Abstract
Spinocerebellar ataxia type 7 (SCA7) is a rare autosomal dominant neurodegenerative disorder characterized by progressive neuronal loss in the cerebellum, brainstem, and retina, leading to cerebellar ataxia and blindness as major symptoms. SCA7 is due to the expansion of a CAG triplet repeat that is translated into a polyglutamine tract in ATXN7. Larger SCA7 expansions are associated with earlier onset of symptoms and more severe and rapid disease progression. Here, we summarize the pathological and genetic aspects of SCA7, compile the current knowledge about ATXN7 functions, and then focus on recent advances in understanding the pathogenesis and in developing biomarkers and therapeutic strategies. ATXN7 is a bona fide subunit of the multiprotein SAGA complex, a transcriptional coactivator harboring chromatin remodeling activities, and plays a role in the differentiation of photoreceptors and Purkinje neurons, two highly vulnerable neuronal cell types in SCA7. Polyglutamine expansion in ATXN7 causes its misfolding and intranuclear accumulation, leading to changes in interactions with native partners and/or partners sequestration in insoluble nuclear inclusions. Studies of cellular and animal models of SCA7 have been crucial to unveil pathomechanistic aspects of the disease, including gene deregulation, mitochondrial and metabolic dysfunctions, cell and non-cell autonomous protein toxicity, loss of neuronal identity, and cell death mechanisms. However, a better understanding of the principal molecular mechanisms by which mutant ATXN7 elicits neurotoxicity, and how interconnected pathogenic cascades lead to neurodegeneration is needed for the development of effective therapies. At present, therapeutic strategies using nucleic acid-based molecules to silence mutant ATXN7 gene expression are under development for SCA7.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00778-5) contains supplementary material, which is available to authorized users.
Keywords: Spinocerebellar ataxia type 7, Ataxin-7, Polyglutamine expansion, Retinal degeneration, Aggregation, SAGA
Introduction
Spinocerebellar ataxia type 7 (SCA7) belongs to the large family of autosomal dominant cerebellar ataxias (ADCAs), a clinically, genetically, and neuropathologically heterogeneous group of disorders defined by variable degrees of cerebellar ataxia, and diverse non-cerebellar symptoms. ADCAs are rare disorders with a prevalence of 0.3 to 2 per 100,000 and are pathologically characterized by prominent atrophy of the cerebellum, brainstem, and spinal cord, together with degeneration at variable level of other brain regions. ADCAs are classified as types I, II, and III. While type I is characterized by cerebellar ataxia associated with variable neurological features and type III by pure cerebellar ataxia, type II or SCA7 is distinguished clinically by the additional presence of a pigmentary maculopathy [1, 2]. SCA7 was estimated to account for 1–11.7% of genetically diagnosed ADCAs in diverse populations worldwide [3–11]. However, the frequency of SCA7 is higher where local founder effects were observed as in Scandinavia, Mexico, South Africa, and Zambia [9, 12–14].
SCA7 is due to the expansion of a polyglutamine (polyQ) tract located in the ataxin-7 (ATXN7). SCA7 is thus pathomechanistically related to the group of CAG/polyQ expansion disorders, which includes other SCAs (types 1–3, 6, and 17), Huntington’s disease (HD), spinal bulbar muscular atrophy (SBMA), and dentatorubro pallidoluysian atrophy (DRPLA) [15]. These diseases are caused by the expansion of a translated CAG triplet repeat in the causative genes, leading to an expanded polyQ tract in the corresponding proteins. SCA7 and other polyQ disorders share a number of common features, including dominant inheritance (except for the X-linked SBMA), adult onset of symptoms and selective brain degeneration responsible for disease-specific symptoms. There is an apparent polyQ length threshold above which the pathology becomes fully penetrant, and the longer the polyQ expansion, the earlier is the disease onset and more severe are symptoms. The pathogenesis of these diseases is complex as polyQ expansion confers toxic gain of function to the mutant protein as well as loss of the normal protein function [15]. Finally, a hallmark of all polyQ diseases is the intracellular accumulation of amyloid-like aggregates containing mutant protein fragments [16]. Except for the presence of a polyQ domain, ATXN7 and the other polyQ proteins do not share any similitude and have different functions. PolyQ proteins are ubiquitously expressed, contrasting with the selective neuronal degeneration. Therefore, it is thought that disease particularities must come from the function of the protein into which the polyQ expansion is embedded.
In the case of SCA7, ATXN7 is a known member of the multiprotein Spt-Ada-Gcn5 acetyltransferase (SAGA) complex [17], a transcriptional coactivator conserved from yeast to human. SAGA harbors chromatin remodeling activities that are essential for RNA polymerase II (RNAPII) transcription [18]. Transcriptional alterations were reported in cellular and animal models of SCA7; however, other cellular perturbations are observed and affect autophagy, mitochondria, as well as cell-cell interaction and maintenance of neuronal differentiation. In this review, we will outline the major clinical, pathological, and genetic aspects of the disease, and then focus on recent advances in understanding the pathogenesis and in developing biomarkers and therapeutic strategy for SCA7.
Genetic Aspects of SCA7
The gene responsible for SCA7 has been localized to chromosome 3p12-21.1 in 1995 [19–21]. A strong anticipation in age of onset in SCA7 families was already well documented at that time, suggesting the involvement of a causative trinucleotide repeat expansion mutation. This hypothesis was later confirmed by the detection of a 130-kDa polyQ-containing protein in cell extracts of SCA7 patients using the specific anti-polyQ 1C2 antibody [22, 23]. Two years later, library screening for CAG repeat sequences led to the identification of the ATXN7 gene [24].
The gene consists of 13 exons spanning 140 kb of genomic DNA and encodes the ATXN7 that harbors a polymorphic polyQ stretch in the amino-terminus [24, 25]. The blood cell genotyping indicated that the wild-type alleles of ATXN7 have 4–35 CAG repeats with 70–80% which carry 10 CAG. Alleles with 28–35 CAG are prone to expand upon paternal transmission and are responsible for rare de novo SCA7 mutation [26–28]. Alleles with 34–36 CAG repeats are associated with reduced penetrance of mild- and late-appearing disease symptoms [27, 29]. In SCA7 patients, mutant alleles have typically 36 or more CAG repeats and can even reach > 460 [30].
CAG expansions are dynamic mutation and show instability in germline and somatic tissues. Among CAG/polyQ disorders, SCA7 CAG repeats show the highest tendency to expand upon transmission, explaining the strong anticipation observed in families (mean 19 ± 13 years [3, 31, 32]). The instability of expanded alleles is significantly greater upon paternal than in maternal transmission [26, 32].
The length of CAG repeats is inversely correlated with the age of onset and the disease duration. The majority of SCA7 alleles have 36–55 CAG repeats and are responsible for the classical adult-onset form, which progresses over several decades until death [32]. Repeats > 70 CAG are typically responsible for juvenile-onset forms with accelerated disease course, while extremely large repeats (> 100 CAG) cause severe infantile forms leading to death within few years or months. The repeat length also relates to symptoms at onset: large repeat expansions associated with early onset cause visual loss before cerebellar ataxia, while ataxia is the first manifestation of shorter expansions associated with later disease onset [3, 4, 26].
Clinical and Neuropathological Features
The classical neurological signs of SCA7 adult form are gait ataxia progressing to limb ataxia and retinal degeneration. Common symptoms include spastic ataxia gait, dysarthria, dysphagia, slow eye movement, ophthalmoplegia, prominent hyperreflexia with crossed supraclavicular, pectoral and hip adductor reflexes, spasticity of the lower limbs, and pyramidal signs [3, 4, 33–35]. SCA7 patients with difficulties in attributing emotions and with cognitive impairment affecting verbal memory and fluency were also reported [36–38]. It is also proposed that extracerebellar component of SCA7 accounts for sudden changes in laryngeal muscle tone, producing instability in the voice [39]. However, as for many neurodegenerative disorders, the accuracy of clinical assessment has limitation due to variation in the age of onset and disease progression. The recent study on a large cohort of 50 patients originating from a founder mutation in Mexico has allowed the specific clinical characterization of adult-onset patients (> 18 years) and early-onset patients (≤ 18 years) and the new observation of frontal-executive deficits and sensory-motor peripheral neuropathy in SCA7 [35, 40]. It is noteworthy that infantile forms are much more severe and cause multisystem disorder including failure to thrive, hypotonia, myoclonic seizures, and non-central nervous systems dysfunctions like congestive heart failure, patent ductus arteriosus, atrial septum defect, renal failure, hepatomegaly, muscle atrophy, capillary leak syndrome, and hemangioma [30, 33, 41–46].
Brain atrophy of SCA7 patients has been characterized in several neuropathological studies using both postmortem tissues and imaging techniques. Postmortem cerebellum of SCA7 patients shows substantial neuronal loss in the Purkinje cell layer, in the dentate nuclei and, to a less extent in the granule cell layer [32, 47–49]. Neuronal loss is also frequently observed in the inferior olivary nuclei and in the basis pontis associated with the atrophy of spinocerebellar and pyramidal tracts [50]. In addition to these structures, postmortem brains of adult- and late-onset patients show widespread neurodegeneration in diverse structures of the telencephalon, diencephalon, midbrain, medulla oblongata, and spinal cord. Atrophy or loss of myelinated fibers was observed in the cerebellar white matter and extracerebellar associated structures including the spinal cord [11, 32, 47–50]. Brain imaging confirmed the atrophy of cerebellum’s grey and white matters and the pons. There is a close relationship between cerebellar volume and the motor impairment in people with SCA7 [51]. Lobules IX and X as well as lobules I, II, and VIIIb of cerebellum appear particularly vulnerable [38]. The unique pattern of cerebellar damages distinguishes SCA7 from other subtypes of SCA [52]. Structural and functional magnetic resonance imaging and fractional anisotropy further confirmed widespread but regio-specific alterations of diverse extracerebellar grey and white matters [38, 51, 53, 54]. Using a new tractography method, the fixel-based analysis, Adanyeguh et al. [55] were able to document longitudinal volumetric changes in the cerebellum and pons as well as in the corpus callosum in SCA7 patients, which might prove useful as disease biomarkers.
Visual impairment in SCA7 is first due to cone photoreceptor degeneration and leads to the decrease of acuity and color vision in the tritan (blue-yellow) axis. The retinopathy then progresses toward a cone-rod dystrophy and complete blindness [11, 33–35, 50, 56–64]. Fundoscopy of SCA7 patient retina shows progressive atrophy of the macula with apparition of granular pigmentation, pale areas with pigmentary atrophy and poor vasculature. Postmortem retinal sections reveal the almost complete loss of photoreceptors and the substantial loss of bipolar and ganglion cells resulting in severe thinning of the nuclear and plexiform layers especially in the foveal and parafoveal regions [32, 48, 57]. In addition, damages in the Bruch’s membrane, hypertrophy or degeneration of the retinal pigmentary epithelium, hypomyelination of the optic nerve, decreased corneal endothelial cell density, and increased corneal thickness were also reported [11, 33, 34, 48, 49, 62, 64].
Overall, the initial disease symptoms in SCA7 correlate well with the primary neuropathology. However, as the disease progresses, the symptoms become more diverse and the degenerative process more widespread.
ATXN7 Protein
ATXN7 Expression and Structure
In situ hybridization and immunohistological analyses showed that ATXN7 gene is widely expressed in neural and non-neural tissues [65–71]. The protein is present in cytoplasm and nuclei of neurons in different ratio depending on the brain region. Various subcellular localizations were even observed in a single neuronal type such as the Purkinje cell, in which ATXN7 can be exclusively cytoplasmic or nuclear or in both compartments [70]. In the retina, ATXN7 is present in all neurons, located in nuclei and inner segments of photoreceptors and absent from their outer segments. The subcellular localization of ATXN7 is thus highly regulated, suggesting different functions. However, there is no apparent correlation between cellular or subcellular localization and the vulnerability of neurons to degenerate in SCA7.
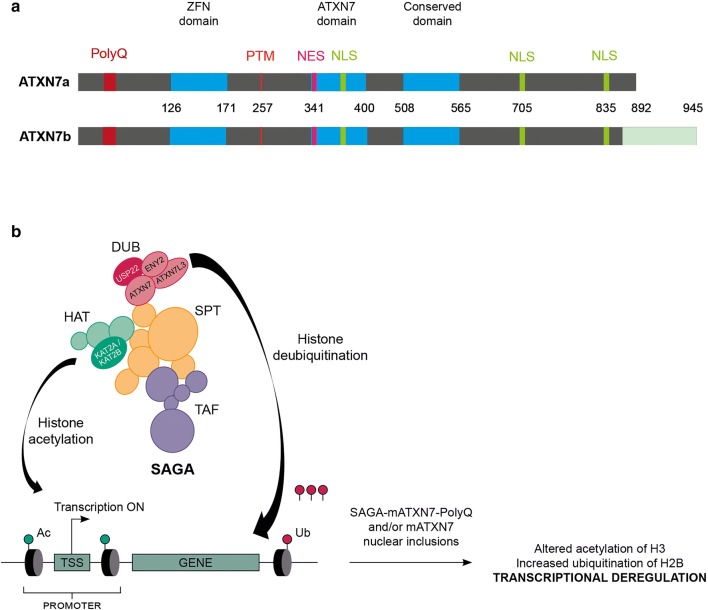
Consistent with the nucleo-cytoplasmic localization, ATXN7 contains three nuclear localization signals (NLS) and one nuclear export signal (NES) (Fig. 1a). The subcellular localization of ATXN7 might also be controlled by alternative splicing of its pre-mRNA, which leads to the production of two different isoforms, ATXN7a and ATXN7b (Fig. 1a). ATXN7b (945 aa) has a longer C-terminus and is found predominantly in the cytoplasm [72], while ATXN7a (892 aa) is predominantly nuclear. The extent to which each isoform contributes to the pathogenesis remains unclear, as most studies have been done so far with ATXN7a.
Fig. 1.
ATXN7 isoforms and SAGA complex. (A) ATXN7a and ATXN7b isoforms corresponding to a wild-type allele with 10 CAG-encoding polyQ tract, are generated by alternative splicing. The conserved domains are indicated as blue boxes: a typical C2H2 zinc-finger (ZNF) motif, an atypical Cys-X9–10–Cys-X5–Cys-X2-His motif known as ATXN7 domain and a third conserved domain. NLS nuclear localization signal, NES nuclear export signal, PTM posttranslation modification site for acetylation and SUMOylation. (B) SAGA is composed of four modules: SPT module forming the core architecture, the TAF module interacts with transcriptional machinery, the HAT module contains histone acetyltransferease KAT2A/KAT2B, and the DUB module contains the deubiquitination enzyme USP22, ATXN7, ATXN7L3, and ENY2. ATXN7 anchors DUB in the core complex. SAGA plays a role in RNAPII transcription by acetylation of histone H3 on gene promoter, deubiquitination of histone H2B on gene body, and interaction with transcriptional machinery. mATXN7 alters SAGA epigenetic activities, following its integration in the complex or by sequestration of SAGA components in nuclear inclusions, leading to transcriptional deregulation.
Sequence analysis also showed that ATXN7 has two domains homologous to the yeast protein Sgf73 [17, 73, 74]. Sgf73 is a component of the multiprotein SAGA complex involved in chromatin remodeling. Biochemical studies confirmed that ATXN7 in human is also a core component of SAGA, initially identified as the TBP-free TAF-containing complex (TFTC) or the SPT3-TAF9–GCN5 (STAGA) complex [17, 75]. Despite limited homology between Sgf73 and ATXN7, the human protein can complement the loss of SGF73 in yeast [76]. Additional sequence analysis led to the identification of three paralogs, ATXN7L1, ATXN7L2, and ATXN7L3, that are consistently present in vertebrates [17, 77]. ATXN7 and its paralogs share the 2 homologous domains [17]. Domain I is a typical C2H2 zinc-finger motif, while domain II has an atypical Cys-X9–10–Cys-X5–Cys-X2-His motif, now known as SCA7 domain (InterPro: IPR013243). A third domain (III) is common to all ATXN7 paralogs, apart for ATXN7L3 and its Sgf11 yeast ortholog. The polyQ motif is unique to ATXN7 and its vertebrate orthologs.
ATXN7 and SAGA Functions
SAGA is composed of four distinct modules forming a core architecture responsible for interaction with transcriptional machinery, and chromatin modifications through histone acetyltransferase (HAT) and deubiquitinase (DUB) activities (Fig. 1b). In the HAT module, the acetyltransferase KAT2A/KAT2B acetylates lysine 9 of histone H3 (H3K9ac) located on genes promoters, allowing chromatin decompaction and accessibility to transcription factors. In the DUB module, the ubiquitin protease USP22 removes monoubiquitin from lysine 120 on histone H2B (H2Bub) on gene bodies to optimize transcriptional initiation and elongation by RNAPII. SAGA complex acts as a bona fide coactivator for all RNAPII transcribed genes [18]. In yeast, SAGA inactivation strongly decreases mRNA synthesis of all active genes transcribed by RNAPII.
