Abstract
Uterine leiomyoma is one of the most common gynaecologic benign tumours, but its genetic basis remains largely unknown. Six previous GWAS identified 33 genetic factors in total. Here, we performed a two-staged GWAS using 13,746 cases and 70,316 controls from the Japanese population, followed by a replication analysis using 3,483 cases and 4,795 controls. The analysis identified 9 significant loci, including a novel locus on 12q23.2 (rs17033114, P = 6.12 × 10−25 with an OR of 1.177 (1.141-1.213), LINC00485). Subgroup analysis indicated that 5 loci (3q26.2, 5p15.33, 10q24.33, 11p15.5, 13q14.11) exhibited a statistically significant effect among multiple leiomyomas, and 2 loci (3q26.2, 10q24.33) exhibited a significant effect among submucous leiomyomas. Pleiotropic analysis indicated that all 9 loci were associated with at least one proliferative disease, suggesting the role of these loci in the common neoplastic pathway. Furthermore, the risk T allele of rs2251795 (3q26.2) was associated with longer telomere length in both normal and tumour tissues. Our findings elucidated the significance of genetic factors in the pathogenesis of leiomyoma.
Subject terms: Computational biology and bioinformatics, Genetics, Molecular biology, Diseases, Health care, Molecular medicine, Pathogenesis, Risk factors
Introduction
Uterine leiomyoma is one of the most common gynaecologic benign tumours. Its estimated lifetime risk is 30-50% in Japan1 and 70-80% in European populations2,3. Although leiomyoma is a benign neoplasm, patients exhibit many types of symptoms, such as vaginal bleeding, pelvic pain, or infertility4. Over 600,000 hysterectomies were performed per year in the USA due to leiomyomas, and 9.4 billion dollars for annual medical expenses were needed in the USA5. Therefore, leiomyomas are gaining much attention in health economics.
Leiomyoma growth is stimulated by sex steroids, such as oestrogen and progesterone, and several aetiological factors, such as age, early age at menarche, obesity, and parity, are linked to an increased leiomyoma risk2,6–11. In addition, there is some evidence for a genetic component of disease predisposition. African American women have a higher risk than European-American women2, and first-degree relatives of affected women have a 2.5-fold greater risk than population average12. We also reported that a positive family history is associated with a higher leiomyoma risk among Japanese individuals (odds ratio of 5.496 (5.061–5.969))13.
There have been six published genome-wide association studies (GWAS) of uterine leiomyoma, and some common single nucleotide polymorphisms (SNPs) associated with leiomyoma risks at 1p36.12 (CDC42/WNT4)14–17, 1q24.3 (DNM3)16, 2p25.1 (GREB1)14,15,17, 2p23.2 (BABAM2)17, 3p24.1 (NEK10)14, 3q26.2 (TERC/LRRC34)15,17, 3q2915, 4q12 (SCFD2/LNX1/PDGFRA)14,15,17, 4q13.3 (SULT1B1/SULT1E1)14,15,17, 4q22.3(PDLIM5)17, 5p15.33 (TERT)14,15,17, 5q35.2 (ZNF346)15,17, 6p21.31(GRM4/HMGA1)17, 6q25.2 (SYNE1/ESR1)14–17, 9p24.33 (KANK1/DMRT1/ANKRD15/LOC105375949)14–17, 10p11.22(ZEB1/ARHGAP12)17, 10q24.33 (OBFC1)14–18, 11p15.5 (SCGB1C1/BET1L/SIRT3/RIC8A)14–18, 11p14.1(FSHB)17, 11p13 (WT1/PDHX/CD44)14–17, 11q22.3 (ATM/C11orf65/KDELC2)14,15,17, 12q13.11 (SLC38A2/LOC100288798)15,17, 12q15(PTPRR)17, 12q24.31 (PITPNM2)17, 13q14.11 (LINC0/FOXO1)14,15,17, 16q12.1 (HEATR3/SALL1)15,16, 17p13.1 (TP53)14–17, 20p12.3 (MCM8)14,17, 20q13.13(LOC105372640)16, 22q13.1 (TNRC6B/CYTH4)14–19, Xq13.1 (TEX11/MED12)15,17, and Xq26.2 (RAP2C)15,17. In addition, uterine leiomyoma harbors genetic alteration of some driver genes including MED12 mutations, biallelic inactivation of FH, and HMGA2 rearrangements20–22. However, only a portion of the genetic variations of uterine leiomyoma could be explained. In a previous study using 1,607 Japanese patients, our group identified three risk loci for uterine leiomyomas on 10q24.33, 11p15.5, and 22q13.1. To gain insights into the genetic causes of uterine leiomyoma, here we performed a GWAS using 5,720 patients of Japanese ancestry.
