Abstract
Polyglutamine expansion disorders, which include Huntington’s disease, have expanded CAG repeats that result in polyglutamine expansions in affected proteins. How this specific feature leads to distinct neuropathies in 11 different diseases is a fascinating area of investigation. Most proteins affected by polyglutamine expansions are ubiquitously expressed, yet their mechanisms of selective neurotoxicity are unknown. Induced pluripotent stem cells have emerged as a valuable tool to model diseases, understand molecular mechanisms, and generate relevant human neural and glia subtypes, cocultures, and organoids. Ideally, this tool will generate specific neuronal populations that faithfully recapitulate specific polyglutamine expansion disorder phenotypes and mimic the selective vulnerability of a given disease. Here, we review how induced pluripotent technology is used to understand the effects of the disease-causing polyglutamine protein on cell function, identify new therapeutic targets, and determine how polyglutamine expansion affects human neurodevelopment and disease. We will discuss ongoing challenges and limitations in our use of induced pluripotent stem cells to model polyglutamine expansion diseases.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00810-8) contains supplementary material, which is available to authorized users.
Key Words: Triplet repeat disorders, neurodegeneration, polyglutamine, Huntington’s disease, induced pluripotent stem cells.
Polyglutamine Expansion Diseases
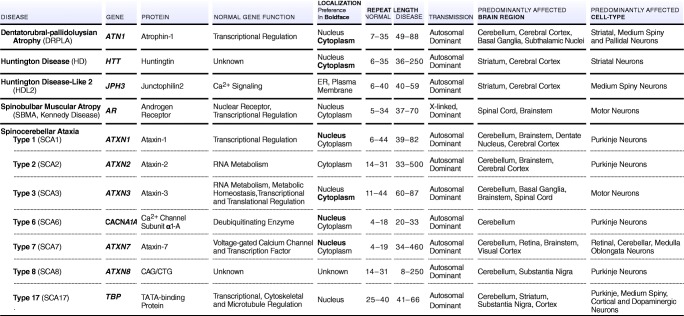
Polyglutamine (polyQ) expansion diseases belong to a large family of repeat expansion disorders characterized by repeating DNA sequences in affected genes, which can result in neurodegenerative disease. More than 40 rare genetic diseases are caused by repeat expansions. In polyQ expansion diseases, cytosine–adenine–guanine (CAG) expansion is localized in coding sequences of the gene (exonic). The disease phenotype manifests when the length of trinucleotide expansion of the mutated gene reaches a threshold which depends upon the disease (Table 1). When the mutated gene is transmitted through each generation, the length of the trinucleotide tract increases. This leads to an earlier age of onset and often a more severe disease phenotype, in a phenomenon known as anticipation [1–3]. The abnormal expansion of CAG repeats within exonic regions expresses a polyQ tract, which typically results in a structurally altered protein. Currently, expansion of CAG repeats is implicated in 11 human neurodegenerative disorders (Table 1): dentatorubral-pallidoluysian atrophy (DRPLA) [4, 5], Huntington’s disease (HD) [6, 7], Huntington’s disease-like 2 (HDL2), spinobulbar muscular atrophy (SBMA, Kennedy disease) [8, 9], and spinocerebellar ataxia types (SCA1, 2, 3, 6, 7, 8, and 17) [10, 11]. PolyQ expansion diseases possess autosomal dominant inheritance, except for the SBMA, which is X-linked.
Table 1.
PolyQ expansion diseases and their characteristics.
Induced pluripotent stem cells (iPSCs) have emerged as a valuable tool for disease modeling and generating relevant human neural subtypes in vitro [12–17]. This new approach has uncovered molecular mechanisms that contribute to neurodegenerative disease onset [18–22]. However, to better understand how expanded polyQ proteins lead to abnormal neuronal function in human tissue, we will need to fully harness iPSCs to generate specific neuronal populations that faithfully recapitulate polyQ expansion disorder phenotypes. Further understanding of the human neurodevelopmental process that leads to the formation of specific brain regions and cell types will allow us to better model these diseases. Cortical and striatal neurons are needed to model HD and DRPLA, whereas motor neurons are relevant for SBMA, and Purkinje cells for SCA1s (Table 1, Fig. 1). Thus, protocols for differentiating iPSCs into relevant central nervous system (CNS) neurons are needed to closely mimic the affected neuronal phenotype. Furthermore, it is essential to determine how faithfully in vitro iPSC models correspond to native disease phenotypes. In this review, we will evaluate how these models, derived from patients with polyQ expansion disease, can shed light on molecular mechanisms related to expanded CAG tracts, selective vulnerability of neurons to the mutant polyQ protein, and the impact of other cells types on neuropathological states. We will discuss opportunities to improve our understanding of these diseases using iPSCs.
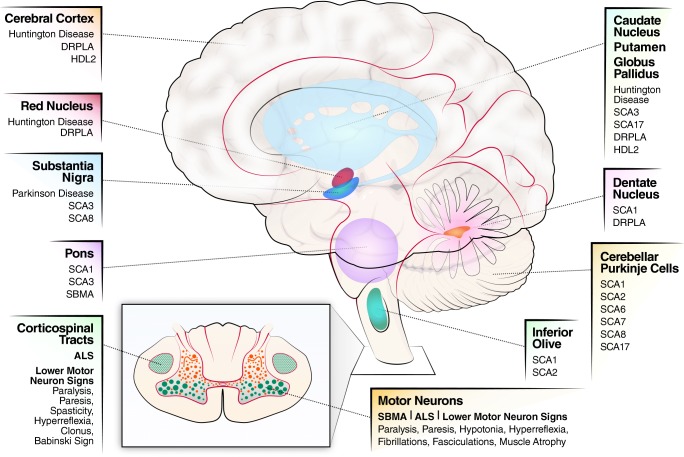
Fig. 1.
Schematic of polyQ expansion diseases and affected areas of the brain.
Heterogeneity Within PolyQ Expansion Diseases: Distinct Protein and CAG Repeat Lengths
The 11 polyQ neurodegenerative disorders illustrate the heterogeneity within polyQ diseases and their affected proteins (see Fig. 1) [23]. Whereas expanded CAG repeats and resultant polyQ sequences broadly drive neurodegeneration, disease-specific features are unique to each protein (Table 1). These disorders feature a range of CAG repeat lengths in both normal and disease alleles. For example, Atrophin-1 (ATN1), the gene involved in DRPLA, contains 7 to 35 repeats in wild-type versus 49 to 88 in disease conditions. The wild-type voltage-dependent calcium channel (CACNA1A) gene, involved in SCA6, contains 4 to 18 repeat expansions, whereas the disease phenotype contains 19 to 33 CAGs. For SCA8, CAG and CTG expansions involve 80 to 250 repeats in the ATXN8 gene (Table 1).
