Abstract
In 2011, a hexanucleotide repeat expansion in the first intron of the C9orf72 gene was identified as the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The proposed disease mechanisms include loss of C9orf72 function and gain of toxicity from the bidirectionally transcribed repeat-containing RNAs. Over the last few years, substantial progress has been made to determine the contribution of loss and gain of function in disease pathogenesis. The extensive body of molecular, cellular, animal, and human neuropathological studies is conflicted, but the predominance of evidence favors gain of toxicity as the main pathogenic mechanism for C9orf72 repeat expansions. Alterations in several downstream cellular functions, such as nucleocytoplasmic transport and autophagy, are implicated. Exciting progress has also been made in therapy development targeting this mutation, such as by antisense oligonucleotide therapies targeting sense transcripts and small molecules targeting nucleocytoplasmic transport, and these are now in phase 1 clinical trials.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00797-2) contains supplementary material, which is available to authorized users.
Key Words: C9orf72, amyotrophic lateral sclerosis, frontotemporal dementia, antisense oligonucleotide, RNA foci, dipeptide repeat proteins, nucleocytoplasmic transport
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are two devastating, adult-onset neurodegenerative disorders with distinct clinical presentations. ALS, also known as Lou Gehrig’s disease, is the most common motor neuron disease and is invariably fatal. Its rapid progression is caused by the loss of upper and lower motor neurons and leads to muscle weakness and paralysis [1]. FTD is the second common dementia in people under 65 years of age. Only Alzheimer’s disease is more prevalent. In FTD, the degeneration of neurons in the frontal and temporal lobes leads to behavioral and personality changes, deficits in executive functions, and language impairments [2]. It is now increasingly recognized that ALS and FTD belong to a continuous disease spectrum with clinical, pathological, and genetic overlaps. In clinic, about 15% of FTD patients show symptoms of ALS, and up to 50% of ALS patients have detectable cognitive impairment [3]. Pathologically, more than 97% of ALS and about 50% of FTD cases accumulate inclusions of the RNA-binding protein TAR DNA–binding protein 43 (TDP-43) in neuron and glia. Genetically, mutations in several genes cause either ALS or FTD [4]. In 2011, a hexanucleotide repeat expansion in the C9orf72 (abbreviated from its genomic loci in chromosome 9 open reading frame 72) was identified as the most common genetic cause of both ALS and FTD [5–7], and accounts for 40% of familial ALS, 5–10% of sporadic ALS, 12–25% of familial FTD, and 6–7% of sporadic FTD cases [8].
Identification of the C9orf72 locus added ALS/FTD (referred to as c9ALS/FTD thereafter) to the increasing number of repeat expansion disorders (e.g., myotonic dystrophy, Huntington’s disease, and several spinocerebellar ataxias) [9]. Based on lessons learned from these other repeat expansion disorders and initial pathological characterization of C9orf72 postmortem tissues, three disease mechanisms have been proposed: 1) loss of C9orf72 function; 2) toxicity from the bidirectionally transcribed repeat-containing RNAs, such as mediated through sequestration of key RNA-binding proteins (RBPs) into RNA foci; and 3) toxicity from production of dipeptide repeat (DPR) proteins through a noncanonical repeat-associated non-AUG-dependent (RAN) translation. Although the C9orf72 mutation was only identified 8 years ago, research progress has been rapid, and interestingly, insights from studying C9orf72 are significantly influencing the way we think about neurodegeneration and therapy. In this review, we will discuss the current understanding of the pathogenic mechanisms caused by the C9orf72 repeat expansions and therapy development.
C9orf72 Gene Structure, Repeat Sizes, and Somatic Mosaicism
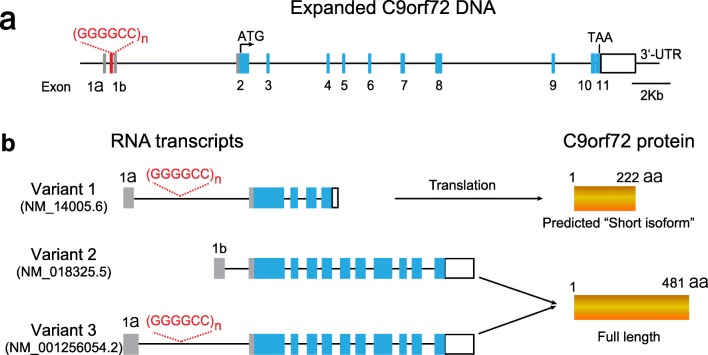
The hexanucleotide repeat expansion is localized in the first intron of the C9orf72 gene (Fig. 1a). The actual sizes of the repeats vary dramatically among C9orf72 patients. The threshold pathogenic repeat size is unknown, but 30 is used in most publications. Healthy individuals have typically fewer than 11 repeats, whereas most patients have several hundreds to thousands [6, 10–14]. A small percentage of patients also have shorter expansions, 45–80 repeats [15]. Unlike Huntington’s disease and other CAG repeat expansion disorders where a phenomenon called anticipation causes increased size and worse phenotype in offspring, increased C9orf72 repeat size in offspring is only reported in a few families [16, 17]. The correlation of the repeat size and clinical and pathological characteristics has also been explored, and the results are variable, depending on patient size, ethnic background, and tissues in which expansion size was measured [10, 13, 15, 18–22]. Longer repeat sizes seem to be associated with a survival disadvantage [19] and an earlier onset of disease [10, 13, 15]. Interestingly, individuals with an intermediate repeat length in blood or fibroblast DNAs have a significant increase of repeat size (repeat instability) and variation in repeat size (mosaicism) in different regions of the central nervous system (CNS) [15, 18, 19]. The mechanisms of such instability and somatic mosaicism are not well understood but need to be considered when using repeat size from peripheral blood and fibroblast cells for genetic counseling.
Fig. 1.
C9orf72 gene structure, RNA transcripts, and proteins. (a) Schematic representation of the human C9orf72 gene with expanded GGGGCC hexanucleotide repeats. (b) Three RNA transcripts can be produced from the C9orf72 gene. Variant 1 is predicted to result in a “short isoform” C9ORF72 protein of 222 amino acids, whereas variants 2 and 3 encode a long C9ORF72 protein of 481 amino acids
Pathogenic Mechanisms of c9ALS/FTD: Loss of Function Versus Gain of Toxicity
Our current understanding of the contributions of loss of function and gain of toxicity to c9ALS/FTD pathogenesis is driven by complementary studies of human postmortem tissues and experimental model systems in vitro and in vivo. When interpreting these data, one should keep in mind the advantages and shortcomings of each system. Patient postmortem studies are more disease-relevant but are anatomically complex and generally capture the later stages of the diseases. Published experimental model systems, on the other hand, often use repeat sizes significantly shorter than those found in patients, mainly due to the technical difficulties in obtaining pure and long GGGGCC repeats. They also frequently rely on high overexpression of a single potential toxic species that may not reflect the endogenous expression in patients. Despite these challenges, a picture of pathogenic mechanisms leading to c9ALS/FTD is emerging.
Loss of C9orf72 Function Alone Is Insufficient to Cause ALS/FTD
Reduced C9orf72 Expression in c9ALS/FTD Patients
In humans, C9orf72 is transcribed into three major RNA isoforms (Fig. 1b). In variants 1 (NM_145005.6) and 3 (NM_001256054.2), the expanded GGGGCC repeat is located in an intron between two alternatively spliced exons, and in variant 2 (NM_018325.5), the repeat is located in the promoter region. Variant 2 expresses at higher level than variants 1 and 3 in CNS tissues [23]. The presence of an enormous GGGGCC repeat expansion in the intron region may cause early abortion during transcription, leading to the increased transcripts containing upstream of the repeats and the decreased transcripts containing downstream of the repeats in C9orf72 patients [24]. In addition, cytosine hypermethylation of CG dinucleotides in CpG islands (regions with significantly higher frequency of the CG sequence) is an important epigenetic modification that could lead to gene silencing. There is a CpG island at the 5′ of GGGGCC repeats, and the repeat expansion itself also provides many more CpG islands. In C9orf72 patients, the CpG island 5′ of the GGGGCC repeats is hypermethylated [25], as is the repeat itself [26]. Finally, trimethylation of histones H3 and H4 at the C9orf72 locus was detected in C9orf72 patient blood, suggesting another mechanism for transcription regulation [27]. All of these could contribute to downregulation of C9orf72 gene expression and loss of C9orf72 protein function (Fig. 2a). Many independent groups have shown reduced one or more of the C9orf72 RNA transcripts in induced pluripotent stem cell (iPSC)-derived neurons [28–30] and patient tissues, including lymphoblasts, frontal cortex, and cerebellum [6, 7, 31]. Demonstrating reduced C9orf72 protein levels has been more challenging, due to the low abundance of C9orf72 protein and a lack of good antibodies for its detection. However, several studies suggest that the expression levels of C9orf72 proteins are reduced in the frontal and temporal cortices in patients [32–34].
Fig. 2.