Results
Genome-wide association study of uterine leiomyoma
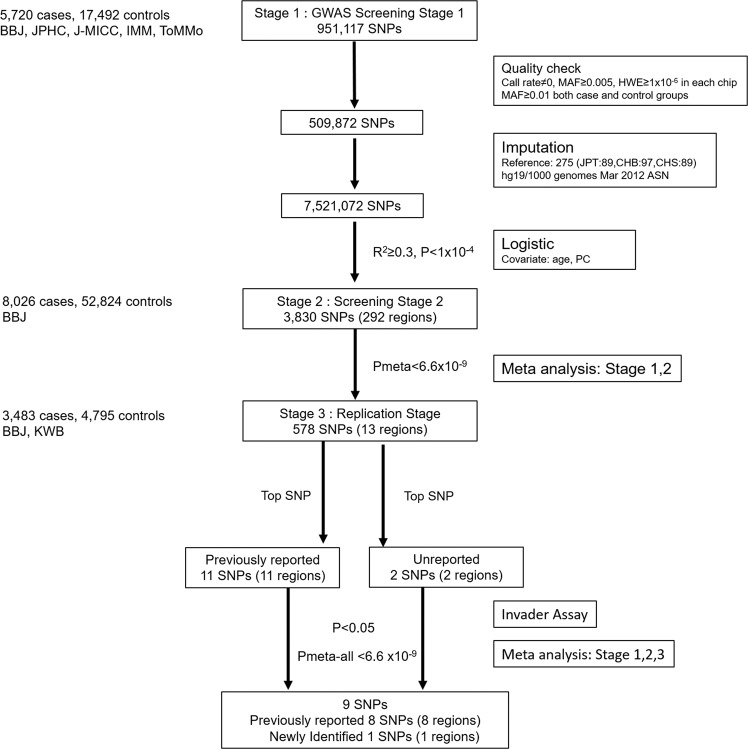
A total of DNA samples from 5,720 uterine leiomyoma patients and 17,492 controls were analysed in stage 1 (GWAS screening stage 1). All samples were genotyped using the Illumina OmniExpressExome or OmniExpress and HumanExome BeadChip in a previous analysis23. After performing a standard quality-control procedure, we conducted genome-wide imputation and obtained the genotyping results of 7,521,072 SNPs. A Manhattan plot of stage 1 analysis is shown in Supplemental Fig. S1. We found four loci (1p36.12, 10q24.33, 11p15.5, 22q13.1) with a genome-wide significance threshold. The genomic inflation factor λ was calculated to be 1.18 and 1.02 (λ1000) (Supplemental Fig. S2)24.
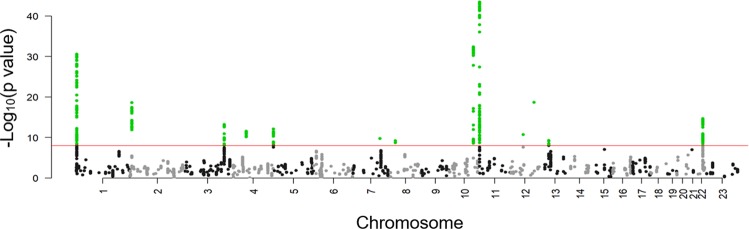
We selected 3,830 SNPs in 292 genomic regions with a suggestive association (P < 1.0 × 10−4) for further analysis in screening stage 2 (Fig. 1). DNA samples from 8,026 uterine leiomyoma patients and 52,824 controls that were genotyped in a previous analysis25 were used in stage 2. A Manhattan plot of the meta-analysis (Stage1 and 2) is shown in Fig. 2. As a result, 578 SNPs in 13 regions showed a significant association after Bonferroni correction (P < 0.05/7,521,072 = 6.6 × 10−9) (Supplemental Table S1).
Figure 1.
Study design of a GWAS of uterine leiomyoma. The meta-analysis comprised three stages.
Figure 2.
Manhattan plot of the meta-analysis of GWAS screening stage 1 and screening stage 2. The red horizontal line represents the genome-wide significance threshold of P = 6.6 × 10−9.
We performed further replication analysis of SNPs in 13 regions. We selected 13 SNPs from 578 SNPs in the replication analysis using the top SNPs in each region (Fig. 1). Thirteen SNPs were successfully genotyped by the invader assay, using 3,483 cases and 4,795 controls (Supplemental Table S2). We validated the association of nine SNPs in nine genomic regions (P < 0.05) with the same risk allele as those in the GWAS (Supplemental Fig. S3).
Meta-analysis of all three stages revealed nine SNPs associated with leiomyoma risk at the genome-wide significance level (P < 6.6 × 10−9) (Table 1 and Supplemental Table S3). These SNPs included rs3820282 (1p36.12), rs124793436 (2p25.1), rs2251795 (3q26.2), rs2242652 (5p15.33), rs75228775 (10q24.33), rs2280543 (11p15.5), rs17033114 (12q23.2), rs7989971 (13q14.11) and rs12484776 (22q13.1). A locus on 12q23.2 (rs17033114, P = 6.12 × 10−25 with an OR of 1.177 (1.141-1.213)) was newly identified. Except for rs12484776 at 22q13.1, no apparent heterogeneity among the three cohorts was found (Table 1 and Supplemental Table S3). Regional plots of the nine loci are shown in Supplemental Fig. S4.
Table 1.