PolyQ Disease Proteins: Dysregulation of Subcellular Localization
The protein product in each of these neurodegenerative disorders is unique with specific subcellular localization and function (Table 1). Cellular localization of conformationally altered polyQ protein is generally associated with neurotoxicity [6, 24–27]. Many proteins involved in polyQ expansion disease have nuclear localization and export signals that guide cross-compartmental interactions with binding partners during disease progression. For example, SBMA is caused by CAG repeat expansion in the androgen receptor (AR) gene. Under basal conditions, the inactive form of AR is localized in the cytoplasm. Upon activation by androgen, AR undergoes posttranslational modification that results in translocation to the nucleus, where the protein acts as a nuclear receptor by binding to chromatin and regulating the expression of androgen-responsive genes [28–30]. The polyQ-expanded AR has reduced intranuclear mobility, nuclear export rate, and transcriptional activity [31]. Another example is SCA1, which is caused by an abnormal number of CAG repeats in Ataxin-1 (ATXN1). The ATXN1 protein is often localized within the nucleus, where it is involved in transcriptional regulation and RNA (ribonucleic acid) metabolism [32]. Irwin et al. [33] showed that polyQ expansion prevents ataxin-1 from exiting the nucleus, leading to accumulation and subsequent neurotoxicity in SCA1. SCA1 mice expressing ataxin-1 with a nonfunctional nuclear localization signal do not develop the disease, suggesting that nuclear translocation is important in mutant ataxin-1-mediated neurotoxicity. This principle of improper subcellular localization can also affect cytoplasmic proteins; the accumulation of mutant ataxin-2 in the cytoplasm and stress granules correlates with toxicity in SCA2 [34, 35].
PolyQ Expansion Diseases Affect Different Areas of the Brain and Neuronal Cell Types
The unique protein for each disease and its biology determine disease-specific features. Although proteins structurally altered by polyQ expansion are broadly expressed throughout different tissues and organs, their primary effect is neuronal dysfunction characterized by synaptic loss, atrophy of dendritic arborizations, axonal swelling, and loss of neuronal markers. Each CAG repeat disorder targets a different region in the brain (Fig. 1, Table 1) and neuronal subpopulations, leading to specific clinical and neuropathologic features, as reviewed by Lieberman et al. [36]. For example, SBMA neuropathogenesis is associated with loss of motor neurons in the brainstem and spinal cord as well as muscle atrophy (Table 1, Fig. 1). In SCA7, degeneration caused by polyQ-expanded ATXN7 is characterized by the loss of Purkinje neurons and rapidly progressing cerebellar and retinal degeneration.
Common Pathogenic Mechanisms of PolyQ Diseases
An enduring question in the field of polyQ expansion diseases is how the translation of CAG repeats into polyQ tracts at the molecular level can lead to neurodegeneration in different polyQ diseases. Numerous studies have identified a common pathogenic mechanism underlying all polyQ expansion diseases: mutant protein misfolding leads to aberrant interactions with key binding partners, protein aggregation, formation of nuclear and cytoplasmic inclusions, toxic fragment generation via mutant protein cleavage, transcriptional dysregulation, autophagy, repeat-associated non-ATG (RAN) translation, RNA toxicity, DNA damage, and mitochondrial dysfunction [37–39]. These mechanisms are carefully reviewed in accompanying articles of this special edition [40–46]. Our review covers a subset of the 11 diseases, as patient-derived iPSC models have not been characterized for all polyQ diseases. Many groups are generating patient-derived polyQ expansion disease iPSCs, but these have not yet been characterized with respect to disease modeling [47–52], and thus will not be included in our review. We will focus on research efforts directed at using iPSCs to model these diseases, starting with HD.
Huntington’s Disease
HD is a rare, progressive neurodegenerative disease that affects 1 in every 10,000 individuals [53]. HD is characterized by involuntary movements, emotional disturbances, and cognitive decline as the disease progresses [54]. HD arises from a mutation in exon 1 of the huntingtin (HTT) gene, resulting in CAG triplet repeat expansion [6]. CAG repeat length inversely correlates with disease severity and age of onset. However, CAG repeat length is not the only factor that determines age of onset, as other genetic and environmental modifiers account for ~ 35% of the variance. HD is associated with progressive neuronal loss in the striatum, cortex, and globus pallidus, resulting in a degenerative disease of the brain that shares some features with other polyQ diseases [55]. Neuronal dysfunction and cell death in HD are attributed to both “a toxic gain of function” of mutant HTT (mHTT) and “a loss of function” of normal HTT. Although monogenic in nature, HD pathogenesis is incredibly complex, and despite the current state-of-the-field, there is a need to determine the exact disease mechanism of the polyQ expansion in HTT in human-relevant models. We will consider advances in HD patient-derived iPSCs to model HD pathogenesis. Common themes from these studies are that neurodevelopmental changes occur due to repeat expansion in the HTT protein and that identification of new molecular pathways highlight opportunities for therapeutic intervention in HD [56–61].
History of HD Embryonic Stem Cells
Prior to using iPSCs to model HD, embryonic stem cells (ESCs), derived from the inner cell mass of donated blastocyst-stage embryos, were used as models. ESCs are pluripotent stem cells that can self-renew and give rise to all cell types in the body [62]. Although ESCs can be used to model HD and for the development of treatments for HD, they have several limitations, including limited access to preimplantation HD embryos and ethical concerns associated with the process of obtaining human ESCs. Additionally, as a treatment model, ESC-derived allogeneic transplantation substantially increases the risk of immune rejection and tumor development. Despite these limitations, the studies described below established methods for creating cells similar to medium spiny neurons (MSNs), created human models for HD, and validated the efficacy of stem cell transplantation.
Aubry et al. [15] differentiated hESCs into gamma aminobutyric acid-, dopamine-, and cAMP-regulated neuronal phosphoprotein (GABAergic DARPP-32) striatal projection-like neurons. The therapeutic potential of these hESC-derived striatal progenitors was evaluated by performing xenograft transplantations in vitro at 5 arbitrary stages of neuronal maturation in the quinolinate-lesioned right striatum of immunotolerant nude rats. All transplanted animals had surviving grafts that labeled positively for human nuclear antigen (HNA). Early-stage xenografts maximally differentiated into structures resembling neural rosettes and resulted in non-neuroectodermal teratoma-like regions. Whereas xenografts transplanted at later, progressively more mature stages of neuronal differentiation did not display graft overgrowth, they had limited capacity to produce mature DARPP-32-positive striatal neurons, depending on the precise in vitro stage of transplantation.
As hESC therapy evolved, improved protocols circumvented the issue of teratoma formation. Shin et al. [63] developed a robust hESC monolayer differentiation method that yielded functional GABAergic neurons that expressed putative glutamate receptors, formed functional inhibitory and excitatory synapses, and recapitulated electrophysiological properties relevant to the neuronal cell type. Upon in vivo transplantation, the graft-derived neurons exhibited a striatal phenotype with over 50% of the NeuN+ (neuronal nuclei) cells coexpressing DARPP-32, suggesting that they were MSNs.
