Abstract
Bud dormancy is one of the most important defensive mechanisms through which plants resist cold stress during harsh winter weather. DAM, Dof, and WRKY have been reported to be involved in many biological processes, including bud dormancy. In the present study, grapevine (Vitis vinifera) and other thirteen plants (six woody plants and seven herbaceous plants) were analyzed for the quantity, sequence structure, and evolution patterns of their DAM, Dof, and WRKY gene family members. Moreover, the expression of VvDAM, VvDof, and VvWRKY genes was also investigated. Thus, 51 DAM, 1,205 WRKY, and 489 Dof genes were isolated from selected genomes, while 5 DAM, 114 WRKY, and 50 Dof duplicate gene pairs were identified in 10 genomes. Moreover, WGD and segmental duplication events were associated with the majority of the expansions of Dof and WRKY gene families. The VvDAM, VvDof, and VvWRKY genes significantly differentially expressed throughout bud dormancy outnumbered those significantly differentially expressed throughout fruit development or under abiotic stresses. Interestingly, multiple stress responsive genes were identified, such as VvDAM (VIT_00s0313g00070), two VvDof genes (VIT_18s0001g11310 and VIT_02s0025g02250), and two VvWRKY genes (VIT_07s0031g01710 and VIT_11s0052g00450). These data provide candidate genes for molecular biology research investigating bud dormancy and responses to abiotic stresses (namely salt, drought, copper, and waterlogging).
Electronic supplementary material
The online version of this article (10.1007/s13205-019-2039-3) contains supplementary material, which is available to authorized users.
Keywords: Vitis vinifera, Bud dormancy, DAM, Dof, WRKY, 14 species
Introduction
Defense mechanisms have evolved in plants under various challenging conditions over deep evolutionary timescales. Dormancy mechanisms have evolved in plants as a mechanism of survival against the low temperatures and short photoperiods that occur during harsh winter seasons (Ríos et al. 2014). Plant growth and developmental cycles generally pause during dormancy periods and occur even in seeds and buds. Bud dormancy can be classified into three different categories (Lang 1987): (1) ecodormancy, when growth is prevented by environmental conditions, such as low temperatures, water shortage, and nutrient deficiency; (2) paradormancy, when growth is inhibited by distal organs, such as via apical dominance, and photoperiodic responses; (3) endodormancy, when growth is prevented by internal bud signaling, such as chilling responses, and photoperiodic responses. However, unfortunately, if there are too few accumulated chilling hours, flowering quality and uniformity can be affected, which causes a drastic reduction in fruit production in the subsequent season. Accordingly, endodormancy may play an important role in fruit production.
Much research has been conducted to investigate the mechanisms underlying dormancy period over recent decades, aside from investigations of its economic impacts. From the beginning of the twentieth century to the present, dormancy studies have gone through three stages: observation of phenomena, cellular modification research, and metabolic analysis (Beauvieux et al. 2018). In particular, this has been highlighted by genetic and genomics studies. Recent advancements in transcriptomic technology have led to the identification of molecular pathways controlling bud dormancy in perennial trees, including Populus trichocarpa (Rohde et al. 2007; Ruttink et al. 2007), Prunus mume (Yamane et al. 2008; Zhong et al. 2013), Prunus persica (Leida et al. 2010), Pyrus bretschneideri (Liu et al. 2012), Pyrus pyrifolia (Bai et al. 2013), Prunus pseudocerasus (Zhu et al. 2015), Cunninghamia lanceolata (Xu et al. 2016), Quercus petraea (Ueno et al. 2013), and Vitis vinifera (Fennell et al. 2015; Khalil-Ur-Rehman et al. 2017; Min et al. 2017; Sreekantan et al. 2010). Specific gene expression patterns suggest that bud dormancy is associated with stress responses, hormone signaling, chromatin modification, and carbon metabolism (Regier et al. 2010; Rios et al. 2014; Saito et al. 2015; Wen et al. 2016; Wisniewski et al. 2015).
Among the well-known pathways that have thus far been characterized in plants, dormancy-associated MADS-box (DAM), DNA-binding one zinc finger (Dof), and WRKY genes have been identified as being involved in dormancy regulation. In peach, six PpDAM genes show distinctive seasonal expression patterns in the top buds, and moreover, PpDAM1, PpDAM2, and PpDAM4 appear to be closely linked with terminal bud formation (Bielenberg et al. 2008; Li et al. 2009). Pear DAM1 could regulate bud dormancy transition under the direct activation of PpHB22 in ‘Suli’ pear (Yang et al. 2018). Overexpression of PmDAM6 could inhibit growth, repress bud break competency of dormant buds and delays bud outgrowth in apple plants (Yamane et al. 2019). Similar regulation models have been demonstrated in Euphorbia esula (Horvath et al. 2010), Pyrus pyrifolia (Ito et al. 2015; Saito et al. 2015; Ubi et al. 2010), Malus × domestica (Mimida et al. 2015), Pyrus bretschneideri (Niu et al. 2015), and Prunus mume (Sasaki et al. 2011). Dof genes have been implicated in bud dormancy transitional stage development in Prunus persica and Fagus sylvatica (Chen et al. 2017; Lesur et al. 2015). In peach, five PpDofs were up-regulated in the transitional stage, may be involved in dormancy; and one PpDof was highly expressed at the end (5 February) of dormancy, may play a role in bud flush (Chen et al. 2017). Pear Dof9.2 could repress flowering time by promoting the levels of PbTFL1a and PbTFL1b (Liu et al. 2019). The dormancy status in Retama raetam was accompanied by the transcription level of one WRKY (Pnueli et al. 2002). In Arabidopsis, WRKY41 controls seed dormancy via direct regulation the expression level of ABI3 (Ding et al. 2014). Moreover, at least six peach WRKY genes have been proven to have higher expression levels in endodormancy and lower expression levels in ecodormancy, which indicates that they may play a key role in dormancy regulation (Chen et al. 2016).
In the present study, the number, sequence structure, and evolution of DAM, Dof, and WRKY gene families were analyzed by comparisons between these genes in Vitis vinifera (woody plant) and 13 other plant species, namely, 6 woody plant species (Actinidia chinensis, Citrus sinensis, Malus domestica, Populus trichocarpa, Prunus persica, and Pyrus bretschneideri) and 7 herbaceous plant species (Ananas comosus, Arabidopsis thaliana, Carica papaya, Fragaria vesca, Musa acuminate, Oryza sativa, and Solanum lycopersicum). Additionally, we analyzed the expression profiles of VvDAM, VvDof, and VvWRKY genes in V. vinifera during bud dormancy, fruit development, and different abiotic stresses to identify candidate genes for further study (Table 1).
Table 1.
The RT-qPCR primer sequences of VvDAMs, VvDofs, and VvWRKYs