Results for newly identified and previously reported risk variants associated with uterine leiomyoma at P < 6.6 × 10−9 in Japanese populations.
| SNP | Locus | Nearest Gene | Non Reference Allele | Reference Allele | GWAS Screening Stage 1 | Screening Stage 2 | Replication stage | Combined | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | Heterogeneity | ||||||
| Qa | I2 b | |||||||||||||||||
| Newly Identified Locus | ||||||||||||||||||
| rs17033114 | 12q23.2 | LINC00485 | T | C | 1.147 | 1.084-1.214 | 2.62E-06 | 1.185 | 1.135-1.237 | 2.25E-14 | 1.200 | 1.118-1.288 | 4.52E-07 | 1.177 | 1.141-1.213 | 6.12E-25 | 0.556 | 0.000 |
| Reported Loci | ||||||||||||||||||
| rs3820282 | 1p36.12 | WNT4 | C | T | 0.854 | 0.816-0.893 | 3.19E-12 | 0.853 | 0.825-0.882 | 5.42E-20 | 0.870 | 0.817-0.927 | 1.58E-05 | 0.856 | 0.835-0.877 | 3.49E-35 | 0.856 | 0.000 |
| rs12479436 | 2p25.1 | GREB1 | G | T | 1.094 | 1.046-1.144 | 6.45E-05 | 1.144 | 1.107-1.183 | 5.06E-15 | 1.072 | 1.006-1.141 | 3.09E-02 | 1.118 | 1.090-1.145 | 9.50E-19 | 0.109 | 54.970 |
| rs2251795 | 3q26.2 | MYNN | A | T | 0.882 | 0.843-0.923 | 5.89E-08 | 0.910 | 0.878-0.943 | 9.08E-08 | 0.933 | 0.874-0.996 | 3.83E-02 | 0.904 | 0.882-0.928 | 1.24E-14 | 0.333 | 8.950 |
| rs2242652 | 5p15.33 | TERT | G | A | 0.891 | 0.847-0.938 | 1.03E-05 | 0.900 | 0.865-0.936 | 1.70E-07 | 0.885 | 0.823-0.952 | 9.64E-04 | 0.895 | 0.870-0.921 | 2.42E-14 | 0.905 | 0.000 |
| rs75228775 | 10q24.33 | SLK | C | T | 0.755 | 0.698-0.817 | 1.28E-12 | 0.751 | 0.708-0.796 | 2.98E-21 | 0.813 | 0.726-0.910 | 3.22E-04 | 0.761 | 0.729-0.795 | 7.45E-35 | 0.465 | 0.000 |
| rs2280543 | 11p15.5 | SIRT3 | C | T | 1.393 | 1.296-1.498 | 3.99E-19 | 1.337 | 1.266-1.412 | 2.15E-25 | 1.279 | 1.157-1.414 | 1.54E-06 | 1.344 | 1.296-1.394 | 2.05E-47 | 0.384 | 0.000 |
| rs7989971 | 13q14.11 | FLJ42392 | C | G | 1.122 | 1.073-1.174 | 5.14E-07 | 1.067 | 1.030-1.105 | 2.23E-04 | 1.071 | 1.005-1.140 | 3.39E-02 | 1.085 | 1.058-1.113 | 3.33E-10 | 0.207 | 36.590 |
| rs12484776 | 22q13.1 | TNRC6B | A | G | 0.837 | 0.800-0.876 | 4.16E-15 | 0.937 | 0.905-0.971 | 2.11E-04 | 0.870 | 0.816-0.927 | 2.03E-05 | 0.893 | 0.871-0.916 | 4.63E-18 | 0.000 | 87.260 |
aQ for heterogeneity across all studies was calculated using Cochran’s Q test.
bI2, heterogeneity index.
Association of previously reported uterine leiomyoma loci in the Japanese population
We also analysed previously reported uterine leiomyoma loci in our screening dataset. We obtained the results of 45 SNPs among 70 SNPs in 33 genomic regions (Supplemental Table S4). We found that 10 SNPs in four regions showed genome-wide significance (P < 5.0 × 10−8) with the same risk allele as in previous studies. In addition, 34 SNPs showed a suggestive association (P < 0.05). 35 SNPs showed same risk allele with the previous study. These results indicate that multiple variations are associated with uterine leiomyoma risk irrespective of the ethnic background.
Association of previously reported somatic mutations in uterine leiomyoma
We also analysed SNPs on driver genes such as MED12, FH, and HMGA2 which harbor somatic mutations in uterine leiomyoma. We obtained the results of 270 SNPs in 3 genomic regions (Supplemental Table S5). Rs4360450 nearby MED12 at Xq13.1 was reported to be a novel candidate SNP, but this SNP is not polymorphic in the Japanese population17. Moreover, rs5937008 at Xq13.1 was not associated with any disease risks in our samples (P = 1.2 × 10−1)15. In addition, 19 SNPs on MED12, 36 SNPs on FH, and 215 SNPs on HMGA2 (lowest p-value of 0.236, 0.005013, and 0.001735, respectively) did not show significant association with leiomyoma. Thus, neither of these loci were associated with uterine leiomyoma in Japanese population.
Subgroup analysis of 9 identified loci
We conducted subgroup analysis using samples from KWB on these nine SNPs (Supplemental Fig. S5). Rs2251795, rs2242652, rs75228775, rs2280543, and rs7989971 exhibited statistically significant effects among multiple leiomyomas patients compared with single leiomyoma patients. Submucous leiomyoma causes serious symptoms such as infertility, abnormal genital bleeding, or hypermenorrhea26, and rs2251795 and rs75228775 were also associated with submucous leiomyoma compared with other two subtypes. Rs2280543 on 11p15.5 was associated with intramural leiomyoma27, but that trend was not validated in this study.