To overcome limitations posed by genetic heterogeneity between control and disease hESCs, Lu et al. [64] generated an allelic series of HD hESCs on an isogenic background by stable transfection of cDNA constructs encoding the HTT exon1 fragment with varying polyQ repeat lengths (Q23, Q73, and Q145). The hESC-derived neurons showed a quantitative relationship between neurodegeneration and soluble monomeric mHTT levels. The authors further confirmed that siRNA knockdown of HTT and guanosine triphosphatase Ras homolog enriched in the striatum (Rhes), a HD toxicity modifier, rescued the neurodegeneration phenotype. mHTT reduction of 10% was sufficient to prevent toxicity, whereas a 90% reduction of wild-type HTT was safely tolerated in these neurons. Thus, hESC-derived neurons could be valuable tools for high-throughput screening of compounds targeting either mHTT levels or toxicity. A limitation of this system is that the HTT protein contains only exon 1 fragment and it is overexpressed. Another study of hESC lines carrying CAG37 and CAG51 repeat expansions [65] revealed no discernible differences in pluripotent gene expression, mitochondrial activity, or forebrain neuronal specification, compared with healthy hESC controls. However, elevated glutamate-induced perturbations were observed only in terminally differentiated HD CAG51 neurons [66]. Interestingly, models derived from HD patient iPSCs have deficits in neurodevelopmental pathways and numerous phenotypes prior to differentiation into GABAergic neurons [56–61], which we will describe below.
History of HD iPSC
First introduced in 2006, genetic reprogramming of easily accessible adult somatic cells, including skin fibroblasts or blood cells, to pluripotency created new possibilities for modeling hereditary diseases [67–69]. Similar to ESCs, iPSCs can be expanded without limits in culture and can potentially be differentiated into any somatic cell type. iPSC technology also obviates many ethical and resource-related concerns posed by ESCs, thus representing a superior alternative for basic science research and medical applications. The discovery of iPSCs has been so influential that the 2012 Nobel Prize in Physiology or Medicine was awarded to its creators Drs. Gurdon and Yamanaka. This technology continues to fuel the development of autologous iPSC-derived therapies using a patient’s own cells in which the pathologic allele has been genetically corrected. In the context of HD, iPSCs facilitate the selection of donors with discrete genotypical and phenotypical associations (e.g., similar CAG repeat lengths but different ages of clinical onset) that can contribute to the identification of modifier genes.
Park et al. [70] in 2008 first demonstrated the feasibility of reprogramming HD patient fibroblasts into iPSCs. The HD iPSC cell line was generated from fibroblasts obtained from a 20-year-old female patient using lentiviral transduction of doxycycline-inducible human octamer-binding transcription factor 4 (OCT4), sex-determining region Y (SRY) box 2 (SOX2), avian myelocytomatosis virus oncogene cellular homolog (c-MYC), Kruppel-like factor 4 (KLF4), and NANOG cDNA vectors. The resulting iPSCs retained expanded CAG triplet repeat sequences in 1 allele, and 19 repeats in the other, were karyotypically normal, and exhibited elevated expression of pluripotency markers compared with their somatic cell controls. Upon further differentiation, iPSCs gave rise to all 3 germ layers (i.e., endoderm, mesoderm, and ectoderm). In a separate study, Zhang et al. [71] showed they maintained putative ESC markers (OCT4, NANOG, SOX2, and stage-specific embryonic antigen-4 (SSEA4)) in HD iPSCs after extended culture, and could differentiate iPSCs into neuronal stem cells (NSCs) that were positive for neuroectoderm stem cell marker (Nestin), SRY-box 1 (SOX1), and paired box 6 (PAX6). They also demonstrated that iPSCs can be differentiated even further into striatal-like neurons that expressed neuronal markers βIII-tubulin, GABA, calbindin, and, at later stages of differentiation, the MSN marker DARPP-32. HD NSCs had higher levels of caspase-3 activity than normal NSCs, especially under conditions of growth factor withdrawal.
In 2012, an HD iPSC consortium reported the derivation and characterization of a panel of 14 iPSC lines: 8 from HD iPSC clones derived from 6 patients and 6 normal iPSC clones derived from 4 unaffected individuals, including 1 HD patient sibling [72]. The iPSC lines were karyotypically normal and were differentiated into NSCs. Hierarchical clustering analysis and protein profiling revealed that both HD and control NSCs segregated into distinct clusters with specific gene and protein expression patterns. Transcriptomic analysis revealed that HD NSCs exhibited CAG expanded repeat–associated changes in actin cytoskeleton, cell–cell adhesion, and energy metabolism. Furthermore, compared with normal NSC-derived neurons, HD NSC-derived neurons displayed altered electrophysiological properties as well as increased vulnerability and cell death under the trophic stress of BDNF withdrawal, glutamate-induced excitotoxic stress, or addition of cellular stressors.
Although the HD consortium-generated iPSC lines serve as excellent models to study HD pathogenesis at the molecular and cellular levels, diverse genetic backgrounds between disease and control lines can potentially create interference by exaggerating or diminishing HTT-specific effects [73, 74]. To circumvent this issue, the first isogenically corrected HD iPSC line was developed by the Ellerby lab in 2012 [56]. The patient-derived HD iPSC line generated by Park et al. [70] was genetically corrected by replacing the expanded CAG repeat (72Q/19Q) with a normal repeat using targeted homologous recombination (HR) [56]. The corrected “C116 iPSCs” (21Q/19Q) retained characteristics of pluripotent stem cells. The merit of this approach is that differences in gene expression between the 2 lines can be solely attributed to mutant HTT-specific effects. Isogenic correction, which can ameliorate relevant HD disease phenotypes, also represents a powerful tool for patient-specific modeling of HD. CAG repeat contraction in C116 iPSCs corrected cadherin and transforming growth factor beta (TGF-β) signaling pathways. Additionally, upon transplantation, C116 iPSC-derived NSCs populated the striatum in the R6/2 HD mouse model and underwent further differentiation into DARPP-32-expressing MSN-like neurons.
Another study that emphasized the importance of using isogenic controls for disease modeling and evaluation of HD-specific effects employed a piggyBac transposon-based approach, which allowed excision of the selection cassette without a DNA scar [75]. Removal of the selection cassette in a previous work left several extra nucleotides in the sequence [23]. The expanded polyQ tract in a CAG180 HD human (h)iPSC line was edited by homologous recombination (HR) to yield 3 corrected clones (HD-C#1, HD-C#2, and HD-C#3) with a nonisogenic CAG33 hiPSC line as control. Removal of the selection cassette from the genomic locus of the HTT gene after targeting ensured optimal disease modeling. HD and isogenically corrected iPSC lines maintained pluripotency and normal karyotype, and could be further differentiated into synaptically active neurons. Evaluation of the isogenically corrected lines showed the rescue of characteristic phenotypic abnormalities associated with HD, including impaired neural rosette formation, increased susceptibility to growth factor withdrawal, and deficits in mitochondrial respiration. However, a study by Hamilton et al. [76] using human neurons derived from HD iPSCs did not find mitochondrial dysfunction. Further investigations comparing conditions of culture and differentiation are required to understand the differences in these studies.