| Gene ID | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| VIT_00s0313g00070 | CAACAATCACTCCAGGTTGAGC | TTGGACACCCTTTGACTGAAGA |
| VIT_15s0107g00120 | AGACTCAGAGGCTGAGGCAAAT | TCCTGCAGTAGGTCTCCCTTTC |
| VIT_18s0001g07460 | GCTCAGCTGATGGAAGAGAACA | TGAGGAGGACCACTACTGCAAC |
| VIT_00s0218g00040 | CCTTGTTTCAGCATCGATCAAC | TGCTGTAATTTCCAGTGCAGGT |
| VIT_00s0253g00060 | TATGATTGGATCCCACATGACC | TTGTTGGTGCTGTTGAGTTTGA |
| VIT_00s0652g00010 | CAGTTGGAGCCATTGAAGTGTC | GCAGATGTCTTGGGCCTCTTAT |
| VIT_01s0026g02580 | CCAAAGACACTGAGGATTGACG | CACCGGAGACATTTCAGCTATG |
| VIT_02s0025g02250 | CACAACTGGGTTTCCTATGCAG | CCCAGTTGAATCTCCTTGCTCT |
| VIT_03s0063g01350 | GCCACTTCTGCAAATCATGC | GTTCAGCAACCTGGTGAAACTC |
| VIT_06s0004g03420 | GGGTGCAGGAAGAACAAGAGAT | TACCCTAACTGCCCACCAGTTT |
| VIT_06s0004g04520 | GCTCGATTAGACCTGGTTCGAT | ACCTCCTCTCGTCCAGTATCGT |
| VIT_07s0255g00020 | CCACCAATCATGCCACATCTAT | CAAATCCCATGTCCTCGAAAC |
| VIT_08s0007g00180 | GAGGTTTAGAGCAGTGGCGATT | AGCCACCTGACTTAACCCAGAC |
| VIT_08s0056g01230 | ACCACGATTCAGACCCAGATTT | ACTTGAGTTTGCTGCTCCTCCT |
| VIT_08s0105g00170 | ATATTTATGGGCTGCCACCACT | AATCAAATGGGCTCCAGTTACC |
| VIT_09s0002g02490 | TCTACGTCGTCGGTGTCTTCTC | CGGATTCAGACTCTCCAACAAG |
| VIT_10s0003g00030 | CCTTTCTCAGCCTCGCTACTTC | CGGAGATAAGTTGGCTGGTTCT |
| VIT_10s0003g00040 | CTTCCGAGCTCAATCTTCCATT | TTCGGTAATCATCAGTGCGAGT |
| VIT_10s0003g01260 | GGGTTCTTGGAGAGTTGCTTGT | CAAGTTTGACCTCCCGATGAG |
| VIT_13s0019g01410 | AATTCCCGATTGGTAGCTGTTC | AACTCGGTGGCTCAGTACCTTC |
| VIT_14s0108g00980 | ATGCGTTTCCTCCTCAGATACC | TAAGCAGCTGCAGGGTAGAATG |
| VIT_15s0046g00150 | AGAAGCCAAGGACATGGAAGAG | TTGCAGAAGTACCTTGGCTGAG |
| VIT_16s0098g01420 | TGCTCAAGAGTACTGGGATTGC | CTCCCATACCCACTTTGAAACC |
| VIT_17s0000g04850 | GCAAAAGATGTCCGATGAGTTG | AACGTTTCCAGGAAGATCGAAG |
| VIT_17s0000g06310 | AAAGCTGCCAGAGGTATTGGAC | CAATTCGAACAGTTTGGAGTGC |
| VIT_17s0000g08290 | CTTCTTCACCTTCCGGAGACAT | TTGCAGAAGTACCTTGGCTGAG |
| VIT_18s0001g11310 | TGGTACTCGCAAGAACTCCAAG | CAGACACATCCCCATTCAGACT |
| VIT_18s0001g15730 | CTTGAAGGCCACACACAGTTG | TCACTGGCTGCCATGTAAAGTT |
| VIT_00s0463g00010 | ATCCTGATTCAAAGCGAAGGAG | GGATTGCCTTTCACCACTTTCT |
| VIT_01s0010g03930 | CTTTTCAAACCAGGAGCCAAGT | ATAGGCGTTGAACCTGCTTCTT |
| VIT_01s0011g00220 | TAAGCCTGCTGATGATGGCTAC | TTGTTGGGTAGTGGTGCTTGAT |
| VIT_01s0011g00720 | GCTATTACCGGTGCACAAACAG | AACTTCACGGTGGAACGACATA |
| VIT_01s0026g01730 | AGAAAGCAGGTGCAAAGATGTG | GCAGTAGAGGAGGTAGCGAAGG |
| VIT_02s0025g00420 | GTAACAGCCGAGGGTCTGTCTT | GGCTGTGTTCAGCTGTGTAGGT |
| VIT_02s0025g01280 | TCCGATTCAAACTGCTTCTCAC | GGAAACTGGGATCAAAGTCCAC |
| VIT_02s0154g00210 | AAAGGATCTCCACATCCAAGGT | GATGATCCAAGATGCAACAAGC |
| VIT_04s0008g01470 | GAAACAGTGAGAGTGGGCAGAA | CCGTACTTCCTCCATTTGAACC |
| VIT_04s0008g05750 | GAAGGAGCACACAACCATGAAC | TCCCGGAAAGAGTTAGATCGAG |
| VIT_04s0008g05760 | ACCCTTCTCCCAGAGCCTACTT | AGCTCGGTTGAAGCCTAATGAC |
| VIT_04s0008g06600 | AGGAGGAAATTGGTGATGTGGT | GGGAGCTGAACATGCTAAACCT |
| VIT_04s0023g00470 | TCAGCTCTTGGATCCACACTTC | GAGCGCACTTACAGCAGATGTT |
| VIT_04s0069g00920 | GCAGTGTTTCTAACGGAAAGCA | TTCTTCACCCGAGATCTCCTTC |
| VIT_04s0069g00970 | TGGAGACTGCCAAGTGAAGAAG | TGGGAGGAAGCTTGTAAAGTCC |
| VIT_04s0069g00980 | ATTGAGGACAAGATCGGAGGAG | CCTCTTCTTCACTTGGCATCCT |
| VIT_05s0077g00730 | GGAGGAAGAGCAAGATCAGGAG | AAAGGGCTGTTCTTCACAGCTT |
| VIT_06s0004g00230 | TTGCTGAGAAGCCAAAAGACAC | GAAGCTTCATTCATGGCTGCTA |
| VIT_06s0004g07500 | CCTTCTCCGACAACAGGAACTT | TGTCCTCTCAAGGCTTGTTCAG |
| VIT_07s0005g01520 | ATCAACCACAACAACCCATCAC | GCTGCTCATCAAATACGACACC |
| VIT_07s0005g01710 | TGAAGAAACGAGTGGAGAGGTG | CATCGTTGAAAGAGGGATGTTG |
| VIT_07s0005g02570 | TCTTCCACCTCGTTTCCTTCTC | GTGTTGCCATAACTGCTCCTTG |
| VIT_07s0031g00080 | CCGGCAATAAGCATGAAAAT | ATCAGCATCTTTGGGTCGTC |
| VIT_07s0031g01710 | CAGTGAAGAACAGCCCCAAT | CATCAAAGGAACCTGGTCGT |
| VIT_07s0031g01840 | TGCCAACAATCCCTAAAAGC | TGCTCAAGCACTCATTCACC |
| VIT_07s0141g00680 | GTTTGGGAAACCCATCATTG | CTATTGCTCCGCCGATACAT |
| VIT_08s0007g00570 | TCCCTTCACCATCCAAGAAG | ATCTTCCAGCAACCCTTGTG |
| VIT_08s0040g03070 | ATGGCCAGAAGGTTGTGAAG | TTCAGAGGTTGCTGCATTTG |
| VIT_08s0058g00690 | CCTTTTCCAGTCCCAAACAA | TTTGGATTGAAGCGATTTCC |
| VIT_08s0058g01390 | GGTTATGCCTGGCGAAAATA | TGACTTCAGCATGCTTTTGC |
| VIT_09s0018g00240 | GAACGCAGAAAACAGGAAGC | AGTTCCCAACGACTCCATTG |
| VIT_10s0003g01600 | GCAATTTTCTCAAGCCAAGC | CGATTCATCCTCTTCCCTCA |
| VIT_10s0003g02810 | CAGAAGTGCACGGTGAAGAA | CTCCCATCTGTCCTGGTGTT |
| VIT_10s0003g05740 | GGAGATGGGAAACGTTGAGA | GCTGTTGTTCCCTGTCCAAT |
| VIT_10s0116g01200 | CAATTGCAAGTGGAGCTTGA | TTGGGCCTAGATCGATGAAC |
| VIT_11s0037g00150 | GGGATCTCAGGTGATGGCTA | AGGCATGTCATGGTCGTGTA |
| VIT_11s0052g00450 | AGTCAAGCCCATTTCATTCG | GAGGTGCGAACAGAGGAGTC |
| VIT_12s0028g00270 | GGCTTCCCATTCTTTTGTGA | CTGTACCCTTTCCCGAATCA |
| VIT_12s0055g00340 | AGCGATGGCTTCAACAACTT | CTAATTGGGCGGATGAGTGT |
| VIT_12s0057g00550 | CCAACCTGCTATTGCAGTCA | CGGTTCACCACTTGGCTTAT |
| VIT_12s0059g00880 | CCAAGCCACCCACTACAGAT | GCTTGAAGAACCACCAGCTC |
| VIT_13s0067g03130 | AAGCGCTCCAAGCACAGTAT | CCAATTCTAGGCGCTGCTAC |
| VIT_13s0067g03140 | TGTCAGCTGAGGATCTGGTG | TTTTGCCCATATTTCCTCCA |
| VIT_14s0068g01770 | TGGAGGCTTTGGTAGGTCTG | TGCCTTTTGACCGTACTTCC |
| VIT_14s0081g00560 | TCCCACGATGTCATCCACTA | GCTCCCATCTTCTCCCCTAC |
| VIT_14s0108g00120 | TGCCATTGATGAAGAATCCA | TTGGACGCAAACTCTGTCAC |
| VIT_14s0108g01280 | CACCGATGCAACTACACACC | CTCCCTTTGACCTGCTTCTG |
| VIT_15s0021g01310 | GAGGAGAACCAGGTGCTGAG | CATCGTTGGCATTACCAGTG |
| VIT_15s0046g01140 | GGCCACATCTGAATCCAACT | TCCAGCGGGAAACTGTAGTC |
| VIT_15s0046g02150 | CAGCGACCTTTACCCAAGAG | TTCCTCCAGCGATATCCATC |
| VIT_15s0046g02190 | CCAAATTCAGCAGCAACAGA | AACCCCTCAGCTGGTATGTG |
| VIT_16s0050g01480 | CAAGACAACCCCGAGGAGTA | CGTCTTCAAACATTGGCTCA |
| VIT_16s0050g02510 | TGGGAAGTTGGGAACAAAAG | CGCTACACCTCAGCAATGAA |
| VIT_17s0000g01280 | ATCAAAATCACCCGCAAATC | TGCTTTCTGCCCATATTTCC |
| VIT_17s0000g05810 | TATCCTCCCCCAAGATTTCC | AGGATCTGGTAGGGGCTGTT |
| VIT_18s0001g10030 | TCTGCTCCCAACTCTTTCGT | AAAAGATGATCCGCACTTGG |
| VIT_19s0015g01870 | CATTCAAGCCTGTTGCAGAA | TGCCTGAACATGGAATGTGT |
| VIT_19s0090g00840 | CATCACAACCATCCCCTACC | GCCTCTGGTGCTGTAAGAGG |
| VIT_19s0090g01720 | TAATAGCCATGACGCAGCAG | TTAGTGCGGGTTTGGGTTAG |
Results
Identification of DAM, WRKY, and Dof gene family members in the selected genomes
A total number of 51 (DAM), 1205 (WRKY), and 489 (Dof) gene family members were identified from the selected genome (Table 2, detailed in Tables S1–S3). Most species had only two or three DAMs, while at least five DAMs were found in Populus trichocarpa (9), Fragaria vesca (6), Malus domestica (6), and Prunus persica (5). The total number of WRKY gene family members in the selected genomes ranged from 49 to 153, and the three species with the most WRKY genes were Musa acuminate (152), Malus domestica (141), and Pyrus bretschneideri (112). The total number of Dof gene family members in the selected genomes ranged from 20 to 73, and the three species with the most Dof TFs were Musa acuminate (73), Malus domestica (59), and Pyrus bretschneideri (46).
Table 2.