Pleiotropic association of 9 identified loci
To further investigate the role of these loci in another hormone-related uterine hyperplastic disease (endometriosis) and other malignant tumours (endometrial cancer, ovarian cancer, breast cancer, oesophageal cancer, gastric cancer, colorectal cancer, lung cancer and prostate cancer), we analysed these SNPs in other diseases (Supplemental Fig. S6). Rs3820282, rs12479436, rs75228775, rs2280543, and rs17033114 were associated with endometriosis (P < 0.05). In addition, rs2251795 at 3q26.2 (breast, oesophageal, colorectal, lung and prostate cancer), rs2242652 at 5p15.33 (breast, gastric, lung and prostate cancer), rs75228775 at 10q24.33 (lung and prostate cancer), and rs7989971 at 13q14.11 (oesophageal and colorectal cancer) were associated with multiple cancers. Thus, six among nine loci were associated with at least one malignant disease, suggesting the role of these loci in the common neoplastic pathway.
Association of identified loci with telomere length
The overall telomere length was significantly shorter in tumours than in adjacent matched myometrium (P = 2.35 × 10−5, Supplementary Fig. S7(a)), as previously reported15,28,29. Three regions among nine (3q26.2, 5p15.33, 10q24.33) were shown to be related to telomere length30,31. To evaluate the role of these genetic variations on telomere maintenance in uterine tissues, we analysed the association between variations in 3q26.2, 5p15.33, 10q24.33 and telomere length (Supplemental Table S6 and Supplemental Fig. S7(b,c)). As a result, the risk T allele of rs2251795 was significantly associated with longer relative telomere length in both normal and tumour tissues.
Discussion
Here, we reported 9 regions that were significantly associated with uterine leiomyoma in the Japanese population. In this study, we identified 12q23.2 as a novel locus.
Although six uterine leiomyoma GWAS have been reported14–19, recent studies were conducted in European or African American populations. Our group previously reported a GWAS of uterine leiomyoma18, but more than 17,000 cases and 75,000 controls were analysed in this study. This is one of the most comprehensive uterine leiomyoma studies performed in an Asian population.
MAF of rs17033114 is about 5% and 23% in European and Japanese population. In addition, our previous study of Japanese samples only used a small sample size of 1,607 at the screening stage18. These differences may lead to negative association of 12q23.2 in the previous studies. rs17033114 have been reported to be related with fetal birth weight32, suggesting the role of this SNP in cell growth or hormonal environment in uterine. The further study is needed to clarify the molecular mechanisms how this SNP regulates fetal birth weight and uterine leiomyoma risk.
Uterine leiomyoma is known to be an oestrogen- or progesterone-dependent tumour6,7. Furthermore, the WNT/β-catenin signalling pathway is considered to play an important role in tumorigenesis33–35. On 1p36.12, rs3820282 lies in the first intron of the WNT4. The risk T allele of rs3820282 was shown to strengthen the binding of ERα36. This possible ERα/WNT4 signalling is supported by the association of the same region with endometriosis, which is an oestrogen-responsive disease. Additionally, some intronic SNPs may change the secondary structure of RNAs and DNAs, and these changes may influence gene expression and protein binding, and leads to tumorigenesis37,38.
Both TERT and TERC play important roles in tumorigenesis and could be involved in various malignant tumours through telomere maintainance14. Rs2242652 at 5p15.33 is located on TERT, and rs2251795 at 3q26.2 is adjacent to TERC (telomerase RNA component). Our findings reveal that these two SNPs were associated with some malignancies, such as breast cancer. In addition, the T allele of rs2251795 is associated with longer relative telomere length in both tumour and normal tissues (P = 0.0225 and 0.0359). Although rs2251795 has not been reported in a GWAS study, rs12638862, rs12696304, rs2293607, rs10936599, rs1317082, and rs10936601 on 3p26.2 were reported to be correlated with telomere length39–42. These six SNPs exhibit strong linkage disequilibrium (LD) (R2 > 0.71) with rs2251795 in JPT. This result is concordant with the previous studies that indicated the association of rs2293607 (T allele) and rs10936599 (C allele) (absolute linkage equilibrium with T allele of rs2251795 in JPT) with longer leukocyte telomere length30,43. On the other hand, the previous study reported that the risk allele of rs2736100 on TERT (5p15.33) was associated with shorter telomere length, while TERC loci (rs10936600 on 3q26.2) was not associated with telomere length in leiomyoma tissues15. Although the effect of uterine leiomyoma risk allele on telomere length is inverted between TERT and TERC loci, the relationship between telomere length and neoplastic disease risk is contradictory44,45. In addition, different SNPs on TERC and TERT loci were analysed in our and previous studies by using relatively small (less than 50) samples15. These results indicated the important roles of telomere pathway in the pathogenesis of uterine leiomyoma. However, further analysis is necessary to fully elucidate the functional roles of these variations on uterine leiomyoma risk.
We also observed associations of rs2251795, rs2242652, rs75228775, rs2280543 and rs7989971 with multiple leiomyomas. In addition, rs2251795 and rs75228775 were significantly related to submucous leiomyoma. Although uterine leiomyoma is benign and has many symptoms, submucous leiomyoma especially causes infertility, abnormal genital bleeding, or hypermenorrhea26. Collecting the information of these variants in advance may enable clinical application and personalized medicine.