Use of iPSC-Derived NSCs and Medium Spiny Neurons to Model HD
Normal and mutant HTT have important roles in pre- and postnatal development as established in HD models in vitro and in vivo. Therefore, elucidating HD pathogenesis along the differentiation axis of ESC or iPSC to NSC to mature neurons will help decipher cellular mechanisms relevant to developmental deficits that ultimately contribute to disease progression [61, 77]. Additionally, these studies will reveal important insights into the role of normal HTT during development [78].
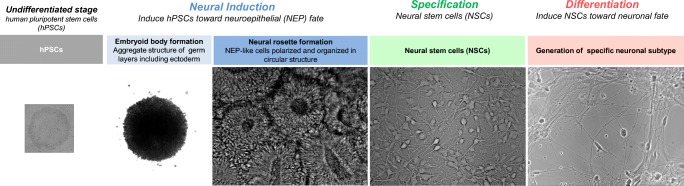
Differentiation of MSNs from iPSCs typically comprises 3 steps: neural induction, regional patterning toward a lateral ganglionic eminence (LGE) identity, and terminal differentiation [79] (Fig. 2). Developmentally, the LGE of the fetal telencephalon gives rise to MSNs. Approaches used to differentiate iPSCs into MSNs are based on mimicking brain development.
Fig. 2.
Schematic of steps involved in making neural stem cells and differentiated neurons.
Neural induction of iPSCs into primitive neural cells or NSCs is typically driven by dual inhibition of SMAD (homologies to the Caenorhabditis elegans “small” worm phenotype, or SMA, and Drosophila Mothers Against Decapentaplegic, or MAD, signaling) [80] using TGF-β receptor 1/Activin A inhibitor (SB431542) in combination with either bone morphogenetic protein (BMP) antagonist noggin [80], BMP inhibitor dorsomorphin, or its analogue LDN193189 [81–83] (Fig. 2). Dual SMAD inhibition facilitates neural induction by driving the formation of neural rosettes [84], thereby favoring a neuroectodermal fate and suppressing other fates [80].
Morphogens, including sonic hedgehog (SHH), or its small-molecule agonist purmorphamine, determine dorso-ventral patterning and induce specific neuronal identities within the neural tube [85, 86]. Dickkopf (DKK1), used in several differentiation protocols, inhibits wingless Int-1 (WNT) signaling [87, 88]. A combination of SHH and DKK1 is reported to favor GABAergic neuron generation, including MSNs [89]. Additionally, Activin A treatment can be used to prepattern iPSC- or ESC-derived NSCs selectively toward a DARPP-32-positive MSN fate through LGE induction in a SHH-independent manner [90]. The rationale for using Activin A is based on the reported presence of activin receptors in the developing striatum [91].
Terminal differentiation commonly involves supplementing the differentiation medium with neurotrophic growth factors, including brain-derived neurotrophic factor (BDNF) [15, 59, 60, 72, 89, 92–97] and/or glial cell-derived neurotrophic factor (GDNF) [74, 89, 90, 92, 96], required for neuronal maturation and survival [98–102]. Various protocols also call for additional chemical and recombinant protein supplements, including valproic acid, cyclic adenosine monophosphate (cAMP), vitamin A, insulin, and insulin-like growth factor 1 (IGF-1) [90, 92, 94, 103, 104]. Kemp et al. [97] formulated a pair of enhanced defined media, termed Synaptojuice, which synchronizes neurogenesis in prepatterned, iPSC/ESC-derived NSCs by modulating GABA, cAMP response element-binding protein (CREB), and WNT signaling, then accelerating synaptogenesis and neuronal maturation by WNT and tyrosine kinase receptor B (TrkB) signaling modulation. Neurons generated using this protocol expressed synaptic markers and exhibited electrophysiological properties typical of GABAergic inhibitory neurons. Another method uses inducible expression of Gsx2 and Ebf1 in ESCs to make MSNs [105].
Alternatively, somatic cells, such as fibroblasts, can be transdifferentiated into MSNs by forced expression of COUP-TF-interacting protein 2 (CTIP2), distal-less homeobox 1 (DLX1), distal-less homeobox 2 (DLX2), myelin transcription factor 1 like (MYT1L), miR-9/9, and miR-124 [106–108].
Several MSN differentiation protocols add recombinant proteins in the differentiation medium. However, using a recombinant protein is not desirable for manufacturing clinical-grade MSNs. The distinct advantages conferred by small molecules over recombinant proteins include better cell permeability, lack of immunogenicity, lower cost, and rapid, reversible biological effects. Therefore, Wu et al. [109] developed a small-molecule-based chemical recipe [110, 111]. The chemical recipe used in this study included small molecules for dual SMAD inhibition, and all patterning morphogens were replaced by small molecules, such as smoothened agonist (SAG) for SHH signaling [112, 113] and tankyrase inhibitor XAV939 for canonical WNT signaling inhibition [114]. Neurons derived using this methodology expressed putative MSN markers and demonstrated electrophysiological properties consistent with those of inhibitory GABAergic neurons. Upon transplantation into brains of neonatal mice or HD mouse models, the neurons exhibited efficacy as seen by correction of motor deficits. Thus, this study outlines a method to generate clinical-grade MSNs that facilitate industrial production and clinical use.
Consistent with observations in postmortem brain slices of HD patients [115], terminal differentiation of iPSCs into GABA medium spiny-like neurons (GMSLNs) recapitulates and models nuclear abnormalities, such as altered envelope integrity and increased levels of nuclear indentations in HD GMSLNs [95], compared with healthy GMSLNs. Premature neuronal aging mimicked by proteasome system inhibition (MG132 treatment) further increased the nuclear irregularity index in HD GMSLNs. HD GMSLNs also displayed perturbations in calcium homeostasis [116], increased number of lysosomes/autophagosomes, and enhanced neuronal cell death as a function of aging. These disease phenotypes were rescued by treatment with the quinazoline derivative EVP4593. Another study revealed decreased mitochondrial density and impaired trafficking in HD GMSLNs, and that these deficits were aggravated as a function of neuronal aging [117].
Although these methods allow us to model HD in a human “MSN”-like neuron, challenges still remain. We do not know if MSNs have similar identities to those formed during human development, nor have we developed conditions to make pure subpopulations of MSNs (e.g., striatonigral and striatopalidal, striosome, and matrix subpopulations). We also do not know if these models represent MSNs found in the human adult brain. A recent study has shown that hPSC-derived medial ganglionic eminence (MGE)/caudal ganglionic eminence (CGE)-like progenitors can differentiate into functional striatal interneurons [118] which will allow modeling in this cell type. With recent advances in single-cell technology, such as RNAseq and ATACseq, that can further our understanding of MSN profiling and development, we can expect rapid advances in disease modeling for HD.