Proposed pathogenic mechanisms in C9orf72 ALS/FTD. (a) The presence of expanded GGGGCC repeats potentially causes abortive transcription from exon 1a and hypermethylation of both DNA and histones to reduce C9orf72 RNA transcription, leading to a loss of C9orf72 protein function. (b) Bidirectionally transcribed repeat-containing RNAs cause neuronal toxicity by sequestration of RNA-binding proteins into RNA foci or production of at least 5 aberrant dipeptide repeat proteins [poly(GA), poly(GP), poly(GR), poly(PR), and poly(PA)] through a novel repeat-associated non-AUG-dependent translation mechanism
Physiological Function of C9orf72 Protein
Transcript variants 2 and 3 encode the full-length C9orf72 protein with 481 amino acids, and variant 1 produces a predicted “short isoform” protein of 222 amino acids with a unique lysine residue at the C-terminus(Fig. 1b). Whether the predicted short isoform C9orf72 is expressed in nature is controversial. Xiao et al. [35] developed an antibody that selectively detects the predicted short isoform C9orf72 protein and showed that its expression levels on nuclear membranes were lower in c9ALS cases than in controls. However, using an antibody against the N-terminal region of C9orf72 that could recognize both the full-length and the predicted short isoform C9orf72, we demonstrated that C9orf72 is predominantly, if not exclusively, expressed as the full-length protein in human and mouse tissues [34]. This is further confirmed by another study using novel knockout-validated C9orf72 monoclonal rat and mouse antibodies [33].
Nevertheless, the physiological function of the full-length C9orf72 protein is emerging. Bioinformatic analysis shows that C9orf72 protein has high homology with the differentially expressed in normal and neoplastic cell (DENN) protein family, which functions as guanine nucleotide exchange factor (GEF) proteins to activate RAB GTPase and regulate membrane trafficking [36, 37]. In agreement with this, C9orf72 interacts with Smith–Magenis syndrome chromosomal region candidate gene 8 (SMCR8) and WD repeat-containing protein 41 (WDR41), and this complex possesses GEF activity for different Rab proteins and plays a role in multiple steps of autophagy/lysosomal pathways [38–45]. Several Rab proteins are substrates of C9orf72, including Rab1, Rab3, Rab5, Rab7, Rab8a, Rab11, and Rab39 [40, 41, 43–45]. Knockdown C9orf72 in human cell lines and primary neurons inhibits autophagy induction, leading to accumulation of P62 [40, 45]. Accumulation of autophagy substrates, including P62, is also observed in spleens of C9orf72 knockout mice and in iPSC–derived neurons from C9orf72 patients [45, 46]. iPS neurons from C9orf72 patients also showed increased sensitivity to autophagy inhibitions, suggesting that reductions in C9orf72 levels contribute to cellular stress [29]. In agreement with this, C9orf72 protein is necessary for the formation of stress granules [47].
Loss of C9orf72 Function in ALS/FTD Pathogenesis
Is reduced C9orf72 function as the main disease driver for ALS/FTD caused by the C9orf72 repeat expansions? In human, only one sporadic ALS patient has ever been found to carry a splice site mutation potentially causing a heterozygous loss of function, and it is unclear whether this mutation is the cause of disease or variant of undetermined significance [48, 49]. In addition, one would expect that patients homozygous for C9orf72 repeat expansions (and thus with less C9orf72 than heterozygotes) will have more severe symptoms of disease, but this is not the case [50]. Although not conclusive, these human studies do not support a loss of function as the disease-driving mechanism.
Studies in lower species model systems suggest that C9orf72 is critical for the development of motor systems and motor functions. In zebrafish, blocking C9orf72 translation or splicing with antisense morpholino oligonucleotides resulted in axonopathy and reduced mobility [51]. Overexpressing of C9orf72 that lacks the complete DENN protein domain also phenocopied the morpholino experiment, suggesting the indispensability of the complete DENN protein domain [52]. Similarly, deletion of C9orf72 orthologue in Caenorhabditis elegans led to age-dependent motility deficits, paralysis, and increased susceptibility to environmental osmotic stress [53]. Primary hippocampal neurons cultured from C9orf72 knockout mice have reduced dendritic arborization and spinal density, suggesting its role in neuronal morphogenesis [54]. However, sustained reduction of mouse C9orf72 to 30–40% of its normal level throughout the CNS using antisense oligonucleotides (ASOs) is well tolerated, producing no behavioral or pathological features characteristic of ALS/FTD [55]. Several ubiquitous or neuronal/glialcell–specific C9orf72 knockout mice have also been characterized. None of these mice produced any ALS/FTD-like pathological or behavioral phenotypes [41, 42, 56–60], indicating that C9orf72 loss-of-function alone is not the main ALS/FTD disease driver in mice. Interestingly, global C9orf72 knockout mice develop a proinflammatory/autoimmune phenotype with enlarged spleen and lymph nodes, consistent with the observation that C9orf72 ALS/FTD patients also show an increased prevalence of autoimmune disease [61]. These findings are particularly noteworthy, given growing appreciation of the role of the immune system in a variety of neurodegenerative diseases [62].
Overall, current available data suggest that C9orf72 loss of function is insufficient to precipitate disease. Instead, hypermethylation of the mutant C9orf72 allele (one of the mechanisms leading to reduced C9orf72 expression) might actually be protective, potentially by decreasing the transcription of toxic repeat-containing C9orf72 RNAs [63–65]. However, given the role of C9orf72 in autophagy and microglia, pathways implicated in ALS and FTD, haploinsufficiency might contribute to the disease process in synergy with the gain-of-toxicity mechanism. This will be discussed in the section “Synergistic Mechanisms.”
Gain of Toxicity Plays a Central Role in Disease Pathogenesis
RNAs containing sense GGGGCC and antisense CCCCGG repeats are bidirectionally transcribed from the C9orf72 mutation. These repeat-containing RNAs can exert toxic gain-of-function and drive neurodegeneration (Fig. 2b). First, they may form complicated secondary structures, such as hairpins, i-motifs, and G-quadruplexes [66–68], which, in turn, sequester RBPs into intranuclear RNA foci that can be experimentally visualized using fluorescence in situ hybridization with probes against the repeats. These RNA foci lead to depletion of the normal function of the sequestered RBPs and neuronal death. The RBP sequestration model is best demonstrated in the neuromuscular disease myotonic dystrophy 1, where the muscleblind family proteins are sequestered by the CTG repeats in the DMPK gene and cause downstream splicing changes [69]. Secondly, despite being in an intronic region, the repeat of the C9orf72 RNAs can remarkably be translated through an unusual mechanism (RAN translation) in every reading frame to produce five different DPR proteins that might be detrimental to neurons. More specifically, poly(GA) and poly(GR) are uniquely translated from the sense GGGGCC RNA; poly(PR) and poly(PA) are uniquely from the antisense CCCCGG RNA, and poly(GP) is from either direction.
In c9ALS/FTD patients, both sense and antisense RNA foci were detected in fibroblast, lymphoblast, iPSC-derived neurons, and several CNS regions of c9ALS/FTD patients [28, 55, 70–73]. RNA foci are most abundant in neurons but also occur in astrocytes, microglia, and oligodendrocytes [55, 71, 72]. Similarly, accumulations of DPR proteins have been detected using either immunohistochemical assays for DPR inclusions or immunoblotting and ELISA-based assays for the soluble form [71, 73–76]. DPR inclusions are widely distributed throughout the brains of c9ALS/FTD and, interestingly, are less frequent in the spinal cord [76–80]. Based on the staining patterns, they can be categorized into neuronal cytoplasmic inclusions (NCIs), intranuclear inclusions (INIs), dystrophic neurites (DNs), and diffuse dendritic or cytoplasmic staining. So far, no DPR inclusions have been reported in astrocyte, microglia, and oligodendrocytes. Of the different types, the sense-strand RNA-encoded poly(GA), poly(GP), and poly(GR) inclusions are far more frequent than the antisense-strand RNA-encoded poly(PA) and poly(PR), and they can co-occur in the same neuron [74]. SDS-soluble poly(GA) and poly(GP) can also be detected in CSF and blood by immunoassay [81–83]. This will be further discussed in the section “Biomarkers.”
To determine the gain-of-toxicity mechanism from the C9orf72 repeats, several studies have overexpressed GGGGCC repeats with variable sizes and demonstrated deleterious phenotypes in cell culture [84], C. elegans [85], zebrafish [86], and Drosophila [87–89]. The gain-of-toxicity mechanism is also supported by studies expressing individual DPR proteins (discussed below), as well as studies using iPSC-derived neurons. Short or interrupted repeats are generally used, given the technical difficulties to obtain long and pure repeats. Adeno-associated viruses expressing 66 or 149 GGGGCC repeats were injected intraventrically into newborn mice to induce somatic transgenesis. These mice accumulated nuclear RNA foci and inclusions of DPR proteins and consequently exhibited hyperactivity, anxiety, antisocial behavior, and motor deficits [90, 91]. Four different groups, including ours, also generated transgenic mice expressing a bacterial artificial chromosome (BAC) that can accommodate genomic DNA from c9ALS/FTD patients to express long repeats (up to ~1000 repeats) in the context of the human gene. All these mouse models produced sense and antisense RNA foci, as well as DPR proteins from the sense strand. Two transgenic models failed to produce neurodegeneration [92, 93], and our mice, expressing exons 1–5 of the human C9orf72 gene and ~450 repeats, developed increased age-dependent anxiety and cognitive deficits, accompanied by the loss of hippocampal neurons, but no unusual motor phenotype [94]. In the last model expressing the full-length C9orf72 gene and ~500 repeats, a subset of female mice developed an acute motor phenotype, including paralysis and decreased survival. Other females and males showed slower progression [95]. The reasons for differences in these mouse models are unknown. They feature different genetic backgrounds, repeat sizes, transgene insertion sites, and expression levels. An in-depth comparison of all BAC models especially the accumulation of RNA foci and DPR proteins will provide insights to the toxic species leading to motor and/or cortical neuron death.
In summary, expressing GGGGCC repeats in model systems clearly recapitulates both cellular pathology (RNA foci and DPR proteins) and neuronal degeneration as in patients, even though unphysiologically high levels of expression were used. It is also challenging to dissect the relative contributions of RNA foci or DPR proteins to disease pathogenesis since these repeats will produce both RNA foci and DPR proteins, and the relative contributions of sense and antisense repeat–containing RNAs remain unclear.
