Abstract
Huntington’s disease (HD) is an autosomal dominant disorder caused by an expansion in the trinucleotide CAG repeat in exon-1 in the huntingtin gene, located on chromosome 4. When the number of trinucleotide CAG exceeds 40 repeats, disease invariably is manifested, characterized by motor, cognitive, and psychiatric symptoms. The huntingtin (Htt) protein and its mutant form (mutant huntingtin, mHtt) are ubiquitously expressed but although multiple brain regions are affected, the most vulnerable brain region is the striatum. Striatal medium-sized spiny neurons (MSNs) preferentially degenerate, followed by the cortical pyramidal neurons located in layers V and VI. Proposed HD pathogenic mechanisms include, but are not restricted to, excitotoxicity, neurotrophic support deficits, collapse of the protein degradation mechanisms, mitochondrial dysfunction, transcriptional alterations, and disorders of myelin. Studies performed in cell type-specific and regionally selective HD mouse models implicate both MSN cell-autonomous properties and cell–cell interactions, particularly corticostriatal but also with non-neuronal cell types. Here, we review the intrinsic properties of MSNs that contribute to their selective vulnerability and in addition, we discuss how astrocytes, microglia, and oligodendrocytes, together with aberrant corticostriatal connectivity, contribute to HD pathophysiology. In addition, mHtt causes cell-autonomous dysfunction in cell types other than MSNs. These findings have implications in terms of therapeutic strategies aimed at preventing neuronal dysfunction and degeneration.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00782-9) contains supplementary material, which is available to authorized users.
Key Words: Cell-autonomous, non-cell-autonomous, glia, medium spiny neurons, corticostriatal
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder characterized by motor deficits and involuntary movements, most commonly chorea at early stages, cognitive dysfunction, and psychiatric manifestations [1]. HD is caused by an expansion in the trinucleotide CAG repeat in exon-1 in the huntingtin gene, located on chromosome 4. When the number of trinucleotide CAG repeats exceeds 40, HD is fully manifested [2], as the polyglutamine (polyQ) tract confers neurotoxicity apparently via both gain- and loss-of-function. An inverse correlation exists between the CAG repeat length and age of onset [3]. Mutant huntingtin (mHtt) is expressed in all cells [4], but the gamma-aminobutyric acid (GABA)ergic striatal projection medium spiny neuron (MSNs) is the most, or differentially, vulnerable to dysfunction and neurodegeneration [5, 6]. Cortical pyramidal neurons in layers V and VI are also highly perturbed by the expression of mHtt [7]. A long-standing question in understanding HD is whether MSN vulnerability is cell-autonomous or non-cell-autonomous [8], a question which now extends to cortical neurons and non-neuronal cell types.
mHtt causes dysfunction of protein degradation systems, including autophagy and the ubiquitin proteasome system [9, 10], leading to altered proteostasis. A major hallmark of HD is the presence of mHtt aggregates, and although not necessarily toxic on their own, Htt is a very large protein with the potential to interact and bind with many other proteins, altering their function [11]. Pre-deposition, oligomeric mHtt species may also comprise a principal component of mHtt toxicity [12, 13], similar to amyloid in Alzheimer’s disease [14]. Transcriptional alterations in HD are partly attributed to mHtt sequestration of transcription factors [15–18]. Through interaction with mitochondrial proteins, mHtt perturbs mitochondrial function and induces bioenergetic deficits and calcium buffering abnormalities [19]. Finally, mHtt induces presynaptic neurotrophin deficits and postsynaptic signaling deficits [20, 21] and glutamate-induced excitotoxicity likely has pre- and postsynaptic etiologies [22]. These processes cannot be fully attributed to restricted expression of mHtt in MSNs and may not even be restricted to the corticostriatal system as alterations in the dopamine system are also implicated in HD [23]. It is impossible to cover all the possible interactions, but this review is focused on cell-autonomous and non-cell-autonomous mHtt-induced alterations in MSNs, corticostriatal communication, and other non-neuronal contributions to MSN dysfunction and neurodegeneration. It is therefore also of course impossible to create sections that do not overlap. For example, in discussing excitotoxicity, there is obvious overlap with the role of astrocytes.
HD Mouse “Models” to Explore Cell-Specific Vulnerability
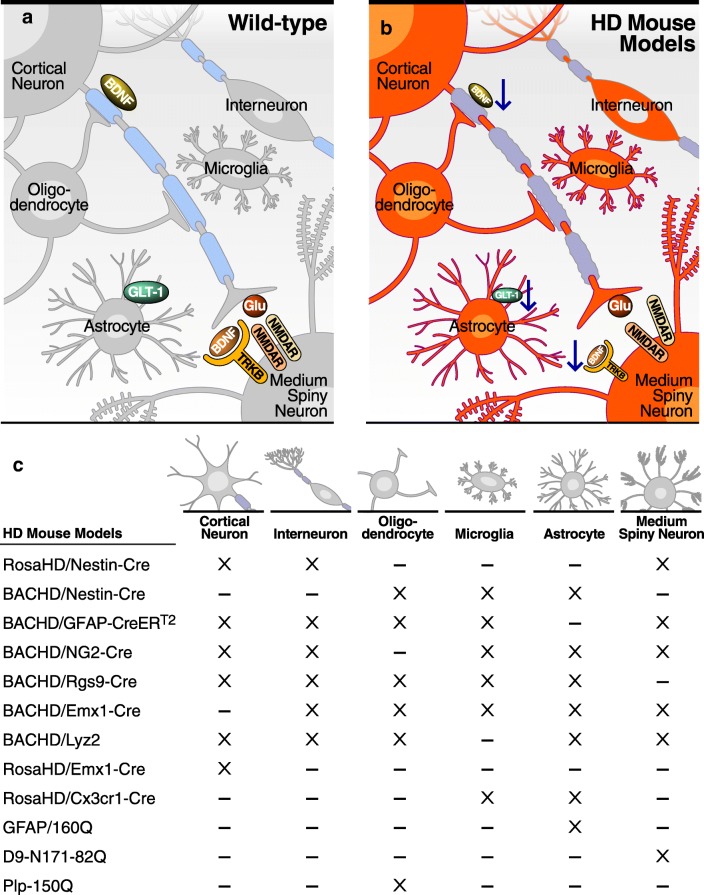
In order to decipher cell-autonomous and non-cell-autonomous pathogenic mechanisms driving HD pathophysiology, novel mouse lines have been generated with cell-specific expression or deletion of mHtt. Phenotypes of animal models discussed herein are summarized in Table 1. Except for knock-in (KI) models, comparisons are hampered by differences in promoter, level of expression of transgene, and importantly, length of Htt, i.e., fragment versus full length (FL) (Fig. 1).
Table 1.
Highlighted mouse lines contributing to study of cell-autonomous and non-cell-autonomous impact of mHtt on HD-like phenotypes
| Pan-cellular | ||||||||
| Motor deficits | Inclusions | Neuronal loss | Abnormal white matter | Microglial/astroglial activation | Transcriptional abnormalities | Mitochondrial | Corticostriatal synapse abnormalities | |
| R6 [24] | Yes | Yes | Absent [25] | Yes | Yes | Yes | Yes | Yes [26] |
| YAC [27] | Yes | Yes | Yes | Yes [28] | Yes [29] | Yes [30] | Yes [31] | Yes |
| BAC [32] | Yes | Yes | Presence of dark neurons | Yes | Not evaluated | Yes | Not evaluated | Yes [33] |
| Pan-neuronal | ||||||||
| Motor deficits | Inclusions | Neuronal loss | Abnormal white matter | Microglial/astroglial activation | Transcriptional abnormalities | Mitochondrial | Corticostriatal synapse abnormalities | |
| Prion [34] | Yes | Yes | Yes | Not evaluated | Astrogliosis | Yes | Yes | Not evaluated |
| RosaHD/Nestin-Cre [35] | Yes | Yes | Dark neurons | Not evaluated | Astrogliosis | Not evaluated | Not evaluated | Yes |
| Neuronal subtype-specific | ||||||||
| Motor deficits | Inclusions | Neuronal loss | Abnormal white matter | Microglial/astroglial activation | Transcriptional abnormalities | Mitochondrial | Corticostriatal synapse abnormalities | |
| D9-N171-82Q [36] | Yes | Yes | No striatal volume loss | Not evaluated | Not evaluated (no astrogliosis) | Yes | Yes [37] | Yes [37] |
| RosaHD/Emx1-Cre [35] | Absent | Yes | Absent (only degeneration vacuoles) | Not evaluated | Not evaluated (no astrogliosis) | Not evaluated | Not evaluated | No |
| mHtt neuronal subtype-specific deletions | ||||||||
| Motor deficits | Inclusions | Neuronal loss | Abnormal white matter | Microglial/astroglial activation | Transcriptional abnormalities | Mitochondrial | Corticostriatal synapse abnormalities | |
| BACHD/Rgs9-Cre [38] | Yes | Yes | Not evaluated | Not evaluated | Not evaluated | Not evaluated | Not evaluated | Yes? |
| BACHD/Emx1-Cre [38] | Improved | Not evaluated | Not evaluated | Not evaluated | Not evaluated | Not evaluated | Not evaluated | No |
| BACHD/Nestin-Cre [35] | Improved | Not evaluated | Recovered striatal volume | Not evaluated | Yes | Not evaluated | Not evaluated | Not evaluated |
| Non-neuronal subtype-specific deletions | ||||||||
| Motor deficits | Inclusions | Neuronal loss | Abnormal white matter | Microglial activation | Transcriptional abnormalities | Mitochondrial | Corticostriatal synapse abnormalities | |
| BACHD/GFAP-CreERT2 [39] | Improved | Not evaluated | Partial recovery of striatal volume | Not evaluated | Not evaluated | Not evaluated | Not evaluated | No |
| BACHD/Lyz2 [40] | Yes | Not evaluated | No recovery of striatal volume | Not evaluated | Absent | Not evaluated | Not evaluated | Not evaluated |
| BACHD/NG2-Cre [41] | Improved | Not evaluated | No recovery striatal volume | No | Not evaluated | Improved | Not evaluated | Not evaluated |
| Non-neuronal specific overexpressers | ||||||||
| Motor deficits | Inclusions | Neuronal loss | Abnormal white matter | Microglial activation | Transcriptional abnormalities | Mitochondrial | Corticostriatal synapse abnormalities | |
| GFAP/160Q [42] | Yes | Yes | No | Not evaluated | Not evaluated | qPCR for specific astroglial genes | Not evaluated | Not evaluated |
| RosaHD/Cx3cr1-Cre [43] | Not evaluated | Not evaluated | Not evaluated | Not evaluated | Yes | Yes | Not evaluated | Not evaluated |
| Plp-150Q [44] | Yes | Yes | Not evaluated | Yes | Not evaluated | Yes | Not evaluated | Not evaluated |
Fig. 1.
Schematic representation of cell-specific contribution to striatal and corticostriatal function in (A) wild-type and (B) HD pancellular context. (C) Cell-type-specific mHtt overexpressers and deleters mouse models. “X”: mHtt expression; “-”: no mHtt expression
Pan-cellular HD Models
Creation of the original HD mouse models, and later KI models, resulted in pan-cellular models with either overexpression or endogenous levels of mHtt. R6 mice, generated by Bates’ group in 1996 [24], were the first HD transgenic mouse models. Both R6/1 and R6/2 mouse lines express 67 amino acids of the N-terminal human protein, driven by the human HTT promoter, and a single copy of the transgene with either 115 CAG repeats or three copies with 144 repeats, respectively [24, 45]. Both lines display weight loss, enlarged cerebral ventricles, neuronal atrophy, progressive increase of mHtt aggregates, motor alterations, and cognitive deficits. Cell death is not replicated in these lines [25]. Later, full-length mHtt transgenic mice, via yeast or bacterial artificial chromosomes (YAC and BAC), were generated [27, 32]. They express full-length mHtt and contain all the introns, exons, and regulatory sequences, with 72 or 128 CAG repeats in the YAC and 97 CAG/CAA in the BACHD. Insertion sites are unknown. Both models develop motor deficits, neuronal degeneration, corticostriatal synaptic dysfunction, and mHtt aggregates. Contemporaneously, mHtt KI mice were generated [46–48], in which exon 1 of the mouse gene was replaced by the human HTT exon 1 with an expanded number of CAG repeats, resulting in the expression of the HD mutation in its appropriate genomic context. They recapitulate much of the HD phenotype although with a slower progression than the over-expressers, and with less neuronal loss than is seen in human HD. None of these pan-cellular models, however, is an effective tool to investigate specific cell subtype-specific contribution to HD physiopathology, without further manipulations, discussed below.
Pan-neuronal mHtt Fragment Transgenic Models
The Prion promoter was used to produce a mouse line, Prp-N171-82Q, which expresses the N-terminal 171 amino acids of mHtt with a CAG repeat of 82, principally in neurons [49]. This model has been valuable for unraveling the contribution of neuronal versus non-neuronal cells in HD. The Nestin-Cre mouse was used for this purpose as well in a line in which an Exon1 mHtt fragment was inserted into the Rosa26 locus and mHtt expression was entirely dependent on Cre-mediated excision [35]. It should be noted that the Nestin promoter also directs expression to glial progenitors. The Prp-N171-82Q mice develop motor deficits, mHtt aggregates, astrogliosis, decreased lifespan, and neuronal degeneration, as demonstrated with cleaved caspase-3 immunostaining [34, 49]. The Nestin-Cre Exon1 mice driven by the Rosa26 promoter present age-dependent motor deficits, mHtt aggregates, gliosis, degenerating “dark neurons” and reduced spontaneous IPSC frequency [35]. These data imply that these deficits are neuronal cell-autonomous.
Neuronal Subtype-Specific mHtt Fragment and Full-Length Mice
Two models were created to determine if there are cell-autonomous effects of mHtt in MSNs. The RosaHD-Dlx5/6-Cre mouse [50] expresses Exon1 mHtt only in MSNs and in a subset of cortical and olfactory interneurons during embryonic development. Although RosaHD-Dlx5/6-Cre mice accumulate mHtt aggregates and develop minor glutamatergic NMDA receptor alterations, they do not develop motor deficits. We generated the D9-N171-82Q line, which expresses a similar mHtt fragment as does the prion promoter mouse, but driven by select regulatory regions of the Ppp1r1b/DARPP-32 gene, restricting transgene expression to MSNs and Purkinje cells [36]. Comparisons between pan-cellular and pan-neuronal models demonstrated that mHtt expression in neurons, and selectively in MSNs in the forebrain, is sufficient to induce certain aspects of the murine HD molecular and behavioral phenotypes. Inclusions developed and comparison of striatal gene expression to other models revealed that transcriptomic abnormalities are induced largely, intrinsically by mHtt, independent of cortical or glial expression [51], as is complex II mitochondrial dysfunction [37]. Corticostriatal synaptic dysfunction, albeit present, is minimal [51].
To further investigate the corticostriatal connection, cell-autonomous, and cell–cell interaction mechanisms in HD, Yang and colleagues [35] used the Emx1-Cre mouse crossed with the Exon1/Rosa26 mouse described above, thereby restricting mHtt expression to cortical pyramidal neurons. Although Emx-1-Cre HD mice developed mHtt aggregation in cortical neurons, they did not exhibit electrophysiologic or motor deficits, implying a role for interactions between cortical interneurons and pyramidal neurons in mHtt-induced cortical dysfunction [35]. Unfortunately, not all neuronal subtype-specific models were analyzed for microgliosis, astrogliosis, or transcriptomic changes.
