4 kb pHV2 plasmid (Hartman et al. 2010). It has a stable genome when compared with other extreme halophiles such as Halobacterium salinarum, which undergoes frequent IS-mediated rearrangement (López-García et al. 1995; Sapienza et al. 1982). One of the major incentives to exploit H. volcanii in biotechnology is it has an extensive genetic toolbox to perform a plethora of genetic manipulations. Transformation of DNA into H. volcanii is achieved using the PEG protocol (Cline et al. 1989). The method was further improved by generating strains deleted for the mrr restriction endonuclease gene, which reduces the transformation efficiency by cleaving the incoming foreign DNA at methylated sites (Allers et al. 2010). The mrr deficient H. volcanii strain allows direct and efficient transformation, eliminating the need to passage the foreign DNA in an Escherichia coli dam mutant. There are multiple shuttle vectors available for use, which are based on pHK2, pHV2 and pHV1 origins (Allers and Mevarech 2005; Allers et al. 2004). The ability of H. volcanii to grow in minimal and synthetic media has led to the development of multiple successful auxotrophic selection markers, where genes involved in amino acid or nucleotide biosynthetic pathways are used to complement chromosomal mutations. Uracil (pyrE2), leucine (leuB), thymidine (hdrB) and tryptophan (trpA) based selection markers have been used extensively for performing gene deletion/complementation in H. volcanii (Allers et al. 2004; Bitan-Banin et al. 2003; Leigh et al. 2011). A CRISPRi toolkit is also available for gene interference based on the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system (Maier et al. 2019). Gene expression reporters based on β-galactosidase and green fluorescent protein (GFP) are available (Holmes and Dyall-Smith 2000; Reuter and Maupin-Furlow 2005). For protein production, both inducible and constitutive promoter systems have been developed (Haque et al. 2019; Large et al. 2007), and metal based protein purification is commonly used owing to purification tags in the expression vectors (Allers et al. 2010). Given these genetic tools and a wide range of salt tolerance, it is not surprising that H. volcanii has been used as a heterologous host to study genes from other halophiles. For example, gas vesicles produced by the closely related species Haloferax mediterranei have been characterized using H. volcanii as a heterologous host, which is naturally devoid of gas vesicle genes (Pfeifer et al. 2002).
Biotechnological potential of H. volcanii
Due to their remarkable properties such as antioxidants, colouring and light absorption, carotenoids are of great interest in biotechnology and medicine (Eggersdorfer and Wyss 2018; Fiedor and Burda 2014; Zhang et al. 2014a). Recently, there has been growing interest in naturally derived carotenoids owing to the increasing demand for bioactive compounds of natural origin with higher bioaccessibility and environmentally benign bioprocessing methods (Giani et al. 2019). The carotenoid biosynthesis pathway in H. volcanii begins with a condensation of two C20 molecules into C40 phytoene, a reaction catalysed by phytoene synthase, encoded by the crtB (HVO_2524) gene. Subsequently, phytoene is converted to lycopene by the phytoene desaturase enzyme encoded by crtI (HVO_2528). In H. volcanii, the lye (HVO_2527) gene encoded lycopene elongase subsequently converts lycopene to bacterioruberin. H. volcanii predominantly produces the C50 carotenoid bacterioruberin (82%) and its derivatives such as monoanhydrobacterioruberin (7%), bisanhydrobacterioruberin (2%) and dihydromonoanhydrobacterioruberin (2%), along with small amount of lycopene (0.3%) (Rønnekleiv 1995). A recent study has shown that bacterioruberin produced by H. volcanii is three times more effective as an antioxidant compared with β-carotene and improves the cell viability, motility and velocities of ram sperm cells (Zalazar et al. 2019). HvLON3 strain, a conditional H. volcanii mutant for Lon protease produces 10–15 times more bacterioruberin than the parental strain given that Lon protease degrades the phytoene synthase, the rate limiting enzyme in carotenoid biosynthesis (Cerletti et al. 2018). H. volcanii is incapable of naturally producing β-carotene since the lycopene cyclase (encoded by crtY) is absent, whereas other halophiles such as Haloarcula marismortui and Halobacterium salinarum possess crtY (Baliga et al. 2004; Peck et al. 2002). However, the carotenoid biosynthetic pathway in H. volcanii can be genetically manipulated to produce β-carotene through the heterologous expression of crtY from Haloarcula marismortui (Smith and Ron 2018). Compared with other halophilic archaea, H. volcanii is better suited for heterologous pathway engineering due to its faster growth, readily available genetic toolkit and stable genome (Leigh et al. 2011). Given the absence of a rigid cell wall, H. volcanii affords a cheap and simple protocol for carotenoid extraction compared with the lengthy and complicated extraction procedure from pink shrimp or carrot, which involves the use of expensive equipment (Mezzomo et al. 2011; Rawson et al. 2011; Zalazar et al. 2019). Hence, by remodelling the carotenoid biosynthetic pathway through genetic engineering, production and extraction of β-carotene using H. volcanii may provide a more effective, efficient and affordable method of natural carotenoid synthesis than those currently offered by chemical synthesis route or by extraction route from other microbes, plants and algae.
Enzyme mediated biocatalysis offers a more cost-effective and environmentally sustainable way of generating biotechnologically valuable chemicals when compared with traditional chemical syntheses (Hughes and Lewis 2018). H. volcanii possesses alcohol dehydrogenase (ADH) enzymes, which catalyse the production of important building blocks in fine chemical, flavour and fragrance industries such as chiral alcohols (Chapuis and Jacoby 2001; Schmid et al. 2001). It encodes two haloalkaliphilic and thermoactive ADHs: HvADH1 (HVO_2428) and HvADH2 (HVO_B0071), which catalyse the reversible oxidation of primary and secondary alcohols into aldehydes and ketones, and the reduction of aldehydes and ketones into their corresponding alcohols in the presence of NADPH/NADH cofactors (Timpson et al. 2013). HvADH2 is industrially more attractive compared with HvADH1 due to greater enzyme activity, dual cofactor and broad substrate scope, stability in organic solvents and halophilic and thermophilic conditions (Alsafadi and Paradisi 2013; Timpson et al. 2013). Given its activity in high temperature (> 80 °C), it is described as the most thermoactive haloarchaeal ADH to date (Cao et al. 2008; Timpson et al. 2012; Timpson et al. 2013). It accepts a broad range of substrates ranging from medium chain alcohols to branched and aromatic alcohols for the oxidative reaction. For the reduction reaction, it accepts a range of aromatic ketone substrates such as alkyl, methyl and alpha-halo aryl ketones (Alsafadi et al. 2017). Immobilization within epoxy resin and metal organic framework material further improves the enzyme activity over a broader pH range and temperature and enhances enzyme stability in organic solvents (Alsafadi and Paradisi 2014; Carucci et al. 2018). It is also possible to improve the substrate scope of HvADH2 for biocatalysis through rational in silico enzyme design (Cassidy et al. 2017). H. volcanii expression systems can be used to overexpress and characterize industrially attractive enzymes from other halophiles (e.g. HmADH12 from extreme halophile Haloarcula marismortui and HsADH2 from the halophilic archaeon Halobacterium salinarum NRC-1) using the tryptophan inducible expression system (Liliensiek et al. 2013; Timpson et al. 2012). The same expression system has also been successfully used to produce haloarchaeal transaminase capable of producing chiral amines (Kelly et al. 2019). In this work, the authors overexpressed a haloarchaeal transaminase from a Triassic period halite deposit that showed halophilic activity, thermo and organic solvent tolerance profile.