Use of iPSC-Derived Cortical Neurons to Model HD
Cortical neurons are a significant neuronal subtype affected in HD. There are now several methods to differentiate iPSCs into excitatory and inhibitory cortical neurons. Forced expression of neurogenin-2 (NGN2) transcription factor in iPSCs differentiates cells into a population of almost 100% pure excitatory cortical-like neurons with excitable membranes and synaptic function. NGN2-induced neurons have been used to model amyloid β and tau pathologies, which feature prominently in the cortex of human brains with Alzheimer disease and frontotemporal dementia [119, 120]. For forebrain cholinergic neurons, Ortiz-Virumbrales et al. [121] developed a method that includes adding dual SMAD inhibitors then a transition into BrainPhys medium supplemented with B27 and fluorescence-activated cell sorting (FACS) of p75+ neural precursor cells for purification. In another method, Mariani et al. [122] generated cortical neurons from iPSCs in ~ 1 month. Cortical neurons are identified by immunostaining with antibodies against T-box, brain transcription factor 1 (TBR1), and special AT-rich sequence-binding protein 2 (SATB2). Mehta et al. [123] differentiated HD patient-derived iPSCs into all classes of cerebral cortical projection neurons using a method described by Shi et al. [124]. As described below and consistent with other transcriptomic analysis, altered developmental pathways and corticogenesis were identified. The next step is then to prepare defined subpopulations of cortical neurons to elucidate cell-specific mechanisms of vulnerability for HD.
HD Astrocyte and Microvascular Endothelial Models Derived from iPSCs
HD is characterized by both autonomous and nonautonomous cellular mechanisms. Other cell types, such as astrocytes, contribute to aberrant corticostriatal connectivity found in HD [40]. Astrocytes are the most abundant CNS glial cell type and are robustly activated in postmortem HD brains [125]. They are essential for maintaining the blood–brain barrier (BBB) and have important roles in synapse formation, maintenance, and pruning, as well as providing trophic support to neurons [126]. Similar to neurons, astrocytes are of neuroectodermal origin. Thus, several in vitro differentiation protocols for astrocytes involve generating neural rosettes that give rise to NSCs that can be further differentiated into astrocytes by adding specific combinations of selection factors, including ciliary neurotrophic factor (CNTF), fibroblast growth factor 2 (FGF2), BMP2, and IGF-1 [126].
Juoperri et al. reported the presence of cytoplasmic, electron-clear vacuoles in iPSC-derived astrocytes generated from a father with late-onset HD (50 CAG repeats) and his daughter with juvenile HD (109 CAG repeats) under basal conditions. The disease phenotype was significantly more pronounced in juvenile HD astrocytes [127]. Additionally, treatment with chloroquine, an autophagy inhibitor, exacerbated the vacuolation disease phenotype. Most vacuoles were found to be autophagosomes, identified by positive immunoreactivity for the microtubule-associated proteins 1A/1B light chain 3B (LC3) marker. This is consistent with cytoplasmic vacuolation observed in lymphoblasts harvested from HD patients in which CAG repeat length correlated with cytoplasmic vacuole number [128, 129]. Further studies by Garcia et al. [130] showed that HD iPSC-derived astrocytes have impaired inward-rectifying K+ currents and altered Ca2+ signaling, and that HD astrocytes do not protect neurons exposed to glutamate. These findings underscore how iPSC-derived astrocytes can be used for successful in vitro modeling of HD and pave the way for high-throughput therapeutic screens.
BBB is impaired in HD patients [131]. Lim et al. [132] advanced the field with an HD iPSC-derived brain microvascular endothelial model. This iPSC-derived blood brain model showed intrinsic abnormalities in angiogenesis and barrier properties using transcriptomic analysis. This model will allow the dissection of BBB disruption in HD and provide a model for evaluating potential therapeutics.
Use of iPSC-Derived Microglia to Model HD
Microglia, resident macrophages of the brain, have important roles in normal brain development, CNS surveillance, and maintenance of homeostasis in both healthy and pathologic brains [133, 134]. They also act as key players in neuroinflammation, which is implicated in the pathogenesis of several neurodegenerative diseases, including HD [134]. Autonomous microglia activation in the presence of mHTT initiates sterile inflammation. Therapeutic strategies that dampen neuronal inflammation slow disease progression in HD mouse models [135, 136]. Traditionally, most research in microglia used either primary cells from postmortem brain tissue or cultured fetal or adult CNS tissue. These cellular models do not represent microglia in the native brain environment, and additionally lose the microglial transcriptome signature within the first few hours of ex vivo culture [137]. These findings demonstrate a need for more HD studies using iPSC-derived microglia. Several protocols have described the generation of iPSC-derived microglia. Some utilize complex coculture systems mimicking the brain microenvironment to promote precursor iPSC differentiation into microglia [138–140], whereas other methods rely entirely on cocktails of specific cytokines and growth factors to steer differentiation toward microglia [141–143]. Disease-relevant phenotyping can be performed in microglia monocultures in response to specific cytokine stimuli or drug treatments, or by using microglia neuron cocultures [144]. In addition, models of 2D and 3D cocultures, including organoids, allow us to assay neurotoxicity at higher levels of complexity [141, 144].
Importance of Oligodendrocytes for HD Modeling
Oligodendrocytes are myelinating cells of the CNS, and impaired remyelination has been reported in HD mouse models [145–147] as well as in the postmortem HD brain [148–150]. Oligodendrocyte defects are directly mediated by mutant HTT [151, 152]. Thus, generations of iPSC-derived oligodendrocytes [153, 154] may facilitate a deep exploration of demyelination in the HD.
Brain Organoid Models for HD Disease Modeling
Recently, several studies reported protocols that generate cerebral organoids derived from human iPSCs to model brain development and neurological disorders [155–158]. The term cerebral organoid was coined to emphasize the ability of this 3-dimensional (3D) tissue to develop a variety of regional brain identities that can model different aspects of neurogenesis, neuronal migration, and maturation. The self-patterning and self-organization processes that occur during organoid generation display regionalization patterns similar to early stages of human brain development, such as those of the forebrain, hindbrain, and cortex. Moreover, Paşca et al. [158] generated a novel organoid model to enable both neurogenesis and gliogenesis, including nonreactive astrocyte generation. Currently, several laboratories are working on HD organoid models to understand developmental issues with mHTT and to identify novel targets for the disease. The first HD organoid system was developed by Conforti et al. [159] using HD iPSCs. As reported in earlier studies on HD iPSCs, there were obvious deficits in neural development. Using the Lancaster protocol for cerebral organoids, Conforti et al. demonstrated that patient HD iPSCs of different CAG repeat lengths had abnormal cell organization and neuronal identities in cerebral organoids. Transcriptomic analysis demonstrated control organoids overlapped with mature human fetal cortical areas whereas HD organoids had overlap with the immature ventrical zone/subventrical zone. Using an ADAM10 inhibitor during HD organoid formation rescued some of these alterations [159].
