Abstract
Purpose
Alterations to mismatch repair (MMR) pathways are a known cause of cancer, particularly colorectal and endometrial carcinomas. Recently, checkpoint inhibitors have been approved for use in MMR-deficient cancers of any type (Prasad et al. in JAMA Oncol 4:157–158, 2018). Functional studies in breast cancer have shown associations between MMR loss, resistance to aromatase inhibitors and sensitivity to palbociclib (Haricharan et al. in Cancer Discov 7:1168–1183, 2017). Herein, we investigate the clinical meaning of MMR deficiency in breast cancer by immunohistochemical assessment of MSH2, MSH6, MLH1 and PMS2 on a large series of breast cancers linked to detailed biomarker and long-term outcome data.
Methods
Cases were classified as MMR intact when all four markers expressed nuclear reactivity, but MMR-deficient when at least one of the four biomarkers displayed loss of nuclear staining in the presence of positive internal stromal controls on the tissue microarray core.
Results
Among the 1635 cases with interpretable staining, we identified 31 (1.9%) as MMR-deficient. In our cohort, MMR deficiency was present across all major breast cancer subtypes, and was associated with high-grade, low-progesterone receptor expression and high tumor-infiltrating lymphocyte counts. MMR deficiency is significantly associated with inferior overall (HR 2.29, 95% CI 1.02–5.17, p = 0.040) and disease-specific survival (HR 2.71, 95% CI 1.00–7.35, p = 0.042) in the 431 estrogen receptor-positive patients who were uniformly treated with tamoxifen as their sole adjuvant systemic therapy.
Conclusion
Overall, this study supports the concept that breast cancer patients with MMR deficiency as assessed by immunohistochemistry may be good candidates for alternative treatment approaches such as immune checkpoint or CDK4 inhibitors.
Electronic supplementary material
The online version of this article (10.1007/s10549-019-05438-y) contains supplementary material, which is available to authorized users.
Keywords: Mismatch repair protein, Breast cancer, Tissue microarray, Tumor-infiltrating lymphocytes
Introduction
Mismatch repair is a highly conserved mechanism that maintains replication fidelity and mediates DNA damage signaling [1–4]. Key players in this pathway include EXO1, DNA-binding protein RPA, DNA polymerase, and four major proteins that form heterodimeric complexes: MutS—mutS homolog 2 (MSH2) and mutS homolog 6 (MSH6), MutL—mutL homolog 1 (MLH1) and postmeiotic segregation increased 2 (PMS2) [3, 5]. MutS recognizes and attaches to abnormal DNA whereas MutL enhances recognition and facilitates the formation of a repair complex [5, 6]. The MMR pathway not only corrects base pair mismatches and insertion or deletion loops commonly found in microsatellite regions, it is also involved in cell cycle checkpoints and apoptosis [2–4]. Consequently, deficiencies in MMR pathways promote oncogenesis.
The clinical management of MMR is well-established in colorectal and endometrial cancers. Currently, universal testing in colorectal cancer is recommended by the National Comprehensive Cancer Network and the Evaluation of Genomic Applications in Practice and Prevention Working Group [7] as neither Amsterdam criteria nor Bethesda guidelines [8] can entirely identify all mutation carriers [9]. Likewise, supporting evidence is growing for systematic screening of MMR in endometrial cancers, reflecting the similar rates of Lynch Syndrome in patients presenting with endometrial carcinoma and colorectal carcinoma [11–12].
New treatment strategies have recently become available to MMR-deficient breast cancer patients [13]. In 2017, the United States Food and Drug Administration approved the checkpoint inhibitor pembrolizumab for use in advanced MMR-deficient solid tumors of any tissue type [1]. Furthermore, MMR-deficient breast cancers (specifically those with loss of MutL [14]) have been shown to be resistant to aromatase inhibitors but sensitive to palbociclib (a CDK4/6 inhibitor) [14–16].
The MMR DNA damage repair pathway is likely to hold significant clinical relevance since high mutational load tumors correlate with poor survival [17] and endocrine therapy resistance in ER+ breast cancer patients [18]. However, genomic studies have suggested MMR loss is rare in breast cancer (1–2%) [19], making it difficult to assemble sufficient numbers of cases to power meaningful associative or survival studies. To understand the clinical meaning of MMR deficiency in breast cancer, we assessed a large breast cancer tissue microarray series linked to detailed biomarker and long-term outcome data for immunohistochemically determined loss of MSH2, MSH6, MLH1 or PMS2.
