Abstract
Ovarian cancer (OC) accounts for more than 150,000 deaths worldwide every year. Patients are often diagnosed at an advanced stage with metastatic dissemination. Although platinum- and taxane-based chemotherapies are effective treatment options, they are rarely curative and eventually, the disease will progress due to acquired resistance. Emerging evidence suggests a crucial role of long non-coding RNAs (lncRNAs) in the response to therapy in OC. Transcriptome profiling studies using high throughput approaches have identified differential expression patterns of lncRNAs associated with disease recurrence. Furthermore, several aberrantly expressed lncRNAs in resistant OC cells have been related to increased cell division, improved DNA repair, up-regulation of drug transporters or reduced susceptibility to apoptotic stimuli, supporting their involvement in acquired resistance. In this review, we will discuss the key aspects of lncRNAs associated with the development of resistance to platinum- and taxane-based chemotherapy in OC. The molecular landscape of OC will be introduced, to provide a background for understanding the role of lncRNAs in the acquisition of malignant properties. We will focus on the interplay between lncRNAs and molecular pathways affecting drug response to evaluate their impact on treatment resistance. Additionally, we will discuss the prospects of using lncRNAs as biomarkers or targets for precision medicine in OC. Although there is still plenty to learn about lncRNAs and technical challenges to be solved, the evidence of their involvement in OC and the development of acquired resistance are compelling and warrant further investigation for clinical applications.
Keywords: ovarian cancer, lncRNA, drug resistance, chemotherapy, precision medicine
Introduction
Ovarian cancer (OC) is the fifth most lethal cancer in women and accounts for more than 150,000 deaths annually worldwide (1). According to molecular and pathological features, epithelial OCs are stratified into type I or type II (2). Type I OC's (including endometrioid, clear cell, mucinous, and low-grade serous carcinomas) are genetically stable with frequent mutations in KRAS, BRAF, CTNNB1, and PTEN. In contrast, type II (mainly HGSC) comprises more aggressive tumors with high-grade and propensity for invasion and metastasis leading to high mortality rates (3). These tumors are genetically unstable, presenting a high frequency of TP53 mutations and BRCA1/2 alterations. Originally HGSC was thought to arise from the squamous epithelial cell layer of the ovary. However, recent findings demonstrate that the molecular profile of HGSCs has a closer resemblance to the epithelium of the distal fallopian tube, suggesting that this tissue is an alternative site of origin (4, 5). HGSC is the most common and deadliest type of OC and will be the main focus of this review.
Due to the aggressive and invasive nature of HGSC around 70% of the patients have metastatic disease (FIGO stage III-IV) at the time of diagnosis. Surgery combined with chemotherapy is the primary treatment. Platinum-based chemotherapy is the cornerstone of chemotherapeutic treatment, namely cisplatin or carboplatin, combined with a taxane, such as paclitaxel or docetaxel (6). Initially, most patients respond well to the treatment; however, the majority of them will eventually acquire resistance and experience relapse (7, 8). To improve the prognosis, targeted therapies can be applied either as adjuvant or second-line treatments. Bevacizumab, an inhibitor or of vascular endothelial growth factor (VEGF) can be administered as first-line treatment in combination with carboplatin and paclitaxel. Inhibitors of Poly (ADP-ribose) polymerase (PARP) proteins are often used as second-line treatment for recurrent disease, mainly in patients with BRCA mutations. A recent randomized phase 3 trial performed in patients with a germline BRCA mutation has shown that the addition of oral PARP inhibitor (Olaparib) as maintenance therapy after chemotherapy prolongs the median progression free survival (PFS) by at least 3 years (9).
Despite the comprehensive combination of chemotherapy and maintenance treatment with targeted therapies, most patients develop resistance to treatment. Consequently, patients with disseminated HGSC have an extremely poor prognosis with a 5-year survival rate of only ~20% (10). The knowledge of the underlying molecular mechanisms involved in the development of resistance to chemotherapy is crucial for treatment decisions and the discovery of novel anticancer drug targets.
Advances in sequencing technologies and large-scale genomic projects such as Encyclopedia of DNA elements (ENCODE) (11) and The Cancer Genome Atlas Program (TCGA) (12) have opened avenues to improve our understanding of the mechanisms of response to treatment, development of therapeutic resistance and cancer progression (13–15). Initial studies focused on describing the small percentage of DNA transcribed into RNA encoding for proteins, whereas the non-coding RNA (ncRNA) was regarded as irrelevant and with unknown function for cellular health and disease. However, compelling evidence now reveals the involvement of these transcripts in the regulation of several cellular processes (16, 17). Furthermore, several cancer types have been associated with dysregulated expression of lncRNAs (18).
LncRNAs in Cancer
NcRNA comprises several different classes of molecules involved in gene regulation and chromatin modification. MicroRNA (miRNA), endogenous small interfering RNA (endo-siRNA) and piwi-interacting RNA (piRNA) are different classes of small ncRNAs involved in heterochromatin formation, histone modification, DNA methylation targeting, and gene silencing. Long non-coding RNAs (lncRNAs) are a subclass of non-translated RNA-sequences defined by an arbitrary length of more than 200 base pairs. These structurally complex RNA molecules interact directly with both DNA, RNA, and proteins affecting various cellular processes including genomic imprinting, gene transcription, mRNA splicing and protein activity (19–21). We are only beginning to understand how these molecules regulate cellular function, and how dysregulation can lead to malignant transformation.
The majority of lncRNAs are physically located in the proximity of protein-coding genes. Furthermore, lncRNAs are often classified according to their position relative to those genes as sense, antisense, intronic, intergenic, and bidirectional (22). Their expression levels are usually low and often compartmentalized to the cytoplasm or nucleus (23). Many lncRNAs exhibit low inter-species homology, and their expression signatures are often tissue-specific, indicating the importance of lncRNAs in cellular differentiation and embryonic development (24–26).
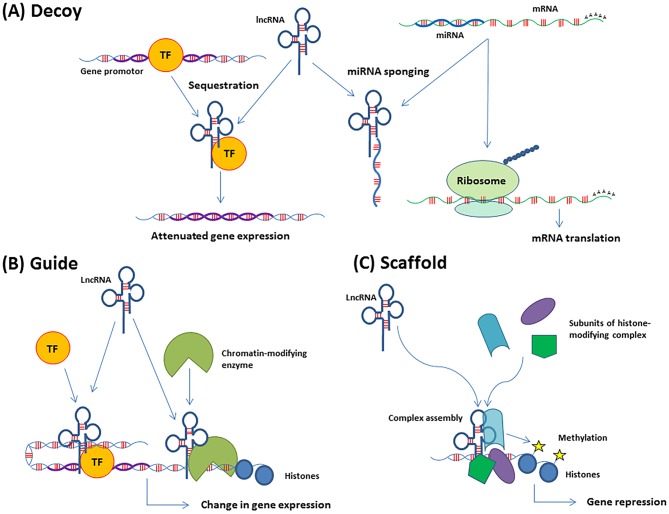
The lncRNA mechanisms of action usually fall into three categories, decoys, guides, or scaffolds (Figure 1). The decoys function as competing endogenous RNAs (ceRNAs) and modulate gene expression by sequestering transcription factors or miRNAs (also called sponging). Consequently, the availability of the targeted molecule is limited and the downstream effect reduced. The guide lncRNAs help to localize transcription factors or chromatin modifiers to specific areas of the genome, whereby transcription can be modulated. Dynamic scaffolds support transient assembly of protein complexes that bind genomic regions to affect chromatin structure (27, 28). The functions are not mutually exclusive, and many lncRNAs have more than one function. The single-stranded circular RNA (circRNA) is a sub-group of lncRNA recently discovered (29). Although the function of circRNA is still poorly understood, evidence indicates a role in miRNA regulation by sponging and intracellular transportation. LncRNAs are also stratified into cis- and trans-acting regulators, where the cis-regulators, exert their effect on neighboring genes on the allele from which they are transcribed, and the trans-regulators control gene expression at distant genomic sites.
Figure 1.
Functions of lncRNAs in gene regulation. (A) Decoys can sequester transcription factors (TF) or complementary RNA transcripts, such as miRNAs (also called miRNA sponging). The consequence of TF sequestration is attenuated expression of the genes regulated by that TF. The effect of miRNA sponging is the release of the molecule e.g., mRNA which is targeted by that miRNA. The mRNA is then translated. (B) Guides recruit molecules, such as TFs or chromatin-modifying enzymes to their target areas of the genome, which leads to the regulation of gene expression. (C) Scaffolds support transient assembly of protein complexes at genomic regions, which can promote histone modifications and DNA methylation.
Unsurprisingly, aberrant expression of lncRNAs has been associated with several diseases, including cancer. Dysregulated lncRNAs can exert oncogenic or tumor suppressor functions through transcriptional regulation impacting cellular proliferation, differentiation, invasiveness, apoptosis, and metabolism (30).
Many cancer-associated lncRNAs display similar expression patterns in different cancer types. Overexpression of Hox transcript antisense RNA (HOTAIR) was first described in breast cancer, where it was associated with increased invasiveness and metastasis (31). Subsequent studies revealed an association of increased expression of HOTAIR with disease progression and poor prognosis in colorectal (32), non-small cell lung (33), hepatocellular (34–36), gastric (37, 38), pancreatic (39), and ovarian (40) carcinomas. Single nucleotide polymorphisms (SNPs) in HOTAIR were recently correlated with increased susceptibility to develop OC in a Chinese population (41, 42). In ovarian cancer, HOTAIR overexpression was associated with poor differentiation, advanced FIGO stage and lymph node metastasis (40, 43).
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is another lncRNA widely overexpressed in various solid tumor types (44, 45), including OC (46–49). Several studies on OC cell lines showed that depletion of MALAT1 suppresses viability, proliferation, migration, and invasion (46, 47, 50). MALAT1 is highly conserved among mammals and is primarily known to localize to nuclear splicing speckles, where it interacts with splicing factors to regulate alternative splicing (51). In OC, MALAT1 was demonstrated to suppress alternative splicing of pro-apoptotic factors, causing apoptotic and anoikis resistance (50).
HOTAIR and MALAT1 are examples of widely expressed lncRNAs with oncogenic potential. Several other well-studied lncRNAs are found to be involved in the regulation of cellular processes such as proliferation, genomic stability, metabolism, and apoptosis to ensure homeostasis. These functions are executed through the lncRNAs directly or indirectly influence on the transcription of various proteins, which can lead to context-dependent oncogenic or tumor-suppressive properties. For a comprehensive overview of lncRNA's involved in cancer see (52–54).
A better understanding of the interplay between coding- and non-coding RNA and the integration of more molecular markers could potentially improve the predictive value of the molecular subtypes and provide a stronger tool for personalized therapeutic approaches.