Lignin and its derivatives are considered as serious contaminants in paper and pulp industries, tanneries and textile mills due to their poor biodegradability and intense colour (Costa et al. 2017; Raghukumar et al. 2008). Given that waste streams in these industries contain high concentrations of recycled salt, halophilic enzymes capable of lignin degradation could play a pivotal role in biopulping and biobleaching processes (Amoozegar et al. 2019). Laccase is a multicopper oxidase enzyme that couples oxidation of phenolic compounds to the four-electron reduction of molecular oxygen to water (Solomon et al. 1996). It has received attention in the biofuel industry due to its catalytic properties and helps to break down lignocellulose materials, prominent feedstocks for advanced biofuel production (Kudanga and Le Roes-Hill 2014). It has also been implicated in the detoxification of water and soil, in bilirubin level determination in serum and for use in biosensors and bioreactors (Sakurai and Kataoka 2007). H. volcanii encodes LccA (HVO_B0205), a rare archaeal laccase, which is secreted into the culture medium as a highly stable glycoprotein that is active at high salt concentrations (0.1 to 1.4 M) and temperature (55 °C). It accepts a broad range of organic substrates such as bilirubin, syringaldazine (SGZ), 2,2-azino-bis-(3-ethylben- zothiazoline-6-sulfonic acid) (ABTS) and dimethoxyphenol (DMP) and retains its activity in organic solvents such as dimethyl sulfoxide (DMSO) and dimethylformamide (DMF) (Uthandi et al. 2009). Hence, the LccA enzyme could serve as valuable biotechnological tool for the removal of phenolic contaminants in phenol-laden saline wastewater derived from various industrial settings (Moussavi et al. 2010). Despite the potential of LccA in biofuel industry, its direct in vivo lignolytic activity on lignin has still to be experimentally verified (Brink et al. 2019).
As can be seen from the above examples, enzymes from H. volcanii show polyextremophilicity, meaning that they are not only active in high salt environments but also are functional at increased temperature and in organic solvents. It has been established that enzymes from halophiles show greater activity and stability in organic solvents compared with their non-halophilic counterparts (Margesin and Schinner 2001). This unusual ability to be active in environments with low water activity (high salt, solvent and/or desiccation) has made H. volcanii and its enzymes ideal candidates for biotechnology advances.
Halocins are antimicrobial compounds naturally produced by halophilic archaea and bacteria living in hypersaline environments (Shand and Leyva 2007). These compounds are believed to confer selective advantages to the organism for survival in a competitive environment, including against other halophiles (Atanasova et al. 2013). Due to the emergence of antimicrobial resistant pathogens, there is a dire need to screen and characterize novel antimicrobials (Baker 2015). This has included the search for halocins that possess a broad range antimicrobial spectrum. H. volcanii KPS1 strain produces halocin that is active at high temperatures (up to 80 °C) and over a broad range of pH (3.0–9.0) and salinity (Kavitha et al. 2011). Most importantly, it shows antimicrobial activity against both Gram-positive and Gram-negative bacteria. Large amounts of salts are used in tanning processes, which provides a favourable environment for the growth of halophiles. Given its anti-halophilic activity, halocins can be exploited in the textile industry to inhibit the unwanted growth of halophiles that could be damaging for the product (Birbir et al. 2004). The ability of H. volcanii to degrade aromatic hydrocarbons such as anthracene, naphthalene, phenanthrene, pyrene and benzoanthracene could also be beneficial for bioremediation in hypersaline environments (Bonfa et al. 2011).
Polyhydroxyalkanoates (PHA) are polyesters composed of hydroxy fatty acids, which serve as intracellular storage material of carbon source and energy (Rehm 2007). They are found in a variety of microbes including haloarcheal species belonging to the genera Haloarcula, Haloferax, Halococcus, Haloquadratum, Halobacterium, Haloterrigena and Natronobacterium (Koller 2019). These water-insoluble nanosized cytoplasmic compounds crystallize after solvent extraction and most importantly, exhibit thermo-plastic and elastomeric properties (Poli et al. 2011). Therefore, PHA compounds have received global attention due to their importance as natural, bio-based, biocompatible and biodegradable alternatives to plastics of petrochemical origin. Poly 3-hydroxybutyrate (PHB) is the best known polyhydroxyalkanoate (PHA). Polymerization of 3-hydroxybutyryl-CoA results in PHA formation through PHA synthase activity (Rehm 2003). H. volcanii produces PHB up to 7% of its cell dry weight when grown on media containing 250 g/L salts, 10 g/L glucose and 1 g/L yeast extract (Fernandez-Castillo et al. 1986). An additional report for H. volcanii mediated PHA accumulation from sugarcane bagasse substrate, a fibrous leftover by-product of the sugarcane industry, was recently published (Salgaonkar and Bragança 2017). In contrast, another study reported no detection of PHB from H. volcanii (Legat et al. 2010). H. volcanii has been successfully used as a heterologous host to generate PHA through the expression of PHA synthase genes from the halophile Haloarcula hispanica (Han et al. 2009).
Alongside facile genetics, at least three other traits make H. volcanii an attractive host for bioprocessing. Firstly, industrially desirable quasi-sterile conditions are readily achieved, since few other microbes cannot grow in the molar concentrations of salt present in growth media for H. volcanii. Secondly, the cost involved in product extraction is significantly reduced since it is easy to lyse H. volcanii cells through addition of water and/or the use of simple chelating agents to disrupt the outer S-layer. Finally, the potential for LPS (lipopolysaccharide) mediated toxicity caused by bacterial hosts is eliminated by the use of H. volcanii. These properties would substantially facilitate downstream processing in economic and environmental terms during bioprocessing.
The S-layer of H. volcanii is a two-dimensional array of protein or glycoprotein subunits, which form part of the cell envelope components (Tamir and Eichler 2017). The ability of isolated archaeal S-layer proteins to assemble into regular arrays in liquid or on surfaces such as metals, polymers or interfaces such as lipid films and liposomes makes them attractive for potential applications in nanobiotechnology. These arrays show reproducible physiochemical properties down to the nanometre scale (Debabov 2004). The pores in S-layers are identical in size and morphology, and functional groups such as carboxylic acid, amine and hydroxyl groups on the protein lattices and pore areas are aligned in defined positions and orientations. These combined features have led to their applications in the production of isoporous ultrafiltration membranes, bioanalytical sensors, biomimetics, affinity membranes, immunotherapy, drug delivery, vaccine development and immobilization matrices for binding of functional molecules such as enzymes and antibodies (Ilk et al. 2008; Sleytr et al. 2007). Mammalian olfactory receptor from rat has been heterologously expressed in the lipid bilayer of the H. volcanii (Lobasso et al. 2015). Therefore, the lipid matrix of H. volcanii could serve as an ideal biomaterial for the future development of nanovesicle based hybrid biosensor devices.
Challenges of applying H. volcanii in biotechnology
Halophiles adapt to high salt environment using two alternative strategies (Oren 2008). The first strategy, termed ‘salt-out’, is mainly adopted by halotolerant bacteria and eukaryotes. Excess salt is excluded from halotolerant bacteria and eukaryotes, with an increase in compatible organic solutes such as glycerol or glycine betaine in the cytoplasm to maintain an osmotic balance. These compatible solutes do not interfere with intracellular enzymatic activity. The second strategy, termed ‘salt-in’, is adopted mainly by haloarchaea including H. volcanii. Equimolar concentrations of salt, predominantly KCl, are accumulated in the cytoplasm to maintain the osmotic balance (Oren 2013). Since biosynthesis of organic osmotic solutes in the ‘salt-out’ strategy is energetically costly compared with the ‘salt-in’ strategy, it is haloarchaea such as H. volcanii that thrive in hypersaline conditions (Oren 1999). Halophilic proteins from H. volcanii have evolved to function in the presence of molar concentration of salt, they have an overall increase in the number of highly positively charged acidic residues on surface to make the proteins soluble in high salt environment, along with a decrease in overall hydrophobicity through the substitution of large hydrophobic residues on the surface with small hydrophilic ones (Danson and Hough 1997; Mevarech et al. 2000; Wright et al. 2002).