Methods
Study cohort
This large tissue microarray series linked to clinical outcomes was built from formalin-fixed paraffin-embedded previously frozen tissues using 0.6 mm cores. Material collection was approved by the Clinical Research Ethics Board of the University of British Columbia (H17-00509), and the characteristics of this cohort have been published [20–22]. Briefly, this cohort comprises 3992 female patients from the province of British Columbia referred to the British Columbia Cancer Agency and diagnosed with primary invasive breast cancer from 1986 to 1992. Patients were treated according to the provincial guidelines in place during the study era [23], and the median follow-up time was 12.5 years. Data for comparative biomarkers on this series have been published: ER [23, 24], PR [23, 25], HER2 [23, 26], CK5/6 [23], EGFR [23], Ki67 [27], PD-1 and PD-L1 [28] (Refer to Supplemental Table A for antibody clones and scoring criteria) and tumor-infiltrating lymphocytes (as assessed on hematoxylin and eosin slides using standardized methods[20].
Immunohistochemistry
Array sections at 4 μm were mounted on charged glass slides and baked for an hour at 60 °C to prepare for staining on the Ventana Discovery automated stainer (Ventana Medical Systems, Tuscon, AZ). Protocols were adapted from Nordic immunohistochemical Quality Control (NordiQC) [29]. Slides were processed according to manufacturer’s protocol with proprietary reagents. Cell Conditioning 1, heat-induced antigen retrieval, and the discovery anti-HQ HRP detection kit from Ventana were used on all slides; only the dilution and incubation time varied with each biomarker.
Slides were incubated with MSH2 (mouse monoclonal G219-1129; Cell Marque: CMQ-286M14, 1:200 dilution), MSH6 (rabbit monoclonal EP49; Epitomics: AC-0047, 1:50 dilution), MLH1 (mouse monoclonal ES05; Leica Biosystems: NCL-L-MLH1, 1:50 dilution), or PMS2 (rabbit monoclonal EP51; Epitomics: AC-0049, 1:20 dilution). Tonsil tissues, as recommended by NordiQC, were included in each run as an external positive control.
Prior to application to the study cohort, the immunohistochemistry (IHC) protocols optimized for the four MMR biomarkers—MSH2, MSH6, MLH1 and PMS2—were run on independent formalin-fixed paraffin-embedded breast cancer tissue microarrays from a smaller training series to confirm staining interpretability in epithelium and stroma. Furthermore, staining patterns from our optimized protocols were comparable to staining patterns of clinically validated protocols from the Vancouver General Hospital on a colorectal cancer resection control, which was IHC confirmed with MLH1 and PMS2 deletions.
Scoring
Stained slides were scanned with the Olympus BLISS system. MMR protein expression was scored by a pathologist blinded to the associated outcome data. According to published guidelines, only cores with absent nuclear staining in all carcinoma cells in the concurrent presence of positive stromal cell internal controls on the same tissue microarray core were categorized as deficient [30, 31]. Cores with nuclear staining in carcinoma cells and stromal controls were categorized as intact. Biomarkers were reported following REMARK guidelines [32].
Data analysis
Statistical analysis was performed with IBM SPSS software (version 25.0). Each biomarker was first dichotomized as intact or deficient. Then, MMR status was assessed. A case was either “MMR intact” when all four biomarkers expressed nuclear positivity, or “MMR-deficient” when nuclear positivity was absent from any of the four biomarkers tested, with respective internal stromal controls. Only cases with interpretable results from all four biomarkers were included in the correlative and survival analyses for MMR deficiency (Refer to Supplemental Figure A for case distribution of the entire cohort and Supplemental Table B for distribution of uninterpretable staining).
Correlations of MMR status with key breast cancer biomarkers were assessed by Fisher’s exact test: ER, PR and HER2 as standard subtype biomarkers; CK5/6 and EGFR for basal-like subtype; Ki67 to examine proliferation for Luminal A vs Luminal B subtype; and immune markers PD-1 and PD-L1. Survival analyses were performed by Kaplan–Meier plot with log-rank test.