The BACHD mouse described above [32] is engineered with loxP sites flanking Exon1, allowing for cell-type specific and regional deletion, and simultaneously therefore the generation in many cases of cell-type specific overexpression models. Striatal (Rgs9) or cortical (Emx-1) Cre recombinase mice were crossed with the BACHD mouse to delete mHtt from MSN or cortical projection neurons, respectively [38]. These studies, discussed in more detail below, demonstrated the requirement for mHtt expression in projection neurons for a more representative and complete HD phenotype in MSNs, most notably including typical electrophysiologic abnormalities. It is useful also to consider these mice relative to those in which mHtt in non-neuronal cells has been investigated, as Rgs9-Cre-mediated deletion, for example, leaves striatal glial mHtt expression intact.
Deletion and Overexpression of mHtt in Non-neuronal Cells
The BACHD mouse and cell type-specific promoters have allowed for the creation of mice with mHtt either specifically deleted in non-neuronal subtypes on a pan-cellular mHtt background, or selectively overexpressed on an otherwise wild-type background.
Astrocytes
There are astrocytic changes in HD and in many pan-cellular and pan-neuronal HD mouse lines, including morphological changes, e.g., thicker processes and larger soma containing mHtt aggregates, and impaired glutamate metabolism and potassium homeostasis [52], discussed in detail below. The GFAP-CreERT2 transgenic mouse was used to conditionally delete mHtt in astrocytes in the BACHD mouse [39], and the GFAP promoter was used to drive mHtt Exon1 expression in astrocytes [42]. Astrocytic mHtt contributes to the BACHD phenotype as its deletion improved motor, psychiatric, pathological, and electrophysiological abnormalities [39]. Overexpression of mHtt in astrocytes led to decreased lifespan and body weight, as well as motor and transcriptional dysfunction [42], specifically implicating cell-autonomous and non-cell-autonomous effects of mHtt in astrocytes.
Microglia
Microgliosis is also observed in HD brains, and mHtt has therefore been deleted or expressed selectively in microglial cells in the BACHD mouse [40], and in this setting, no contribution to phenotype was observed with either modulation. Conversely, however, mHtt transgene expression driven with a microglial-specific promoter led to cell-autonomous microglial activation and heightened inflammatory response to LPS in Q175 KI mice, thereby suggesting cell-autonomous effects of mHtt on microglia. However, it is not clear that there was otherwise a phenotypic effect without additional stimuli [43]. The possible role of neuroinflammation in HD is discussed further below.
Oligodendroglia
Finally, because myelin alterations are also present in human HD brain and mouse models of HD [28, 53–55], mHtt has been selectively expressed and repressed in oligodendrocytes. The Plp promoter was used to express N-terminal mHtt with 150 glutamines in murine oligodendroglia [44] and, the Ng2-Cre mouse line was used to delete oligodendroglial mHtt expression in BACHD mice [41]. In brief, with additional details provided below, both cell-autonomous effects in oligodendrocytes and non-cell-autonomous effects on neurons arise from mHtt expression in oligodendrocytes. These include transcriptional abnormalities that potentially explain the myelin deficit in HD and which could play a pivotal role in axonal integrity and associated motor coordination deficits.
Intrinsic Vulnerability of MSNs
The cell-autonomous nature of some HD phenotypes in MSNs and their selective vulnerability raises the question as to which MSN characteristics may induce this dysfunction. MSNs are unique in that they receive a massive glutamatergic, excitatory input from the cortex in addition to all being dopaminoceptive. Much effort has been expended in determining whether vulnerability is due to this combination of afferents, and/or MSN-enriched gene expression patterns uniquely sensitive to mHtt. In addition to being selectively vulnerable to mHtt expression, MSNs are also highly sensitive to quinolinic acid [56] or 3-nitropropionic acid (3-NP; [57]), which were utilized as the first models of HD prior to identification of the gene and its mutation [2]. Lesions from these toxins are due to excitotoxicity or inhibition of the citric acid cycle, respectively, yielding clues to cell intrinsic properties of MSNs leading to vulnerability in HD. Proteomic and transcriptomic studies have identified proteins selectively enriched in striatal neurons which might account for MSN-specific vulnerability to a toxic stimulus [58–66] and many of the candidates are involved in the pathways mediating neurotransmission and energy utilization. Unsurprisingly, there are multiple candidates, e.g., Ptpn5, Rhes, Pde10A, Foxp2, and Bcl11b, some of which we will specifically discuss. Individual validation is required for each gene.
Ptpn5, which encodes for STEP (striatal-enriched protein tyrosine phosphatase), is an example in which a targeted validation approach has been used to assay the role of a striatal-enriched protein that is not known to directly interact with Htt. STEP protein regulates the endocytosis of NMDA receptor subunits [67] and via dephosphorylation modulates intracellular signaling pathways, including those requiring mitogen activated protein kinase (MAPK) or Fyn [68, 69]. In R6/1 and several other mouse models, phosphorylation of STEP is increased, and STEP is thereby inactivated, hypothetically exerting a compensatory, neuroprotective role inducing resistance to excitotoxicity [70]. Genetic and pharmacologic inhibition of STEP in the R6/1 mouse delays the appearance of motor dysfunction, improves cognitive function, and decreases the size of mHtt aggregates [71]. Contrary to these data, in a full-length HD model, STEP activity is actually increased, hypothetically increasing pathogenic, extrasynaptic NMDA receptor localization, which is reduced by inactivation of STEP [72]. These studies highlight the need to look at multiple models, including fragment and full-length, and at different disease stages.
PDE10A is a second example of a protein that does not interact with mHtt protein but has a relevant role in HD pathology. This phosphodiesterase directly impacts cyclic AMP levels and consequently cAMP response element-binding protein (CREB), which modulates gene expression. Pde10A expression is increased in R6/2 MSNs [73], and its inhibition improves motor function, prevents striatal atrophy, reduces microgliosis, and restores brain-derived neurotrophic factor (BDNF) and active CREB protein levels in the R6/2 model [73–75]. The basic function of PDE10A in fact makes it a therapeutic target in multiple hyperkinetic movement disorders [76].
Amongst the many Htt interacting proteins, Ras homologue enriched in striatum (Rhes) is selectively expressed within the striatum and increases mHtt toxicity via sumoylation [77]. However, using different models, there are again discrepant reports regarding the role of this protein in HD. Rhes deletion is neuroprotective in a 3-NP model, ameliorating motor dysfunction and preventing striatal degeneration [78]. Silencing of Rhes in HD genetic mouse models, however, exacerbated both striatal pathology and the “psychiatric” phenotype [79] and, in agreement with these data, its overexpression improved motor function and brain pathology [80].
Two well-characterized transcription factors (TFs) with enriched expression in the striatum, Bcl11b/Ctip2 and Foxp2, are decreased in some HD models, although Foxp2 is not decreased in patients. Both appear to interact with mHtt, as determined by co-immunoprecipitation [81, 82]. Bcl11b overexpression in STHdhQ111 cells rescues mitochondrial metabolic activity [81], but only Foxp2 contribution to HD pathology has been evaluated in vivo. Interestingly, Foxp2 overexpression in the BACHD model rescues motor coordination deficits, whereas knockdown in wild-type mice leads to symptoms overlapping with those of HD, both unsurprisingly implying cell-autonomous effects [82]. Much work, however, remains to determine whether any individual striatal-enriched gene, or their combination, actually accounts for MSN vulnerability.
Differential Vulnerability of Striatal Neuronal Subtypes
MSNs are morphologically homogeneous, but are sub-grouped based on molecular properties, and origins and destinations of afferent and efferent projections. The striatum also contains heterogeneous interneuron subtypes, all of which are considered relatively resistant to neurodegeneration in HD, although they may be dysfunctional. Amongst MSN subtypes, there is some evidence of further differential vulnerability. MSNs are divided approximated 50/50 amongst direct pathway MSNs (dMSNs), which project to the substantia nigra and express the dopamine type 1 receptor (Drd1/D1R), substance P and dynorphin [83] and the apparently more vulnerable indirect pathway MSNs (iMSNs), which project to the globus pallidus pars externa and express D2-type dopamine receptors (Drd2/D2R) and enkephalin, [84, 85]. Because dMSNs and iMSNs express similar levels of mHtt [86], other mechanisms must account for their different responses in HD. Physiologically, iMSNs are more excitable than dMSNs, perhaps due to higher expression of NMDA receptors [87]. dMSNs also display lower input and membrane resistance, higher branching complexity, along with increased whole-cell capacitance relative to iMSNs, phenomena that may also contribute to differences in excitability [88]. As already inferred, iMSNs and dMSNs also differ in their transcriptomes, and a selectively expressed gene could cell autonomously confer either vulnerability or protection. For example, iMSNs express higher levels of the potassium channel Kir2.3 subunit that might contribute to relative excitability [89], and higher levels of the BDNF receptor, TrkB [90, 91], suggesting greater susceptibility to a BDNF deficit. Déglon and colleagues [92] identified additional genes selectively expressed in dMSNs and specifically downregulated in Hdh KI mouse striatum, e.g., glutathione peroxidase Gpx6, interferon alpha-inducible protein 27 like 2B (Ifi27l2b) and protein tyrosine phosphatase Ptpn7. To date, only Gpx6 has been experimentally validated, in addition to having been identified by others as modulating the HD phenotype using a synthetic lethal screen. Overexpression in R6/2 HD mice improved motor function and ameliorated biochemical abnormalities [93], validating this approach, and again pointing to cell-autonomous actions of neuronal genes altered in HD.
Animal models poorly recapitulate the early loss of iMSNs observed in HD patients, but electrophysiological assays of iMSNs and dMSNs at pre- and postsymptomatic stages support differential vulnerability, including of their output structures [94, 95], although not always in the same direction as in human samples. Morphologically, corticostriatal inputs to dMSNs are altered whereas inputs to iMSNs are spared in HD140Q KI mice [96]. Similarly, dMSNs obtained from Q175 heterozygous symptomatic KI mice display reduced action potential amplitude and decreased rheobase, along with selective spine loss, more so than do iMSNs [97]. These are further examples of how mouse models, although very valuable in unraveling pathophysiology in HD, do not perfectly recapitulate human disease.
Generation of iPSCs to Unravel Cell-Autonomous Versus Non-cell-Autonomous Mechanisms in HD
Human-induced pluripotent stem cells (hiPSCs), somatic cells reprogrammed to a pluripotent state, are now a critical and unique cellular model to decipher pathogenic mechanisms of HD and are particularly useful in the study of cell-autonomous and non-cell-autonomous mechanisms, based on their differentiation into single or multiple cell types, including organoids. In less than a decade, many hiPSC lines have been generated by multiple groups, with different numbers of CAG repeats [98–106]. The mutant locus may be corrected by either homozygous recombination [107] or more commonly now, CrispR [108–110], creating control lines in isogenic backgrounds, thereby limiting the experimental variables. In all cases, when differentiated into “MSNs”, these cell lines recapitulate cell-autonomous transcriptional alterations, demonstrate increased cell death with excitotoxicity and/or neurotrophin deficiency including in a CAG repeat-dependent manner, and have bioenergetic deficits, all likely HD pathophysiologic mechanisms that are discussed below. iPSC-induced neural stem cells (NSCs) also recapitulate aspects of the HD phenotype, including altered cholesterol levels and increased levels of reactive oxygen species [111]. In fact, a highly novel aspect emerging from research using iPSCs is the revelation of CAG-dependent abnormalities very early in neuronal development, lending support to the notion that HD has a strong developmental component [112, 113].
Notably, cortical neurons differentiated from HD iPSCs also demonstrate altered transcriptional profiles and morphology, revealing cell-autonomous and developmental dysfunction in neuronal subtypes other than MSNs [114, 115]. With the rapid technical progress in the iPSC field, reports are soon likely to emerge characterizing glial subtypes and organoids derived from these lines.
Pathogenic Mechanisms: Cell-Autonomous, Non-cell-Autonomous, or Both
As current knowledge regarding intrinsic properties of MSNs does not adequately explain their selective vulnerability in HD, there is great interest in non-cell-autonomous and circuit-based pathogenic mechanisms in HD, including the role of non-neuronal cells. This is similar to the thought processes for other neurodegenerative disorders, e.g., amyotrophic lateral sclerosis [116–118] and Parkinson’s disease [119, 120]. Here, we will review the proposed HD pathogenic mechanisms in the context of cell-autonomous versus non-cell-autonomous etiology.
Aberrant Corticostriatal Communication
The MSNs receive major glutamatergic input from the cortex [121, 122], and these afferents also carry much of their neurotrophic support in the form of BDNF [123, 124], highlighting the direct impact of cortical projection neurons on MSN function and survival. Corticostriatal communication deficits are detectable in early stages of HD [125], and within the cortex, human pyramidal cortical neurons from layers V and VI are particularly vulnerable, with a 30% reduction in number [126]. As in the striatum, cortical interneurons are largely spared from degeneration [127], although data from a mouse model suggest that interneuron-pyramidal interactions are required for pyramidal neuron dysfunction [35]. Evidence from HD mouse models exists suggesting that cortical alterations, involving mainly cortical layers II/III, may actually precede striatal pathology [128, 129]. For example, there are dendritic alterations including a reduced number of spines in cortical pyramidal neurons in HdhQ7/Q111 KI and R6/2 mice, prior to striatal structural changes [130, 131]. The physiologic status of cortical neurons in HD models is highly complex and may change with disease stage. Functionally, cortical neurons in R6/1 mice have, at various stages of disease, altered resting membrane potential, input resistance and cell capacitance [26, 132] along with abnormal firing rate and an imbalance between excitatory and inhibitory inputs [26, 125].
We have already noted that the D9-N171-82Q mouse did not display the full range of MSN electrophysiologic alterations seen in models with more widely expressed mHtt [37], and the expression of an Htt fragment rather than full-length likely also impacts the phenotype. The BACHD model allowed for more extensive analysis of the corticostriatal pathway in mice in which full-length mHtt expression is restricted to select neuronal populations [35, 38, 50]. Mice with knockdown of cortical expression showed improvement of the motor and psychiatric alterations relative to mice with expression in both striatum and cortex, but suppression of mHtt in cortical afferents did not prevent striatal atrophy. This result is complementary to what is seen in Emx1-Cre-Exon1-103Q Htt mice, an animal model that only expresses mHtt in cortical neurons and does not display striatal pathology [35], and also confirms the existence of MSN cell-autonomous pathology.
Pan-cellular HD mouse models have decreased levels of pre- and postsynaptic proteins at the corticostriatal synapse, including VGlut1 [133, 134], rabphilin3A [135], PSD-95, and SAP-97 [136]. Furthermore, in HD corticostriatal co-cultures from Q140 KI mice, and in Q175 KI mice in vivo, there is a decreased number of corticostriatal contacts [33, 137]. Suppression of mHtt expression in the striatum in the BACHD mouse restores the levels of postsynaptic, but not presynaptic, proteins as well as preventing MSN degeneration. Interestingly, reduction of mHtt expression only in the cortical output neurons of BACHD mice restores the levels of presynaptic proteins and also exerts benefits at the postsynaptic level in the MSNs, but does not prevent neurodegeneration. Furthermore, in the BACHD mice with reduced mHtt expression in cortical output neurons, there is an amelioration of striatal evoked excitatory and inhibitory postsynaptic current (EPSCs/IPSCs) imbalance [38] and improvement in bursting properties, but not firing rate, of MSNs in vivo [138]. These data indicate the requirement for corticostriatal cell–cell communication for the full pathophysiologic state to develop.