This peculiarity poses a challenge for the expression of active halophilic proteins in industrially attractive heterologous hosts such as E. coli, since these proteins often misfold and aggregate in low ionic strength environment. However, there have been a few reports of heterologous expression of halophilic proteins in E. coli (Cao et al. 2008; Cendrin et al. 1993; Domenech and Ferrer 2006; Johnsen and Schönheit 2004), which rely on solubilization of inclusion bodies followed by laborious solubilization and refolding of insoluble proteins in a hypersaline environment (Connaris et al. 1999; Singh and Panda 2005). An additional caveat is the requirement for prior knowledge of the target protein characteristics. In any case, this approach is not universally effective, as many enzymes remain inactive. For example, enzymatically active protein could not be recovered from the heterologously overexpressed alcohol dehydrogenase (HmADH12) of the extreme halophile Haloarcula marismortui, despite repeated attempts to solubilize the inclusion bodies in varying salt and pH conditions (Timpson et al. 2012). In another study, no protein expression was observed following attempts of overexpression in E. coli of two halophilic alcohol dehydrogenases (ADH/D1 and ADH/A1) from the deep Red Sea brine pools (Strillinger et al. 2016). These drawbacks of heterologous expression have led to the development of halophilic systems to generate biotechnologically important proteins that are functionally active.
One of the major obstacles in exploiting extremozymes is the low yield or activity of enzymes when cultivated on a large bioreactor scale, as required for industrial applications (Elleuche et al. 2014; Sarmiento et al. 2015). For example, it is essential to produce active alcohol dehydrogenase (HvADH), laccase (LccA) enzymes and bacterioruberin and halocins proteins (Table 1) from H. volcanii on large scale to realize their true biotechnological potential. It is challenging to ensure expression and purification of bulk quantities of highly active proteins in a halophilic environment. Co-purification of unwanted non-specific proteins with the target protein is often encountered by the researchers following affinity tagged protein purification (Kimple et al. 2013). A bottleneck of large-scale protein overexpression in H. volcanii host is their intrinsic ability to form biofilms (Chimileski et al. 2014; Frols et al. 2012), which can interfere with expensive sensors present in the bioreactor during the fermentation process and may also alter the characteristics of expressed proteins. Molar concentrations of salt required to culture H. volcanii can rapidly corrode stainless-steel bioreactors. This necessitates the use of reactors made of alternative materials rather than the readily available stainless-steel.
Table 1.
Biotechnologically important components encoded by H. volcanii
| Component | Potential applications | References |
|---|---|---|
| Phytoene synthase (HVO_2524), phytoene desaturase (HVO_2528) and lycopene elongase (HVO_2527) | Carotenoid biosynthesis | (Smith and Ron 2018; Zalazar et al. 2019) |
| Alcohol dehydrogenases-HvADH1 (HVO_2428) and HvADH2 (HVO_B0071) | Fine chemical production through biocatalysis | (Timpson et al. 2013; Alsafadi et al. 2017; Haque et al. 2019) |
| Laccase (HVO_B0205) | Biopulping, biobleaching, biofuel production | (Uthandi et al. 2009) |
| Halocin | Antimicrobials, bioremediation | (Kavitha et al. 2011) |
| Polyhydroxyalkanoate | Bioplastic production | (Koller 2019) |
| Outer surface layer (S-layer) | Nanobiotechnology | (Ilk et al. 2008; Sleytr et al. 2007; Lobasso et al. 2015) |
Tools for H. volcanii applications in biotechnology
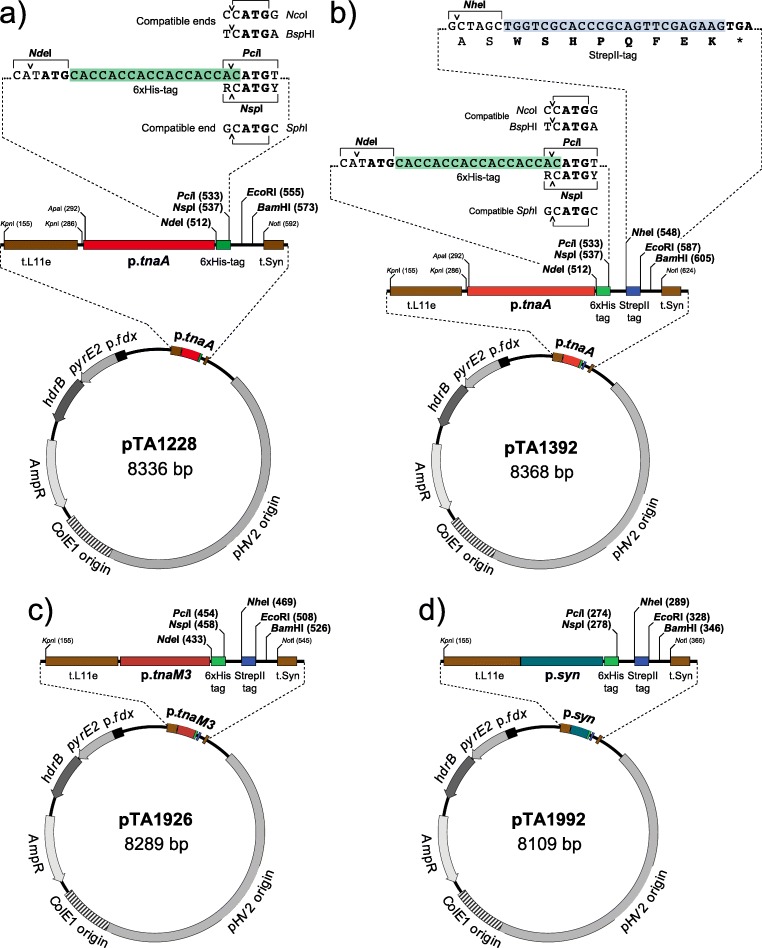
For protein overexpression, a modified strain H. volcanii host strain H1424 (ΔpyrE2 ΔhdrB Δmrr Nph-pitA cdc48d-Ct) is available along with its corresponding plasmid pTA1228 (Fig. 1a) (Brendel et al. 2014; Stroud et al. 2012). Regulated gene expression systems enable the tailoring of expression of biotechnologically important enzymes/proteins according to the industrial need. The pTA1228 plasmid (Fig. 1a) contains the tryptophan inducible promoter p.tnaA for protein expression (Large et al. 2007), which is active in the presence of >1 mM tryptophan. The plasmid pTA1228 also encodes an N-terminal hexahistidine (6xHis) tag to facilitate protein purification using immobilized metal-based affinity chromatography (IMAC), since this affinity tag is compatible with high salt media. The host strain H1424 is deleted for the histidine-rich PitA and Cdc48d proteins, major contaminants of the His-tagged proteins purified from H. volcanii using IMAC (Bab-Dinitz et al. 2006). Given the essential nature of the pitA and cdc48d, it was not possible to delete these genes completely; therefore, pitA was replaced with an orthologue (Nph-pitA) from the haloalkaliphile Natronomonas pharaonis that does not contain a high number of histidine residues, and a truncated version of cdc48d coding for a protein (Cdc48d-Ct) lacking the histidine rich C terminus was engineered (Allers et al. 2010; Stroud et al. 2012). The expression cassette in pTA1228 is flanked by two transcriptional terminators (t.L11e and t.Syn) to prevent read through transcription (Fig. 1a), and the pHV2 replication origin maintains the plasmid at a copy number of 6 per genome (Allers et al. 2004). Incorporation of pyrE2 and hdrB marker genes in pTA1228 enables selection on media lacking uracil and thymidine, respectively; the latter is lacking in rich media (Hv-YPC), facilitating maximal cell growth and protein overexpression (Allers et al. 2004; Bitan-Banin et al. 2003).
Fig. 1.