ATXN7/Sgf73 belongs to the DUB module and anchors DUB to SAGA [78]. Domain I of ATXN7/Sgf73 mediates interactions between the three other DUB components, the yeast Ubp8, Sgf11, and Sus1 (respectively, USP22, ATXN7L3, and ENY2 orthologs in human) [79, 80] (Fig. 1b). Deletion of SGF73 in yeast disrupts the integrity of DUB and as a consequence, the genome level of H2Bub increases [81]. In fly and human cells, ATXN7 inhibition also leads to dissociation of DUB from SAGA, and in contrast to yeast, a DUB activity seems to persist in the absence of ATXN7 and the level of H2Bub decreases [82]. ATXN7 can also bind nucleosomes through its SCA7 domain [83]. This binding is thought to fine-tune the deubiquitination activity of SAGA through optimal positioning of DUB relative to its substrate. Therefore, ATXN7/Sgf73 appears to be a functional molecular scaffold of SAGA that plays a role in the regulation of H2B ubiquitin levels.
SAGA is involved in diverse cellular processes including cell growth and survival, genomic integrity, and cancer. Interestingly, it was shown that yeast lacking the DUB components SGF73, SGF11, and UBP8 are exceptionally long-lived [84]. This new function of DUB in aging depends on sirtuin pathway. Furthermore, chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) revealed that Sgf73 binds to promoter of genes encoding ribosomal proteins involved in aging regulation in yeast [85]. ATXN7, therefore, may also play a similar role in aging, which might be relevant to the pathogenesis of SCA7.
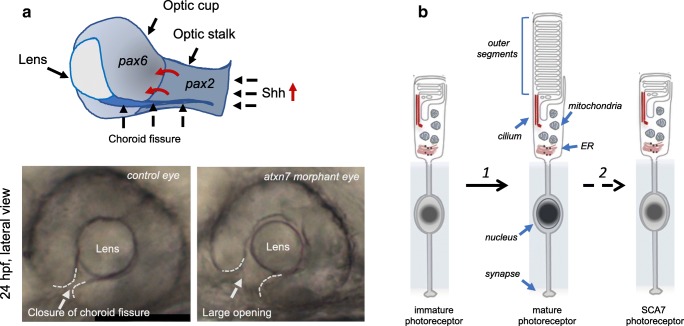
In metazoans, SAGA functions appear critical for diverse neural development processes [86]. Interestingly, subunits of the DUB module play a role in the proper development of the visual system in flies. For instance, Drosophila mutants for Nonstop or Sgf11 (the ortholog of USP22 and ATXN7L3, respectively) cause a loss of glial cells in the lamina plexus of the optic lobe, resulting in misprojection of photoreceptor axons into the medulla [87–89]. While flies with loss of Atxn7 ortholog die at prepupation, a small number of “escapers” were observed with lesions throughout neural and retinal structures [82]. Consistently, targeted RNAi knock down of Atxn7 expression within the retina and lamina leads to a progressive age-dependent retinal degeneration [82]. In contrast to fly, inactivation of zebrafish atxn7 primarily results in ocular coloboma [77], a structural malformation responsible for visual impairment in about 1/5000 live birth in human (Fig. 2a). The lack of Atxn7 leads to elevated Hedgehog signaling in the forebrain, which alters the expression domain of pax6 and pax2 genes and causes an alteration of proximo-distal patterning of the optic vesicle during early eye development of zebrafish, and hence coloboma. The role of Atxn7 in the regulation of Hh signaling is consistent with previous work demonstrating that defective mouse KAT2A (also named GCN5), another component of SAGA, causes alteration of Shh signaling and brain malformation [90]. Interestingly, at later developmental stages of zebrafish, atxn7 inactivation leads to malformations of the outer segment of photoreceptor. The differentiation of immature photoreceptors into mature ones is under the control of the transcription factor Crx, and it ends with the formation and elongation of outer segments, which contains the photopigment rhodopsin and cone opsins. Fish lacking Atxn7 show reduced expression of crx and rhodopsin genes [77, 91]. Thus, the observation in zebrafish indicates that Atxn7 is involved in the proximo-distal patterning during early eye development and in photoreceptor full differentiation during late eye development [77] (Fig. 2). Interestingly, Yanicosta et al. [91] showed that zebrafish Atxn7 is also required for full differentiation of Purkinje neurons of the cerebellum, another neuronal type highly affected in SCA7.
Fig. 2.
ATXN7 function in zebrafish eye morphogenesis and photoreceptor differentiation and dysfunction in SCA7 photoreceptor differentiation. (A) Inactivation of zebrafish atxn7, through antisense morpholinos and crispr/cas9 approach, leads to coloboma, an incomplete closure of the eye cup. During early eye morphogenesis, the closure of the choroid fissure is controlled by Shh signaling from the forebrain, which regulates the precise proximo-distal patterning of pax6 expression in the optic cup and pax2 in the optic stalk. When atxn7 is inactivated in zebrafish, the level of Shh is increased (red arrow) and the expression pattern of pax2 is extended in the optic cup (curved red arrows), leading to much larger opening of the choroid fissure at 28-h postfertilization (hpf) compared to control (lower panels) and later to coloboma (bright field images of control and atxn7 morphant eyes; YT, unpublished data). (B) The differentiation of immature photoreceptors into mature ones involves to the formation of outer segments, a structure entirely filled with discs of folded double membranes where phototransduction takes place, and is associated with chromatin compaction. In mature photoreceptors, outer segments are dynamic structures undergoing daily partial shedding at the apical side and resynthesis at the base. Depletion of zebrafish Atxn7 leads to incomplete formation of outer segments, indicating that Atxn7 is involved in full photoreceptor differentiation [1]. In SCA7, mATXN7 toxicity leads to down-regulation of photoreceptor genes involved in outer segment formation and function, and hence photoreceptor outer segments are progressively lost [2] due to a lack of renewal. SCA7 photoreceptors also show chromatin decompaction.
Despite the abundance of ATXN7 in the cytoplasm, only few studies have looked into its cytoplasmic function. Nakamura et al. [92] showed that overexpressed ATXN7 associates with microtubules and may play a role in their stabilization. Using yeast two-hybrid screen, Kahle et al. [93] found that ATXN7 interacts with several cytoplasmic proteins, including the CEP70 and CEP72 which localize at centrosomes and ciliary microtubules. ATXN7 also localizes to centrosome and cilia in mammalian cells (YT, unpublished data). Recently, it was shown that KAT2A/KAT2B are also associated to centrosome and acetylate several cytoplasmic targets [94]. The physiological role of ATXN7 in the cytoplasm thus deserves further investigation, as it might help to understand pathomechanisms underlying SCA7.
Mutant ATXN7 Misfolding and Accumulation
The wild-type ATXN7 is expressed at low level in most tissues. A major consequence of polyQ expansion in the mutant ATXN7 (mATXN7) is the progressive accumulation of misfolded forms of the protein in the brain. Over time, misfolded mATXN7 forms large aggregates in cell nuclei, the so-called nuclear inclusions (NIs) [95, 96]. In postmortem brains, NIs are widely distributed in degenerating and spared brain regions [47], indicating that NIs per se might be poor indicator of mATXN7 toxicity. However, in SCA7 knock-in (KI) and transgenic (tg) mouse models (see Table 1), misfolded mATXN7 accumulates faster in nuclei of vulnerable neurons such as photoreceptors and Purkinje cells [95–97], indicating a close link between accumulation and toxicity. Furthermore, NIs sequester many cellular proteins including proteasomes, chaperones, RNA binding proteins, transcription activators such as the CREB-binding protein (CBP), and subunits of SAGA complex (see Table 2), which may therefore lose their biological functions and contribute to the pathogenesis.
Table 1.
List of SCA7 mouse models
| Model* | Design** | Pathology*** | Ref. |
|---|---|---|---|
| P7E (tg) | Pcp-2 (ATXN7 90Q) (Purkinje cells) | Motor and cerebellar pathology (onset 11 months) | [97] |
| B7E2 (tg) | PDGF-ß (ATXN7 128Q) (ubiquitous) | Motor and cerebellar pathology (onset 3-5 months) | [95] |
| R7E (tg) | Rhodopsin (ATXN7 90Q) (rod photoreceptors) | Retinopathy (onset 5 weeks) | [97] |
| Prp SCA7-c92Q (tg) | Murine prion (ATXN7 92Q) (brain except Purkinje cells; retina) | Motor and cerebellar pathology (onset 13-15 months); Retinopathy (onset 11 weeks) | [98] |
| moPRP-Flag-SCA7-92Q-myc (tg) | Murine prion (Flag-ATXN7 92Q-myc) (brain except Purkinje cells; retina) | Motor and cerebellar pathology (onset 13-15 months) | [99] |
| Gfa2-SCA7-92Q (tg) | Human Gfa2 (ATXN7 92Q) (Bergmann glia) | Cerebellar pathology (9-12 months) | [100] |
| PrP-floxed-SCA7-92Q BAC (tg) | BAC murine prion (Floxed ATXN7 92Q) (whole brain) | Motor and cerebellar pathology (onset 21 weeks) | [101] |
| PrP-Ataxin-7-92Q-D266N | Murine prion (ATXN7 92Q with D266N mutation) (brain except Purkinje cells; retina) | No motor and cerebellar phenotype; no retinopathy; die prematurely at 40-63 week-old | [102] |
| Ataxin-7-Q52 (tg) | PDGF-β (ATXN7 52Q) (whole brain) | Motor and cerebellar pathology (onset 9 months) | [103] |
| SCA7266Q/5Q (KI) | Mouse Atxn7 266Q | Motor, cerebellar and hippocampal pathology ; Retinopathy (onset 5 weeks) | [96] |
| SCA7100Q/100Q (KI) | Mouse Atxn7 100Q | Motor, cerebellar and hippocampal pathology; Retinopathy (onset 11 months) | [104] |
*Transgenic (tg); knock-in (KI)
**Promoter (ATXN7 protein and polyQ length) (tissue or cell expression)
***Major pathological features (onset of pathology when known)
Table 2.
List of proteins found in ATXN7 nuclear inclusions (NIs)
| Symbol | Description | Function | SCA7 model | Ref. |
|---|---|---|---|---|
| TAFII30 | TATA-box binding protein associated factor 10 | Subunit of TFIID, transcription initiation factor | B7E2 Tg mice | [95] |
| TFIIEα | Transcription initiation factor IIE subunit alpha | Component of basal transcriptional machinery | B7E2 Tg mice | [95] |
| TFIIFβ | Transcription initiation factor IIF subunit beta | Component of basal transcriptional machinery | B7E2 Tg mice | [95] |
| XPB (p89) | TFIIH basal transcription factor complex helicase XPB subunit | Subunit of TFIIH; nucleotide excision repair | B7E2 Tg mice | [95] |
| KAT2A (GCN5) | Lysine acetyltransferase 2A | Acetyltransferase, transcriptional activator; SAGA HAT component | human astrocytes | [105] |
| USP22 | Ubiquitin-specific peptidase 22 | Deubiquitination of histones H2A and H2B; transcriptional activator; SAGA DUB component | HEK293T cells | [106] |
| ATXN7L3 | Ataxin-7-like protein 3 | SAGA DUB component; USP22 and ENY2 recrutment into SAGA | human astrocytes | [107] |
| HDAC3 | Histone deacetylase 3 | Histone deacetylase activity; transcription repression | HEK293T cells | [108] |
| CBP (KAT3A) | CREB binding protein | Transriptional co-activator; Acetyltransferase |
SCA7 patient brain SCA7266Q/5Q KI mice |
[96, 109, 110] |
| CRX | Cone-rod homeobox | PR-specific transcription factor | HEK293T cells | [98] |
| P53* | Tumor protein P53 | Transcriptional factor, cellular stress response regulation | PC12 cells | [111, 112] |
| PML | Promyelocytic leukemia protein | Transcriptional regulation, apoptosis, senescence, DNA damage response, and viral defense mechanisms | SCA7 patient brain | [109] |
| SNF5 | SWI/SNF-related matrix-associated protein | Component of SWI/SNF remodelling complex | B7E2 Tg mice | [95] |
| FUS/TLS | FUS RNA-binding protein | DNA and RNA metabolism, regulation of transcription, RNA splicing, RNA export to the cytoplasm | LV-mouse model of SCA7 | [113] |
| Phosphorylated TDP-43 | TAR DNA-binding protein 43 | DNA and RNA binding protein; regulation of transcription and splicing | LV-mouse model of SCA7 | [113] |
| MBNL1 | Muscleblind-like splicing regulator 1 | Splicing regulation | LV-mouse model of SCA7 | [113] |
| HDJ-2 (DNAJ1) | DnaJ heat shock protein family (Hsp40) member A1 | Chaperon protein |
SCA7 patient brain B7E2, R7E, P7E Tg mice HEK293 and SH-SY5Y cells |
[95, 97, 109, 110] |
| HSC70 (HSPA8) | Heat shock protein family A (Hsp70) member 8 | Chaperon protein | SCA7266Q/5Q KI mice | [95, 96] |
| HSP70 (HSPA7) | Heat shock protein family A (Hsp70) member 4 | Protein folding, cellular stress control | Neurons in Drosophila expressing mATXN7T COS-7 cells | [114–116] |
| SUMO 1, 2, 3 | Small ubiquitin-like modifier 1, 2, and 3 | Nuclear transport, transcriptional regulation, apoptosis, and protein stability |
human cortex SCA7266Q/5Q KI mice Atxn7100Q/5Q KI mice HeLa cells |
[114, 117] |
| RNF4 | E3 ubiquitin-protein ligase RNF4 | SUMO-dependent ubiquitin ligase; proteasomal degradation | HeLa cells | [117] |
| UB | Ubiquitin | Protein degradation |
SCA7 patient brain HEK293, SH-SY5Y, PC12, and COS-7 cells SCA7 266Q/5Q KI mice R7E, P7E Tg mice Neurons in Drosophila expressing mATXN7T |
[96, 97, 110, 114, 116, 118, 119] |
| P62 | Ubiquitin-binding protein P62 | Autophagy receptor |
SCA7 patient brain SCA7 266Q/5Q KI |
[118] |
| S4 (MTS-2) | Proteasome 26S subunit, ATPase 1 | 19S regulator unit of proteasome; ATP-dependent degradation of ubiquitinated proteins | R7E, P7E Tg mice | [97] |
| S7 (MSS-1) | Proteasome 26S subunit, ATPase 2 | 19S regulator unit of proteasome; ATP-dependent degradation of ubiquitinated proteins |
SCA7 patient brain R7E, P7E Tg mice Neurons in Drosophila expressing mATXN7T |
[97, 109, 110, 116] |
| RPN10 | Proteasome 26S subunit, non-ATPase 4 | 19S regulator unit of proteasome | COS-7 cells | [114] |
| α1,2,3,5,6,7; β2 | Proteasome subunit alpha 1,2,3,5,6,7; beta 2 | 20S core unit of proteasome; ATP-dependent degradation of ubiquitinated proteins |
R7E, P7E Tg mice HEK293, and SH-SY5Y cells |
[97, 110, 120] |
| CASP-7 | Caspase 7 | Endopeptidase, apoptosis cascade |
COS-7 cells Prp-SCA7 Tg mice |
[121] |
| CASP-3 | Caspase 3 | Activation cascade of caspases involved in apoptosis | SCA7 patient brainHEK293, SH-SY5Y, COS-7 cells | [110, 114, 120] |
| N-terminus of APP, APLP1, and APLP2 | Amyloid beta-protein precursor (APP) family | Synaptic functions, glucose homeostasis |
PC12 cells P7E Tg mice |
[119] |
Data come from coimmunofluorescence studies. All cell culture systems are based of ATXN7 overexpression
Tg transgenic, KI knockin
*Based on filter retardation results
NIs in different SCA7 mouse models are majorly formed by an amino-terminal fragment of mATXN7 containing the polyQ expansion [97, 122, 123]. The mATXN7 fragment is SDS-insoluble and formic acid-soluble and can be analyzed by western blot [123]. Young et al. [121] showed that caspase-7 can cleave mATXN7 and release a fragment of similar size to the one that accumulates in neurons. In HEK293T cells, the caspase-7 fragment of mATXN7 is more toxic than the full-length mATXN7 [121]. Interestingly, PrP-Ataxin-7-92Q-D266N tg mice expressing a mATXN7 form resistant to caspase-7 cleavage show reduced neurodegeneration, improved visual and motor performance and prolonged lifespan, when compared to SCA7 tg mice which express a mATXN7 that can be cleaved by caspase-7 [102]. These results suggest that the caspase-7 cleavage is a critical step in the pathogenesis and could be a major target for therapy development. The caspase-7 fragment of mATXN7 is short enough to diffuse passively through nuclear pore complexes, but it accumulates in the nucleus, indicating a specific retention mechanism in this compartment [121]. In the nucleus, mATXN7 fragment may cause toxicity by replacing the full-length protein in the SAGA complex or through aggregation and sequestration of SAGA components and other proteins [106, 107].