The Molecular Landscape of HGSC
The most prominent molecular feature of HGSC is high genomic instability (55), possibly initiated by TP53 dysregulation and its associated effects in DNA damage repair (56). Mutations of TP53 were reported in up to 96% of HGSC cases, mostly missense mutations (70.4%), which can result in a dominant-negative effect, gain or loss of protein function. Frameshift (12%), non-sense (8.67%), and splice site (5%) mutations, leading to loss of protein function have also been described (57). Only a few other genes were reported as commonly mutated in HGSC, including BRCA1 (12.5%) and BRCA2 (11.5%) (58).
Genetic predisposition is recognized in a minority of the patients with HGSC, with around 70% of familial cases presenting inherited pathogenic mutations in BRCA1 and BRCA2 (59). These mutations contribute to an increased risk of developing ovarian cancer (44% for BRCA1 and 27% for BRCA2 carriers), compared to the normal population. Mutations in other genes with low penetrance also have an important role in ovarian cancer development. The increased lifetime risk for women harboring mutations in genes involved in the DNA damage repair by homologous recombination (HR), such as BRIP1 (5.8%) (60), RAD51C (5.2%), and RAD51D (12%) have been reported in OC (61). Alterations in genes involved in DNA mismatch repair associated with Lynch syndrome (MSH2, MLH1, PMS2, and MSH6), in rare cases prompt HGSC development (59, 62, 63).
The deficiency in DNA damage repair pathways is compatible with the high genomic instability observed in epithelial OC, with copy number alterations (CNA) affecting a significant fraction of the genome. Recurrent focal amplification of CCNE1, MYC, and MECOM genes are frequently identified in the TCGA cohort (58). Cases showing CCNE1 amplification are mutually exclusive with BRCA mutated cases suggesting the involvement of different pathways in the tumorigenesis of HGSC (58). Deficiency of the HR pathway was described in around 50% of HGSC cases, which has been associated with BRCA1 (20% of cases) and BRCA2 (5%) germline or somatic mutations and, BRCA1 promoter hypermethylation (10%). Genomic alterations in other genes involved in the HR repair pathway, such as amplification or mutation of EMSY (8%), focal deletion or mutation of PTEN (7%), hypermethylation of RAD51C (3%), mutation of ATM/ATR (2%), and mutation of Fanconi Anemia genes (58) have also been reported.
Some sporadic ovarian tumors share the phenotypic traits with tumors harboring germline mutations in BRCA1/2 (BRCAness phenotype), which may reflect molecular similarities. The BRCAness phenotype predicts responsiveness to platinum-based chemotherapy (64) and PARP inhibitors (65). In a population-based study that evaluated the mutational profile of HR genes, a better overall survival in BRCAness patients was described (66). Another approach to identifying HR deficiency was performed based on scores of the CNA profile of tumors, named “genomic scars,” which was very high in HGSC, and also correlated to PARP inhibitors or platinum-based chemotherapy sensitivity (67).
The integrative analysis of CNA, mutations, and gene expression alterations of HGSC identified RB1 and PI3K/Ras pathways deregulated in 67% and 45% of the cases, respectively (58). Amplification of PIK3CA, PIK3CB, and PIK3K4 was correlated to the decreased overall survival of OC patients. The analysis of PIK3CA protein product p110α and p-Akt confirmed the involvement of the PIK3/AKT pathway in survival (68). The PIK3A/AKT/mTOR pathway was also shown to be implicated in therapy resistance. Advanced OC patients who did not respond to subsequent chemotherapy presented activation of the pathway compared to responsive patients (69). Besides, GAB2, a signaling intermediate of PI3K and MAPK pathways, was reported as amplified in 44% of ovarian cancer samples (70). Although HGSC rarely exhibits mutations in KRAS or BRAF, the main activators of the MAPK pathways, almost half of tumors display an expression of active downstream MAPKs (71).
In addition to the specific pathways and genes altered in HGSC, distinct molecular subtypes were identified based on the differential expression profiles (72). The expression analysis of 489 tumors performed by TCGA and compared to an external cohort revealed four HGSC subtypes: proliferative, mesenchymal, immunoreactive, and differentiated (58). The proliferative subtype was characterized by low expression of ovarian tumor markers and high expression of transcription factors and proliferation markers. The mesenchymal subtype presented a high expression of HOX genes and markers suggestive of increased stromal components. The immunoreactive subtype was characterized by T-cell chemokine ligands, CXCL11 and CXCL10, and the receptor, CXCR3. The differentiated subtype was related to high expression of MUC16, MUC1, and SLPI (the secretory fallopian tube maker), suggesting a more mature stage of development (58). Patients with the HGSC immunoreactive subtype presented better prognosis, while patients with the mesenchymal or proliferative subtypes showed worse overall survival (73, 74).
LncRNAs Signature of Ovarian Cancer
Lately, the predictive value of differentially expressed lncRNAs has received increased attention due to their presence in liquid biopsies and potential as biomarkers for therapeutic response and prognosis (49, 75, 76). A meta-analysis including 1,333 OC patients established that altered lncRNAs are, in general, associated with decreased overall survival (76). In this analysis, 11 lncRNAs (HOTAIR, TC010441, AB073614, ANRIL, MALAT1, NEAT1, CCAT2, UCA1, HOXA11-AS, SPRY4-IT1, and ZFAS1) were identified with significantly increased expression in OC patients (76). Additional data have been reported to support the role of lncRNAs in OC. Eight lncRNAs were significantly correlated with overall survival, in a comprehensive analysis of lncRNA expression profiles of 544 OC patients from TCGA (75). Six of them (RP4-799P18.3, RP11-57P19.1, RP11-307C12.11, RP11-254I22.1, RP1-223E5.4, and GACAT3) were positively correlated with overall survival, while the last two (PTPRD-AS1 and RP11-80H5.7) were inversely correlated. The eight-lncRNA signature showed prognostic value and was able to stratify patients according to clinical outcome into high- and low-risk groups. Furthermore, this signature demonstrated predictive value for the response to platinum-based chemotherapy (75). A prognostic signature was identified for recurrent disease based on datasets extracted from the Gene Expression Omnibus (GEO). The signature comprised four well-known cancer-related lncRNAs, RUNX1-IT1, MALAT1, H19, and HOTAIRM1, and two less well-described transcripts LOC100190986 and AL132709.8. These lncRNAs were confirmed as differentially expressed in validation cohorts independently of tumor stage, tumor grade and histology type (49).
In silico analysis of RNA sequencing data derived from 391 patients retrieved from TCGA revealed three additional lncRNAs (NBR2, ZNF883, and WT1-AS) associated with recurrent OC (77). Based on the results, two interesting interactions were predicted; WT1-AS-miR-375-RBPMS and WT1-AS-miR-27b-TP53, suggesting that WT1-AS regulates two important tumor suppressors RBPMS and TP53, through miRNA sponging (77).
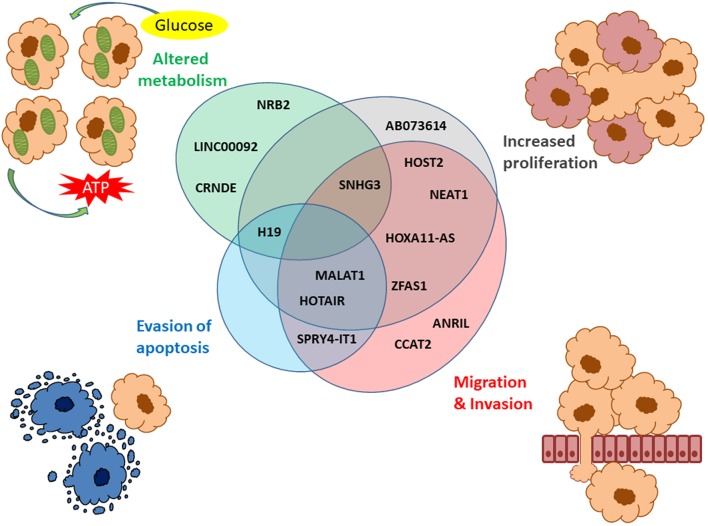
Several lncRNAs identified in large-scale studies have also been validated individually (46, 78–82), and across different cancer types (83). Also, functional studies have revealed that many of the dysregulated lncRNAs associated with OC are involved in one or several hallmarks of cancer such as increased proliferation, altered metabolism, evasion of apoptosis, migration or invasion (Figure 2) (79, 84–86).
Figure 2.
LncRNAs described in OC associated with the hallmarks of cancer. The hallmarks of cancer include increased proliferation (gray), migration/invasion (red), evasion of apoptosis (blue), and altered metabolism (green). The Venn diagram shows the lncRNAs associated with these four hallmarks with several of them involved in more than one hallmark of cancer.
Two lncRNAs are described specifically in ovarian cancer, ovarian adenocarcinoma amplified (OVAL) (87) and human ovarian cancer-specific transcript 2 (HOST2) (88). An intergenic region encompassing the full OVAL gene was found amplified in higher frequency in OC patients in comparison to other cancers. OVAL amplification and its increased expression suggest an oncogenic function in OC (87). So far no mechanisms or functional interactions have been described for OVAL. The expression of HOST2 is dramatically increased in OC tissues and cell lines, compared to normal ovarian tissues and non-ovarian cell lines (88). Furthermore, HOST2 was associated with increased proliferation, migration and invasion in OC. The mechanism of action of this potential driver is suggested to be through sequestration of miRNA let-7b, which is known to promote the expression of several oncogenes (89).
The involvement of dysregulated lncRNAs in the development of OC is well-documented. Considering the described oncogenic and tumor suppressor functions of lncRNA, their role in the development of resistance to therapy is expected (90–92). Differential lncRNA expression profiles were demonstrated in cisplatin-resistant and cisplatin sensitive OC, supporting their role in acquired resistance to chemotherapy (93). However, the mechanism by which lncRNAs contribute to acquired resistance remains incompletely understood. More detailed insights might lead to discoveries of new biomarkers or therapeutic targets. Signatures with the potential to predict therapeutic resistance would be valuable tools for clinicians, aiding the selection of the optimal treatment strategies for individual patients. In the following section, the involvement of lncRNAs in the development of therapeutic resistance will be highlighted, with a focus on platinum and taxane-based treatment regimens.
LncRNAs Involved in Platinum-Resistance
The development of anti-cancer drug resistance is often complex and multifactorial, depending on the specific drug and the histological subtype of cancer. Carboplatin and cisplatin are the most commonly used drugs for the first-line treatment of advanced stages of HGSC. These platinum-based agents interact with DNA forming mono adducts or interstrand, intrastrand, and protein crosslinks mainly at guanine. The crosslinking affects DNA repair and synthesis and leads to the accumulation of single and double-strand breaks, which results in cell cycle arrest and apoptosis (94, 95). Moreover, the release of reactive oxygen species that activates inflammatory pathways may also contribute to the cytotoxic effects of these compounds (96).