Regulated and constitutive protein expression system in H. volcanii.a pTA1228. The p.tnaA promoter is induced by L-tryptophan and allows for regulated protein expression. An N-terminal histidine (His) tag is available for immobilized metal-based affinity chromatography (IMAC) protein purification. A multiple cloning site is located after His tag. At the 5′ end of the halophilic gene, the ATG start codon is replaced with an NcoI, BspHI, PciI or SphI site (depending on the coding sequence) and inserted in-frame with the His tag at the PciI or SphI site; at the 3′ end of the gene, an EcoRI or BamHI site is used. The cloning cassette is flanked by L11e and t.Syn terminators to prevent read-through transcription. b pTA1392. A derivative of pTA1228. A C-terminal streptadivin II (StrepII) tag is available along with the His tag. At the 3′ end of the gene, the stop codon is replaced with an NheI site and inserted in-frame with the StrepII tag. c pTA1926. A 50% reduced strength tryptophan inducible promoter, when compared with p.tnaA. Both His and Strep II tags are available. d) pTA1992. A strong p.syn promoter for constitutive protein expression with both His and Strep II tags
To give researchers more options for protein purification, another plasmid pTA1392 (Fig. 1b) is available that encodes a C-terminal Streptavidin II tag (Strep II) for protein purification, alongside the established N-terminal His-tag (Braun et al. 2019); both tags are compatible with high salt concentrations. For protein expression at lower levels, for example if the protein is toxic to the host cells, the pTA1926 plasmid (Fig. 1c) contains the p.tnaM3 promoter that shows 50% reduced activity compared with p.tnaA promoter present in pTA1228 and pTA1392 (Braun et al. 2019). For constitutive overexpression of proteins that are not harmful to the host, the pTA1992 plasmid (Fig. 1d) contains the highly active p.syn promoter (Haque et al. 2019; Large et al. 2007). This plasmid has the potential to make biotechnological processes cost effective since its usage alleviates the need for repeated tryptophan supplementation into H. volcanii cultures, which is an expensive amino acid.
A simple tryptophan-inducible protein overexpression system was reported that allows halophilic protein overexpression using a stirred tank bioreactor system (Strillinger et al. 2016). This study overexpressed two thermohalophilic alcohol dehydrogenases (ADH/D1 and ADH/A1), which showed about 28 fold more enzyme activity when compared with the uncontrolled shaker flask cultivations. The use of bioreactors has inherent advantages such as control of pH, temperature, supply of oxygens and nutrient media during the fermentation process. To prevent biofilm formation inside bioreactors, a H. volcanii strain (H1895) that is deleted for the two essential genes for biofilm formation: an integral membrane protein (HVO_1033) and ATPase subunit (HVO_1034) of the archaeal pilus biosynthesis machinery could be exploited in biotechnology given its inability to form biofilm during protein overexpression (Strillinger et al. 2016; Tripepi et al. 2013; Tripepi et al. 2012).
Alternative bioreactors other than corrosion prone stainless-steel are available for large scale halophilic protein production. A stirred and aerated bioreactor made of corrosion resistant materials such as polyetherether ketone (PEEK), borosilicate glass or silicon nitrite ceramics has been successfully used to generate poly-L-glutamic acid (PGA) or poly -β-hydroxy butyric acid (PHB) from the cultures of extreme halophilic archaeon Natrialba spp. (Hezayen et al. 2000). Another extreme halophile H. mediterranei has been cultured using a bioreactor with parts made of PEEK and borosilicate glass (Lorantfy et al. 2014). However, these materials are costly and more work is necessary in designing more energy efficient, corrosion-resistant and cost-effective alternative bioreactors for culturing H. volcanii.
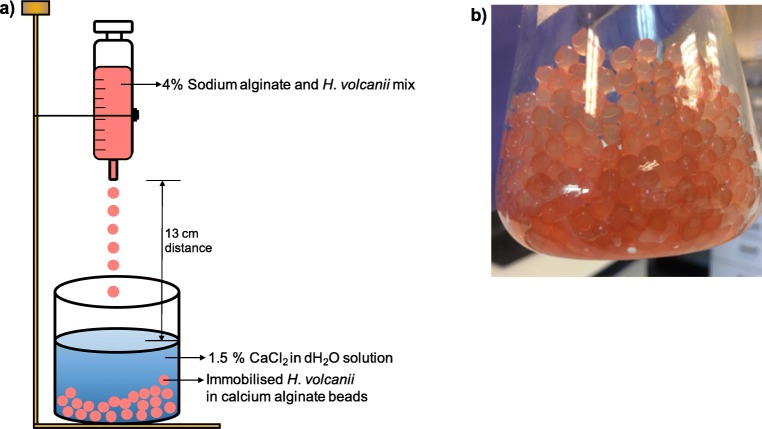
Recent work has shown H. volcanii can be immobilized in inexpensive, non-toxic and highly porous calcium alginate beads following a simple method and used as whole cell biocatalyst (Fig. 2) (Haque et al. 2019). It offers halophilic enzyme expression with high product yield and reusability in a hypersaline environment. Furthermore, it provides data that calcium alginate beads with once immobilized H. volcanii can be used repeatedly for 12 successive biocatalytic cycles without compromising the bead morphology. Product yield from a batch of immobilized beads with H. volcanii that had been stored at room temperature for a month was very similar to freshly prepared beads. This system is particularly cost-effective with enzymatic processes that require expensive NADPH/NADH cofactor supplementation, since in situ cofactors generated by H. volcanii are used for the catalytic reaction, thus making the process self-sufficient. In this reported system, 1.1 g dry weight (from 120 ml broth culture) of immobilized H. volcanii whole cell biocatalyst showed high product yield range of 96.4–100% in the presence of 5 mM acetophenone substrate after 24 h. In contrast, a yield range of 48–94% was found from 500 ml of Yarrowia lipolytica in presence of 3.3 mM of the same substrate after 48 h (Janeczko et al. 2015). In another work, a product yield range of 53–63% was found with 5 g of the fungus, Aspergillus niger whole cell biocatalyst in the presence of 1 mM acetophenone substrate after 24 h (Kurbanoglu et al. 2007). Therefore, the performance of this immobilized H. volcanii whole cell biocatalyst is superior compared with the reported above two studies.
Fig. 2.
Simple method for immobilization of H. volcanii within the calcium alginate hydrogels. a Calcium alginate beads with encapsulated H. volcanii is formed instantaneously following the dropwise addition of resuspended H. volcanii cells in 4% sodium alginate solution into 1.5% CaCl2 solution. b Formation of pink beads due to the presence of high carotenoid pigment confirms entrapment of H. volcanii within the calcium alginate beads
Immobilized H. volcanii within the calcium alginate beads offer multiple advantages for biotechnological applications (Fig. 2). Firstly, it stabilizes the whole cells within the calcium alginate bead matrix leading to higher cell density and specific productivity (Strand et al. 2004). Secondly, the porous alginate bead matrix eases the product separation and flow of material to the H. volcanii cells. Thirdly, this type of highly defined and reproducible encapsulation within the natural alginate biopolymer ensures biocompatible microenvironment for the maintenance of viable H. volcanii cells (Melvik and Dornish 2004). Finally, immobilized nature of the whole cells means it is ideal for their applications in bioreactors without the problem of cell wash out (Obradovic et al. 2004). Such simple system can easily be scaled up by packing the beads with immobilized H. volcanii inside a stirred tank fermenter (Strillinger et al. 2016). Organism such as Alcaligenes faecalis immobilized within the calcium alginate beads has been successfully used in packed bed bioreactor as whole cell biocatalyst (Xue et al. 2013; Zhang et al. 2014b). Intermittent or continuous substrate feeding approach using flow biocatalysis can be combined with in situ product removal strategy to further increase product yield and alleviate toxicity mediated by the presence of excessive substrate/product in the immobilized system (Freeman et al. 1993; Tamborini et al. 2018)). Therefore, if the biotechnological process involves supplementation or generation of a toxic compound, use of immobilized H. volcanii whole cell approach would be preferable over the bioreactor scale fermentation since it would offer more protection to the cells.
Interest in halophiles such as H. volcanii has led to the development of curated bioinformatic databases such as HaloWeb, HProtDB and HaloDom. These tools greatly accelerate bioprospecting through genome mining. HaloWeb (http://halo4.umbi.umd.edu) provides the complete genome sequences for all haloarchaeal genomes, along with BLAST options against various genomes and genomic maps (DasSarma et al. 2010b). HProtDB (http://webapp.cabgrid.res.in/protein/) contains information about physical and biochemical properties of halophilic proteins for 21 strains of Archaea and Bacteria (Sharma et al. 2014). HaloDom catalogues all the halophilic species studied to date across all three domains of life, Archaea, Bacteria and Eukarya (http://halodom.bio.auth.gr/). It also provides a ‘Halopredictor’ function to predict the signatures of halophilic adaptation in the FASTA sequence of a novel protein (Loukas et al. 2018). The ‘Halohandbook’ is a manual containing detailed laboratory protocols for studying halophiles including H. volcanii (Dyall-Smith 2009). It has been compiled with input from leading halophile researchers.