There is a limitation to this study. Patients in stage 2 and stage 3 from BBJ and controls in all stages were based on medical history obtained by questionnaire or medical records. Therefore, control samples could potentially have undiagnosed leiomyoma. However, four previous GWAS studies of uterine leiomyoma also used similar criteria for control samples14,17,18. This possible under-diagnosis of the controls may weaken the power of the GWAS, but this should not lead to false-positive results. Because many of previously reported loci were validated in our study, we considered that our case-control samples and statistical methods/results are reliable and valid.
In conclusion, we identified nine loci, including a novel locus in the Japanese population. Our study provides evidence for the possible role of telomeres in the aetiology of leiomyoma. Our findings contribute to the elucidation of the molecular pathogenesis of uterine leiomyoma.
Materials and Methods
Study participants
The characteristics of each cohort are shown in Table 2. Stage 1 included 5720 leiomyoma patients from the BioBank Japan (BBJ) cohort13,46. Stage 2 included 8,026 patients with a history of leiomyoma from BBJ, and stage 3 included 2,582 unrelated patients with a history of leiomyoma from BBJ and 901 leiomyoma patients from Keio Women’s Health Biobank (KWB). The diagnosis of uterine leiomyoma in stage 1 was confirmed by physicians. Patients in stage 2 and stage 3 from BBJ were based on medical history obtained by questionnaire or medical records. The diagnosis of uterine leiomyoma from KWB in stage 3 was based on ultrasound examination and/or magnetic resonance imaging (MRI). The 17,492 female controls for stage 1 were from three population-based cohorts, including the JPHC (Japan Public Health Center-based Prospective) Study47, the J-MICC (Japan Multi-Institutional Collaborative Cohort) Study48, IMM (Iwate Tohoku Medical Megabank Organization) and ToMMo (Tohoku University Tohoku Medical Megabank Organization)49. Controls for stage 2 and 3 were from BBJ. A total of 52,824 and 4,795 female controls without a history of leiomyoma and malignancies from BBJ were used as controls for stages 2 and 3, respectively. There were no exclusion criteria, and we used all available samples at the time of the experiment. Genomic DNA was extracted from peripheral blood leukocytes using a standard method. All participants provided informed consent, and this project was approved by the ethics committees at University of Tokyo, Keio University School of Medicine, RIKEN Center for Integrative Medical Sciences, Tohoku University, Iwate Medical University, Nagoya University Graduate School of Medicine and National Cancer Center. This study was conducted in accordance with the Declaration of Helsinki.
Table 2.
Selected characteristics of cases and controls in this study. Sample size after quality control and genotyping platforms in each of the studies included in this study.
| Stage | Sample Type | Source | Number | Age Mean ± Std | Genotyping Platform |
|---|---|---|---|---|---|
| Stage 1: GWAS screening Stage 1 | |||||
| case | BBJa | 5,720 | 44.8 ± 9.6 | OmniExpressExome BeadChip/OmniExpress and HumanExome | |
| control | JPHCb, J-MICCc, IMMd, ToMMoe | 17,492 | 55.9 ± 10.0 | OmniExpressExome | |
| Stage 2: Screening Stage 2 | |||||
| case | BBJa | 8,026 | 64.9 ± 10.5 | OmniExpressExome BeadChip/OmniExpress and HumanExome | |
| control | BBJa | 52,824 | 63.6 ± 15.7 | ||
| Stage 3: Replication Stage | |||||
| case | BBJa, KWBf | 3,483 | 63.8 ± 14.4 | Invader Assay | |
| control | BBJa | 4,795 | 65.9 ± 14.1 | ||
aBBJ; BioBank Japan, bJPHC; The Japan Public Health Center-based Prospective Study, cJ-MICC; Japan Multi-Institutional Collaborative Cohort Study, dIMM; Iwate Tohoku Medical Megabank Organization, eToMMo; Tohoku University Tohoku Medical Megabank Organization, fKWB; Keio Women's Health Biobank.
Genotyping and statistical analysis
In stage 1 and stage 2, all samples were genotyped for 951,117 SNPs with the Illumina HumanOmniExpressExome BeadChip or a combination of the Illumina HumanOmniExpress and HumanExome BeadChips (Table 2) in a previous study23. In stage 3, samples were genotyped by the multiplex PCR-based Invader assay (Third Wave Technologies)50 (Table 2). In stages 1 and 2, we excluded (i) samples with a call rate < 0.98, (ii) samples from closely related individuals identified by identity-by-descent analysis, (iii) sex-mismatched samples with a lack of information, and (iv) samples from non–East Asian outliers identified by principal component analysis of the studied samples and the three major reference populations (Africans, Europeans, and East Asians) in the International HapMap Project51. We then applied standard quality-control criteria for variants, excluding those with (i) SNP call rate < 0.99, (ii) minor allele frequency < 1%, and (iii) Hardy–Weinberg equilibrium P value < 1.0 × 10−6. We prephased the genotypes with MACH52 and imputed dosages with minimac and the 1000 Genomes Project Phase 1 (version 3) East Asian reference haplotypes53. Imputed SNPs with an imputation quality R2 < 0.3 were excluded from the subsequent association analysis.