Results
MMR deficiency is rare in breast cancer
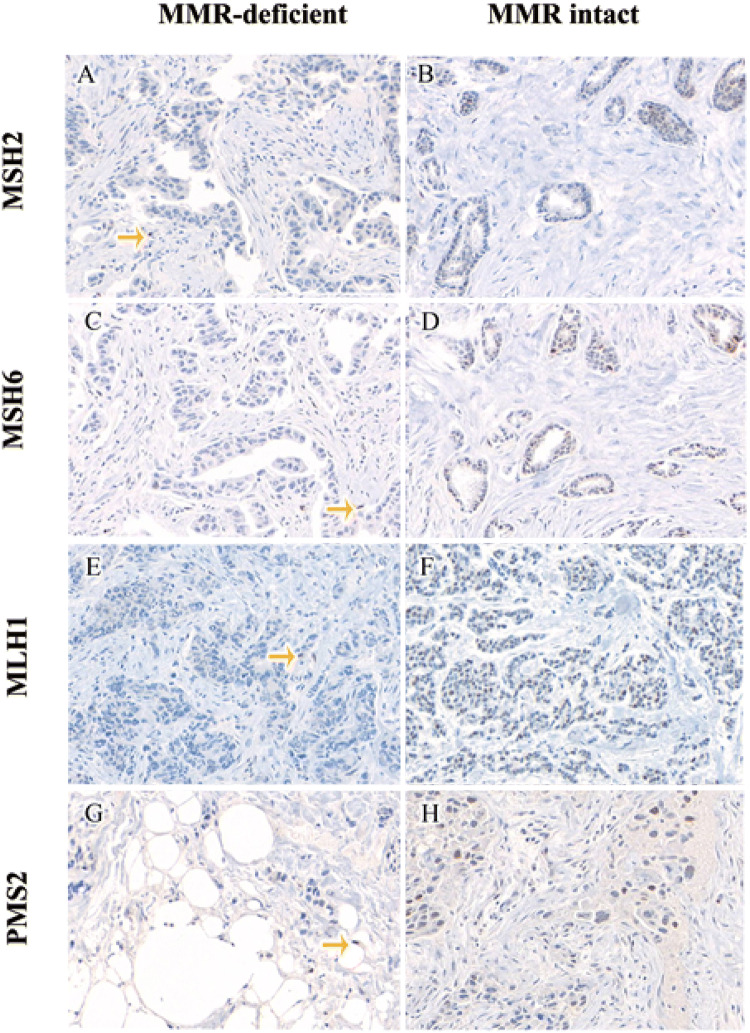
Interpretable staining for each individual MMR biomarker (MSH2, MSH6, MLH1 and PMS2) is summarized in Table 1. Of the 1635 cases interpretable for all four MMR biomarkers, we identified 31 cases as MMR-deficient (1.9%). Twenty-five cases had loss of a single MMR biomarker and six cases had paired losses. Four cases had paired MLH1 and PMS2 losses, and two cases had MSH2 and MSH6 losses. Eleven cases had PMS2 loss only, ten cases had MLH1 loss only, three cases had MSH6 loss only and one case had MSH2 loss only (Fig. 1). Figure 2 illustrates examples of MMR-deficient and MMR intact cases.
Table 1.
Summary of interpretable staining in each biomarker
| Interpretable staining | MMR intact | MMR loss | |
|---|---|---|---|
| MSH2 | 2399 (60.1%) | 2363 (98.5%) | 36 (1.5%) |
| MSH6 | 2488 (62.3%) | 2440 (98.1%) | 48 (1.9%) |
| MLH1 | 1930 (48.3%) | 1891 (98.0%) | 39 (2.0%) |
| PMS2 | 2159 (54.1%) | 2115 (98.0%) | 44 (2.0%) |
Uninterpretable staining includes lack of viable cancer cells, core dropouts from staining or sectioning, insufficient tumour cells and technical fails (apparent loss but without internal positive controls). Refer to Supplemental Table B for detailed distribution
Fig. 1.
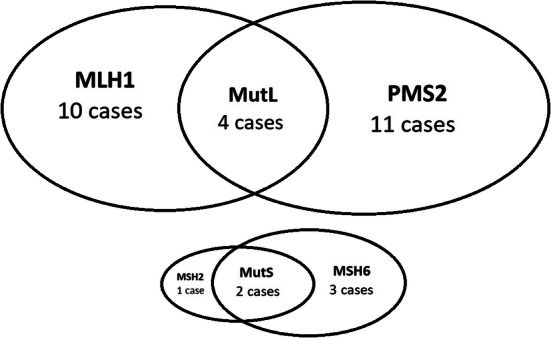
Distribution of MMR-deficient cases
Fig. 2.