Number of DAM, Dof, and WRKY in each species
| Actinidia chinensis | Ananas comosus | Arabidopsis thaliana | Carica papaya | Fragaria vesca | Musa acuminata | Oryza sativa | Solanum lycopersicum | Citrus sinensis | Malus domestica | Populus trichocarpa | Prunus persica | Pyrus bretschneideri | Vitis vinifera | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAM | 2 | 3 | 2 | 2 | 6 | 3 | 3 | 2 | 2 | 6 | 9 | 5 | 3 | 3 | 51 |
| Dof | 25 | 26 | 36 | 20 | 23 | 73 | 30 | 32 | 24 | 59 | 45 | 25 | 46 | 25 | 489 |
| WRKY | 106 | 56 | 72 | 49 | 58 | 152 | 101 | 81 | 53 | 140 | 105 | 61 | 112 | 59 | 1205 |
Sequence characteristics and genomic locations of DAM, WRKY, and Dof gene family members
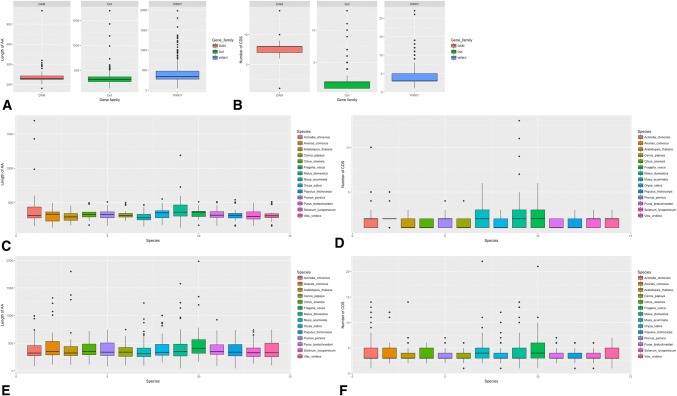
First, the inferred protein sequences and coding sequences (CDS) of all three genes were characterized (Fig. 1). The average DAM protein sequences were shorter than those of WRKY and Dof protein sequences (Fig. 1a). The average Dof protein sequence in apple was longer than those in the others, while the average WRKY protein sequence in strawberry was longer than those in the other species (Fig. 1c, e). The average DAM gene CDS was significantly longer than those of WRKY and Dof genes (Fig. 1b). Most Dof genes contained two CDS regions in each species, while most WRKY genes contained three CDS regions in each species. Moreover, Dof genes in Ananas comosus and WRKY genes in Arabidopsis thaliana had a high level of agreement with each other, while Dof genes in Musa acuminate and WRKY genes in Fragaria vesca differed in the number of CDS regions (Fig. 1d, f).
Fig. 1.
The overview of length and CDS quantity of DAM, Dof, and WRKY in selected genomes. a The length of DAM, Dof, and WRKY in selected genomes. b The CDS quantity of DAM, Dof, and WRKY in selected genomes. c The boxplot of length of Dof in selected genomes. d The boxplot of CDS quantity of Dof in selected genomes. e The boxplot of length of WRKY in selected genomes. f The boxplot of CDS quantity of WRKY in selected genomes
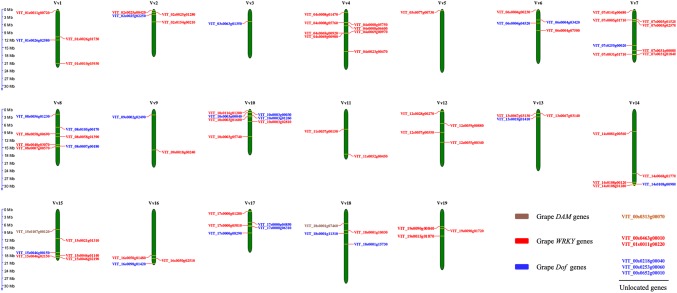
The physical location of grapevine DAM, WRKY, and Dof gene family members were assigned to chromosomes 1–19, except for one DAM (VIT_00s0313g00070), two WRKY (VIT_00s0463g00010 and VIT_01s0011g00220), and three Dof (VIT_00s0218g00040, VIT_00s0253g00060, and VIT_00s0652g00010) genes, which were assigned to unmapped chromosomes (Fig. 2, Tables S1–S3). No WRKY gene family member was assigned to grapevine chromosome 3, while no Dof gene family member was assigned to chromosomes 4, 5, 11, 12, or 19. The WRKY and Dof gene family members in the other plants shared a genomic distribution pattern similar to that observed in grapevine (Tables S2–S3).
Fig. 2.
The distribution of grapevine DAMs, Dofs, and WRKYs on chromosomes
Protein sequence alignment, phylogenetic tree, and domain analysis of DAM, WRKY, and Dof gene family members
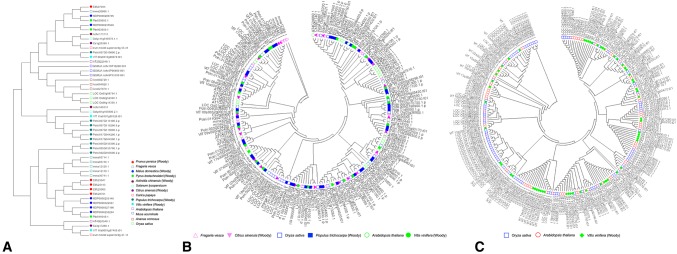
All the protein sequences of DAM genes were aligned using MEGA, and woody species, such as Vitis vinifera, Malus domestica, and Prunus persica were separated from herbaceous species (e.g., Arabidopsis thaliana, Oryza sativa, and Fragaria vesca (Fig. 3a). Based on the large size of the Dof and WRKY families, Dof protein sequences from six plants and WRKY protein sequences from three plants were also aligned using MEGA, respectively. The NJ phylogenetic trees showed that Dof and WRKY protein sequences from woody plants were respectively separated from those from herbaceous plant species (Fig. 3b, c).
Fig. 3.
Phylogenetic tree of DAMs (a), Dofs (b), and WRKYs (c) in selected genomes. The species were represented by different icons
SMART analysis indicated that all the DAM members contained one MADS domain and one K-box domain, except Achn340131, LOC_Os06g11330.1, and MDP0000527190 (Fig. S1a). Achn340131 had two K-box domains and one MADS domain located between the two K-box domains. LOC_Os06g11330.1 and MDP0000527190 contained only one MADS domains. Fourteen domain models represented 489 Dof genes from 14 species (Fig. S1b). Four hundred and eighty-seven Dof members contained one zf-Dof domain, while two Dof genes contained two zf-Dof domains (Achn386081 and Achn321981). Fifteen other domains were also found in inferred Dof protein sequences. Some domains were annotated to have transcription functions (e.g., EF1_GNE and E2F_TDF), while others were annotated to be related to nucleic acid binding, DNA binding, and protein binding function (e.g., 35EXOc, AT_hook, and PHD) or signaling function (e.g., Efh and Jas). Only 9.5% of WRKY family members contained two WRKY domains, although most of them contained a single WRKY domain (Fig. S1c). Moreover, more than 30 other domains were also found in inferred WRKY protein sequences. WRKY proteins have been found to have extensive functions in plant development and stress responses. In this study, some domains were annotated to be involved in stress responses (e.g., LRR and LRR_CC), while others were annotated to have multiple binding functions (e.g., RRM, TIR, WD40, ZnF_BED, and PUA) or to be associated with proteins of unknown function (e.g., DUF1664, DUF4413, DUF863, and DUF2985).
Duplication events and divergence rates of DAM, WRKY, and Dof gene families
Owing to the inadequate annotations for pear, apple, peach, and papaya, these species were excluded from duplication event and divergence rate analyses, leaving the ten remaining species. Five duplicate gene pairs were found within the DAM gene family using MCScanX. Among these, one strawberry gene pair (mrna12119-mrna12120) and two Populus gene pairs (Potri.007G115000-Potri.007G115100, Potri.007G115100-Potri.007G115200) were identified as tandem repeats, while another two DAM gene pairs (LOC_Os06g11330-LOC_Os02g52340, Potri.017G044200-Potri.007G115000) were identified as segmental duplicate gene pairs (Table 3). The dates of three Populus DAM duplication events were projected to have occurred between 4.89 and 23.71 million years ago (MYA). One gene pair was under positive selection (Potri.007G115000-Potri.007G115100), while the others were found to be under purifying selection. Notably, the date of the rice DAM duplication event was predicted to have occurred 109.57 MYA, which was much earlier than that in poplar.
Table 3.
Ka/Ks analysis and estimation of the absolute dates of the duplication events between the duplicated rice, populus, strawberry DAM homologues
| Duplicated pair | Duplicate type | Ka | Ks | Ka/Ks | Purifying selection | Date (million years) |
|---|---|---|---|---|---|---|
| LOC_Os06g11330-LOC_Os02g52340 | Segmental | 0.0956 | 1.4244 | 0.0671 | Yes | 109.5715 |
| mrna12119-mrna12120 | Tandem | 0.2459 | 0.7261 | 0.3387 | Yes | NA |
| Potri.007G115000-Potri.007G115100 | Tandem | 0.1817 | 0.1643 | 1.1059 | No | 9.0266 |
| Potri.007G115100-Potri.007G115200 | Tandem | 0.0724 | 0.0890 | 0.8128 | Yes | 4.8921 |
| Potri.017G044200-Potri.007G115000 | Segmental | 0.2126 | 0.4314 | 0.4927 | Yes | 23.7055 |
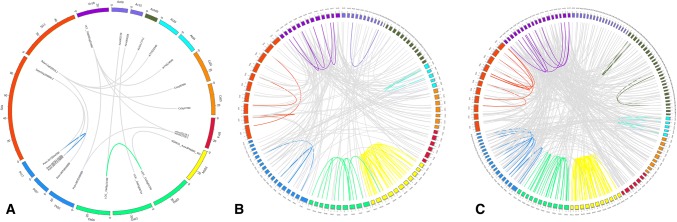
We also performed a synteny analysis with sequences from the remaining ten species (including grapevine), for which fourteen gene pairs were isolated (Table 4). Three of these gene pairs belonged to rice, one pair belonged to Populus, six pairs belonged to tomato, and four pairs belonged to grapevine. Notably, LOC_Os06g11330 and LOC_Os02g52340 were inferred to be segmental duplicate gene pairs in rice, which were inferred to be orthologs of a pineapple gene (Aco002729) and a banana gene (GSMUA_Achr3P06800_001), respectively. One tomato DAM gene (Solyc01g105800) was orthologous to two Populus genes (Potri.007G115200 and Potri.017G044200), while one tomato DAM gene (Solyc11g010570) was orthologous to one gene each in kiwifruit (Achn171711), Arabidopsis (AT2G22540), sweet orange (Cs1g20360), and Populus (Potri.007G010800), respectively. One grapevine DAM gene (VIT_18s0001g07460) was also orthologous to four DAM genes in four species. The Ka/Ks ratios of these fourteen duplicate gene pair were significantly lower than those among three Populus duplicate gene pairs (0.4927–1.1059). All the duplicate gene pairs, except tandem gene pairs, were also visualized using Circos software (Fig. 4a).