In stage 1, we conducted a GWAS by a logistic regression model using PLINK and incorporating the top ten principal components as covariates. A total of 3,830 SNPs (P < 1 × 10−4) from stage 1 were evaluated using 60,850 samples in stage 2. In stage 2, a logistic regression model incorporating age and the top two principal components was used. In stage 3, we selected 13 SNPs within 13 regions in the following two steps (Fig. 1). First, we selected the top eleven SNPs for the previously reported eleven loci. For one SNP, it was difficult to make a probe for the Invader assay, and we selected a SNP with high LD (r2 > 0.8) as a substitute. Second, we selected two top SNPs within the newly identified two loci.
The meta-analysis was conducted using PLINK. Heterogeneity between studies was examined using Cochran’s Q test54. The threshold of heterogeneity was P < 0.05/13. We calculated the genomic inflation factor λGenomic Control (GC) in R. λGC adjusted to a sample size of 1,000 (λ1,000) was calculated using the following formula24, as large sample sizes cause inflated λGC values55: λ1,000 = 1 + (1−λobs) × (1/ncases + 1/ncontrols)/(1/1,000cases + 1/1,000controls). A quantile-quantile plot was drawn using R. A Manhattan plot of the associations was constructed by plotting −log10 (P values) against chromosome position using R. We generated regional plots with LocusZoom (v. 1.3)56. A forest plot was drawn using R.
Subgroup analysis
We used the subjects from KWB in stage 3 for the subgroup analysis. This study analysed the clinical factors: localization, number of tumours, and recurrence. Localization and number of uterine leiomyomas were assessed using ultrasound or MRI. Leiomyomas are classified into three subgroups based on their location within the layers of the uterus. Leiomyomas are classified as subserosal (found just below the uterine serosa), intramural (within the myometrium), or submucous (located just beneath the endometrium). The numbers of leiomyomas divided into two groups: one or more than two tumours. Each subgroup was analysed using a logistic regression model with an adjustment for age. Control samples were the same as those used in stage 3. Then, comparisons between subgroups were performed using a logistic regression model with an adjustment for age.
Pleiotropy association analysis
The association of 9 SNPs with other diseases was evaluated in the pleiotropy analysis. GWAS results in BioBank Japan for another hormone-related uterine disease (endometriosis (n = 705)) and malignant tumours (endometrial cancer (n = 931), ovarian cancer (n = 681), breast cancer (n = 5,272), oesophageal cancer (n = 1,225), colorectal cancer (n = 6,692), gastric cancer (n = 6,171), lung cancer (n = 3,874) and prostate cancer (n = 5,088))57–59 were used in this study.
Tumour specimens and nucleic acid extraction
We evaluated 16 pairs of uterine leiomyoma and corresponding normal myometrium tissue, 2 normal myometrium samples and 27 uterine leiomyoma samples. These samples were collected between 2016 and 2017 at the Keio University Hospital, Tokyo, at hysterectomy or myomectomy. All tumour tissues were examined histopathologically. Tissues were preserved in RNA later solution (QIAGEN) at 4 °C until RNA extraction. Collected tissues were homogenized in QIAzol lysis reagent (QIAGEN) using Precellys 24 (Bertin Corporation). Total RNA was extracted from the tissue samples with an AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN).
Measurement of relative telomere length by real‐time quantitative PCR (qPCR)
Relative telomere length was determined by the quantitative PCR method described by Cawthon60. The relative telomere length was calculated as the ratio of telomere repeats to a single-copy gene (T/S ratio). The single-copy gene refers to the 36B4 gene, which encodes the acid ribosomal phosphoprotein. For each sample, the quantity of telomere repeats and the quantity of the single-copy gene were determined by comparison to a reference. The normal sample that exhibited the highest copy numbers of telomere DNA among 18 normal samples was used as a reference in Telomere length qPCR analysis. The primers used for the telomere repeat copy number and the single-copy gene copy number amplification were as follows (written 5′ → 3′): telomere forward, CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT; telomere reverse, GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT; 36B4 forward, CAGCAAGTGGGAAGGTGTAATCC; 36B4 reverse, CCCATTCTATCATCAACGGGTACAA. The relative telomere length was calculated using the comparative 2-ΔΔCt method: ΔΔCt = ΔCt sample - ΔCtreference sample and ΔCtsample = CtTelomere - Ct36B4. The relationship between genotype and relative telomere length was examined by a linear regression analysis.
Supplementary information
Acknowledgements
We thank all the participants in this study. We are grateful for the staff of BioBank Japan, Tohoku Medical Megabank, Iwate Tohoku Medical Megabank, J-MICC, JPHC, and KWB for their outstanding assistance. We also thank Satoyo Oda and Akane Sei for their technical assistance. This study was partially supported by the BioBank Japan project and the Tohoku Medical Megabank project, which is supported by the Ministry of Education, Culture, Sports, Sciences and Technology Japan and the Japan Agency for Medical Research and Development. The JPHC Study has been supported by the National Cancer Center Research and Development Fund since 2011 and was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan from 1989 to 2010. The J-MICC Study was supported by Grants-in-Aid for Scientific Research for Priority Areas of Cancer (17015018) and Innovative Areas (221S0001) and the JSPS KAKENHI Grant (16H06277) from the Japan Ministry of Education, Science, Sports, Culture and Technology.