Staining patterns of MMR biomarker: MSH2 loss (a), MSH2 intact (b), MSH6 loss (c), MSH6 intact (d), MLH1 loss (e), MLH1 intact (f), PMS2 loss (g), PMS2 intact (h). Stromal cell internal positive control in MMR-deficient cases are indicated by gold arrows
MMR-deficient breast cancers are likely high grade with low-progesterone receptor expression and high-TIL counts
Among the clinical parameters examined (age, tumor grade, tumor size, lymphovascular invasion, nodal and menstrual status), MMR deficiency was significantly associated with Grade 3 histology (Table 2). We also evaluated associations between MMR-deficient and other important biomarkers previously assessed on this British Columbia cohort (Table 3). Among additional biomarkers with data available for evaluation, MMR deficiency correlated with low-progesterone receptor (PR) expression: 75% (21 cases) of MMR-deficient cases were negative for PR compared to less than half of MMR intact cases. Additionally, cases displaying MMR deficiency had significantly higher TIL counts (median of 5, interquartile range 1–10) compared to MMR intact cases (median of 1, p = 0.009 by Mann–Whitney test).
Table 2.
Association of MMR status with demographic and pathological features
| Parameters | MMR loss n = 31 |
MMR intact n = 1604 |
p value |
|---|---|---|---|
| Age at diagnosis (years) | |||
| < 50 | 11 | 490 | N.S. |
| ≥ 50 | 20 | 1114 | |
| Tumour grade | |||
| 1 and 2 | 6 | 636 | 0.015 |
| 3 | 25 | 905 | |
| Unknown | 63 | ||
| Tumour size (cm) | |||
| ≤ 2 | 17 | 782 | N.S. |
| > 2 | 14 | 812 | |
| Unknown | 10 | ||
| Lymphovascular invasion | |||
| Negative | 12 | 812 | N.S. |
| Positive | 17 | 727 | |
| Unknown | 2 | 65 | |
| Nodal status | |||
| Negative | 12 | 870 | N.S. |
| Positive | 19 | 730 | |
| Unknown | 4 | ||
| Menstrual status | |||
| Premenopausal | 12 | 504 | N.S. |
| Postmenopausal | 19 | 1067 | |
| Unknown | 33 | ||
N.S. not significant (p value > 0.05)
Table 3.
Association of MMR status with biomarkers
| Parameters | MMR loss n = 31 |
MMR intact n = 1604 |
p value |
|---|---|---|---|
| ER | N.S. | ||
| Negative | 9 | 478 | |
| Positive | 22 | 1125 | |
| Unknown | 1 | ||
| PR | 0.004 | ||
| Negative (< 1%) | 21 | 718 | |
| Positive (≥ 1%) | 7 | 825 | |
| Unknown | 3 | 61 | |
| Her2 (erbb2) | N.S. | ||
| Negative | 25 | 1325 | |
| Positive | 5 | 248 | |
| Unknown | 1 | 31 | |
| Krt5/6 (CK5/6) | N.S. | ||
| Negative | 22 | 1308 | |
| Positive | 4 | 160 | |
| Unknown | 5 | 136 | |
| EGFR | N.S. | ||
| Negative | 26 | 1233 | |
| Positive | 2 | 250 | |
| Unknown | 3 | 121 | |
| Ki67 | N.S. | ||
| Negative (< 14%) | 12 | 754 | |
| Positive (≥ 14%) | 15 | 764 | |
| Unknown | 4 | 86 | |
| Subtypes | |||
| Luminal A | 10 | 616 | N.S. |
| Luminal B | 12 | 494 | |
| HER2E | 2 | 134 | |
| Triple negative/basal | 4 | 275 | |
| Indeterminate | 3 | 85 | |
| PD-1 | |||
| Intra-epithelial TIL | N.S. | ||
| 0 | 28 | 1357 | |
| ≥ 1% | 2 | 184 | |