RNA Foci-Mediated Toxicity
Few neuropathological studies have attempted to correlate the abundance and distribution of RNA foci with c9ALS/FTD clinical features. Mizielinska et al. [72] showed that patients with more sense RNA foci had an earlier age of symptom onset in a small number of C9orf72 FTD patients. Antisense foci showed a similar trend [72]. In agreement with this, antisense RNA foci are associated with nucleoli and mislocalization of TDP-43 [70, 96]. However, another study with larger patient population reported a contradictory observation. Patients with more antisense RNA foci in middle frontal gyrus (cortical layers 3–6) neurons showed a later age of onset [97].
Expressing C9orf72 GGGGCC repeats in primary neurons [84], Drosophila [87, 98], and zebrafish [86] suggests that the toxicity arises from RNA foci because no DPR proteins were detected. However, the inability to detect DPR proteins in these studies is insufficient to exclude a role for these proteins. Others argue against the toxicity from RNA foci. In two important studies, researchers generated Drosophila expressing either pure GGGGCC repeats or interrupted repeats of similar size, but with stop codons inserted in each reading frame to prevent the translation of the repeats into DPR proteins [88, 99]. Both pure and interrupted repeats form similar levels of RNA foci. Expression of pure repeats caused toxicity and early lethality whereas the interrupted ones had no effect. In another study, Drosophila expressing 160 GGGGCC repeats in the intron formed abundant sense RNA foci in the nucleus but produced little DPR protein and no neurodegeneration, again suggesting that nuclear sense RNA foci are not sufficient to drive neurodegeneration in this model [100]. There are, however, some caveats for interpreting these experiments, including whether the interrupted repeat RNAs have similar secondary structures to sequester the same key RBPs as the pure repeats, and whether RNA foci in humans may sequester RBPs that are not expressed in Drosophila.
A key to validating RNA foci–mediated toxicity is to identify the RBP(s) that are sequestered and thus lose function. Many proteins are proposed to interact with short, synthesized C9orf72 repeat RNAs in vitro, including hnRNPA1, hnRNPA3, hnRNP-H, hnRNP-H1/F, nucleolin, Pur-α, double-stranded RNA-specific editase B2 (ADARB2), RanGAP1, THO complex subunit 4 (ALYREF), serine/arginine-rich splicing factor 1 (SRSF1), SRSF2, and Zfp106 [24, 28, 87, 101–105]. In c9ALS/FTD brain tissues, sense and antisense RNA foci colocalize with hnRNPA1, hnRNP-H/F, SRSF2, and ALYREF [101, 102, 106]. But most of these RNA-binding proteins appear to colocalize with RNA foci at a low frequency; only hnRNP-H colocalizes with 70% of all sense foci detected [102]. Nevertheless, key data to support RNA foci–mediated toxicity are missing (i.e., demonstrating that the loss of function of any of these RBPs leads to C9orf72 diseases and upregulation of their functions can rescue phenotypes caused by C9orf72 repeat expansions).
DPR Protein-Mediated Toxicity
To gain insights for whether and/or which DPR proteins are the primary culprit of c9ALS/FTD, several clinicopathological studies have been carried out. Most of these studies raised suspicions about the pathogenicity of DPR inclusions, including 1) regions with the highest burden of DPR inclusions are cerebellum, hippocampus, and neocortex, where no significant loss of neurons occurs; 2) DPR inclusions do not differ in the neuroanatomical distributions in the brain between FTD and ALS cases; 3) DPR inclusions are rarely observed in lower motor neurons in the spinal cord; and most importantly, 4) no obvious correlation between the abundance levels of DPR inclusions with neurodegeneration [77, 78, 107, 108]. However, a few neuropathological studies support DPR protein–mediated toxicity. Mackenzie et al. showed moderate associations for the amount of poly(GA)-positive DNs with local degeneration in the frontal cortex [77]. In another study, more abundant poly(GA) NCIs in cortical, hippocampal, and motor regions were associated with earlier age at symptom onset [107]. Poly(GR) inclusions, but not the other DPR inclusions, correlated with areas of neurodegeneration in C9orf72 ALS and were unique in colocalizing with TDP-43 pathology in a small sample of brains that were obtained shortly after death [34]. In a larger cohort of 40 c9ALS/FTD cases, middle frontal gyrus poly(GR) inclusions were strongly correlated with neurodegeneration in this brain region [109]. Notably, these studies detect only aggregated, insoluble proteins, and it is possible that soluble species may mediate toxicity. Alternatively, CNS regions that have extensive DPR pathology but are unaffected by neurodegeneration might contain protective factors.
To move pathology to pathogenesis, several groups have expressed individual DPR proteins in isolation in yeast [110], mammalian cells [84, 111–117], Drosophila [84, 88, 89, 112, 118, 119], zebrafish [86, 120, 121], and mice [122–127]. Of all the DPR proteins, poly(GR) and poly(PR) are most toxic. Synthetic 20-mer poly(GR) and poly(PR) can cause rapid death of U2OS cells and cultured human astrocytes when added exogenously [128]. These DPR proteins also consistently show toxicity when overexpressed in several different cell lines, such as HEK293T cells, NSC-34 cells, and iPSC-derived neurons [84, 111, 112]. In Drosophila, targeted expression of poly(GR) or (PR) resulted in eye degeneration, motor deficits, and reduced survival [84, 88, 89, 112, 118, 119]. Finally, AAV-mediated or transgenic expression of poly(GR) and poly(PR) in mice caused age-dependent neurodegeneration, brain atrophy, and motor/memory deficits [124–127]. Poly(GA) also exerts toxicity. Synthetic poly(GA) exogenously applied to human cells or primary neurons is toxic [129]. Poly(GA) overexpression in cultured cells [115, 116], primary neurons [115, 116], zebrafish [120, 121], Drosophila [88], or mouse brains [122, 123] leads to toxicity. The remaining DPR proteins [poly(PA) and poly(GP)] are less likely to be the toxic species. These studies provide compelling evidence that certain DPR proteins are toxic. However, since high overexpression levels of individual DPR proteins were used, a key question remains to determine whether (which) DPR proteins are the main toxic species in c9ALS/FTD when expressed at the endogenous levels in patients. Another important issue is whether different DPR proteins, and repeat RNA and DPR proteins act synergistically, potentially in conjunction with the loss of function of C9orf72, to elicit downstream effects.
Synergistic Mechanisms
In c9ALS/FTD patients, pathological evidence exists for both loss of C9orf72 function and gain of toxicity. Whether loss of C9orf72 function synergizes with gain of toxicity to exacerbate ALS/FTD disease is not known. Mice with loss of C9orf72 do not develop ALS/FTD-related behavioral deficits, but additional stressors might be required as second hits to induce neurodegeneration in these mouse models. Shi et al. reported that loss of C9orf72 in human iPSC–derived motor neurons impacts lysosomal biogenesis and vesicular trafficking and exacerbates toxicity to poly(GR) and poly(PR) exposure [30]. Similarly, loss of C9orf72 protein sensitized cells to toxicity induced by expression of 30 and 60 GGGGCC repeats [47]. Shao et al. [130] bred C9orf72 knockout mice with one BAC transgenic model expressing the repeat-expanded full-length human C9orf72 gene to determine the synergism. One would doubt such strategy since transgene overexpression would compensate for loss of the mouse C9orf72 endogenous protein. But surprisingly, the transgenic mouse line does not express full-length C9orf72 proteins from the transgene and C9orf72 haploinsufficiency exacerbate motor behavior deficits in a dose-dependent manner [130]. It is premature to say that C9orf72 loss of function induces neurodegeneration together with toxic RNA and DPR proteins. Further investigations crossing C9orf72 knockout mice with transgenic mice that do not express the full-length C9orf72 proteins or injecting AAV virus expressing GGGGCC repeats into C9orf72 knockout mice are needed to address this question. In addition, poly(GA) recruits poly(GR) into inclusions and reduces poly(PR) toxicity when co-expressed in human cells and Drosophila [117, 118]. Whether interaction between individual DPR proteins enhances or suppresses their toxicity is thus a point of interest. Poly(GA) can also induce nuclear RNA foci in cells expressing 80 GGGGCC repeats and in patient fibroblast cells, suggesting a positive feedback loop between RNA foci and DPR proteins [131]. Finally, translational frameshifting has been suggested to occur in cells expressing short C9orf72 repeats [132] and other repeat expansions [133], which might produce chimeric repeat peptides. However, it is unknown whether such a frameshift occurs in C9orf72 mutation carriers and, if it does, the contribution of chimeric repeat peptides to disease pathogenesis.
Downstream Molecular Pathway Dysfunctions
In addition to identifying the major toxic species, research has focused on determining cellular dysfunctions that result from the C9orf72 repeat expansions. These events provide a basic understanding of disease pathophysiology and new therapeutic targets (Fig. 3).
Fig. 3.