Conclusions
Characteristics such as facile genetics, adaptation to low water environment and high salt tolerance have great potential in making H. volcanii a valuable resource for biotechnology. Of particular interest is the use of H. volcanii as a heterologous host to study important biotechnological enzymes from other halophiles, ranging from moderate to extreme. However, challenges still remain in applying H. volcanii in a broad range of biotechnological applications. With the discovery of novel biotechnologically important proteins, expansion of genetic toolkits and rapidly advanced technologies, the potential for exploiting H. volcanii in biotechnology can only grow.
Author contributions
All authors contributed equally to writing the manuscript.
Funding information
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/P002536/1]. The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
Compliance with ethical standards
Competing interests
The authors declare that there are no competing interests.
Ethical approval
Not applicable, since the work does not involve any study with human participants or animals.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allers T. Overexpression and purification of halophilic proteins in Haloferax volcanii. Bioeng Bugs. 2010;1(4):288–290. doi: 10.4161/bbug.1.4.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Mevarech M. Archaeal genetics — the third way. Nat Rev Genet. 2005;6(1):58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- Allers T, Ngo H-P, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70(2):943–953. doi: 10.1128/aem.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Barak S, Liddell S, Wardell K, Mevarech M. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol. 2010;76(6):1759–1769. doi: 10.1128/aem.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsafadi D, Paradisi F. Effect of organic solvents on the activity and stability of halophilic alcohol dehydrogenase (ADH2) from Haloferax volcanii. Extremophiles. 2013;17(1):115–122. doi: 10.1007/s00792-012-0498-0. [DOI] [PubMed] [Google Scholar]
- Alsafadi D, Paradisi F. Covalent immobilization of alcohol dehydrogenase (ADH2) from Haloferax volcanii: how to maximize activity and optimize performance of halophilic enzymes. Mol Biotechnol. 2014;56(3):240–247. doi: 10.1007/s12033-013-9701-5. [DOI] [PubMed] [Google Scholar]
- Alsafadi D, Alsalman S, Paradisi F. Extreme halophilic alcohol dehydrogenase mediated highly efficient syntheses of enantiopure aromatic alcohols. Org Biomol Chem. 2017;15(43):9169–9175. doi: 10.1039/C7OB02299A. [DOI] [PubMed] [Google Scholar]
- Amoozegar MA, Siroosi M, Atashgahi S, Smidt H, Ventosa A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology. 2017;163(5):623–645. doi: 10.1099/mic.0.000463. [DOI] [PubMed] [Google Scholar]
- Amoozegar MA, Safarpour A, Noghabi KA, Bakhtiary T, Ventosa A. Halophiles and their vast potential in biofuel production. Front Microbiol. 2019;10:1895. doi: 10.3389/fmicb.2019.01895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A, Simões MF, Grötzinger SW, Eppinger J, Bragança J, Bajic VB. Bioprospecting Archaea: focus on extreme halophiles. In: Paterson R, Lima N, editors. Bioprospecting: success, potential and constraints. Cham: Springer International Publishing; 2017. pp. 81–112. [Google Scholar]
- Atanasova NS, Pietilä MK, Oksanen HM. Diverse antimicrobial interactions of halophilic archaea and bacteria extend over geographical distances and cross the domain barrier. Microbiologyopen. 2013;2(5):811–825. doi: 10.1002/mbo3.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bab-Dinitz E, Shmuely H, Maupin-Furlow J, Eichler J, Shaanan B. Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics. 2006;22(6):671–675. doi: 10.1093/bioinformatics/btk043. [DOI] [PubMed] [Google Scholar]
- Baker S. A return to the pre-antimicrobial era? Science. 2015;347(6226):1064. doi: 10.1126/science.aaa2868. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, Hung P, Date SV, Marcotte E, Hood L, Ng WV. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004;14(11):2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbir M, Eryilmaz S, Ogan A. Prevention of halophilic microbial damage on brine cured hides by extremely halophilic halocin producer strains. J Soc Leath Tech Ch. 2004;88:99–104. [Google Scholar]
- Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185(3):772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfa MR, Grossman MJ, Mellado E, Durrant LR. Biodegradation of aromatic hydrocarbons by Haloarchaea and their use for the reduction of the chemical oxygen demand of hypersaline petroleum produced water. Chemosphere. 2011;84(11):1671–1676. doi: 10.1016/j.chemosphere.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Braun F, Thomalla L, van der Does C, Quax TEF, Allers T, Kaever V, Albers S-V. Cyclic nucleotides in archaea: cyclic di-AMP in the archaeon Haloferax volcanii and its putative role. Microbiologyopen. 2019;8(9):e00829. doi: 10.1002/mbo3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel J, Stoll B, Lange SJ, Sharma K, Lenz C, Stachler AE, Maier LK, Richter H, Nickel L, Schmitz RA, Randau L, Allers T, Urlaub H, Backofen R, Marchfelder A. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J Biol Chem. 2014;289(10):7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink DP, Ravi K, Lidén G, Gorwa-Grauslund MF. Mapping the diversity of microbial lignin catabolism: experiences from the eLignin database. Appl Microbiol Biotechnol. 2019;103(10):3979–4002. doi: 10.1007/s00253-019-09692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liao L, Xu XW, Oren A, Wang C, Zhu XF, Wu M. Characterization of alcohol dehydrogenase from the haloalkaliphilic archaeon Natronomonas pharaonis. Extremophiles. 2008;12(3):471–476. doi: 10.1007/s00792-007-0133-7. [DOI] [PubMed] [Google Scholar]
- Carucci C, Bruen L, Gascón V, Paradisi F, Magner E. Significant enhancement of structural stability of the hyperhalophilic ADH from Haloferax volcanii via entrapment on metal organic framework support. Langmuir. 2018;34(28):8274–8280. doi: 10.1021/acs.langmuir.8b01037. [DOI] [PubMed] [Google Scholar]
- Cassidy J, Bruen L, Rosini E, Molla G, Pollegioni L, Paradisi F. Engineering substrate promiscuity in halophilic alcohol dehydrogenase (HvADH2) by in silico design. PLoS One. 2017;12(11):e0187482. doi: 10.1371/journal.pone.0187482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendrin F, Chroboczek J, Zaccai G, Eisenberg H, Mevarech M. Cloning, sequencing, and expression in Escherichia coli of the gene coding for malate dehydrogenase of the extremely halophilic archaebacterium Haloarcula marismortui. Biochemistry. 1993;32(16):4308–4313. doi: 10.1021/bi00067a020. [DOI] [PubMed] [Google Scholar]
- Cerletti M, Paggi R, Troetschel C, Ferrari MC, Guevara CR, Albaum S, Poetsch A, De Castro R. LonB protease is a novel regulator of carotenogenesis controlling degradation of phytoene synthase in Haloferax volcanii. J Proteome Res. 2018;17(3):1158–1171. doi: 10.1021/acs.jproteome.7b00809. [DOI] [PubMed] [Google Scholar]
- Chapuis C, Jacoby D. Catalysis in the preparation of fragrances and flavours. Appl Catal A. 2001;221:93–117. doi: 10.1016/S0926-860X(01)00798-0. [DOI] [Google Scholar]