| Unknown | 1 | 63 | |
| PD-L1 | |||
| Negative (< 1%) | 20 | 1356 | 0.059 |
| Positive (≥ 1%) | 6 | 165 | |
| Unknown | 5 | 83 | |
| H&E sTILs count (%) | |||
| Median | 5 | 1 | 0.009a |
| Interquartile range | 1–10 | 1–5 | |
| Unknown | 3 | 97 | |
N.S. not significant (p value > 0.05)
aMann–Whitney test
MMR deficiency is present at similar frequencies across all major subtypes
As basal-like breast cancers are known to have a higher mutational burden than other intrinsic subtypes, and mismatch repair deficiency has been shown to be associated with genomic instability, we analyzed the distribution of MMR-deficient cases by subtype. Results demonstrated no significant differences in MMR deficiency across all the major intrinsic subtypes of breast cancer, as determined by immunohistochemistry (Table 3). Briefly, cases with ER or PR positivity (≥ 1%) and Her2 negativity and low Ki67 (< 14%) were classified as Luminal A; cases with ER+/PR+/HER2−/high Ki67 or ER+/PR+/HER2+ were classified as Luminal B; cases with HER2 positivity with ER and PR negativity ***cases were classified as HER2 enriched; ER−/PR−/HER2− were classified as triple negative or basal if CK5/6+ or EGFR+. The frequency of MMR-deficient cases ranged from 0.5% in basal breast cancers to 2.4% in Luminal B.
Univariable survival analysis
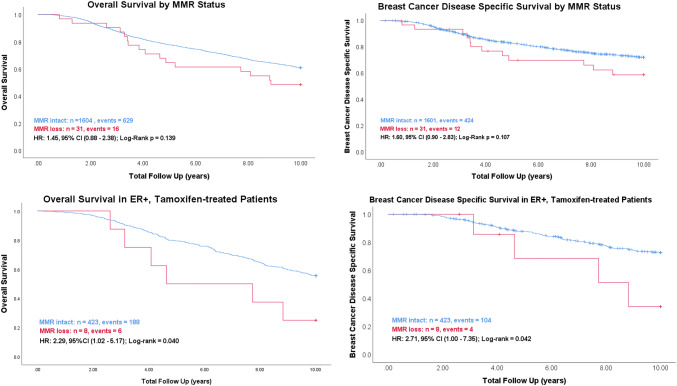
As this large breast cancer series had clinical outcome data, we analyzed the prognostic value of MMR deficiency (Fig. 3). Among the whole cohort, overall survival (HR 1.45, p = 0.139, n = 1635) and breast cancer disease-specific survival (HR 1.60, p = 0.107, n = 1632) displayed a non-significant decreasing trend with MMR deficiency. The separation of curves was similar for relapse-free survival (HR 1.30, p = 0.355, n = 1635, Supplemental Figure B). In survival analyses stratified by treatment (Fig. 3), MMR deficiency was associated with significantly shorter survival within the cohort of ER-positive patients treated with tamoxifen as their sole adjuvant therapy (OS: HR 2.29, p = 0.040, n = 431; DSS: HR 2.71, p = 0.042, n = 431). Due to the low number of ER-negative cases with MMR deficiency, survival analyses in this population were not conducted.
Fig. 3.
Overall survival (a) and breast cancer disease-specific survival (b) in the whole cohort. Overall survival (c) and breast cancer disease-specific survival (d) in ER-positive, tamoxifen-treated patients
Discussion
We present the largest series to date assessing mismatch repair protein deficiency in breast cancer, as determined by immunohistochemistry and linked to survival outcomes. Using our assay, we identified 31 MMR-deficient cases out of 1635 that had data for all four MMR biomarkers (MSH2, MSH6, MLH1 and PMS2). Despite its relative rarity in breast cancer, this population is important as MMR-deficient cancers have been highly responsive to immune therapies such as PD-1 or CDK4/6 checkpoint inhibitors [1, 13, 33–35].
