Abstract
β-1,4-N-Acetyl-Galactosaminyltransferase 1 (B4GALNT1) encodes the key enzyme B4GALNT1 to generate gangliosides GM2/GD2. GM2/GD2 gangliosides are surface glycolipids mainly found on brain neurons as well as peripheral nerves and skin melanocytes and are reported to exacerbate the malignant potential of melanomas. In order to elucidate the mechanism, we performed functional analyses of B4GALNT1-overexpressing cells. We analyzed ganglioside pattern on four melanoma and two neuroblastoma cell lines by high performance liquid chromatography (HPLC). We overexpressed B4GALNT1 in GM2/GD2-negative human melanoma cell line (SH4) and confirmed production of GM2/GD2 by HPLC. They showed higher anchorage independence growth (AIG) in colony formation assay, and exhibited augmented motility. In vitro, cell proliferation was not affected by GM2/GD2 expression. In vivo, GM2/GD2-positive SH4 clones showed significantly higher tumorigenesis in NOD/Scid/IL2Rγ-null mice, and immunostaining of mouse CD31 revealed that GM2/GD2 induced remarkable angiogenesis. No differences were seen in melanoma stem cell and Epithelial-Mesenchymal Transition markers between GM2/GD2-positive and -negative SH4 cells. We therefore concluded that B4GALNT1, and consequently GM2/GD2, enhanced tumorigenesis via induction of angiogenesis, AIG, and cell motility. RNA-Seq suggested periostin as a potential key factor for angiogenesis and AIG. These findings may lead to development of novel therapy for refractory melanoma.
Subject terms: Melanoma, Glycobiology, Cell migration, Cancer genetics, Tumour angiogenesis
Introduction
Malignant melanoma is the most common and lethal skin cancer1,2. It is a cancer with one of the biggest rise in incidence3,4, and the overall 5-year survival rate is less than 10% for patients with stage IV disease5,6. There have been major advances in the treatment of advanced melanoma including Ipilimumab, an antibody to cytotoxic T-lymphocyte-associated-antigen-4 (CTLA-4), and BRAF inhibitor7–9. However, the anti-CTLA-4 antibody shows benefit in less than 50% of patients10. While BRAF inhibitors increased survival compared to other chemotherapies, its indication is limited to about half of patients with BRAF V600 mutations, and almost all patients develop resistance to these inhibitors11. While the combination of Nivolumab (monoclonal antibody against programmed death 1, PD-1) and Ipilumumab has demonstrated an impressive 2-year overall survival rate of 63.8% in stage III-IV patients12, further improvement of therapy is still needed for the treatment of advanced melanoma patients.
β-1,4-N-Acetyl-Galactosaminyltransferase 1 (B4GALNT1) encodes B4GALNT1 (GM2/GD2 synthase), and it works as the key enzyme which transfers a N-acetylgalactosamine (GalNAc) to GM3/GD3, yielding gangliosides GM2/GD2 as part of their stepwise synthesis (Fig. 1A). Gangliosides, including GM2 or GD2, belong to the family of glycosphingolipids (GSL) and contain one or more sialic acids, N-acetyl derivatives of neuraminic acid, in their hydrophilic oligosaccharide chain.13 Gangliosides are sialic acid-containing glycosphingolipids that are most abundant in the nervous system, especially brain neurons14. They also exist in peripheral nerves and skin melanocytes15,16. These molecules are reported to have important biological functions, such as intercellular communication, cell cycling, cell growth, adhesion, differentiation, and cell motility17–19. Gangliosides are not only detected at high levels in tumors of neuroectodermal cell origin but also related to the biological and clinical behavior of many kinds of tumors20. Recently, some analysis revealed that patients with higher expression of B4GALNT1 and GM2/GD2 correlated with poorer prognosis in renal cell carcinoma (TCGA data set; Human Protein Atlas), neuroblastoma21, and melanoma22. Thus, B4GALNT1 gene is considered to be key tumor-associated antigens23–27, indicating that their expression is a meaningful marker for metastatic condition and are potential therapeutic targets for melanoma.
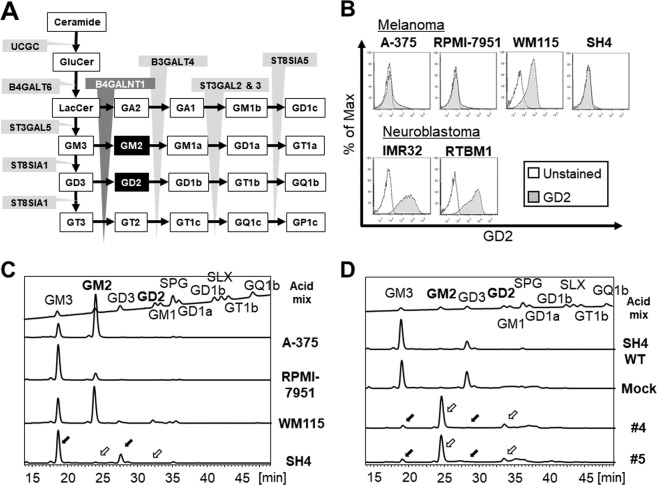
Figure 1.
Schemes of ganglioside synthesis and analyses of gangliosides in the cells. (A) Glycosylation sequences for biosynthesis of GM2/GD2. B4GALNT1 (β-1,4-N-Acetyl-Galactosaminyltransferase 1) is the critical enzyme for the GM2/GD2 synthesis. (B) Flow cytometry Analysis of GD2 on the cultured cells; four human melanoma cell lines (A375, RPMI-7951, WM-115 and SH4), and two human neuroblastoma cell lines (IMR32 and RTBM1). (C) Detail analysis of acidic gangliosides on the four melanoma cell lines. The surface expression of gangliosides was analyzed by HPLC. Y-axis indicated intensity of fluorescence. (D) HPLC-based analysis of acidic gangliosides on SH4 cell line before and after B4GALNT1 overexpression. Mock is SH4 with pcDNA3.1(+) expression vector alone. #4 and #5 single cells are isolated SH4 clones with B4GALNT1 overexpression. Black arrows; GM3/GD3, white arrows; GM2/GD2.
Our findings indicate the involvement of B4GALNT1 and GM2/GD2 in tumor establishment and progression as well as a potential direction of therapeutic approach via controlling B4GALNT1, and consequently GM2/GD2 expression in cancers such as melanoma.
Results
GM2/GD2 expression status in melanoma and neuroblastoma cell lines
To assess the GM2/GD2 expression level, four melanoma (A-375, RPMI-7951, WM115 and SH4) and two neuroblastoma cell lines (IMR32 and RTBM1) were measured by flow cytometry. One melanoma (WM115) and both of two neuroblastoma cell lines expressed high level of GM2/GD2 (Fig. 1B).
Because gangliosides including GM2/GD2 require stepwise synthesis reactions (Fig. 1A), a model for induced expression of GM2/GD2 on cell surface via overexpression of B4GALNT1 needs the following conditions; 1) both GM3 and GD3 are positive, and 2) both GM2 and GD2 are negative. To evaluate these conditions accurately in the six cell lines, HPLC-based high-specificity analysis of gangliosides was performed (Fig. 1C). Being that SH4 melanoma cell line showed high expression of both GD3 and GM3 (black arrows) and no expression of GD2 and GM2 (white arrows), SH4 fulfilled the aforementioned conditions and was used in the following study. Other results of neuroblastoma cells were shown in Fig. S1.