Drivers of Cre recombinase are rarely entirely cell type or region specific, and recombination is usually below 100%, so there are caveats in the interpretation of these experiments. For example, Emx1 directs expression to pallial precursors, including glia [139], so the knockdown in the BAC mice may include glia along with excitatory cortical neurons, leaving open the possibility that glia may be contributing to the improvement. In addition, although there was a comprehensive phenotyping of the BACHD variants, mitochondrial function and the transcriptome were not assessed, and none of the studies has determined the transneuronal effect of cortical mHtt on striatal gene expression. Despite any caveats, however, these results from multiple mouse models demonstrate that mHtt acts cell autonomously in both MSNs and cortical neurons, so that expression in either neuronal set causes a phenotype. However, as would be expected, expression in the cortical projection neurons has a greater impact on the striatum than vice versa, but amelioration of symptoms likely requires “treatment” of both compartments.
To further examine the cortical and striatal interactions in vitro, microfluidic chambers were introduced to reconstitute the network, and these represent an improved strategy over co-culture of striatal and cortical neurons. In this system, presynaptic mHtt expression in HD cortical neurons is necessary and sufficient to produce abnormalities of the corticostriatal network as determined by single cell calcium imaging, using GCaMP6f as a calcium sensor. Conversely, HD MSNs receiving WT cortical inputs were normal in terms of synaptic function as determined by the same assay [33]. These results support the hypothesis that the origin of specific, and most, aspects of corticostriatal synaptic dysfunction lies in the cortical neurons. A caveat to this conclusion is that cultured neurons, like iPSCs, are relatively immature, and other aspects of MSN dysfunction were not assayed.
Finally, in addition to perturbation of corticostriatal glutamatergic projections, there is an early reduction of thalamostriatal glutamatergic projections in HD (reviewed by Reiner and Deng, [140]). Thalamic inputs to MSNs are reduced in HD140Q KI mice in presymptomatic stages, prior to corticostriatal disconnection [141]. Notably, thalamocortical abnormalities do not worsen with disease progression, suggesting that most probably developmental alterations are selectively affecting this synapse [141]. Experimental, isolated manipulation of mHtt in thalamus has not been performed.
Contribution of Glial Cells to HD Pathophysiology
The focus of the study of mHtt pathogenic mechanisms was long restricted to neurons, but the last decade has witnessed an explosion of knowledge as to how non-neuronal central nervous system cells may contribute to neurodegeneration. Notably, Htt expression level, and therefore of course its mutant form, is similar between astrocytes, microglia, and oligodendrocytes [142]. Furthermore, mHtt inclusions develop in all three cell types, although in different proportions [142], which may contribute to, or protect from, certain aspects of mHtt toxicity. We noted above that there are cell-autonomous effects of mHtt in all three major glial types, but we will now discuss cell–cell interactions and non-cell-autonomous effects arising from non-neuronal cells in more detail.
Astrocytes and Neuronal Support
Neurodegenerative disorders are usually accompanied by astrogliosis in the most affected brain regions. In fact, the Vonsattel scale commonly used for grading of human HD neuropathology is partly based on the presence of glial cells in the putamen, which accumulate during disease progression [143]. HD astrocytes express higher levels of GFAP relative to WT, have altered arborization, and increased soma size [143]. Adding to the complexity of non-neuronal cells in neurodegenerative disease is the recent identification of the role of microglia in inducing a toxic astrocyte phenotype detrimental to neurons [144, 145], so that non-cell-autonomous actions of a glial subtype may be exerted on another glial subtype and not solely on neurons.
In mouse models, expression of mHtt restricted to astrocytes induced their activation, and expression restricted to neurons altered glial gene expression [42, 146], but in several pan-cellular models, astrogliosis is poorly recapitulated. For example, astrogliosis is undetectable in symptomatic R6/2 mice [147], with the most aggressive phenotype amongst HD mouse models. Despite the lack of gliosis, however, astrocytes are dysfunctional in full-length and fragment models of HD. Overall, they demonstrate decreased efficiency in glutamate and potassium buffering, due to altered expression of glutamate transporters, Glt-1, and potassium channels, Kir2.1, Kir2.3, Kir4.1, and Kv2.1 [52]. Electrophysiological parameters in striatal HD astrocytes of pan-cellular models show mHtt-induced alterations, including increased depolarization and membrane resistance, along with lower membrane conductance [147]. Restricted expression of mHtt in astrocytes recapitulates some of these phenotypes, including loss of glutamate transporters [42], indicating that this deficit is cell-autonomous. Astrocytes generated from human HD iPSCs display vacuolization in a CAG repeat-length dependent manner [148] and importantly, bipotential glial progenitor cells derived from human cells do not mature normally after engraftment and differentiation in vivo. Furthermore, HD embryonic stem cell (ESC)–derived glial progenitor cells (GPCs) have lower morphological fiber network complexity relative to WT, accompanied by transcriptional abnormalities with prominent downregulation of glial differentiation factors [149].
Restricted expression of mHtt in astrocytes also appears to lead to non-cell-autonomous neuronal deficits. Thus, transgenic or viral-induced expression of mHtt in astrocytes leads to motor deficits, although this may not require that there be neuronal abnormalities [149, 150], and expression in astrocytes exacerbates neuronal dysfunction via non-cell-autonomous mechanisms, with both cell types contributing to alterations in the glutamate cycle. In another study, there was an increased response to LPS in mice that expressed mHtt in both astrocytes and neurons relative to neurons only, which was ascribed to Iκb activity, supporting the non-cell-autonomous exacerbation of an HD neuronal phenotype by astrocytes [150].
In microfluidic chamber experiments [33] in which striatal astrocytes were included with mHtt-expessing MSNs, but without cortical neuron mHtt expression, corticostriatal connectivity alterations were absent. However, in vivo, deletion of mHtt in astrocytes BACHD mice restores the levels of postsynaptic proteins and the amplitude of NMDA currents in MSNs [39]. These results again suggest a non-cell-autonomous effect of astrocytic mHtt on MSN synaptic structure and function. Most recently, Benraiss and colleagues [151] generated HD glial chimeras by injecting mHtt-expressing human glial progenitor cells (HD-hGPCs) into the striatum of immunodeficient mice within 24 h of birth. These mice developed motor deficits accompanied by hyperexcitability of striatal neurons. Conversely, transplanted healthy astroglia improved motor function, prolonged life span, and normalized MSN excitability in R6/2 mice. These results demonstrate a clear contribution of HD glia to MSN dysfunction, in a non-cell-autonomous manner. Please note that the role of astrocytes and glutamate uptake in HD is discussed below.
Microglia/Neuroinflammation
Reactive microglia and molecular evidence of neuroinflammation are associated with most neurodegenerative disorders, but the role of microglia in HD pathophysiology is much less definitive than in, for example, Alzheimer’s disease. In addition, again similar to other neurodegenerative diseases, the answer to the question as to whether microglia play a pathogenic or protective role remains elusive. Microglial cells express high levels of mHtt [152] and there is an increased number of microglial cells prior to the onset of neuronal degeneration, as presymptomatic HD patients contain activated microglia within the striatum [153]. Microglia progressively increase in number, correlating with HD severity [154]. There is also an increase of pro-inflammatory cytokines in HD human striatum [155–157]. Using published RNA seq datasets, Botas and colleagues identified over 300 differentially expressed genes that overlap between early human HD brain and mouse models, and these included many genes related to immune function which, when assayed in a Drosophila HD model, were deemed to be pathogenic and driving disease via the NFkB pathway [65].
This last study drew heavily from previously published databases from HD mouse models with transcriptional profiles associated with the striatal inflammatory response [158–160]. Morphologically, some pan-cellular HD models, e.g., R6/2, display an increased number of Iba1 positive cells in the striatum, particularly as compared to neuronal models, suggesting that MSN mHtt does not by itself induce microgliosis [29, 161, 162]. Characterization of microglia in activated states has rapidly advanced in the last few years using RNA seq, and the classification into the traditional M1 and M2 phenotypes, releasing pro-inflammatory or anti-inflammatory cytokines, respectively, now appears too simplistic. In older studies, however, the “M1” microglial phenotype was detected in symptomatic HD mice, based on an activated morphology with retracted processes, relative to the longer processes in quiescent microglia, and pro-inflammatory cytokine and lymphokine production [163]. Moreover, some therapies proven effective in ameliorating HD phenotype, e.g., the modulation of mHtt sumoylation, via knockdown of the HTT-selective E3 SUMO ligase Pias1 or Pde10A inhibition, are linked with decreased microgliosis [74, 164]. Together, these human and mouse data might suggest that reactive microglia exacerbate HD pathology rather than exerting a neuroprotective role.
As with astrocytes, mouse models in which mHtt is selectively expressed or repressed in microglial cells have been derived and phenotyped to differentiate between cell-autonomous and non-cell-autonomous microglial functions in HD. First, microglial cells in HD mice were proposed to be more sensitive to sterile inflammation [163] but Crotti and colleagues [43] demonstrated that mHtt expression restricted to microglia induces a transcriptional pro-inflammatory response. These same mice had increased striatal neuronal death relative to WT after a peripheral pro-inflammatory challenge with LPS, raising the specter of a possible non-cell-autonomous effect on neurons from microglia expressing mHtt. Furthermore, microglial-derived interleukin-34 contributes to HD pathology in a biolistic brain slice HD model, suggesting a non-cell-autonomous effect of microglia on mHtt neurons [165]. In vitro, WT microglia can be activated by mHtt-expressing neurons [166], suggesting a non-cell-autonomous effect in the other direction, i.e., of neuronal mHtt on microglia. These data are supported by experiments utilizing co-culture of mHtt-expressing microglia with WT neurons, in which cell death was induced ex vivo and subsequently, in vivo (after sterile inflammation) [43], but mice were not assayed for motor deficits or neuropathology. In summary, although there is convincing evidence of microglial dysfunction in HD, the gaps in knowledge along with the absence of phenotype in the in vivo selective expression experiments discussed above [40] leave the field with an unclear picture as to the role of microglia in HD. Likewise, the cross-talk between mHtt expression astrocytes and microglia requires more investigation, including in the absence of neuronal expression.
Oligodendrocytes/Myelination/Dysmyelination
As with neuroinflammation, white matter abnormalities are prevalent in several neurodegenerative diseases. Abnormalities in myelin-related proteins together with axonal myelination deficits are present in several brain regions in HD, including the corpus callosum and posterior white matter tracts [53, 54, 167] and occur early in disease [168–170] and in some, but not all, animal models [55, 171]. MHtt aggregates in oligodendrocytes are present in multiple models, including R6/2 and HD140Q KI and zQ175 mice [142]. Analysis of oligodendroglial mHtt expression was enhanced by crossing a HD140Q KI mouse with a Plp-GFP line, allowing the visualization of the cell body and also oligodendrocyte processes, which are reduced in this mouse relative to WT [44].
The cell-autonomous and non-cell-autonomous contributions of oligodendrocytes to the HD phenotype are being scrutinized using similar methods as for other cell subtypes, i.e., selective expression and knockdown in vivo, and assays of iPSC-derived oligodendroglial lineage cells. Expression of the N-terminal fragment of mHtt with either 23 or 150 CAG repeats driven by the Plp promoter leads to compromised oligodendrocyte process length and myelination, but not cell viability, with 150CAG relative to 23CAG [44]. Plp-150Q mice displayed decreased expression of myelin-related genes, motor deficits, and decreased life span. Interestingly, there was also axonal degeneration, suggesting that mHtt could act non-cell autonomously on neurons, although the axonal degeneration presented after demyelination was already present, so it may be secondary and not directly due to mHtt [44]. Selective suppression of mHtt in the NG2+ oligodendrocyte progenitor cell population in BACHD mice prevented the development of white matter abnormalities and ameliorated some motor alterations [41]. In summary, there appear to be primary cell-autonomous and non-cell-autonomous effects of mHtt in oligodendroglia, with a significant contribution to motor dysfunction [41, 44].
Dysregulation in the expression of myelin-related proteins is commonly observed in HD models. Huang et al. [44] demonstrated that mHtt expression in oligodendrocytes is directly responsible for myelin gene expression deficits, because it has the ability to bind myelin regulatory factor, perturbing its function as a transcriptional activator. However, these microarray analyses could reflect changes in gene expression caused by demyelination and axonal degeneration secondary to some other process. In vitro, profound transcriptional abnormalities were detected in purified HD-hGPCs, affecting principally genes related to glial differentiation and function, and also regulation of synaptic function [151], implying a primary, cell-autonomous deficit. Importantly, specific oligodendrocyte- and myelin-related genes were altered. Transplantation of HD-hGPCs cells demonstrated a myelination defect due to altered oligodendrocyte differentiation and myelin production. These data strongly support the presence of a cell-autonomous detrimental effect of mHtt in the oligodendroglial lineage.
Neurotrophic Supply/BDNF Deficit
BDNF is synthesized in the cortex and is anterogradely transported to the striatum, in which it binds to tyrosine receptor kinase B (TrkB) which is highly expressed in MSNs. BDNF production [172], transport [173] and intracellular signaling pathways [174] are altered in HD. First, mHtt compromises BDNF at a transcriptional level through binding to transcription factors that regulate its expression [172]. Second, Htt indirectly binds to kinesin and dynein/dynactin [175, 176], and the HD mutation thereby compromises vesicular transport, directly affecting BDNF supply to the MSNs. Because MSNs do not produce BDNF [123, 177], lack of cortical production of BDNF results in a non-cell-autonomous negative effect on MSNs. In addition, however, there are cell-autonomous impacts on this system within MSNs, as there is some evidence, albeit contradictory, showing decreased TrkB levels in striatal neurons of HD patients and in some HD models [178–180]. It is also possible that TrkB localization in MSN dendrites is compromised, along with its signal transduction pathway [181].
The striatum does not develop normally in the absence of BDNF or TrkB [90, 182–184], but the survival of MSNs is not entirely dependent on BDNF. BDNF withdrawal induces increased cell death in HD iPSC-derived neural progenitor cells (NPCs) and more differentiated iPSC-derived MSNs relative to controls [98, 102, 185], perhaps due to increased excitotoxicity. Reducing cortical expression of BDNF worsens HD pathology in vivo. Overexpression or ectopic supply of BDNF ameliorates HD pathology in R6/1 and YAC128 mice and also in an HD rat model [186–190].
Corticostriatal co-cultures in microfluidic chambers again provide an ideal system in which to evaluate the impact of mHtt on both sides of the corticostriatal connection. MHtt-expressing cortical neurons from HD140Q KI and BACHD mice displayed cell-autonomous deficits in anterograde, and also retrograde in the latter, transport of BDNF [33, 191, 192], that could be pharmacologically corrected [192]. Restoration of BDNF transport by targeting TriC subunits, a subtype of cytosolic chaperonins, prevented striatal atrophy in BACHD cultured neurons [192]. The velocity of BDNF transportation in BACHD cortical neurons was slower than in WT neurons.