Table 4.
Ka/Ks analysis of duplicated DAM homologues in selected genomes
| Duplicate pair | Ka | Ks | Ka/Ks | Purifying selection |
|---|---|---|---|---|
| LOC_Os02g52340-GSMUA_Achr3P06800_001 | 0.2525 | 1.5375 | 0.1642 | Yes |
| LOC_Os03g08754-Aco004028 | 0.2825 | 2.0331 | 0.1390 | Yes |
| LOC_Os06g11330-Aco002729 | 0.1990 | 0.7448 | 0.2672 | Yes |
| Potri.002G105600-mrna12119 | 0.3443 | 1.4492 | 0.2376 | Yes |
| Solyc01g105800-Potri.007G115200 | 0.3771 | 2.4953 | 0.1511 | Yes |
| Solyc01g105800-Potri.017G044200 | 0.3659 | 1.4135 | 0.2589 | Yes |
| Solyc11g010570-Achn171711 | 0.1392 | 3.7684 | 0.0369 | Yes |
| Solyc11g010570-AT2G22540 | 0.2080 | 3.7418 | 0.0556 | Yes |
| Solyc11g010570-Cs1g20360 | 0.1549 | 3.7352 | 0.0415 | Yes |
| Solyc11g010570-Potri.007G010800 | 0.1355 | 1.5937 | 0.0850 | Yes |
| VIT_18s0001g07460-AT4G24540 | 0.2373 | 3.6573 | 0.0649 | Yes |
| VIT_18s0001g07460-Cs3g17260 | 0.1468 | 1.5855 | 0.0926 | Yes |
| VIT_18s0001g07460-mrna12120 | 0.2717 | 1.1965 | 0.2271 | Yes |
| VIT_18s0001g07460-Potri.002G105600 | 0.1548 | 1.9183 | 0.0807 | Yes |
Fig. 4.
Synteny analysis of DAMs (a), Dofs (b), and WRKY (c) among selected species
Fifty duplicate Dof gene pairs and 114 duplicate WRKY gene pairs were identified in each species, and all of them were under purifying selection. Moreover, 96% of Dof gene pairs and 89.47% WRKY gene pairs were inferred to have arisen through segmental duplication. Additionally, 72% and 68.42% of gene pairs belonged to banana and rice, respectively (Tables S4–S5).
The Populus WRKY duplication events were inferred to have occurred first, between 8.9075 and 102.8418 MYA (24.6086 MYA mean), followed by the duplication of Arabidopsis WRKY genes (6.7098–73.3367 MYA, 36.9943 MYA mean), tomato WRKY genes (3.0886–283.8233 MYA, 99.6510 MYA mean), grapevine WRKY genes (58.9438–376.3792 MYA, 173.1244 MYA mean), and rice WRKY genes (6.9985–297.8946 MYA, 185.9315 MYA mean). For Dof duplicate gene pairs, mean inferred duplication events within each species occurred in the following order: tomato (136.9214 MYA), grapevine (129.7212 MYA), rice (125.7269 MYA), Arabidopsis (29.9528 MYA), and Populus (14.7926 MYA).
One hundred and fifty-eight duplicate Dof gene pairs and 377 duplicate WRKY gene pairs were identified, with more than 70% of gene pairs having grapevine WRKY or Dof members (Tables S6–S7). All the duplicate gene pairs, except tandem gene pairs, were also visualized using Circos software (Fig. 4b, c).
The expression patterns of grapevine DAM, WRKY, and Dof gene family members during bud dormancy
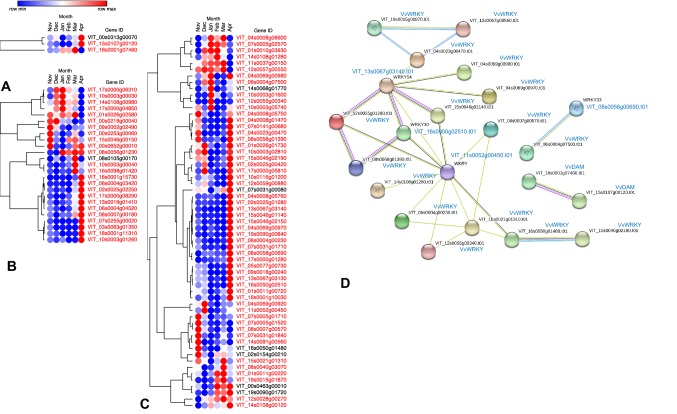
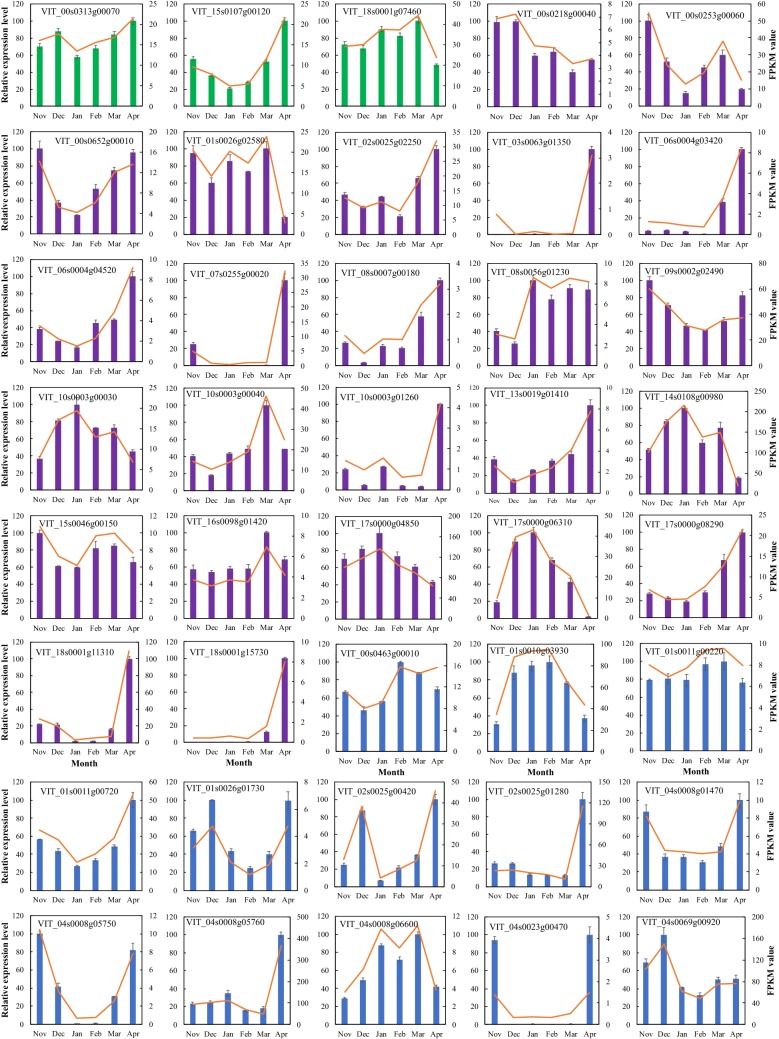
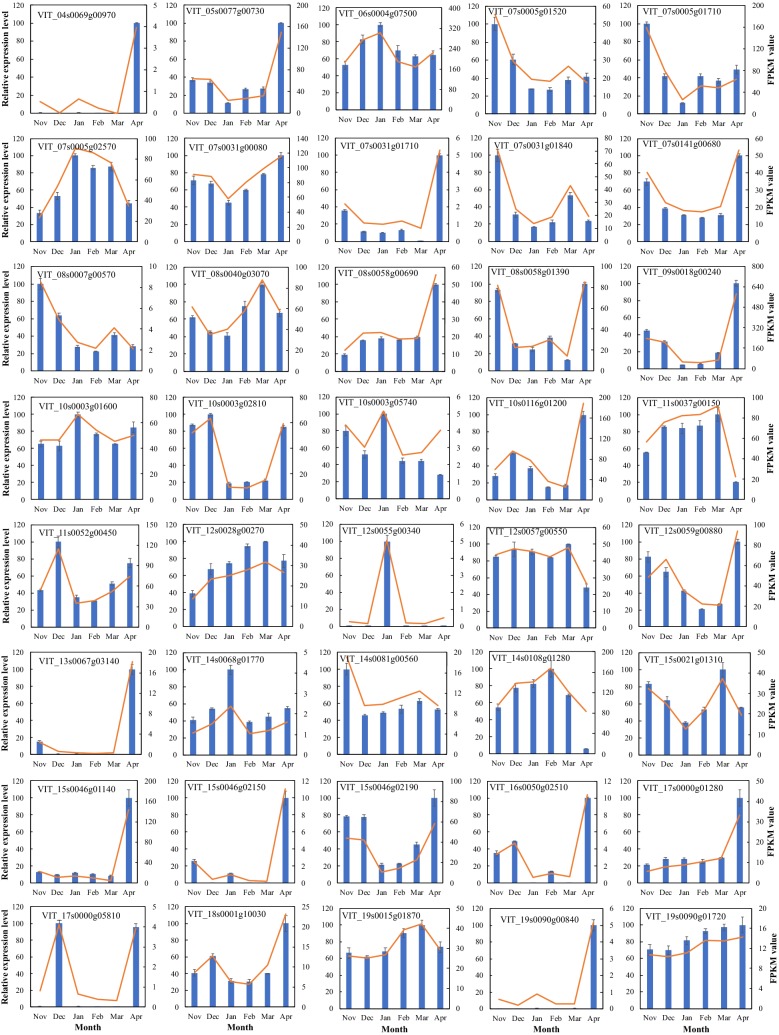
The expression profiles of grapevine DAM, WRKY, and Dof gene family members were obtained from our unpublished RNA-seq data (Fig. 5 and Table S8) and RT-qPCR results (Fig. 6). Here, we also marked the differentially expressed genes (DEGs) in red according their expression levels. Two VvDAM genes (VIT_15s0107g00120 and VIT_18s0001g07460) were significantly expressed during bud dormancy, and three VvDAM genes had their highest expression levels in April or March. Twenty-four out of 25 VvDof genes (except VIT_08S0105g00170) were significantly expressed during bud dormancy. These 24 VvDof genes were classified into 2 subgroups according to the highest expression level of each gene. Ten VvDof genes (VIT_17s0000g06310, VIT_10s0003g00030, VIT_14s0108g00980, VIT_17s0000g04850, VIT_01s0026g02580, VIT_00s0218g00040, VIT_09s0002g02490, VIT_00s0253g00060, VIT_15s0046g00150, VIT_00s0652g00010, and VIT_08s0056g01230) had their highest expression levels in November, December, or January. The remaining 14 VvDof genes had their highest expression levels in March or April, with relatively lower expression levels in November to January. Forty-nine out of 55 VvWRKY genes were significantly expressed during bud dormancy, and over half of them (33 VvWRKY genes) had their highest expression levels in April or March. The remaining VvWRKY genes had their highest expression levels in November or January. In total, nearly 90% of VvDof genes and VvWRKY genes were significantly differentially expressed during bud dormancy. Furthermore, we also performed a protein–protein interaction (PPI) analysis of the proteins encoded by differentially expressed VvDAM, VvDof, and VvWRKY genes in bud dormancy using the STRING database, inferring that 20 VvWRKY, 2 VvDAM, and 0 VvDof proteins interact. VvWRKY54 (VIT_13s0067g03140), VvWRKY30 (VIT_16s0050g02510), and VvWRKY (VIT_11s0052g00450) were also detected as part of the network core in the PPI analysis (Fig. 5d).