Generation of GM2/GD2-positive SH4 melanoma clones
The SH4 cells were transfected with expression vectors with or without B4GALNT1 gene cassette, to establish GM2/GD2-positive and -negative SH4 clones. Two GM2/GD2-high clones were selected by single cell isolation (#4 and #5, Fig. S2A). These two clones showed significant expression of GD2, whereas Mock (pcDNA3.1(+) alone) and two clones showed no GD2 expression. The expressions of B4GALNT1 in mRNA level were in correspondence with those by flow cytometry (Fig. S2B). Additionally, HPLC revealed that the clones #4 and #5 expressed GM2/GD2 at high level (Fig. 1D). The reason that GD2 level in the transfected clones is very low compared to the GD3 level in the parental cells was interpreted that B4GALNT1 and ST8Sia1 competes GM3 as a substrate. It is known that GD2 is not synthesized from GM228.
Induction of morphological change, anchorage independence growth, and cell motility
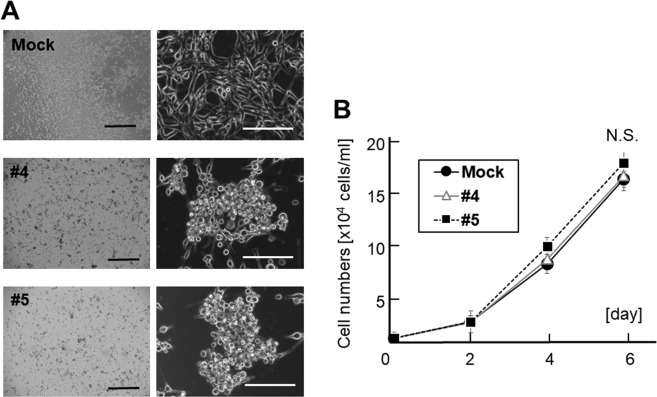
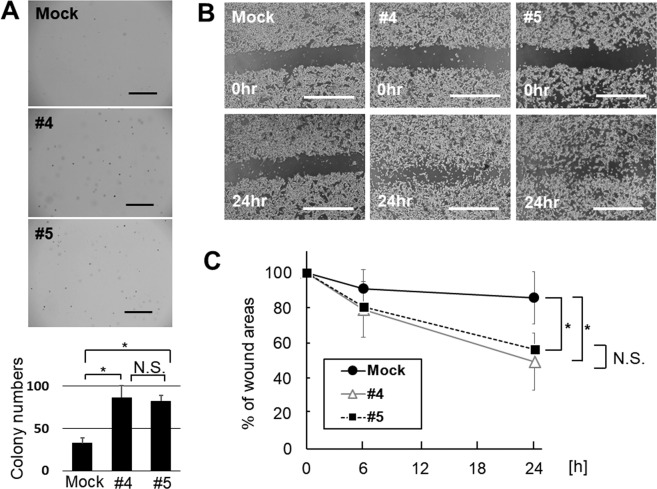
The SH4 clones overexpressing GM2/GD2, #4 and #5, exhibited a distinct morphological appearance compared to SH4 Wild type (WT) or the mock transduced cells. The cells were round and formed aggregation. More than half of them were detached from the bottom of flask, but still capable of survival and proliferation after detachment (Fig. 2A). No significant difference was seen between the proliferation of GM2/GD2-positive SH4 clones and control (Fig. 2B). A soft agar colony formation assay demonstrated that GM2/GD2-positive SH4 clones formed larger and greater number of colonies than GM2/GD2-negative cells (#4; 86.6 ± 13.9, #5; 82.5 ± 6.5, Mock; 32.7 ± 6.6, #4 vs Mock; p < 0.0001, #5 vs Mock; p < 0.0001, Fig. 3A). There was no significant difference between the two GM2/GD2-positive SH4 clones (#4 vs #5; P = 0.15). In addition, faster wound closure was observed in the GM2/GD2-positive SH4 clones than the control cells (#4: 49.7 ± 16.4 vs #5: 56.5 ± 25.3 vs Mock: 85.9 ± 14.8, #4 vs Mock: p < 0.0001, #5 vs Mock: p < 0.0001, #4 vs #5: p = 0.98, Fig. 3B,C), indicating enhanced motility.
Figure 2.
Effect of B4GALNT1 overexpression on cell morphology and growth. (A) Morphological changes in SH4 after B4GALNT1 overexpression. Scale bar, 1,000 μm (left) and, 200 μm (right). (B) Cell growth of SH4 with or without GM2/GD2-expression 6 days later. Results represent the means s.d. from three independent experiments.
Figure 3.
Effect of B4GALNT1 overexpression to cell behavior. (A) Photographs of colonies of SH4 cells with and without B4GALNT1 overexpression 14 days later. Scale bar, 1,000 μm. Results represent the means ∓ s.d. of three independent experiments. (B) Light microscopic images of SH4 cells that were scratched and compared the wound width 6 and 24 h later. Scale bar, 1,000 μm. (C) Average wound widths, expressed as a percent of the original width, obtained from 30 measurements in each photo. Results represent the means ∓ s.d. of three independent experiments. *P < 0.01 compared with Mock. N.S.; Not Significant.
Enhancement of tumor incidence and growth speed in vivo
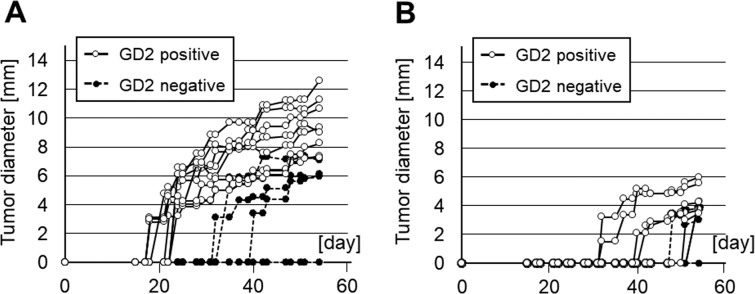
To assess the in vivo effect of GM2/GD2 induced by B4GALNT1 overexpression, the two GM2/GD2-positive SH4 clones and Mock were inoculated in NOD/Scid/IL2Rγ-null (NSG) mice to assess tumor initiation and growth. After transplanting 2 × 106 GM2/GD2-positive and -negative SH4 cells, all mice receiving #4 and #5 cells developed tumors, whereas only three out of six mice injected with Mock cells developed tumors (P = 0.038, Fig. 4A, Table 1). In the NSG mice transplanted with lower number (2 × 105) of GM2/GD2-positive or -negative SH4 cells, seven out of eight mice injected with #4 and #5 cells developed tumors, whereas two out of six mice injected with Mock cells developed tumors (P = 0.038, Fig. 4B, Table 1). Moreover, in the NSG mice transplanted with 2 × 106 GM2/GD2-positive or -negative SH4 cells, tumors derived from GM2/GD2-positive cells grew to be approximately three times larger than the GM2/GD2-negative Mock at day 54 (9.4 ± 1.9 vs 3.2 ± 3.5 mm, P = 0.003, Fig. 4A). In the NSG mice transplanted with 2 × 105 GM2/GD2-positive or -negative SH4 cells, tumors derived from GM2/GD2-positive cells grew to be over three times as large as those from GM2/GD2- negative Mock in 54 days (4.0 ± 1.8 vs 1.1 ± 1.8 mm, P = 0.026, Fig. 4B). Every tumor was solid, firm, and partially fibrotic.
Figure 4.
Effects of B4GALNT1-overexpressing on the proliferation of SH4 cells in xenograft models. (A) 2 × 106, (B) 2 × 105 SH4 cell were inoculated into lower flank of NSG mice subcutaneously.
Table 1.