Like BDNF, TrkB receptor plays a pivotal role in the development, survival, and function of mature MSNs, and its early deletion [90] or knockdown [182] leads to neuronal loss. Post-mitotic TrkB deletion restricted to MSNs induces specific alterations in their gene expression, some of which overlap with genes altered in HD [193]. As alluded to above, retrograde and anterograde TrkB transport in striatal axons also depends on Htt, which participates in the transport of selective cargos, such as BDNF, APP, and TrkB. Htt knockdown retards TrkB movement in striatal axons [181], possibly due to alterations in the binding of TrkB-loaded vesicles to the cytoskeleton [33, 181]. Cultures in microfluidic chambers showed that retrograde transport of TrkB is specifically altered in HD striatal dendrites compromising dendrite-to-nucleus signaling [181, 194]. In conclusion, mHtt disrupts the BDNF/TrkB system cell autonomously in both cortical and striatal neurons, and non-cell autonomously via the MSN requirement for BDNF delivered by cortical afferents.
Notably, although most of the striatal BDNF is supplied by the cortex, cortical and striatal astrocytes are also important for BDNF production and release, particularly after injury [195, 196], adding complexity to understanding the mechanism of deficient neurotrophic supply in HD. In fact, although BDNF is strongly decreased in pan-cellular models of HD and in the putamen of HD patients [197–199], cortical BDNF is not decreased in an animal model in which mHtt is expressed only in neuronal cells (PrP-N171-82Q) [34] or specifically in MSNs (D9-N171-98Q) [36]. Unfortunately, BDNF has not been assayed in mice in which mHtt is selectively increased in astrocytes [146], but the induction of BDNF synthesis and release from astrocytes has been demonstrated to be beneficial in different models of HD [189].
In vitro, transduction of mHtt into cultured astrocytes with adenovirus reduced BDNF transcription and secretion by disrupting the function of transcription factors CREB-binding protein (CBP) and Sp1, and impairing clathrin and the Golgi complex, respectively [200]. However, in a HD140Q KI mouse, transcription of BDNF is not decreased in astrocytes [201]. This divergence could arise due to higher levels of mHtt after viral transduction than in the HD140Q KI mouse. BDNF exocytosis, however, is reduced in the HD140Q KI astrocytes, due to disruption of the conversion of Rab3a-GTP to Rab3a-GDP, highlighting the multiplicity of pathogenic mechanisms via which mHtt disrupts the BDNF/TrkB system.
Mitochondrial/Energy Dysfunction: Respiratory Chains and ATP Production
The generation of truncated N-terminal and full-length transgenic and then KI HD mouse models largely replaced the use of toxin models, including 3-NP, a drug that specifically targets the mitochondrial respiratory chain in striatal neurons, resulting in lesions resembling HD [202]. Alterations in structure, function, and dynamics of mitochondria exist in multiple HD models and in patients. In fact, mHtt has been shown to directly or indirectly alter mitochondrial dynamics (biogenesis, fission, fusion, trafficking, and degradation), calcium buffering capacity, transcription, membrane potential, ATP production, and protein import [203, 204]. Mitochondria produce the high level of ATP required to maintain MSNs in a hyperpolarized state, and for calcium buffering, and both processes are dysregulated in HD. The use of 3-NP as a toxin model for HD demonstrated that striatal neurons are vulnerable to inhibition of respiratory chain complex II, succinate dehydrogenase. Lentiviral or transgenic expression of mHtt in cultured neurons or MSNs in vivo, respectively, decreases components of complex II and/or its function, and importantly, overexpression of complex II components confers neuroprotection [31, 37].
Most of the studies regarding mitochondrial alterations in HD were performed in the striatum or in neuronal primary cultures. Striatal mitochondria are selectively more vulnerable to the expression of mHtt possibility due to intrinsic properties [205]. First, neuronal and astrocytic striatal mitochondria are less efficient in buffering calcium than their cortical counterparts [205]. Second, cortical neurons have higher maximal respiration in comparison with striatal neurons [206]. Third, PGC-1α, a crucial transcription factor in mitochondria, is decreased in HD models [207, 208]. This protein regulates mitochondrial/nuclear DNA (mtDNA/nDNA) ratio and citrate synthase activity. Interestingly, PGC-1α transcription is equally altered in both HD striatum and cortex, but only the striatum manifests decreased mtDNA/nDNA ratio [209]. These data suggest that selective striatal vulnerability in HD may be mediated, in part, by the intrinsic properties of striatal neuronal mitochondria. Surprisingly, however, some full-length models, e.g., YAC128 [206] and HD iPSC-derived neural cells [210] do not display the mitochondrial respiratory chain activity alterations that are present in pan-neuronal (R6/1 and R6/2) and MSN-specific (D9-N171-98Q) truncated models [37, 51].
The discrepancy in the effects of mHtt on mitochondrial function in full-length and truncated models may arise from multiple molecular mechanisms. For instance, aggregated forms of mHtt may account for the mitochondrial dysfunction, because truncated models generate larger and more numerous mHtt aggregates than do full-length. Consistent with this explanation, mitochondria are not dysfunctional in heart tissue from R6/2 mice, in which mHtt is expressed but does not aggregate, whereas in quadriceps muscle in R6/2 mice, aggregates are present and respiratory chain complex activities are decreased [211]. Interestingly, aggregates and mitochondria are frequently closely apposed [212].
Another process mediated by mitochondria which is perturbed in HD striatum is energy metabolism, with both cell-autonomous and non-cell-autonomous etiologies. In co-cultures of WT and BACHD neurons and astrocytes, the inclusion of mHtt-expressing astrocytes impairs glucose uptake in WT neurons but importantly, neuronal genotype does not alter metabolism [213]. Mass spectrometry studies in R6/2 mice reveal altered expression of proteins related to energy metabolism in astrocytes, pointing again to an astrocytic pathogenic role in HD [214]. On the other hand, HD iPSC-derived neural cells have decreased glycolytic capacity relative to corrected cells, attributed mainly to glycolytic deficits contributing to ATP depletion [210]. These results suggest that mHtt cell autonomously compromises metabolism in neurons. Future work should include assays of mitochondrial function in animal models in which mHtt expression is restricted to single cell types, e.g., astrocytes and microglia.
Finally, mHtt also disrupts the levels and activity of proteins that mediate mitochondrial fission and fusion. Both events regulate mitochondrial morphology, number, size, and functionality. In general, fusion proteins are downregulated and fission is upregulated in HD striatum [215–219]. Striatal neurons are intrinsically more sensitive to the presence of mHtt increasing fission than are cortical neurons. In fact, the activity of Drp1, a GTPase that mediates mitochondrial fission, is strongly upregulated in the striatum compared with cortex [216]. Furthermore, the non-aggregated form of mHtt disrupts striatal mitochondrial transport [220], whereas in cortical neurons mitochondrial transport is only affected in the presence of mHtt aggregates [221]. In total, data support a large component of cell-autonomous factors impacting on MSN mitochondria in the presence of mHtt.
Huntingtin Aggregation
Generation of mHtt aggregates is a hallmark of HD and occurs in all cell types discussed herein. Full-length mHtt has a lower tendency to aggregate than do N-terminal fragments [222], accounting for the differences in detectable aggregates between full-length and fragment mouse models. Post-translational modifications (PTMs) differentially affect mHtt toxicity and aggregation, particularly those on the N-terminal 17 amino acids [223]. Thus, BACHD mice lacking the N17 domain have a more severe phenotype than does the original BACHD mouse [224], although they express lower levels of mHtt. There are of course PTMs outside the N17 terminus, including on serine 421, which is phosphorylated via Akt and GSK, thereby protecting against NMDA-mediated toxicity and regulating mHtt toxicity and clearance [225]. Remarkably, there are lower levels of serine 421 phosphorylation in the striatum than in the cortex or cerebellum [226], suggesting that striatal neurons could be selectively vulnerable in a cell-autonomous manner to mHtt aggregation and its possible downstream effects due to decreased phosphorylation on serine 421. Along with these data, imbalance in kinase/phosphatase levels and activity participates in neuronal viability and dysfunction in HD [227, 228]. In fact, Akt activation and level inversely correlate with the most affected brain regions in HD [229].
Striatal neurons clear mHtt more slowly than do cortical neurons [230], adding to the cell-autonomous, intrinsic mechanisms contributing to striatal neuronal vulnerability in HD. Furthermore, astrocytes degrade mHtt more rapidly than neurons, which is in line with the higher ubiquitin proteasome system (UPS) activity detected in astrocytes compared with neurons [231]. The non-cell-autonomous mechanism via which astrocytes can modulate aggregation in HD neurons is being unraveled with the use of cellular specific models. First, astrocytic exosomes reduce mHtt aggregation in HD140Q KI mice [232], suggesting that these exosomes carry cargo which participates in prevention of mHtt aggregation. The authors specifically suggest that αB-crystallin, a small heat shock protein that reduces mHtt aggregation and toxicity, participates in this exosome secretion because its overexpression in HD astrocytes prevents these deficits. However, the direct link between the restoration of αB-crystallin astrocytic protein and mHtt aggregation has not been established, although reduction of mHtt in astrocytes is known to redistribute mHtt neuronal aggregates [39].
Excitotoxicity and Potassium and Calcium Buffering
The use of quinolinic acid (QA) as a toxin-induced model of HD revealed that MSNs demonstrate greater vulnerability to glutamate-mediated excitotoxicity than do other neuronal subtypes [233, 234]. Presymptomatic and symptomatic HD models exhibit different levels of sensitivity to QA-induced excitotoxicity. Increased resistance to excitotoxicity at late stages of HD may be attributed to the redistribution of NMDA receptors within the neuron, and the differential activation of signaling pathways dependent on the complexed proteins, e.g., PSD-95 [235] or regulatory element antagonist modulator (DREAM) [236], and/or their subcellular localization [237, 238]. Young D9-N171-98Q mice respond to QA similarly to their WT counterparts, but pan-neuronal HD models present increased sensitivity to excitotoxicity at presymptomatic stages, which suggests a non-cell-autonomous mechanism. Once symptomatic, both D9-N171-98Q and pan-cellular HD models respond similarly, suggesting that at advanced stages this non-cell-autonomous mechanism is lost and a specific MSN quality confers resistance to excitotoxicity. Under physiologic conditions glutamate clearance is accelerated in R6/2 mice [239], possibly accounting for the resistance to excitotoxicity at this stage. At later stages, the number of functional excitatory synapses is reduced, compromising glutamate release.
Finally, MSN vulnerability to glutamate excitotoxicity in the presence of mHtt is partly attributed to neurodevelopmental alterations, because expression of mHtt restricted to the developmental period recapitulates this selective vulnerability at early stages of disease [112]. These data lend further support to the notion that has evolved from iPSC-based studies of HD as a neurodevelopmental disorder [112, 113] and it has been suggested that epigenetic-based, HDAC-associated therapies could reverse presymptomatic alterations [240].
Vulnerability to excitotoxicity in HD is also determined by the cell-autonomous effects of MSN mHtt on the activation, localization, and stabilization of glutamate receptors. The NMDA receptor mislocalization hypothesis has emerged as a major contributor to the increased sensitivity of HD MSNs to excitotoxicity. Briefly, stimulation of synaptic NMDA receptor activity is neuroprotective, whereas extrasynaptic NMDA receptor stimulation can lead to cell death [238]. Previously discussed results regarding the role of STEP highlight the relevance of synaptic versus extrasynaptic NMDA receptor imbalance at early stages of HD. Other findings include the enhanced GluN2B extrasynaptic expression in YAC128 MSNs [241], which may be further enhanced by the increased extrasynaptic localization of PSD-95, which acts as scaffolding protein for GluN2B. MHtt also upregulates GluN3A subunit expression [242], which possibly participates in GluN2B localization [243]. GluN2B cleavage is induced, but its phosphorylation is reduced, contributing to extrasynaptic GluN2B localization [72].
Deciphering the role of excitotoxicity in HD increasingly includes the study of the contribution of astrocytes. An increased level of glutamate at HD synapses, which may arise due to altered clearance, would represent a source of excitotoxicity. There are discrepant reports regarding the functionality and level of glutamate transporters in HD [244–249]. The astrocytic glutamate transporter SlcA12, aka Glt-1 or EAAT2, that regulates glutamate uptake [214, 250], is decreased in astrocytes transduced with viral vectors expressing mHtt [143]. Although the BACHD mouse has normal levels of Glt-1 mRNA [39], other HD mouse models including R6/1 [251], R6/2 [147, 250–253], YAC128 [254] and Q175 homozygotes [147] express decreased levels of Glt-1 in striatal lysates, including at corticostriatal synapses. These data suggest an addition non-cell-autonomous mechanism via which mHtt expression in astrocytes could increase QA vulnerability of MSNs at initial disease stages. HD striatal astrocytes also display alterations in glutamate buffering and consequently aberrant calcium signaling that further impacts MSNs [255]. Intrinsically, mHtt expression in cultured astrocytes enhances calcium-dependent glutamate release, possibly due to increased levels of pyruvate carboxylase, an enzyme which facilitates glutamate synthesis [256]. Conversely, silencing of mHtt in HD astrocytes generated from monkey iPSCs restored Glt-1 levels and glutamate uptake ability [257], implying that cell-autonomous effects may be overwhelmed when astrocytes are in their natural milieu with intact cell–cell communication.
Although it is easy to speculate about how inadequate glutamate buffering might increase QA susceptibility, there is no evidence yet of early dysregulation of Glt-1 expression in astrocytes in HD models. In addition, HD pathology is not worsened by Glt-1 deletion [250], and presymptomatic knockdown of Glt-1 does not affect glutamate clearance in the YAC128 model [239]. On the other hand, virally mediated overexpression of Glt-1 prevents the appearance of motor deficits in a Q175 KI mouse model and normalizes neuronal striatal firing frequency [258]. Finally, R6/2 mice treated with ceftriaxone, an antibiotic that increases striatal expression of Glt-1, improves motor function in the early symptomatic stage [252]. Further work is required to resolve the study of GLT-1 status, perhaps with a focus on genetically accurate models.
In addition to the glutamate transporter, the astrocytic Kir4.1 potassium channel is also altered in HD. This channel maintains low levels of potassium in the extracellular space, maintaining neurons in a hyperpolarized state and regulating MSN excitability. R6/2 striatal neurons are in fact hyperexcitable and striatal interstitial potassium levels are increased in this animal model [151]. Engraftment of healthy glia into R6/2 mice normalized interstitial potassium levels and MSN excitability, implying a non-cell-autonomous mechanism. In addition, although mHtt suppression in astrocytes does not restore Kir4.1 expression [39], viral-mediated restoration of Kir4.1 levels in HD astrocytes ameliorates pathology in R6/2 mice, reducing MSN excitability [147]. These data are similar to those for Glt-1. However, the improvement of the HD phenotype by the potentiation of Kir4.1 expression cannot be attributed solely to potassium levels, because overexpression of Kir4.1 increases the level of Glt-1. Finally, although reduced Kir4.1 protein levels were detected in R6/2 and Q175 striatal homogenates, not all striatal astrocytes have decreased levels of Kir4.1 [147], and thus how mHtt reduces Kir4.1 level is not entirely clear.