Fig. 5.
Heatmaps and PPI analysis of VvDAMs, VvDofs, and VvWRKYs during grapevine bud dormancy. The expression value was represented by the colors, and the significantly expressed genes were marked as red font. a VvDAMs expression during grapevine bud dormancy. b VvDofs expression during grapevine bud dormancy. c VvWRKYs expression during grapevine bud dormancy. d Protein–protein interaction of VvDAMs, VvDofs, and VvWRKYs
Fig. 6.
RT-qPCR results of VvDAMs (green), VvDofs (purple), and VvWRKYs (blue) in grapevine bud dormancy
The expression patterns of DAM, WRKY, and Dof gene family members in Arabidopsis seed and potato tuber dormancy release
Three AtDof gene transcripts and one AtWRKY gene transcript were identified by Cadman et al. (2006) to be significantly expressed during Arabidopsis seed dormancy. In that study, among them, one AtDof gene (AT5G39660) had expression levels at least twofold higher in all five dormant states compared to both after-ripened states, while two AtDof genes (AT5G60200 and AT5G62940) and one AtWRKY gene (AT1G13960) had showed higher expression levels that were at least twofold higher in both after-ripened states compared to all five dormant states (Table S9). Previous research revealed that 7 StDof, 35 StWRKY, and zero StDAM genes were identified as differentially expressed throughout potato tuber dormancy release (Liu et al. 2015). Notably, most StWRKY genes were down-regulated, whereas most StDof genes were up-regulated during potato tuber dormancy release (Table S9).
The expression patterns of grapevine DAM, WRKY, and Dof genes throughout fruit development
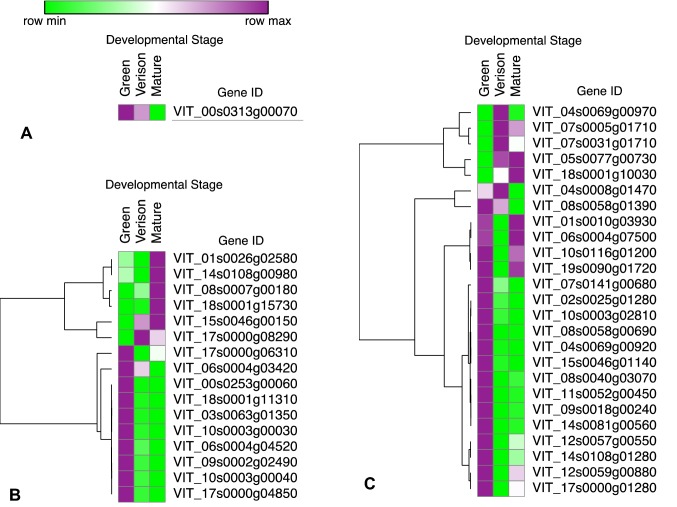
Fruit development is one of the most important metabolic processes in plants. One VvDAM, 16 VvDof, and 25 VvWRKY genes were identified as differentially expressed during fruit developmental stages (Fig. 7, Table S10). Remarkably, the number of differentially expressed genes (DEGs) across fruit development (1 DAM, 16 Dof, and 25 WRKY genes) was much lower than those across bud dormancy (2 DAM, 24 Dof, and 53 WRKY genes). Based on the expression profile data, all VvDAM DEGs, 62.5% of VvDof DEGs, and 68% of VvWRKY DEGs were highly expressed during the green stage, with expression thereafter reduced throughout the fruit maturation process.
Fig. 7.
Heatmaps of significantly differential expressed VvDAMs (a), VvDofs (b), and VvWRKYs (c) expression during grapevine fruit development. The expression value was represented by the colors
The expression patterns of grapevine DAM, WRKY, and Dof genes under abiotic stress
To determine stress-responsive VvDAM, VvDof, and VvWRKY genes, the RNA-seq data from grapevine plants under drought, salt, waterlogging, and copper stresses were also used in this study. Under salt stress, two VvDAM, four VvDof, and nine VvWRKY genes were identified as differentially expressed. Except for three VvDof DEGs, the expression most of these DEGs were inhibited under salt stress. Two VvDAM, five VvDof, and eleven VvWRKY genes were identified as DEGs under copper stress, with most of these DEGs induced by copper stress. Four VvDof and twenty-three VvWRKY genes were identified as drought stress DEGs, with most of these DEGs repressed under drought stress. Two VvDAM, five VvDof, and twenty-three VvWRKY genes were identified as DEGs under waterlogging stress, and most of them were repressed under waterlogging stress. Some of the identified DEGs responded to multiple stresses. For example, one VvDAM (VIT_00s0313g00070) responded to copper, salt, and waterlogging stresses. One VvDof gene (VIT_18s0001g11310) responded to waterlogging and drought stresses, and two VvDof genes (VIT_02s0025g02250 and VIT_06s0004g03420) responded to salt, drought, and copper stresses. One VvWRKY gene (VIT_07s0031g01710) responded to salt, copper, drought, and waterlogging stresses, and six VvWRKY genes (VIT_01s0010g03930, VIT_07s0031g00080, VIT_10s0003g01600, VIT_10s0116g01200, VIT_11s0052g00450, and VIT_14s0108g01280) responded to at least three kinds of abiotic stress (Table S11, Fig. S2–S5).
Discussion
DAM, Dof, and WRKY gene family members were broadly distributed across the investigated plants species: Arabidopsis, rice, Populus, grapevine, and peach. Increasing evidence has indicated that these genes play an important role in the regulation of bud dormancy. However, these three gene families in woody plants and herbaceous plants had not been compared previously. In this study, seven woody plants and seven herbaceous plants were used as experimental materials to analyze the distribution of gene family members, sequence structure, and evolution of the DAM, Dof, and WRKY gene families.
The identification and evolution of DAM, Dof, and WRKY gene families in selected genomes
The abundance of duplicate genes in plant genomes was inferred to have mostly been caused by ancient duplication events, with a higher retention rate of extant pairs among duplicated genes (Panchy et al. 2016). As Panchy et al. (2016) reported, apple had the highest rate of duplication and retention (84.4%) among 41 sequenced land plant genomes. We also found that apple had the second most copies of DAM, Dof, and WRKY genes among the 14 species investigated (6 DAM, 59 Dof, and 140 WRKY gene family members, Table 2). Moreover, ten other species were also investigated in this work, and they were classified into two groups based on the inferred rounds of whole genome duplication (WGD) events that had produced them according to our results and work by Panchy et al. (2016). Among the ten species, three rounds of WGD events (two duplications and one triplication) took place in Arabidopsis thaliana, Carica papaya, Citrus sinensis, Fragaria vesca, Prunus persica, and Vitis vinifera, with the number of DAM, Dof, and WRKY gene copies ranging from 2 to 6, 20 to 36, and 49 to 72, respectively. Four or five rounds of WGD events took place in Solanum lycopersicum (two duplications and two triplications), Oryza sativa (five duplications), Malus domestica (three duplications and one triplication), and Populus trichocarpa (three duplications and one triplication), with the numbers of DAM, Dof, and WRKY gene copies ranging from 2 to 9, 30 to 59, and 81 to 140, respectively (Table 2). It is also clear that 40% of DAM, 96% of Dof, and 89.5% of WRKY duplicate gene pairs were associated with segmental duplications (Table 3, Table S4, and S5). Most of the DAM, Dof, and WRKY gene copies were derived from WGD or segmental duplication events, which has also been previously reported for soybean WRKY (Yin et al. 2013; Yu et al. 2016), peanut WRKY (Song et al. 2016), Chinese cabbage WRKY (Tang et al. 2014), Chinese white pear WRKY (Huang et al. 2015), Musa WRKY(Goel et al. 2016), Populus WRKY (He et al. 2012; Ma et al. 2015b), Chinese cabbage Dof (Ma et al. 2015a), and tomato Dof (Cai et al. 2013) gene families.