Tumor incidence in vivo.
| GM2/GD2 | p-value | |||
|---|---|---|---|---|
| positive | negative | |||
| cell | 2 × 106 | 8/8 | 3/6 | P < 0.05 |
| number | 2 × 105 | 7/8 | 2/6 | P < 0.05 |
No evident difference in major cancer stem cell markers
To evaluate the possibility that B4GALNT1 overexpression might enhance tumor incidence via induction of stemness, several melanoma stem cell markers were analyzed in GM2/GD2- positive and -negative cells by flow cytometry. Previous reports indicated that CD13329–31, CD16629, CD27132, Nestin30,and ABCB533 are potential melanoma stem cell markers. Aldehyde dehydrogenase (ALDH) activity has also been reported as a potential marker of melanoma stem cell34. However, B4GALNT1 overexpression did not induce any meaningful change in any of these markers (Fig. S3A–F), there was no significant difference in CD133, CD166, and ABCB5 between GM2/GD2-positive and -negative cells. There was a small decrease in the expression of CD271 and Nestin in the #5 GM2/GD2-positive cell line, but this alone is unlikely to be the cause of enhanced tumor growth. That indicated that B4GALNT1 was unlikely to affect the stemness in the SH4 melanoma cell line.
Promotion of angiogenesis in vivo
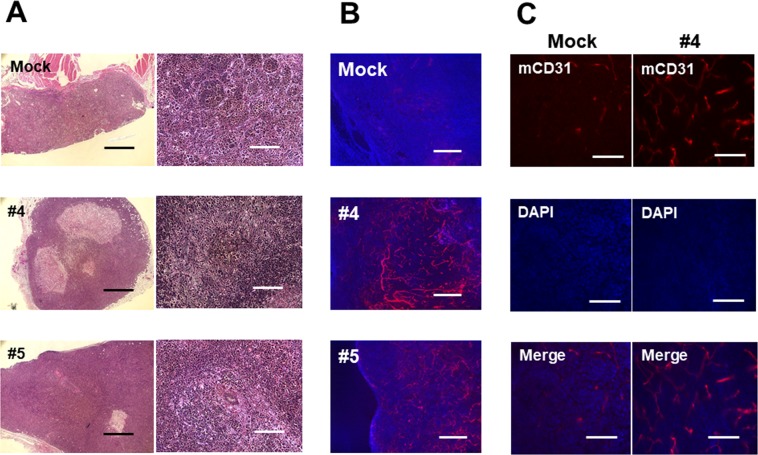
We hypothesized that B4GALNT1 may enhance tumor induction and growth by increasing tumor vascularization. Tumors of similar size from each group were stained by hematoxylin and eosin (H&E) and immunofluorescence for murine-CD31. In H&E staining, the tumors induced by GM2/GD2-positive or -negative SH4 cells did not differ in any characteristics examined: cell shape, number of giant cells, nuclear-to-cytoplasm volume ratio reversal, and hyperchromatism (Fig. 5A). The major blood vessel in the GM2/GD2-positive tumors were more prominent than the one supplying the Mock tumor. Although some tumors showed evidence of local invasion, none of the tumor-bearing mice developed metastasis.
Figure 5.
Histological analyses of the tumors. (A) Images of tissue sections of SH4 (H&E). (B,C) Immunostaining with murine CD31. Scale bar, 1,000 μm (A; left), 200 μm (A; right), 400 μm (B), and 100 μm (C), respectively.
Immunofluorescence staining in tumors derived from GM2/GD2-positive clones by anti-mouse CD31 Ab exhibited many well-structured vessels, while GM2/GD2-negative Mock tumors had much less (Fig. 5B,C). Immunofluorescence performed using selective anti-human-CD31 antibody (non-reactive to mouse CD31) as a negative control did not show any staining of blood vessels (data not shown).
RNA-Seq revealed potential key molecule downstream of B4GALNT1
To identify the difference of the transcriptional profile between GM2/GD2-positive and -negative cells, we compared them by RNA-Seq and analyzed using edgeR. There were a total of 26,484 genes detected in the individual libraries (Table S1), and 472 genes showed over +/− two-fold change between the two groups (Table S2). The heat map was shown in Fig. S4A. Among the 472 genes, 117 genes were up-regulated and 351 genes were down-regulated by B4GALNT1 overexpression. There was no significant difference between the two Mock clones (Fig. S4B), as well as between two GM2/GD2-positive samples (#4 and #5 clones; Fig. S4C). Genes exhibiting the Top 10 largest fold changes are listed in Table 2. The top up-regulated gene, PTPRD, is a member of the protein tyrosine phosphatase (PTP) family, and is known to be a signaling molecule that regulates a variety of cellular processes including cell growth, differentiation as a tumor suppressor35, which was often down-regulated in a variety of tumors. The second highest was B4GALNT1, suggesting that our overexpression of the gene in SH4 had succeeded. The third highest, POSTN (periostin), functions as a ligand for alpha-V/beta-3 and alpha-V/beta-5 integrins to support adhesion and migration36–38. In addition, it is known to increase angiogenesis. Furthermore, CPVL (Carboxypeptidase Vitellogenic Like), the top down-regulated gene was related to maturation of monocytes into macrophages39. Carboxypeptidases are a large class of proteases that act to cleave a single amino acid from the carboxy termini of proteins or peptides. The exact function of this protein, however, has not been determined. The second and third downregulated genes were melanoma-associated antigen 12 (MAGEA1240) and chondrosarcoma associated Gene 1 (CSAG141) which are oncogenes, supposed to be overexpressed in tumors, but in this case they were down-regulated upon B4GALNT1 overexpression.
Table 2.
Top 10 of the greatest genes expression changes in SH4 with vs without GM2/GD2.
| Symbol | Gene name | Expr Fold change | p-value |
|---|---|---|---|
| Up-regulation | |||
| PTPRD | Protein tyrosine phosphatase receptor type D | 735.0563 | 0.005722 |
| B4GALNT1 | Beta-1,4-N-acetyl- galactosaminyltransferase 1 | 359.3801 | 1.4E-150 |
| POSTN | Periostin | 286.3453 | 4.95E-26 |
| SERPINB2 | Serpin family B member 2 | 240.9988 | 7.94E-06 |
| HLA-DRB1 | human leukocyte antigen, class II, DR beta 1 | 228.4819 | 1.07E-05 |
| IL13RA2 | Interleukin 13 receptor alpha 2 | 191.2431 | 0.000616 |
| CSMD1 | CUB and sushi multiple domains 1 | 188.313 | 7.75E-05 |
| MYBPC1 | Myosin binding protein C type 1 | 182.0119 | 0.000805 |
| ADGRL2 | Adhesion G protein- coupled receptor L2 | 158.3556 | 4.73E-92 |
| TNFRSF14 | TNF receptor superfamily member 14 | 142.9792 | 0.011144 |
| Down-regulation | |||
| CPVL | Carboxypeptidase vitellogenic like | −1370.3 | 3.27E-37 |
| MAGEA12 | Melanoma-associated antigen 12 | −1143.23 | 9.84E-16 |
| CSAG1 | Chondrosarcoma associated gene 1 | −676.619 | 8.41E-12 |
| GIPC3 | PDZ domain containing family Member 3 | −535.495 | 1.85E-20 |
| CPM | Carboxypeptidase M | −484.607 | 3.42E-15 |
| ZXDA | Zinc finger, X-linked, duplicated A | −457.61 | 9.68E-09 |
| FAM83H | Family with sequence similarity 83 | −412.334 | 1.45E-08 |
| PRAME | Preferentially expressed antigen in melanoma | −411.078 | 0.000851 |
| GABRQ | Gamma-aminobutyric acid type A receptor theta subunit | −402.747 | 1.57E-21 |
| LDB2 | LIM domain binding 2 | −371.463 | 0.000011 |
To further elucidate the role of the 472 genes, we categorized them by function related to cancer malignancy using Ingenuity Pathway Analysis (IPA). This included several genes that functioned as melanoma incidence, proliferation, mobility and colony formation. Some genes were up- or down-regulated as the past reports and others were not (Table 3; “*” means that the fold changes went the opposite direction compared with previous findings). Some of them changed in the opposite direction compared to past reports in melanoma carcinogenesis. These genes may not function as downstream of B4GALNT1, and may have shown a change opposite of other literature due to negative feedback. Pathway analysis was attempted, but no known pathway was found that could fully explain our expression profile suggesting that the revelation of the pathway downstream of glycolipids is not complete.