Like excitotoxicity, a biphasic alteration in dopamine release exists, with an initial increase, participating in the characteristic hyperkinetic phenotype, to a decrease at later stages [23, 259]. Dopamine (DA) signaling non-cell autonomously contributes to NMDA receptor modulation, possibly playing a role in excitotoxicity, and dopamine also trans-synaptically regulates specific MSN transcripts known to be dysregulated in HD, e.g., neuropeptides. One study ascribes early changes in dopamine release in the pan-cellular R6/1 model to transcriptional dysregulation of SK3 channels [260], and its mislocalization, but there is little known as to whether DA release dysregulation is cell-autonomous due to substantia nigra DAergic neuron dysfunction, or non-cell-autonomous, due to feedback from MSNs.
Transcriptional Alterations
New data are constantly emerging regarding transcriptional dysregulation in HD, both from human brain and from mouse models [30, 58, 261–267]. Although as discussed above, specific transcription factors (TFs) are implicated, e.g., Foxp2 [82], it is unlikely that HD transcriptional pathology can be attributed to any single TF. The question of causation versus compensation for specific genes has also only been approached for a limited number of candidates, e.g., PGC1α, CBP in vitro, and Foxp2 [82, 207, 208], with a larger screen in Drosophila [61]. This question persists because although regional deep sequencing from Hdh KI mice revealed that there are more dysregulated genes in the striatum than in the cortex [58]. The cerebellum, which is frequently used as a “control” as it is relatively resistant in HD, displays gene expression changes that qualitatively highly overlap with the striatum [268–270].
There are three main mechanisms proposed to account for the alterations in the HD transcriptome. First, mHtt sequesters TFs thru interactions of the polyQ tracts, impeding their function and compromising the transcription of selective genes [11, 271–273]. MHtt directly interacts with repressor element-1 silencing transcription factor (REST), retaining it in the nucleus and thereby repressing the expression of neuronal-related genes [274, 275]. Second, the presence of mHtt is associated with histone hypoacetylation [276–278], that in some cases, may arise due to TF sequestration. One example is acetyltransferase CBP, which is found in mHtt aggregates thereby reducing its function [18, 279]. Third, there is likely reduced trans-synaptic regulation of transcription, specifically that by BDNF [172]. This last possibility, which would be non-cell-autonomous, has not been directly addressed.
In all studies using bulk sequencing, neuronal transcriptomic changes could easily mask glial transcriptional alterations, both in older microarray experiments and in RNA seq assays. To date, in most cases, studies of the cell-specific transcriptome in mouse lines in which mHtt is expressed exclusively in non-neuronal cells are lacking. There are, however, isolated reports examining cell-autonomous and non-cell-autonomous transcriptional regulation of specific genes. For example, as discussed above, viral-mediated expression of mHtt in MSNs leads to dysregulation of the glutamate transporters Glast and Glt-1 in astrocytes [143]. Also as previously noted, mHtt expression restricted to microglia promotes a pro-inflammatory profile, but unfortunately the neuronal transcriptome was not assayed in these studies. There are almost certainly sequencing studies in process from cell subtype-specific mice expressing mHtt which will tell us much more about transcriptional dysregulation and the translatome in neuronal subtypes, e.g., dMSNs and iMSNs, non-neuronal cells, single cells, and their interactions.
Other approaches are being used to unravel selective cell-type gene expression patterns in adult mouse brain. To avoid the generation of genetically modified mice to perform translational profiling, Merienne et al. [92] developed an approach in which direct and indirect MSNs, astrocytes, and microglia are identified with selective transcriptomic signatures. Briefly, laser capture microdissection was performed in mice expressing Drd2-eGFP (iMSNs), Glt-1-eGFP-eGFP (astrocytes), Cx3cr1-eGFP (microglia), and Drd1-Tomato (dMSNs). Comparison between existing transcriptional data bases of HD models and data bases generated from different striatal cell types, revealed that most upregulated genes observed in HD are highly represented in glia, and conversely, most of the downregulated transcripts, in neurons. These data suggest that mHtt induces transcriptional alterations in a cell type-specific manner, but do not speak to whether they are cell-autonomous.
As iPSC differentiation methods evolve, transcriptome assays in more mature cells will be able to significantly contribute to such studies. As noted, these experiments have already revealed specific gene expression networks selectively altered during development and have recapitulated epigenetic alterations in iPSC-derived neurons and glial cells. There is always some heterogeneity in the differentiated cells, but these transcriptional changes are largely considered to be cell-autonomous in neuronal preparations.
In summary, although the striatal MSN is the most vulnerable neuronal subtype in HD, it has long been a question as to whether their dysfunction and degeneration is cell-autonomous or non-cell-autonomous, the latter particularly due to the corticostriatal connection. Definitive data have emerged demonstrating that both mechanisms are at work, but that the corticostriatal connection is required for the full picture of striatal pathology in HD. Additional work is therefore required to identify the major pathogenic mechanisms in cortical neurons, which may differ from those in MSNs due to intrinsic differences between the two neuronal subtypes, and to further characterize the mechanisms via which the cortical neurons impact the MSNs, e.g., trans-synaptic transcriptional dysregulation. Additional tools have been developed to more carefully examine these questions in cortical and MSN neuronal subtypes, for example, differences in vulnerability between direct and indirect pathway, and striosome and matrix, MSNs. The question of the effects of mHtt on cell types and/or their interactions has increased in relevance based on enormous strides in identifying the roles of non-neuronal cells in HD pathophysiology. Much of this is due to advances in the derivation of mouse lines with cell subtype mHtt expression in vivo, and the use of HD iPSC-derived neurons and non-neuronal cells, which has also led to an important notion of neurodevelopmental abnormalities in HD, suggesting that early treatment may be required.
Electronic supplementary material
(PDF 431 kb)
Acknowledgments
This work was supported by R01NS100529. Thank you to John Carroll for graphics in Figure 1.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 2.HDCRG A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 4.Marques Sousa C, Humbert S. Huntingtin: here, there, everywhere! J Huntingtons Dis. 2013;2:395–403. doi: 10.3233/JHD-130082. [DOI] [PubMed] [Google Scholar]
- 5.Kassubek J, Bernhard Landwehrmeyer G, Ecker D, et al. Global cerebral atrophy in early stages of Huntington’s disease: quantitative MRI study. Neuroreport. 2004;15:363–365. doi: 10.1097/00001756-200402090-00030. [DOI] [PubMed] [Google Scholar]
- 6.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Rosas HD, Koroshetz WJ, Chen YI, et al. Evidence for more widespread cerebral pathology in early HD: an MRI-based morphometric analysis. Neurology. 2003;60:1615–1620. doi: 10.1212/01.wnl.0000065888.88988.6e. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich ME. Huntington’s disease and the striatal medium spiny neuron: cell-autonomous and non-cell-autonomous mechanisms of disease. Neurotherapeutics. 2012;9:270–284. doi: 10.1007/s13311-012-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding RJ, Tong Y-F. Proteostasis in Huntington’s disease: disease mechanisms and therapeutic opportunities. Acta Pharmacol Sin. 2018;39:754–769. doi: 10.1038/aps.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega Z, Lucas JJ. Ubiquitin-proteasome system involvement in Huntington’s disease. Front Mol Neurosci. 2014;7:77. doi: 10.3389/fnmol.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Lajoie P, Snapp EL. Formation and toxicity of soluble polyglutamine oligomers in living cells. PLoS One 2010;5. [DOI] [PMC free article] [PubMed]
- 13.Takahashi T, Kikuchi S, Katada S, et al. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta U, Nilson AN, Kayed R. The role of amyloid-beta oligomers in toxicity, propagation, and immunotherapy. EBioMedicine. 2016;6:42–49. doi: 10.1016/j.ebiom.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Roon-Mom WMC, Reid SJ, Jones AL, et al. Insoluble TATA-binding protein accumulation in Huntington’s disease cortex. Mol Brain Res. 2002;109:1–10. doi: 10.1016/s0169-328x(02)00450-3. [DOI] [PubMed] [Google Scholar]
- 16.Schaffar G, Breuer P, Boteva R, et al. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 17.Suhr ST, Senut MC, Whitelegge JP, et al. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J Cell Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steffan JS, Kazantsev A, Spasic-Boskovic O, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci. 2002;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira JMA. Mitochondrial bioenergetics and dynamics in Huntington’s disease: tripartite synapses and selective striatal degeneration. J Bioenerg Biomembr. 2010;42:227–234. doi: 10.1007/s10863-010-9287-6. [DOI] [PubMed] [Google Scholar]
- 20.Alberch J, Perez-Navarro E, Canals JM. Neurotrophic factors in Huntington’s disease. Prog Brain Res. 2004;146:195–229. doi: 10.1016/s0079-6123(03)46014-7. [DOI] [PubMed] [Google Scholar]
- 21.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Raymond LA. Excitotoxicity in Huntington disease. Clin Neurosci Res. 2003;3:121–128. [Google Scholar]
- 23.Cepeda C, Murphy KPS, Parent M, et al. The role of dopamine in Huntington’s disease. Prog Brain Res. 2014;211:235–254. doi: 10.1016/B978-0-444-63425-2.00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD Gene with an Expanded. 1996;87:493–506. [DOI] [PubMed]
- 25.Francelle L, Galvan L, Brouillet E. Possible involvement of self-defense mechanisms in the preferential vulnerability of the striatum in Huntington’s disease. Front Cell Neurosci. 2014;8:1–13. doi: 10.3389/fncel.2014.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummings DM, Andre VM, Uzgil BO, et al. Alterations in cortical excitation and inhibition in genetic mouse models of Huntington’s disease. J Neurosci. 2009;29:10371–10386. doi: 10.1523/JNEUROSCI.1592-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slow EJ, van Raamsdonk J, Rogers D, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 28.Teo RTY, Hong X, Yu-Taeger L, et al. Structural and molecular myelination deficits occur prior to neuronal loss in the YAC128 and BACHD models of Huntington disease. Hum Mol Genet. 2016;25:2621–2632. doi: 10.1093/hmg/ddw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmons DA, Belichenko NP, Yang T, et al. A small molecule TrkB ligand reduces motor impairment and neuropathology in R6/2 and BACHD mouse models of Huntington’s disease. J Neurosci. 2013;33:18712–18727. doi: 10.1523/JNEUROSCI.1310-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labadorf A, Hoss AG, Lagomarsino V, et al. RNA sequence analysis of human huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS One. 2015;10:e0143563. doi: 10.1371/journal.pone.0143563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damiano M, Diguet E, Malgorn C, et al. A role of mitochondrial complex II defects in genetic models of Huntington’s disease expressing N-terminal fragments of mutant huntingtin. Hum Mol Genet. 2013;22:3869–3882. doi: 10.1093/hmg/ddt242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray M, Shirasaki DI, Cepeda C, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virlogeux A, Moutaux E, Christaller W, et al. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington’s disease. Cell Rep. 2018;22:110–122. doi: 10.1016/j.celrep.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Yu Z-X, Li S-H, Evans J, et al. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu X, Li C, Wei W, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Brown TB, Bogush AI, Ehrlich ME. Neocortical expression of mutant huntingtin is not required for alterations in striatal gene expression or motor dysfunction in a transgenic mouse. Hum Mol Genet. 2008;17:3095–3104. doi: 10.1093/hmg/ddn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SH, Thomas CA, Andre VM, et al. Forebrain striatal-specific expression of mutant huntingtin protein in vivo induces cell-autonomous age-dependent alterations in sensitivity to excitotoxicity and mitochondrial function. ASN Neuro. 2011;3:e00060. doi: 10.1042/AN20110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang N, Gray M, Lu XH, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat Med. 2014;20:536–541. doi: 10.1038/nm.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood TE, Barry J, Yang Z, et al. Mutant huntingtin reduction in astrocytes slows disease progression in the BACHD conditional Huntington’s disease mouse model. Hum Mol Genet. 2019;28:487–500. doi: 10.1093/hmg/ddy363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petkau TL, Hill A, Connolly C, et al. Mutant huntingtin expression in microglia is neither required nor sufficient to cause the Huntington’s disease-like phenotype in BACHD mice. Hum Mol Genet. 2019;00:1–10. doi: 10.1093/hmg/ddz009. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari C, Garcia-miralles M, Caron NS, et al. Intrinsic mutant HTT-mediated defects in oligodendroglia cause myelination deficits and behavioral abnormalities in Huntington disease. 2019;1–6. [DOI] [PMC free article] [PubMed]
- 42.Bradford J, Shin J-Y, Roberts M, et al. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci. 2009;106:22480–22485. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crotti A, Benner C, Kerman BE, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. 2014;17:513–521. doi: 10.1038/nn.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang B, Wei WJ, Wang G, et al. Mutant huntingtin downregulates myelin regulatory factor-mediated myelin gene expression and affects mature oligodendrocytes. Neuron. 2015;85:1212–1226. doi: 10.1016/j.neuron.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies SW, Turmaine M, Cozens BA, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 46.Wheeler VC, White JK, Gutekunst CA, et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 47.Menalled LB, Sison JD, Wu Y, et al. Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington’s disease knock-in mice. J Neurosci. 2002;22:8266–8276. doi: 10.1523/JNEUROSCI.22-18-08266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CH, Tallaksen-Greene S, Chien WM, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 49.Schilling G, Becher MW, Sharp AH, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 50.Gu X, Andre VM, Cepeda C, et al. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington’s disease. Mol Neurodegener. 2007;2:8. doi: 10.1186/1750-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas EA, Coppola G, Tang B, et al. In vivo cell-autonomous transcriptional abnormalities revealed in mice expressing mutant huntingtin in striatal but not cortical neurons. Hum Mol Genet. 2011;20:1049–1060. doi: 10.1093/hmg/ddq548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khakh BS, Beaumont V, Cachope R, et al. Unravelling and exploiting astrocyte dysfunction in Huntington’s disease. Trends Neurosci. 2017;40:422–437. doi: 10.1016/j.tins.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faria AV, Ratnanather JT, Tward DJ, et al. Linking white matter and deep gray matter alterations in premanifest Huntington disease. NeuroImage Clin. 2016;11:450–460. doi: 10.1016/j.nicl.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tabrizi SJ, Scahill RI, Durr A, et al. Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: the 12-month longitudinal analysis. Lancet Neurol. 2011;10:31–42. doi: 10.1016/S1474-4422(10)70276-3. [DOI] [PubMed] [Google Scholar]