The average divergence times of DAM, Dof, and WRKY gene families across the species were also notable. The average divergence time of Populus DAM genes (12.5414 MYA), Dof genes (14.77926 MYA), and WRKY genes (24.6086 MYA) was substantially later than those in rice (109.5715 MYA in DAM, 125.7269 MYA in Dof, and 185.9315 MYA in WRKY genes), Arabidopsis (29.9528 MYA in Dof and 36.9943 MYA in WRKY genes), and tomato (136.9214 MYA in Dof and 99.6510 in WRKY genes). Otherwise, unlike Populus, grapevine (a woody plant) has an earlier divergence time among its Dof (129.7213 MYA) and WRKY (173.1244 MYA) gene families relative to those of Populus (Tables S4–S5). This may explain the high number of orthologs between grapevine and other species (Tables S6–S7).
VvDAM, VvDof, and VvWRKY expression profiles during fruit development and under abiotic stresses
A considerable body of research has indicated that Dof and WRKY genes are involved in fruit development, including in tomato (Cai et al. 2013; Rohrmann et al. 2011), banana (Feng et al. 2016; Wang et al. 2012a), and grapevine (Fernandez et al. 2007; Haider et al. 2017; Terrier et al. 2005). With the help of high-throughput sequencing data, VvDAM, VvDof, and VvWRKY, the expression profiles during fruit development were obtained. One VvDAM DEG, 10 out of 16 VvDof DEGs, and 17 out of 25 VvWRKY DEGs were highly expressed at the green stage (Fig. 7). Other VvDof and VvWRKY DEGs exhibited contrasting expression profiles, with higher expression levels at veraison (a key fruit development stage in grapevine indicating the onset of ripening) or mature stages. This indicates that VvDAM, VvDof, and VvWRKY genes might play a significant role in grapevine fruit development through interactions with other proteins. Here, we also identified three VvDof (VIT_00s0652g00010, VIT_06s0004g03420, and VIT_02s0025g02250) genes and one VvWRKY gene (VIT_07s0031g01710) that were responsive to multiple stresses (Fig. S2–S5). In plants, various Dof and WRKY genes were determined to be responsive to abiotic stress. Overexpression of the homologs SlCDF1 and SlCDF3 in Arabidopsis increased salt and drought stresses tolerance, and various stress-responsive genes were activated in the overexpression lines (Corrales et al. 2014). AtWRKY6 is involved in responses to low-phosphorus stress through regulating PHOSPHATE (PHO1) expression (Chen et al. 2009). In rice, 13 OsWRKY genes differentially responded to salt, drought, cold, or heat stresses (Qiu et al. 2004). Among 14 wheat WRKY genes, eight were responsive to low temperature, high temperature, salt, or drought stresses (Wu et al. 2008). However, overexpression of VvWRKY30 in Arabidopsis increased resistance to salt stress (Zhu et al. 2019). Meanwhile the expression levels of GmWRKY13, GmWRKY21, and GmWRKY54 were induced by salt and drought stresses in soybean (Zhou et al. 2008). Moreover, numerous studies have demonstrated that a single transcriptional factor may be responsive to multiple stresses. For example, the expression levels of AtWRKY25 and AtWRKY33 both respond to salt, cold, and heat stresses (Li et al. 2011; Jiang and Deyholos 2009). Interestingly, one VvDAM (VIT_00s0313g00070) was identified as a significantly expressed gene under multiple abiotic stresses, e.g., copper, salt, and waterlogging stresses (Fig. S2, S3, and S5). Most recent reports on DAM gene family members have focused on dormancy (Li et al. 2009; Mimida et al. 2015; Niu et al. 2015). These data have isolated one VvDAM, two VvDof, and seven VvWRKY genes as major genes that respond to abiotic stress, which can function as regulators of several different processes and may also mediate the crosstalk between different signaling pathways.
VvDAM, VvDof, and VvWRKY expression during bud dormancy
Bud dormancy is an essential biological process for plant survival in cold temperatures. Many reports have revealed the involvement of DAM, Dof, and WRKY genes in metabolic activities. For example, DAM1 and the ABA metabolism and signaling pathway (NCED1 and AREB1) function through a feedback mechanism to regulate pear bud dormancy (Tuan et al. 2017). In hybrid aspen, the SVP ortholog SVL acts downstream of ABA in dormancy regulation and promotes dormancy by suppressing the growth-promoting gibberellic acid pathway (Singh et al. 2018). In addition to this, Dof and WRKY genes were found to be differentially expressed during bud dormancy in peach (Song et al. 2016), oak (Derory et al. 2006), and pear (Liu et al. 2012). The numbers of VvDAM, VvDof, and VvWRKY DEGs were 2, 24, and 49, respectively. In general, over 90% of VvDof and VvWRKY genes were differentially expressed throughout grapevine bud dormancy. Notably, there were substantially more VvDAM, VvDof, and VvWRKY genes associated with bud dormancy than with fruit development or abiotic stress. This suggests that bud dormancy was shaped by evolution of the VvDof and VvWRKY gene families, such that the Dof and WRKY DEGs associated with Arabidopsis seed and potato tuber dormancy release processes were much fewer in number. Intriguingly, multiple abiotic stress-responsive VvDof and VvWRKY genes were also differentially expressed across bud dormancy. For example, two VvWRKY genes (VIT_11s0052g00450 and VIT_14s0108g01280) responded to bud dormancy and multiple abiotic stresses, which should accordingly be treated as candidate genes for further study. However, the single multiple abiotic stress-responsive VvDAM gene (VIT_00s0313g00070) was not differentially expressed across bud dormancy. One VvDAM gene (VIT_18s0001g07460) is a possible candidate gene for grapevine bud dormancy regulation. This suggests that a differential regulation mechanism functions between bud dormancy and other biological processes.
The analysis of bud dormancy expression profiles of tandem duplicated genes revealed high expression divergence between two VvWRKY pairs (VIT_07s0005g01710-VIT_05s0077g00730 and VIT_08s0058g00690-VIT_06s0004g07500). The highest expression value of VIT_07s0005g01710 was in November, while that of VIT_05s0077g00730 was in March. The highest expression value of VIT_08s0058g00690 was in March, while that of VIT_06s0004g07500 was in January. This also indicated that the gene family has evolved different functions across environmental stresses and throughout development, with evolutionary divergence occurring among gene families. Theses phenomena have been observed in the glutathione S-transferase (GST) family in sorghum, rice, and Arabidopsis (Chi et al. 2010). This study systematically analyzed DAM, Dof, and WRKY expression in grapevine under different environmental stresses and across different developmental stages, thus providing candidate genes for further functional analysis.
Conclusion
This study has investigated the distribution, sequence characteristics, evolution, and expression profile of grapevine DAM, Dof, and WRKY gene family members, with comparisons to those from other species. These three gene families in each species differed in number, but WGD and segmental duplication events had a predominant effect on the expansion of Dof and WRKY gene families. More DAM, Dof, and WRKY genes were associated with bud dormancy than with fruit development or abiotic stresses. One, two, and seven candidate stress-response genes were identified among the DAM, Dof, and WRKY genes investigated. This study provides the foundation for further analyses of grapevine abiotic stress responses.
Methods
Plant materials
Three-year-old Vitis vinifera cv. ‘Rosario Bianco’ (‘Rosaki’ × ‘Muscat of Alexandria’) grapevines were grown in a greenhouse at Jiangsu Agricultural Expo Garden, Jiangsu, China (N32°0′41.99″, E119°15′7.11″) with permission from Jiangsu Vocational College of Agriculture and Forestry. The dormancy period began in November and ended by April, when bud break occurred. Grapevine buds were divided into three groups depending on their position along the stem: the bottom group (the third, fourth, and fifth buds from the cordon), the center group (the eighth, ninth, and tenth buds from the cordon), and the top group (the fourteenth, fifteenth, and sixteenth buds from the cordon). At least 30 buds from each group were sampled every month between November and April (of the following year). Samples were frozen in liquid nitrogen, and then stored at − 80 °C for further analysis.
Sequence retrieval
Protein, mRNA, and genome annotation files were downloaded from online databases. Citrus sinensis datasets were retrieved from the Citrus sinensis annotation project database (V2, https://citrus.hzau.edu.cn/orange/). Actinidia chinensis data were retrieved from the Kiwifruit Genome database (V1, https://bioinfo.bti.cornell.edu/kiwi). Pyrus bretschneideri data were retrieved from the Pear Genome Project archives (V1, https://peargenome.njau.edu.cn). Multiple datasets for Arabidopsis thaliana (TAIR10), Musa acuminate (MA1), Prunus persica (Prupe1.0), Vitis vinifera, and Solanum lycopersicum (SL2.50), (IGGP_12 ×) were downloaded from the Ensembl Plants database (https://plants.ensembl.org/). Multiple datasets for Ananas comosus (V3), Carica papaya (ASGPBv0.4), Fragaria vesca (V1.1), Malus domestica (V1.0), Oryza sativa (v7.0), and Populus trichocarpa (V3.1) were downloaded from JGI archives (https://genome.jgi.doe.gov/portal/).
Identification of DAM, Dof, and WRKY gene family members
To identify DAM, Dof, and WRKY gene family members from the 14 genomes, 2 search tools were employed: Blast and HMMER. (1) Blast searches were performed using the nucleotide sequences of target genes (9 PpDAM, 2 AtDAM, 2 SlDAM, 3 OsDAM genes; 36 AtDof, 30 OsDof genes; 72 AtWRKY, and 101 OsWRKY genes; detailed in Tables S1–S3) against the peptide databases of 14 species. (2) Protein sequences of target DAM genes (9 PpDAM, 2 AtDAM, 2 SlDAM, and 3 OsDAM genes, detailed in Table S1) were used to build the HMMER file. The Dof pfam file (PF02701) and WRKY pfam file (PF03106) were downloaded from the pfam database (https://pfam.xfam.org). DAM HMMER, Dof pfam, and WRKY pfam files were used to search the peptide databases of 14 species, respectively. (3) The results from the above two methods were combined, and redundant sequences were removed, leaving all the putative DAM, Dof, and WRKY gene family members from the 14 species.