Resistant clones can arise through a clonal selection of cells able to prevent, repair, or withstand DNA damage. The tumor suppressor p53 and its related nuclear transcription factors are important mediators of the cytotoxic effects of platinum therapy. DNA damage normally leads to a p53-dependent release of pro-apoptotic factors. Consequently, reduced activity of p53 or the related pathways is associated with platinum-resistance (97). Platinum-induced DNA damage can also be repaired by HR, and hence, the activity of the BRCA1/2 genes reduces the responsiveness to platinum therapy (98). In accordance, reversions of BRCA1/2 germline mutations have previously been reported as a mechanism of resistance to therapy (8). The mismatch repair system is another mechanism by which the cell can detect platinum induced lesions. Loss of mismatch repair-related genes such as MLH1 and MSH2 prevents the cells from recognizing the damage caused by platinum therapy. Consequently, apoptosis is not initiated and the cells are therefore less sensitive to the treatment (99, 100). Furthermore, platinum resistance was associated with epithelial to mesenchymal transition (EMT) (101), implying a range of molecular alterations promoting invasive properties and resistance to oncogene-induced senescence. The major pathways involved in EMT are TGF-β, HIF1-α, Wnt/β-catenin, and Notch, which have also been associated with platinum resistance (102, 103). General resistance mechanisms include reprogramming of metabolism and mitochondrial dysregulation, suppression of apoptotic mediators, up-regulated efflux pumps, reduced drug uptake, or intracellular detoxification (104). The molecular alterations leading to the platinum-resistant phenotype rarely include single nucleotide mutations in the known driver or resistance genes. Instead, it appears that the resistance arises from a highly patient-specific and adaptable pattern of altered methylation, gene amplifications, reversion of BRCA1/2 mutations, promotor fusion, and translocation, and differential expression of ncRNAs (8, 105, 106).
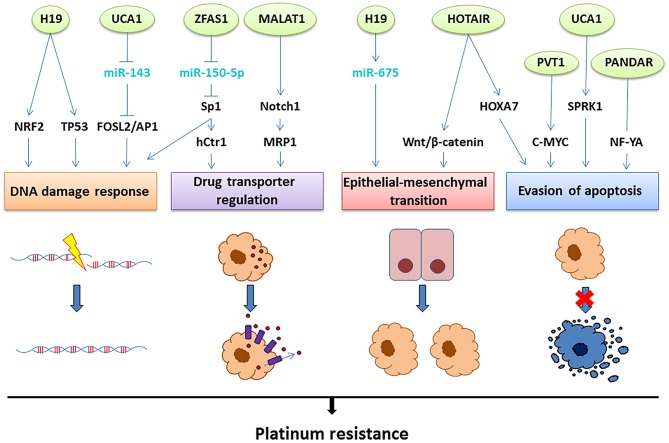
Although several lncRNAs have been associated with platinum resistance in OC (Table 1), the resistance-associated molecular mechanisms have been elucidated in only a few of them (Figure 3). Next, we will present the current knowledge of the functions and roles of a set of lncRNAs associated with platinum resistance in OC.
Table 1.
List of lncRNAs associated with platinum-resistance in ovarian cancer.
| lncRNA | Category | Expression in OC tissue* | Expression in platinum-resistant cell lines** | Mechanisms of resistance*** | References |
|---|---|---|---|---|---|
| HOTAIR | Antisense | ↑ (cisplatin resistance or treated with carboplatin) | ↑ (cisplatin/carboplatin) | ↑ HOXA7 | (107–110) |
| MALAT1 | Intergenic | ↑ | ↑ (cisplatin) | ↑ Notch1 → ↑ abcc1 | (111, 112) |
| H19 | Intergenic | ↑ (recurrent disease) | ↑ (cisplatin) | ↑ GSH pathway | (113) |
| ZFAS1 | Antisense | ↑ (platinum resistance) | ↑ (cisplatin) | ↓ miR-150-5p → ↑ SP1 | (80, 114) |
| UCA1 | Intergenic | ↑ (cisplatin resistance) | ↑ (cisplatin) | ↑ SRPK1 ↓ miR-143 → ↑ FOSL2 | (115–117) |
| PANDAR | Antisense | ↑ (disease recurrence + wt-p53) | ↑ (cisplatin) | ↓ NF-YA ↑ SFRS2 - ↓ p53 | (118) |
| PVT1 | Intergenic circRNA | ↑ (cisplatin resistance) | ↑ (cisplatin) | ↑ c-MYC | (119) |
| ZBED3-AS1 | Antisense | ↑ (platinum resistance) | ↑ (cisplatin/carboplatin) | N/A | (120) |
| F11-AS1 | Antisense | ↓ (platinum resistance) | ↓ (cisplatin/carboplatin) | N/A | (120) |
| GAS5 | Intergenic | ↓ (platinum resistance) | ↓ (cisplatin/carboplatin) | N/A | (120) |
| BC200 | Intergenic | ↓ | ↓ (carboplatin) | N/A | (121) |
| LINC00312 | Intergenic | ↓ (cisplatin+paclitaxel resistance) | ↓ (cisplatin) | ↑ Bcl-2/Caspase-3 pathway | (122) |
| BX641110 | N/A | ↑ (cisplatin) | N/A | (123) | |
| CRNDE | Intergenic | N/A | ↑ (cisplatin) | N/A | (123) |
| HOXC-AS3 | Antisense | N/A | ↑ (cisplatin) | N/A | (123) |
| RP11-384P7.7 | Intergenic | N/A | ↑ (cisplatin) | N/A | (123) |
| PLAC2 | Intronic | N/A | ↑ (cisplatin) | N/A | (123) |
| RP11-6N17 | Intergenic | N/A | ↑ (cisplatin) | N/A | (123) |
| RP11-65J3.1-002 | Intergenic | N/A | ↓ (cisplatin) | N/A | (123) |
| AC141928.1 | Intergenic | N/A | ↓ (cisplatin) | N/A | (123) |
| GS1-600G8.5 | Intergenic | N/A | ↓ (cisplatin) | N/A | (123) |
| SNHG15 | Intergenic | ↑ | ↑ (cisplatin) | N/A | (124) |
| EBIC | Processed pseudogene | ↑ | ↑ (cisplatin) | ↑ Wnt/β-catenin pathway | (125) |
The order of the lncRNAs corresponds to the appearance of the individual descriptions in the manuscript. Only the first 7 lncRNAs are described in detail in the manuscript, and are selected based on substantiating evidence in the literature.
The expression of the lncRNAs in OC tissue is indicated by arrows, ↑ for higher and ↓ for lower expression in platinum-resistant patients (patient characteristics indicated in parenthesis), compared to expression in platinum-sensitive patients. If no patient characteristics are indicated, the expression was determined in ovarian cancer tissue from patients with unspecified sensitivity to platinum drugs and normalized to adjacent or normal ovarian tissue.
The expression in resistant OC cell lines is indicated by arrows; ↑ for higher and ↓ for lower expression in platinum-resistant, compared to expression in platinum-sensitive cell lines. The drug the cell lines are resistant to is indicated in parenthesis.
The effect of lncRNAs on associated pathways, miRNAs, genes or transcription factors involved in resistance mechanisms are indicated by arrows: ↑ induction and ↓ repression.
N/A, information not available.
Figure 3.
The lncRNAs involved in platinum resistance in OC. Aberrant expression of the lncRNAs depicted on top (green circles) leads to platinum resistance through four main mechanisms: improved DNA damage response, upregulation of drug transporters leading to efflux of the drug, epithelial-mesenchymal transition or evasion of apoptosis. The molecular mechanisms suggested linking the lncRNAs to these resistance mechanisms involve interactions with miRNAs (light blue) and direct or indirect regulation of transcription factors and signaling pathways (black), as illustrated above.
HOTAIR located within the HOXC gene cluster (mapped on12q13.13) was previously introduced due to its involvement in OC. HOTAIR recruits lysine-specific demethylase 1 (LSD1) and Polycomb Repressive Complex 2 (PRC2) and guide them to promote epigenetic silencing of HOXD genes (126). Additionally, HOTAIR regulates other HOX genes including HOXA7, which is consistently overexpressed in several tumor types (40, 127, 128). The knockdown of HOTAIR led to reduced expression of HOXA7, which increased susceptibility to apoptosis and restored cisplatin sensitivity in resistant OC cells (107). In general, HOTAIR is more abundant in advanced OC tissues and was also overexpressed in cisplatin-resistant OC cell lines, compared to sensitive controls (108). Furthermore, HOTAIR expression was correlated with poor survival in patients who received carboplatin compared with untreated patients (109). Knockdown of HOTAIR in a mouse xenograft model, enhanced the effect of treatment with cisplatin, suggesting its potential as a target to re-sensitize ovarian cancer cells to platinum treatment. This effect has been attributed to reduced activation of the Wnt/β-catenin pathway, which is known to promote excess stem cell renewal and EMT (110). Thus, overexpression of HOTAIR might contribute to platinum resistance by increased transcription of HOXA7 and Wnt/β-catenin dependent induction of EMT. Overexpression of three additional lncRNAs DNM3OS, MEG3, and MIAT have been associated with EMT in ovarian cancer (129). However, the direct link between dysregulation of these transcripts and the development of platinum resistance is unexplored.
As previously mentioned, MALAT1 plays an important oncogenic role in multiple cancers (44, 45). Recently, MALAT1 has also been associated with resistance to therapy (130). In OC, MALAT1 knockdown increased cell death during treatment with cisplatin, indicating its potential involvement in resistance (111). MALAT1 was demonstrated to correlate with NOTCH1 expression, which is also up-regulated during platinum resistance in OC (112). NOTCH1 knockdown attenuates cisplatin resistance by directly down-regulating the expression of the multidrug resistance-associated protein 1 (ABCC1) in OC (131). ABCC1 encodes a transporter of molecules across cellular membranes, including the efflux of a range of drugs (132). In lung adenocarcinoma and colorectal cancer, MALAT1 promotes resistance to taxane- and platinum-based drugs, respectively. In these cases, EMT was identified as a mechanism of resistance (130, 133); however, this effect has not yet been investigated in OC.
The imprinted maternally expressed transcript H19 gene is located in a well-conserved gene cluster also containing the insulin-like growth factor 2 (IGF2). Both genes are regulated by genomic imprinting, and H19 is only transcribed from the maternal allele, whereupon it plays an important role in embryonic development and growth control (134). Aberrant expression of H19 has been demonstrated in several different cancers (135, 136); although its exact carcinogenic role is still under debate (137). The understanding of its function is challenged by the variety of transcriptional products deriving from the H19 gene locus and its complex regulation. As an example, the miR-675 is transcribed from the first exon of H19 (138) and has been associated with EMT and metastatic progression in colorectal and pancreatic cancers (139, 140). Furthermore, H19 is directly induced by the c-Myc oncogene (141) and its expression has been associated with the hypoxic stress response, involving p53 and hypoxia-inducible factor 1-α (HIF1-α) (142). Transcriptome analysis revealed differential expression of H19 in cisplatin-resistant OC cells compared to their sensitive progenitors. The involvement of H19 in platinum resistance was validated in tissues from 41 cases of HGSC treated with either cisplatin or carboplatin. The patients were divided into two groups according to their recurrence-free survival (threshold of 12 months), where H19 expression was shown to be significantly higher in patients with early recurrence (113). The role of H19 in cisplatin resistance was related to oxidative stress and induction of the glutathione (GSH) pathway, where H19 regulates several targets (GSR, G6PD, GCLC, GCLM, GSTP1, and NQO1) of the nuclear factor erythroid 2 (NRF2), an important factor in the antioxidant defense (113). The glutathione pathway has been suggested as a detoxifying mechanism to platinum-induced oxidative toxicity and is often up-regulated during the development of resistance (143, 144). H19 overexpression has also been correlated with cisplatin resistance in other cancers, including seminomas (145) and non-small cell lung cancer (146), where it was associated with evasion of apoptosis.