- Chimileski S, Franklin MJ, Papke RT. Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 2014;12:65–65. doi: 10.1186/s12915-014-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989;35(1):148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- Cohen S, Shilo M, Kessel M. Nature of the salt dependence of the envelope of a Dead Sea archaebacterium, Haloferax volcanii. Arch Microbiol. 1991;156(3):198–203. doi: 10.1007/BF00249115. [DOI] [Google Scholar]
- Connaris H, Chaudhuri JB, Danson MJ, Hough DW. Expression, reactivation, and purification of enzymes from Haloferax volcanii in Escherichia coli. Biotechnol Bioeng. 1999;64(1):38–45. doi: 10.1002/(SICI)1097-0290(19990705)64:1<38::AID-BIT4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Costa S, Dedola D, Pellizzari S, Blo R, Rugiero I, Pedrini P, Tamburini E. Lignin biodegradation in pulp-and-paper mill wastewater by selected white rot fungi. Water. 2017;9:935. doi: 10.3390/w9120935. [DOI] [Google Scholar]
- Danson MJ, Hough DW. The structural basis of protein halophilicity. Comp Biochem Physiol Part A Physiol. 1997;117(3):307–312. doi: 10.1016/S0300-9629(96)00268-X. [DOI] [Google Scholar]
- DasSarma P, Coker J, Huse V, Das Sarma S (2010a) Halophiles, industrial applications. In Encyclopedia of industrial biotechnology: bioprocess, bioseparation and cell technology. Hoboken: John Wiley & Sons, Inc.2009
- DasSarma SL, Capes MD, DasSarma P, DasSarma S. HaloWeb: the haloarchaeal genomes database. Saline Systems. 2010;6:12–12. doi: 10.1186/1746-1448-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lourdes MM, Pérez D, García MT, Mellado E. Halophilic bacteria as a source of novel hydrolytic enzymes. Life. 2013;3(1):38–51. doi: 10.3390/life3010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debabov VG. Bacterial and archaeal S-layers as object of bionanotechnology. Mol Biol (Mosk) 2004;38(4):578–591. doi: 10.1023/B:MBIL.0000036999.77762.6a. [DOI] [PubMed] [Google Scholar]
- Domenech J, Ferrer J. A new D-2-hydroxyacid dehydrogenase with dual coenzyme-specificity from Haloferax mediterranei, sequence analysis and heterologous overexpression. Biochim Biophys Acta. 2006;1760(11):1667–1674. doi: 10.1016/j.bbagen.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Dyall-Smith M (2009) The Halohandbook- protocols for haloarchaeal genetics. https://haloarchaea.com/wp-content/uploads/2018/10/Halohandbook_2009_v7.3mds.pdf
- Eggersdorfer M, Wyss A. Carotenoids in human nutrition and health. Arch Biochem Biophys. 2018;652:18–26. doi: 10.1016/j.abb.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Elazari-Volcani B(W) Bacteria in the bottom sediments of the Dead Sea. Nature. 1943;152(3853):274–275. doi: 10.1038/152274c0. [DOI] [Google Scholar]
- Elleuche S, Schröder C, Sahm K, Antranikian G. Extremozymes—biocatalysts with unique properties from extremophilic microorganisms. Curr Opin Biotechnol. 2014;29:116–123. doi: 10.1016/j.copbio.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Fernandez-Castillo R, Rodriguez-Valera F, Gonzalez-Ramos J, Ruiz-Berraquero F. Accumulation of poly (beta-hydroxybutyrate) by halobacteria. Appl Environ Microbiol. 1986;51(1):214–216. doi: 10.1128/AEM.51.1.214-216.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedor J, Burda K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients. 2014;6(2):466–488. doi: 10.3390/nu6020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Woodley JM, Lilly MD. In situ product removal as a tool for bioprocessing. Biotechnology (NY) 1993;11(9):1007–1012. doi: 10.1038/nbt0993-1007. [DOI] [PubMed] [Google Scholar]
- Frols S, Dyall-Smith M, Pfeifer F. Biofilm formation by haloarchaea. Environ Microbiol. 2012;14(12):3159–3174. doi: 10.1111/j.1462-2920.2012.02895.x. [DOI] [PubMed] [Google Scholar]
- Giani M, Garbayo I, Vílchez C, Martínez-Espinosa RM. Haloarchaeal carotenoids: healthy novel compounds from extreme environments. Mar Drugs. 2019;17(9):524. doi: 10.3390/md17090524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lu Q, Zhou L, Liu H, Xiang H. Identification of the polyhydroxyalkanoate (PHA)-specific acetoacetyl coenzyme a reductase among multiple FabG paralogs in Haloarcula hispanica and reconstruction of the PHA biosynthetic pathway in Haloferax volcanii. Appl Environ Microbiol. 2009;75(19):6168–6175. doi: 10.1128/aem.00938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque RU, Paradisi F, Allers T. Haloferax volcanii as immobilised whole cell biocatalyst: new applications for halophilic systems. Appl Microbiol Biotechnol. 2019;103(9):3807–3817. doi: 10.1007/s00253-019-09725-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM, Maupin-Furlow J, Pohlschroder M, Daniels C, Pfeiffer F, Allers T, Eisen JA. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5(3):e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezayen FF, Rehm BH, Eberhardt R, Steinbuchel A. Polymer production by two newly isolated extremely halophilic archaea: application of a novel corrosion-resistant bioreactor. Appl Microbiol Biotechnol. 2000;54(3):319–325. doi: 10.1007/s002530000394. [DOI] [PubMed] [Google Scholar]
- Holmes ML, Dyall-Smith ML. Sequence and expression of a halobacterial beta-galactosidase gene. Mol Microbiol. 2000;36(1):114–122. doi: 10.1046/j.1365-2958.2000.01832.x. [DOI] [PubMed] [Google Scholar]
- Hughes G, Lewis JC. Introduction: biocatalysis in industry. Chem Rev. 2018;118(1):1–3. doi: 10.1021/acs.chemrev.7b00741. [DOI] [PubMed] [Google Scholar]
- Ilk N, Egelseer EM, Ferner-Ortner J, Küpcü S, Pum D, Schuster B, Sleytr UB. Surfaces functionalized with self-assembling S-layer fusion proteins for nanobiotechnological applications. Colloid Surf A Physicochem Eng Asp. 2008;321(1):163–167. doi: 10.1016/j.colsurfa.2007.12.038. [DOI] [Google Scholar]
- Janeczko T, Bąkowski W, Walczak E, Robak M, Dmochowska-Gładysz J, Kostrzewa-Susłow E. Biotransformation of acetophenone and its halogen derivatives by Yarrowia lipolytica strains. Ann Microbiol. 2015;65(2):1097–1107. doi: 10.1007/s13213-014-0955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen U, Schönheit P. Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui. J Bacteriol. 2004;186(18):6198–6207. doi: 10.1128/JB.186.18.6198-6207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamekura M. Diversity of extremely halophilic bacteria. Extremophiles. 1998;2(3):289–295. doi: 10.1007/s007920050071. [DOI] [PubMed] [Google Scholar]
- Kavitha PK, Lipton A, S AR, MS A. Growth characteristics and halocin production by a new isolate, Haloferax volcanii KPS1 from Kovalam solar saltern (India) Res J Biol Sci. 2011;6:257–262. doi: 10.3923/rjbsci.2011.257.262. [DOI] [Google Scholar]
- Kelly SA, Magill DJ, Megaw J, Skvortsov T, Allers T, McGrath JW, Allen CCR, Moody TS, Gilmore BF. Characterisation of a solvent-tolerant haloarchaeal (R)-selective transaminase isolated from a Triassic period salt mine. Appl Microbiol Biotechnol. 2019;103(14):5727–5737. doi: 10.1007/s00253-019-09806-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple ME, Brill AL, Pasker RL (2013) Overview of affinity tags for protein purification. Curr Protoc Protein Sci:9.9.1–9.9.23. 10.1002/0471140864.ps0909s73 [DOI] [PMC free article] [PubMed]
- Koller Polyhydroxyalkanoate Biosynthesis at the Edge of Water Activitiy-Haloarchaea as Biopolyester Factories. Bioengineering. 2019;6(2):34. doi: 10.3390/bioengineering6020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudanga T, Le Roes-Hill M. Laccase applications in biofuels production: current status and future prospects. Appl Microbiol Biotechnol. 2014;98(15):6525–6542. doi: 10.1007/s00253-014-5810-8. [DOI] [PubMed] [Google Scholar]