Isogenic HD Allelic Series for hESC-Modeled Neurons, Hepatocytes, and Myotubes
Despite being a neurocentric disease, systemic effects are frequently observed in HD but are often overlooked [160–162]. Thus, Ooi et al. [163] used genome-editing techniques to generate a selection of cassette-free, isogenic HD (IsoHD) hESC allelic series with varying CAG repeats (30, 45, 65, and 81 CAGs) ranging from normal to juvenile onset. Each of these lines was further differentiated into NSCs, mature neurons, hepatocytes, and myotubes to investigate the transcriptional landscape of HD. Transcriptomic profiling revealed that changes in differentially expressed genes did not necessarily have a linear correlation across different CAG sizes. However, measurements of phenotypic abnormalities (e.g., mitochondrial respiration deficits, expression of DNA damage markers, and cell death) largely correlated with CAG repeat size with effects more readily observed in 81Q NSCs, followed by 65Q and 45Q NSCs. Interestingly, hepatocytes, which are considered not to be involved in HD pathology, were found to have the largest transcriptional response, with significant differential expression across CAG lengths in 36% of the expressed genes. Overall, the study outlined how research expanding on range of CAG sizes may help uncover both linear and complex CAG-dependent relationships.
Altered Signaling Pathways and Potential Therapeutic Targets
Previous characterization of HD lines by differential gene expression and pathway analysis in our lab revealed that 4466 genes were differentially expressed in HD NSCs, in contrast to only 370 genes in HD iPSCs [58]. These findings indicated that HD-associated cellular and molecular phenotypes were evident only in differentiated neural-specific HD NSCs, but not in precursor HD iPSCs. This makes NSCs an attractive cell type for HD stem cell modeling. Further genomic analysis revealed the disruption of striatal neural development and identified new therapeutic targets for the disease. The study found that TGF-β and Netrin-1 were among the most dysregulated pathways in HD NSCs. Treatment with exogenous TGF-β and Netrin-1 rescued caspase-3-mediated apoptosis and mitochondrial respiration deficits in HD NSCs. Another study from our lab showed that HD NSCs displayed dysregulated or even elevated matrix metalloproteinase (MMP) expression, contributing to increased mHTT cleavage and generation of toxic fragments [57]. TGF-β treatment upregulated expression of the endogenous MMP inhibitor, inhibitor of metalloproteinase-1 (TIMP-1), thereby suppressing MMP activity and the associated production of toxic N-terminal fragments. Thus, upregulation of TIMP-1 and correction of the dysregulated MMP/TIMP axis represent an important mechanism of TGF-β-conferred neuroprotection in HD.
Unbiased omics analysis performed on neural cells from HD patient-derived iPSC lines with juvenile onset (CAG 60 and 109 repeats) and control iPSC lines (CAG 21 to 33 repeats) [59] identified 1869 differentially expressed genes (DEGs) and revealed altered neurodevelopmental pathways and synaptic homeostasis in HD lines. Dysregulated pathways relevant to neuronal differentiation and maturation included axonal guidance, Wnt signaling, Ca2+ signaling, neuronal CREB signaling, and glutamate and GABA receptor signaling. RNAseq analysis comparing DEGs with 1647 mouse ortholog genes suggests that differentiated HD lines either mature more slowly than healthy control lines or abruptly shut off gene expression in earlier stages of development. HD-related histone marks strongly correlated with gene expression and peak profiles of the dysregulated genes, representing an altered epigenetic program. The omics-based approach used in this study also helped identify an existing small molecule, isozaxole-9 (Isx-9), that could target affected pathways. Isx-9 restored expression of select dysregulated genes, abrogated neuronal vulnerability in response to stress induced by BDNF withdrawal, and corrected Ca2+ signaling deficits. Furthermore, Isx-9 treatment improved cognition and rescued synaptic pathology in the R6/2 HD model.
Proteomic and metabolomic studies have also been carried out on HD iPSCs to identify altered signaling pathways and points for therapeutic intervention in HD. Kedaigle et al. [164] found reduced ATP levels in HD cells compared with controls as well as lower expression of glycolytic enzymes, suggesting that the addition of pyruvate or other metabolites may have therapeutic benefit in HD. Another study discovered the therapeutic target ubiquitin protein ligase (UBR5) in HD iPSC-derived models [165]. A quantitative proteomics analysis of HD iPSCs suggested that ataxia telangiectasia mutated (ATM) and p53 are therapeutic targets for HD [166].
Gene Editing for HD: CRISPR-Cas9 In Vitro and In Vivo
Genetic correction of HD iPSCs using targeted HR in combination with antibiotic selection to generate an isogenically corrected “C116” iPSC line represented the first example of gene editing in patient-derived HD stem cells [56]. However, using traditional HR is inefficient. Although this proof-of-concept study demonstrates a feasible approach to edit expanded polyQ tracts, new advances in genome editing technology, such as CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats), greatly enhance the ability to develop engineered lines with increased efficiency [167]. CRISPR-Cas9 can also be used to genetically correct the HTT gene or lower HTT levels in vivo. The Cas9 protein is an RNA-guided nuclease belonging to the CRISPR system that can be targeted to a specific gene locus using a 20-nucleotide guide RNA (gRNA) with homology to the target region, followed by a 5′-NGG protospacer adjacent motif (PAM). The complexed Cas9 then cleaves DNA upstream of PAM [168]. CRISPR-Cas9 thus enhances the rate of HR between genomic and donor DNA [169–172]. HR accuracy can be refined by using the mutant Cas9 D10A. The mutation changes wild-type Cas9, which normally creates double-stranded DNA breaks, into a nickase that produces single-stranded breaks, thereby favoring HR over nonhomologous end joining at the target site [173] and improving target sequence specificity [174–176].
As an extension of their previous work [56], An et al. [177] corrected HD-iPSCs with both CRISPR/Cas9 and a rapid antibody-based screening approach. The 12% recombination rate observed with Cas9 and HTT gRNA was remarkably higher than the 1% rate reported at this locus using traditional HR [56]. Xu et al. [74] used a piggyBac transposon-based approach for seamless cassette excision from the HTT genomic locus after Cas9 targeting. Dabrowska et al. [176] demonstrated that, in patient-derived HD fibroblasts transfected with Cas9 D10A nickase and 2 gRNAs with upstream and downstream polyQ tract targets, excision of expanded CAG repeats in at least 1 allele resulted in a 68 to 82% decrease in HTT mRNA and protein expression. Nonallele-specific editing was performed in this study. In an alternative approach, the presence of SNPs specific to the mutant CAG expanded allele was exploited to selectively inactivate the mutant allele without impacting the normal allele [178, 179].
gRNA/Cas9 complexes administered by viral delivery are also effective in HD mouse models in vivo. Yang et al. [180] found that CRISPR/Cas9-mediated reduction of endogenous mouse mHTT expression in heterozygous 140Q knock-in HD mice effectively depleted striatal mHTT aggregates, attenuated early HD-associated neuropathology, and significantly improved grip strength and performance on rotarod and balance beam tests, alleviating typical HD motor deficits. CRISPR/Cas9-mediated gene editing also attenuated reactive astrogliosis in the HD brain. Another study showed a 40% reduction in human mHTT expression in a transgenic HD mouse model using SNP-dependent mutant allele-specific targeting [179].