Author contributions
K.S. contributed in the conceptualisation, formal analysis, methodology, interpretation of the results and writing the original draft. C.Ta. contributed in the conceptualisation, formal analysis, methodology, interpretation of the results and supervision. These authors equally contributed to the paper. A.H., T.C., W.Y., F.K. and N.Su. contributed in methodology and sample collection. C.Te., Y.K., A.T., Y.M., M.H. and M.K. contributed in methodology and assisted with data analysis. N.F., T.T., A.S., A.F., A.K. and K.A. contributed in methodology, sample collection and data analysis. H.I., K.W., T.Y., N.Sa., M.I. and S.T. contributed in methodology. D.A. contributed in conceptualisation and supervised the project. K.M. supervised the project and critically revised the original manuscript. All authors reviewed and approved the manuscript.
Data availability
The genotyping results are available in National Bioscience Database center (JGAS00000000114 / https://humandbs.biosciencedbc.jp/). Clinical information are available from Biobank Japan (https://biobankjp.org/english/index.html) on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kensuke Sakai and Chizu Tanikawa.
Supplementary information
is available for this paper at 10.1038/s41598-020-58066-8.
References
- 1.Kawamura S, et al. Prevalence of uterine myoma detected by ultrasound examination in the atomic bomb survivors. Radiat. Res. 1997;147:753–758. doi: 10.2307/3579491. [DOI] [PubMed] [Google Scholar]
- 2.Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am. J. Obstet. Gynecol. 2003;188:100–107. doi: 10.1067/mob.2003.99. [DOI] [PubMed] [Google Scholar]
- 3.Catherino WH, Parrott E, Segars J. Proceedings from theNational Institute of Child Health and Human Development conference on the Uterine Fibroid Research Update Workshop. Fertil. Steril. 2011;95:9–12. doi: 10.1016/j.fertnstert.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta S, Jose J, Manyonda I. Clinical presentation of fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2008;22:615–626. doi: 10.1016/j.bpobgyn.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Cardozo ER, et al. The estimated annual cost of uterine leiomyomata in the United States. Am. J. Obstet. Gynecol. 2012;206:211.e1–9. doi: 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh EE, Bulun SE. Steroid hormones and leiomyomas. Obstet. Gynecol. Clin. North Am. 2006;33:59–67. doi: 10.1016/j.ogc.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa H, et al. Progesterone is essential for maintenance and growth of uterine leiomyoma. Endocrinology. 2010;151:2433–2442. doi: 10.1210/en.2009-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore AB, et al. Association of race, age and body mass index with gross pathology of uterine fibroids. J. Reprod. Med. 2008;53:90–96. [PubMed] [Google Scholar]
- 9.Dragomir AD, et al. Potential risk factors associated with subtypes of uterine leiomyomata. Reprod. Sci. 2010;17:1029–1035. doi: 10.1177/1933719110376979. [DOI] [PubMed] [Google Scholar]
- 10.Baird DD, Dunson DB. Why is parity protective for uterine fibroids? Epidemiology. 2003;14:247–250. doi: 10.1097/01.EDE.0000054360.61254.27. [DOI] [PubMed] [Google Scholar]
- 11.Laughlin SK, Hartmann KE, Baird DD. Postpartum factors and natural fibroid regression. Am. J. Obstet. Gynecol. 2011;204:496.e1–6. doi: 10.1016/j.ajog.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vikhlyaeva EM, Khodzhaeva ZS, Fantschenko ND. Familial predisposition to uterine leiomyomas. Int. J. Gynaecol. Obstet. 1995;51:127–131. doi: 10.1016/0020-7292(95)02533-I. [DOI] [PubMed] [Google Scholar]
- 13.Hirata M, et al. Cross-sectional analysis of BioBank Japan clinical data: A large cohort of 200,000 patients with 47 common diseases. J. Epidemiol. 2017;27:S9–S21. doi: 10.1016/j.je.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rafnar T, et al. Variants associating with uterine leiomyoma highlight genetic background shared by various cancers and hormone-related traits. Nat. Commun. 2018;9:3636. doi: 10.1038/s41467-018-05428-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Välimäki, N. et al. Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability. Elife7, (2018). [DOI] [PMC free article] [PubMed]
- 16.Edwards TL, et al. A Trans-Ethnic Genome-Wide Association Study of Uterine Fibroids. Front. Genet. 2019;10:511. doi: 10.3389/fgene.2019.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher CS, et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 2019;10:4857. doi: 10.1038/s41467-019-12536-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cha P-C, et al. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat. Genet. 2011;43:447–450. doi: 10.1038/ng.805. [DOI] [PubMed] [Google Scholar]
- 19.Hellwege JN, et al. A multi-stage genome-wide association study of uterine fibroids in African Americans. Hum. Genet. 2017;136:1363–1373. doi: 10.1007/s00439-017-1836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mäkinen N, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 21.Lehtonen R, et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am. J. Pathol. 2004;164:17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 23.Akiyama M, et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017;49:1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 24.Freedman ML, et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 25.Tanikawa C, et al. Novel Risk Loci Identified in a Genome-Wide Association Study of Urolithiasis in a Japanese Population. J. Am. Soc. Nephrol. 2019;30:855–864. doi: 10.1681/ASN.2018090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J-H, et al. Impact of submucous myoma on the severity of anemia. Fertil. Steril. 2011;95:1769–72.e1. doi: 10.1016/j.fertnstert.2011.01.142. [DOI] [PubMed] [Google Scholar]