Table 3.
Gene expression characterestics.
| Symbol | Expr Fold change | findings | Symbol | Expr Fold change | findings |
|---|---|---|---|---|---|
| Up-regulatation | Down-regulated | ||||
| Invasion of tumor | |||||
| POSTN | 286.345 | Increases | EDN3 | −248.929 | Decreases |
| NRP1 | 117.774 | Increases | VCAN* | −243.495 | Increases |
| IGFBP5* | 26.687 | Decreases | IL24 | −15.763 | Decreases |
| MMP1 | 20.329 | Increases | SERPINE1* | −9.371 | Increases |
| TGFBR2* | 6.627 | Decreases | SFRP1 | −6.729 | Decreases |
| ITGA1 | 4.868 | Increases | MMP2* | −6.046 | Increases |
| TBX2 | 2.561 | Increases | GPC1* | −4.435 | Increases |
| HMGB3* | 2.021 | Decreases | CEACAM1* | −4.131 | Increases |
| SPHK1 | 2.012 | Increases | EGF* | −3.683 | Increases |
| CTGF* | −2.077 | Decreases | CSF1* | −2.879 | Increases |
| Migration of melanoma cell | |||||
| NRP1* | 117.774 | Decreases | VCAN | −243.495 | Affects |
| SDC2 | 11.904 | Increases | L1CAM* | −20.041 | Increases |
| TNC | 2.217 | Increases | SERPINA5* | −10.969 | Increases |
| SPHK1 | 2.012 | Increases | SERPINE1* | −9.371 | Increases |
| MMP2* | −6.046 | Increases | |||
| EGF* | −3.683 | Increases | |||
| FN1* | −2.15 | Increases | |||
| Cell proliferation of tumor cell lines | |||||
| POSTN | 286.345 | Increases | PRAME* | −411.078 | Increases |
| SERPINB2* | 240.999 | Decreases | VCAN* | −243.495 | Increases |
| IL13RA2 | 191.243 | Affects | EMILIN2 | −124.892 | Decreases |
| NRP1 | 117.774 | Affects | TRPM2* | −94.449 | Increases |
| HTN1 | 60.939 | Increases | SYNM* | −26.275 | Increases |
| MAF* | 50.99 | Decreases | PDGFB* | −22.149 | Increases |
| POU4F1 | 50.781 | Increases | L1CAM* | −20.041 | Increases |
| VIP | 39.551 | Increases | CARD10* | −16.191 | Increases |
| CDK14 | 30.195 | Increases | IL24 | −15.763 | Decreases |
| IGFBP5 | 26.687 | Increases | S100A4* | −15.312 | Increases |
| Colony formation | |||||
| POSTN | 286.345 | Affects | VCAN* | −243.495 | Increases |
| HTN3* | 98.45 | Decreases | NUPR1 | −31.659 | Decreases |
| VIP | 39.551 | Increases | PDGFB* | −22.149 | Increases |
| TP63 | 8.144 | Increases | IL24 | −15.763 | Decreases |
| CDCA7L | 4.871 | Increases | S100A4* | −15.312 | Increases |
| ITGA1 | 4.868 | Increases | SERPINE1 | −9.371 | Decreases |
| ALDH1A1 | 4.81 | Affects | TBX3* | −7.463 | Increases |
| HAS2 | 4.155 | Increases | DUSP5* | −6.896 | Increases |
| TNC | 2.217 | Affects | SFRP1* | −6.729 | Increases |
| LIMA1* | 2.133 | Decreases | PTGES* | −6.519 | Increases |
*means that the fold changes went the opposite direction compared with previous findings.
Discussion
In this study, we analyzed a variety of changes in the SH4 melanoma cell line upon overexpression of GM2/GD2 by transfection of B4GALNT1 gene. One of remarkable findings in vitro is that GM2/GD2-positive SH4 cells showed significant difference of AIG compared to Mock (Fig. 3A), interestingly, Mahata et al. also revealed that GM2/GD2 is associated with AIG by knocking out GM2/GD2 synthase42. AIG is often reported as a critical factor for tumorigenesis or exacerbation of malignancy42–44. Although we initially expected contribution of Epithelial-Mesenchymal Transition (EMT), neither CDH1 (E-cadhelin) nor VIM (vimentin), the major EMT markers, showed change at the mRNA level (data not shown). On the other hand, our RNA-Seq result showed that the expression of POSTN in B4GALNT1-overexpressing cells increased by almost 300 times than Mock cells. Periostin is generally known as a cancer suppresser45 and it also help to migration in neuronal cell development46. While periostin is involved in numerous biological processes, it sometimes contributes to tumorigenesis by promoting cancer cell survival, invasion, and metastasis actively36–38. It is also known that high expression of periostin protein and/or mRNA is detected in variety of solid tumors38,47. Kudo et al. showed that periostin overexpression promoted invasion in head and neck squamous cell carcinoma cells48 and to explore the genes that are coordinately expressed with periostin, they performed microarray analysis. Among the genes changed in their study, SULF1 was upregulated clearly in our result as well (9.30-fold; Table S2). On top of that, Kotbuki et al. directly revealed that periostin increased cell proliferation and invasion in melanoma cell in vitro and in vivo using overexpression system49, and Fukuda et al. showed that periostin was a key factor in promoting melanoma cell metastasis using shRNA50. We therefore speculated that the findings support our conclusion that periostin and its downstream gene overexpression promoted migration induced by GM2/GD2. Furthermore, Bao et al. demonstrated that periostin activated the downstream Akt/PKB pathway via αvβ3 integrin, by in which they observed phosphorylation of Akt1/PKBα on Ser473 to promote cellular survival in colon cancer51. Their phosphorylation level, not the total amount, would therefore be contributing to the downstream effect of GM2/GD2. This may explain why our RNA-Seq result did not show a remarkable fold change in Akt/PKB pathway (Table S1).
In our observation, B4GALNT1 overexpression did not affect cell proliferation in vitro, while multiple genes related to tumor cell proliferation-promoting, such as POSTN, IL13RA2, NRP1 were up-regulated, and EMILIN2, which is a proliferation-suppressing gene, was down-regulated notably in mRNA level (Fig. 2B, Table 3). The relationship between ganglioside and cell proliferation is still controversial; some research suggest promotion of proliferation by gangliosides52–54, while others show inhibition55. These discrepancies may be explained by the difference in the ratio of GD2 + 3 vs GM2 + 3, as well as GD2 vs GD3, which may contribute to differences in cell proliferation. The report by Shibuya et al. which induced GM2/GD2 like ours showed enhanced cell migration55, similar to what we observed, supporting that B4GALNT1 and consequently GM2/GD2 intensified migration of SH4 cell (Fig. 3B). Periostin is known to promote motility of several different kinds of cells56–58. While our IPA analysis did not include the category of melanoma cell migration, RNA-Seq indicated that SDC2 and VCAN might have led to the motility (Table 3).