ZNFX1 antisense RNA 1 (ZFAS1) is transcribed from the antisense strand close to the protein-coding gene ZNFX1, including three C/D box small nucleolar RNAs (Snord12, Snord12b, and Snord12c) (147). The role of the lncRNA ZFAS1 (zinc finger antisense 1) varies among human cancers. ZFAS1 is downregulated in breast tumors compared to normal mammary tissue (147), whereas it is overexpressed in colorectal cancer (148), indicating tissue-specific functions. In OC, ZFAS1 overexpression was identified as part of an eight-lncRNA expression signature predictive of platinum-sensitivity, based on transcriptome data from 258 patients with HGSCs (114). The authors also found increased ZFAS1 expression in cisplatin-treated OC cell lines. Functional studies in OC cell lines revealed that the ZFAS1 knockdown resulted in increased sensitivity to cisplatin. This effect was shown to involve sequestration of miR-150-5p, which prevents binding of the transcription factor specific protein 1 (SP1) (80). SP1 has been appointed an important oncogene and a potential therapeutic target in several tumor types (149). Additionally, SP1 is involved in DNA damage response (150), and regulation of a copper transporter (hCtr1), which is associated with platinum drug transport (151).
Urothelial carcinoma associated 1 (UCA1) is expressed during embryonic development and subsequently abolished in most tissues, including ovarian epithelia. In OC tissue and several other cancers, UCA1 is re-activated and overexpressed (115, 152). In cancer tissues, UCA1 is regulated by HIF1-α, indicating its involvement in the response to hypoxia (153). Through sponging of miR-143, UCA1 prevents the repression of FOSL2, a subunit of the Activator protein 1 (AP-1) also involved in the hypoxic regulation. Consequently, UCA1 overexpression might lead to the up-regulation of the hypoxic response involving AP-1. Hypoxia has been shown to promote cisplatin resistance, through HIF1-α and p53 activation (154). A significant UCA1 overexpression in OC tissues from cisplatin-resistant patients (116) was reported. In vitro assays revealed that stable transfection of OC cells with UCA1 promotes resistance toward cisplatin (115). In addition, UCA1 was shown to affect the activity of the serine/arginine-rich protein-specific kinase 1 (SRPK1), an oncogene that suppresses apoptotic factors (115).
UCA1 is also associated with resistance to Paclitaxel in OC (155, 156), which can be reverted by UCA1 knockdown in cell lines (156). This effect was related to reduced sponging of miR-129 and, subsequently down-regulation of the ABCB1 gene that encodes an efflux pump previously correlated with multidrug resistance in cancer (156, 157). These findings suggested that the mechanism of resistance involving UCA1 is multifactorial, and could include improved response to DNA damage, reduced activation of apoptotic factors and increased efflux of the drugs.
The promoter of CDKN1A Antisense DNA damage Activated RNA (PANDAR) is a widely acknowledged oncogene mainly involved in regulating the response to DNA damage (158). The transcription of PANDAR is p53-dependent and promotes cell survival by impeding apoptosis through sequestering of the NF-YA transcription factor (159). In OC cell lines, an inverse relationship was demonstrated between PANDAR expression and cisplatin sensitivity. This effect involved interaction between PANDAR and the splicing factor arginine/serine-rich 2 (SFRS2), which led to negative feedback regulation of TP53 (118). Patients with wild type TP53 showed increased expression of PANDAR and SFRS2 at disease recurrence, compared to the time of diagnosis (118). The PANDAR-dependent suppression of p53 in resistant cells prevents the normal DNA damage response, whereby the cells can evade apoptosis. Since HGSCs have a very high occurrence of inactivating TP53 mutations, the role of PANDAR in platinum resistance remains to be elucidated.
Plasmacytoma variant translocation 1 (PVT1) is a well-established oncogene in OC (160, 161), as well as other cancers such as gastric (162) and breast (163). PVT1 is located in proximity to the MYC locus, where it encodes several alternative splice isoforms. In addition, the PVT1 locus contains a cluster of at least six miRNAs (miR-1204,−1205,−1206,−1207-3p,−1207-5p, and−1208) (164). Transcription of PVT1 can be regulated by p53 through a canonical binding site, indicating its involvement in the response to DNA damage. PVT1 is often co-expressed with MYC, with which it interacts and stabilizes to potentiate its activity (165). MYC has been suggested as a potential therapeutic target in platinum-resistant OC, as its overexpression confers resistance toward cisplatin (166). The role of PVT1 in the development of therapeutic resistance in OC is ambiguous since it was both demonstrated to promote cisplatin resistance by suppressing apoptotic factors (119), but also to be an effector in the cytotoxic response to treatment with carboplatin and docetaxel, by activating p53 and potentially promoting apoptosis (167). However, since p53 is often affected by the loss of function mutations in HGSC, the cytotoxic effect of PVT1 in response to carboplatin and docetaxel might be blunted in these cases. Studies in other cancers support the involvement of PVT1 in cisplatin resistance (168, 169). The opposing effects described for PVT1 could be due to the differences in the mechanisms of action of the two treatment regimens and underlines the need for further investigation.
Other mechanisms of resistance than the ones reported here have been suggested to involve lncRNAs. As an example, the lncRNAs MPRL (170), LINC00312 (122), and SNHG3 (84) are involved in mitochondrial function and altered expression of these have been associated with platinum resistance, either through effects on energy metabolism or mitochondrial-dependent apoptosis. In general, the interactions between lncRNA and the mitochondrial genome is not yet well-understood and should be further investigated.
LncRNAs Involved in the Taxane Resistance
Taxanes are microtubule-stabilizing agents that bind to the β-subunit of tubulin dimers to promote and stabilize polymerization. This mechanism inhibits microtubule disassembly that is a necessary event in mitosis; consequently, mitotic arrest and eventually apoptosis are promoted (171, 172). Paclitaxel is most often used in combination with the platinum-based chemotherapy as a first-line treatment, or as a single agent in platinum-resistant OC patients. Unfortunately, repeated exposure often leads to acquired resistance. Docetaxel, a second-generation taxane, can be used in some cases; however, shared resistance mechanisms result in low response rates. In vitro experiments have demonstrated that an inverse relationship exists between resistance to platins and taxanes, suggesting separate resistance mechanisms and emphasizing the benefits of combined treatments (173). The most common resistance mechanisms to taxanes comprise structural changes in the β-tubulin target region, altered expression of apoptotic and mitotic factors and overexpression of the multidrug resistance genes (ABC-transporters) (174, 175).
Since the taxanes are rarely used as a single agent in the treatment of OC, only a few studies have investigated the role of lncRNAs in the development of paclitaxel resistance in tissues. A combined analysis of two expression datasets comparing (1) patients with complete and incomplete response to chemotherapy and (2) two OC cell lines with paclitaxel resistance with two sensitive OC cell lines was performed. The combined analysis identified a panel of seven lncRNAs (XR_948297, XR_947831, XR_938728, XR_938392, NR_103801, NR_073113, and NR_036503) differentially expressed in both cell lines and tissues, and had predictive value for resistance to therapeutic regimens containing paclitaxel (176). However, the signature described in this study needs further validation and the functional implications for differential expression of the selected lncRNAs should be explored.
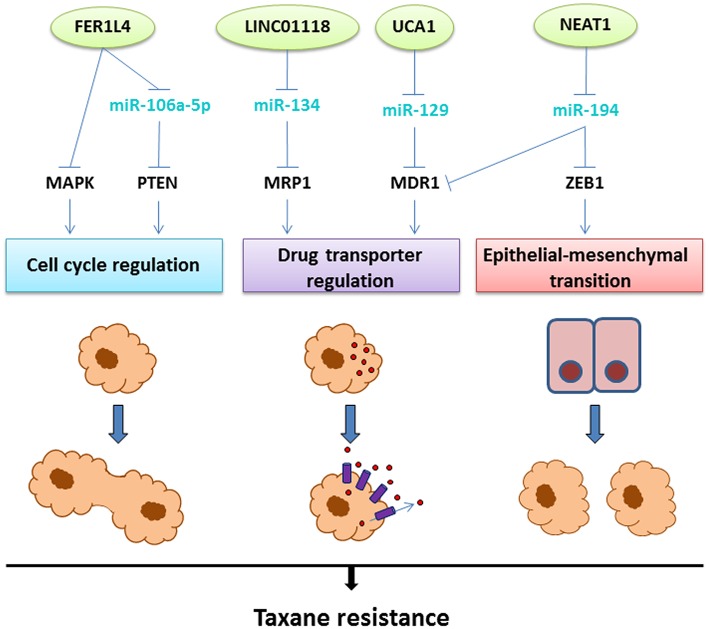
A list of lncRNAs associated with taxane resistance in OC is detailed in Table 2. Few of these lncRNAs have well-described functions and will be presented below. The interplay between the described lncRNAs, their molecular pathways, and resistance mechanisms are illustrated in Figure 4.
Table 2.
List of lncRNAs associated with taxane-resistance in ovarian cancer.
| lncRNA | Category | Expression in OC tissue* | Expression in paclitaxel-resistant cell lines** | Mechanisms of resistance*** | Reference |
|---|---|---|---|---|---|
| UCA1 | Intergenic | N/A | ↑ | ↓ miR-129 → ↑ abcb1 | (155, 156) |
| FER1L4 | Pseudogene | ↓ | ↓ | MAPK | (177) |
| LINC01118 | Intergenic | ↑ | ↑ | ↓ miR-134 → ↑ abcc1 | (178) |
| NEAT1 | Intergenic | ↑ (paclitaxel resistance) | ↑ | ↓ miR-194 → ↑ ZEB1 | (179) |
| Xist | Intergenic | ↓ (recurrent disease) | ↓ | N/A | (180) |
| KB-1471A8.2 | Antisense | ↓ | ↓ | ↓ CDK4 | (181) |
| OIP5-AS1 | Antisense | N/A | ↓ | N/A | (182) |
The order of the lncRNAs corresponds to the appearance of the individual descriptions in the manuscript. Only the first 4 lncRNAs are described in detail in the manuscript and are selected based on substantiating evidence in the literature.