Interaction of ATXN7 with the histone deacetylase 3 (HDAC3) was shown to stabilize the protein in a deacetylase-independent manner [108]. HDAC3 is increased in the cerebellum of moPrp-Flag-SCA7-92Q-myc tg mice and specially localizes in Purkinje cells and Bergmann glia, two cell types involved in SCA7 cerebellar pathology. Further studies shall elucidate the functional nature of this interaction and whether increased HDAC3 level contributes to the SCA7 pathogenesis.
Posttranslational modifications of lysine-257 (K257) also play a critical role in the turnover and accumulation of mATXN7. The accumulation of the mATXN7 fragment is associated with an increased acetylation of K257, which is located close to the caspase-7 cleavage site [99]. In the absence of lysine acetylation, the fragment is readily degraded by autophagy, whereas K257 acetylation seems to prevent the clearance of the mATXN7 fragment. Interestingly, the same K257 residue was shown to be a specific site for ATXN7 SUMOylation [114]. Marinello et al. showed that mATXN7 is more SUMOylated than the wild-type form, and that addition of poly-SUMO2/3 chains to mATXN7 promotes its recruitment to clastosomes [117]. The clastosomes are specialized nuclear bodies formed by the promyelocytic leukemia (PML) protein and containing active proteasomes for protein degradation. In PML clastosomes, poly-SUMOylated mATXN7 is ubiquitinated by the SUMO-dependent ubiquitin ligase RNF4 for its final degradation by the proteasome. Treatment with interferon beta increases SUMO pathway enzymes and clastosome formation by increasing PML expression [117], promoting further poly-SUMOylation of mATXN7 and enhanced clearance. Moreover, interferon beta increases the survival of rat primary Purkinje neurons overexpressing mATXN7 [124, 125]. Importantly, in the brain of SCA7 patients, mATXN7 aggregates often colocalize with PML nuclear bodies [109, 126]. Endogenous clastosomes might prevent the accumulation of mATXN7 for several decades before onset of aggregation. Over time, the degradative activity of clastosomes might be overwhelmed by the aggregation process.
SCA7 Pathology in Animal Models
Several KI and tg mouse models have been generated over the past years, reproducing several aspects of the disease including retinopathy, motor defects, cerebellar pathology and premature death, and have provided important insights into the nature of SCA7 neurodegeneration (Table 1) ( also see reference (127) for review on SCA7 mouse models).
Progressive Loss of Neuron Identity
The retina—as a simple and accessible central nervous system tissue—is extremely helpful to delineate close correlations between functional, morphological, and molecular alterations during pathological process. In SCA7 mouse models, the retina develops normally before showing progressive electroretinograph dysfunction and thinning of the retinal layers [96–98]. Cone photoreceptors are affected before rods, as in patients, and photoreceptor dysfunction is followed by cell death [96, 98, 123, 128]. The loss of photoreceptor function correlates with the early and progressive reduction of expression of photoreceptor-specific genes important for the phototransduction and outer segment formation [96, 98, 129, 130].
In mouse retina, about 75% of all cells are photoreceptors, and 97% of these are rods. The R7E tg mice, which express mATXN7 only in rods, have allowed the long-term study of mATXN7 toxicity in a single type of neuron and provided critical information about the nature of photoreceptor dysfunction [97]. R7E rods primarily lose their outer segments [123], a structure entirely filled with discs of folded double membranes where phototransduction takes place (Fig. 2b). In mature photoreceptors, outer segments are dynamic structures undergoing daily partial shedding at the apical side and re-synthesis at the base. Thus, outer segments renewal necessitates a high biosynthetic activity and specific gene expression programs [131]. Expression profile analysis of R7E retina unveiled the alteration of genetic programs controlling the maintenance of mature photoreceptor identity. This is evidenced on the one hand by the downregulation of photoreceptors specific transcription factor coding genes (Crx, Nrl (neural retina leucine zipper protein), and Nr2e3 (Nuclear Receptor Subfamily 2, Group E, Member 3)) as well as many of their target genes involved in outer segment renewal and function, and on the other hand by the re-activation of genes (Stat3 and Hes5) that normally inhibit the premature differentiation of photoreceptors during development [129]. After the loss of outer segments, R7E rods progressively lose their polarity, and relapse to round cell shape, consistent with the loss of photoreceptor differentiation identity [123]. As zebrafish study showed that Atxn7 is involved in the full differentiation of photoreceptors during development, it is proposed that one of the primary toxic effects of polyQ expansion is to alter the function of human ATXN7 in the maintenance of differentiated photoreceptors (Fig. 2b).
Whether other neurons vulnerable in SCA7 also progressively lose their differentiation identity remains to be investigated. Purkinje neurons are excellent candidate cells to study, as zebrafish Atxn7 was shown to be involved in their differentiation during development [91]. SCA7 mouse models show motor dysfunction and signs of Purkinje cell pathology. The expression profile of the whole cerebellum of SCA7266Q/5Q KI mice (in which an expansion of 266 CAG repeats was introduced in one allele of mouse Atxn7) and of different tg models revealed gene deregulations affecting specific neuronal pathways including synaptic transmission, axonal transport, and neuronal differentiation [103, 132, 133]. However, the current approaches have been insufficient to provide a deep understanding of gene deregulation at a single cerebellar neuronal-type level. Nevertheless, one interesting finding is the downregulation of a set of myelin-associated genes (Cnp, Mag, Mbp, Mog, Mobp, and Plp1) and of their regulators, the transcription factor OLIG1 and transferrin in the Ataxin-7-Q52 tg mice, which express mATXN7 under PDGF promoter [103]. This is consistent with the loose and poorly compacted myelin sheaths observed in the cerebellar white matter of these mice, and with the myelin pallor and loss of myelinated fibers reported in the cerebellar white matter of SCA7 patients [47]. mATXN7 toxicity might compromise genetic programs controlling oligodendrocyte maturation and myelin sheath integrity and function.
Cell Autonomous and Non-cell Autonomous Toxicity of mATXN7
Mouse studies showed that mATXN7 toxicity is cell autonomous and non-cell autonomous [97, 101, 122]. In R7E tg mice, the degeneration affects rods, which express mATXN7, as well as cones and bipolar neurons, which do not express the mutant protein [97, 129]. In the P7E tg mice, which express mATXN7 only in Purkinje cells, the cerebellar pathology is very mild [97]. In contrast, in the PrP-SCA7-c92Q tg mouse model [122], Purkinje neurons show signs of degeneration despite the fact the mouse prion promoter (MoPrP) used in this model drives the expression of mATXN7 in all cerebellar cells, except for Purkinje cells. In PrP-SCA7-c92Q tg mice, Purkinje cell pathology might result in part from dysfunction of Bergmann glia cells [100]. Indeed, Bergmann glia cells, which express mATXN7, have low levels of the glia-specific glutamate transporter GLAST, and hence a decreased glutamate uptake function. This might in turn cause glutamate accumulation and excitotoxicity in Purkinje neurons [122]. Indeed, when mATXN7 is expressed only in Bergmann glia cells, as in Gfa2-SCA7-92Q tg mice [100], the level of GLAST expression is also reduced and Purkinje cells degenerate, although at a milder severity than in PrP-SCA7-c92Q tg mice.
An even more complex cell-cell interaction between Bergmann glia, Purkinje and inferior olive neurons in the SCA7 cerebellar dysfunction was revealed using the PrP-floxed-SCA7-92Q BAC tg mice. In this model, mATXN7 cDNA is expressed in the whole cerebellum and can be deleted in specific cell types upon crossing with mice expressing Cre recombinase under cell-specific promoters [101]. Deletion of mATXN7 from Bergmann glia has mild beneficial effects on the pathology and does not prevent Bergmann glia degeneration, which thus appears to be also non-cell autonomous. In contrast, deletion of mATXN7 from Purkinje and inferior olive neurons improves motor performance and corrects Bergmann glia pathology. Finally, deletion of mATXN7 in the three cell types is more effective to prevent the pathology. Thus, the SCA7 cerebellar pathology results from dysfunctions of different cell types and of their close cell-cell interactions.
Pathomechanisms Underlying SCA7 Neurodegeneration
Deregulation of Gene Expression
Several studies performed in cultured cells and mouse have shown that deregulation of gene expression is central in the pathogenesis. In SCA7 mouse retina [96, 98, 123, 129, 134], the level of messenger RNA (mRNA) coding for photoreceptor proteins progressively decreases over time, and correlates with photoreceptor dysfunction and loss of differentiation [123]. The decreased mRNA level results from transcriptional deficit, as ChIP-qPCR analysis showed a reduced occupancy of RNAPII on promoter of photoreceptor genes [130]. Yet, the molecular mechanisms underlying selective transcription alterations in SCA7 are not fully understood.
mATXN7 was shown to affect SAGA functions through different mechanisms, including sequestration of SAGA components in NIs [106, 107], alteration of SAGA integrity [75, 76], and aberrant interaction with SAGA component [135] or with promoter-bound transcription factors [98, 136]. There is also evidence that epigenetic marks regulated by SAGA, histone H3 acetylation and H2Bub deubiquitination, are altered in SCA7 [75, 76, 105–107, 130]. In the case of SCA7 mouse photoreceptors, ChIP-qPCR analysis of histone H3 acetylation on promoter of photoreceptor-specific genes has led to contradictory results. Histone H3 is hypoacetylated on photoreceptor gene promoters in the PrP-SCA7-c92Q tg model [75] and hyperacetylated in the R7E tg mice [130]. The discrepancy between these two studies remains unclear. The issue is likely to be solved by a genome-wide study of histone H3 acetylation using ChIP-seq. In the PrP-SCA7-c92Q tg mice, it was shown that in combination to histone hypoacetylation, mATXN7 in SAGA suppresses CRX transactivation activity on photoreceptor gene promoter, which may explain in part the loss of photoreceptor gene expression [98, 136]. Monoubiquitination of H2B is globally increased in cultured cells expressing mATXN7 [105–107]. This is likely due to sequestration of DUB components, ATXN7L3 and USP22, in mATXN7 aggregates. Increase of H2Bub was also reported in the cerebellum of SCA7100Q/5Q KI mice [107]. Although the current data support that SAGA dysfunction accounts for SCA7 transcriptional dysregulations, it remains to be determined how the dysfunction of a general co-activator complex like SAGA, which is involved in the expression of all RNAPII-regulated genes, would only affect specific subsets of genes in SCA7 affected tissues. Interestingly, while normal rod nuclei contain a large central region of heterochromatin, rod nuclei in SCA7266Q/5Q KI and R7E tg mice showed severe decondensation of heterochromatin that increases rod nuclear volume [130, 134]. Heterochromatin decondensation in R7E retina could result from H3 hyperacetylation and/or from a low expression of histone H1 [137], and might account for major changes in the expression level of photoreceptor-specific genes, which necessitate high expression to renew the outer segments.
The retina and cerebellum are highly affected in SCA7; however, the mechanism underlying tissue-specificity of the disease is unclear, especially as ATXN7 and SAGA components are widely expressed. Interestingly, Tan et al. [138] showed that SAGA can regulate the expression of miR-124, a microRNA highly expressed in the cerebellum and retina. Moreover, miR-124 can regulate the amount of ATXN7 transcripts. In SCA7 KI mice, miR-124 was found expressed at low level, and in turn the Atxn7 transcripts were high. Therefore, posttranscriptional upregulation of mATXN7 transcripts in the retina and cerebellum might account for the tissue specificity of SCA7.
Besides SAGA dysfunction, other mechanisms are proposed to contribute to transcriptional alterations observed in SCA7. mATXN7 aggregates sequester CBP [95], a histone acetyltransferase, and impair CBP-mediated transcription in SK-N-SH neuroblastoma cells [139]. CBP is sequestered in aggregates in other polyQ diseases and is thought to be involved in epigenetic alterations found in HD [140]. In SK-N-SH cells, mATXN7 also alters the transcription mediated by the retinoid-related orphan nuclear receptor RORA [139]. This finding is interesting because Rora gene is deleted in Staggerer mice, causing defect in Purkinje cells differentiation and congenital ataxia. Furthermore, in SCA1 mice, RORA forms a complex with ATXN1 and RORA-target genes are highly deregulated [141]. Finally, the activity of HDAC3, which interacts with ATXN7, appears to be increased in the retina of moPrP-Flag-SCA7-92Q-myc tg mice [108]. HDAC3 activity is also affected in other polyQ disorders and its specific inhibition improves disease in HD [142, 143]. The potential contribution of CBP, RORA, and HDAC3 to the SCA7 pathogenesis thus deserves further investigations, using genetic or pharmacological approaches in SCA7 mice. mATXN7 aggregation was also shown to induce a stress response involving the JNK/c-JUN signaling pathway in R7E tg retina [115]. Activation of c-JUN in R7E retina plays a role in the repression of the Nrl coding for photoreceptor specific transcription factor [144]. Interestingly, the use of genetic approach to inhibit c-JUN activation in R7E mice delays retinal degeneration.
Most interesting, a study by Ajayi et al. [145] made a link between metabolic defect in SCA7 and transcriptional alterations. Indeed, early studies reported mitochondrial abnormalities in liver and skeletal muscle biopsies from SCA7 patients [146–149]. Abnormal mitochondria were also observed in the retina of R7E tg mice [123]. Furthermore, reduced electron transport chain activity and metabolic acidosis were reported in muscle biopsy of patients [147]. Using PC12 cells expressing mATXN7, Ajayi et al. [145] showed that p53 is sequestered in mATXN7 aggregates and that p53 transcriptional activity is reduced, leading to dysregulation of metabolic proteins, such as TIGAR, AIF, and NOX1. These alterations result in a reduced respiratory capacity of cells, associated with an increased reliance on glycolysis for energy production and a subsequent reduction of ATP. Investigation of these transcriptional and metabolic pathways in SCA7 mice is thus warranted, in particular because loss of AIF in mice results in neurodegeneration of cerebellar and retinal neurons.
Mitochondrial Dysfunction and Metabolic Impairment
A recent study in SCA7 patients and in mice provided additional insight into the impairment of mitochondrial function [150]. 31P magnetic resonance spectroscopy (MRS) revealed the inability of SCA7 patients to increase ATP production during completion of a visual task. When SCA7266Q/5Q KI mice were housed in metabolic cages to measure indirect calorimetry, they presented deficits in oxygen consumption rate and respiratory exchange. Furthermore, SCA7 mouse Purkinje neurons show increased fragmentation of the mitochondrial network and significant enlargement of mitochondria.
To investigate further the mitochondrial function in SCA7, Ward et al. [150] generated induced pluripotent stem cells (IPSCs) from patients (with 50Q, 65Q, or 70Q) and related controls, as well as isogenic IPSC clones in which endogenous ATXN7 was knockout and cDNAs for wild type (10Q) or mutant (113Q) ATXN7 were overexpressed. Interestingly, after differentiation into neural progenitor cells (NPCs), the mitochondrial network also shows increased fragmentation in SCA7 patient NPCs 70Q and isogenic NPCs ATXN7-113Q, in comparison to their control NPCs. Isogenic NPCs ATXN7-113Q have impairment in mitochondrial bioenergetics as observed by extracellular flux analysis. Moreover, the authors provided evidence suggesting that mitochondrial dysfunction is related to deficit in production of nicotinamide adenine dinucleotide (NAD+), which represents a substrate for NADH production, the key electron donor involved in oxidative phosphorylation. For instance, both SCA7 mice and isogenic NPCs ATXN7-113Q show reduced expression of NMNAT1, which catalyzes the synthesis of NAD+ in the salvage pathway. Furthermore, the level of NAD+ is significantly decreased in nuclei and mitochondria of patient NPCs. Finally, metabolomic analysis of SCA7 patients revealed altered tryptophan-kynurenine pathway upstream of NAD+ precursor de novo synthesis. Therefore, these findings have therapeutic implications for SCA7, as numerous strategies are being developed to allow NAD+ repletion in age-related diseases.