Cellular processes impaired by the C9orf72 repeat expansions and potential therapeutic interventions. A wide range of cellular pathways have been implicated in c9ALS/FTD, including DNA damage, nucleolar stress, nucleocytoplasmic transport deficits, ER stress, autophagy dysfunction, translational inhibition, proteasome inhibition, and altered stress granule dynamics. Therapies targeting these deficits, as well as directly targeting repeat expanded C9orf72 DNA/RNA and DPR proteins, are also highlighted
Nucleocytoplasmic Transport Deficits
Active transport of proteins and RNAs through nuclear pore complexes (NPCs), a process called nucleocytoplasmic transport (NCT), is essential for cellular functions [134]. Zhang et al. [98] showed that RanGAP1 binds to GGGGCC repeat RNA in vitro and is sequestered into sense RNA foci. RanGAP1 activates Ran small GTPase, which plays an important role in regulating the interactions between cargo molecules and transport receptors, such as the importin and exportin family of proteins [134]. In both Drosophila overexpressing 30 GGGGCC repeats and c9ALS iPSC–derived neurons, the nucleocytoplasmic Ran gradient is decreased, and the nuclear import of proteins and nuclear export of RNAs are compromised. In addition, the repeat toxicity can be readily suppressed by increased activity of RanGAP1, suggesting that RNA foci–mediated NCT deficit is a fundamental pathway in c9ALS/FTD [98]. In parallel, unbiased genetic screenings for phenotypic modifiers of Drosophila expressing GGGGCC repeats [89] or poly(PR) [119], yeast [110], and human cells expressing poly(GR) or poly(PR) [135] identified that many proteins involved in NCT mitigate or exacerbate disease phenotypes. Mechanistically, poly(GR) and poly(PR) interact with nucleopore proteins (Nups), including the central channel of the nucleopore, to reduce trafficking [136]. Poly(GA) also sequesters nucleocytoplasmic transport proteins, such as HR23, and induces an abnormal distribution of RanGAP1 in mice [122]. The primary cause for the NCT dysfunction in c9ALS/FTD is unknown. NCT defects have also been reported in other neurodegenerative disorders, such as Alzheimer’s disease [137] and Huntington’s disease [138, 139], suggesting that it may play a more global role in the neurodegenerative process.
Nucleolar Dysfunction
Nucleolus plays an important role in ribosomal RNA biogenesis. When applied exogenously to astrocyte cultures, poly(GR) and poly(PR) 20-mers accumulate in nucleolus, leading to splicing changes and impaired ribosomal RNA maturation [128]. Overexpressed poly(GR) and poly(PR) also colocalize with nucleoli in cultured cells, primary neurons, iPSC-derived neurons, and Drosophila, leading to abnormal nucleolar morphology [84]. However, poly(GR) and poly(PR) do not localize within the nucleolus in C9orf72 patient brains. Instead, sense RNA foci can bind nucleolin, a principle component of the nucleolus, and aberrant subcellular localization of nucleolin was observed in C9orf72 iPSC–derived motor neurons and patient motor cortex [24]. Contrary to this, antisense RNA foci associated with nucleoli were identified neuropathologically to be associated with disease [96]. Nucleolin mislocalization was also found in C9orf72 BAC transgenic mice without significant changes in ribosomal RNA processing or splicing [92]. Interestingly, C9orf72 patient brains exhibit bidirectional nucleolar volume changes, with smaller nucleoli overall but enlarged nucleoli in neurons containing poly(GR) inclusions [140].
DNA Damage
Nucleolar stress induces DNA damage, and postmitotic neurons are particularly susceptible to DNA damage. An age-dependent increase in DNA damage and oxidative stress was reported in iPSC-derived motor neurons of patients with C9orf72 repeat expansions [141]. DNA damage is also increased in spinal cord neurons of C9orf72 ALS patients [142]. Such DNA damage is partially caused by poly(GR) since expression of poly(GR) in iPSC-derived control neurons, neuronal cell lines or in mice in vivo is sufficient to induce DNA damage and toxicity [126]. Mechanistically, poly(GR) binds to Atp5a1 and compromises mitochondrial function, leading to increased oxidative stress and an overactivated Ku80-dependent DNA repair pathway [126, 143]. Introducing ectopic Atp5a1 expression or reduction in oxidative stress by treatment with antioxidants partially rescues DNA damage in motor neurons from C9orf72 carriers and neurons expressing poly(GR). As a result of DNA damage, the levels of phosphorylated ATM and P53 and other downstream proapoptotic proteins, such as PUMA, Bax, and cleaved caspase-3, are significantly increased in C9orf72 patient neurons. Partial loss of Ku80 function in these neurons through CRISPR/Cas9-mediated ablation or small RNA-mediated knockdown suppresses the apoptotic pathway. Thus, poly(GR)-mediated DNA damage contributes to ALS/FTD pathogenesis, and partial inhibition of overactivated Ku80-dependent DNA repair pathway is a promising therapeutic target.
Alteration in Stress Granules
Eukaryotic cells have evolved sophisticated strategies to combat unexpected cellular stress. Cytoplasmic ribonucleoprotein (RNP) granules, such as processing bodies (P-bodies) and stress granules (SGs), assemble quickly under stress to sequester messenger RNAs (mRNAs), translation initiation factors, 40S ribosomes, and many other RNA-binding proteins. In this way, only essential proteins needed for survival are produced. These granular structures can either dissemble upon release of cellular stress or be degraded by the autophagy pathway. Dysregulated formation or clearance of SGs has been proposed as a key pathogenic mechanism for ALS/FTD [144]. In recent years, groundbreaking work from several groups showed that SG-associated RNA-binding proteins, such as FUS and hnRNP1/2, undergo liquid–liquid-phase separation (LLPS) to form droplets in vitro and have the propensity to further fibrilize into irreversible hydrogel or even insoluble amyloid structures [145–148]. One characteristic of these RBPs is that they all contain prion-like, intrinsically disordered low-complexity domains (LCDs). Poly(GR) and poly(PR) interact with these LCD proteins, impair LLPS, and disrupt the dynamics of stress granules [149]. Consistent with this, primary cortical neurons overexpressing poly(PR) showed a reduction in cytoplasmic P-bodies with larger sizes and an increase in stress granule formation [84], and knockdown of several of the LCD proteins modifies the eye degeneration phenotype in Drosophila expressing poly(GR) [112]. Activation of the SG response or exposure to either poly(GR) or poly(PR) was also sufficient to disrupt nucleocytoplasmic transport by promoting the sequestration of several nuclear pore proteins into SGs [150]. Interestingly, poly(GR) and poly(PR) themselves undergo LLPS at high concentrations, and GGGGCC repeat RNA also undergoes gel transition itself [151] and promotes the phase transition of RNA granule proteins in vitro and in cells [152].
Translation Inhibition and Ubiquitin Proteasome System
In addition to regulating stress granule dynamics and ribosomal RNA synthesis, overexpression of poly(GR) and (PR) blocked global translation in an in vitro translation assay and in cell lines [114]. This was attributed to direct binding of poly(GR) and poly(PR) to mRNA, thereby blocking access to the translational machinery. Poly(PR) and poly(GR) also interact with translation initiation and elongation factors and ribosome subunits in pull-down assays [114]. In addition, GGGCC repeat–containing RNAs sequester ribosomal subunits, trigger stress granule formation, and inhibit translation [152, 153]. Interestingly, 26S proteasome complexes, components of the ubiquitin proteasome system (UPS), are sequestered within poly(GA) aggregates in cells as revealed by protein cryo-electron tomography technology, which allows 3D imaging of the cell interior in close-to-native conditions [154]. These results provide new insight into the mechanism by which poly(GA) deposition promotes UPS impairment and regulate protein homeostasis.
Therapeutics
Currently, no cure is available for ALS or FTD. The identification of C9orf72 as the most common genetic cause and fast progress in research unveiling disease mechanisms has inspired multiple therapeutic interventions for patients carrying this mutation, the prospects of which are particularly exciting (Fig. 3).
Targeting C9orf72 Repeat-Expanded RNA/DNA
Broad evidence supports a gain of toxicity from which the C9orf72 repeat RNAs play a central role in ALS/FTD pathogenesis. Thus, inhibiting the transcription or selectively reducing repeat-containing RNAs is a promising strategy with antisense oligonucleotide therapy being the most advanced in terms of clinical development. Indeed, unraveling the exact contributions from RNA toxicity and DPR protein toxicity or the exact cellular pathways may not be necessary for such an approach to be successful. However, unraveling the exact contributions from sense and antisense strand transcripts is necessary, since these must be targeted separately.
Antisense Oligonucleotide Therapy
ASOs are designed synthetic oligonucleotides or oligonucleotide analogs that bind to target RNAs. Depending on its chemical modifications, ASOs selectively degrade mRNAs through endonuclease RNase H recruitment or prevent the interaction of RNAs with RBPs, thereby modulating its splicing/processing without degradation [155]. In recent years, ASO therapy for neurological disorders has gained significant interest, with the FDA approval of nusinersen (Spinraza) for spinal muscular atrophy (SMA) in 2016. Nusinersen increases the expression of survival motor neuron (SMN) protein by modulating splicing of the SMN2 pre-mRNAs [156]. The ability of ASOs to selectively degrade mRNAs is also advantageous if the pathogenesis of neurodegeneration is caused by gain of toxicity from mutant RNA/proteins. Phase I trials have been completed with ASOs targeting superoxide dismutase 1 (SOD1) in familiar ALS [157] and mutant huntingtin gene in Huntington’s disease [158]. For C9orf72 diseases, early studies showed that ASOs targeting within or immediately upstream of the sense repeats reduced sense RNA foci, increased survival from glutamate excitotoxicity, and abrogated aberrant gene expression patterns in fibroblast and iPSC-derived neurons [28, 55, 101]. These observations were further extended to in vivo mouse models. We demonstrated that a single-dose, intraventricular administration of ASOs targeting the sense repeat–containing RNAs led to sustained reduction of sense RNA foci and DPR proteins and mitigated the behavioral and cognitive deficits in transgenic mice expressing 450 repeats [94]. Importantly, transcript variants 1 (NM_145005.6) and 3 (NM_001256054.2) that carry the repeat expansion can be specifically targeted by ASOs without reducing variant 2 (NM_018325.5) expression. Since variant 2 is expressed at much higher levels than variants 1 and 3, overall C9orf72 abundance remained relatively intact [94]. Thus, even if loss of function is important in pathogenesis, toxic transcripts can be reduced without further exacerbating loss. Based on these encouraging results, a phase I clinical trial of ASOs targeting the sense strand for C9ALS patients has started by Ionis Pharmaceuticals and its partner Biogen Inc. in September 2018 (NCT03626012).