- Kurbanoglu EB, Zilbeyaz K, Kurbanoglu NI, Kilic H. Enantioselective reduction of substituted acetophenones by Aspergillus niger. Tetrahedron Asymmetry. 2007;18(10):1159–1162. doi: 10.1016/j.tetasy.2007.05.017. [DOI] [Google Scholar]
- Large A, Stamme C, Lange C, Duan Z, Allers T, Soppa J, Lund PA. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol Microbiol. 2007;66(5):1092–1106. doi: 10.1111/j.1365-2958.2007.05980.x. [DOI] [PubMed] [Google Scholar]
- Le Borgne S, Paniagua D, Vazquez-Duhalt R. Biodegradation of organic pollutants by halophilic bacteria and archaea. J Mol Microbiol Biotechnol. 2008;15(2–3):74–92. doi: 10.1159/000121323. [DOI] [PubMed] [Google Scholar]
- Lee HS. Diversity of halophilic archaea in fermented foods and human intestines and their application. J Microbiol Biotechnol. 2013;23(12):1645–1653. doi: 10.4014/jmb.1308.08015. [DOI] [PubMed] [Google Scholar]
- Legat A, Gruber C, Zangger K, Wanner G, Stan-Lotter H. Identification of polyhydroxyalkanoates in Halococcus and other haloarchaeal species. Appl Microbiol Biotechnol. 2010;87(3):1119–1127. doi: 10.1007/s00253-010-2611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35(4):577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- Liliensiek AK, Cassidy J, Gucciardo G, Whitely C, Paradisi F. Heterologous overexpression, purification and characterisation of an alcohol dehydrogenase (ADH2) from Halobacterium sp. NRC-1. Mol Biotechnol. 2013;55(2):143–149. doi: 10.1007/s12033-013-9666-4. [DOI] [PubMed] [Google Scholar]
- Lobasso S, Vitale R, Lopalco P, Corcelli A. Haloferax volcanii, as a novel tool for producing mammalian olfactory receptors embedded in archaeal lipid bilayer. Life (Basel) 2015;5(1):770–782. doi: 10.3390/life5010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, St Jean A, Amils R, Charlebois RL. Genomic stability in the archaeae Haloferax volcanii and Haloferax mediterranei. J Bacteriol. 1995;177(5):1405–1408. doi: 10.1128/jb.177.5.1405-1408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorantfy B, Seyer B, Herwig C. Stoichiometric and kinetic analysis of extreme halophilic Archaea on various substrates in a corrosion resistant bioreactor. New Biotechnol. 2014;31(1):80–89. doi: 10.1016/j.nbt.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Loukas A, Kappas I, Abatzopoulos TJ. HaloDom: a new database of halophiles across all life domains. J Biol Res (Thessalon) 2018;25:2–2. doi: 10.1186/s40709-017-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L-K, Stachler A-E, Brendel J, Stoll B, Fischer S, Haas KA, Schwarz TS, Alkhnbashi OS, Sharma K, Urlaub H, Backofen R, Gophna U, Marchfelder A. The nuts and bolts of the Haloferax CRISPR-Cas system I-B. RNA Biol. 2019;16(4):469–480. doi: 10.1080/15476286.2018.1460994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5(2):73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- Melvik JE, Dornish M. Alginate as a carrier for cell immobilisation. In: Nedović V, Willaert R, editors. Fundamentals of cell immobilisation biotechnology. Dordrecht: Springer Netherlands; 2004. pp. 33–51. [Google Scholar]
- Mevarech M, Werczberger R. Genetic transfer in Halobacterium volcanii. J Bacteriol. 1985;162(1):461–462. doi: 10.1128/JB.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevarech M, Frolow F, Gloss LM. Halophilic enzymes: proteins with a grain of salt. Biophys Chem. 2000;86(2–3):155–164. doi: 10.1016/s0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- Mezzomo N, Maestri B, dos Santos RL, Maraschin M, Ferreira SRS. Pink shrimp (P. brasiliensis and P. paulensis) residue: influence of extraction method on carotenoid concentration. Talanta. 2011;85(3):1383–1391. doi: 10.1016/j.talanta.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Mohammadipanah F, Hamedi J, Dehhaghi M. Halophilic bacteria: potentials and applications in biotechnology. In: Maheshwari DK, Saraf M, editors. Halophiles: biodiversity and sustainable exploitation. Cham: Springer International Publishing; 2015. pp. 277–321. [Google Scholar]
- Moussavi G, Barikbin B, Mahmoudi M. The removal of high concentrations of phenol from saline wastewater using aerobic granular SBR. Chem Eng J. 2010;158(3):498–504. doi: 10.1016/j.cej.2010.01.038. [DOI] [Google Scholar]
- Mullakhanbhai MF, Larsen H. Halobacterium volcanii spec. Nov., a Dead Sea halobacterium with a moderate salt requirement. Arch Microbiol. 1975;104(1):207–214. doi: 10.1007/bf00447326. [DOI] [PubMed] [Google Scholar]
- Obradovic B, Nedović VA, Bugarski B, Willaert RG, Vunjak-Novakovic G. Immobilised cell bioreactors. In: Nedović V, Willaert R, editors. Fundamentals of cell immobilisation biotechnology. Dordrecht: Springer Netherlands; 2004. pp. 411–436. [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63(2):334–348. doi: 10.1128/MMBR.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems. 2008;4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Industrial and environmental applications of halophilic microorganisms. Environ Technol. 2010;31(8–9):825–834. doi: 10.1080/09593330903370026. [DOI] [PubMed] [Google Scholar]
- Oren A (2013) Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front Microbiol 4(315). 10.3389/fmicb.2013.00315 [DOI] [PMC free article] [PubMed]
- Oren A. Halophilic microbial communities and their environments. Curr Opin Biotechnol. 2015;33:119–124. doi: 10.1016/j.copbio.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Oren A, Arahal DR, Ventosa A. Emended descriptions of genera of the family Halobacteriaceae. Int J Syst Evol Microbiol. 2009;59(3):637–642. doi: 10.1099/ijs.0.008904-0. [DOI] [PubMed] [Google Scholar]
- Patel S, Saraf M. Perspectives and application of halophilic enzymes. In: Maheshwari DK, Saraf M, editors. Halophiles: biodiversity and sustainable exploitation. Cham: Springer International Publishing; 2015. pp. 403–419. [Google Scholar]
- Peck RF, Johnson EA, Krebs MP. Identification of a lycopene beta-cyclase required for bacteriorhodopsin biogenesis in the archaeon Halobacterium salinarum. J Bacteriol. 2002;184(11):2889–2897. doi: 10.1128/jb.184.11.2889-2897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F, Gregor D, Hofacker A, Plosser P, Zimmermann P. Regulation of gas vesicle formation in halophilic archaea. J Mol Microbiol Biotechnol. 2002;4(3):175–181. [PubMed] [Google Scholar]
- Poli A, Di Donato P, Abbamondi GR, Nicolaus B. Synthesis, production, and biotechnological applications of exopolysaccharides and polyhydroxyalkanoates by archaea. Archaea. 2011;2011:1–13. doi: 10.1155/2011/693253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghukumar C, D'Souza-Ticlo D, Verma AK. Treatment of colored effluents with lignin-degrading enzymes: an emerging role of marine-derived fungi. Crit Rev Microbiol. 2008;34(3–4):189–206. doi: 10.1080/10408410802526044. [DOI] [PubMed] [Google Scholar]
- Rawson A, Tiwari BK, Tuohy MG, O’Donnell CP, Brunton N. Effect of ultrasound and blanching pretreatments on polyacetylene and carotenoid content of hot air and freeze dried carrot discs. Ultrason Sonochem. 2011;18(5):1172–1179. doi: 10.1016/j.ultsonch.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Rehm BH. Polyester synthases: natural catalysts for plastics. Biochem J. 2003;376(Pt 1):15–33. doi: 10.1042/bj20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BH. Biogenesis of microbial polyhydroxyalkanoate granules: a platform technology for the production of tailor-made bioparticles. Curr Issues Mol Biol. 2007;9(1):41–62. [PubMed] [Google Scholar]