The 1.9% frequency of MMR deficiency in our study corroborates well with the genomic findings reported from the Sanger Centre, who on the basis of mutational signatures derived from whole genome sequencing of 640 cases, identified 11 cases (1.7%) as MMR-deficient [19]. In addition, another study that sequenced MMR genes in 12,019 cancers comprising 32 cancer types reported less than 2% frequency of MMR deficiency in breast cancer [36]. Furthermore, a recent study in 94 HER2-positive luminal B breast cancer patients showed that, although 13.5% of cases had a germline mutation (V384D) in the MLH1 gene, only 3 cases (3.2%) were MLH1-deficient by IHC [37]. In contrast, a recently published cohort from Italy reported a ten-times higher frequency (17%, 75 out of 444 cases) of homogenous MMR loss by immunohistochemistry [30]; although when further investigated by microsatellite instability assay, all but seven of these were negative (meaning only 1.6% of cases overall were MSI positive). The discrepancy in their reported frequency of MMR-deficient cases in breast cancer by IHC could be due to the inclusion of cases which lack an internal positive control. As this requirement was not mentioned in the reported methods, assessments may be vulnerable to technical false-negative MMR staining, especially when working with clinical cases acquired outside of strict research protocols where variabilities in pre-analytical specimen handling are unavoidable [38]. This discrepancy reiterates the need for IHC assays, which are much less expensive and more widely available than MSI or mutational signature assays, to be standardized and validated with appropriate quality control programs if they are to be put into clinical use [38, 39].
Our results suggest that MMR-deficient cases are associated with poor prognostic factors such as high-grade and high-TIL counts. MMR deficiency is also present across all major breast cancer subtypes by immunohistochemistry in our cohort (1.6% in Luminal A, 2.4% in Luminal B, 1.5% in HER2 enriched, 0.5% in basal), illustrating that MMR deficiency testing is potentially relevant in all major breast cancer subtypes. Poor survival in the ER-positive, tamoxifen-treated cohort suggests MMR status can potentially identify a subpopulation of ER-positive patients that may benefit from treatments beyond endocrine therapy alone.
Our study does have several limitations. Although we examined a large TMA set with extensive published data, each case is represented by a single 0.6 mm core. To avoid overestimating MMR deficiency rates, we only included cases where all four tested MMR proteins had interpretable data not only for carcinoma cells but also for positive stromal cell controls. As cases with apparent MMR biomarker loss without internal positive controls ranged from 230 cases (5.8%) for MSH6 to 523 (13.1%) for MLH1 (Supplemental Table B), the frequency of MMR deficiency we report could be an underestimation, although it does agree quite closely with genomic findings as described above [19, 36]. The biomarker patterns in MMR-deficient cases were not always coherent with expected mismatch repair biology (i.e. some cases showed MLH1 loss with intact PMS2, or MSH2 loss with intact MSH6) [38, 40], a result that may arise from false negatives from tumor heterogeneity that is not adequately assessed using tissue microarrays [41]. Unfortunately, in our series, sequencing data were not available for the great majority of the 1635 cases with MMR IHC results. Although eight positive cases had panel sequencing data available for 83 genes [42], this data was not sufficient to confidently infer MMR genotypic status.
Another limitation of our study is that the determination of MMR loss by IHC is based on its strong correlation with the functionality of MMR rather than direct assessment of DNA mutational patterns. We are aware of the potential misrepresentation, but have found many sources supporting the robustness of IHC compared to genomic methods [19, 30]. Some reports suggest there is a tradeoff of higher specificity achieved through curated genomic methods versus a higher sensitivity using protein expression detected through IHC methods[19]. We opted for IHC because MMR deficiency in breast cancer patients is very rarely hereditary [37, 43]. Additionally, IHC is well-established in colorectal cancer and endometrial cancer, inexpensive and readily available [40, 44].
Our initial cohort comprises 3992 patients. After accounting for inevitable tissue loss from IHC handling, insufficient tumor sampling due to core depletions from sectioning, and exclusion of data from cores without concurrent positive internal stromal controls, we were left with 1635 cases with all four interpretable MMR biomarkers. Since the nature of our TMA resources is one core per case, we lost data on cases that failed to meet the strict MMR-deficient criteria. In the future, one may include additional replicate cores per case to increase the probability of having interpretable stromal controls. Nonetheless, to the best of our knowledge, 1635 cases remains the largest cohort of breast cancer patients examined for mismatch repair protein expression linked to long-term survival data.