- 55.Jin J, Peng Q, Hou Z, et al. Early white matter abnormalities, progressive brain pathology and motor deficits in a novel knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2015;24:2508–2527. doi: 10.1093/hmg/ddv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beal MF, Ferrante RJ, Swartz KJ, et al. Chronic quinolinic acid lesions in rats closely resemble Huntington’s disease. J Neurosci. 1991;11:1649–1659. doi: 10.1523/JNEUROSCI.11-06-01649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouary S, Bizat N, Altairac S, et al. Major strain differences in response to chronic systemic administration of the mitochondrial toxin 3-nitropropionic acid in rats: Implications for neuroprotection studies. Neuroscience. 2000;97:521–530. doi: 10.1016/s0306-4522(00)00020-8. [DOI] [PubMed] [Google Scholar]
- 58.Langfelder P, Cantle JP, Chatzopoulou D, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19:623–633. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vodicka P, Mo S, Tousley A, et al. Mass spectrometry analysis of wild-type and knock-in Q140/Q140 Huntington’s disease mouse brains reveals changes in glycerophospholipids including alterations in phosphatidic acid and lyso-phosphatidic acid. J Huntingtons Dis. 2015;4:187–201. doi: 10.3233/JHD-150149. [DOI] [PubMed] [Google Scholar]
- 60.Ament SA, Pearl JR, Cantle JP, et al. Transcriptional regulatory networks underlying gene expression changes in Huntington’s disease. Mol Syst Biol. 2018;14:e7435. doi: 10.15252/msb.20167435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin J, Cheng Y, Zhang Y, et al. Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J Neurochem. 2012;123:477–490. doi: 10.1111/j.1471-4159.2012.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wegrzynowicz M, Holt HK, Friedman DB, et al. Changes in the striatal proteome of YAC128Q mice exhibit gene-environment interactions between mutant huntingtin and manganese. J Proteome Res. 2012;11:1118–1132. doi: 10.1021/pr200839d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choudhury KR, Das S, Bhattacharyya NP. Differential proteomic and genomic profiling of mouse striatal cell model of Huntington’s disease and control; probable implications to the disease biology. J Proteome. 2016;132:155–166. doi: 10.1016/j.jprot.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 64.Ring KL, An MC, Zhang N, et al. Genomic analysis reveals disruption of striatal neuronal development and therapeutic targets in human Huntington’s disease neural stem cells. Stem Cell Rep. 2015;5:1023–1038. doi: 10.1016/j.stemcr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Ramahi I, Lu B, Di Paola S, et al. High-throughput functional analysis distinguishes pathogenic, nonpathogenic, and compensatory transcriptional changes in neurodegeneration. Cell Syst. 2018;7:28–40.e4. doi: 10.1016/j.cels.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazarei G, Neal SJ, Becanovic K, et al. Expression analysis of novel striatal-enriched genes in Huntington disease. 2010;19:609–622. [DOI] [PMC free article] [PubMed]
- 67.Snyder EM, Nong Y, Almeida CG, et al. Regulation of NMDA receptor trafficking by amyloid-β. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 68.Lombroso PJ, Ogren M, Kurup P, et al. Molecular underpinnings of neurodegenerative disorders: striatal-enriched protein tyrosine phosphatase signaling and synaptic plasticity. F1000Research. 2016;5. [DOI] [PMC free article] [PubMed]
- 69.Xu J, Kurup P, Foscue E, et al. Striatal-enriched protein tyrosine phosphatase regulates the PTPalpha/Fyn signaling pathway. J Neurochem. 2015;134:629–641. doi: 10.1111/jnc.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saavedra A, Giralt A, Rue L, et al. Striatal-enriched protein tyrosine phosphatase expression and activity in Huntington’s disease: a STEP in the resistance to excitotoxicity. J Neurosci. 2011;31:8150–8162. doi: 10.1523/JNEUROSCI.3446-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Forn M, Martinez-Torres S, Garcia-Diaz Barriga G, et al. Pharmacogenetic modulation of STEP improves motor and cognitive function in a mouse model of Huntington’s disease. Neurobiol Dis. 2018;120:88–97. doi: 10.1016/j.nbd.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 72.Gladding CM, Sepers MD, Xu J, et al. Calpain and STriatal-Enriched protein tyrosine phosphatase (STEP) activation contribute to extrasynaptic NMDA receptor localization in a Huntington’s disease mouse model. Hum Mol Genet. 2012;21:3739–3752. doi: 10.1093/hmg/dds154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leuti A, Laurenti D, Giampa C, et al. Phosphodiesterase 10A (PDE10A) localization in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2013;52:104–116. doi: 10.1016/j.nbd.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 74.Giampa C, Laurenti D, Anzilotti S, et al. Inhibition of the striatal specific phosphodiesterase PDE10A ameliorates striatal and cortical pathology in R6/2 mouse model of Huntington’s disease. PLoS One. 2010;5:e13417. doi: 10.1371/journal.pone.0013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beaumont V, Zhong S, Lin H, et al. Phosphodiesterase 10A inhibition improves cortico-basal ganglia function in Huntington’s disease models. Neuron. 2016;92:1220–1237. doi: 10.1016/j.neuron.2016.10.064. [DOI] [PubMed] [Google Scholar]
- 76.Whiteley EL, Tejeda GS, Baillie GS, et al. PDE10A mutations help to unwrap the neurobiology of hyperkinetic disorders. Cell Signal. 2019;60:31–38. doi: 10.1016/j.cellsig.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Subramaniam S, Snyder SH. Huntington’s Disease is a disorder of the corpus striatum: Focus on Rhes (Ras homologue enriched in the striatum) Neuropharmacology. 2011;60:1187–1192. doi: 10.1016/j.neuropharm.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 78.Mealer RG, Subramaniam S, Snyder SH. Rhes deletion is neuroprotective in the 3-nitropropionic acid model of Huntington’s disease. J Neurosci. 2013;33:4206–4210. doi: 10.1523/JNEUROSCI.3730-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee JH, Sowada MJ, Boudreau RL, et al. Rhes suppression enhances disease phenotypes in Huntington’s disease mice. J Huntingtons Dis. 2014;3:65–71. doi: 10.3233/JHD-140094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JH, Tecedor L, Chen YH, et al. Reinstating aberrant mTORC1 activity in huntington’s disease mice improves disease phenotypes. Neuron. 2015;85:303–315. doi: 10.1016/j.neuron.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Desplats PA, Lambert JR, Thomas EA. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington’s disease. Neurobiol Dis. 2008;31:298–308. doi: 10.1016/j.nbd.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hachigian LJ, Carmona V, Fenster RJ, et al. Control of Huntington’s disease-associated phenotypes by the striatum-enriched transcription factor Foxp2. Cell Rep. 2017;21:2688–2695. doi: 10.1016/j.celrep.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 83.Deng YP, Albin RL, Penney JB, et al. Differential loss of striatal projection systems in Huntington’s disease: a quantitative immunohistochemical study. J Chem Neuroanat. 2004;27:143–164. doi: 10.1016/j.jchemneu.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 84.Richfield EK, Maguire-Zeiss KA, Vonkeman HE, et al. Preferential loss of preproenkephalin versus preprotachykinin neurons from the striatum of Huntington’s disease patients. Ann Neurol. 1995;38:852–861. doi: 10.1002/ana.410380605. [DOI] [PubMed] [Google Scholar]
- 85.Sapp E, Ge P, Aizawa H, et al. Evidence for a preferential loss of enkephalin immunoreactivity in the external globus pallidus in low grade Huntington’s disease using high resolution image analysis. Neuroscience. 1995;64:397–404. doi: 10.1016/0306-4522(94)00427-7. [DOI] [PubMed] [Google Scholar]
- 86.Fusco FR, Chen Q, Lamoreaux WJ, et al. Cellular localization of huntingtin in striatal and cortical neurons in rats: lack of correlation with neuronal vulnerability in Huntington’s disease. J Neurosci. 1999;19:1189–1202. doi: 10.1523/JNEUROSCI.19-04-01189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kreitzer AC, Malenka RC. Endocannabinoid-mediated rescue of striatal LTD and motor deficits in Parkinson’s disease models. Nature. 2007;445:643–647. doi: 10.1038/nature05506. [DOI] [PubMed] [Google Scholar]
- 88.Gertler TS, Chan CS, Surmeier DJ. Dichotomous anatomical properties of adult striatal medium spiny neurons. J Neurosci. 2008;28:10814–10824. doi: 10.1523/JNEUROSCI.2660-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen W, Tian X, Day M, et al. Cholinergic modulation of Kir2 channels selectively elevates dendritic excitability in striatopallidal neurons. Nat Neurosci. 2007;10:1458–1466. doi: 10.1038/nn1972. [DOI] [PubMed] [Google Scholar]
- 90.Baydyuk M, Russell T, Liao G-Y, et al. TrkB receptor controls striatal formation by regulating the number of newborn striatal neurons. Proc Natl Acad Sci. 2011;108:1669–1674. doi: 10.1073/pnas.1004744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Besusso D, Geibel M, Kramer D, et al. BDNF-TrkB signaling in striatopallidal neurons controls inhibition of locomotor behavior. Nat Commun. 2013;4:1–12. doi: 10.1038/ncomms3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merienne N, Meunier C, Schneider A, et al. Cell-type-specific gene expression profiling in adult mouse brain reveals normal and disease-state signatures. Cell Rep. 2019;26:2477–2493.e9. doi: 10.1016/j.celrep.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 93.Shema R, Kulicke R, Cowley GS, et al. Synthetic lethal screening in the mammalian central nervous system identifies Gpx6 as a modulator of Huntington’s disease. Proc Natl Acad Sci. 2014;112:268–272. doi: 10.1073/pnas.1417231112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.André VM, Fisher YE, Levine MS. Altered balance of activity in the striatal direct and indirect pathways in mouse models of Huntington’s disease. Front Syst Neurosci. 2011;5:1–11. doi: 10.3389/fnsys.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barry J, Akopian G, Cepeda XC, et al. Striatal direct and indirect pathway output structures are differentially altered in mouse models of Huntington’s disease. J Neurosci 2018;38:4678–4694. [DOI] [PMC free article] [PubMed]
- 96.Deng Y-P, Wong T, Wan JY, et al. Differential loss of thalamostriatal and corticostriatal input to striatal projection neuron types prior to overt motor symptoms in the Q140 knock-in mouse model of Huntington’s disease. Front Syst Neurosci. 2014;8:1–15. doi: 10.3389/fnsys.2014.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goodliffe JW, Song H, Rubakovic A, et al. Differential changes to D1 and D2 medium spiny neurons in the 12-month-old Q175+/− mouse model of Huntington’s Disease. PLoS One. 2018;13:1–28. doi: 10.1371/journal.pone.0200626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mattis VB, Tom C, Akimov S, et al. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum Mol Genet. 2014;24:3257–3271. doi: 10.1093/hmg/ddv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim RG, Quan C, Reyes-Ortiz AM, et al. Huntington’s disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep. 2017;19:1365–1377. doi: 10.1016/j.celrep.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lim RG, Salazar LL, Wilton DK, et al. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci. 2017;20:648–660. doi: 10.1038/nn.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ward JM, La Spada AR. The expanding world of stem cell modeling of Huntington’s disease: creating tools with a promising future. Genome Med. 2012;4:68. doi: 10.1186/gm369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.An MC, Zhang N, Scott G, et al. Genetic correction of huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Castiglioni V, Onorati M, Rochon C, et al. Induced pluripotent stem cell lines from Huntington’s disease mice undergo neuronal differentiation while showing alterations in the lysosomal pathway. Neurobiol Dis. 2012;46:30–40. doi: 10.1016/j.nbd.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 104.Jeon I, Lee N, Li J-Y, et al. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30:2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 105.Kaye JA, Finkbeiner S. Modeling Huntington’s disease with induced pluripotent stem cells. Mol Cell Neurosci. 2013;56:50–64. doi: 10.1016/j.mcn.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vachey G, Deglon N. CRISPR/Cas9-mediated genome editing for Huntington’s disease. Methods Mol Biol. 1780;2018:463–481. doi: 10.1007/978-1-4939-7825-0_21. [DOI] [PubMed] [Google Scholar]
- 107.An MC, O’Brien RN, Zhang N, et al. Polyglutamine disease modeling: epitope based screen for homologous recombination using CRISPR/Cas9 system. PLoS Curr 2014;6. [DOI] [PMC free article] [PubMed]
- 108.Shin JW, Kim K-H, Chao MJ, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25:4566–4576. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Monteys AM, Ebanks SA, Keiser MS, et al. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu X, Tay Y, Sim B, et al. Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in huntington disease patient-derived induced pluripotent stem cells. Stem Cell Rep. 2017;8:619–633. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ritch JJ, Valencia A, Alexander J, et al. Multiple phenotypes in Huntington disease mouse neural stem cells. Mol Cell Neurosci. 2012;50:70–81. doi: 10.1016/j.mcn.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Molero AE, Arteaga-Bracho EE, Chen CH, et al. Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington’s disease. Proc Natl Acad Sci. 2016;113:5736–5741. doi: 10.1073/pnas.1603871113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Humbert S. Is Huntington disease a developmental disorder? EMBO Rep. 2010;11:899–899. doi: 10.1038/embor.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barnat M, Le Friec J, Benstaali C, et al. Huntingtin-mediated multipolar-bipolar transition of newborn cortical neurons is critical for their postnatal neuronal morphology. Neuron. 2017;93:99–114. doi: 10.1016/j.neuron.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 115.Mehta SR, Tom CM, Wang Y, et al. Human Huntington’s disease iPSC-derived cortical neurons display altered transcriptomics, morphology, and maturation. Cell Rep. 2018;25:1081–1096.e6. doi: 10.1016/j.celrep.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 116.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen H, Kankel MW, Su SC, et al. Exploring the genetics and non-cell autonomous mechanisms underlying ALS/FTLD. Cell Death Differ. 2018;25:646–660. doi: 10.1038/s41418-018-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ferraiuolo L. The non-cell-autonomous component of ALS: new in vitro models and future challenges. Biochem Soc Trans. 2014;42:1270–1274. doi: 10.1042/BST20140168. [DOI] [PubMed] [Google Scholar]
- 119.Dawson TM. Non-autonomous cell death in Parkinson’s disease. Lancet Neurol Engl; 2008. p. 474–475. [DOI] [PMC free article] [PubMed]
- 120.Chai M, Kohyama J. Non-cell-autonomous neurotoxicity in Parkinson’s disease mediated by Astroglial alpha-Synuclein. Stem Cell Rep. 2019;12:183–185. doi: 10.1016/j.stemcr.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yeterian EH, Van Hoesen GW. Cortico-striate projections in the rhesus monkey: the organization of certain cortico-caudate connections. Brain Res. 1978;139:43–63. doi: 10.1016/0006-8993(78)90059-8. [DOI] [PubMed] [Google Scholar]
- 122.Selemon LD, Goldman-Rakic PS. Longitudinal topography and interdigitation of corticostriatal projections in the rhesus monkey. J Neurosci. 1985;5:776–794. doi: 10.1523/JNEUROSCI.05-03-00776.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Altar CA, Cai N, Bliven T, et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 124.Kokaia Z, Andsberg G, Yan Q, et al. Rapid alterations of BDNF protein levels in the rat brain after focal ischemia: evidence for increased synthesis and anterograde axonal transport. Exp Neurol. 1998;154:289–301. doi: 10.1006/exnr.1998.6888. [DOI] [PubMed] [Google Scholar]