RNA Interference Strategy
RNA interference (RNAi) represents another alternative approach against RNA/protein-mediated gain of toxicity. RNAi is a biological process in which certain double-stranded RNA molecules inhibit the expression of target genes by destroying the messenger RNAs in the cytoplasm mediated through the RNA-induced silencing complex (RISC) [159]. The three most common RNAi approaches include short interfering RNAs (siRNAs), shRNAs, or artificial microRNAs (miRNAs). Given the recent progress of using adeno-associated virus (AAV) for therapeutic gene delivery [160], RNAi therapy becomes even more attractive to treat neurodegenerative disorders. The major challenge for C9orf72 ALS/FTD is to target the repeat-containing C9orf72 transcripts in the nucleus. Indeed, one study showed that siRNA significantly reduced C9orf72 mRNA in patient fibroblast but did not affect nuclear RNA foci abundance [55]. However, another study suggested that single-strand silencing RNAs reduced both sense and antisense RNA foci by reducing mutant RNA transcript via RNAi and sterically blocking RBP binding to RNAs [161]. Designed double-strand RNA targeting the repeat region can also block RNA foci formation [162]. In addition, AAV5-driving miRNAs can silence C9orf72 and reduce RNA foci in both the nucleus and cytoplasm in iPSC-derived motor neurons and in an ALS mouse model [163, 164]. Further studies are needed to demonstrate the efficacy of RNAi to mitigate behavioral deficits in transgenic mice.
Small Molecules
Small molecules offer an attractive approach for targeting C9orf72 repeat DNA/RNAs given their pharmacological advantages, such as a small molecular mass, which is important for blood–brain barrier permeability. TMPyP4, a known G-quadruplex binder, binds (GGGGCC)8 RNA in vitro, ablates the sequestration of RBPs [165], and rescues transport defect pathologies and neurodegeneration in Drosophila overexpressing 30 repeats [98]. Recent progress has also been made in the design of nucleotide structure–specific small molecules. Several compounds bind the hairpin structure of GGGGCC repeats and significantly decrease foci formation and poly(GP) accumulation in cultured cells overexpressing 66 repeats and patient iPSC-derived neurons [83, 166]. It is unknown whether these compounds also reduce poly(GR) and poly(PR), two DPR proteins that are most toxic in model systems. Further experiments are also needed to demonstrate that they are beneficial in reducing cellular toxicity both in vitro and in vivo.
Reduce Repeat-Containing RNA Transcription
The complicated secondary structure formed by GC-rich C9orf72 repeat expansions suggest that specialized transcription machinery may be required to facilitate RNA polymerase II through such long repeats. DRB sensitivity-inducing factor (DSIF) complex and the RNA polymerase II associated factor 1 complex (PAF1C) are highly conserved and have nonredundant roles in activating RNA polymerase II during elongation [167]. SUPT4H and SUPT5H (components of DSIF) and PAF1C play critical roles in the transcriptional elongation of C9orf72 repeat expansions [168, 169]. Targeting SUPT4H reduces levels of sense and antisense C9orf72 repeat transcripts, as well as accumulation of RNA foci and DPR products, and ameliorates neurodegeneration in C9orf72 iPSC-derived neurons and in Drosophila [168]. Similarly, in C9orf72 patients, the expression levels of PAF1 and LEO1 (components of PAF1C) are upregulated, and their expression correlates positively with the expression of repeat-containing transcripts. Depletion of PAF1C reduces RNA and poly(GR) dipeptide production in the Drosophila-expressing GGGGCC transgene [169]. Although exciting, careful examination of this therapeutic strategy is needed since depletion of SUPT4H causes a global RNA reduction [170].
CRISPR
Recent advances in genome editing with CRISPR/Cas technology have made it possible to target the mutant gene at the genomic level. CRISPR/Cas is an abbreviation for clustered regularly interspaced short palindromic repeats and CRISPR-associated protein. It was adapted from a naturally occurring genome-editing system in bacteria [171]. CRISR/Cas9 can be designed to cut the C9orf72 repeat expansions, following gene repair through nonhomologous end joining. As a proof of concept, genetic correction of C9orf72 repeat expansions in patient iPSCs has been achieved [30, 126]. In addition, Pinto et al. [172] targeted enzymatically dead Cas9 to directly bind repeat-expanded DNAs, such as CTG expansion in DM1, CTGG expansion in DM2, and GGGGCC expansion in C9ALS/FTD. This approach sterically impedes repeat RNA transcription and leads to a drastic decrease in the production of aberrant proteins produced from RAN translation [172]. Similar to ASOs, RNA-targeting Cas9 with RNA endonuclease can degrade repeat expanded RNAs and reduce RNA foci and DPR protein levels in cell lines [173]. The clinical use of CRISPR/Cas technology in ALS/FTD is still in its infancy. Ethical concerns, adequate methods of delivery and cell targeting, off-target and safety measures, and treatment efficacy are the main issues that need to be addressed before applying this method to patients.
Targeting DPR Proteins and Downstream Mechanisms
Overall, ASO, RNAi, and other abovementioned approaches target the upstream of disease mechanism and also positively affect different downstream pathways. For example, Zhang et al. [98] showed that ASOs targeting C9orf72 sense RNA or TMPyP4 treatment to inhibit RBPs bindings to repeat RNAs can rescue NCT deficits. Nevertheless, strategies directly targeting DPR proteins and downstream mechanisms have also been pursued.
Target DPR Proteins
Aggregates of DPR proteins are a pathological hallmark of C9orf72 ALS/FTD, and DPR proteins seem to exert toxicity in model systems. In addition to inhibiting RAN translation as we learn more about its mechanisms [153, 174, 175], one way to directly mitigate DPR protein–mediated toxicity is to increase toxic protein turnover rate. To this end, overexpressing the small heat shock protein HSPB8 facilitated the autophagy-mediated disposal of a large variety of classical misfolded aggregation-prone proteins and significantly decreased the accumulation of most DPR insoluble species [176]. Antibody immunization against DPR proteins represents another novel therapeutic approach. In other neurodegenerative disorders, active and passive immunization have been explored to target toxic proteins that transmit from cells to cells, such as amyloid-β, tau, and α-synuclein [177]. The prion-like propagation of misfolded/aggregated proteins is also a pathogenic principle in ALS/FTD [178, 179]. Using coculture cell assays, poly(GA) was shown to possess cell-to-cell transmission properties [180] and so do poly(GP) and poly(PA) [131]. Interestingly, poly(GA) antibodies reduce intracellular poly(GA) aggregations and seeding activity of C9orf72 patient brain extracts [131]. Since poly(GR) and poly(PR) are more toxic than poly(GA), poly(GP), and poly(PA) in experimental models, further studies are needed to determine which DPR proteins and conformations to target. The specificity of antibodies against DPR proteins is also a concern as similar short repetitive sequences are present in human proteome.
Target Downstream Mechanisms
Other therapeutic approaches aim to correct downstream cellular pathways affected by C9orf72 repeat expansions. Reducing nuclear export by targeting the nuclear export factors SRSF1 or exportin 1 ameliorates the toxicity mediated by C9orf72 repeat expansion in Drosophila [98]. This could be due to the reduced export of toxic repeat-containing RNA to the cytoplasm and thus less DPR proteins [181] or through a more general mechanism to reverse alterations in NCT and consequent mislocalization of RBPs, such as TDP-43 [98]. In addition to the genetic tools, selective inhibitors of nuclear export (SINEs) mitigate NCT deficits and neurodegeneration in a Drosophila model [98]. SINE compounds also improve primary neuron survival and partially improve motor function in rats overexpressing mutant TDP-43 [182]. Currently, Karyopharm Therapeutics and its partner Biogen Inc. are taking one of the SINE compounds, KPT-350, into clinical trials. Zhang et al. [150] also showed that inhibition of stress granule assembly by using ASOs targeting ataxin 2 is sufficient to abrogate NCT dysfunction and neurodegeneration in patient-derived neurons and in vivo. As reducing ataxin 2 significantly extend the lifespan of TDP-43 mice, this strategy holds great promise [183].
Biomarkers
One of the critical themes that has emerged over the last two decades of clinical trials in ALS relates to biomarkers. There is no clear diagnostic biomarker for diagnosis, tracking disease progression, or determining if a putative therapy engages its therapeutic target. In C9orf72-associated diseases, effective biomarkers could also identify phenoconversion of asymptomatic genetic carriers. Clinical, neurophysiological, and imaging biomarkers are difficult to employ and/or insensitive. Recently, progress has emerged for biomarkers in the blood and CSF. Neurofilament proteins herald the onset of disease in presymptomatic carriers and appear to track with disease progression, although these biomarkers are nonspecific to C9orf72 diseases and are sensitive to damage caused by free radicals [184]. Significantly, the DPR protein poly(GP) was detected in CSF and in peripheral blood mononuclear cells from c9ALS patients. Thus, tracking poly(GP) in CSF provides a direct readout of target engagement for ASO and other clinical trials [82].