The expression of the lncRNAs in OC tissue is indicated by arrows, ↑ for higher and ↓ for lower expression in resistant patients (patient characteristics are indicated in parenthesis), compared to expression in sensitive patients. If no patient characteristics are indicated, the expression was determined in ovarian cancer tissue from patients with unspecified sensitivity to platinum drugs and normalized to adjacent or normal ovarian tissue.
The expression in paclitaxel-resistant OC cell lines is indicated by arrows; ↑ for higher and ↓ for lower expression and the drug they are resistant to is indicated in parenthesis.
The effect of lncRNAs on associated pathways, miRNAs, genes or transcription factors involved in resistance mechanisms are indicated by arrows: ↑ induction and ↓ repression.
N/A, information not available.
Figure 4.
The lncRNAs involved in taxane resistance in OC. Aberrant expression of the lncRNAs depicted on top (green circles) leads to taxane resistance through three main mechanisms: cell cycle regulation, upregulation of drug transporters leading to efflux of the drug, or epithelial-mesenchymal transition. The molecular mechanisms linking the lncRNAs to these resistance mechanisms involve interactions with miRNAs (light blue) and regulation of transcription factors or signaling pathways (black), as illustrated above.
The Fer-1-like family member 4 (FER1L4) pseudogene is a lncRNA associated with tumor-suppressive properties in cancer (183). FER1L4 acts as a decoy for miR-106a-5p which also interacts with the tumor suppressor PTEN (184). In OC, FER1L4 is expressed at low levels compared to normal ovarian epithelial cells and, at even lower levels in paclitaxel-resistant cell lines. Transfection with FER1L4 led to MAPK pathway suppression and restored the sensitivity to paclitaxel, indicating an important role in the development of resistance (177). Overall, PTEN-AKT-mTOR and MAPK are major cancer driver pathways deeply involved in resistance to chemotherapy (including paclitaxel) in several cancers (185).
Long Intergenic Non-Coding RNA 1118 (LINC01118) was recently identified as overexpressed in paclitaxel and cisplatin-resistant cell lines and OC compared with normal and benign tissues (178). These findings were supported by in vitro studies showing that knockdown conferred increased sensitivity to paclitaxel, whereas overexpression led to resistance. MiR-134 was predicted as a direct target, and functional assays showed that LINC01118 was able to regulate the ABCC1 gene through miR-134 repression (178). As previously described, ABCC1 upregulation is associated with multidrug resistance in cancers (132). No additional studies have been performed correlating LINC01118 with drug resistance or even cancer, warranting further investigations of this lncRNA.
Nuclear paraspeckle assembly transcript 1 (NEAT1) is transcribed from the MEN1 (familial tumor syndrome multiple endocrine neoplasia type 1) locus on chromosome 11 and is a well-described oncogene (186). NEAT1 is overexpressed in OC tissue and cell lines accordingly and is correlated with metastatic potential and poor prognosis (79, 187). In paclitaxel-resistant OC cells, NEAT1 acts as a decoy for miR-194, promoting upregulation of ZEB1 (zinc finger E-box-binding homeobox 1) (179). ZEB1 is an important transcription factor and mediator of EMT and was previously associated with drug resistance (188). Besides, paclitaxel resistance was attenuated by NEAT1 knockdown, which was associated with suppression of the efflux pump P-glycoprotein encoded by the ABCB1 gene. This effect was also related to the interaction between NEAT1 and miR-194, since the suppression of the efflux pump was rescued by miR-194 knockdown. The involvement of NEAT1 in resistance to paclitaxel was validated in OC xenografts in mice, where knockdown restored paclitaxel sensitivity (179). These results substantiate the role of NEAT1 in paclitaxel resistance and indicate the potential for therapeutic targeting.
Perspectives and Future Directions
Collectively, the present review provides compelling evidence of the association between lncRNA expression pattern and therapeutic response in OC, indicating that the etiology of acquired resistance is more complex than originally described. We are just beginning to understand the biological role and function of some of these lncRNAs, and how they can be exploited for clinical purposes.
Two of the most obvious applications for lncRNAs in OC is the establishment of biomarker panels with predictive value for prognosis and/or drug response, or for therapeutic targeting to prevent or reverse resistance to chemotherapy. The presence of circulating lncRNAs in body fluids such as blood and urine at detectable levels, suggests that they could represent excellent biomarkers (189, 190). Several lncRNA signatures with predictive value for platinum-sensitivity in OC have recently been identified (49, 114, 120). However, further studies are needed to determine their clinical applicability.
The oncogenic behavior of some lncRNAs, combined with their tissue specificity and content of targetable residues, emphasize their potential as targets for therapeutic intervention. Furthermore, lncRNA targeting is one of the only therapeutic approaches to upregulate tumor suppressors in a locus-specific manner (191). Artificially synthesized polymers of nucleic acids, known as peptide nucleic acids (PNAs) are thermally stable, not affected by nucleases and can be modified for in vivo administration. Also, PNAs specific for RNA targets is much easier to design and synthesize than small-molecule oncogene inhibitors (192). For example, HOTAIR was targeted with a PNA, designed to prevent the interaction between HOTAIR and the EZH2 subunit of PRC2, in mice with platinum-resistant ovarian tumor xenografts. The treatment reduced HOTAIR expression and re-sensitized the tumors to treatment with cisplatin, which resulted in prolonged survival. The study provided proof of concept for targeting oncogenic lncRNAs as a strategy for precision medicine (193). However, more conclusive evidence of the complex molecular interactions of individual lncRNAs is paramount to determine the physiological impact of targeted treatments before clinical testing.
So far, the molecular profiling of lncRNAs and the identification of functional interactions have proved to be difficult to replicate in different studies. The main platforms for high throughput analysis of lncRNAs, such as RNA sequencing and expression arrays, offer different advantages and drawbacks, and the downstream bioinformatics is not yet standardized. Although RNA sequencing offers the advantage of including all potential lncRNA transcripts, the complexity of the following sequence assembly often hampers the correct annotation (194). In contrast, array-based methods provide a more standardized workflow and a much simpler downstream analysis but are limited to a selection of annotated transcripts. Several studies revise old data sets from publically available sources to perform in silico investigations. However, the experimental setup behind these data sets was rarely designed for the identification of lncRNAs. The low expression of lncRNA transcripts requires specific methodological considerations for optimal results. Furthermore, computational prediction of functional interactions should always be validated experimentally in the specific tissue of interest.
The studies investigating lncRNAs are increasing exponentially and both, experimental and bioinformatic methods are constantly improving. Several lncRNA-targeting therapeutics are already in the clinical pipeline (191), and some have reached clinical trials (195, 196). GENCODE (23), a spin-off from ENCODE is currently attempting to annotate all non-coding transcripts of the entire human genome. Complete annotation of human lncRNAs, standardization of experimental procedures and bioinformatic analysis combined with improved insights into the functional roles of lncRNAs in the development of resistance, will provide a novel paradigm for biomarker discovery and precision medicine in OC.
Author Contributions
CA and SR conceived and designed the study. CA drafted the manuscript and designed the figures. LD and KS participated in drafting the molecular and clinical information, respectively. SR revised and edited the full content of the manuscript. All authors have read, revised critically and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The preparation of this manuscript was financially supported by the Region of Southern Denmark Research Fund.
References
- 1.Reid F. The world ovarian cancer coalition atlas -global trends in incidence mortality and survival. In: Coalition WO, editor. The Every Woman Study (The World Ovarian Cancer Coalition: ) (2018). p. 1–39. [Google Scholar]
- 2.Kurman RJ, Shih Ie M. Pathogenesis of ovarian cancer: lessons from morphology and molecular biology and their clinical implications. Int J Gynecol Pathol. (2008) 27:151–60. 10.1097/PGP.0b013e318161e4f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int. (2014) 2014:934261. 10.1155/2014/934261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducie J, Dao F, Considine M, Olvera N, Shaw PA, Kurman RJ, et al. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat Commun. (2017) 8:990. 10.1038/s41467-017-01217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. (2017) 8:1093. 10.1038/s41467-017-00962-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd LR, Muggia FM. Carboplatin/paclitaxel induction in ovarian cancer: the finer points. Oncology. (2018) 32:418–20, 422–4. [PubMed] [Google Scholar]
- 7.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. (2014) 384:1376–88. 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 8.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. (2015) 521:489–94. 10.1038/nature14410 [DOI] [PubMed] [Google Scholar]
- 9.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2018) 379:2495–505. 10.1056/NEJMoa1810858 [DOI] [PubMed] [Google Scholar]
- 10.Howlader NA, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, et al. editors. SEER Cancer Statistics Review, 1975-2016. Bethesda, MD: National Cancer Institute; (2019). Available online at: https://seer.cancer.gov/csr/1975_2016/ [Google Scholar]
- 11.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. (2018) 46:D794–801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Institute T. N. C The Cancer Genome Atlas. Available online at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
- 13.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. (2005) 205:275–92. 10.1002/path.1706 [DOI] [PubMed] [Google Scholar]
- 14.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. (2011) 17:297–303. 10.1038/nm.2323 [DOI] [PubMed] [Google Scholar]
- 15.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. (2015) 19:A68–77. 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. (2013) 152:1298–307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. (2011) 21:354–61. 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 19.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. (2016) 539:452–55. 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zampetaki A, Albrecht A, Steinhofel K. Long non-coding RNA structure and function: is there a link? Front Physiol. (2018) 9:1201. 10.3389/fphys.2018.01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. (2012) 482:339–46. 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. (2017) 1008:1–46. 10.1007/978-981-10-5203-3_1 [DOI] [PubMed] [Google Scholar]