Autophagy
Today, autophagy is recognized as a major player of intracellular homeostasis by degrading and clearing most long-lived proteins, damaged mitochondria and aggregate-prone proteins, including those causing neurodegenerative diseases. Defects in the autophagy pathway have been observed in several neurodegenerative conditions. Furthermore, it is suggested that mutation in the autophagic protein ATG5 is responsible for the symptoms of childhood ataxia [151].
Several lines of evidence suggest that the autophagy/lysosome process is affected in SCA7. Firstly, the autophagic flux is impaired in PC12 and HEK293 cells overexpressing mATXN7 [111, 118]. Secondly, there is selective accumulation of various autophagy and endosome/lysosome markers in cerebellar neurons, but not spared striatal neurons of SCA7 patients and KI mice [118], suggesting specific perturbations of autophagic process in vulnerable neurons. Interestingly, transcriptome of SCA7 peripheral blood mononuclear cells (PBMCs) indicated that the upregulated genes were significantly associated with lysosomal machinery function [118]. Furthermore, the expression of the early autophagy-associated gene ATG12, was increased in these patients’ cells in relation with disease severity. The mechanism whereby mATXN7 may inhibit autophagy was investigated in cultured cells (PC12 and HEK293) expressing mATXN7 [111]. Autophagy induction in mammalian cells is regulated by a complex formed by ULK1, FIP200, ATG13, and ATG101. However, in SCA7 cells, the level of ULK1 and FIP200 is reduced, consistent with decreased autophagic activity. It is proposed that aberrant interaction of p53 with FIP200 and their sequestration in mATXN7 aggregates result in decreased soluble FIP200 levels and subsequent destabilization of ULK1. Interestingly, treatment with inhibitor of polyQ aggregation or with a p53 inhibitor that prevents interaction with FIP200, both restored the soluble levels of FIP200 and ULK1, and increased the autophagic activity and cell survival [111].
In summary, these results suggest that the autophagy/lysosome pathway is impaired in cells undergoing neurodegeneration in SCA7 and can be considered as target in therapeutic strategies. Additionally, the changed transcriptional profiling of easily available patient material (PBMC) paves a way to explore potential biomarkers associated to autophagy.
Cell Death Mechanisms
SCA7 patients loose neuronal cells in the cerebellum, retina and other brain regions [48]. Notably, postmortem retina show an almost complete disappearance of photoreceptors [32]. One recurrent neuropathological observation in mouse models of polyQ disorders is the presence in the brain of dark degenerating neurons. Dark neurons are also observed in postmortem brain of HD patients [152]. Dark degeneration occurs in the retina of R7E tg mice and the cerebellum of Prp-SCA7-c92Q tg mice [100, 123]. Dark photoreceptors show atrophic nuclei with atypical scalloped shape and clumping of chromatin, but no fragmentation or blebs associated to apoptotic-like mechanisms. Dark photoreceptors are readily observed in early-onset stage in the SCA7 mice [123].
Interestingly, apoptosis was also observed in the retina of several SCA7 mouse models [96, 98, 123]. In R7E retina, apoptosis occurs for a short time window during early disease stages [123]. Recently, Lebon et al. [153] reported the necessity to use a modified TUNEL protocol to efficiently detect apoptotic photoreceptor nuclei in R7E tg model. The need a dephosphorylation step for TUNEL staining suggests that the DNase activity involved in this cell death exposes 3’P ends, pointing out toward the involvement of the LEI/L-DNase II and AIF endonucleases. LEI/L-DNase II and AIF were indeed translocated in the nucleus of TUNEL-positive photoreceptors. Their implication during apoptosis of SCA7 photoreceptors death has been confirmed by intravitreous injections of specific siRNAs. When either LEI or AIF were downregulated, the number of apoptotic photoreceptors was significantly decreased [153]. Moreover, the results suggest that AIF and LEI activation depend on calpain-1, the protease responsible for AIF release from mitochondria. Accordingly, the injection of calpain inhibitor VI decreased the number of dying retinal cells in SCA7.
Concomitant with the apoptotic wave in R7E mouse retina, stealthy cells expressing proliferation markers were observed and expressed photoreceptor-specific genes afterwards, suggesting that new cells might be produced to replace dead photoreceptors at early disease stages [123]. From these observations, it appears that R7E photoreceptors go through different cell fates as a response to mATXN7 toxicity such as loss of neuronal identity, apoptosis, dark degeneration, and cell proliferation. Different cellular responses may be triggered by different mATXN7 toxic species, as observed by biochemical approach. Indeed, the relative amount of full-length mATXN7, proteolytic fragments, SDS-soluble and insoluble aggregates varies considerably from early to late disease stages and might influence the way how individual neurons respond to proteotoxic stress [123].
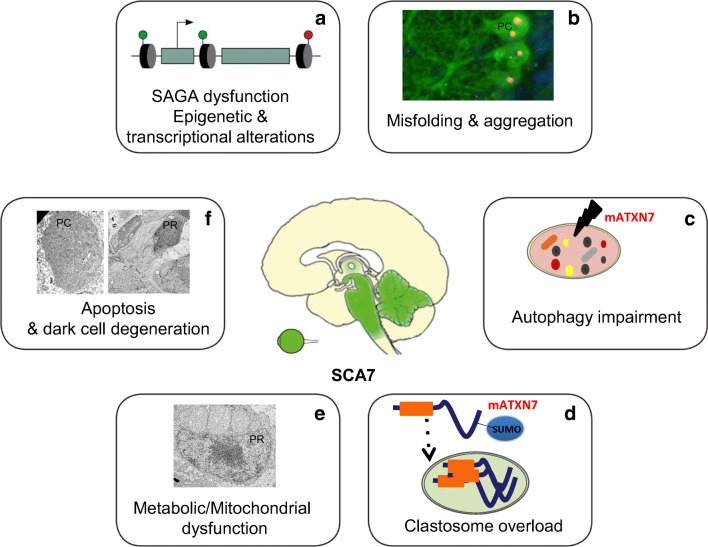
In summary, studies in cellular and mouse models of SCA7 have identified several mechanisms whereby mutant ATXN7 toxicity triggers multiple interconnected pathogenic cascades that cause cellular dysfunctions and lead to cell death (Fig. 3). At the same time, they have provided promising molecular targets to potential treatment of the disease.
Fig. 3.
Overview of pathomechanisms occurring in SCA7. SCA7 pathogenesis triggers multiple pathogenic cascades that cause cellular dysfunctions and lead to cell death. (A) Mutant ATXN7 causes gene deregulation by altering transcription and epigenetics through different mechanisms, including sequestration of SAGA components in nuclear inclusions, alteration of SAGA integrity, and aberrant interaction with SAGA components or with promoter-bound transcription factor such as CRX. (B) Mutant ATXN7 misfolds, accumulates in nucleus, and forms aggregates that sequester a large number of cellular proteins, which may therefore lose their biological functions and contribute to the pathogenesis (fluorescent image of section of SCA7 mouse cerebellum with mATXN7 aggregates (red) in the nuclei of Purkinje cell (green); ANC, unpublished data). (C) Mutant ATXN7 can be degraded by autophagy; however, studies of SCA7 cells, mice and patient’s brains indicate that autophagy is impaired in the disease. (D) Mutant ATXN7 is also degraded by nuclear clastosomes, which might prevent its accumulation for several decades before onset of aggregation. Over time, the degradative activity of clastosomes might be overwhelmed by the aggregation process. (E) SCA7 mitochondria show morphological enlargement and dysfunction, and several lines of evidence indicate metabolic deficits in SCA7 (electron micrograph showing large mitochondira in photoreceptor (PR) of SCA7 mouse; YT, unpublished data). (F) SCA7 patients loose neurons in retina, cerebellum, brainstem and other brain structures. In SCA7 mice, neuronal loss occurs through apoptosis and dark cell degeneration (electron micrographs showing dark degeneration of Purkinje cell (PC) and photoreceptor (PR): YT, unpublished data)
Therapeutic Perspectives
SCA7 and the other polyQ diseases are currently untreatable. Only some symptoms of disease can be partially alleviated using pharmacological and non-pharmacological measures [154]. Interestingly, it was recently shown that physical training significantly improves some clinical features and biochemical parameters of SCA7 [155]. Studies on SCA7 pathomechanisms have provided several directions for therapeutic development aimed at preventing or reversing SCA7 symptomatology.
Preventing Purkinje Cell Degeneration
Purkinje cells are among the most vulnerable neurons in SCA7 and many other SCAs, and as the sole output neurons from the cerebellar cortex, play a critical role in information processing of the cerebellum. Given that excess of glutamate might make SCA7 Purkinje cells vulnerable to excitotoxicity-induced degeneration, strategies diminishing glutamate levels are of high interest. Accordingly, the ceftriaxone, a β-lactamic antibiotic is a promising compound for SCA7 and other SCAs showing excitotoxic Purkinje cell degeneration. Ceftriaxone induces GLT-1 expression and hence promotes glutamate clearance [156]. Administration of ceftriaxone in presymptomatic SCA28 mice (Afg3l2+/− mouse model) protects Purkinje cells from excitotoxicity-mediated dark degeneration and prevents the onset of ataxia [157]. Treatment at postsymptomatic stage is also efficient to stop disease progression. Another potential therapeutic strategy is to provide factors with neurotrophic effects on Purkinje cells. Hepatocyte growth factor (HGF) plays a neurotrophic role in the cerebellum [158]. Overexpression of HGF was shown to provide beneficial effect in ALS mice by maintaining GLT-1 levels [159], and to restore GLT-1 and GLAST levels in SCA7266Q/5Q KI mice, preventing Purkinje cell shrinkage and motor dysfunction [160]. HGF is currently under consideration for therapeutic development of a number of human pathologies including brain injury. It is unclear if approaches aiming at the protection of Purkinje cells would also have beneficial effect on other degenerating neurons in SCA7.
Preventing mATXN7 Accumulation
One of the most significant therapeutic target is the intracellular accumulation of misfolded mATXN7, which positively correlates with SCA7 progression. Several strategies can be considered, such as the inhibition of the production of aggregation-prone mATXN7 fragment, the slowdown of neo-synthesis or the increase of protein clearance. The inhibition of caspase-7 activity, through pharmacological approach or genetic intervention, could be explored as caspase-7 cleavage was shown to be an early step in protein accumulation in SCA7 mice [102]. To increase the clearance of toxic misfolded proteins, several strategies have been implemented to enhance autophagy, a major clearance route for intracytoplasmic aggregates. However, in SCA7 patient brains, mATXN7 mostly accumulates in cell nuclei [48]. Therefore, the use interferon beta, which fosters mATXN7 clearance over the wild-type form through the induction of nuclear PML-clastosomes [124, 125], is a promising avenue. As a first preclinical trial, Sittler and colleagues [124] performed intraperitoneal injection of interferon beta three times a week in 5-week-old presymptomatic SCA7266Q /5Q KI mice. They observed a significant decrease of mATXN7 aggregation and improved motor functions. However, the treatment was not efficient enough to protect against weight loss and premature death at 11 weeks. The partial beneficial effects are likely explained by the very severe phenotype and its rapid progression due to an expansion of 266 CAG repeats, responsible for infantile forms of SCA7 in humans. Since interferon beta has been used for many years in the treatment of multiple sclerosis, it may hold promise as a potential treatment to delay motor symptoms in SCA7 patients.
To improve protein clearance of polyQ proteins, it is proposed to use pharmacological modulation autophagy and ubiquitin-proteasome system or induction of molecular chaperones to assist the protein folding and degradation [161–163]. However, Helmlinger et al. [162] showed that overexpression of heat shock proteins HSP40 and HSP70 is not sufficient to prevent the retinopathy of R7E tg mice. Other strategies aim at inhibiting polyQ aggregation using peptides, antibodies recognizing aggregation-prone polyQ protein conformations or small chemical molecules [163–165]. Most of these approaches show promising results in both cell-based and animal models of polyQ disorders, to inhibit protein aggregation, to restore altered cellular functions, to alleviate behavior deficits and many of them await to be translated in clinical trials [166]. Preventing protein aggregation with small molecules is a therapeutically relevant strategy, as shown with the case of familial amyloid polyneuropathy, where Tafamidis strongly binds transthyretin and prevent its aggregation [167]. However, in contrast to transthyretin, polyglutamine domains are intrinsically disordered and adopt many transient conformations upon aggregation, which are difficult to target efficiently with a single compound. To overcome this limitation, new strategies are been developed to combine molecules targeting distinct steps or subpathways of the aggregation cascade to provide stronger inhibitory effects [168–170]. Strategies that aims at the inhibition of mATXN7 aggregation still need to be evaluated in SCA7 mouse models.
The therapeutic potential of many molecules remains unexplored, because of their intrinsic properties (poor water solubility, poor pharmacokinetic properties), and difficulty to penetrate the blood–brain barrier. There is thus a growing necessity of developing drug delivery system, such as nanoparticules, to improve target specificity and increase bioavailability of a drug in the brain [171, 172].
Silencing Gene Expression
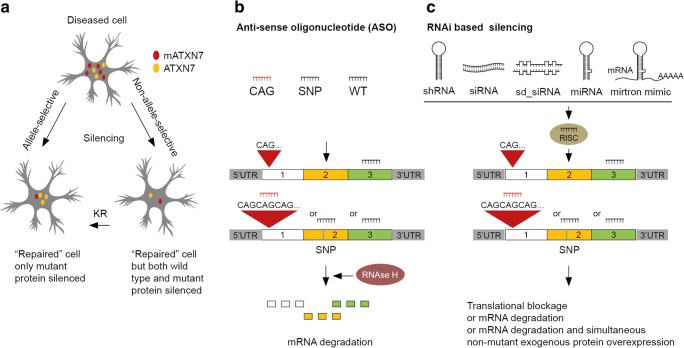
One emerging therapeutic approach in polyQ neurodegenerative disorders is to prevent the expression of toxic proteins using gene silencing. Several different strategies are available to target RNA or DNA of the polyQ-associated genes, and the pros and cons are being evaluated (see [173] for critical review). The current prevailing strategy is transcript targeting with oligonucleotide (ON)-based molecules (Fig. 4). ON-based molecules form duplex with their target RNA and induce RNA degradation or prevent translation through diverse cellular mechanisms, thereby reducing protein production [173]. Preclinical testing of ON-based tools in rodent models of neurodegenerative disorders is well advanced, and small-scale clinical tests are ongoing. For patients affected by polyQ disorders, with typically one normal and one mutant allele, important issues are the potential deleterious effects that could result from concomitant wild-type mRNA silencing. In the case of SCA7, studies in zebrafish and fly indicate that inhibition of the wild-type ATXN7 affects the differentiation of photoreceptor and Purkinje neurons, two highly vulnerable neurons in SCA7 [77, 82, 91]. Taking this concern into account, ON-based strategies, specific to the expanded CAG repeat or with non-allele selectivity, are being developed for SCA7.
Fig. 4.
Potential genetic therapies for SCA7 and other polyQ SCAs. (A) Different gene silencing strategies. Oligonucleotide (ON)-based molecules can be designed in a non-allele-selective manner to lower both the wild type and mutant ATXN7, or in an allele-selective manner to preferentially lower the toxic mutant. A third strategy, named knockdown-replacement (KR), combines a non-allele selective protein lowering with simultaneous delivery of a functional copy of the wild type gene to preserve the protein physiological function. (B) Anti-sense oligonucleotide (ASO) can be designed to develop non-allele-selective approach for ATXN7 silencing (ASO WT), or to preferentially target the mutant allele by the SNP-targeting strategy (ASO SNP) or by CAG-targeting (ASO CAG). The formation of an RNA–DNA hybrid activates RNase H and leads to transcript degradation. (C) Similarly, non-allele-selective and allele-selective downregulation of target transcripts can also be achieved through shRNA, siRNA, sd-siRNA, miRNA, and mirtron. miRNA is generally designed with a mismatch, resulting in translational inhibition. All these molecules act through the RISC pathway: shRNA, siRNA and mirtron lead to target mRNA degradation; sd-siRNA causes translational blockage. miRNA microRNA, mRNA messenger RNA, RISC RNA-induced silencing complex, sd-siRNA self-duplexing siRNA, shRNA short hairpin RNA, siRNA small interfering RNA, SNP single nucleotide polymorphism
Ramachandran et al. [174] were the first to assess an ON-based strategy using the Prp-floxedSCA7-92Q BAC mice, which express human mATXN7 cDNA with 92 CAG repeats, in addition to the endogenous mouse protein. In this study adeno-associated viral vectors were used to introduce miRNA targeting Atxn7 transcripts in the deep cerebellar nucleus of mutant mice. This strategy lowered wild type and mutated ATXN7 by about 35–50% and significantly improved motor functions and cerebellar status compared to untreated mice.