Overall, our study reports a low frequency of MMR loss in breast cancer, as determined by IHC, which is present across all major subtypes. Frequencies agree with genomic data, but the IHC approach we used facilitated the examination of large numbers of cases with long-term follow-up, increasing the power to assess the clinical relevance of the relatively rare state of MMR deficiency in breast cancer. There appears to be an association of MMR loss with grade 3, low PR and high-TIL count tumors, as well as with worse survival among ER-positive patients treated with tamoxifen as their sole adjuvant systemic therapy, supporting the concept that patients with such tumors may be good candidates for alternative treatment approaches such as checkpoint or CDK4 inhibitors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
This study was supported by the Canadian Cancer Society (Grant #705463) to TON.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
Torsten O. Nielsen and Matthew J. Ellis have a proprietary interest (Bioclassifer LLC, Nanostring Technologies) in the PAM50 subtype classifier, not used in this study. The other authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in this study was approved by the Clinical Research Ethics Board of the University of British Columbia (H17-00509).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/20/2020
In the original publication of the article, the funding statement was published incompletely. The corrected funding statement should read as below:
References
- 1.Prasad V, Kaestner V, Mailankody S. Cancer drugs approved based on biomarkers and not tumor type-FDA approval of pembrolizumab for mismatch repair-deficient solid cancers. JAMA Oncol. 2018;4:157–158. doi: 10.1001/jamaoncol.2017.4182. [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negureanu L, Salsbury FR. The molecular origin of the MMR-dependent apoptosis pathway from dynamics analysis of MutSα-DNA complexes. J Biomol Struct Dyn. 2012;30:347–361. doi: 10.1080/07391102.2012.680034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 6.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 7.Group EoGAiPaPEW Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SA, Laurino M, Bowen DJ, et al. Initiation of universal tumor screening for Lynch syndrome in colorectal cancer patients as a model for the implementation of genetic information into clinical oncology practice. Cancer. 2016;122:393–401. doi: 10.1002/cncr.29758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Kong JK, Yang W, et al. DNA mismatch repair protein immunohistochemistry and MLH1 promotor methylation testing for practical molecular classification and the prediction of prognosis in endometrial cancer. Cancers. 2018;10:279. doi: 10.3390/cancers10090279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins JC, Yang EJ, Muto MG, et al. Universal screening for mismatch-repair deficiency in endometrial cancers to identify patients with Lynch syndrome and Lynch-like syndrome. Int J Gynecol Pathol. 2017;36:115–127. doi: 10.1097/PGP.0000000000000312. [DOI] [PubMed] [Google Scholar]
- 12.Snowsill T, Coelho H, Huxley N, et al. Molecular testing for Lynch syndrome in people with colorectal cancer: systematic reviews and economic evaluation. Health Technol Assess. 2017;21:1–238. doi: 10.3310/hta21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Peng Y, Peng G. Mismatch repair-based stratification for immune checkpoint blockade therapy. Am J Cancer Res. 2018;8:1977–1988. [PMC free article] [PubMed] [Google Scholar]
- 14.Haricharan S, Punturi N, Singh P, et al. Loss of MutL disrupts CHK2-dependent cell-cycle control through CDK4/6 to promote intrinsic endocrine therapy resistance in primary breast cancer. Cancer Discov. 2017;7:1168–1183. doi: 10.1158/2159-8290.CD-16-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anurag M, Punturi N, Hoog J, et al. Comprehensive profiling of DNA repair defects in breast cancer identifies a novel class of endocrine therapy resistance drivers. Clin Cancer Res. 2018;24:4887–4899. doi: 10.1158/1078-0432.CCR-17-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anurag M, Ellis MJ, Haricharan S. DNA damage repair defects as a new class of endocrine treatment resistance driver. Oncotarget. 2018;9:36252–36253. doi: 10.18632/oncotarget.26363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haricharan S, Bainbridge MN, Scheet P, Brown PH. Somatic mutation load of estrogen receptor-positive breast tumors predicts overall survival: an analysis of genome sequence data. Breast Cancer Res Treat. 2014;146:211–220. doi: 10.1007/s10549-014-2991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies H, Morganella S, Purdie CA, et al. Whole-genome sequencing reveals breast cancers with mismatch repair deficiency. Cancer Res. 2017;77:4755–4762. doi: 10.1158/0008-5472.CAN-17-1083. [DOI] [PubMed] [Google Scholar]