Conclusions and Future Perspectives
Since the discovery of C9orf72 repeat expansions, an impressive effort has been made to investigate disease mechanisms. Many disease models were developed to determine the loss of function and/or gain of toxicity and downstream cellular function alterations. Therapeutic approaches and biomarkers have also been explored. Excitingly, antisense oligonucleotide therapy to selectively degrade repeat-containing RNAs has gone into a phase I clinical trial for C9orf72 ALS patients. With a continuing fast pace of research on these devastating diseases, a promising therapy should soon be on the horizon.
Electronic supplementary material
(PDF 506 kb)
(PDF 515 kb)
Acknowledgements
JR is supported by grants from ALS Association (5356S3), Target ALS (20134792), the National Institute of Neurological Diseases and Stroke (NIH R01NS088578 and NS047101), and Pam Golden and is involved in Biogen-sponsored clinical trials. JJ is a recipient of career development grant from Muscular Dystrophy Association (479769).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jie Jiang, Email: jie.jiang@emory.edu.
John Ravits, Email: jravits@ucsd.edu.
References
- 1.Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olney NT, Spina S, Miller BL. Frontotemporal Dementia. Neurol Clin. 2017;35(2):339–74. doi: 10.1016/j.ncl.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng AS, Rademakers R, Miller BL. Frontotemporal dementia: a bridge between dementia and neuromuscular disease. Ann N Y Acad Sci. 2015;1338:71–93. doi: 10.1111/nyas.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurology. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen HP, Van Broeckhoven C, van der Zee J. ALS Genes in the Genomic Era and their Implications for FTD. Trends Genet. 2018;34(6):404–23. doi: 10.1016/j.tig.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 9.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature Reviews Genetics. 2010;11(4):247–58. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. American Journal of Human Genetics. 2013;92(3):345–53. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchman VL, Cooper-Knock J, Connor-Robson N, Higginbottom A, Kirby J, Razinskaya OD, et al. Simultaneous and independent detection of C9ORF72 alleles with low and high number of GGGGCC repeats using an optimised protocol of Southern blot hybridisation. Molecular Neurodegeneration. 2013;8:12. doi: 10.1186/1750-1326-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobson-Stone C, Hallupp M, Loy CT, Thompson EM, Haan E, Sue CM, et al. C9ORF72 repeat expansion in Australian and Spanish frontotemporal dementia patients. PLoS One. 2013;8(2):e56899. doi: 10.1371/journal.pone.0056899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubers A, Marroquin N, Schmoll B, Vielhaber S, Just M, Mayer B, et al. Polymerase chain reaction and Southern blot-based analysis of the C9orf72 hexanucleotide repeat in different motor neuron diseases. Neurobiology of Aging. 2014;35(5):1214.e1–6. doi: 10.1016/j.neurobiolaging.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 14.Ishiura H, Takahashi Y, Mitsui J, Yoshida S, Kihira T, Kokubo Y, et al. C9ORF72 repeat expansion in amyotrophic lateral sclerosis in the Kii peninsula of Japan. Archives of Neurology. 2012;69(9):1154–8. doi: 10.1001/archneurol.2012.1219. [DOI] [PubMed] [Google Scholar]
- 15.Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J, et al. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Molecular Psychiatry. 2016;21(8):1112–24. doi: 10.1038/mp.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proudfoot M, Gutowski NJ, Edbauer D, Hilton DA, Stephens M, Rankin J, et al. Early dipeptide repeat pathology in a frontotemporal dementia kindred with C9ORF72 mutation and intellectual disability. Acta Neuropathologica. 2014;127(3):451–8. doi: 10.1007/s00401-014-1245-7. [DOI] [PubMed] [Google Scholar]
- 17.Van Mossevelde S, van der Zee J, Gijselinck I, Sleegers K, De Bleecker J, Sieben A, et al. Clinical Evidence of Disease Anticipation in Families Segregating a C9orf72 Repeat Expansion. JAMA Neurology. 2017;74(4):445–52. doi: 10.1001/jamaneurol.2016.4847. [DOI] [PubMed] [Google Scholar]
- 18.Nordin A, Akimoto C, Wuolikainen A, Alstermark H, Jonsson P, Birve A, et al. Extensive size variability of the GGGGCC expansion in C9orf72 in both neuronal and non-neuronal tissues in 18 patients with ALS or FTD. Human Molecular Genetics. 2015;24(11):3133–42. doi: 10.1093/hmg/ddv064. [DOI] [PubMed] [Google Scholar]
- 19.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurology. 2013;12(10):978–88. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benussi L, Rossi G, Glionna M, Tonoli E, Piccoli E, Fostinelli S, et al. C9ORF72 hexanucleotide repeat number in frontotemporal lobar degeneration: a genotype-phenotype correlation study. Journal of Alzheimer’s Disease: JAD. 2014;38(4):799–808. doi: 10.3233/JAD-131028. [DOI] [PubMed] [Google Scholar]
- 21.Dols-Icardo O, Garcia-Redondo A, Rojas-Garcia R, Sanchez-Valle R, Noguera A, Gomez-Tortosa E, et al. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Human Molecular Genetics. 2014;23(3):749–54. doi: 10.1093/hmg/ddt460. [DOI] [PubMed] [Google Scholar]
- 22.Suh E, Lee EB, Neal D, Wood EM, Toledo JB, Rennert L, et al. Semi-automated quantification of C9orf72 expansion size reveals inverse correlation between hexanucleotide repeat number and disease duration in frontotemporal degeneration. Acta Neuropathologica. 2015;130(3):363–72. doi: 10.1007/s00401-015-1445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzu P, Blauwendraat C, Heetveld S, Lynes EM, Castillo-Lizardo M, Dhingra A, et al. C9orf72 is differentially expressed in the central nervous system and myeloid cells and consistently reduced in C9orf72, MAPT and GRN mutation carriers. Acta Neuropathologica Communications. 2016;4(1):37. doi: 10.1186/s40478-016-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. American Journal of Human Genetics. 2013;92(6):981–9. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xi Z, Zhang M, Bruni AC, Maletta RG, Colao R, Fratta P, et al. The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathologica. 2015;129(5):715–27. doi: 10.1007/s00401-015-1401-8. [DOI] [PubMed] [Google Scholar]
- 27.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathologica. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathologica. 2013;126(3):385–99. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nature Medicine. 2018;24(3):313–25. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, et al. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathologica. 2015;130(6):863–76. doi: 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiology of Aging. 2014;35(7):1779 e5- e13. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frick P, Sellier C, Mackenzie IRA, Cheng CY, Tahraoui-Bories J, Martinat C, et al. Novel antibodies reveal presynaptic localization of C9orf72 protein and reduced protein levels in C9orf72 mutation carriers. Acta Neuropathologica Communications. 2018;6(1):72. doi: 10.1186/s40478-018-0579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, et al. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathologica. 2018;135(3):459–74. doi: 10.1007/s00401-017-1793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao S, MacNair L, McGoldrick P, McKeever PM, McLean JR, Zhang M, et al. Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Annals of Neurology. 2015;78(4):568–83. doi: 10.1002/ana.24469. [DOI] [PubMed] [Google Scholar]
- 36.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics (Oxford, England) 2013;29(4):499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Frontiers in Genetics. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amick J, Roczniak-Ferguson A, Ferguson SM. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Molecular Biology of the Cell. 2016;27(20):3040–51. doi: 10.1091/mbc.E16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung J, Nayak A, Schaeffer V, Starzetz T, Kirsch AK, Muller S, et al. Multiplex image-based autophagy RNAi screening identifies SMCR8 as ULK1 kinase activity and gene expression regulator. eLife. 2017;6. [DOI] [PMC free article] [PubMed]
- 40.Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. The EMBO Journal. 2016;35(12):1276–97. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathologica Communications. 2016;4(1):51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, et al. Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genetics. 2016;12(11):e1006443. doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Science Advances. 2016;2(9):e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Human Molecular Genetics. 2014;23(13):3579–95. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. The EMBO Journal. 2016;35(15):1656–76. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoki Y, Manzano R, Lee Y, Dafinca R, Aoki M, Douglas AGL, et al. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain: a journal of neurology. 2017;140(4):887–97. doi: 10.1093/brain/awx024. [DOI] [PubMed] [Google Scholar]
- 47.Maharjan N, Kunzli C, Buthey K, Saxena S. C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly. Hypersensitizing Cells to Stress. Molecular Neurobiology. 2017;54(4):3062–77. doi: 10.1007/s12035-016-9850-1. [DOI] [PubMed] [Google Scholar]
- 48.Harms MB, Cady J, Zaidman C, Cooper P, Bali T, Allred P, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiology of Aging. 2013;34(9):2234 e13–9. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu F, Liu Q, Lu CX, Cui B, Guo XN, Wang RR, et al. Identification of a novel loss-of-function C9orf72 splice site mutation in a patient with amyotrophic lateral sclerosis. Neurobiology of Aging. 2016;47:219.e1-.e5. doi: 10.1016/j.neurobiolaging.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 50.Fratta P, Poulter M, Lashley T, Rohrer JD, Polke JM, Beck J, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathologica. 2013;126(3):401–9. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Annals of Neurology. 2013;74(2):180–7. doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 52.Yeh TH, Liu HF, Li YW, Lu CS, Shih HY, Chiu CC, et al. C9orf72 is essential for neurodevelopment and motility mediated by Cyclin G1. Experimental Neurology. 2018;304:114–24. doi: 10.1016/j.expneurol.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Therrien M, Rouleau GA, Dion PA, Parker JA. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One. 2013;8(12):e83450. doi: 10.1371/journal.pone.0083450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho WY, Tai YK, Chang JC, Liang J, Tyan SH, Chen S, et al. The ALS-FTD-linked gene product, C9orf72, regulates neuronal morphogenesis via autophagy. Autophagy. 2019;15(5):827–42. doi: 10.1080/15548627.2019.1569441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(47):E4530–9. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA. Vieira de Sa R, et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Annals of Neurology. 2015;78(3):426–38. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee HC, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Scientific Reports. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sudria-Lopez E, Koppers M, de Wit M, van der Meer C, Westeneng HJ, Zundel CA, et al. Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathologica. 2016;132(1):145–7. doi: 10.1007/s00401-016-1581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science (New York, NY) 2016;351(6279):1324–9. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Science Translational Medicine. 2016;8(347):347–93. doi: 10.1126/scitranslmed.aaf6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller ZA, Sturm VE, Camsari GB, Karydas A, Yokoyama JS, Grinberg LT, et al. Increased prevalence of autoimmune disease within C9 and FTD/MND cohorts: Completing the picture. Neurology(R) Neuroimmunology & Neuroinflammation. 2016;3(6):e301. doi: 10.1212/NXI.0000000000000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molteni M, Rossetti C. Neurodegenerative diseases: The immunological perspective. J Neuroimmunol. 2017;313:109–15. doi: 10.1016/j.jneuroim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, et al. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathologica. 2014;128(4):525–41. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMillan CT, Russ J, Wood EM, Irwin DJ, Grossman M, McCluskey L, et al. C9orf72 promoter hypermethylation is neuroprotective: Neuroimaging and Neuropathologic Evidence. Neurology. 2015;84(16):1622–30. doi: 10.1212/WNL.0000000000001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russ J, Liu EY, Wu K, Neal D, Suh E, Irwin DJ, et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathologica. 2015;129(1):39–52. doi: 10.1007/s00401-014-1365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Scientific Reports. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. The Journal of Biological Chemistry. 2013;288(14):9860–6. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovanda A, Zalar M, Sket P, Plavec J, Rogelj B. Anti-sense DNA d(GGCCCC)n expansions in C9ORF72 form i-motifs and protonated hairpins. Scientific Reports. 2015;5:17944. doi: 10.1038/srep17944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahimov F, Kunkel LM. The cell biology of disease: cellular and molecular mechanisms underlying muscular dystrophy. The Journal of Cell Biology. 2013;201(4):499–510. doi: 10.1083/jcb.201212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cooper-Knock J, Higginbottom A, Stopford MJ, Highley JR, Ince PG, Wharton SB, et al. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathologica. 2015;130(1):63–75. doi: 10.1007/s00401-015-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathologica. 2013;126(6):829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathologica. 2013;126(6):845–57. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(51):E4968–77. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathologica. 2013;126(6):881–93. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 75.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science (New York, NY). 2013;339(6125):1335–8. [DOI] [PubMed]
- 76.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathologica. 2015;130(6):845–61. doi: 10.1007/s00401-015-1476-2. [DOI] [PubMed] [Google Scholar]
- 78.Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathologica. 2015;130(4):537–55. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez-Deza J, Lee YB, Troakes C, Nolan M, Al-Sarraj S, Gallo JM, et al. Dipeptide repeat protein inclusions are rare in the spinal cord and almost absent from motor neurons in C9ORF72 mutant amyotrophic lateral sclerosis and are unlikely to cause their degeneration. Acta Neuropathologica Communications. 2015;3:38. doi: 10.1186/s40478-015-0218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schipper LJ, Raaphorst J, Aronica E, Baas F, de Haan R, de Visser M, et al. Prevalence of brain and spinal cord inclusions, including dipeptide repeat proteins, in patients with the C9ORF72 hexanucleotide repeat expansion: a systematic neuropathological review. Neuropathology and Applied Neurobiology. 2016;42(6):547–60. doi: 10.1111/nan.12284. [DOI] [PubMed] [Google Scholar]
- 81.Lehmer C, Oeckl P, Weishaupt JH, Volk AE, Diehl-Schmid J, Schroeter ML, et al. Poly-GP in cerebrospinal fluid links C9orf72-associated dipeptide repeat expression to the asymptomatic phase of ALS/FTD. EMBO Mol Med. 2017;9(7):859–68. doi: 10.15252/emmm.201607486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, Prudencio M, et al. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Science Translational Medicine. 2017;9(383). [DOI] [PMC free article] [PubMed]
- 83.Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83(5):1043–50. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84(6):1213–25. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Hao L, Saur T, Joyal K, Zhao Y, Zhai D, et al. Forward Genetic Screen in Caenorhabditis elegans Suggests F57A10.2 and acp-4 As Suppressors of C9ORF72 Related Phenotypes. Front Mol Neurosci. 2016;9:113. [DOI] [PMC free article] [PubMed]
- 86.Swinnen B, Bento-Abreu A, Gendron TF, Boeynaems S, Bogaert E, Nuyts R, et al. A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathologica. 2018;135(3):427–43. doi: 10.1007/s00401-017-1796-5. [DOI] [PubMed] [Google Scholar]
- 87.Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science (New York, NY) 2014;345(6201):1192–4. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525(7567):129–33. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science (New York, NY) 2015;348(6239):1151–4. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chew J, Cook C, Gendron TF, Jansen-West K, Del Rosso G, Daughrity LM, et al. Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Molecular Neurodegeneration. 2019;14(1):9. doi: 10.1186/s13024-019-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Rourke JG, Bogdanik L, Muhammad A, Gendron TF, Kim KJ, Austin A, et al. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron. 2015;88(5):892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, et al. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron. 2015;88(5):902–9. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron. 2016;90(3):535–50. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, et al. C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron. 2016;90(3):521–34. doi: 10.1016/j.neuron.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 96.Aladesuyi Arogundade O, Stauffer JE, Saberi S, Diaz-Garcia S, Malik S, Basilim H, et al. Antisense RNA foci are associated with nucleoli and TDP-43 mislocalization in C9orf72-ALS/FTD: a quantitative study. Acta Neuropathologica. 2019;137(3):527–30. doi: 10.1007/s00401-018-01955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DeJesus-Hernandez M, Finch NA, Wang X, Gendron TF, Bieniek KF, Heckman MG, et al. In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathologica. 2017;134(2):255–69. doi: 10.1007/s00401-017-1725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moens TG, Mizielinska S, Niccoli T, Mitchell JS, Thoeng A, Ridler CE, et al. Sense and antisense RNA are not toxic in Drosophila models of C9orf72-associated ALS/FTD. Acta Neuropathologica. 2018;135(3):445–57. doi: 10.1007/s00401-017-1798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, et al. Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS. Neuron. 2015;87(6):1207–14. doi: 10.1016/j.neuron.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Science Translational Medicine. 2013;5(208):208–149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Reports. 2013;5(5):1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathologica. 2013;125(3):413–23. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 104.Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, et al. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife. 2016;5. [DOI] [PMC free article] [PubMed]
- 105.Celona B, Dollen JV, Vatsavayai SC, Kashima R, Johnson JR, Tang AA, et al. Suppression of C9orf72 RNA repeat-induced neurotoxicity by the ALS-associated RNA-binding protein Zfp106. eLife. 2017;6. [DOI] [PMC free article] [PubMed]
- 106.Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain: a journal of neurology. 2014;137(Pt 7):2040–51. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, et al. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathologica Communications. 2014;2:70. doi: 10.1186/2051-5960-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathologica. 2013;126(6):859–79. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- 109.Sakae N, Bieniek KF, Zhang YJ, Ross K, Gendron TF, Murray ME, et al. Poly-GR dipeptide repeat polymers correlate with neurodegeneration and Clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathologica Communications. 2018;6(1):63. doi: 10.1186/s40478-018-0564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nature Neuroscience. 2015;18(9):1226–9. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Human Molecular Genetics. 2015;24(9):2426–41. doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- 112.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167(3):774–88. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):260–5. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, et al. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Human Molecular Genetics. 2016;25(9):1803–13. doi: 10.1093/hmg/ddw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, et al. Aggregation-pronec9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathologica. 2014;128(4):505–24. [DOI] [PMC free article] [PubMed]
- 116.May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, et al. C9orf72 FTLD/ALS-associatedGly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathologica. 2014;128(4):485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Human Molecular Genetics. 2015;24(6):1630–45. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- 118.Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathologica. 2015;130(4):525–35. doi: 10.1007/s00401-015-1448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovicic A, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Scientific Reports. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Swaminathan A, Bouffard M, Liao M, Ryan S, Callister JB, Pickering-Brown SM, et al. Expression of C9orf72-related dipeptides impairs motor function in a vertebrate model. Human Molecular Genetics. 2018;27(10):1754–62. doi: 10.1093/hmg/ddy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ohki Y, Wenninger-Weinzierl A, Hruscha A, Asakawa K, Kawakami K, Haass C, et al. Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Molecular Neurodegeneration. 2017;12(1):6. doi: 10.1186/s13024-016-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nature Neuroscience. 2016;19(5):668–77. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, et al. Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathologica. 2017;134(2):241–54. doi: 10.1007/s00401-017-1711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nature Medicine. 2018;24(8):1136–42. doi: 10.1038/s41591-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang YJ, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, et al. Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science (New York, NY). 2019;363(6428). [DOI] [PMC free article] [PubMed]