These studies suggest that mHTT allele-specific CRISPR/Cas9-mediated gene editing could be used to efficiently and permanently eliminate polyQ expansion-mediated neuronal toxicity in the adult brain. Combining gene editing to correct existing diseased cells in the host striatum with isogenically corrected NSCs or MSN transplantation to replace lost neurons may facilitate optimal recovery and rescue HD progression, especially in patients with late-stage HD.
Modeling Other PolyQ Diseases with iPSCs
In this section, we will discuss recent advances in polyQ-based iPSC models for SBMA and SCAs. We will review the progress made in the differentiation and characterization of iPSC-derived neuronal cells relevant for each disorder. The models described here take advantage of opportunities offered by iPSC technology to study diseases during different stages of development. iPSC-generated patient disease models generally include the following steps: (i) iPSC generation using reprogrammed adult cells, (ii) iPSC differentiation toward NSC fate, and (iii) NSC differentiation toward the desired neuronal fate.
SBMA or Kennedy’s Disease
SBMA, or Kennedy’s disease, is an X-linked dominant neuromuscular disease characterized by late onset and progressive degeneration of lower motor neurons in the spinal cord and brainstem, as well as skeletal muscle atrophy [8, 9]. SBMA is caused by CAG repeat expansion in the coding region of the AR gene (Table 1). Normal AR is localized in the cytoplasm and is activated by ligand binding, after which the AR is translocated to the nucleus, where it acts as a nuclear receptor. SBMA neuropathogenesis is complex as it results from both loss and gain of function in the AR protein with polyQ tract. The toxic gain of function is likely in part due to altered function in the nucleus, resulting in aberrant interaction with other nuclear factors and transcriptional dysregulation. However, how polyQ expansions in AR lead to motor neuron vulnerability and degeneration remains unclear.
Characterization of iPSCs Derived from SBMA patients
Nihei et al. [181] generated iPSC lines from an SBMA patient and a healthy control using retroviral transduction of 5 factors (OCT4, SOX2, KLF4, LIN28, and NANOG). Neither CAG repeat expansions nor contractions were observed during reprogramming and long passaging of SBMA iPSC models. These observations were consistent with results from Cortes et al. [182], in which SBMA iPSCs generated from 3 SBMA patients showed that CAG repeats in the AR gene were unchanged. On the other hand, Grunseich et al. [183] generated SBMA iPSCs from 6 patients and 3 controls using 3 reprogramming approaches: lentivirus with OCT4, SOX2, KLF4 and c-MYC, Sendai virus, and episomal vectors. Notably, they reported both expansion and contraction in stretches of CAG repeats when comparing SBMA iPSC clones with parental fibroblasts, but no variations were observed in the control lines. This CAG instability might be caused by mosaicism within the parental fibroblast population. Nuclear inclusions generally observed in patient tissue were not detected in the SBMA iPSC model. At the transcriptional level, AR mRNAs were similar in the SBMA iPSC and control lines, but AR protein levels were lower in SBMA iPSCs when compared with controls. Overall, Grunseich et al. [183] found that SBMA iPSCs expressed polyQ-expanded AR and that its nuclear translocation was ligand-dependent.
Characterization of NSCs Derived from SBMA iPSC
SBMA NSCs recapitulate disease-specific phenotypes related to SBMA, including accumulation of mutant polyQ-AR aggregates and decreased mitochondrial membrane potential [182]. Furthermore, autophagy dysregulation was evident in SBMA NSCs. PolyQ expansion prevented mutant AR from physically interacting with the transcription factor EB (TFEB), a key regulator of the autophagy-lysosome pathway. Interestingly, the induction of TFEB activity in SBMA NSCs rescued autophagy and metabolic defects. SBMA NSCs thus appear to mimic SBMA disease hallmarks and can elucidate signaling pathways relevant to disease pathogenesis, such as TFEB, autophagy, and metabolic dysregulation [182].
Characterization of Motor Neurons Derived from SBMA NSCs
For motor neuron differentiation, Nihei et al. [181] induced a neural fate by growing NSCs in N2 medium. To identify homeobox 9 (Hb9)-positive neurons, NSCs were transduced with a lentivirus expressing Hb9, a transcription factor specific to mature motor neurons. Positive NSCs for Hb9 expression were then selected and cultured for 2 weeks in motor neuron medium supplemented with N2, B27, retinoic acid (RA), and SHH. In these cultures, treatment with AR agonist led to increased AR protein levels in differentiated neurons. In mutant polyQ-expanded AR, AR ligand activation increased mutant AR accumulation in motor neurons derived from SBMA NSCs, compared with control lines. However, the authors were unable to show formation of nuclear inclusions, one of the key hallmarks of SBMA [181].
Grunseich et al. [184] used an established protocol to differentiate motor neurons from SBMA NSCs. Neural induction was performed during the EB stage in N2 medium supplemented with SMAD inhibitors LDN-193189 and SB431542. Motor neuron specification was carried out in N2 medium containing BDNF, RA, and SHH. For the final maturation stage, cells were cultured with N2, B27, BDNF, GDNF, CNTF, SHH, and RA. Overall, protocol duration was about 40 days. Grunseich et al. [183] showed motor neuron differentiation efficiency was similar between control and SBMA iPSC patient cells, and both conditions were positive for SMI-32, β-tubulin, and neurofilament markers. These observations illustrate that mutant AR does not interfere with the differentiation potential of SBMA NSCs. Notably, motor neurons from patients with CAG expansions of over 60 repeats have more acetylated α-tubulin than control lines, which may be correlated with reduced histone deacetylase 6 (HDAC6) activity. The authors also demonstrated that SBMA-derived motor neurons possess alterations in lysosomal localization, as illustrated by increased glycosylation of LAMP1, a lysosomal marker. Because HDAC6 is important for trafficking misfolded protein to the aggresome, Grunseich et al. [183] proposed that decreased HDAC6 activity reduces autophagic flux with changes in protein trafficking and lysosomal function. As SBMA also has neuromuscular symptoms, other relevant cell types should be investigated to model this disease with iPSCs.
SCAs
Spinocerebellar ataxias (SCAs) belong to a large family of autosomal dominant cerebellar ataxias (ADCAs), and are characterized by progressive neurodegeneration of the cerebellum and its connections [185, 186]. The clinical phenotype is principally related to progressive ataxia, dysarthria, and oculomotor deficits. SCAs refer to a heterogeneous group of disorders classified according to their genetic mutations, such as those caused by nucleotide repeat expansions in coding or noncoding sequences, or by point mutations [11]. SCAs are chronologically numbered with each newly identified disease-causing gene, and 47 SCAs have been identified thus far. SCAs caused by unstable expansion of CAG repeats encoding for polyQ tracts are composed of 7 subtypes: SCA1, SCA2, SCA3, SCA6, SCA7, SCA8, and SCA17 (Table 1). Here, we will focus on studies that characterized iPSC-based disease modeling for SCAs. Currently, only SCA2, SCA3, SCA6, and SCA7 have been subjected to iPSC-based disease modeling.
iPSC-Based Disease Modeling for SCA2
SCA2 is an, progressive cerebellar ataxia characterized by severe loss of cerebellar Purkinje cells. The pons, medulla oblongata, and spinal cord are also affected [187]. SCA2 is caused by an unstable CAG repeat expansion in the coding region of the ataxin-2 (ATXN2) gene (Table 1). The normal polyQ-ATXN2 contains 14 to 31 repeats, but the mutant protein contains 33 to 500 repeats [188, 189]. ATXN2 is broadly expressed throughout the body and in the CNS, with high levels of expression in Purkinje cells [35, 189]. In the normal brain, ATXN2 protein is localized mainly in the cytoplasm. Notably, histopathological studies of the brain from SCA2 patients showed predominantly cytoplasmic, instead of nuclear, inclusion bodies [35, 189]. Although the normal function of ATXN2 and its role in SCA2 neuropathogenesis are not fully understood, the protein may be involved in RNA metabolism and metabolic pathways [190, 191].