- 20.Burugu S, Gao D, Leung S, et al. TIM-3 expression in breast cancer. Oncoimmunology. 2018;7:e1502128. doi: 10.1080/2162402X.2018.1502128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Lachapelle J, Leung S, et al. CD8 + lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14:R48. doi: 10.1186/bcr3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 24.Cheang MC, Treaba DO, Speers CH, et al. Immunohistochemical detection using the new rabbit monoclonal antibody SP1 of estrogen receptor in breast cancer is superior to mouse monoclonal antibody 1D5 in predicting survival. J Clin Oncol. 2006;24:5637–5644. doi: 10.1200/JCO.2005.05.4155. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Chia SK, Mehl E, et al. Progesterone receptor is a significant factor associated with clinical outcomes and effect of adjuvant tamoxifen therapy in breast cancer patients. Breast Cancer Res Treat. 2010;119:53–61. doi: 10.1007/s10549-009-0318-0. [DOI] [PubMed] [Google Scholar]
- 26.Chia S, Norris B, Speers C, et al. Human epidermal growth factor receptor 2 overexpression as a prognostic factor in a large tissue microarray series of node-negative breast cancers. J Clin Oncol. 2008;26:5697–5704. doi: 10.1200/JCO.2007.15.8659. [DOI] [PubMed] [Google Scholar]
- 27.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burugu S, Gao D, Leung S, et al. LAG-3 + tumor infiltrating lymphocytes in breast cancer: clinical correlates and association with PD-1/PD-L1 + tumors. Ann Oncol. 2017;28:2977–2984. doi: 10.1093/annonc/mdx557. [DOI] [PubMed] [Google Scholar]
- 29.Vyberg M, Nielsen S. Proficiency testing in immunohistochemistry—experiences from Nordic Immunohistochemical Quality Control (NordiQC) Virchows Arch. 2016;468(1):19–29. doi: 10.1007/s00428-015-1829-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusco N, Lopez G, Corti C, et al. Mismatch Repair Protein Loss as a Prognostic and Predictive Biomarker in Breast Cancers Regardless of Microsatellite Instability. JNCI Cancer Spectr. 2018;2:pky056. doi: 10.1093/jncics/pky056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kheirelseid EA, Miller N, Chang KH, et al. Mismatch repair protein expression in colorectal cancer. J Gastrointest Oncol. 2013;4:397–408. doi: 10.3978/j.issn.2078-6891.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fremd C, Hlevnjak M, Zapatka M, et al. Mismatch repair deficiency drives durable complete remission by targeting programmed death receptor 1 in a metastatic luminal breast cancer patient. Breast Care. 2019;14:53–59. doi: 10.1159/000492580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills AM, Dill EA, Moskaluk CA, et al. The relationship between mismatch repair deficiency and PD-L1 expression in breast carcinoma. Am J Surg Pathol. 2018;42:183–191. doi: 10.1097/PAS.0000000000000949. [DOI] [PubMed] [Google Scholar]
- 36.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SE, Lee HS, Kim KY, et al. High prevalence of the MLH1 V384D germline mutation in patients with HER2-positive luminal B breast cancer. Sci Rep. 2019;9:10966. doi: 10.1038/s41598-019-47439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McConechy MK, Talhouk A, Li-Chang HH, et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol Oncol. 2015;137:306–310. doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
- 40.Niu BT, Hammond RFL, Leen SLS, et al. Two versus four immunostains for Lynch syndrome screening in endometrial carcinoma. Histopathology. 2019;75:442–445. doi: 10.1111/his.13898. [DOI] [PubMed] [Google Scholar]
- 41.Joost P, Veurink N, Holck S, et al. Heterogenous mismatch-repair status in colorectal cancer. Diagn Pathol. 2014;9:126. doi: 10.1186/1746-1596-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffith OL, Spies NC, Anurag M, et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat Commun. 2018;9:3476. doi: 10.1038/s41467-018-05914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorscher S. Rationale for evaluating breast cancers of Lynch syndrome patients for mismatch repair gene expression. Breast Cancer Res Treat. 2019 doi: 10.1007/s10549-019-05394-7. [DOI] [PubMed] [Google Scholar]
- 44.Mojtahed A, Schrijver I, Ford JM, et al. A two-antibody mismatch repair protein immunohistochemistry screening approach for colorectal carcinomas, skin sebaceous tumors, and gynecologic tract carcinomas. Mod Pathol. 2011;24:1004–1014. doi: 10.1038/modpathol.2011.55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.