Antisense oligonucleotide (ASO) therapy is also a very promising therapeutic tool. Clinical trials using ASO are ongoing for several neurodegenerative diseases. For instance, IONIS-HTTRx (also known as RG6042), a non-selective antisense drug candidate, delivered using lumbar puncture successfully passed the proof of concept phase I/II trial in which it lowered mutant protein levels by 40–60% in the cerebrospinal fluids [175, 176]. It has recently been reported that ASO-mediated knockdown of Atxn7 might be an effective treatment for SCA7 retinal degeneration [177]. Preclinical treatments using intravitreal injection of Atxn7 ASO were evaluated in SCA7266Q/5Q knockin mice, a genetically accurate model with wild type (5Q) and mutant (266Q) forms of mouse ATXN7. Vitreous humor injections leads to efficient delivery of ASOs into retinal neurons including photoreceptors, and more than 40% knockdown of Atxn7 mRNA expression even 6 weeks after injection. A single-dose injection of Atxn7 ASO at presymptomatic stage resulted in marked reduction of mATXN7 aggregates, and preservation of the photoreceptor outer and inner segments and other histological parameters affected in eyes of untreated mice. Further analysis indicated an amelioration of transcriptional deficits of photoreceptor-specific genes as well as rescue of altered histone H2B deubiquitination and histone H3 acetylation on photoreceptor gene promoters. Finally, and most importantly, ERG of cone and rod photoreceptors measured the improvement of visual function of SCA7266Q/5Q KI mice treated with Atxn7 ASO several weeks after injection. No adverse effect of Atxn7 ASO was observed in wild-type mice. Atxn7 ASO treatment was also efficient after the onset of visual symptom, leading to a marked reduction in mATXN7 aggregation and an increase of cone and rod ERG activities 6 weeks after intravitreal injection. Preservation of rod function but not of cones was still observed 9 weeks after injection. Thus, this study represents an important proof of concept of ASO-based therapy for SCA7 retinal degeneration. It also provides the groundwork for the assessment of ASO’s innocuity in long-term treatment and efficiency against cerebellar degeneration.
Considering that treatment in patients will last decades, selective suppression of mutant proteins is thought to have superior therapeutic outcomes and safety profiles. Allele-selective silencing strategies using ASOs directed against single nucleotide polymorphisms (SNPs) or single-strand short interfering RNAs (ss-siRNA) targeting the expanded CAG repeat on the mutant transcript have provided promising results in HD [178, 179]. Two antisense drug candidates, WVE-120101 and WVE-120102, target frequent SNP variants located in expanded allele of mutant Huntingtin [175]. A SNP linked to the SCA7 mutation was identified in 50% of SCA7 patients in the South African population which shows a strong founder effect [180]. Using short-hairpin RNA (shRNA) targeting this SNP, allele-specific mATXN7 suppression was achieved in patients’ cells [181]. However, targeting disease-linked SNPs will apply to a subset of patients, since they do not all bear targetable SNPs. ASO specifically targeting the expanded CAG repeat tract of the mATXN7 transcript was recently tested [177]. CAG-ASO provided significant knockdown of expanded ATXN7 expression in patient fibroblasts. However, when injected in the eye of SCA7266Q/5Q KI mice, the efficacy of CAG-ASO was only transient and to a lesser degree than the Atxn7 ASO. Krzyzosiak and colleagues [182, 183] have developed atypical ss-siRNA composed of self-complementary CUG repeats, containing chemical modifications and a single base mutation to facilitate guide-strand only self-duplex (sd) formation of guide. The sd-siRNAs efficiently form base mismatch complex with their CAG repeat targets, which results in selective inhibition of mutant mRNA translation [182]. When tested in SCA7 patient fibroblasts, sd-siRNAs showed a high efficiency and allele selectivity for silencing the mATXN7 protein (by about 75%) with the simultaneous upregulation of wild type ATXN7 [184]. Although targeting the expansion carries a promising application potential, the (non)protein-coding transcriptional off-target silencing should be carefully assessed in more advanced animal models.
An alternative to allele selective targeting was recently developed using specific knockdown-replacement strategy, which combines a non-allele specific gene silencing with simultaneous delivery of a functional copy of the wild-type gene to preserve the protein physiological function [185]. In this strategy, the authors used artificial mirtrons targeting ATXN7 transcript, and a mirtron resistant ATXN7 transgene carrying silent mutations [185]. Mirtrons are a class of intronic miRNAs that form premicroRNA hairpins after splicing. Mirtron maturation is thus independent of DROSHA and avoids overloading this processing machinery. The efficacy of the approach was significant in cells, despite potential limitation due to the splicing efficiency of mirtrons [185]. The development of knockdown-replacement strategy in vivo is challenging as both the silencing agent and the expression vector for the normal allele must be efficiently delivered, and the exogenous protein has to be expressed at an appropriate level.
Together, these studies provide the first proofs of efficacy of ON-based strategy to lower mATXN7 expression. However, additional studies are required to investigate the off-targets, the inflammatory responses, the effects of long-term treatments of chronic neurodegenerative pathology. Also, the brain or retina delivery approach need to be met. Synthetic ON-based strategies require regular delivery of the drug in multiple applications, which might be inconvenient. For this reason, research groups and companies also work on development of strategies, where the viral vectors could be used to permanently silence protein of interest in single drug application. For example, a viral vector miRNA-based strategy is now investigated for HD [175]. In any case, for siRNA, ASO or viral vector delivery, safety and efficacy issues related to biodistribution and durability still need to be evaluated [186, 187]. The recent years have seen the development of a powerful technologies to target and edit specific disease genes, such as TALEN, Zinc finger endonucleases, and CRISPR/Cas9 systems [188–190]. Currently under development for the most frequent polyQ diseases, CRISPR/Cas9 strategies have the potential to have great success in clinical trials for all polyQ neurodegenerative disorders.
Biomarkers for Clinical Trials
In rare inherited neurodegenerative disorders such as SCA7 and other ataxias, the lack of sensitive biomarkers and the small number of patients is a real challenge in clinical trials for getting achievable statistical sample size. Currently, ataxias are evaluated using diverse clinical scales for cerebellar and non-cerebellar phenotypical signs, i.e., Assessment and Rating of Ataxia (SARA), Brief Ataxia Rating Scale (BARS), semiquantitative non-ataxia scale (INAS), Composite Cerebellar Functional Severity Score (CCFS) (see [191] for review). The disease severity of SCA7 was estimated slower than that of other polyQ SCAs using SARA scores [192]. At present, neuroimaging biomarkers are considered powerful alternatives to subjective clinical scores for the detection of disease progression. For instance, SCA1, SCA2, SCA3, and SCA7 patients show larger longitudinal effect for brain volumetry (> 1.2) compared to standard clinical scores (< 0.8) [55, 193]. Proton magnetic resonance spectroscopy (1H MRS) using a 3T system was also used to determine neurometabolites levels in the cerebellar vermis and pons of patients with SCA1, SCA2, SCA3, and SCA7 [194]. This approach revealed early metabolic changes in SCA patients including lower level of neuronal N-acetylaspartate and N-acetylaspartylglutamate metabolites (tNAA), higher level of glial marker myo-inositol (myo-Ins) and elevated level of total creatinine (tCr). Severe phenotype evaluated using SARA scoring was associated with lower levels of tNAA in the vermis and pons of SCA7 patients, which may indicate neuronal integrity loss as well as CNS myelination disturbances.
Identification of disease-specific body fluids biomarkers is another alternative for disease progression measures in clinical trials. Recent studies have demonstrated the presence of miRNAs in human blood and cerebrospinal fluid (CSF) [195–197]. MicroRNA expression was found to be deregulated in models of polyQ SCA [198]. Several miRNAs were recently found upregulated in the plasma of SCA7 patients in comparison to healthy controls [199]. Four of them showed diagnostic value to differentiate healthy controls from patients. Furthermore, computational analysis on differentially expressed miRNAs in adult-onset and early-onset patients identified another group of four miRNAs with potential prognosis value. Importantly, the expression of prognosis-related miRNAs showed significant correlation with SARA and INAS rating scales.
Assessment of oxidative stress deriving from protein misfolding, neuroinflammation, and mitochondrial dysfunction might be another good indicator of neurodegeneration processes. Emerging evidence based on patient’s material indicates that oxidative stress might play a critical role in neurodegeneration occurring in several polyQ disorders [200–202]. mATXN7 expression induces oxidative stress in PC12 cells [145]. Clinical studies performed on SCA7 patients and matched healthy subjects indicate impairments in peripheral levels of different oxidative stress markers. SCA7 patients exhibit higher levels of lipid hydroperoxides and malondialdehyde and protein carbonyls [203]. The physiological response to oxidative stress result in induction of antioxidant enzymes such as glutathione reductase, glutathione peroxidase, and paraoxonase. The cupric reducing antioxidant capacity is also significantly increased in SCA7. These results suggest the activation of the antioxidant defense system. Interestingly, a more severe oxidative stress was observed in early-onset patients compared to adult-onset patients. A positive correlation between oxidative stress markers and disease severity evaluated by clinical scores was also observed.
Final Remarks
While biochemical approaches and the characterization of cellular and animal models of SCA7 have greatly advanced our understanding of disease pathogenesis in SCA7, much more needs to be learned before we get a solid comprehension of the pathogenic mechanisms underlying neuronal specific dysfunction and neuronal cell loss. Several of the therapeutic strategies against SCA7 are promising and can be proposed to clinical trials. Nevertheless, further fundamental investigations are required to identify new molecular targets for SCA7. Since SCA7 shares many common pathological features with other degenerative disorders affecting the cerebellum and the retina, identification of therapeutics in SCA7 or in any of the related diseases is likely to be cross-beneficial.
Electronic Supplementary Material
(PDF 498 kb)
Acknowledgments
We are grateful to Nathalie Daigle for critical reading of the manuscript, and Samantha Carrillo-Rosas for images in Fig. 2a.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Funding Information
YT was supported by funding from Institut National de la Santé et de la Recherche Médicale (INSERM); Centre National de la Recherche Scientifique (CNRS); University of Strasbourg; Agence Nationale de la Recherche (ANR) Ciliataxia-ANR-13-BSV1-0016-01 and ANR-10-LABX-0030-INRT under the frame programme Investissements d’Avenir labelled ANR-10-IDEX-0002-02; ANR-15-JPWG-0008-03 from the Joint Program for Neurodegenerative diseases research entitled Model-PolyQ: Advanced models of polyglutamine disorders -HD, SCA2, SCA3, SCA7; the Fondation pour la Recherche Médicale (FRM) [DVS20131228917]; Association RETINA-FRANCE; Connaître les Syndrômes Cérébelleux (CSC).
Compliance with ethical standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Invited review: Molecular Pathogenesis and Advances in Therapeutics of Repeat Expansion Disorders, guest edited by Dr. Lisa Ellerby
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harding AE. The clinical features and classification of the late onset autosomal dominant cerebellar ataxias. A study of 11 families, including descendants of the 'the Drew family of Walworth'. Brain : a journal of neurology. 1982;105(Pt 1):1–28. doi: 10.1093/brain/105.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Harding AE. Clinical features and classification of inherited ataxias. Adv Neurol. 1993;61:1–14. [PubMed] [Google Scholar]
- 3.David G, Durr A, Stevanin G, Cancel G, Abbas N, Benomar A, et al. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7) Human molecular genetics. 1998;7(2):165–70. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- 4.Johansson J, Forsgren L, Sandgren O, Brice A, Holmgren G, Holmberg M. Expanded CAG repeats in Swedish spinocerebellar ataxia type 7 (SCA7) patients: effect of CAG repeat length on the clinical manifestation. Human molecular genetics. 1998;7(2):171–6. doi: 10.1093/hmg/7.2.171. [DOI] [PubMed] [Google Scholar]
- 5.Bryer A, Krause A, Bill P, Davids V, Bryant D, Butler J, et al. The hereditary adult-onset ataxias in South Africa. Journal of the neurological sciences. 2003;216(1):47–54. doi: 10.1016/s0022-510x(03)00209-0. [DOI] [PubMed] [Google Scholar]
- 6.Gu W, Wang Y, Liu X, Zhou B, Zhou Y, Wang G. Molecular and clinical study of spinocerebellar ataxia type 7 in Chinese kindreds. Arch Neurol. 2000;57(10):1513–8. doi: 10.1001/archneur.57.10.1513. [DOI] [PubMed] [Google Scholar]
- 7.Jardim LB, Silveira I, Pereira ML, Ferro A, Alonso I. do Ceu Moreira M, et al. A survey of spinocerebellar ataxia in South Brazil - 66 new cases with Machado-Joseph disease, SCA7, SCA8, or unidentified disease-causing mutations. Journal of neurology. 2001;248(10):870–6. doi: 10.1007/s004150170072. [DOI] [PubMed] [Google Scholar]
- 8.Storey E, du Sart D, Shaw JH, Lorentzos P, Kelly L, McKinley Gardner RJ, et al. Frequency of spinocerebellar ataxia types 1, 2, 3, 6, and 7 in Australian patients with spinocerebellar ataxia. Am J Med Genet. 2000;95(4):351–7. doi: 10.1002/1096-8628(20001211)95:4<351::aid-ajmg10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 9.Magana JJ, Gomez R, Maldonado-Rodriguez M, Velazquez-Perez L, Tapia-Guerrero YS, Cortes H, et al. Origin of the spinocerebellar ataxia type 7 gene mutation in Mexican population. Cerebellum. 2013;12(6):902–5. doi: 10.1007/s12311-013-0505-8. [DOI] [PubMed] [Google Scholar]
- 10.Bang OY, Huh K, Lee PH, Kim HJ. Clinical and neuroradiological features of patients with spinocerebellar ataxias from Korean kindreds. Arch Neurol. 2003;60(11):1566–74. doi: 10.1001/archneur.60.11.1566. [DOI] [PubMed] [Google Scholar]
- 11.Martin JJ. Spinocerebellar ataxia type 7. Handb Clin Neurol. 2012;103:475–91. doi: 10.1016/B978-0-444-51892-7.00030-9. [DOI] [PubMed] [Google Scholar]
- 12.Jonasson J, Juvonen V, Sistonen P, Ignatius J, Johansson D, Bjorck EJ, et al. Evidence for a common Spinocerebellar ataxia type 7 (SCA7) founder mutation in Scandinavia. Eur J Hum Genet. 2000;8(12):918–22. doi: 10.1038/sj.ejhg.5200557. [DOI] [PubMed] [Google Scholar]
- 13.Magana JJ, Tapia-Guerrero YS, Velazquez-Perez L, Cerecedo-Zapata CM, Maldonado-Rodriguez M, Jano-Ito JS, et al. Analysis of CAG repeats in five SCA loci in Mexican population: epidemiological evidence of a SCA7 founder effect. Clinical genetics. 2014;85(2):159–65. doi: 10.1111/cge.12114. [DOI] [PubMed] [Google Scholar]
- 14.Smith DC, Atadzhanov M, Mwaba M, Greenberg LJ. Evidence for a common founder effect amongst South African and Zambian individuals with Spinocerebellar ataxia type 7. Journal of the neurological sciences. 2015;354(1-2):75–8. doi: 10.1016/j.jns.2015.04.053. [DOI] [PubMed] [Google Scholar]