- 27.Edwards TL, Hartmann KE, Velez Edwards DR. Variants in BET1L and TNRC6B associate with increasing fibroid volume and fibroid type among European Americans. Hum. Genet. 2013;132:1361–1369. doi: 10.1007/s00439-013-1340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogalla P, et al. Telomere repeat fragment sizes do not limit the growth potential of uterine leiomyomas. Biochem. Biophys. Res. Commun. 1995;211:175–182. doi: 10.1006/bbrc.1995.1793. [DOI] [PubMed] [Google Scholar]
- 29.Bonatz G, et al. Telomere shortening in uterine leiomyomas. Am. J. Obstet. Gynecol. 1998;179:591–596. doi: 10.1016/S0002-9378(98)70050-X. [DOI] [PubMed] [Google Scholar]
- 30.Codd, V. et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45, 422–7, 427e1–2 (2013). [DOI] [PMC free article] [PubMed]
- 31.Liu Y, et al. A genome-wide association study identifies a locus on TERT for mean telomere length in Han Chinese. PLoS One. 2014;9:e85043. doi: 10.1371/journal.pone.0085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warrington NM, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, et al. Paracrine activation of WNT/β-catenin pathway in uterine leiomyoma stem cells promotes tumor growth. Proc. Natl. Acad. Sci. USA. 2013;110:17053–17058. doi: 10.1073/pnas.1313650110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono M, et al. Inhibition of canonical WNT signaling attenuates human leiomyoma cell growth. Fertil. Steril. 2014;101:1441–1449. doi: 10.1016/j.fertnstert.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono M, Bulun SE, Maruyama T. Tissue-specific stem cells in the myometrium and tumor-initiating cells in leiomyoma. Biol. Reprod. 2014;91:149. doi: 10.1095/biolreprod.114.123794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, et al. Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. N. Engl. J. Med. 2017;377:1156–1167. doi: 10.1056/NEJMoa1612665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu L, et al. Functional study of risk loci of stem cell-associated gene lin-28B and associations with disease survival outcomes in epithelial ovarian cancer. Carcinogenesis. 2012;33:2119–2125. doi: 10.1093/carcin/bgs243. [DOI] [PubMed] [Google Scholar]
- 38.Lu L, et al. An insulin-like growth factor-II intronic variant affects local DNA conformation and ovarian cancer survival. Carcinogenesis. 2013;34:2024–2030. doi: 10.1093/carcin/bgt168. [DOI] [PubMed] [Google Scholar]
- 39.Codd V, et al. Common variants near TERC are associated with mean telomere length. Nat. Genet. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangino M, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012;21:5385–5394. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pooley KA, et al. A genome-wide association scan (GWAS) for mean telomere length within the COGS project: identified loci show little association with hormone-related cancer risk. Hum. Mol. Genet. 2013;22:5056–5064. doi: 10.1093/hmg/ddt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prescott J, et al. Genome-wide association study of relative telomere length. PLoS One. 2011;6:e19635. doi: 10.1371/journal.pone.0019635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorajoo R, et al. Loci for human leukocyte telomere length in the Singaporean Chinese population and trans-ethnic genetic studies. Nat. Commun. 2019;10:2491. doi: 10.1038/s41467-019-10443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baird DM. Variation at the TERT locus and predisposition for cancer. Expert Rev. Mol. Med. 2010;12:e16. doi: 10.1017/S146239941000147X. [DOI] [PubMed] [Google Scholar]
- 45.Walsh KM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat. Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagai A, et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol. 2017;27:S2–S8. doi: 10.1016/j.je.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsugane S, Sawada N. The JPHC study: design and some findings on the typical Japanese diet. Jpn. J. Clin. Oncol. 2014;44:777–782. doi: 10.1093/jjco/hyu096. [DOI] [PubMed] [Google Scholar]
- 48.Hamajima N. & J-MICC Study Group. The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pac. J. Cancer Prev. 2007;8:317–323. [PubMed] [Google Scholar]
- 49.Kuriyama S, et al. The Tohoku Medical Megabank Project: Design and Mission. J. Epidemiol. 2016;26:493–511. doi: 10.2188/jea.JE20150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohnishi Y, et al. A high-throughput SNP typing system for genome-wide association studies. J. Hum. Genet. 2001;46:471–477. doi: 10.1007/s100380170047. [DOI] [PubMed] [Google Scholar]
- 51.International HapMap 3 Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Genet. Epidemiol. 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes; pp. 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breslow, N. E. & Day, N. E. Statistical methods in cancer research. Volume II–The design and analysis of cohort studies. IARC Sci. Publ. 1–406 (1987). [PubMed]
- 55.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanikawa C, et al. GWAS identifies two novel colorectal cancer loci at 16q24.1 and 20q13.12. Carcinogenesis. 2018;39:652–660. doi: 10.1093/carcin/bgy026. [DOI] [PubMed] [Google Scholar]
- 58.Tanikawa C, et al. Genome-wide association study identifies gastric cancer susceptibility loci at 12q24.11-12 and 20q11.21. Cancer Sci. 2018;109:4015–4024. doi: 10.1111/cas.13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanai M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 60.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotyping results are available in National Bioscience Database center (JGAS00000000114 / https://humandbs.biosciencedbc.jp/). Clinical information are available from Biobank Japan (https://biobankjp.org/english/index.html) on reasonable request.