- 23.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. (2014) 24:616–28. 10.1101/gr.165035.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouckenheimer J, Assou S, Riquier S, Hou C, Philippe N, Sansac C, et al. Long non-coding RNAs in human early embryonic development and their potential in ART. Hum Reprod Update. (2016) 23:19–40. 10.1093/humupd/dmw035 [DOI] [PubMed] [Google Scholar]
- 26.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. (2007) 29:288–99. 10.1002/bies.20544 [DOI] [PubMed] [Google Scholar]
- 27.Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. (2018) 3:108–17. 10.1016/j.ncrna.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. (2014) 15:7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 29.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. (2012) 7:e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. (2012) 9:703–19. 10.4161/rna.20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. (2011) 71:6320–6. 10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- 33.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. (2013) 13:464. 10.1186/1471-2407-13-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. (2011) 18:1243–50. 10.1245/s10434-011-1581-y [DOI] [PubMed] [Google Scholar]
- 35.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. (2013) 29:946–50. 10.3892/or.2012.2219 [DOI] [PubMed] [Google Scholar]
- 36.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. (2011) 39:2119–28. 10.1177/147323001103900608 [DOI] [PubMed] [Google Scholar]
- 37.Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE. (2013) 8:e77070. 10.1371/journal.pone.0077070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. (2013) 30:670. 10.1007/s12032-013-0670-0 [DOI] [PubMed] [Google Scholar]
- 39.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. (2013) 32:1616–25. 10.1038/onc.2012.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui L, Xie XY, Wang H, Chen XL, Liu SL, Hu LN. [Expression of long non-coding RNA HOTAIR mRNA in ovarian cancer]. Sichuan Da Xue Xue Bao Yi Xue Ban. (2013) 44:57–9. [PubMed] [Google Scholar]
- 41.Qiu H, Wang X, Guo R, Liu Q, Wang Y, Yuan Z, et al. HOTAIR rs920778 polymorphism is associated with ovarian cancer susceptibility and poor prognosis in a Chinese population. Future Oncol. (2017) 13:347–55. 10.2217/fon-2016-0290 [DOI] [PubMed] [Google Scholar]
- 42.Wu H, Shang X, Shi Y, Yang Z, Zhao J, Yang M, et al. Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget. (2016) 7:41047–52. 10.18632/oncotarget.8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. (2015) 333:238–48. 10.1016/j.yexcr.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Xu AM, Zhang JY, He XM, Pan YS, Cheng G, et al. Prognostic significance of long non-coding RNA MALAT-1 in various human carcinomas: a meta-analysis. Genet Mol Res. (2016) 15:1–12. 10.4238/gmr.15017433 [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B, et al. The long noncoding RNA MALAT-1 is a novel biomarker in various cancers: a meta-analysis based on the GEO database and literature. J Cancer. (2016) 7:991–1001. 10.7150/jca.14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou A, Liu R, Wu X. Long non-coding RNA MALAT1 is up-regulated in ovarian cancer tissue and promotes SK-OV-3 cell proliferation and invasion. Neoplasma. (2016) 63:865–72. 10.4149/neo_2016_605 [DOI] [PubMed] [Google Scholar]
- 47.Lin Q, Guan W, Ren W, Zhang L, Zhang J, Xu G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol Rep. (2018) 39:2644–52. 10.3892/or.2018.6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, et al. The long noncoding RNA MALAT-1 Is highly expressed in ovarian cancer and induces cell growth and migration. PLoS ONE. (2016) 11:e0155250. 10.1371/journal.pone.0155250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang K, Hou Y, Li A, Li Z, Wang W, Xie H, et al. Identification of a six-lncRNA signature associated with recurrence of ovarian cancer. Sci Rep. (2017) 7:752 10.1038/s41598-017-00763-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. (2019) 58:196–205. 10.1002/mc.22919 [DOI] [PubMed] [Google Scholar]
- 51.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. (2010) 39:925–38. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. (2019) 9:1354–66. [PMC free article] [PubMed] [Google Scholar]
- 53.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. (2015) 21:1253–61. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 54.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. (2018) 109:2093–100. 10.1111/cas.13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohn EC, Ivy SP. Whence high-grade serous ovarian cancer. Am Soc Clin Oncol Educ Book. (2017) 37:443–8. 10.14694/EDBK_174718 [DOI] [PubMed] [Google Scholar]
- 56.Testa U, Petrucci E, Pasquini L, Castelli G, Pelosi E. Ovarian cancers: genetic abnormalities, tumor heterogeneity and progression, clonal evolution and cancer stem cells. Medicines. (2018) 5:1–74. 10.3390/medicines5010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Cao L, Nguyen D, Lu H. TP53 mutations in epithelial ovarian cancer. Transl Cancer Res. (2016) 5:650–63. 10.21037/tcr.2016.08.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cancer cGenome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. (2011) 474:609–15. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bewtra C, Watson P, Conway T, Read-Hippee C, Lynch HT. Hereditary ovarian cancer: a clinicopathological study. Int J Gynecol Pathol. (1992) 11:180–7. 10.1097/00004347-199207000-00003 [DOI] [PubMed] [Google Scholar]
- 60.Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. (2015) 107:1–6. 10.1093/jnci/djv214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. (2015) 33:2901–7. 10.1200/JCO.2015.61.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Folkins AK, Longacre TA. Hereditary gynaecological malignancies: advances in screening and treatment. Histopathology. (2013) 62:2–30. 10.1111/his.12028 [DOI] [PubMed] [Google Scholar]
- 63.Song H, Cicek MS, Dicks E, Harrington P, Ramus SJ, Cunningham JM, et al. The contribution of deleterious germline mutations in BRCA1, BRCA2 and the mismatch repair genes to ovarian cancer in the population. Hum Mol Genet. (2014) 23:4703–9. 10.1093/hmg/ddu172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alsop K, Fereday S, Meldrum C, A., deFazio, Emmanuel C, George J, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. (2012) 30:2654–63. 10.1200/JCO.2011.39.8545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ledermann JA, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. (2016) 17:1579–89. 10.1016/S1470-2045(16)30376-X [DOI] [PubMed] [Google Scholar]
- 66.Hjortkjaer M, Malik Aagaard Jorgensen M, Waldstrom M, Ornskov D, Sogaard-Andersen E, Jakobsen A, et al. The clinical importance of BRCAness in a population-based cohort of Danish epithelial ovarian cancer. Int J Gynecol Cancer. (2019) 29:166–73. 10.1136/ijgc-2018-000017 [DOI] [PubMed] [Google Scholar]
- 67.Marquard AM, Eklund AC, Joshi T, Krzystanek M, Favero F, Wang ZC, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res. (2015) 3:9. 10.1186/s40364-015-0033-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang J, Zhang L, Greshock J, Colligon TA, Wang Y, Ward R, et al. Frequent genetic abnormalities of the PI3K/AKT pathway in primary ovarian cancer predict patient outcome. Genes Chromosomes Cancer. (2011) 50:606–18. 10.1002/gcc.20883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. (2005) 11:2875–8. 10.1158/1078-0432.CCR-04-2142 [DOI] [PubMed] [Google Scholar]
- 70.Dunn GP, Cheung HW, Agarwalla PK, Thomas S, Zektser Y, Karst AM, et al. In vivo multiplexed interrogation of amplified genes identifies GAB2 as an ovarian cancer oncogene. Proc Natl Acad Sci USA. (2014) 111:1102–7. 10.1073/pnas.1311909111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu CY, Bristow R, Cha MS, Wang BG, Ho CL, Kurman RJ, et al. Characterization of active mitogen-activated protein kinase in ovarian serous carcinomas. Clin Cancer Res. (2004) 10:6432–6. 10.1158/1078-0432.CCR-04-0893 [DOI] [PubMed] [Google Scholar]
- 72.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. (2008) 14:5198–208. 10.1158/1078-0432.CCR-08-0196 [DOI] [PubMed] [Google Scholar]
- 73.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. (2013) 123:517–25. 10.1172/JCI65833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. (2014) 106:1–8. 10.1093/jnci/dju249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao H, et al. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget. (2016) 7:32433–48. 10.18632/oncotarget.8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ning L, Hu YC, Wang S, Lang JH. Altered long noncoding RNAs and survival outcomes in ovarian cancer: a systematic review and meta-analysis (PRISMA Compliant). Medicine. (2018) 97:e11481. 10.1097/MD.0000000000011481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, Han L, Zhou L, Wang L, Zhang LM. Prediction of candidate RNA signatures for recurrent ovarian cancer prognosis by the construction of an integrated competing endogenous RNA network. Oncol Rep. (2018) 40:2659–73. 10.3892/or.2018.6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem. (2018) 49:1289–303. 10.1159/000493408 [DOI] [PubMed] [Google Scholar]
- 79.Liu Y, Wang Y, Fu X, Lu Z. Long non-coding RNA NEAT1 promoted ovarian cancer cells' metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. (2018) 109:2188–98. 10.1111/cas.13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia B, Hou Y, Chen H, Yang S, Liu T, Lin M, et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget. (2017) 8:19534–46. 10.18632/oncotarget.14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hong L, Chen W, Wu D, Wang Y. Upregulation of SNHG3 expression associated with poor prognosis and enhances malignant progression of ovarian cancer. Cancer Biomark. (2018) 22:367–74. 10.3233/CBM-170710 [DOI] [PubMed] [Google Scholar]
- 82.Szafron LM, Balcerak A, Grzybowska EA, Pienkowska-Grela B, Podgorska A, Zub R, et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget. (2015) 6:43897–910. 10.18632/oncotarget.6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chi Y, Wang D, Wang J, Yu W, Yang J. Long Non-coding RNA in the pathogenesis of cancers. Cells. (2019) 8:1–44. 10.3390/cells8091015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. (2018) 150:343–54. 10.1016/j.ygyno.2018.06.013 [DOI] [PubMed] [Google Scholar]
- 85.Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. (2017) 21:3176–84. [PubMed] [Google Scholar]
- 86.Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. (2015) 46:2497–505. 10.3892/ijo.2015.2943 [DOI] [PubMed] [Google Scholar]
- 87.Akrami R, Jacobsen A, Hoell J, Schultz N, Sander C, Larsson E. Comprehensive analysis of long non-coding RNAs in ovarian cancer reveals global patterns and targeted DNA amplification. PLoS ONE. (2013) 8:e80306. 10.1371/journal.pone.0080306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rangel LB, Sherman-Baust CA, Wernyj RP, Schwartz DR, Cho KR, Morin PJ. Characterization of novel human ovarian cancer-specific transcripts (HOSTs) identified by serial analysis of gene expression. Oncogene. (2003) 22:7225–32. 10.1038/sj.onc.1207008 [DOI] [PubMed] [Google Scholar]