- 15.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–47. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 16.Matos CA, Almeida LP, Nobrega C. Proteolytic Cleavage of Polyglutamine Disease-Causing Proteins: Revisiting the Toxic Fragment Hypothesis. Curr Pharm Des. 2017;23(5):753–75. doi: 10.2174/1381612822666161227121912. [DOI] [PubMed] [Google Scholar]
- 17.Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Human molecular genetics. 2004;13(12):1257–65. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 18.Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, et al. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes & development. 2014;28(18):1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmberg M, Johansson J, Forsgren L, Heijbel J, Sandgren O, Holmgren G. Localization of autosomal dominant cerebellar ataxia associated with retinal degeneration and anticipation to chromosome 3p12-p21.1. Human molecular genetics. 1995;4(8):1441–5. doi: 10.1093/hmg/4.8.1441. [DOI] [PubMed] [Google Scholar]
- 20.Benomar A, KROLS L, STEVANIN G, CANCEL G, LEGUERN E, DAVID G, et al. The Gene for Autosomal-Dominant Cerebellar-Ataxia with Pigmentary Macular Dystrophy Maps to Chromosome 3p12-P21.1. Nature genetics. 1995;10(1):84–8. doi: 10.1038/ng0595-84. [DOI] [PubMed] [Google Scholar]
- 21.Gouw LG, Kaplan CD, Haines JH, Digre KB, Rutledge SL, Matilla A, et al. Retinal Degeneration Characterizes a Spinocerebellar Ataxia Mapping to Chromosome 3p. Nature genetics. 1995;10(1):89–93. doi: 10.1038/ng0595-89. [DOI] [PubMed] [Google Scholar]
- 22.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, et al. Polyglutamine expansion as a pathological epitope in Huntington's disease and four dominant cerebellar ataxias. Nature. 1995;378(6555):403–6. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 23.Stevanin G, Trottier Y, Cancel G, Durr A, David G, Didierjean O, et al. Screening for proteins with polyglutamine expansions in autosomal dominant cerebellar ataxias. Human molecular genetics. 1996;5(12):1887–92. doi: 10.1093/hmg/5.12.1887. [DOI] [PubMed] [Google Scholar]
- 24.David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nature genetics. 1997;17(1):65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- 25.Michalik A, Del-Favero J, Mauger C, Lofgren A, Van Broeckhoven C. Genomic organisation of the spinocerebellar ataxia type 7 (SCA7) gene responsible for autosomal dominant cerebellar ataxia with retinal degeneration. Human genetics. 1999;105(5):410–7. doi: 10.1007/s004390051123. [DOI] [PubMed] [Google Scholar]
- 26.Giunti P, Stevanin G, Worth PF, David G, Brice A, Wood NW. Molecular and clinical study of 18 families with ADCA type II: evidence for genetic heterogeneity and de novo mutation. American journal of human genetics. 1999;64(6):1594–603. doi: 10.1086/302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevanin G, Giunti P, Belal GD, Durr A, Ruberg M, Wood N, et al. De novo expansion of intermediate alleles in spinocerebellar ataxia 7. Human molecular genetics. 1998;7(11):1809–13. doi: 10.1093/hmg/7.11.1809. [DOI] [PubMed] [Google Scholar]
- 28.Mittal U, Roy S, Jain S, Srivastava AK, Mukerji M. Post-zygotic de novo trinucleotide repeat expansion at spinocerebellar ataxia type 7 locus: evidence from an Indian family. J Hum Genet. 2005;50(3):155–7. doi: 10.1007/s10038-005-0233-0. [DOI] [PubMed] [Google Scholar]
- 29.Nardacchione A, Orsi L, Brusco A, Franco A, Grosso E, Dragone E, et al. Definition of the smallest pathological CAG expansion in SCA7. Clinical genetics. 1999;56(3):232–4. doi: 10.1034/j.1399-0004.1999.560309.x. [DOI] [PubMed] [Google Scholar]
- 30.van de Warrenburg BP, Frenken CW, Ausems MG, Kleefstra T, Sinke RJ, Knoers NV, et al. Striking anticipation in spinocerebellar ataxia type 7: the infantile phenotype. Journal of neurology. 2001;248(10):911–4. doi: 10.1007/s004150170082. [DOI] [PubMed] [Google Scholar]
- 31.Monckton DG, Cayuela ML, Gould FK, Brock GJ, Silva R, Ashizawa T. Very large (CAG)(n) DNA repeat expansions in the sperm of two spinocerebellar ataxia type 7 males. Human molecular genetics. 1999;8(13):2473–8. doi: 10.1093/hmg/8.13.2473. [DOI] [PubMed] [Google Scholar]
- 32.Michalik A, Martin JJ, Van Broeckhoven C. Spinocerebellar ataxia type 7 associated with pigmentary retinal dystrophy. Eur J Hum Genet. 2004;12(1):2–15. doi: 10.1038/sj.ejhg.5201108. [DOI] [PubMed] [Google Scholar]
- 33.Enevoldson TP, Sanders MD, Harding AE. Autosomal dominant cerebellar ataxia with pigmentary macular dystrophy. A clinical and genetic study of eight families. Brain : a journal of neurology. 1994;117(Pt 3):445–60. doi: 10.1093/brain/117.3.445. [DOI] [PubMed] [Google Scholar]
- 34.Gouw LG, Digre KB, Harris CP, Haines JH, Ptacek LJ. Autosomal dominant cerebellar ataxia with retinal degeneration: clinical, neuropathologic, and genetic analysis of a large kindred. Neurology. 1994;44(8):1441–7. doi: 10.1212/wnl.44.8.1441. [DOI] [PubMed] [Google Scholar]
- 35.Velazquez-Perez L, Cerecedo-Zapata CM, Hernandez-Hernandez O, Martinez-Cruz E, Tapia-Guerrero YS, Gonzalez-Pina R, et al. A comprehensive clinical and genetic study of a large Mexican population with spinocerebellar ataxia type 7. Neurogenetics. 2015;16(1):11–21. doi: 10.1007/s10048-014-0424-y. [DOI] [PubMed] [Google Scholar]
- 36.Sokolovsky N, Cook A, Hunt H, Giunti P, Cipolotti L. A preliminary characterisation of cognition and social cognition in spinocerebellar ataxia types 2, 1, and 7. Behav Neurol. 2010;23(1-2):17–29. doi: 10.3233/BEN-2010-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Agata F, Caroppo P, Baudino B, Caglio M, Croce M, Bergui M, et al. The recognition of facial emotions in spinocerebellar ataxia patients. Cerebellum. 2011;10(3):600–10. doi: 10.1007/s12311-011-0276-z. [DOI] [PubMed] [Google Scholar]
- 38.Chirino A, Hernandez-Castillo CR, Galvez V, Contreras A, Diaz R, Beltran-Parrazal L, et al. Motor and cognitive impairments in spinocerebellar ataxia type 7 and its correlations with cortical volumes. The European journal of neuroscience. 2018;48(10):3199–211. doi: 10.1111/ejn.14148. [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Coello A, Valadez-Jimenez VM, Cisneros B, Carrillo-Mora P, Parra-Cardenas M, Hernandez-Hernandez O, et al. Voice Alterations in Patients With Spinocerebellar Ataxia Type 7 (SCA7): Clinical-Genetic Correlations. J Voice. 2017;31(1):123 e1- e5. doi: 10.1016/j.jvoice.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Salas-Vargas J, Mancera-Gervacio J, Velazquez-Perez L, Rodrigez-Labrada R, Martinez-Cruz E, Magana JJ, et al. Spinocerebellar ataxia type 7: a neurodegenerative disorder with peripheral neuropathy. Eur Neurol. 2015;73(3-4):173–8. doi: 10.1159/000370239. [DOI] [PubMed] [Google Scholar]
- 41.Benton CS, de Silva R, Rutledge SL, Bohlega S, Ashizawa T, Zoghbi HY. Molecular and clinical studies in SCA-7 define a broad clinical spectrum and the infantile phenotype. Neurology. 1998;51(4):1081–6. doi: 10.1212/wnl.51.4.1081. [DOI] [PubMed] [Google Scholar]
- 42.Whitney A, Lim M, Kanabar D, Lin JP. Massive SCA7 expansion detected in a 7-month-old male with hypotonia, cardiomegaly, and renal compromise. Dev Med Child Neurol. 2007;49(2):140–3. doi: 10.1111/j.1469-8749.2007.00140.x. [DOI] [PubMed] [Google Scholar]
- 43.Ansorge O, Giunti P, Michalik A, Van Broeckhoven C, Harding B, Wood N, et al. Ataxin-7 aggregation and ubiquitination in infantile SCA7 with 180 CAG repeats. Annals of neurology. 2004;56(3):448–52. doi: 10.1002/ana.20230. [DOI] [PubMed] [Google Scholar]
- 44.Trang H, Stanley SY, Thorner P, Faghfoury H, Schulze A, Hawkins C, et al. Massive CAG Repeat Expansion and Somatic Instability in Maternally Transmitted Infantile Spinocerebellar Ataxia Type 7. JAMA neurology. 2014. [DOI] [PubMed]
- 45.Donis KC, Mattos EP, Silva AA, Furtado GV, Saraiva-Pereira ML, Jardim LB, et al. Infantile spinocerebellar ataxia type 7: Case report and a review of the literature. Journal of the neurological sciences. 2015;354(1-2):118–21. doi: 10.1016/j.jns.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 46.Gousse G, Patural H, Touraine R, Chabrier S, Rolland E, Antoine JC, et al. Lethal form of spinocerebellar ataxia type 7 with early onset in childhood. Arch Pediatr. 2018;25(1):42–4. doi: 10.1016/j.arcped.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 47.Rub U, Brunt ER, Gierga K, Seidel K, Schultz C, Schols L, et al. Spinocerebellar ataxia type 7 (SCA7): first report of a systematic neuropathological study of the brain of a patient with a very short expanded CAG-repeat. Brain pathology. 2005;15(4):287–95. doi: 10.1111/j.1750-3639.2005.tb00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rub U, Brunt ER, Seidel K, Gierga K, Mooy CM, Kettner M, et al. Spinocerebellar ataxia type 7 (SCA7): widespread brain damage in an adult-onset patient with progressive visual impairments in comparison with an adult-onset patient without visual impairments. Neuropathol Appl Neurobiol. 2008;34(2):155–68. doi: 10.1111/j.1365-2990.2007.00882.x. [DOI] [PubMed] [Google Scholar]
- 49.Horton LC, Frosch MP, Vangel MG, Weigel-DiFranco C, Berson EL, Schmahmann JD. Spinocerebellar ataxia type 7: clinical course, phenotype-genotype correlations, and neuropathology. Cerebellum. 2013;12(2):176–93. doi: 10.1007/s12311-012-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin JJ, Van Regemorter N, Krols L, Brucher JM, de Barsy T, Szliwowski H, et al. On an autosomal dominant form of retinal-cerebellar degeneration: an autopsy study of five patients in one family. Acta Neuropathol (Berl). 1994;88(4):277–86. doi: 10.1007/BF00310370. [DOI] [PubMed] [Google Scholar]
- 51.Hernandez-Castillo CR, Galvez V, Diaz R, Fernandez-Ruiz J. Specific cerebellar and cortical degeneration correlates with ataxia severity in spinocerebellar ataxia type 7. Brain Imaging Behav. 2016;10(1):252–7. doi: 10.1007/s11682-015-9389-1. [DOI] [PubMed] [Google Scholar]
- 52.Hernandez-Castillo CR, King M, Diedrichsen J, Fernandez-Ruiz J. Unique degeneration signatures in the cerebellar cortex for spinocerebellar ataxias 2, 3, and 7. Neuroimage Clin. 2018;20:931–8. doi: 10.1016/j.nicl.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernandez-Castillo CR, Alcauter S, Galvez V, Barrios FA, Yescas P, Ochoa A, et al. Disruption of visual and motor connectivity in spinocerebellar ataxia type 7. Mov Disord. 2013;28(12):1708–16. doi: 10.1002/mds.25618. [DOI] [PubMed] [Google Scholar]
- 54.Hernandez-Castillo CR, Galvez V, Morgado-Valle C, Fernandez-Ruiz J. Whole-brain connectivity analysis and classification of spinocerebellar ataxia type 7 by functional MRI. Cerebellum Ataxias. 2014;1:2. doi: 10.1186/2053-8871-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adanyeguh IM, Perlbarg V, Henry PG, Rinaldi D, Petit E, Valabregue R, et al. Autosomal dominant cerebellar ataxias: Imaging biomarkers with high effect sizes. Neuroimage Clin. 2018;19:858–67. doi: 10.1016/j.nicl.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benomar A, Leguern E, Durr A, Ouhabi H, Stevanin G, Yahyaoui M, et al. Autosomal-Dominant Cerebellar-Ataxia with Retinal Degeneration (Adca Type-Ii) Is Genetically Different from Adca Type-I. Annals of neurology. 1994;35(4):439–44. doi: 10.1002/ana.410350411. [DOI] [PubMed] [Google Scholar]
- 57.Aleman TS, Cideciyan AV, Volpe NJ, Stevanin G, Brice A, Jacobson SG. Spinocerebellar ataxia type 7 (SCA7) shows a cone-rod dystrophy phenotype. Exp Eye Res. 2002;74(6):737–45. doi: 10.1006/exer.2002.1169. [DOI] [PubMed] [Google Scholar]
- 58.Abe T, Tsuda T, Yoshida M, Wada Y, Kano T, Itoyama Y, et al. Macular degeneration associated with aberrant expansion of trinucleotide repeat of the SCA7 gene in 2 Japanese families. Arch Ophthalmol. 2000;118(10):1415–21. doi: 10.1001/archopht.118.10.1415. [DOI] [PubMed] [Google Scholar]
- 59.Inaba H, Yabe I, Yashima M, Soma H, Nakamura Y, Houzen H, et al. Unusual retinal phenotypes in an SCA7 family. Intern Med. 2009;48(16):1461–4. doi: 10.2169/internalmedicine.48.2072. [DOI] [PubMed] [Google Scholar]
- 60.Thurtell MJ, Fraser JA, Bala E, Tomsak RL, Biousse V, Leigh RJ, et al. Two patients with spinocerebellar ataxia type 7 presenting with profound binocular visual loss yet minimal ophthalmoscopic findings. J Neuroophthalmol. 2009;29(3):187–91. doi: 10.1097/WNO.0b013e3181b41764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hugosson T, Granse L, Ponjavic V, Andreasson S. Macular dysfunction and morphology in spinocerebellar ataxia type 7 (SCA 7) Ophthalmic Genet. 2009;30(1):1–6. doi: 10.1080/13816810802454081. [DOI] [PubMed] [Google Scholar]
- 62.Manrique RK, Noval S, Aguilar-Amat MJ, Arpa J, Rosa I, Contreras I. Ophthalmic features of spinocerebellar ataxia type 7. J Neuroophthalmol. 2009;29(3):174–9. doi: 10.1097/WNO.0b013e3181b2828e. [DOI] [PubMed] [Google Scholar]
- 63.Azevedo PB, Rocha AG, Keim LMN, Lavinsky D, Furtado GV, de Mattos EP, et al. Ophthalmological and Neurologic Manifestations in Pre-clinical and Clinical Phases of Spinocerebellar Ataxia Type 7. Cerebellum. 2019. [DOI] [PubMed]
- 64.Campos-Romo A, Graue-Hernandez EO, Pedro-Aguilar L, Hernandez-Camarena JC, Rivera-De la Parra D, Galvez V, et al. Ophthalmic features of spinocerebellar ataxia type 7. Eye (Lond). 2018;32(1):120–7. doi: 10.1038/eye.2017.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindenberg KS, Yvert G, Muller K, Landwehrmeyer GB. Expression analysis of ataxin-7 mRNA and protein in human brain: evidence for a widespread distribution and focal protein accumulation. Brain pathology. 2000;10(3):385–94. doi: 10.1111/j.1750-3639.2000.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cancel G, Duyckaerts C, Holmberg M, Zander C, Yvert G, Lebre AS, et al. Distribution of ataxin-7 in normal human brain and retina. Brain : a journal of neurology. 2000;123(Pt 12):2519–30. doi: 10.1093/brain/123.12.2519. [DOI] [PubMed] [Google Scholar]
- 67.Mauger C, Del-Favero J, Ceuterick C, Lubke U, van Broeckhoven C, Martin J. Identification and localization of ataxin-7 in brain and retina of a patient with cerebellar ataxia type II using anti-peptide antibody. Brain Res Mol Brain Res. 1999;74(1-2):35–43. doi: 10.1016/s0169-328x(99)00256-9. [DOI] [PubMed] [Google Scholar]
- 68.Jonasson J, Strom AL, Hart P, Brannstrom T, Forsgren L, Holmberg M. Expression of ataxin-7 in CNS and non-CNS tissue of normal and SCA7 individuals. Acta neuropathologica. 2002;104(1):29–37. doi: 10.1007/s00401-001-0514-4. [DOI] [PubMed] [Google Scholar]
- 69.Einum DD, Gouw L, MAtilla A, Townsend J, Fu YH, Ptacek LJ. Expression analysis of ataxin-7 reveals restricted localization of an alternate isoform. American journal of human genetics. 1999;65(4):A104-A. [Google Scholar]
- 70.Strom AL, Jonasson J, Hart P, Brannstrom T, Forsgren L, Holmberg M. Cloning and expression analysis of the murine homolog of the spinocerebellar ataxia type 7 (SCA7) gene. Gene. 2002;285(1-2):91–9. doi: 10.1016/s0378-1119(02)00399-2. [DOI] [PubMed] [Google Scholar]
- 71.Einum DD, Townsend JJ, Ptacek LJ, Fu YH. Ataxin-7 expression analysis in controls and spinocerebellar ataxia type 7 patients. Neurogenetics. 2001;3(2):83–90. doi: 10.1007/s100480000100. [DOI] [PubMed] [Google Scholar]
- 72.Einum DD, Clark AM, Townsend JJ, Ptacek LJ, Fu YH. A novel central nervous system-enriched spinocerebellar ataxia type 7 gene product. Arch Neurol. 2003;60(1):97–103. doi: 10.1001/archneur.60.1.97. [DOI] [PubMed] [Google Scholar]
- 73.Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Human molecular genetics. 2003;12(21):2845–52. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- 74.Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Molecular and cellular biology. 2002;22(13):4723–38. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(24):8472–7. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(24):8478–82. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carrillo-Rosas S, Weber C, Fievet L, Messaddeq N, Karam A, Trottier Y. Loss of zebrafish Ataxin-7, a SAGA subunit responsible for SCA7 retinopathy, causes ocular coloboma and malformation of photoreceptors. Human molecular genetics. 2018. [DOI] [PubMed]