Another finding was that GM2/GD2 strongly induced angiogenesis. The effect of B4GALNT1 for tumor incidence in vivo was assessed by injecting GM2/GD2-positive SH4 cells into NSG mice. As shown in Fig. 4A,B, the tumors injected GM2/GD2-positive cells showed a higher tumor establishment rate. This result corresponds with the fact that B4GALNT1 is a clinical marker for advanced melanoma59. Liu Y et al. revealed that gangliosides accelerate tumor angiogenesis in murine cells and demonstrated that GM2/GD2-negative cells formed much smaller tumors, using GM3 synthase and GM2 synthase double knockout low ganglioside tumor model60. We assessed murine-CD31 expression in the tumors derived from GM2/GD2-positive cell by immunofluorescence staining and observed that B4GALNT1-overexpressing clones induced many CD31 positive endothelial cells and well-developed vessels. In addition, the surface of the GM2/GD2-positive tumors were better vascularized than that of Mock by observation of the recovered tumor with eyes. Tumor progression requires endothelial cells to be activated for the formation of a vascular system. Lang Z et al. found that the enrichment of human umbilical vein endothelial cell (HUVEC) membranes with ganglioside results in amplified VEGF-induced signaling that is important for angiogenesis, and concluded that ganglioside enhances VEGF-induced endothelial cell proliferation61. Liu Y et al. reported that reduction of gangliosides depleted vascularization, while addition of wild type gangliosides restored angiogenesis of ganglioside-poor tumor60. To clarify how gangliosides induce blood vessels, we assessed the relations of GM2/GD2 and VEGF. Some other reports indicate that ganglioside enhances VEGF and induces endothelial cell proliferation61,62. In addition, Liu Y et al. also reported that periostin induces angiogenesis via Erk/VEGF pathway63. However, in our result of RNA-Seq and real-time RT-PCR, the expression of VEGF did not show significant correlation with GM2/GD2 level (Table S1, Fig. S5). There is a possibility that interaction between periostin and integrins directly promoted angiogenesis64 or GM2/GD2 lowered the threshold for cytokine stimulation60,65.
While GM2 and GD2 were reported to be increased greatly in cancer stem cells in breast cancer66,67, our data ruled out the possibility that GM2/GD2 enhanced tumor incidence via induction of cell stemness. We assessed some melanoma stem cell markers, such as CD133, CD166, CD271, Nestin, ABCB5, and ALDH activity by flow cytometry, and there was no evidence indicative of GM2/GD2 involvement in stemness (Fig. S3A–F).
In summary, our findings demonstrated that in the SH4 melanoma cell line, overexpression of B4GALNT1 as well as its main products GM2/GD2 promotes AIG and cell migration in vitro and enhances tumor incidence by inducing angiogenesis in vivo. To our best knowledge, this is the first time that RNA-Seq was performed to elucidate the influence of B4GALNT1. This result indicates that GM2/GD2 or B4GALNT1 is upstream regulator of periostin, and it might cause some change of characters related to tumorigenesis mentioned above in melanoma cell line. In this study, we have not only shown how GM2/GD2 exacerbates tumors’ malignant characters by using B4GALNT1 artificial expression system, but also reconfirmed RNA-Seq is useful tool to find novel potential target in cancer.
Materials and Methods
All experiments were performed in accordance with relevant guidelines and regulations.
Cell lines
Human melanoma cell lines (A375, RPMI-7951, SH4 and WM115) were purchased from American Type Culture Collection (Manassas, USA). Human neuroblastoma cell lines (IMR32 and RTBM1) were provided by Dr. Hajime Hosoi (Kyoto Prefectural University of Medicine, Japan). The melanoma cell lines were maintained in Dulbecco’s modified Eagle’s high-glucose medium (DMEM, Corning, USA) and neuroblastoma cell lines were maintained in Eagle’s minimal essential medium (EMEM, Corning) supplemented with 10% FBS, 100 U/ml penicillin and 10 mg/ml streptomycin (Corning). HUVEC was maintained in Endothelial Cell Growth Media (Sigma-Aldrich, USA). All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2.
Construction of a cDNA expression vector, gene transfection and selection
Human B4GALNT1 cDNA was cloned from IMR32 with the primers listed in Table S3. The fragment was first inserted into Topo vector using Zero Blunt TOPO PCR Cloning Kit (Invitrogen, USA). After confirmation of sequence, the cDNA cut out by BamHI and NotI was inserted into the cDNA3.1(+) expression vector (Invitrogen). SH4 cells were plated in a 60-mm plastic plate (Corning) and then transfected with the plasmids by using Superfect (Qiagen, Germany). Stable transfectants were isolated in the presence of 600 μg/ml G418 (Roche, Germany).
Flow cytometric analysis
Cells were trypsinized and washed twice with flow cytometry buffer (FCB, PBS supplemented with 1% FBS and 0.02% sodium azide (Sigma-Aldrich)). Cells were incubated with the anti-hGD2 mAbs (MAB2052, Millipore, USA) and anti-hABCB5 mAbs (MA5-17026, Thermo Fischer Scientific, USA (1:100, 100 μl/106 cells) for 1 h and then washed in FCB. The cells were subsequently incubated with FITC-labeled anti-mouse goat IgG (sc-2010; 1:1,000, 1 ml/106 cells, Santa Cruz Biotechnology, USA) for 40 min, and washed twice with FCB. Cells were incubated in PE-conjugated anti-hCD133/2 mAbs (#130-090-853, Miltenyi Biotec, Germany), PE-conjugated anti-hALCAM/CD166 mAbs (#105902, R&D systems, USA) and FITC-conjugated anti-hCD271/NGFR antibody (#345103, BioLegend, USA), (1:200, 1 ml/106 cells) for 15 min and then washed with FCB. With PE-conjugated anti-hNestin mAbs (#196908, R&D systems), cells (1 × 106 cells/200 μl) were fixed with 200 μl of cold 4% paraformaldehyde (Sigma-Aldrich) at room temperature for 30 min, and then washed in FCB twice. After incubation with 500 μl 0.1% Triton X-100 (Sigma-Aldrich) for 10 min at room temperature, cells were washed with FCB twice and stained with the antibody (1:200, 1 ml/106 cells) on ice for 30 min in the dark, and subsequently washed with FCB. ALDH activity was determined by ALDEFLUOR Kit (Stemcell Technologies, Canada) according to the manufacturer’s instructions. All procedures were performed at 4 °C. The samples were immediately analyzed using FACS Canto II flow cytometer (BD Bioscience, USA). In each sample at least 1 × 104 events were collected. The data was analyzed with FlowJo software (FlowJo, LLC, USA).
High-performance liquid chromatography (HPLC)
HPLC was carried out as described previously68–70. Briefly, the acidic glycosphingolipids were extracted from each melanoma and neuroblastoma cell line (1 × 106 cells) and digested with recombinant endoglycoceramidase II from Rhodococcus sp. (Takara Bio, Japan). The released oligosaccharides were labeled with 2-aminopyridine and separated using a HPLC system equipped with a fluorescence detector. Normal-phase HPLC was performed on a TSK gel Amide-80 column (Tosoh, Japan). The molecular size of each PA-oligosaccharide is given in glucose units (Gu) based on the elution times of PA-isomaltooligosaccharides. Reversed-phase HPLC was performed on a TSK gel ODS-80Ts column (Tosoh). The retention time of each PA-oligosaccharide is given in glucose units based on the elution times of PA-isomaltooligosaccharides. Thus, a given compound on these two columns provides a unique set of Gu (amide) and Gu (ODS) values, which correspond to coordinates of the 2-D map. PA-oligosaccharides were analyzed using LC⁄ ESI MS⁄ MS. Standard PA-oligosaccharides, PA-GM1 and PA-GD1a, were purchased from Takara Bio and PA-LST-a and PA-SPG were obtained from our previous study71.