- 89.Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. (2015) 24:841–52. 10.1093/hmg/ddu502 [DOI] [PubMed] [Google Scholar]
- 90.Huang P, Li F, Li L, You Y, Luo S, Dong Z, et al. lncRNA profile study reveals the mRNAs and lncRNAs associated with docetaxel resistance in breast cancer cells. Sci Rep. (2018) 8:17970. 10.1038/s41598-018-36231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L, et al. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS ONE. (2014) 9:e108133. 10.1371/journal.pone.0108133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu Y, Zhu QN, Deng JL, Li ZX, Wang G, Zhu YS. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther. (2018) 11:3185–94. 10.2147/OTT.S158104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Q, Zhang J, Zhou J, Yang B, Liu P, Cao L, et al. lncRNAs are novel biomarkers for differentiating between cisplatin-resistant and cisplatin-sensitive ovarian cancer. Oncol Lett. (2018) 15:8363–70. 10.3892/ol.2018.8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rosenberg B, VanCamp L, Trosko JE, Mansour VH. Platinum compounds: a new class of potent antitumour agents. Nature. (1969) 222:385–6. 10.1038/222385a0 [DOI] [PubMed] [Google Scholar]
- 95.Hah SS, Stivers KM, de Vere White R.W., Henderson PT. Kinetics of carboplatin-DNA binding in genomic DNA and bladder cancer cells as determined by accelerator mass spectrometry. Chem Res Toxicol. (2006) 19:622–6. 10.1021/tx060058c [DOI] [PubMed] [Google Scholar]
- 96.Ferreira JA, Peixoto A, Neves M, Gaiteiro C, Reis CA, Assaraf YG, et al. Mechanisms of cisplatin resistance and targeting of cancer stem cells: adding glycosylation to the equation. Drug Resist Updat. (2016) 24:34–54. 10.1016/j.drup.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 97.Righetti SC, Della Torre G, Pilotti S, Menard S, Ottone F, Colnaghi MI, et al. A comparative study of p53 gene mutations, protein accumulation, and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Cancer Res. (1996) 56:689–93. [PubMed] [Google Scholar]
- 98.Turner NC, Tutt AN. Platinum chemotherapy for BRCA1-related breast cancer: do we need more evidence? Breast Cancer Res. (2012) 14:115. 10.1186/bcr3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. (1996) 56:4881–6. [PubMed] [Google Scholar]
- 100.Henkels KM, Turchi JJ. Cisplatin-induced apoptosis proceeds by caspase-3-dependent and -independent pathways in cisplatin-resistant and -sensitive human ovarian cancer cell lines. Cancer Res. (1999) 59:3077–83. [PubMed] [Google Scholar]
- 101.Marchini S, Fruscio R, Clivio L, Beltrame L, Porcu L, Fuso Nerini I, et al. Resistance to platinum-based chemotherapy is associated with epithelial to mesenchymal transition in epithelial ovarian cancer. Eur J Cancer. (2013) 49:520–30. 10.1016/j.ejca.2012.06.026 [DOI] [PubMed] [Google Scholar]
- 102.Talbot LJ, Bhattacharya SD, Kuo PC. Epithelial-mesenchymal transition, the tumor microenvironment, and metastatic behavior of epithelial malignancies. Int J Biochem Mol Biol. (2012) 3:117–36. [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang W, Shi X, Peng Y, Wu M, Zhang P, Xie R, et al. HIF-1alpha Promotes Epithelial-Mesenchymal Transition and Metastasis through Direct Regulation of ZEB1 in Colorectal Cancer. PLoS ONE. (2015) 10:e0129603. 10.1371/journal.pone.0129603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Norouzi-Barough L, Sarookhani MR, Sharifi M, Moghbelinejad S, Jangjoo S, Salehi R. Molecular mechanisms of drug resistance in ovarian cancer. J Cell Physiol. (2018) 233:4546–62. 10.1002/jcp.26289 [DOI] [PubMed] [Google Scholar]
- 105.Bashashati A, Ha G, Tone A, Ding J, Prentice LM, Roth A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. (2013) 231:21–34. 10.1002/path.4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fang F, Cardenas H, Huang H, Jiang G, Perkins SM, Zhang C, et al. Genomic and epigenomic signatures in ovarian cancer associated with re-sensitization to platinum drugs. Cancer Res. (2018) 78:631–44. 10.1158/0008-5472.CAN-17-1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu S, Lei H, Luo F, Li Y, Xie L. The effect of lncRNA HOTAIR on chemoresistance of ovarian cancer through regulation of HOXA7. Biol Chem. (2018) 399:485–97. 10.1515/hsz-2017-0274 [DOI] [PubMed] [Google Scholar]
- 108.Wang Y, Wang H, Song T, Zou Y, Jiang J, Fang L, et al. HOTAIR is a potential target for the treatment of cisplatinresistant ovarian cancer. Mol Med Rep. (2015) 12:2211–6. 10.3892/mmr.2015.3562 [DOI] [PubMed] [Google Scholar]
- 109.Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. (2015) 7:108. 10.1186/s13073-015-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J, Yang S, Su N, Wang Y, Yu J, Qiu H, et al. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/beta-catenin pathway in human ovarian cancer. Tumour Biol. (2016) 37:2057–65. 10.1007/s13277-015-3998-6 [DOI] [PubMed] [Google Scholar]
- 111.Wang P, Zhang Y, Wang Q, Wu S, Zeng X, Zhou Y, et al. Down-modulation of MALAT1 inhibits proliferation, metastasis, invasion and sensitizes chemosensitivity to cisplatin in ovarian cancer cells. Int J Clin Exp Med. (2018). 11:13158–68. [Google Scholar]
- 112.Bai L, Wang A, Zhang Y, Xu X, Zhang X. Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp Cell Res. (2018) 366:161–71. 10.1016/j.yexcr.2018.03.014 [DOI] [PubMed] [Google Scholar]
- 113.Zheng ZG, Xu H, Suo SS, Xu XL, Ni MW, Gu LH, et al. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep. (2016) 6:26093. 10.1038/srep26093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu R, Zeng Y, Zhou CF, Wang Y, Li X, Liu ZQ, et al. Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients. Sci Rep. (2017) 7:18. 10.1038/s41598-017-00050-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang F, Zhou J, Xie X, Hu J, Chen L, Hu Q, et al. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. (2015) 62:432–8. 10.4149/neo_2015_051 [DOI] [PubMed] [Google Scholar]
- 116.Li Z, Niu H, Qin Q, Yang S, Wang Q, Yu C, et al. lncRNA UCA1 mediates resistance to cisplatin by regulating the miR-143/FOSL2-signaling pathway in ovarian cancer. Mol Ther Nucleic Acids. (2019) 17:92–101. 10.1016/j.omtn.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang L, Cao X, Zhang L, Zhang X, Sheng H, Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemother Pharmacol. (2016) 77:629–34. 10.1007/s00280-016-2963-4 [DOI] [PubMed] [Google Scholar]
- 118.Wang H, Fang L, Jiang J, Kuang Y, Wang B, Shang X, et al. The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis. (2018) 9:1103. 10.1038/s41419-018-1148-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu E, Liu Z, Zhou Y, Mi R, Wang D. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med. (2015) 8:20565–72. [PMC free article] [PubMed] [Google Scholar]
- 120.Song J, Zhang W, Wang S, Liu K, Song F, Ran L. A panel of 7 prognosis-related long non-coding RNAs to improve platinum-based chemoresistance prediction in ovarian cancer. Int J Oncol. (2018) 53:866–76. 10.3892/ijo.2018.4403 [DOI] [PubMed] [Google Scholar]
- 121.Wu DI, Wang T, Ren C, Liu L, Kong D, Jin X, et al. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol Lett. (2016) 11:1189–94. 10.3892/ol.2015.3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang C, Wang M, Shi C, Shi F, Pei C. Long non-coding RNA Linc00312 modulates the sensitivity of ovarian cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway. Biosci Trends. (2018) 12:309–16. 10.5582/bst.2018.01052 [DOI] [PubMed] [Google Scholar]
- 123.Vera O, Rodriguez-Antolin C, de Castro J, Karreth FA, Sellers TA, Ibanez de Caceres I.. An epigenomic approach to identifying differential overlapping and cis-acting lncRNAs in cisplatin-resistant cancer cells. Epigenetics. (2018) 13:251–63. 10.1080/15592294.2018.1436364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qu C, Dai C, Guo Y, Qin R, Liu J. Long noncoding RNA SNHG15 serves as an oncogene and predicts poor prognosis in epithelial ovarian cancer. Onco Targets Ther. (2019) 12:101–11. 10.2147/OTT.S182657 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Xu QF, Tang YX, Wang X. LncRNA EBIC promoted proliferation, metastasis and cisplatin resistance of ovarian cancer cells and predicted poor survival in ovarian cancer patients. Eur Rev Med Pharmacol Sci. (2018) 22:4440–7. 10.26355/eurrev_201807_15495 [DOI] [PubMed] [Google Scholar]
- 126.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Naora H, Montz FJ, Chai CY, Roden RB. Aberrant expression of homeobox gene HOXA7 is associated with mullerian-like differentiation of epithelial ovarian tumors and the generation of a specific autologous antibody response. Proc Natl Acad Sci USA. (2001) 98:15209–14. 10.1073/pnas.011503998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y, Cheng JC, Huang HF, Leung PC. Homeobox A7 stimulates breast cancer cell proliferation by up-regulating estrogen receptor-alpha. Biochem Biophys Res Commun. (2013) 440:652–7. 10.1016/j.bbrc.2013.09.121 [DOI] [PubMed] [Google Scholar]
- 129.Mitra R, Chen X, Greenawalt EJ, Maulik U, Jiang W, Zhao Z, et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun. (2017) 8:1604. 10.1038/s41467-017-01781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li P, Zhang X, Wang H, Wang L, Liu T, Du L, et al. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. (2017) 16:739–51. 10.1158/1535-7163.MCT-16-0591 [DOI] [PubMed] [Google Scholar]
- 131.Zhou Y, Chen Q, Qin R, Zhang K, Li H. MicroRNA-449a reduces cell survival and enhances cisplatin-induced cytotoxicity via downregulation of NOTCH1 in ovarian cancer cells. Tumour Biol. (2014) 35:12369–78. 10.1007/s13277-014-2551-3 [DOI] [PubMed] [Google Scholar]
- 132.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. (1992) 258:1650–4. 10.1126/science.1360704 [DOI] [PubMed] [Google Scholar]
- 133.Chen J, Liu X, Xu Y, Zhang K, Huang J, Pan B, et al. TFAP2C-Activated MALAT1 modulates the chemoresistance of docetaxel-resistant lung adenocarcinoma cells. Mol Ther Nucleic Acids. (2019) 14:567–82. 10.1016/j.omtn.2019.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. (2010) 32:473–80. 10.1002/bies.200900170 [DOI] [PubMed] [Google Scholar]
- 135.Esteves LI, Javaroni AC, Nishimoto IN, Magrin J, Squire JA, Kowalski LP, et al. DNA methylation in the CTCF-binding site I and the expression pattern of the H19 gene: does positive expression predict poor prognosis in early stage head and neck carcinomas? Mol Carcinog. (2005) 44:102–10. 10.1002/mc.20126 [DOI] [PubMed] [Google Scholar]
- 136.Rainho CA, Pontes A, Rogatto SR. Expression and imprinting of insulin-like growth factor II (IGF2) and H19 genes in uterine leiomyomas. Gynecol Oncol. (1999) 74:375–80. 10.1006/gyno.1999.5483 [DOI] [PubMed] [Google Scholar]
- 137.Yoshimura H, Matsuda Y, Yamamoto M, Kamiya S, Ishiwata T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front Biosci. (2018) 23:614–25. 10.2741/4608 [DOI] [PubMed] [Google Scholar]