Bioinformatic analysis
The protein sequence lengths and CDS numbers of DAM, Dof, and WRKY gene family members in each species were calculated using an in-house Perl script and visualized using the ggplot2 package in the R statistical computing environment. Protein sequence domain analysis, protein sequence alignment, phylogenetic analysis, gene distribution analysis, and gene structure visualization were conducted using the Simple Modular Architecture Research Tool (https://smart.embl-heidelberg.de/), the MEGA software, the MapGene2Chro online tool (https://mg2c.iask.in/mg2c_v2.0/), and the GSDS online tool (https://gsds.cbi.pku.edu.cn/index.php), respectively. Identical types and pairs were analyzed using MCScanX and visualized using Circos (Krzywinski et al. 2009; Wang et al. 2012b). ParaAT and KaKs_Calculator were used to estimate the number of non-synonymous substitutions per non-synonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) (Zhang et al. 2006, 2012). The dates of duplication events were estimated using the equation T = Ks/2λ, where λ = 6.5 × 10–9 for grapes and rice (Gaut et al. 1996; Yu et al. 2005), 1.5 × 10–8 for Arabidopsis(Blanc and Wolfe 2004), 9.1 × 10–9 for Populus (Lynch and Conery 2000) and 6.96 × 10–9 for tomato (Cheng et al. 2009). Gene expression was analyzed using NCBI online datasets (GSE77218, SRP070475, SRP074162, and SRP159132) and published supplemental datasets (Leng et al. 2015). A heatmap of the data was drawn using Morpheus (https://software.broadinstitute.org/morpheus/). Protein–protein interaction (PPI) analysis was performed using the STRING database (https://string-db.org/).
RNA isolation and RT-qPCR
Extraction of total RNA was conducted using the SDS-phenol method (Zhang et al. 2010). The Revert Aid™ First-Strand cDNA Synthesis Kit was used for first-strand cDNA synthesis (Fermentas, Glen Burnie, MD, USA). The cDNAs were diluted in double-distilled water by 30-fold. Then, cDNA templates were pooled with EvaGreen 2 × qPCR MasterMix-ROX (ABM, Richmond, BC, Canada) to perform RT-qPCR (real time-quantitative PCR) using an Applied Biosystems® 7500 Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA). The housekeeping gene Actin (AB073011, forward primer GGAAGCTGCGGGAATTCATGAG, reverse primer CCTTGATCTTCATGCTGCTGGG) was used as an internal control to quantify mRNA levels. The primer sequences used in the present study are given in Table 1. The 20-μl PCR volumes contained 500 ng of cDNA, 10 μl of EvaGreen 2 × qPCR MasterMix, 0.6 μl of forward primer, 0.6 μl of reverse primer, and 6.8 μl of nuclease-free H2O. PCR was performed using the following cycling conditions: 10 min (95 °C), followed by 35 cycles of 15 s at 95 °C and 1 min at 62 °C, with a final cooling to 4 °C.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- DAM
Dormancy associated MADS-box
- Dof
DNA-binding one zinc finger
- CDS
Coding sequence
- MYA
Million years ago
- WGD
Whole genome duplication
Authors’ contributions
Conceived and designed the experiments: LFSG and JGF. Analyzed the data: LFSG, XF, KKZ, and MXC. RT-qPCR: MXC, ZQX, and TZ. Wrote the paper: LFSG, XF, and JGF. Revised the paper: LFSG, JGF, YFP, and MXC.
Funding
This work was supported by the National Natural Science Foundation of China (31772283), the Fundamental Research Funds for the Central Universities (KYZ201837).
Availability of data and material
Gene expression was analyzed using NCBI online datasets (GSE77218, SRP070475, SRP074162, and SRP159132) or Leng et al.’s supplemental datasets.
Compliance with ethical standards
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Lingfei Shangguan and Mengxia Chen contributed equally to this work.
References
- Bai S, Saito T, Sakamoto D, Ito A, Fujii H, Moriguchi T. Transcriptome analysis of Japanese pear Pyrus pyrifolia Nakai flower buds transitioning through endodormancy. Plant Cell Physiol. 2013;54(7):1132–1151. doi: 10.1093/pcp/pct067. [DOI] [PubMed] [Google Scholar]
- Beauvieux R, Wenden B, Dirlewanger E. Bud dormancy in perennial fruit tree species: a pivotal role for oxidative cues. Front Plant Sci. 2018;9:657. doi: 10.3389/fpls.2018.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielenberg DG, Wang YE, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genom. 2008;4(3):495–507. [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell Online. 2004;16(7):1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46(5):805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol. 2013;55(6):552–566. doi: 10.1111/jipb.12043. [DOI] [PubMed] [Google Scholar]
- Chen Y-F, Li L-Q, Xu Q, Kong Y-H, Wang H, Wu W-H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell. 2009;21(11):3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tan Q, Sun M, Li D, Fu X, Chen X, Xiao W, Li L, Gao D. Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Mol Genet Genom. 2016;291(3):1319–1332. doi: 10.1007/s00438-016-1171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Liu X, Huan L, Sun M, Liu L, Chen X, Gao D, Li L. Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach Prunus persica. Sci Hortic. 2017;214:18–26. [Google Scholar]
- Cheng X, Zhang D, Cheng Z, Keller B, Ling H-Q. A new family of Ty1-copia-like retrotransposon originated in the tomato genomes by a recent horizontal transfer event. Genetics. 2009;181(4):1183–1193. doi: 10.1534/genetics.108.099150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Cheng Y, Vanitha J, Kumar N, Ramamoorthy R, Ramachandran S, Jiang S-Y. Expansion mechanisms and functional divergence of the glutathione S-transferase family in sorghum and other higher plants. DNA Res. 2010;18(1):1–16. doi: 10.1093/dnares/dsq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales A-R, Nebauer SG, Carrillo L, Fernández-Nohales P, Marqués J, Renau-Morata B, Granell A, Pollmann S, Vicente-Carbajosa J, Molina R-V. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J Exp Bot. 2014;65(4):995–1012. doi: 10.1093/jxb/ert451. [DOI] [PubMed] [Google Scholar]
- Derory J, Léger P, Garcia V, Schaeffer J, Hauser MT, Salin F, Luschnig C, Plomion C, Glössl J, Kremer A. Transcriptome analysis of bud burst in sessile oak Quercus petraea. New Phytol. 2006;170(4):723–738. doi: 10.1111/j.1469-8137.2006.01721.x. [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ. WRKY 41 controls Arabidopsis seed dormancy via direct regulation of ABI 3 transcript levels not downstream of ABA. Plant J. 2014;79(5):810–823. doi: 10.1111/tpj.12597. [DOI] [PubMed] [Google Scholar]
- Feng B-h, Han Y-c, Xiao Y-y, Kuang J-f, Fan Z-q, Chen J-y, Lu W-j. The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J Exp Bot. 2016;67(8):2263–2275. doi: 10.1093/jxb/erw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell AY, Schlauch KA, Gouthu S, Deluc LG, Khadka V, Sreekantan L, Grimplet J, Cramer GR, Mathiason KL. Short day transcriptomic programming during induction of dormancy in grapevine. Front Plant Sci. 2015;6:834. doi: 10.3389/fpls.2015.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Torregrosa L, Terrier N, Sreekantan L, Grimplet J, Davies C, Thomas MR, Romieu C, Ageorges A. Identification of genes associated with flesh morphogenesis during grapevine fruit development. Plant Mol Biol. 2007;63(3):307–323. doi: 10.1007/s11103-006-9090-2. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci. 1996;93(19):10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R, Pandey A, Trivedi PK, Asif MH. Genome-wide analysis of the Musa WRKY gene family: evolution and differential expression during development and stress. Front Plant Sci. 2016;7:299. doi: 10.3389/fpls.2016.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider MS, Kurjogi MM, Khalil-Ur-Rehman M, Fiaz M, Pervaiz T, Jiu S, Haifeng J, Chen W, Fang J. Grapevine immune signaling network in response to drought stress as revealed by transcriptomic analysis. Plant Physiol Biochem. 2017;121:187–195. doi: 10.1016/j.plaphy.2017.10.026. [DOI] [PubMed] [Google Scholar]
- He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31(7):1199–1217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Sung S, Kim D, Chao W, Anderson J. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol Biol. 2010;73(1–2):169–179. doi: 10.1007/s11103-009-9596-5. [DOI] [PubMed] [Google Scholar]
- Huang X, Li K, Xu X, Yao Z, Jin C, Zhang S. Genome-wide analysis of WRKY transcription factors in white pear Pyrus bretschneideri reveals evolution and patterns under drought stress. BMC Genomics. 2015;16(1):1104. doi: 10.1186/s12864-015-2233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Saito T, Sakamoto D, Sugiura T, Bai S, Moriguchi T. Physiological differences between bud breaking and flowering after dormancy completion revealed by DAM and FT/TFL1 expression in Japanese pear Pyrus pyrifolia. Tree Physiol. 2015;36(1):109–120. doi: 10.1093/treephys/tpv115. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol. 2009;69(1–2):91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- Khalil-Ur-Rehman M, Sun L, Li C-X, Faheem M, Wang W, Tao J-M. Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape. BMC Plant Biol. 2017;17(1):18. doi: 10.1186/s12870-016-0960-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. Endo-, para-and ecodormancy: physiological terminology and classification for dormancy research. Hortic Sci. 1987;22:271–277. [Google Scholar]
- Leida C, Terol J, Martí G, Agustí M, Llácer G, Badenes ML, Ríos G. Identification of genes associated with bud dormancy release in Prunus persica by suppression subtractive hybridization. Tree Physiol. 2010;30(5):655–666. doi: 10.1093/treephys/tpq008. [DOI] [PubMed] [Google Scholar]
- Leng X, Jia H, Sun X, Shangguan L, Mu Q, Wang B, Fang J. Comparative transcriptome analysis of grapevine in response to copper stress. Sci Rep. 2015;5:17749. doi: 10.1038/srep17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesur I, Bechade A, Lalanne C, Klopp C, Noirot C, Leplé JC, Kremer A, Plomion C, Le Provost G. A unigene set for European beech Fagus sylvatica L and its use to decipher the molecular mechanisms involved in dormancy regulation. Mol Ecol Resour. 2015;15(5):1192–1204. doi: 10.1111/1755-0998.12373. [DOI] [PubMed] [Google Scholar]