Real time RT–PCR
Total RNA was extracted from a tumor specimen with RNeasy mini kit (Qiagen) and complementary DNA (cDNA) was synthesized by the use of the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions, respectively. The primers used in this experiment are listed in Table S3. Real-time RT-PCR was carried out using LightCycler 480 System (Roche) with SYBR Green (Applied Biosystems, CA) as previously described72. Thermal cycling conditions were: initial denaturation for 10 min at 95 °C, and 40 cycles of 15 sec at 95 °C, and 1 min at 60 °C. Data were analyzed with the Light Cycler software.
Assay for cell proliferation
Cells were seeded at 2 × 104 cells/well in 12-well plate. Every 48 h, cells were dissociated by 0.25% trypsin (Corning) and neutralized by the same volume of 10% DMEM. After that 100 μl 0.5% trypan blue (Sigma-Aldrich) and counted by Cellometer Auto T4 (Nexcelom Bioscience, USA) until days 6.
Anchorage-independent soft agar colony formation assay
Cells were cultured in a two-layer soft agar system73. It consisted of a 1% agarose (RPI, USA) underlayer and a 0.7% agarose overlayer containing 1 × 104 cells in 60-mm dishes (Corning). Colonies were allowed to form for 2 weeks with fresh media added every 3 days. Plates were stained with crystal violet and colonies more than 0.1 mm in diameter were counted.
Wound-healing assay
Wound-healing assays were carried out as described previously74,75. Immediately after scratching (0 h), the plates were photographed and the distance between the edges of the wound area was measured. At 6 h and 24 h after scratching, the plates were photographed and the distance between the edges of the wound region was again measured.
In vivo tumorigenesis
Tumors were induced in 5–6 week old female and male NSG mice (Jackson Laboratory, USA). Each mouse was injected subcutaneously with SH4 cell lines transformed with pcDNA3.1(+) empty vector or the one expressing B4GALNT1 suspended in 0.1 ml of PBS at a single site (2 × 106 cell; left, 2 × 105 cell; right) to the lower flank. Tumor diameter was monitored every 2–3 days on onset of tumor formation. Mice were sacrificed when the largest tumor size reached 16 mm in diameter along with IACUC approved protocol. At the end point of the experiments, tumors were extracted. At least 4 mice were used in each group (Mock, #4 and #5). The animal experiments were performed in accordance with the institutionally approved animal experimental protocol.
Histopathology and immunohistochemistry
Histological specimens were fixed in 10% formalin and routinely processed for embedding in paraffin. The sections were stained with H&E. Some portions of tumors were embedded in OCT and frozen at −20 °C for immunofluorescence analyses. The tumors were sectioned 30-μm-thick by CM 1800 Cryostat (Leica Biosystems, Germany). The sections were fixed by 50 μl of ice-cold acetone for 5 min. After washing the slides in 1 x PBS, they were incubated in blocking buffer (2% bovine serum albumin (BSA) serum in PBS) for 1 h. The sections were incubated with PE-conjugated anti-mouse CD31 antibody (#102507, BioLegend, 1:500 at 4 °C overnight). After washing the slides in 1 x PBS three times, the slides were incubated by 20 μl VECTASHIELD Antifade Mounting Medium with DAPI (H-1200, Vector Laboratories, USA) and coverslipped. The tissues were observed with automated upright microscope System DM5500 B (Leica Biosystems).
RNA sequencing (whole transcriptome shotgun sequencing, WTSS) and analysis
RNAs derived from SH4 with (#4 and #5) or without (Mock; two samples) B4GALNT1 over-expression were analyzed by RNA sequencing as described in ref. 76–78. Quality tested with Bioanalyzer 2100 (Agilent Technologies, USA). Sequencing was accomplished on the MiSeq 500 (Illumina, USA). 50 bp FastQ paired-end reads (n = 23.6 Million per sample) were trimmed using Trimmomatic (v 0.33). Quality control checks on raw sequence data were performed with FastQC. Read mapping was performed via Hisat2 (2.1.0) using the Human UCSC genome (hg38) as reference. Differentially expressed genes were identified using the edgeR (Bioconductor, www.bioconductor.org) feature in CLCGWB (Qiagen) using raw read counts. The generated list was filtered based on a minimum 2 x absolute fold change and false discovery rate (FDR) corrected p < 0.05. Pathway analysis was performed in IPA (Qiagen) using fold change and FDR corrected values.
Statistical analysis
Statistical analysis was performed using the unpaired Student’s t-test. A P-value of less than 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This research was partly supported by grants from NIH/NCI (R01CA168448 and R01CA196215 to MY), and the Research Support from Department of Surgery, University of Minnesota. .
Author contributions
Conception and design: H. Yoshida and M. Yamamoto. Development of methodology: H. Yoshida, L. Koodie, Y. Miyamoto, and M. Yamamoto. Acquisition of data (provide): H. Yoshida (All), L. Koodie (Immunostaining), K. Jacobsen (Animal experiments), and K. Hanzawa (Glycobiology). Analysis and interpretation of data: H. Yoshida and M. Yamamoto. Writing review, and/or revision of the manuscript: H. Yoshida and M. Yamamoto. Administrative, technical, or material support: L. Koodie and K. Jacobsen. Study supervision: M. Yamamoto.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57130-2.