- 138.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. (2012) 14:659–65. 10.1038/ncb2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Costa V, Lo Dico A, Rizzo A, Rajata F, Tripodi M, Alessandro R, et al. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget. (2017) 8:24292–302. 10.18632/oncotarget.14464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang J, Zhang Y, Wei H, Zhang X, Wu Y, Gong A, et al. The mir-675-5p regulates the progression and development of pancreatic cancer via the UBQLN1-ZEB1-mir200 axis. Oncotarget. (2017) 8:24978–87. 10.18632/oncotarget.15330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. (2006) 66:5330–7. 10.1158/0008-5472.CAN-06-0037 [DOI] [PubMed] [Google Scholar]
- 142.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. (2010) 1803:443–51. 10.1016/j.bbamcr.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 143.Godwin AK, Meister A, O'Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci USA. (1992) 89:3070–4. 10.1073/pnas.89.7.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Meijer C, Mulder NH, Timmer-Bosscha H, Sluiter WJ, Meersma GJ, de Vries EG. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer Res. (1992) 52:6885–9. [PubMed] [Google Scholar]
- 145.Wei J, Gan Y, Peng D, Jiang X, Kitazawa R, Xiang Y, et al. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med. (2018) 7:6247–57. 10.1002/cam4.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Q, Cheng N, Li X, Pan H, Li C, Ren S, et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. (2017) 8:2558–67. 10.18632/oncotarget.13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. (2011) 17:878–91. 10.1261/rna.2528811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, Mlcochova J, Kretz M, Svoboda M, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. (2016) 7:622–37. 10.18632/oncotarget.5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vizcaino C, Mansilla S, Portugal J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol Ther. (2015) 152:111–24. 10.1016/j.pharmthera.2015.05.008 [DOI] [PubMed] [Google Scholar]
- 150.Olofsson BA, Kelly CM, Kim J, Hornsby SM, Azizkhan-Clifford J. Phosphorylation of Sp1 in response to DNA damage by ataxia telangiectasia-mutated kinase. Mol Cancer Res. (2007) 5:1319–30. 10.1158/1541-7786.MCR-07-0374 [DOI] [PubMed] [Google Scholar]
- 151.Lai YH, Kuo C, Kuo MT, H.Chen HW. Modulating chemosensitivity of tumors to platinum-based antitumor drugs by transcriptional regulation of copper homeostasis. Int J Mol Sci. (2018) 19:E1486. 10.3390/ijms19051486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. (2006) 12:4851–8. 10.1158/1078-0432.CCR-06-0134 [DOI] [PubMed] [Google Scholar]
- 153.Xue M, Li X, Li Z, Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, invasion. Tumour Biol. (2014) 35:6901–12. 10.1007/s13277-014-1925-x [DOI] [PubMed] [Google Scholar]
- 154.Guo Q, Lan F, Yan X, Xiao Z, Wu Y, Zhang Q. Hypoxia exposure induced cisplatin resistance partially via activating p53 and hypoxia inducible factor-1α in non-small cell lung cancer A549 cells. Oncol Lett. (2018) 16:801–8. 10.3892/ol.2018.8767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Horita K, Kurosaki H, Nakatake M, Kuwano N, Oishi T, Itamochi H, et al. lncRNA UCA1-mediated Cdc42 signaling promotes oncolytic vaccinia virus cell-to-cell spread in ovarian cancer. Mol Ther Oncolytics. (2019) 13:35–48. 10.1016/j.omto.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang J, Ye C, Liu J, Hu Y. UCA1 confers paclitaxel resistance to ovarian cancer through miR-129/ABCB1 axis. Biochem Biophys Res Commun. (2018) 501:1034–40. 10.1016/j.bbrc.2018.05.104 [DOI] [PubMed] [Google Scholar]
- 157.Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE, et al. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. (2016) 115:431–41. 10.1038/bjc.2016.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y, et al. PANDAR: a pivotal cancer-related long non-coding RNA in human cancers. Mol Biosyst. (2017) 13:2195–201. 10.1039/C7MB00414A [DOI] [PubMed] [Google Scholar]
- 159.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. (2011) 43:621–9. 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. (2018) 15:238–50. 10.20892/j.issn.2095-3941.2017.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang Q, Yu Y, Sun Z, Pan Y. Long non-coding RNA PVT1 promotes cell proliferation and invasion through regulating miR-133a in ovarian cancer. Biomed Pharmacother. (2018) 106:61–7. 10.1016/j.biopha.2018.06.112 [DOI] [PubMed] [Google Scholar]
- 162.Ding J, Li D, Gong M, Wang J, Huang X, Wu T, et al. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. (2014) 7:1625–30. 10.2147/OTT.S68854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. (2007) 13:5745–55. 10.1158/1078-0432.CCR-06-2882 [DOI] [PubMed] [Google Scholar]
- 164.Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. (2008) 6:212–21. 10.1158/1541-7786.MCR-07-0105 [DOI] [PubMed] [Google Scholar]
- 165.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. (2014) 512:82–6. 10.1038/nature13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Reyes-Gonzalez JM, Armaiz-Pena GN, Mangala LS, Valiyeva F, Ivan C, Pradeep S, et al. Targeting c-MYC in platinum-resistant ovarian cancer. Mol Cancer Ther. (2015) 14:2260–9. 10.1158/1535-7163.MCT-14-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu E, Liu Z, Zhou Y. Carboplatin-docetaxel-induced activity against ovarian cancer is dependent on up-regulated lncRNA PVT1. Int J Clin Exp Pathol. (2015) 8:3803–10. [PMC free article] [PubMed] [Google Scholar]
- 168.Chen L, Han X, Hu Z, Chen L. The PVT1/miR-216b/Beclin-1 regulates cisplatin sensitivity of NSCLC cells via modulating autophagy and apoptosis. Cancer Chemother Pharmacol. (2019) 83:921–31. 10.1007/s00280-019-03808-3 [DOI] [PubMed] [Google Scholar]
- 169.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS ONE. (2016) 11:e0156274. 10.1371/journal.pone.0156274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tian T, Lv X, Pan G, Lu Y, Chen W, He W, et al. Long noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of pre-miRNA processing. Clin Cancer Res. (2019) 25:3673–88. 10.1158/1078-0432.CCR-18-2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rao S, Orr GA, Chaudhary AG, Kingston DG, Horwitz SB. Characterization of the taxol binding site on the microtubule. 2-(m-Azidobenzoyl)taxol photolabels a peptide (amino acids 217-231) of beta-tubulin. J Biol Chem. (1995) 270:20235–8. 10.1074/jbc.270.35.20235 [DOI] [PubMed] [Google Scholar]
- 172.Jordan MA, Wendell K, Gardiner S, Derry WB, Copp H, Wilson L. Mitotic block induced in HeLa cells by low concentrations of paclitaxel (Taxol) results in abnormal mitotic exit and apoptotic cell death. Cancer Res. (1996) 56:816–25. [PubMed] [Google Scholar]
- 173.Stordal B, Pavlakis N, Davey R. A systematic review of platinum and taxane resistance from bench to clinic: an inverse relationship. Cancer Treat Rev. (2007) 33:688–703. 10.1016/j.ctrv.2007.07.013 [DOI] [PubMed] [Google Scholar]
- 174.Yusuf RZ, Duan Z, Lamendola DE, Penson RT, Seiden MV. Paclitaxel resistance: molecular mechanisms and pharmacologic manipulation. Curr Cancer Drug Targets. (2003) 3:1–19. 10.2174/1568009033333754 [DOI] [PubMed] [Google Scholar]
- 175.Parekh H, Wiesen K, Simpkins H. Acquisition of taxol resistance via P-glycoprotein- and non-P-glycoprotein-mediated mechanisms in human ovarian carcinoma cells. Biochem Pharmacol. (1997) 53:461–70. 10.1016/S0006-2952(97)83383-7 [DOI] [PubMed] [Google Scholar]
- 176.Wang L, Hu Y, Xiang X, Qu K, Teng Y. Identification of long non-coding RNA signature for paclitaxel-resistant patients with advanced ovarian cancer. Oncotarget. (2017) 8:64191–202. 10.18632/oncotarget.19828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu S, Zou B, Tian T, Luo X, Mao B, Zhang X, et al. Overexpression of the lncRNA FER1L4 inhibits paclitaxel tolerance of ovarian cancer cells via the regulation of the MAPK signaling pathway. J Cell Biochem. (2018). 10.1002/jcb.28032. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 178.Shi C, Wang M. LINC01118 modulates paclitaxel resistance of epithelial ovarian cancer by regulating miR-134/ABCC1. Med Sci Monit. (2018) 24:8831–9. 10.12659/MSM.910932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.An J, Lv W, Zhang Y. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther. (2017) 10:5377–90. 10.2147/OTT.S147586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Huang KC, Rao PH, Lau CC, Heard E, Ng SK, Brown C, et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. (2002) 1:769–76. [PubMed] [Google Scholar]
- 181.Zhang M, Liu S, Fu C, Wang X, Zhang M, Liu G, et al. LncRNA KB-1471A8.2 Overexpression suppresses cell proliferation and migration and antagonizes the paclitaxel resistance of ovarian cancer cells. Cancer Biother Radiopharm. (2019) 34:316–24. 10.1089/cbr.2018.2698 [DOI] [PubMed] [Google Scholar]
- 182.Xu J, Wu J, Fu C, Teng F, Liu S, Dai C, et al. Multidrug resistant lncRNA profile in chemotherapeutic sensitive and resistant ovarian cancer cells. J Cell Physiol. (2018) 233:5034–43. 10.1002/jcp.26369 [DOI] [PubMed] [Google Scholar]
- 183.Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. (2014) 35:9613–7. 10.1007/s13277-014-2259-4 [DOI] [PubMed] [Google Scholar]
- 184.Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. (2015) 5:13445. 10.1038/srep13445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu Z, Zhu G, Getzenberg RH, Veltri RW. The upregulation of PI3K/Akt and MAP kinase pathways is associated with resistance of microtubule-targeting drugs in prostate cancer. J Cell Biochem. (2015) 116:1341–9. 10.1002/jcb.25091 [DOI] [PubMed] [Google Scholar]
- 186.Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. (2017) 50:1–13. 10.1111/cpr.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. (2016) 20:3373–7. [PubMed] [Google Scholar]
- 188.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. (2015) 14:481–7. 10.1080/15384101.2015.1006048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine–a new perspective for detecting prostate cancer. Eur Urol. (2004) 46:182–6; discussion: 187. 10.1016/j.eururo.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 190.Isin M, Ozgur E, Cetin G, Erten N, Aktan M, Gezer U, et al. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. (2014) 431:255–9. 10.1016/j.cca.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 191.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. (2017) 474:4219–51. 10.1042/BCJ20170079 [DOI] [PubMed] [Google Scholar]
- 192.Ray A, Norden B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. (2000) 14:1041–60. 10.1096/fasebj.14.9.1041 [DOI] [PubMed] [Google Scholar]
- 193.Ozes AR, Wang Y, Zong X, Fang F, Pilrose J, Nephew KP. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci Rep. (2017) 7:894. 10.1038/s41598-017-00966-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. (2018) 19:535–48. 10.1038/s41576-018-0017-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vidaurre S, Fitzpatrick C, Burzio VA, Briones M, Villota C, Villegas J, et al. Down-regulation of the antisense mitochondrial non-coding RNAs (ncRNAs) is a unique vulnerability of cancer cells and a potential target for cancer therapy. J Biol Chem. (2014) 289:27182–98. 10.1074/jbc.M114.558841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Andes Biotechnologies -Clinical Trials and Pipelines Available online at: http://andesbio.com/clinical-trials-pipeline/ (cited June 05, 2019).