- 78.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics & chromatin. 2009;2(1):2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141(4):606–17. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, et al. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328(5981):1025–9. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nature cell biology. 2008;10(6):707–15. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 82.Mohan RD, Dialynas G, Weake VM, Liu J, Martin-Brown S, Florens L, et al. Loss of Drosophila Ataxin-7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev. 2014;28(3):259–72. doi: 10.1101/gad.225151.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonnet J, Wang YH, Spedale G, Atkinson RA, Romier C, Hamiche A, et al. The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep. 2010;11(8):612–8. doi: 10.1038/embor.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCormick MA, Mason AG, Guyenet SJ, Dang W, Garza RM, Ting MK, et al. The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell reports. 2014;8(2):477–86. doi: 10.1016/j.celrep.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mason AG, Garza RM, McCormick MA, Patel B, Kennedy BK, Pillus L, et al. The replicative lifespan-extending deletion of SGF73 results in altered ribosomal gene expression in yeast. Aging Cell. 2017;16(4):785–96. doi: 10.1111/acel.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang L, Dent SY. Functions of SAGA in development and disease. Epigenomics. 2014;6(3):329–39. doi: 10.2217/epi.14.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma J, Brennan KJ, D'Aloia MR, Pascuzzi PE, Weake VM. Transcriptome Profiling Identifies Multiplexin as a Target of SAGA Deubiquitinase Activity in Glia Required for Precise Axon Guidance During Drosophila Visual Development. G3 (Bethesda) 2016;6(8):2435–45. doi: 10.1534/g3.116.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poeck B, Fischer S, Gunning D, Zipursky SL, Salecker I. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29(1):99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- 89.Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, et al. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. The EMBO journal. 2008;27(2):394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilde JJ, Siegenthaler JA, Dent SY, Niswander LA. Diencephalic Size Is Restricted by a Novel Interplay Between GCN5 Acetyltransferase Activity and Retinoic Acid Signaling. J Neurosci. 2017;37(10):2565–79. doi: 10.1523/JNEUROSCI.2121-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yanicostas C, Barbieri E, Hibi M, Brice A, Stevanin G, Soussi-Yanicostas N. Requirement for zebrafish ataxin-7 in differentiation of photoreceptors and cerebellar neurons. PloS one. 2012;7(11):e50705. doi: 10.1371/journal.pone.0050705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakamura Y, Tagawa K, Oka T, Sasabe T, Ito H, Shiwaku H, et al. Ataxin-7 associates with microtubules and stabilizes the cytoskeletal network. Human molecular genetics. 2012;21(5):1099–110. doi: 10.1093/hmg/ddr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kahle JJ, Gulbahce N, Shaw CA, Lim J, Hill DE, Barabasi AL, et al. Comparison of an expanded ataxia interactome with patient medical records reveals a relationship between macular degeneration and ataxia. Human molecular genetics. 2011;20(3):510–27. doi: 10.1093/hmg/ddq496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fournier M, Orpinell M, Grauffel C, Scheer E, Garnier JM, Ye T, et al. KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat Commun. 2016;7:13227. doi: 10.1038/ncomms13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yvert G, Lindenberg KS, Devys D, Helmlinger D, Landwehrmeyer GB, Mandel JL. SCA7 mouse models show selective stabilization of mutant ataxin-7 and similar cellular responses in different neuronal cell types. Human molecular genetics. 2001;10(16):1679–92. doi: 10.1093/hmg/10.16.1679. [DOI] [PubMed] [Google Scholar]
- 96.Yoo SY, Pennesi ME, Weeber EJ, Xu B, Atkinson R, Chen S, et al. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron. 2003;37(3):383–401. doi: 10.1016/s0896-6273(02)01190-x. [DOI] [PubMed] [Google Scholar]
- 97.Yvert G, Lindenberg KS, Picaud S, Landwehrmeyer GB, Sahel JA, Mandel JL. Expanded polyglutamines induce neurodegeneration and trans-neuronal alterations in cerebellum and retina of SCA7 transgenic mice. Human molecular genetics. 2000;9(17):2491–506. doi: 10.1093/hmg/9.17.2491. [DOI] [PubMed] [Google Scholar]
- 98.La Spada AR, Fu YH, Sopher BL, Libby RT, Wang X, Li LY, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31(6):913–27. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- 99.Mookerjee S, Papanikolaou T, Guyenet SJ, Sampath V, Lin A, Vitelli C, et al. Posttranslational modification of ataxin-7 at lysine 257 prevents autophagy-mediated turnover of an N-terminal caspase-7 cleavage fragment. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29(48):15134–44. doi: 10.1523/JNEUROSCI.4720-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Custer SK, Garden GA, Gill N, Rueb U, Libby RT, Schultz C, et al. Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nature neuroscience. 2006;9(10):1302–11. doi: 10.1038/nn1750. [DOI] [PubMed] [Google Scholar]
- 101.Furrer SA, Mohanachandran MS, Waldherr SM, Chang C, Damian VA, Sopher BL, et al. Spinocerebellar ataxia type 7 cerebellar disease requires the coordinated action of mutant ataxin-7 in neurons and glia, and displays non-cell-autonomous bergmann glia degeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(45):16269–78. doi: 10.1523/JNEUROSCI.4000-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guyenet SJ, Mookerjee SS, Lin A, Custer SK, Chen SF, Sopher BL, et al. Proteolytic cleavage of ataxin-7 promotes SCA7 retinal degeneration and neurological dysfunction. Human molecular genetics. 2015;24(14):3908–17. doi: 10.1093/hmg/ddv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chou AH, Chen CY, Chen SY, Chen WJ, Chen YL, Weng YS, et al. Polyglutamine-expanded ataxin-7 causes cerebellar dysfunction by inducing transcriptional dysregulation. Neurochemistry international. 2010;56(2):329–39. doi: 10.1016/j.neuint.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 104.Chen YC, Gatchel JR, Lewis RW, Mao CA, Grant PA, Zoghbi HY, et al. Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Human molecular genetics. 2012;21(2):394–405. doi: 10.1093/hmg/ddr474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCullough SD, Xu X, Dent SY, Bekiranov S, Roeder RG, Grant PA. Reelin is a target of polyglutamine expanded ataxin-7 in human spinocerebellar ataxia type 7 (SCA7) astrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21319–24. doi: 10.1073/pnas.1218331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang H, Liu S, He WT, Zhao J, Jiang LL, Hu HY. Aggregation of Polyglutamine-expanded Ataxin 7 Protein Specifically Sequesters Ubiquitin-specific Protease 22 and Deteriorates Its Deubiquitinating Function in the Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex. The Journal of biological chemistry. 2015;290(36):21996–2004. doi: 10.1074/jbc.M114.631663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lan X, Koutelou E, Schibler AC, Chen YC, Grant PA, Dent SY. Poly(Q) Expansions in ATXN7 Affect Solubility but Not Activity of the SAGA Deubiquitinating Module. Molecular and cellular biology. 2015;35(10):1777–87. doi: 10.1128/MCB.01454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Duncan CE, An MC, Papanikolaou T, Rugani C, Vitelli C, Ellerby LM. Histone deacetylase-3 interacts with ataxin-7 and is altered in a spinocerebellar ataxia type 7 mouse model. Mol Neurodegener. 2013;8:42. doi: 10.1186/1750-1326-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takahashi J, Fujigasaki H, Zander C, El Hachimi KH, Stevanin G, Durr A, et al. Two populations of neuronal intranuclear inclusions in SCA7 differ in size and promyelocytic leukaemia protein content. Brain : a journal of neurology. 2002;125(Pt 7):1534–43. doi: 10.1093/brain/awf154. [DOI] [PubMed] [Google Scholar]
- 110.Zander C, Takahashi J, El Hachimi KH, Fujigasaki H, Albanese V, Lebre AS, et al. Similarities between spinocerebellar ataxia type 7 (SCA7) cell models and human brain: proteins recruited in inclusions and activation of caspase-3. Human molecular genetics. 2001;10(22):2569–79. doi: 10.1093/hmg/10.22.2569. [DOI] [PubMed] [Google Scholar]
- 111.Yu X, Munoz-Alarcon A, Ajayi A, Webling KE, Steinhof A, Langel U, et al. Inhibition of autophagy via p53-mediated disruption of ULK1 in a SCA7 polyglutamine disease model. J Mol Neurosci. 2013;50(3):586–99. doi: 10.1007/s12031-013-0012-x. [DOI] [PubMed] [Google Scholar]
- 112.Ajayi A, Yu X, Wahlo-Svedin C, Tsirigotaki G, Karlstrom V, Strom AL. Altered p53 and NOX1 activity cause bioenergetic defects in a SCA7 polyglutamine disease model. Biochim Biophys Acta. 2015;1847(4-5):418–28. doi: 10.1016/j.bbabio.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 113.Alves S, Marais T, Biferi MG, Furling D, Marinello M, El Hachimi K, et al. Lentiviral vector-mediated overexpression of mutant ataxin-7 recapitulates SCA7 pathology and promotes accumulation of the FUS/TLS and MBNL1 RNA-binding proteins. Mol Neurodegener. 2016;11(1):58. doi: 10.1186/s13024-016-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Janer A, Werner A, Takahashi-Fujigasaki J, Daret A, Fujigasaki H, Takada K, et al. SUMOylation attenuates the aggregation propensity and cellular toxicity of the polyglutamine expanded ataxin-7. Human molecular genetics. 2010;19(1):181–95. doi: 10.1093/hmg/ddp478. [DOI] [PubMed] [Google Scholar]
- 115.Merienne K, Helmlinger D, Perkin GR, Devys D, Trottier Y. Polyglutamine expansion induces a protein-damaging stress connecting heat shock protein 70 to the JNK pathway. The Journal of biological chemistry. 2003;278(19):16957–67. doi: 10.1074/jbc.M212049200. [DOI] [PubMed] [Google Scholar]
- 116.Latouche M, Lasbleiz C, Martin E, Monnier V, Debeir T, Mouatt-Prigent A, et al. A conditional pan-neuronal Drosophila model of spinocerebellar ataxia 7 with a reversible adult phenotype suitable for identifying modifier genes. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(10):2483–92. doi: 10.1523/JNEUROSCI.5453-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marinello M, Werner A, Giannone M, Tahiri K, Alves S, Tesson C, et al. SUMOylation by SUMO2 is implicated in the degradation of misfolded ataxin-7 via RNF4 in SCA7 models. Dis Model Mech. 2019;12(1). [DOI] [PMC free article] [PubMed]
- 118.Alves S, Cormier-Dequaire F, Marinello M, Marais T, Muriel MP, Beaumatin F, et al. The autophagy/lysosome pathway is impaired in SCA7 patients and SCA7 knock-in mice. Acta neuropathologica. 2014;128(5):705–22. doi: 10.1007/s00401-014-1289-8. [DOI] [PubMed] [Google Scholar]
- 119.Takahashi-Fujigasaki J, Breidert T, Fujigasaki H, Duyckaerts C, Camonis JH, Brice A, et al. Amyloid precursor-like protein 2 cleavage contributes to neuronal intranuclear inclusions and cytotoxicity in spinocerebellar ataxia-7 (SCA7) Neurobiology of disease. 2011;41(1):33–42. doi: 10.1016/j.nbd.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 120.Latouche M, Fragner P, Martin E, El Hachimi KH, Zander C, Sittler A, et al. Polyglutamine and polyalanine expansions in ataxin7 result in different types of aggregation and levels of toxicity. Mol Cell Neurosci. 2006;31(3):438–45. doi: 10.1016/j.mcn.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 121.Young JE, Gouw L, Propp S, Sopher BL, Taylor J, Lin A, et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. The Journal of biological chemistry. 2007;282(41):30150–60. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- 122.Garden GA, Libby RT, Fu YH, Kinoshita Y, Huang J, Possin DE, et al. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22(12):4897–905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yefimova MG, Messaddeq N, Karam A, Jacquard C, Weber C, Jonet L, et al. Polyglutamine toxicity induces rod photoreceptor division, morphological transformation or death in Spinocerebellar ataxia 7 mouse retina. Neurobiology of disease. 2010;40(1):311–24. doi: 10.1016/j.nbd.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 124.Chort A, Alves S, Marinello M, Dufresnois B, Dornbierer JG, Tesson C, et al. Interferon beta induces clearance of mutant ataxin 7 and improves locomotion in SCA7 knock-in mice. Brain : a journal of neurology. 2013. [DOI] [PubMed]
- 125.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, et al. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. The Journal of cell biology. 2006;174(1):65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takahashi J, Fujigasaki H, Iwabuchi K, Bruni AC, Uchihara T, El Hachimi KH, et al. PML nuclear bodies and neuronal intranuclear inclusion in polyglutamine diseases. Neurobiology of disease. 2003;13(3):230–7. doi: 10.1016/s0969-9961(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 127.Karam A, Trottier Y. Molecular Mechanisms and Therapeutic Strategies in Spinocerebellar Ataxia Type 7. Advances in experimental medicine and biology. 2018;1049:197–218. doi: 10.1007/978-3-319-71779-1_9. [DOI] [PubMed] [Google Scholar]
- 128.Helmlinger D, Abou-Sleymane G, Yvert G, Rousseau S, Weber C, Trottier Y, et al. Disease progression despite early loss of polyglutamine protein expression in SCA7 mouse model. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24(8):1881–7. doi: 10.1523/JNEUROSCI.4407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abou-Sleymane G, Chalmel F, Helmlinger D, Lardenois A, Thibault C, Weber C, et al. Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Human molecular genetics. 2006;15(5):691–703. doi: 10.1093/hmg/ddi483. [DOI] [PubMed] [Google Scholar]
- 130.Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS biology. 2006;4(3):e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11(4):273–84. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 132.Friedrich B, Euler P, Ziegler R, Kuhn A, Landwehrmeyer BG, Luthi-Carter R, et al. Comparative analyses of Purkinje cell gene expression profiles reveal shared molecular abnormalities in models of different polyglutamine diseases. Brain research. 2012;1481:37–48. doi: 10.1016/j.brainres.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 133.Gatchel JR, Watase K, Thaller C, Carson JP, Jafar-Nejad P, Shaw C, et al. The insulin-like growth factor pathway is altered in spinocerebellar ataxia type 1 and type 7. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(4):1291–6. doi: 10.1073/pnas.0711257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Helmlinger D, Tora L, Devys D. Transcriptional alterations and chromatin remodeling in polyglutamine diseases. Trends Genet. 2006;22(10):562–70. doi: 10.1016/j.tig.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 135.Burke TL, Miller JL, Grant PA. Direct inhibition of Gcn5 protein catalytic activity by polyglutamine-expanded ataxin-7. The Journal of biological chemistry. 2013;288(47):34266–75. doi: 10.1074/jbc.M113.487538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, et al. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Human molecular genetics. 2004;13(1):53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- 137.Kizilyaprak C, Spehner D, Devys D, Schultz P. The linker histone H1C contributes to the SCA7 nuclear phenotype. Nucleus. 2011;2(5):444–54. doi: 10.4161/nucl.2.5.17843. [DOI] [PubMed] [Google Scholar]
- 138.Tan JY, Vance KW, Varela MA, Sirey T, Watson LM, Curtis HJ, et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nature structural & molecular biology. 2014;21(11):955–61. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Strom AL, Forsgren L, Holmberg M. A role for both wild-type and expanded ataxin-7 in transcriptional regulation. Neurobiology of disease. 2005;20(3):646–55. doi: 10.1016/j.nbd.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 140.Achour M, Le Gras S, Keime C, Parmentier F, Lejeune FX, Boutillier AL, et al. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington's disease mice. Human molecular genetics. 2015;24(12):3481–96. doi: 10.1093/hmg/ddv099. [DOI] [PubMed] [Google Scholar]