References
- 1.Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. Journal of Investigative Dermatology. 2009;129:1666–1674. doi: 10.1038/jid.2008.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erdei E, Torres SM. A new understanding in the epidemiology of melanoma. Expert review of anticancer therapy. 2010;10:1811–1823. doi: 10.1586/era.10.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rigel DS, Carucci JA. Malignant melanoma: prevention, early detection, and treatment in the 21st century. CA: a cancer journal for clinicians. 2000;50:215–236. doi: 10.3322/canjclin.50.4.215. [DOI] [PubMed] [Google Scholar]
- 4.Kosary CL, Altekruse SF, Ruhl J, Lee R, Dickie L. Clinical and prognostic factors for melanoma of the skin using SEER registries: collaborative stage data collection system, version 1 and version 2. Cancer. 2014;120:3807–3814. doi: 10.1002/cncr.29050. [DOI] [PubMed] [Google Scholar]
- 5.Ernst DS, et al. Burden of illness for metastatic melanoma in Canada, 2011–2013. Current Oncology. 2016;23:e563. doi: 10.3747/co.23.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szczepaniak Sloane RA, et al. Interaction of molecular alterations with immune response in melanoma. Cancer. 2017;123:2130–2142. doi: 10.1002/cncr.30681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. New England Journal of Medicine. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 8.Spain L, Julve M, Larkin J. Combination dabrafenib and trametinib in the management of advanced melanoma with BRAFV600 mutations. Expert opinion on pharmacotherapy. 2016;17:1031–1038. doi: 10.1517/14656566.2016.1168805. [DOI] [PubMed] [Google Scholar]
- 9.Wood K, Luke JJ. Optimal use of BRAF targeting therapy in the immunotherapy era. Current oncology reports. 2016;18:67. doi: 10.1007/s11912-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 10.Callahan MK, Postow MA, Wolchok JD. Immunomodulatory therapy for melanoma: ipilimumab and beyond. Clinics in dermatology. 2013;31:191–199. doi: 10.1016/j.clindermatol.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ascierto PA, et al. The role of BRAF V600 mutation in melanoma. Journal of translational medicine. 2012;10:85. doi: 10.1186/1479-5876-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodi FS, et al. Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. The Lancet Oncology. 2016;17:1558–1568. doi: 10.1016/S1470-2045(16)30366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kholodenko IV, Kalinovsky DV, Doronin II, Deyev SM, Kholodenko RV. Neuroblastoma origin and therapeutic targets for immunotherapy. Journal of immunology research. 2018;2018:7394268. doi: 10.1155/2018/7394268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolter T. Ganglioside biochemistry. ISRN biochemistry. 2012;2012:506160. doi: 10.5402/2012/506160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robert KY, Nakatani Y, Yanagisawa M. The role of glycosphingolipid metabolism in the developing brain. Journal of lipid research. 2009;50:S440–S445. doi: 10.1194/jlr.R800028-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoon DSB, Irie RF, Cochran AJ. Gangliosides from human melanoma immunomodulate response of T cells to interleukin-2. Cellular immunology. 1988;111:410–419. doi: 10.1016/0008-8749(88)90104-9. [DOI] [PubMed] [Google Scholar]
- 17.Todeschini AR, Hakomori S-I. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochimica et Biophysica Acta (BBA)-General Subjects. 2008;1780:421–433. doi: 10.1016/j.bbagen.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ledeen RW, Wu G. Ganglioside function in calcium homeostasis and signaling. Neurochemical research. 2002;27:637–647. doi: 10.1023/A:1020224016830. [DOI] [PubMed] [Google Scholar]
- 19.Hakomori S-I. Traveling for the glycosphingolipid path. Glycoconjugate journal. 2000;17:627–647. doi: 10.1023/A:1011086929064. [DOI] [PubMed] [Google Scholar]
- 20.Hakomori S-I. Tumor malignancy defined by aberrant glycosylation and sphingo (glyco) lipid metabolism. Cancer research. 1996;56:5309–5318. [PubMed] [Google Scholar]
- 21.Cheung IY, Lo Piccolo MS, Kushner BH, Cheung N-KV. Early molecular response of marrow disease to biologic therapy is highly prognostic in neuroblastoma. Journal of clinical oncology. 2003;21:3853–3858. doi: 10.1200/JCO.2003.11.077. [DOI] [PubMed] [Google Scholar]
- 22.Tringali C, et al. Molecular subtyping of metastatic melanoma based on cell ganglioside metabolism profiles. BMC cancer. 2014;14:560. doi: 10.1186/1471-2407-14-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modak S, Gerald W, Cheung NKV. Disialoganglioside GD2 and a novel tumor antigen: potential targets for immunotherapy of desmoplastic small round cell tumor. Medical and pediatric oncology. 2002;39:547–551. doi: 10.1002/mpo.10151. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchida T, Saxton RE, Morton DL, Irie RF. Gangliosides of human melanoma. JNCI: Journal of the National Cancer Institute. 1987;78:45–54. doi: 10.1093/jnci/78.1.45. [DOI] [PubMed] [Google Scholar]
- 25.Chang HR, Cordon‐Cardo C, Houghton AN, Cheung NKV, Brennan MF. Expression of disialogangliosides GD2 and GD3 on human soft tissue sarcomas. Cancer. 1992;70:633–638. doi: 10.1002/1097-0142(19920801)70:3<633::AID-CNCR2820700315>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Heiner JP, et al. Localization of GD2-specific monoclonal antibody 3F8 in human osteosarcoma. Cancer Research. 1987;47:5377–5381. [PubMed] [Google Scholar]
- 27.Wu Z-L, Schwartz E, Seeger R, Ladisch S. Expression of GD2 ganglioside by untreated primary human neuroblastomas. Cancer research. 1986;46:440–443. [PubMed] [Google Scholar]
- 28.Sasaki K, et al. Expression cloning of a GM3-specific alpha-2, 8-sialyltransferase (GD3 synthase) Journal of Biological Chemistry. 1994;269:15950–15956. [PubMed] [Google Scholar]
- 29.Klein WM, et al. Increased expression of stem cell markers in malignant melanoma. Modern pathology. 2007;20:102–107. doi: 10.1038/modpathol.3800720. [DOI] [PubMed] [Google Scholar]
- 30.Sabet MN, Rakhshan A, Erfani E, Madjd Z. Co-expression of putative cancer stem cell markers, CD133 and Nestin, in skin tumors. Asian Pacific Journal of Cancer Prevention. 2014;15:8161–8169. doi: 10.7314/APJCP.2014.15.19.8161. [DOI] [PubMed] [Google Scholar]
- 31.Roudi R, et al. Comparative gene-expression profiling of CD133+ and CD133-D10 melanoma cells. Future. Oncology. 2015;11:2383–2393. doi: 10.2217/fon.15.174. [DOI] [PubMed] [Google Scholar]
- 32.Redmer T, et al. The role of the cancer stem cell marker CD271 in DNA damage response and drug resistance of melanoma cells. Oncogenesis. 2017;6:e291. doi: 10.1038/oncsis.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson BJ, et al. ABCB5 maintains melanoma-initiating cells through a pro-inflammatory cytokine signaling circuit. Cancer research. 2014;74:4196–4207. doi: 10.1158/0008-5472.CAN-14-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem cells. 2012;30:2100–2113. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veeriah S, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proceedings of the National Academy of Sciences. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu AY, Zheng H, Ouyang G. Periostin, a multifunctional matricellular protein in inflammatory and tumor microenvironments. Matrix biology. 2014;37:150–156. doi: 10.1016/j.matbio.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Q-m, et al. Periostin facilitates the epithelial-mesenchymal transition of endometrial epithelial cells through ILK-Akt signaling pathway. BioMed research international. 2016;2016:9842619. doi: 10.1155/2016/9842619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan K, Bao S, Ouyang G. The multifaceted role of periostin in tumorigenesis. Cellular and molecular life sciences. 2009;66:2219–2230. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahoney JA, Ntolosi B, DaSilva RP, Gordon S, McKnight AJ. Cloning and characterization of CPVL, a novel serine carboxypeptidase, from human macrophages. Genomics. 2001;72:243–251. doi: 10.1006/geno.2000.6484. [DOI] [PubMed] [Google Scholar]
- 40.Zhao G, et al. Overexpression and Implications of Melanoma-associated Antigen A12 in Pathogenesis of Human Cutaneous Squamous Cell Carcinoma. Anticancer research. 2019;39:1849–1857. doi: 10.21873/anticanres.13292. [DOI] [PubMed] [Google Scholar]
- 41.Lin C, et al. Cancer/testis antigen CSAGE is concurrently expressed with MAGE in chondrosarcoma. Gene. 2002;285:269–278. doi: 10.1016/S0378-1119(02)00395-5. [DOI] [PubMed] [Google Scholar]