- 126.Choi SY, Lopez-Gonzalez R, Krishnan G, Phillips HL, Li AN, Seeley WW, et al. C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nature Neuroscience. 2019;22(6):851–62. doi: 10.1038/s41593-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hao Z, Liu L, Tao Z, Wang R, Ren H, Sun H, et al. Motor dysfunction and neurodegeneration in a C9orf72 mouse line expressing poly-PR. Nature Communications. 2019;10(1):2906. doi: 10.1038/s41467-019-10956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science (New York, NY) 2014;345(6201):1139–45. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Flores BN, Dulchavsky ME, Krans A, Sawaya MR, Paulson HL, Todd PK, et al. Distinct C9orf72-Associated Dipeptide Repeat Structures Correlate with Neuronal Toxicity. PLoS One. 2016;11(10):e0165084. doi: 10.1371/journal.pone.0165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shao Q, Liang C, Chang Q, Zhang W, Yang M, Chen JF. C9orf72 deficiency promotes motor deficits of a C9ALS/FTD mouse model in a dose-dependent manner. Acta Neuropathologica Communications. 2019;7(1):32. doi: 10.1186/s40478-019-0685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou Q, Lehmer C, Michaelsen M, Mori K, Alterauge D, Baumjohann D, et al. Antibodies inhibit transmission and aggregation of C9orf72 poly-GA dipeptide repeat proteins. EMBO Mol Med. 2017;9(5):687–702. doi: 10.15252/emmm.201607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nature Communications. 2018;9(1):152. doi: 10.1038/s41467-017-02643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wojciechowska M, Olejniczak M, Galka-Marciniak P, Jazurek M, Krzyzosiak WJ. RAN translation and frameshifting as translational challenges at simple repeats of human neurodegenerative disorders. Nucleic Acids Research. 2014;42(19):11849–64. doi: 10.1093/nar/gku794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Knockenhauer KE, Schwartz TU. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell. 2016;164(6):1162–71. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kramer NJ, Haney MS, Morgens DW, Jovicic A, Couthouis J, Li A, et al. CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nature Genetics. 2018;50(4):603–12. doi: 10.1038/s41588-018-0070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(7):E1111–E7. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eftekharzadeh B, Daigle JG, Kapinos LE, Coyne A, Schiantarelli J, Carlomagno Y, et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron. 2019;101(2):349. doi: 10.1016/j.neuron.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron. 2017;94(1):93–107.e6. doi: 10.1016/j.neuron.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, et al. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron. 2017;94(1):48–57.e4. doi: 10.1016/j.neuron.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mizielinska S, Ridler CE, Balendra R, Thoeng A, Woodling NS, Grasser FA, et al. Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration. Acta Neuropathologica Communications. 2017;5(1):29. doi: 10.1186/s40478-017-0432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, et al. Poly(GR) in C9ORF72-RelatedALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron. 2016;92(2):383–91. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Farg MA, Konopka A, Soo KY, Ito D, Atkin JD. The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Human Molecular Genetics. 2017;26(15):2882–96. doi: 10.1093/hmg/ddx170. [DOI] [PubMed] [Google Scholar]
- 143.Lopez-Gonzalez R, Yang D, Pribadi M, Kim TS, Krishnan G, Choi SY, et al. Partial inhibition of the overactivated Ku80-dependent DNA repair pathway rescues neurodegeneration in C9ORF72-ALS/FTD. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(19):9628–33. doi: 10.1073/pnas.1901313116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. The Journal of Cell Biology. 2013;201(3):361–72. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, et al. ALS/FTDMutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88(4):678–90. [DOI] [PMC free article] [PubMed]
- 147.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162(5):1066–77. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 148.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Molecular Cell. 2017;65(6):1044–55. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, et al. Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell. 2018;173(4):958–71. doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546(7657):243–7. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fay MM, Anderson PJ, Ivanov P. ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Reports. 2017;21(12):3573–84. doi: 10.1016/j.celrep.2017.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nature Communications. 2017;8(1):2005. doi: 10.1038/s41467-017-02200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, et al. In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell. 2018;172(4):696–705. doi: 10.1016/j.cell.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bennett CF, Krainer AR, Cleveland DW. Antisense Oligonucleotide Therapies for Neurodegenerative Diseases. Annual Review of Neuroscience. 2019;42:385–406. doi: 10.1146/annurev-neuro-070918-050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biology. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurology. 2013;12(5):435–42. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. The New England Journal of Medicine. 2019;380(24):2307–16. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 159.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nature Reviews Drug Discovery. 2019;18(5):358–78. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hu J, Rigo F, Prakash TP, Corey DR. Recognition of c9orf72 Mutant RNA by Single-Stranded Silencing RNAs. Nucleic Acid Ther. 2017;27(2):87–94. doi: 10.1089/nat.2016.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu J, Liu J, Li L, Gagnon KT, Corey DR. Engineering Duplex RNAs for Challenging Targets: Recognition of GGGGCC/CCCCGG Repeats at the ALS/FTD C9orf72 Locus. Chem Biol. 2015;22(11):1505–11. doi: 10.1016/j.chembiol.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Martier R, Liefhebber JM, Garcia-Osta A, Miniarikova J, Cuadrado-Tejedor M, Espelosin M, et al. Targeting RNA-Mediated Toxicity in C9orf72 ALS and/or FTD by RNAi-Based Gene Therapy. Mol Ther Nucleic Acids. 2019;16:26–37. doi: 10.1016/j.omtn.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Martier R, Liefhebber JM, Miniarikova J, van der Zon T, Snapper J, Kolder I, et al. Artificial MicroRNAs Targeting C9orf72 Can Reduce Accumulation of Intra-nuclear Transcripts in ALS and FTD Patients. Mol Ther Nucleic Acids. 2019;14:593–608. doi: 10.1016/j.omtn.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zamiri B, Reddy K, Macgregor RB, Jr, Pearson CE. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. The Journal of Biological Chemistry. 2014;289(8):4653–9. doi: 10.1074/jbc.C113.502336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang ZF, Ursu A, Childs-Disney JL, Guertler R, Yang WY, Bernat V, et al. The Hairpin Form of r(G4C2)(exp) in c9ALS/FTD Is Repeat-Associated Non-ATG Translated and a Target for Bioactive Small Molecules. Cell Chemical Biology. 2019;26(2):179–90. doi: 10.1016/j.chembiol.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Van Oss SB, Cucinotta CE, Arndt KM. Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation. Trends Biochem Sci. 2017;42(10):788–98. doi: 10.1016/j.tibs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kramer NJ, Carlomagno Y, Zhang YJ, Almeida S, Cook CN, Gendron TF, et al. Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science (New York, NY) 2016;353(6300):708–12. doi: 10.1126/science.aaf7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Goodman LD, Prudencio M, Kramer NJ, Martinez-Ramirez LF, Srinivasan AR, Lan M, et al. Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nature Neuroscience. 2019;22(6):863–74. doi: 10.1038/s41593-019-0396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Naguib A, Sandmann T, Yi F, Watts RJ, Lewcock JW, Dowdle WE. SUPT4H1 Depletion Leads to a Global Reduction in RNA. Cell Reports. 2019;26(1):45–53. doi: 10.1016/j.celrep.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 171.Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nature Reviews Neuroscience. 2016;17(1):36–44. doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pinto BS, Saxena T, Oliveira R, Mendez-Gomez HR, Cleary JD, Denes LT, et al. Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9. Molecular Cell. 2017;68(3):479–90. doi: 10.1016/j.molcel.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, et al. Elimination of Toxic Microsatellite Repeat Expansion RNA by RNA-Targeting Cas9. Cell. 2017;170(5):899–912. doi: 10.1016/j.cell.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation. Nature Communications. 2018;9(1):51. doi: 10.1038/s41467-017-02495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yamada SB, Gendron TF, Niccoli T, Genuth NR, Grosely R, Shi Y, et al. RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nature Neuroscience. 2019;22(9):1383–8. doi: 10.1038/s41593-019-0455-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cristofani R, Crippa V, Vezzoli G, Rusmini P, Galbiati M, Cicardi ME, et al. The small heat shock protein B8 (HSPB8) efficiently removes aggregating species of dipeptides produced in C9ORF72-related neurodegenerative diseases. Cell Stress & Chaperones. 2018;23(1):1–12. doi: 10.1007/s12192-017-0806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gallardo G, Holtzman DM. Antibody Therapeutics Targeting Abeta and Tau. Cold Spring Harbor Perspectives in Medicine. 2017;7(10). [DOI] [PMC free article] [PubMed]
- 178.Hock EM, Polymenidou M. Prion-like propagation as a pathogenic principle in frontotemporal dementia. Journal of Neurochemistry. 2016;138(Suppl 1):163–83. doi: 10.1111/jnc.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ravits J. Focality, stochasticity and neuroanatomic propagation in ALS pathogenesis. Experimental Neurology. 2014;262(Pt B):121–6. doi: 10.1016/j.expneurol.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 180.Chang YJ, Jeng US, Chiang YL, Hwang IS, Chen YR. The Glycine-Alanine Dipeptide Repeat from C9orf72 Hexanucleotide Expansions Forms Toxic Amyloids Possessing Cell-to-Cell Transmission Properties. The Journal of Biological Chemistry. 2016;291(10):4903–11. doi: 10.1074/jbc.M115.694273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, et al. SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nature Communications. 2017;8:16063. doi: 10.1038/ncomms16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Archbold HC, Jackson KL, Arora A, Weskamp K, Tank EM, Li X, et al. TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Scientific Reports. 2018;8(1):4606. doi: 10.1038/s41598-018-22858-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544(7650):367–71. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Floeter MK, Gendron TF. Biomarkers for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Associated With Hexanucleotide Expansion Mutations in C9orf72. Frontiers in Neurology. 2018;9:1063. doi: 10.3389/fneur.2018.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 506 kb)
(PDF 515 kb)