- Reuter C, Maupin-Furlow J. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl Environ Microbiol. 2005;70:7530–7538. doi: 10.1128/AEM.70.12.7530-7538.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Pyzyna B, Atrasz RG, Henderson CA, Morrill KL, Burd AM, DeSoucy E, Fogleman RE, Naylor JB, Steele SM, Elliott DR, Leyva KJ, Shand RF. Growth kinetics of extremely halophilic archaea (family halobacteriaceae) as revealed by arrhenius plots. J Bacteriol. 2005;187(3):923–929. doi: 10.1128/JB.187.3.923-929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Baños M, Garbayo I, Vílchez C, Bonete MJ, Martínez-Espinosa RM. Carotenoids from haloarchaea and their potential in biotechnology. Mar Drugs. 2015;13(9):5508–5532. doi: 10.3390/md13095508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Oliveira T, Belmok A, Vasconcellos D, Schuster B, Kyaw CM (2017) Archaeal S-layers: overview and current state of the art. Front Microbiol 8(2597). 10.3389/fmicb.2017.02597 [DOI] [PMC free article] [PubMed]
- Rønnekleiv M. Bacterial carotenoids 53∗ C50-carotenoids 23; carotenoids of Haloferax volcanii versus other halophilic bacteria. Biochem Syst Ecol. 1995;23(6):627–634. doi: 10.1016/0305-1978(95)00047-X. [DOI] [Google Scholar]
- Sakurai T, Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem Rec. 2007;7(4):220–229. doi: 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- Salgaonkar BB, Bragança JM. Utilization of sugarcane bagasse by Halogeometricum borinquense strain E3 for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Bioengineering (Basel) 2017;4(2):50. doi: 10.3390/bioengineering4020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C, Rose MR, Doolittle WF. High-frequency genomic rearrangements involving archaebacterial repeat sequence elements. Nature. 1982;299(5879):182–185. doi: 10.1038/299182a0. [DOI] [PubMed] [Google Scholar]
- Sarmiento F, Peralta R, Blamey JM. Cold and hot extremozymes: industrial relevance and current trends. Front Bioeng Biotechnol. 2015;3:148–148. doi: 10.3389/fbioe.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409(6817):258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Shand RF, Leyva KJ. Peptide and protein antibiotics from the domain archaea: halocins and sulfolobicins. In: Riley MA, Chavan MA, editors. Bacteriocins: ecology and evolution. Berlin Heidelberg, Berlin, Heidelberg: Springer; 2007. pp. 93–109. [Google Scholar]
- Sharma N., Farooqi M. S., Chaturvedi K. K., Lal S. B., Grover M., Rai A., Pandey P. The Halophile Protein Database. Database. 2014;2014(0):bau114–bau114. doi: 10.1093/database/bau114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J Biosci Bioeng. 2005;99(4):303–310. doi: 10.1263/jbb.99.303. [DOI] [PubMed] [Google Scholar]
- Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol Lett. 2007;267(2):131–144. doi: 10.1111/j.1574-6968.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- Smith EaP, Ron (2018) Haloferax volcanii for carotenoid production. vol Honours Theses, Paper 899
- Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96(7):2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- Strand BL, Skjåk-Bræk G, Gåserød O. Microcapsule formulation and formation. In: Nedović V, Willaert R, editors. Fundamentals of cell immobilisation biotechnology. Dordrecht: Springer Netherlands; 2004. pp. 165–183. [Google Scholar]
- Strillinger E, Grotzinger SW, Allers T, Eppinger J, Weuster-Botz D. Production of halophilic proteins using Haloferax volcanii H1895 in a stirred-tank bioreactor. Appl Microbiol Biotechnol. 2016;100(3):1183–1195. doi: 10.1007/s00253-015-7007-1. [DOI] [PubMed] [Google Scholar]
- Stroud A, Liddell S, Allers T (2012) Genetic and biochemical identification of a novel single-stranded dna-binding complex in Haloferax volcanii. Front Microbiol 3(224). 10.3389/fmicb.2012.00224 [DOI] [PMC free article] [PubMed]
- Tamborini L, Fernandes P, Paradisi F, Molinari F. Flow bioreactors as complementary tools for biocatalytic process intensification. Trends Biotechnol. 2018;36(1):73–88. doi: 10.1016/j.tibtech.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Tamir A, Eichler J. N-glycosylation is important for proper Haloferax volcanii S-layer stability and function. Appl Environ Microbiol. 2017;83(6):e03152–e03116. doi: 10.1128/AEM.03152-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson LM, Alsafadi D, Mac Donnchadha C, Liddell S, Sharkey MA, Paradisi F. Characterization of alcohol dehydrogenase (ADH12) from Haloarcula marismortui, an extreme halophile from the Dead Sea. Extremophiles. 2012;16(1):57–66. doi: 10.1007/s00792-011-0405-0. [DOI] [PubMed] [Google Scholar]
- Timpson LM, Liliensiek AK, Alsafadi D, Cassidy J, Sharkey MA, Liddell S, Allers T, Paradisi F. A comparison of two novel alcohol dehydrogenase enzymes (ADH1 and ADH2) from the extreme halophile Haloferax volcanii. Appl Microbiol Biotechnol. 2013;97(1):195–203. doi: 10.1007/s00253-012-4074-4. [DOI] [PubMed] [Google Scholar]
- Torregrosa-Crespo J, Pire C, Martínez-Espinosa R (2017) Biocompounds from Haloarchaea and their uses in biotechnology. In: Sghaier, Haitham; Najjari, Afef; Ghedira, Kais (Eds.). Archaea - New Biocatalysts, Novel Pharmaceuticals and Various Biotechnological Applications. InTech, 2017, pp 63–82
- Tripepi M, You J, Temel S, Önder Ö, Brisson D, Pohlschröder M. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J Bacteriol. 2012;194(18):4876–4887. doi: 10.1128/JB.00731-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripepi M, Esquivel RN, Wirth R, Pohlschroder M. Haloferax volcanii cells lacking the flagellin FlgA2 are hypermotile. Microbiology. 2013;159(Pt 11):2249–2258. doi: 10.1099/mic.0.069617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthandi S, Saad B, Humbard M, Maupin-Furlow J. LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl Environ Microbiol. 2009;76:733–743. doi: 10.1128/AEM.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DB, Banks DD, Lohman JR, Hilsenbeck JL, Gloss LM. The effect of salts on the activity and stability of Escherichia coli and Haloferax volcanii dihydrofolate reductases. J Mol Biol. 2002;323(2):327–344. doi: 10.1016/s0022-2836(02)00916-6. [DOI] [PubMed] [Google Scholar]
- Xue Y-P, Xu M, Chen H-S, Liu Z-Q, Wang Y-J, Zheng Y-G. A novel integrated bioprocess for efficient production of (R)-(−)-Mandelic acid with immobilized Alcaligenes faecalis ZJUTB10. Org Process Res Dev. 2013;17(2):213–220. doi: 10.1021/op3001993. [DOI] [Google Scholar]
- Yin J, Chen JC, Wu Q, Chen GQ. Halophiles, coming stars for industrial biotechnology. Biotechnol Adv. 2015;33(7):1433–1442. doi: 10.1016/j.biotechadv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Zalazar L, Pagola P, Miro MV, Churio MS, Cerletti M, Martinez C, Iniesta-Cuerda M, Soler AJ, Cesari A, De Castro R. Bacterioruberin extracts from a genetically modified hyperpigmented Haloferax volcanii strain: antioxidant activity and bioactive properties on sperm cells. J Appl Microbiol. 2019;126(3):796–810. doi: 10.1111/jam.14160. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sun Z, Sun P, Chen T, Chen F. Microalgal carotenoids: beneficial effects and potential in human health. Food Funct. 2014;5(3):413–425. doi: 10.1039/c3fo60607d. [DOI] [PubMed] [Google Scholar]
- Zhang J-F, Liu Z-Q, Zhang X-H, Zheng Y-G. Biotransformation of iminodiacetonitrile to iminodiacetic acid by Alcaligenes faecalis cells immobilized in ACA-membrane liquid-core capsules. Chem Pap. 2014;68(1):53–64. doi: 10.2478/s11696-013-0423-8. [DOI] [Google Scholar]