- 42.Mahata B, Banerjee A, Kundu M, Bandyopadhyay U, Biswas K. TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells. Scientific reports. 2015;5:9048. doi: 10.1038/srep09048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori S, et al. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene. 2009;28:2796–2805. doi: 10.1038/onc.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colburn NH, et al. Correlation of anchorage-independent growth with tumorigenicity of chemically transformed mouse epidermal cells. Cancer research. 1978;38:624–634. [PubMed] [Google Scholar]
- 45.Lai J-P, Sandhu DS, Shire AM, Roberts LR. The tumor suppressor function of human sulfatase 1 (SULF1) in carcinogenesis. Journal of gastrointestinal cancer. 2008;39:149–158. doi: 10.1007/s12029-009-9058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalus I, et al. Sulf1 and Sulf2 differentially modulate heparan sulfate proteoglycan sulfation during postnatal cerebellum development: evidence for neuroprotective and neurite outgrowth promoting functions. PLoS One. 2015;10:e0139853. doi: 10.1371/journal.pone.0139853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conway SJ, et al. The role of periostin in tissue remodeling across health and disease. Cellular and Molecular Life Sciences. 2014;71:1279–1288. doi: 10.1007/s00018-013-1494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kudo Y, et al. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer research. 2006;66:6928–6935. doi: 10.1158/0008-5472.CAN-05-4540. [DOI] [PubMed] [Google Scholar]
- 49.Kotobuki Y, et al. Periostin accelerates human malignant melanoma progression by modifying the melanoma microenvironment. Pigment cell & melanoma research. 2014;27:630–639. doi: 10.1111/pcmr.12245. [DOI] [PubMed] [Google Scholar]
- 50.Fukuda K, et al. Periostin is a key niche component for wound metastasis of melanoma. PloS one. 2015;10:e0129704. doi: 10.1371/journal.pone.0129704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao S, et al. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer cell. 2004;5:329–339. doi: 10.1016/S1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida S, et al. Ganglioside GD2 in small cell lung cancer cell lines: enhancement of cell proliferation and mediation of apoptosis. Cancer research. 2001;61:4244–4252. [PubMed] [Google Scholar]
- 53.Hamamura K, et al. Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proceedings of the National Academy of Sciences. 2005;102:11041–11046. doi: 10.1073/pnas.0503658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, et al. Ganglioside synthase knockout in oncogene-transformed fibroblasts depletes gangliosides and impairs tumor growth. Oncogene. 2010;29:3297–3306. doi: 10.1038/onc.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibuya H, et al. Enhancement of malignant properties of human osteosarcoma cells with disialyl gangliosides GD 2/GD 3. Cancer science. 2012;103:1656–1664. doi: 10.1111/j.1349-7006.2012.02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Y, et al. Periostin promotes migration and osteogenic differentiation of human periodontal ligament mesenchymal stem cells via the Jun amino‐terminal kinases (JNK) pathway under inflammatory conditions. Cell proliferation. 2017;50:e12369. doi: 10.1111/cpr.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gillan L, et al. Periostin secreted by epithelial ovarian carcinoma is a ligand for αVβ3 and αVβ5 integrins and promotes cell motility. Cancer research. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 58.Li G, et al. Periostin mediates vascular smooth muscle cell migration through the integrins ανβ3 and ανβ5 and focal adhesion kinase (FAK) pathway. Atherosclerosis. 2010;208:358–365. doi: 10.1016/j.atherosclerosis.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuo CT, et al. Assessment of messenger RNA of beta 1–>4-N-acetylgalactosaminyl-transferase as a molecular marker for metastatic melanoma. Clinical Cancer Research. 1998;4:411–418. [PubMed] [Google Scholar]
- 60.Liu Y, Wondimu A, Yan S, Bobb D, Ladisch S. Tumor gangliosides accelerate murine tumor angiogenesis. Angiogenesis. 2014;17:563–571. doi: 10.1007/s10456-013-9403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lang Z, Guerrera M, Li R, Ladisch S. Ganglioside GD1a enhances VEGF-induced endothelial cell proliferation and migration. Biochemical and biophysical research communications. 2001;282:1031–1037. doi: 10.1006/bbrc.2001.4630. [DOI] [PubMed] [Google Scholar]
- 62.Chung T-W, et al. Ganglioside GM3 inhibits VEGF/VEGFR-2-mediated angiogenesis: direct interaction of GM3 with VEGFR-2. Glycobiology. 2008;19:229–239. doi: 10.1093/glycob/cwn114. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, et al. Periostin promotes tumor angiogenesis in pancreatic cancer via Erk/VEGF signaling. Oncotarget. 2016;7:40148–40159. doi: 10.18632/oncotarget.9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siriwardena B, et al. Periostin is frequently overexpressed and enhances invasion and angiogenesis in oral cancer. British journal of cancer. 2006;95:1396–1403. doi: 10.1038/sj.bjc.6603431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, McCarthy J, Ladisch S. Membrane ganglioside enrichment lowers the threshold for vascular endothelial cell angiogenic signaling. Cancer research. 2006;66:10408–10414. doi: 10.1158/0008-5472.CAN-06-1572. [DOI] [PubMed] [Google Scholar]
- 66.Liang Y-J, et al. Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells. Proceedings of the National Academy of Sciences. 2013;110:4968–4973. doi: 10.1073/pnas.1302825110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang Y-J, et al. Interaction of glycosphingolipids GD3 and GD2 with growth factor receptors maintains breast cancer stem cell phenotype. Oncotarget. 2017;8:47454–47473. doi: 10.18632/oncotarget.17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nomura M, Shimbo T, Miyamoto Y, Fukuzawa M, Kaneda Y. 13‐Cis retinoic acid can enhance the antitumor activity of non‐replicating S endai virus particle against neuroblastoma. Cancer science. 2013;104:238–244. doi: 10.1111/cas.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawaguchi Y, Miyamoto Y, Inoue T, Kaneda Y. Efficient eradication of hormone‐resistant human prostate cancers by inactivated Sendai virus particle. International journal of cancer. 2009;124:2478–2487. doi: 10.1002/ijc.24234. [DOI] [PubMed] [Google Scholar]
- 70.Hatano K, Miyamoto Y, Nonomura N, Kaneda Y. Expression of gangliosides, GD1a, and sialyl paragloboside is regulated by NF‐κB‐dependent transcriptional control of α2, 3‐sialyltransferase I, II, and VI in human castration‐resistant prostate cancer cells. International journal of cancer. 2011;129:1838–1847. doi: 10.1002/ijc.25860. [DOI] [PubMed] [Google Scholar]
- 71.Korekane H, et al. Novel fucogangliosides found in human colon adenocarcinoma tissues by means of glycomic analysis. Analytical biochemistry. 2007;364:37–50. doi: 10.1016/j.ab.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida H, et al. Post-transcriptional modulation of C/EBPα prompts monocytic differentiation and apoptosis in acute myelomonocytic leukaemia cells. Leukemia research. 2012;36:735–741. doi: 10.1016/j.leukres.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 73.Hamburger AW, et al. Direct cloning of human ovarian carcinoma cells in agar. Cancer research. 1978;38:3438–3444. [PubMed] [Google Scholar]
- 74.Yoshida H, et al. PAX3-NCOA2 fusion gene has a dual role in promoting the proliferation and inhibiting the myogenic differentiation of rhabdomyosarcoma cells. Oncogene. 2014;33:5601–5608. doi: 10.1038/onc.2013.491. [DOI] [PubMed] [Google Scholar]
- 75.Kikuchi K, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochemical and biophysical research communications. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 76.Rathe SK, et al. Using RNA-seq and targeted nucleases to identify mechanisms of drug resistance in acute myeloid leukemia. Scientific reports. 2014;4:6048. doi: 10.1038/srep06048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dauer P, et al. Inactivation of cancer-associated-fibroblasts disrupts oncogenic signaling in pancreatic cancer cells and promotes its regression. Cancer research. 2018;78:1321–1333. doi: 10.1158/0008-5472.CAN-17-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark CR, et al. Transposon mutagenesis screen in mice identifies TM9SF2 as a novel colorectal cancer oncogene. Scientific reports. 2018;8:15327. doi: 10.1038/s41598-018-33527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.