- 141.Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, et al. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127(4):697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 142.Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci. 2006;7(10):784–96. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 143.Suelves N, Kirkham-McCarthy L, Lahue RS, Gines S. A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal CAG repeat expansions in Huntington's disease mice. Sci Rep. 2017;7(1):6082. doi: 10.1038/s41598-017-05125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Merienne K, Friedman J, Akimoto M, Abou-Sleymane G, Weber C, Swaroop A, et al. Preventing polyglutamine-induced activation of c-Jun delays neuronal dysfunction in a mouse model of SCA7 retinopathy. Neurobiology of disease. 2007;25(3):571–81. doi: 10.1016/j.nbd.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ajayi A, Yu X, Lindberg S, Langel U, Strom AL. Expanded ataxin-7 cause toxicity by inducing ROS production from NADPH oxidase complexes in a stable inducible Spinocerebellar ataxia type 7 (SCA7) model. BMC Neurosci. 2012;13:86. doi: 10.1186/1471-2202-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cooles P, Michaud R, Best PV. A dominantly inherited progressive disease in a black family characterised by cerebellar and retinal degeneration, external ophthalmoplegia and abnormal mitochondria. Journal of the neurological sciences. 1988;87(2-3):275–88. doi: 10.1016/0022-510x(88)90252-3. [DOI] [PubMed] [Google Scholar]
- 147.Forsgren L, Libelius R, Holmberg M, von Dobeln U, Wibom R, Heijbel J, et al. Muscle morphology and mitochondrial investigations of a family with autosomal dominant cerebellar ataxia and retinal degeneration mapped to chromosome 3p12-p21.1. Journal of the neurological sciences. 1996;144(1-2):91–8. doi: 10.1016/s0022-510x(96)00187-6. [DOI] [PubMed] [Google Scholar]
- 148.Han Y, Yu L, Zheng HM, Guan YT. Clinical and genetic study of spinocerebellar ataxia type 7 in East Asian population. Chin Med J (Engl). 2010;123(16):2274–8. [PubMed] [Google Scholar]
- 149.Modi G, Modi M, Martinus I, Rodda J, Saffer D. The clinical and genetic characteristics of spinocerebellar ataxia type 7 (SCA 7) in three Black South African families. Acta Neurol Scand. 2000;101(3):177–82. doi: 10.1034/j.1600-0404.2000.101003177.x. [DOI] [PubMed] [Google Scholar]
- 150.Ward JM, Stoyas CA, Switonski PM, Ichou F, Fan W, Collins B, et al. Metabolic and Organelle Morphology Defects in Mice and Human Patients Define Spinocerebellar Ataxia Type 7 as a Mitochondrial Disease. Cell reports. 2019;26(5):1189-202 e6. doi: 10.1016/j.celrep.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016;5. [DOI] [PMC free article] [PubMed]
- 152.Turmaine M, Raza A, Mahal A, Mangiarini L, Bates GP, Davies SW. Nonapoptotic neurodegeneration in a transgenic mouse model of Huntington's disease. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(14):8093–7. doi: 10.1073/pnas.110078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lebon C, Behar-Cohen F, Torriglia A. Cell Death Mechanisms in a Mouse Model of Retinal Degeneration in Spinocerebellar Ataxia 7. Neuroscience. 2019;400:72–84. doi: 10.1016/j.neuroscience.2018.12.051. [DOI] [PubMed] [Google Scholar]
- 154.Klockgether T, Mariotti C, Paulson HL. Spinocerebellar ataxia. Nat Rev Dis Primers. 2019;5(1):24. doi: 10.1038/s41572-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 155.Tercero-Perez K, Cortes H, Torres-Ramos Y, Rodriguez-Labrada R, Cerecedo-Zapata CM, Hernandez-Hernandez O, et al. Effects of Physical Rehabilitation in Patients with Spinocerebellar Ataxia Type 7. Cerebellum. 2019;18(3):397–405. doi: 10.1007/s12311-019-1006-1. [DOI] [PubMed] [Google Scholar]
- 156.Lee SG, Su ZZ, Emdad L, Gupta P, Sarkar D, Borjabad A, et al. Mechanism of ceftriaxone induction of excitatory amino acid transporter-2 expression and glutamate uptake in primary human astrocytes. The Journal of biological chemistry. 2008;283(19):13116–23. doi: 10.1074/jbc.M707697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Maltecca F, Baseggio E, Consolato F, Mazza D, Podini P, Young SM, Jr, et al. Purkinje neuron Ca2+ influx reduction rescues ataxia in SCA28 model. J Clin Invest. 2015;125(1):263–74. doi: 10.1172/JCI74770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ieraci A, Forni PE, Ponzetto C. Viable hypomorphic signaling mutant of the Met receptor reveals a role for hepatocyte growth factor in postnatal cerebellar development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(23):15200–5. doi: 10.1073/pnas.222362099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sun W, Funakoshi H, Nakamura T. Overexpression of HGF retards disease progression and prolongs life span in a transgenic mouse model of ALS. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22(15):6537–48. doi: 10.1523/JNEUROSCI.22-15-06537.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Noma S, Ohya-Shimada W, Kanai M, Ueda K, Nakamura T, Funakoshi H. Overexpression of HGF attenuates the degeneration of Purkinje cells and Bergmann glia in a knockin mouse model of spinocerebellar ataxia type 7. Neuroscience research. 2012;73(2):115–21. doi: 10.1016/j.neures.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 161.Cortes CJ, La Spada AR. Autophagy in polyglutamine disease: Imposing order on disorder or contributing to the chaos? Mol Cell Neurosci. 2015;66(Pt A):53–61. doi: 10.1016/j.mcn.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Helmlinger D, Bonnet J, Mandel JL, Trottier Y, Devys D. Hsp70 and Hsp40 chaperones do not modulate retinal phenotype in SCA7 mice. The Journal of biological chemistry. 2004;279(53):55969–77. doi: 10.1074/jbc.M409062200. [DOI] [PubMed] [Google Scholar]
- 163.Buijsen RAM, Toonen LJA, Gardiner SL, van Roon-Mom WMC. Genetics, Mechanisms, and Therapeutic Progress in Polyglutamine Spinocerebellar Ataxias. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 164.Butler DC, McLear JA, Messer A. Engineered antibody therapies to counteract mutant huntingtin and related toxic intracellular proteins. Prog Neurobiol. 2012;97(2):190–204. doi: 10.1016/j.pneurobio.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Popiel HA, Takeuchi T, Burke JR, Strittmatter WJ, Toda T, Wada K, et al. Inhibition of protein misfolding/aggregation using polyglutamine binding peptide QBP1 as a therapy for the polyglutamine diseases. Neurotherapeutics. 2013;10(3):440–6. doi: 10.1007/s13311-013-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Potkin KT, Potkin SG. New directions in therapeutics for Huntington disease. Future Neurol. 2018;13(2):101–21. doi: 10.2217/fnl-2017-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bulawa CE, Connelly S, Devit M, Wang L, Weigel C, Fleming JA, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(24):9629–34. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Saunders JC, Young LM, Mahood RA, Jackson MP, Revill CH, Foster RJ, et al. An in vivo platform for identifying inhibitors of protein aggregation. Nat Chem Biol. 2016;12(2):94–101. doi: 10.1038/nchembio.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Roberts BE, Duennwald ML, Wang H, Chung C, Lopreiato NP, Sweeny EA, et al. A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat Chem Biol. 2009;5(12):936–46. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Aviolat H, Nomine Y, Gioria S, Bonhoure A, Hoffmann D, Ruhlmann C, et al. SynAggreg: A Multifunctional High-Throughput Technology for Precision Study of Amyloid Aggregation and Systematic Discovery of Synergistic Inhibitor Compounds. Journal of molecular biology. 2018;430(24):5257–79. doi: 10.1016/j.jmb.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 171.Joshi AS, Singh V, Gahane A, Thakur AK. Biodegradable Nanoparticles Containing Mechanism Based Peptide Inhibitors Reduce Polyglutamine Aggregation in Cell Models and Alleviate Motor Symptoms in a Drosophila Model of Huntington's Disease. ACS Chem Neurosci. 2019;10(3):1603–14. doi: 10.1021/acschemneuro.8b00545. [DOI] [PubMed] [Google Scholar]
- 172.Escalona-Rayo O, Fuentes-Vazquez P, Leyva-Gomez G, Cisneros B, Villalobos R, Magana JJ, et al. Nanoparticulate strategies for the treatment of polyglutamine diseases by halting the protein aggregation process. Drug Dev Ind Pharm. 2017;43(6):871–88. doi: 10.1080/03639045.2017.1281949. [DOI] [PubMed] [Google Scholar]
- 173.Fiszer A, Krzyzosiak WJ. Oligonucleotide-based strategies to combat polyglutamine diseases. Nucleic Acids Res. 2014;42(11):6787–810. doi: 10.1093/nar/gku385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ramachandran PS, Boudreau RL, Schaefer KA, La Spada AR, Davidson BL. Nonallele specific silencing of ataxin-7 improves disease phenotypes in a mouse model of SCA7. Mol Ther. 2014;22(9):1635–42. doi: 10.1038/mt.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mullard A. Pioneering antisense drug heads into pivotal trials for Huntington disease. Nat Rev Drug Discov. 2019;18(3):161–3. doi: 10.1038/d41573-019-00018-7. [DOI] [PubMed] [Google Scholar]
- 176.Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, et al. Targeting Huntingtin Expression in Patients with Huntington's Disease. The New England journal of medicine. 2019;380(24):2307–16. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 177.Niu C, Prakash TP, Kim A, Quach JL, Huryn LA, Yang Y, et al. Antisense oligonucleotides targeting mutant Ataxin-7 restore visual function in a mouse model of spinocerebellar ataxia type 7. Sci Transl Med. 2018;10(465). [DOI] [PMC free article] [PubMed]
- 178.Southwell AL, Skotte NH, Kordasiewicz HB, Ostergaard ME, Watt AT, Carroll JB, et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther. 2014;22(12):2093–106. doi: 10.1038/mt.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stanek LM, Sardi SP, Mastis B, Richards AR, Treleaven CM, Taksir T, et al. Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington's disease. Hum Gene Ther. 2014;25(5):461–74. doi: 10.1089/hum.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Greenberg J, Solomon GA, Vorster AA, Heckmann J, Bryer A. Origin of the SCA7 gene mutation in South Africa: implications for molecular diagnostics. Clinical genetics. 2006;70(5):415–7. doi: 10.1111/j.1399-0004.2006.00680.x. [DOI] [PubMed] [Google Scholar]
- 181.Scholefield J, Watson L, Smith D, Greenberg J, Wood MJ. Allele-specific silencing of mutant Ataxin-7 in SCA7 patient-derived fibroblasts. Eur J Hum Genet. 2014;22(12):1369–75. doi: 10.1038/ejhg.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fiszer A, Olejniczak M, Galka-Marciniak P, Mykowska A, Krzyzosiak WJ. Self-duplexing CUG repeats selectively inhibit mutant huntingtin expression. Nucleic Acids Res. 2013;41(22):10426–37. doi: 10.1093/nar/gkt825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fiszer A, Ellison-Klimontowicz ME, Krzyzosiak WJ. Silencing of genes responsible for polyQ diseases using chemically modified single-stranded siRNAs. Acta Biochim Pol. 2016;63(4):759–64. doi: 10.18388/abp.2016_1336. [DOI] [PubMed] [Google Scholar]
- 184.Fiszer A, Wroblewska JP, Nowak BM, Krzyzosiak WJ. Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells. Genes (Basel). 2016;7(12). [DOI] [PMC free article] [PubMed]
- 185.Curtis HJ, Seow Y, Wood MJA, Varela MA. Knockdown and replacement therapy mediated by artificial mirtrons in spinocerebellar ataxia 7. Nucleic Acids Res. 2017;45(13):7870–85. doi: 10.1093/nar/gkx483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121(2):500–7. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Donde A, Wong PC, Chen LL. Challenges and Advances in Gene Therapy Approaches for Neurodegenerative Disorders. Curr Gene Ther. 2017;17(3):187–93. doi: 10.2174/1566523217666171013124150. [DOI] [PubMed] [Google Scholar]
- 188.Bailus B, Zhang N, Ellerby LM. Using Genome Engineering to Understand Huntington's Disease. In: Jaenisch R, Zhang F, Gage F, editors. Genome Editing in Neurosciences. Cham (CH) 2017. pp. 87–101. [Google Scholar]
- 189.Saha SK, Saikot FK, Rahman MS, Jamal M, Rahman SMK, Islam SMR, et al. Programmable Molecular Scissors: Applications of a New Tool for Genome Editing in Biotech. Mol Ther Nucleic Acids. 2019;14:212–38. doi: 10.1016/j.omtn.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Malankhanova TB, Malakhova AA, Medvedev SP, Zakian SM. Modern Genome Editing Technologies in Huntington's Disease Research. Journal of Huntington's disease. 2017;6(1):19–31. doi: 10.3233/JHD-160222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Saute JA, Donis KC, Serrano-Munuera C, Genis D, Ramirez LT, Mazzetti P, et al. Ataxia rating scales--psychometric profiles, natural history and their application in clinical trials. Cerebellum. 2012;11(2):488–504. doi: 10.1007/s12311-011-0316-8. [DOI] [PubMed] [Google Scholar]
- 192.Tezenas du Montcel S, Charles P, Goizet C, Marelli C, Ribai P, Vincitorio C, et al. Factors influencing disease progression in autosomal dominant cerebellar ataxia and spastic paraplegia. Arch Neurol. 2012;69(4):500–8. doi: 10.1001/archneurol.2011.2713. [DOI] [PubMed] [Google Scholar]
- 193.Coarelli G, Brice A, Durr A. Recent advances in understanding dominant spinocerebellar ataxias from clinical and genetic points of view. F1000Res. 2018;7. [DOI] [PMC free article] [PubMed]
- 194.Adanyeguh IM, Henry PG, Nguyen TM, Rinaldi D, Jauffret C, Valabregue R, et al. In vivo neurometabolic profiling in patients with spinocerebellar ataxia types 1, 2, 3, and 7. Mov Disord. 2015;30(5):662–70. doi: 10.1002/mds.26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shi Y, Huang F, Tang B, Li J, Wang J, Shen L, et al. MicroRNA profiling in the serums of SCA3/MJD patients. Int J Neurosci. 2014;124(2):97–101. doi: 10.3109/00207454.2013.827679. [DOI] [PubMed] [Google Scholar]
- 196.Serafin A, Foco L, Zanigni S, Blankenburg H, Picard A, Zanon A, et al. Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology. 2015;84(7):645–53. doi: 10.1212/WNL.0000000000001258. [DOI] [PubMed] [Google Scholar]
- 197.Derkow K, Rossling R, Schipke C, Kruger C, Bauer J, Fahling M, et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer's disease. PloS one. 2018;13(7):e0200602. doi: 10.1371/journal.pone.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Koscianska E, Krzyzosiak WJ. Current understanding of the role of microRNAs in spinocerebellar ataxias. Cerebellum Ataxias. 2014;1:7. doi: 10.1186/2053-8871-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Borgonio-Cuadra VM, Valdez-Vargas C, Romero-Cordoba S, Hidalgo-Miranda A, Tapia-Guerrero Y, Cerecedo-Zapata CM, et al. Wide Profiling of Circulating MicroRNAs in Spinocerebellar Ataxia Type 7. Molecular neurobiology. 2019. [DOI] [PubMed]
- 200.Cornelius N, Wardman JH, Hargreaves IP, Neergheen V, Bie AS, Tumer Z, et al. Evidence of oxidative stress and mitochondrial dysfunction in spinocerebellar ataxia type 2 (SCA2) patient fibroblasts: Effect of coenzyme Q10 supplementation on these parameters. Mitochondrion. 2017;34:103–14. doi: 10.1016/j.mito.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 201.Klepac N, Relja M, Klepac R, Hecimovic S, Babic T, Trkulja V. Oxidative stress parameters in plasma of Huntington's disease patients, asymptomatic Huntington's disease gene carriers and healthy subjects : a cross-sectional study. Journal of neurology. 2007;254(12):1676–83. doi: 10.1007/s00415-007-0611-y. [DOI] [PubMed] [Google Scholar]
- 202.Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45(5):667–78. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 203.Torres-Ramos Y, Montoya-Estrada A, Cisneros B, Tercero-Perez K, Leon-Reyes G, Leyva-Garcia N, et al. Oxidative Stress in Spinocerebellar Ataxia Type 7 Is Associated with Disease Severity. Cerebellum. 2018;17(5):601–9. doi: 10.1007/s12311-018-0947-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 498 kb)