- Li Z, Reighard GL, Abbott AG, Bielenberg DG. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot. 2009;60(12):3521–3530. doi: 10.1093/jxb/erp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233(6):1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- Liu G, Li W, Zheng P, Xu T, Chen L, Liu D, Hussain S, Teng Y. Transcriptomic analysis of ‘Suli’pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC genomics. 2012;13(1):700. doi: 10.1186/1471-2164-13-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhang N, Wen Y, Jin X, Yang J, Si H, Wang D. Transcriptomic changes during tuber dormancy release process revealed by RNA sequencing in potato. J Biotechnol. 2015;198:17–30. doi: 10.1016/j.jbiotec.2015.01.019. [DOI] [PubMed] [Google Scholar]
- Liu X, Liu Z, Hao Z, Chen G, Qi K, Zhang H, Jiao H, Wu X, Zhang S, Wu J (2019) Characterization of Dof family in Pyrus bretschneideri and role of PbDof9. 2 in flowering time regulation. Genomics. 10.1016/j.ygeno.2019.05.005 [DOI] [PubMed]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Ma J, Li M-Y, Wang F, Tang J, Xiong A-S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics. 2015;16(1):33. doi: 10.1186/s12864-015-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lu J, Xu J, Duan B, He X, Liu J (2015b) Genome-wide identification of WRKY genes in the desert poplar Populus euphratica and adaptive evolution of the genes in response to salt stress. Evolutionary Bioinformatics 11:EBO. S22067 [DOI] [PMC free article] [PubMed]
- Mimida N, Saito T, Moriguchi T, Suzuki A, Komori S, Wada M. Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol Plant. 2015;59(2):237–244. [Google Scholar]
- Min Z, Zhao X, Li R, Yang B, Liu M, Fang Y. Comparative transcriptome analysis provides insight into differentially expressed genes related to bud dormancy in grapevine Vitis vinifera. Sci Hortic. 2017;225:213–220. [Google Scholar]
- Niu Q, Li J, Cai D, Qian M, Jia H, Bai S, Hussain S, Liu G, Teng Y, Zheng X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear Pyrus pyrifolia white pear group flower bud. J Exp Bot. 2015;67(1):239–257. doi: 10.1093/jxb/erv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002;31(3):319–330. doi: 10.1046/j.1365-313x.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Jing S, Fu J, Li L, Yu D. Cloning and analysis of expression profile of 13 WRKY genes in rice. Chin Sci Bull. 2004;49(20):2159–2168. [Google Scholar]
- Regier N., Streb S., Zeeman S. C., Frey B. Seasonal changes in starch and sugar content of poplar (Populus deltoides x nigra cv. Dorskamp) and the impact of stem girdling on carbohydrate allocation to roots. Tree Physiology. 2010;30(8):979–987. doi: 10.1093/treephys/tpq047. [DOI] [PubMed] [Google Scholar]
- Rios G, Conejero A, Leida C, Petri C, Burgos L, Badenes M Functional characterization of a SAP protein expressed in dormant buds of peach. In: RGC7: 7th International Rosaceae Genomics Conference, 2014. p 20
- Ríos G, Leida C, Conejero A, Badenes ML. Epigenetic regulation of bud dormancy events in perennial plants. Front Plant Sci. 2014;5:247. doi: 10.3389/fpls.2014.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J Exp Bot. 2007;58(15–16):4047–4060. doi: 10.1093/jxb/erm261. [DOI] [PubMed] [Google Scholar]
- Rohrmann J, Tohge T, Alba R, Osorio S, Caldana C, McQuinn R, Arvidsson S, van der Merwe MJ, Riaño-Pachón DM, Mueller-Roeber B. Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. Plant J. 2011;68(6):999–1013. doi: 10.1111/j.1365-313X.2011.04750.x. [DOI] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell. 2007;19(8):2370–2390. doi: 10.1105/tpc.107.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Bai S, Imai T, Ito A, Nakajima I, Moriguchi T. Histone modification and signalling cascade of the dormancy-associated MADS-box gene, PpMADS 13–1, in Japanese pear Pyrus pyrifolia during endodormancy. Plant Cell Environ. 2015;38(6):1157–1166. doi: 10.1111/pce.12469. [DOI] [PubMed] [Google Scholar]
- Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot Prunus mume. Plant Physiol. 2011;157(1):485–497. doi: 10.1104/pp.111.181982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Miskolczi P, Maurya JP, Bhalerao RP. A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr Biol. 2018;29(1):128–133. doi: 10.1016/j.cub.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Song H, Wang P, Lin J-Y, Zhao C, Bi Y, Wang X. Genome-wide identification and characterization of WRKY gene family in peanut. Front Plant Sci. 2016;7:534. doi: 10.3389/fpls.2016.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekantan L, Mathiason K, Grimplet J, Schlauch K, Dickerson JA, Fennell AY. Differential floral development and gene expression in grapevines during long and short photoperiods suggests a role for floral genes in dormancy transitioning. Plant Mol Biol. 2010;73(1–2):191–205. doi: 10.1007/s11103-010-9611-x. [DOI] [PubMed] [Google Scholar]
- Tang Jun, Wang Feng, Hou Xi-Lin, Wang Zhen, Huang Zhi-Nan. Genome-Wide Fractionation and Identification of WRKY Transcription Factors in Chinese Cabbage (Brassica rapa ssp. pekinensis) Reveals Collinearity and Their Expression Patterns Under Abiotic and Biotic Stresses. Plant Molecular Biology Reporter. 2013;32(4):781–795. [Google Scholar]
- Terrier N, Glissant D, Grimplet J, Barrieu F, Abbal P, Couture C, Ageorges A, Atanassova R, Léon C, Renaudin J-P. Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry Vitis vinifera L development. Planta. 2005;222(5):832–847. doi: 10.1007/s00425-005-0017-y. [DOI] [PubMed] [Google Scholar]
- Tuan PA, Bai S, Saito T, Ito A, Moriguchi T. Dormancy-associated mads-box (dam) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 2017;58(8):1378–1390. doi: 10.1093/pcp/pcx074. [DOI] [PubMed] [Google Scholar]
- Ubi BE, Sakamoto D, Ban Y, Shimada T, Ito A, Nakajima I, Takemura Y, Tamura F, Saito T, Moriguchi T. Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. J Am Soc Hortic Sci. 2010;135(2):174–182. [Google Scholar]
- Ueno S, Klopp C, Leplé JC, Derory J, Noirot C, Léger V, Prince E, Kremer A, Plomion C, Le Provost G. Transcriptional profiling of bud dormancy induction and release in oak by next-generation sequencing. BMC Genomics. 2013;14(1):236. doi: 10.1186/1471-2164-14-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-n, Kuang J-f, Shan W, Chen J, Xie H, Lu W-j, Chen J-w, Chen J-y. Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep. 2012;31(8):1485–1494. doi: 10.1007/s00299-012-1263-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49–e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L. H., Zhong W. J., Huo X. M., Zhuang W. B., Ni Z. J., Gao Z. H. Expression analysis of ABA- and GA-related genes during four stages of bud dormancy in Japanese apricot (Prunus mumeSieb. et Zucc) The Journal of Horticultural Science and Biotechnology. 2016;91(4):362–369. [Google Scholar]
- Wisniewski M, Norelli J, Artlip T. Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front Plant Sci. 2015;6:85. doi: 10.3389/fpls.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ni Z, Yao Y, Guo G, Sun Q. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat Triticum aestivem L. Prog Nat Sci. 2008;18(6):697–705. [Google Scholar]
- Xu H, Cao D, Chen Y, Wei D, Wang Y, Stevenson RA, Zhu Y, Lin J. Gene expression and proteomic analysis of shoot apical meristem transition from dormancy to activation in Cunninghamia lanceolata Lamb Hook. Sci Rep. 2016;6:19938. doi: 10.1038/srep19938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J Am Soc Hortic Sci. 2008;133(5):708–716. [Google Scholar]
- Yamane H, Wada M, Honda C, Matsuura T, Ikeda Y, Hirayama T, Osako Y, Gao-Takai M, Kojima M, Sakakibara H. Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PLoS ONE. 2019;14(4):e0214788. doi: 10.1371/journal.pone.0214788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Niu Q, Li J, Zheng X, Ma Y, Bai S, Teng Y. PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in ‘Suli’pear Pyrus pyrifolia White Pear Group. Plant Physiol Biochem. 2018;127:355–365. doi: 10.1016/j.plaphy.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Yin G, Xu H, Xiao S, Qin Y, Li Y, Yan Y, Hu Y. The large soybean Glycine max WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013;13(1):148. doi: 10.1186/1471-2229-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3(2):e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang N, Hu R, Xiang F. Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus. 2016;5(1):920. doi: 10.1186/s40064-016-2647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinform. 2006;4(4):259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang C, Yu H, Cai B, Fang J. Screening of RNA extraction methods for various grapevine organs and tissues. Acta Agriculturae Boreali-Occidentalis Sinica. 2010;19(11):135–140. [Google Scholar]
- Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, Dai L. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 2012;419(4):779–781. doi: 10.1016/j.bbrc.2012.02.101. [DOI] [PubMed] [Google Scholar]
- Zhong W, Gao Z, Zhuang W, Shi T, Zhang Z, Ni Z. Genome-wide expression profiles of seasonal bud dormancy at four critical stages in Japanese apricot. Plant Mol Biol. 2013;83(3):247–264. doi: 10.1007/s11103-013-0086-4. [DOI] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6(5):486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li Y, Xin D, Chen W, Shao X, Wang Y, Guo W. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry Prunus pseudocerasus. Gene. 2015;555(2):362–376. doi: 10.1016/j.gene.2014.11.032. [DOI] [PubMed] [Google Scholar]
- Zhu D, Hou L, Xiao P, Guo Y, Deyholos MK, Liu X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019;280:132–142. doi: 10.1016/j.plantsci.2018.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression was analyzed using NCBI online datasets (GSE77218, SRP070475, SRP074162, and SRP159132) or Leng et al.’s supplemental datasets.