- 125.Bunner KD, Rebec GV. Corticostriatal dysfunction in Huntington’s disease: the basics. Front Hum Neurosci. 2016;10:1–12. doi: 10.3389/fnhum.2016.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cudkowicz M, Kowall NW. Degeneration of pyramidal projection neurons in Huntington’s disease cortex. Ann Neurol. 1990;27:200–204. doi: 10.1002/ana.410270217. [DOI] [PubMed] [Google Scholar]
- 127.Sotrel A, Paskevich PA, Kiely DK, et al. Morphometric analysis of the prefrontal cortex in Huntington’s disease. Neurology. 1991;41:1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- 128.Cepeda C, Wu N, André VM, et al. The corticostriatal pathway in Huntington’s disease. Prog Neurobiol. 2007;81:253–271. doi: 10.1016/j.pneurobio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rebec GV. Corticostriatal network dysfunction in Huntington’s disease: deficits in neural processing, glutamate transport, and ascorbate release. CNS Neurosci Ther. 2018;24:281–291. doi: 10.1111/cns.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Puigdellívol M, Cherubini M, Brito V, et al. A role for Kalirin-7 in corticostriatal synaptic dysfunction in Huntington’s disease. Hum Mol Genet. 2015;24:7265–7285. doi: 10.1093/hmg/ddv426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Murmu RP, Li W, Holtmaat A, et al. Dendritic spine instability leads to progressive neocortical spine loss in a mouse model of Huntington’s disease. J Neurosci. 2013;33:12997–13009. doi: 10.1523/JNEUROSCI.5284-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cummings DM, Milnerwood AJ, Dallérac GM, et al. Aberrant cortical synaptic plasticity and dopaminergic dysfunction in a mouse model of Huntington’s disease. Hum Mol Genet. 2006;15:2856–2868. doi: 10.1093/hmg/ddl224. [DOI] [PubMed] [Google Scholar]
- 133.Anglada-Huguet M, Xifró X, Giralt A, et al. Prostaglandin E2 EP1 receptor antagonist improves motor deficits and rescues memory decline in R6/1 mouse model of Huntington’s disease. Mol Neurobiol. 2014;49:784–795. doi: 10.1007/s12035-013-8556-x. [DOI] [PubMed] [Google Scholar]
- 134.Anglada-Huguet M, Giralt A, Rué L, et al. Loss of striatal 90-kDa ribosomal S6 kinase (Rsk) is a key factor for motor, synaptic and transcription dysfunction in Huntington’s disease. Biochim Biophys Acta Mol basis Dis. 1862;2016:1255–1266. doi: 10.1016/j.bbadis.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 135.Smith R, Petersen A, Bates GP, et al. Depletion of rabphilin 3A in a transgenic mouse model (R6/1) of Huntington’s disease, a possible culprit in synaptic dysfunction. Neurobiol Dis. 2005;20:673–684. doi: 10.1016/j.nbd.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 136.Torres-Peraza JF, Giralt A, Garcia-Martinez JM, et al. Disruption of striatal glutamatergic transmission induced by mutant huntingtin involves remodeling of both postsynaptic density and NMDA receptor signaling. Neurobiol Dis. 2008;29:409–421. doi: 10.1016/j.nbd.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 137.Hintiryan H, Foster NN, Bowman I, et al. The mouse cortico-striatal projectome. Nat Neurosci. 2016;19:1100–1114. doi: 10.1038/nn.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Estrada-Sanchez AM, Burroughs CL, Cavaliere S, et al. Cortical efferents lacking mutant huntingtin improve striatal neuronal activity and behavior in a conditional mouse model of Huntington’s disease. J Neurosci. 2015;35:4440–4451. doi: 10.1523/JNEUROSCI.2812-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gorski JA, Talley T, Qiu M, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reiner A, Deng Y-P. Disrupted striatal neuron inputs and outputs in Huntington’s disease. CNS Neurosci Ther. 2018;24:250–280. doi: 10.1111/cns.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deng YP, Wong T, Bricker-Anthony C, et al. Loss of corticostriatal and thalamostriatal synaptic terminals precedes striatal projection neuron pathology in heterozygous Q140 Huntington’s disease mice. Neurobiol Dis. 2013;60:89–107. doi: 10.1016/j.nbd.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jansen AHP, van Hal M, op den Kelder IC, et al. Frequency of nuclear mutant huntingtin inclusion formation in neurons and glia is cell-type-specific. Glia. 2017;65:50–61. doi: 10.1002/glia.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Faideau M, Kim J, Cormier K, et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum Mol Genet. 2010;19:3053–3067. doi: 10.1093/hmg/ddq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liddelow SA, Guttenplan KA, Clarke LE, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clarke LE, Liddelow SA, Chakraborty C, et al. Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A. 2018;115:E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meunier C, Merienne N, Jollé C, et al. Astrocytes are key but indirect contributors to the development of the symptomatology and pathophysiology of Huntington’s disease. Glia. 2016;64:1841–1856. doi: 10.1002/glia.23022. [DOI] [PubMed] [Google Scholar]
- 147.Tong X, Ao Y, Faas GC, et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat. Neurosci. 2014;17:694–703. doi: 10.1038/nn.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Juopperi TA, Kim WR, Chiang CH, et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntingtons disease patient cells. Mol Brain. 2012;5:1. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Osipovitch M, Asenjo Martinez A, Mariani JN, et al. Human ESC-derived chimeric mouse models of Huntington’s disease reveal cell-intrinsic defects in glial progenitor cell differentiation. Cell Stem Cell. 2019;24:107–122.e7. doi: 10.1016/j.stem.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hsiao HY, Chen YC, Chen HM, et al. A critical role of astrocyte-mediated nuclear factor-κB-dependent inflammation in huntington’s disease. Hum Mol Genet. 2013;22:1826–1842. doi: 10.1093/hmg/ddt036. [DOI] [PubMed] [Google Scholar]
- 151.Benraiss A, Wang S, Herrlinger S, et al. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat Commun. 2016;7:1–13. doi: 10.1038/ncomms11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ferrante RJ, Gutekunst CA, Persichetti F, et al. Heterogeneous topographic and cellular distribution of huntingtin expression in the normal human neostriatum. J Neurosci. 1997;17:3052–3063. doi: 10.1523/JNEUROSCI.17-09-03052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tai YF, Pavese N, Gerhard A, et al. Microglial activation in presymptomatic Huntington’s disease gene carriers. Brain. 2007;130:1759–1766. doi: 10.1093/brain/awm044. [DOI] [PubMed] [Google Scholar]
- 154.Pavese N, Gerhard A, Tai YF, et al. Microglial activation correlates with severity in Huntington disease: a clinical and PET study. Neurology. 2006;66:1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 155.Politis M, Lahiri N, Niccolini F, et al. Increased central microglial activation associated with peripheral cytokine levels in premanifest Huntington’s disease gene carriers. Neurobiol Dis. 2015;83:115–121. doi: 10.1016/j.nbd.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 156.Silvestroni A, Faull RLM, Strand AD, et al. Distinct neuroinflammatory profile in post-mortem human Huntington’s disease. Neuroreport. 2009;20:1098–1103. doi: 10.1097/WNR.0b013e32832e34ee. [DOI] [PubMed] [Google Scholar]
- 157.Bjorkqvist M, Wild EJ, Thiele J, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sapp E, Kegel KB, Aronin N, et al. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol. 2001;60:161–172. doi: 10.1093/jnen/60.2.161. [DOI] [PubMed] [Google Scholar]
- 159.Labadorf A, Hoss AG, Lagomarsino V, et al. Correction: RNA sequence analysis of human huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS One. 2016;11:1–21. doi: 10.1371/journal.pone.0160295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Singhrao SK, Neal JW, Morgan BP, et al. Increased complement biosynthesis by microglia and complement activation on neurons in Huntington’s disease. Exp Neurol. 1999;159:362–376. doi: 10.1006/exnr.1999.7170. [DOI] [PubMed] [Google Scholar]
- 161.Simmons DA, Casale M, Alcon B, et al. Ferritin accumulation in dystrophic microglia is an early event in the development of Huntington’s disease. Glia. 2007;55:1074–1084. doi: 10.1002/glia.20526. [DOI] [PubMed] [Google Scholar]
- 162.Wacker JL, Huang S-Y, Steele AD, et al. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington’s disease. J Neurosci. 2009;29:9104–9114. doi: 10.1523/JNEUROSCI.2250-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Franciosi S, Ryu JK, Shim Y, et al. Age-dependent neurovascular abnormalities and altered microglial morphology in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2012;45:438–449. doi: 10.1016/j.nbd.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 164.Ochaba J, Monteys AM, O’Rourke JG, et al. PIAS1 regulates mutant Huntingtin accumulation and Huntington’s disease-associated phenotypes in vivo. Neuron. 2016;90:507–520. doi: 10.1016/j.neuron.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Khoshnan A, Sabbaugh A, Calamini B, et al. IKK β and mutant huntingtin interactions regulate the expression of IL-34: implications for microglial-mediated neurodegeneration in HD. Hum Mol Genet. 2017;26:4267–4277. doi: 10.1093/hmg/ddx315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kraft AD, Kaltenbach LS, Lo DC, et al. Activated microglia proliferate at neurites of mutant huntingtin-expressing neurons. Neurobiol Aging. 2012;33:621.e17–33. doi: 10.1016/j.neurobiolaging.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Di Paola M, Luders E, Cherubini A, et al. Multimodal MRI analysis of the corpus callosum reveals white matter differences in presymptomatic and early Huntington’s disease. Cereb Cortex. 2012;22:2858–2866. doi: 10.1093/cercor/bhr360. [DOI] [PubMed] [Google Scholar]
- 168.Reading SAJ, Yassa MA, Bakker A, et al. Regional white matter change in pre-symptomatic Huntington’s disease: a diffusion tensor imaging study. Psychiatry Res. 2005;140:55–62. doi: 10.1016/j.pscychresns.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 169.Rosas HD, Lee SY, Bender AC, et al. Altered white matter microstructure in the corpus callosum in Huntington’s disease: implications for cortical “disconnection”. Neuroimage. 2010;49:2995–3004. doi: 10.1016/j.neuroimage.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McColgan P, Gregory S, Seunarine KK, et al. Brain regions showing white matter loss in Huntington’s disease are enriched for synaptic and metabolic genes. Biol Psychiatry. 2018;83:456–465. doi: 10.1016/j.biopsych.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meng Y, Jiang J, Bachevalier J, et al. Developmental whole brain white matter alterations in transgenic Huntington’s disease monkey. Sci Rep. 2017;7:379. doi: 10.1038/s41598-017-00381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zuccato C, Ciammola A, Rigamonti D, et al. Loss of Huntingtin-mediated BDNF gene transcription huntingtin-mediate in Huntington’s disease. Adv Sci. 2001;293:493–499. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 173.Gauthier LR, Charrin BC, Borrell-Pagès M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 174.Ginés S, Paoletti P, Alberch J. Impaired TrkB-mediated ERK1/2 activation in huntington disease knock-in striatal cells involves reduced p52/p46 Shc expression. J Biol Chem. 2010;285:21537–21548. doi: 10.1074/jbc.M109.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McGuire JR, Rong J, Li S-H, et al. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- 176.Engelender S, Sharp AH, Colomer V, et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 177.Conner JM, Lauterborn JC, Yan Q, et al. Distribution of brain-derived neurotrophic factor (BDNF) protein and mRNA in the normal adult rat CNS: evidence for anterograde axonal transport. J Neurosci. 1997;17:2295–2313. doi: 10.1523/JNEUROSCI.17-07-02295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ginés S, Bosch M, Marco S, et al. Reduced expression of the TrkB receptor in Huntington’s disease mouse models and in human brain. Eur J Neurosci. 2006;23:649–658. doi: 10.1111/j.1460-9568.2006.04590.x. [DOI] [PubMed] [Google Scholar]
- 179.Zuccato C, Marullo M, Conforti P, et al. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 2008;18:225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Brito V, Puigdellívol M, Giralt A, et al. Imbalance of p75NTR/TrkB protein expression in Huntington’s disease: implication for neuroprotective therapies. Cell Death Dis. 2013;4:1–15. doi: 10.1038/cddis.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liot G, Zala D, Pla P, et al. Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J Neurosci. 2013;33:6298–6309. doi: 10.1523/JNEUROSCI.2033-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Baydyuk M, Nguyen MT, Xu B. Chronic deprivation of TrkB signaling leads to selective late-onset nigrostriatal dopaminergic degeneration. 2011;228:118–125. [DOI] [PMC free article] [PubMed]
- 183.Li Y, Yui D, Luikart BW, et al. Conditional ablation of brain-derived neurotrophic factor-TrkB signaling impairs striatal neuron development. Proc Natl Acad Sci U S A. 2012;109:15491–15496. doi: 10.1073/pnas.1212899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rauskolb S, Zagrebelsky M, Dreznjak A, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. J Neurosci. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mattis VB, Tom C, Akimov S, et al. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum Mol Genet. 2015;24:3257–3271. doi: 10.1093/hmg/ddv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xie Y, Hayden MR, Xu B. BDNF overexpression in the forebrain rescues Huntington’s disease phenotypes in YAC128 mice. J Neurosci. 2010;30:14708–14718. doi: 10.1523/JNEUROSCI.1637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.da Fonsêca VS, da Silva Colla AR, de Paula Nascimento-Castro C, et al. Brain-derived neurotrophic factor prevents depressive-like behaviors in early-symptomatic YAC128 Huntington’s disease mice. Mol Neurobiol. 2018;55:7201–7215. doi: 10.1007/s12035-018-0890-6. [DOI] [PubMed] [Google Scholar]
- 188.Connor B, Sun Y, Von Hieber D, et al. AAV 1/2-mediated BDNF gene therapy in a transgenic rat model of Huntington’s disease. Gene Ther. 2016;23:283–295. doi: 10.1038/gt.2015.113. [DOI] [PubMed] [Google Scholar]
- 189.Giralt A, Friedman HC, Caneda-Ferrón B, et al. BDNF regulation under GFAP promoter provides engineered astrocytes as a new approach for long-term protection in Huntington’s disease. Gene Ther. 2010;17:1294–1308. doi: 10.1038/gt.2010.71. [DOI] [PubMed] [Google Scholar]
- 190.Gharami K, Xie Y, An JJ, et al. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J Neurochem. 2008;105:369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yu C, Li CH, Chen S, et al. Decreased BDNF release in cortical neurons of a knock-in mouse model of Huntington’s disease. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-34883-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhao X, Chen X-Q, Han E, et al. TRiC subunits enhance BDNF axonal transport and rescue striatal atrophy in Huntington’s disease. Proc Natl Acad Sci. 2016;113:E5655–E5664. doi: 10.1073/pnas.1603020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Unterwald EM, Page ME, Brown TB, et al. Behavioral and transcriptome alterations in male and female mice with postnatal deletion of TrkB in dorsal striatal medium spiny neurons. 2013;1–14. [DOI] [PMC free article] [PubMed]
- 194.Plotkin JL, Day M, Peterson JD, et al. Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington’s disease. Neuron. 2014;83:178–188. [DOI] [PMC free article] [PubMed]
- 195.Fulmer CG, VonDran MW, Stillman AA, et al. Astrocyte-derived BDNF supports myelin protein synthesis after cuprizone-induced demyelination. J Neurosci. 2014;34:8186–8196. doi: 10.1523/JNEUROSCI.4267-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Poyhonen S, Er S, Domanskyi A, et al. Effects of neurotrophic factors in glial cells in the central nervous system: expression and properties in neurodegeneration and injury. Front Physiol. 2019;10:486. doi: 10.3389/fphys.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zuccato C, Liber D, Ramos C, et al. Progressive loss of BDNF in a mouse model of Huntington’s disease and rescue by BDNF delivery. Pharmacol Res. 2005;52:133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 198.Zuccato C, Marullo M, Vitali B, et al. Brain-derived neurotrophic factor in patients with Huntington’s disease. PLoS One 2011;6. [DOI] [PMC free article] [PubMed]
- 199.Canals JM, Pineda JR, Torres-Peraza JF, et al. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J Neurosci 2004;24:7727–7739. [DOI] [PMC free article] [PubMed]
- 200.Wang L, Lin F, Wang J, et al. Expression of mutant N-terminal huntingtin fragment (htt552-100Q) in astrocytes suppresses the secretion of BDNF. Brain Res. 2012;1449:69–82. doi: 10.1016/j.brainres.2012.01.077. [DOI] [PubMed] [Google Scholar]
- 201.Hong Y, Zhao T, Li X-J, et al. Mutant Huntingtin impairs BDNF release from astrocytes by disrupting conversion of Rab3a-GTP into Rab3a-GDP. J Neurosci. 2016;36:8790–8801. doi: 10.1523/JNEUROSCI.0168-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bossi SR, Simpson JR, Isacson O. Age dependence of striatal neuronal death caused by mitochondrial dysfunction. Neuroreport. 1993;4:73–76. doi: 10.1097/00001756-199301000-00019. [DOI] [PubMed] [Google Scholar]
- 203.Yano H, Baranov SV, Baranova OV, et al. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci. 2014;17:822–831. doi: 10.1038/nn.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zheng J, Winderickx J, Franssens V, et al. A mitochondria-associated oxidative stress perspective on Huntington’s disease. Front Mol Neurosci. 2018;11:329. doi: 10.3389/fnmol.2018.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Oliveira JMA, Gonçalves J. In situ mitochondrial Ca2+ buffering differences of intact neurons and astrocytes from cortex and striatum. J Biol Chem. 2009;284:5010–5020. doi: 10.1074/jbc.M807459200. [DOI] [PubMed] [Google Scholar]
- 206.Hamilton J, Pellman JJ, Brustovetsky T, et al. Oxidative metabolism in YAC128 mouse model of Huntington’s disease. Hum Mol Genet. 2015;24:4862–4878. doi: 10.1093/hmg/ddv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cui L, Jeong H, Borovecki F, et al. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 208.Weydt P, Pineda VV, Torrence AE, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1α in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 209.Hering T, Birth N, Taanman JW, et al. Selective striatal mtDNA depletion in end-stage Huntington’s disease R6/2 mice. Exp Neurol. 2015;266:22–29. doi: 10.1016/j.expneurol.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 210.Kedaigle AJ, Fraenkel E, Atwal RS, et al. Bioenergetic deficits in Huntington's disease iPSC-derived neural cells and rescue with glycolytic metabolites. Hum Mol Genet. 2019 [DOI] [PMC free article] [PubMed]
- 211.Kojer K, Hering T, Bazenet C, et al. Huntingtin aggregates and mitochondrial pathology in skeletal muscle but not heart of late-stage R6/2 mice. J Huntingtons Dis. 2019;8(2):145–15. doi: 10.3233/JHD-180324. [DOI] [PubMed] [Google Scholar]
- 212.Chakraborty J, Rajamma U, Mohanakumar KP. A mitochondrial basis for Huntington’s disease: therapeutic prospects. Mol Cell Biochem. 2014;389:277–291. doi: 10.1007/s11010-013-1951-9. [DOI] [PubMed] [Google Scholar]
- 213.Boussicault L, Hérard AS, Calingasan N, et al. Impaired brain energy metabolism in the BACHD mouse model of Huntington’s disease: Critical role of astrocyte-neuron interactions. J Cereb Blood Flow Metab. 2014;34:1500–1510. doi: 10.1038/jcbfm.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Skotte NH, Andersen JV, Santos A, et al. Integrative characterization of the R6/2 mouse model of Huntington’s disease reveals dysfunctional astrocyte metabolism. Cell Rep. 2018;23:2211–2224. doi: 10.1016/j.celrep.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 215.Reddy PH. Increased mitochondrial fission and neuronal dysfunction in Huntington’s disease: implications for molecular inhibitors of excessive mitochondrial fission. Drug Discov Today. 2014;19:951–955. doi: 10.1016/j.drudis.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shirendeb UP, Calkins MJ, Manczak M, et al. Mutant Huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Song W, Chen J, Petrilli A, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim J, Moody JP, Edgerly CK, et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Costa V, Giacomello M, Hudec R, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Orr AL, Li S, Wang C-E, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chang DTW, Rintoul GL, Pandipati S, et al. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 222.Cooper JK, Schilling G, Peters MF, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 223.DeGuire SM, Ruggeri FS, Fares M-B, et al. N-terminal Huntingtin (Htt) phosphorylation is a molecular switch regulating Htt aggregation, helical conformation, internalization, and nuclear targeting. J Biol Chem. 2018;293:18540–18558. doi: 10.1074/jbc.RA118.004621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gu X, Cantle JP, Greiner ER, et al. N17 modifies mutant huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron. 2015;85:726–741. doi: 10.1016/j.neuron.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Weiberth KF, Tsvetkov AS, Yang XW, et al. Serine 421 regulates mutant huntingtin toxicity and clearance in mice. J Clin Invest. 2016;126:3585–3597. doi: 10.1172/JCI80339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Warby SC, Chan EY, Metzler M, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum Mol Genet. 2005;14:1569–1577. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- 227.Bowles KR, Jones L. Kinase signalling in Huntington’s disease. J Huntingtons Dis. 2014;3:89–123. doi: 10.3233/JHD-140106. [DOI] [PubMed] [Google Scholar]
- 228.Saavedra A, Alberch J, Pérez-Navarro E. Don’t take away my P: phosphatases as therapeutic targets in Huntington’s disease. 2012; In Ersoy Tunali (Ed.), Huntington’s disease - core concepts and current advances. Intech, pp. 465–494 Chapter 20.
- 229.Humbert S, Bryson EA, Cordelières FP, et al. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves huntingtin phosphorylation by Akt. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 230.Zhao T, Hong Y, Li S, et al. Compartment-dependent degradation of mutant huntingtin accounts for its preferential accumulation in neuronal processes. J Neurosci. 2016;36:8317–8328. doi: 10.1523/JNEUROSCI.0806-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tydlacka S, Wang C-E, Wang X, et al. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J Neurosci. 2008;28:13285–13295. doi: 10.1523/JNEUROSCI.4393-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hong Y, Zhao T, Li X-J, et al. Mutant huntingtin inhibits αB-crystallin expression and impairs exosome secretion from astrocytes. J Neurosci. 2017;37:9550–9563. doi: 10.1523/JNEUROSCI.1418-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.McGeer EG, McGeer PL. Duplication of biochemical changes of Huntington’s chorea by intrastriatal injections of glutamic and kainic acids. Nature. 1976;263:517–519. doi: 10.1038/263517a0. [DOI] [PubMed] [Google Scholar]
- 234.Coyle JT, Schwarcz R. Lesion of striatal neurones with kainic acid provides a model for Huntington’s chorea. Nature. 1976;263:244–246. doi: 10.1038/263244a0. [DOI] [PubMed] [Google Scholar]
- 235.Cui H, Hayashi A, Sun H-S, et al. PDZ protein interactions underlying NMDA receptor-mediated excitotoxicity and neuroprotection by PSD-95 inhibitors. J Neurosci. 2007;27:9901–9915. doi: 10.1523/JNEUROSCI.1464-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang Y, Su P, Liang P, et al. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J Neurosci. 2010;30:7575–7586. doi: 10.1523/JNEUROSCI.1312-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Petralia RS. Distribution of extrasynaptic NMDA receptors on neurons. ScientificWorldJournal. 2012;2012:267120. doi: 10.1100/2012/267120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Parsons MP, Vanni MP, Woodard CL, et al. Real-time imaging of glutamate clearance reveals normal striatal uptake in Huntington disease mouse models. Nat Commun. 2016;7:1–12. doi: 10.1038/ncomms11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Siebzehnrübl FA, Raber KA, Urbach YK, et al. Early postnatal behavioral, cellular, and molecular changes in models of Huntington disease are reversible by HDAC inhibition. Proc Natl Acad Sci. 2018;115:E8765–E8774. doi: 10.1073/pnas.1807962115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Milnerwood AJ, Gladding CM, Pouladi MA, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 242.Marco S, Giralt A, Petrovic MM, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med. 2013;19:1030–1038. doi: 10.1038/nm.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Martinez-Turrillas R, Puerta E, Chowdhury D, et al. The NMDA receptor subunit GluN3A protects against 3-nitroproprionic-induced striatal lesions via inhibition of calpain activation. Neurobiol Dis. 2012;48:290–298. doi: 10.1016/j.nbd.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 244.Ali NJ, Levine MS. Changes in expression of N-methyl-D-aspartate receptor subunits occur early in the R6/2 mouse model of Huntington’s disease. Dev Neurosci. 2006;28:230–238. doi: 10.1159/000091921. [DOI] [PubMed] [Google Scholar]
- 245.Cha JH, Frey AS, Alsdorf SA, et al. Altered neurotransmitter receptor expression in transgenic mouse models of Huntington’s disease. Philos. Trans R Soc Lond B Biol Sci. 1999;354:981–989. doi: 10.1098/rstb.1999.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jarabek BR, Yasuda RP, Wolfe BB. Regulation of proteins affecting NMDA receptor-induced excitotoxicity in a Huntington’s mouse model. Brain. 2004;127:505–516. doi: 10.1093/brain/awh058. [DOI] [PubMed] [Google Scholar]
- 247.Li L, Fan M, Icton CD, et al. Role of NR2B-type NMDA receptors in selective neurodegeneration in Huntington disease. Neurobiol Aging. 2003;24:1113–1121. doi: 10.1016/j.neurobiolaging.2003.04.003. [DOI] [PubMed] [Google Scholar]
- 248.Benn CL, Slow EJ, Farrell LA, et al. Glutamate receptor abnormalities in the YAC128 transgenic mouse model of Huntington’s disease. Neuroscience. 2007;147:354–372. doi: 10.1016/j.neuroscience.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ambroziak W, Fourie C, Montgomery JM. SAP97-mediated rescue of NMDA receptor surface distribution in a neuronal model of Huntington’s disease. Hippocampus. 2018;28:707–723. doi: 10.1002/hipo.22995. [DOI] [PubMed] [Google Scholar]
- 250.Petr GT, Schultheis LA, Hussey KC, et al. Decreased expression of GLT-1 in the R6/2 model of Huntington’s disease does not worsen disease progression. Eur J Neurosci. 2013;38:2477–2490. doi: 10.1111/ejn.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lievens JC, Woodman B, Mahal A, et al. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiol Dis. 2001;8:807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- 252.Miller BR, Dorner JL, Shou M, et al. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008;153:329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Behrens PF, Franz P, Woodman B, et al. Impaired glutamate transport and glutamate-glutamine cycling: downstream effects of the Huntington mutation. Brain. 2002;125:1908–1922. doi: 10.1093/brain/awf180. [DOI] [PubMed] [Google Scholar]
- 254.Huang K, Kang MH, Askew C, et al. Palmitoylation and function of glial glutamate transporter-1 is reduced in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2010;40:207–215. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 255.Jiang R, Diaz-Castro B, Looger LL, et al. Dysfunctional calcium and glutamate signaling in striatal astrocytes from Huntington’s disease model mice. J Neurosci. 2016;36:3453–3470. doi: 10.1523/JNEUROSCI.3693-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lee W, Reyes RC, Gottipati MK, et al. Enhanced Ca2+−dependent glutamate release from astrocytes of the BACHD Huntington’s disease mouse model. Neurobiol Dis. 2013;58:192–199. doi: 10.1016/j.nbd.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cho IK, Yang B, Forest C, et al. Amelioration of Huntington’s disease phenotype in astrocytes derived from iPSC-derived neural progenitor cells of Huntington’s disease monkeys. PLoS One. 2019;14:e0214156. doi: 10.1371/journal.pone.0214156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bunner KD, Rangel-Barajas C, McCormick BM, et al. Behavioral and electrophysiological improvements following up-regulation of GLT-1 in the Q175 Huntington’s mouse. Abstr Soc Neurosci 2016;418.09.
- 259.Smith-Dijak AI, Sepers MD, Raymond LA. Alterations in synaptic function and plasticity in Huntington disease. J Neurochem. 2019;150:346–365. [DOI] [PubMed]
- 260.Dallerac GM, Levasseur G, Vatsavayai SC, et al. Dysfunctional dopaminergic neurones in mouse models of Huntington’s disease: A role for SK3 channels. Neurodegener Dis. 2015;15:93–108. doi: 10.1159/000375126. [DOI] [PubMed] [Google Scholar]
- 261.Langfelder P, Gao F, Wang N, et al. MicroRNA signatures of endogenous Huntingtin CAG repeat expansion in mice. PLoS One. 2018;13:1–20. doi: 10.1371/journal.pone.0190550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Novati A, Hentrich T, Wassouf Z, et al. Environment-dependent striatal gene expression in the BACHD rat model for Huntington disease. Sci Rep. 2018;8:5803. doi: 10.1038/s41598-018-24243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Brochier C, Gaillard M-C, Diguet E, et al. Quantitative gene expression profiling of mouse brain regions reveals differential transcripts conserved in human and affected in disease models. Physiol Genomics. 2008;33:170–179. doi: 10.1152/physiolgenomics.00125.2007. [DOI] [PubMed] [Google Scholar]
- 264.Le Gras S, Keime C, Anthony A, et al. Altered enhancer transcription underlies Huntington’s disease striatal transcriptional signature. Sci Rep. 2017;7:42875. doi: 10.1038/srep42875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ament SA, Pearl JR, Grindeland A, et al. High resolution time-course mapping of early transcriptomic, molecular and cellular phenotypes in Huntington’s disease CAG knock-in mice across multiple genetic backgrounds. Hum Mol Genet. 2017;26:913–922. doi: 10.1093/hmg/ddx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hervas-Corpion I, Guiretti D, Alcaraz-Iborra M, et al. Early alteration of epigenetic-related transcription in Huntington’s disease mouse models. Sci Rep. 2018;8:9925. doi: 10.1038/s41598-018-28185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Becanovic K, Pouladi MA, Lim RS, et al. Transcriptional changes in Huntington disease identified using genome-wide expression profiling and cross-platform analysis. Hum Mol Genet. 2010;19:1438–1452. doi: 10.1093/hmg/ddq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Luthi-Carter R, Strand AD, Hanson SA, et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington’s disease mouse models reveal context-independent effects. Hum Mol Genet. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- 269.Fossale E, Seong IS, Coser KR, et al. Differential effects of the Huntington’s disease CAG mutation in striatum and cerebellum are quantitative not qualitative. Hum Mol Genet. 2011;20:4258–4267. doi: 10.1093/hmg/ddr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Neueder A, Bates GP. A common gene expression signature in Huntington’s disease patient brain regions. BMC Med Genet. 2014;7:60. doi: 10.1186/s12920-014-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 272.Yamanaka T, Miyazaki H, Oyama F, et al. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–839. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 274.Zuccato C, Belyaev N, Conforti P, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zuccato C, Tartari M, Crotti A, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 276.Vashishtha M, Ng CW, Yildirim F, et al. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci. 2013;110:E3027–E3036. doi: 10.1073/pnas.1311323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Sadri-Vakili G, Cha J-HJ. Mechanisms of disease: histone modifications in Huntington’s disease. Nat Clin Pract Neurol. 2006;2:330–338. doi: 10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 278.Gray SG. Targeting Huntington’s disease through histone deacetylases. Clin Epigenetics. 2011;2:257–277. doi: 10.1007/s13148-011-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.McCampbell A, Taylor JP, Taye AA, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 431 kb)