Characterization of iPSCs Derived from SCA2 patients
Xia et al. [192] generated iPSC lines from a SCA2 patient and healthy control using a retrovirus for each of the 4 factors OCT4, SOX2, KLF4, and c-MYC. They found that CAG repeats remained stable during reprogramming and long passaging of SCA2 iPSC models. Chuang et al. [193] generated SCA2 iPSCs from 4 SCA2 patients using a retrovirus with the same 4 factors. However, the authors did not investigate CAG repeat stability in ATXN2 gene.
Characterization of NSCs Derived from SCA2 iPSC
To generate NSCs, Xia et al. used an embryoid body (EB) approach that allowed subsequent formation of neural rosettes. However, they observed that during the rosette formation step, instead of forming rosette-like structures in normal conditions, SCA2 EB formed a cyst-like structure. Nevertheless, this did not prevent EB differentiation into NSCs. Furthermore, they showed that SCA2 NSCs express less ataxin-2 protein compared with normal conditions.
Characterization of Neurons Derived from SCA2 NSCs
Notably, the neural differentiation protocol used by Xia et al. [192] and Chuang et al. [193] was not designed to generate a cerebellar neuronal population, which are the cells predominantly affected in SCA2. Using time-lapse imaging, Xia et al. showed that, compared with normal conditions, SCA2-neural cells were short-lived. Chuang et al. found that neurons derived from SCA2 iPSCs expressed Purkinje cell markers glutamate decarboxylase 67 (GAD67), LIM homeobox 5 (LHX5), and calbindin 1 (CALB1). They demonstrated that SCA2 neurons recapitulated disease-specific phenotypes related to SCA2, including accumulation of mutant polyQ-ATXN2 into soluble aggregates in the nucleus and occasionally the cytoplasm, as well as the distortion of mitochondrial microstructures. Furthermore, they showed that glutamate associated pathways may be preferentially affected in SCA2 iPSC-derived neurons. Interestingly, glutamate treatment of SCA2-derived neurons exacerbated SCA2-associated pathologic phenotypes in relation to cell death and mitochondrial dysfunction [193]. Those effects were not observed in neurons derived from control iPSCs. The authors hypothesized that the effect of glutamate on SCA2 phenotype was mediated through the calcium signaling pathway. The vulnerability of SCA2 neurons to glutamate could be alleviated by antiglutamate or calcium flux drugs which reduced cell death and improved mitochondrial function. Although neither cytoplasmic nor intranuclear inclusion body formation was observed in either study, iPSC disease modeling for SCA2 appears to be a powerful tool for recapitulating SCA2 disease hallmarks and for deciphering signaling pathways relevant to disease pathogenesis. Notably, the model developed by Chuang et al. illustrates the possibility to use this system as a platform for screening novel therapeutics.
iPSC-Based Disease Modeling for SCA3
SCA3, or Machado–Joseph disease, is the most common autosomal dominant, progressive cerebellar ataxia, and is characterized by neurodegeneration of the spinocerebellar tract, basal ganglia, dentate nuclei, substantia nigra, and spinal cord (Fig. 1, Table 1). Unlike the other SCAs, SCA3 only presents with a mild loss of Purkinje cells, and the inferior olives are typically spared [194]. SCA3 is caused by abnormal CAG repeat expansion in the coding region of the Ataxin-3 (ATXN3) gene (Table 1). The CAG repeats within normal alleles contain only 11 to 44 repeats, whereas disease allele repeats have 60 to 87 [195]. Although ATXN3 is characterized as a deubiquitinating enzyme (DUB) [196], its role in SCA3 neuropathogenesis remains unclear.
Characterization of iPSCs Derived from SCA3 patients
Koch et al. [197] generated iPSC lines from 4 SCA3 patients and 2 healthy controls using retroviral transduction with factors OCT4, SOX2, KLF4, and c-MYC. They observed no expansion of CAG repeats during reprogramming and long passaging of SCA3 iPSC models, consistent with other studies [198–200]. Ou et al. [199] and Hansen et al. [198] used a nonintegrating method to generate iPSC lines from 1 and 2 SCA3 patients, respectively. Both normal and mutant polyQ-ATXN3 were expressed in SCA3 fibroblasts and iPSCs [199].
Characterization of Neurons Derived from SCA3 NSCs
To generate mature neurons derived from SCA3 NSCs, Koch et al. [201] differentiated iPSCs toward long-term, self-renewing neuroepithelial-like stem (lt-NES) cells. lt-NES cells retain neuro- and gliogenic potential after long-term proliferation. The authors found expression of normal and expanded ATXN3 proteins in control and SCA3 lt-NES cells, respectively [197]. Both control and SCA3 lt-NES cells, in the absence of growth factor in a maturation medium, gave rise to a rich dominant fraction of β-III-tubulin neurons and a smaller fraction of glial fibrillary acid protein (GFAP) glia, suggesting that the mutant ATXN3 protein does not interfere with the neuronal differentiation process [197]. Because SCA3 neuropathogenesis affects the hindbrain, Hansen et al. [202] used a neuronal differentiation protocol developed by Yan et al., which gives rise to NSCs that express hindbrain genes, including HOXA2 and HOXB2. Briefly, NSCs are cultured in neuronal maturation medium (N2 and B27) in the presence of BDNF, GDNF, ascorbic acid, and db-cAMP for 28 days. In addition to generating iPSC-derived neurons expressing HOXA2, HOXB2, and gastrulation brain homeobox 2 (GBX2), Hansen et al. demonstrated the presence of forebrain markers forkhead box G1 (FOXG1) and LIM homeobox protein 2 (LHX2). Interestingly, neuronal subtype analysis indicated the presence of cholinergic neurons of hindbrain origin with low expression of GABAergic and dopaminergic markers. However, further studies are needed to quantify the percentage of neurons with hindbrain identity [199]. Chuang et al. [193] also generated mature neurons from SCA3 iPSCs and showed that neuronal cultures expressed Purkinje cell markers GAD67, LHX5, and CALB1.
Functional analysis using whole-cell patch-clamp revealed no functional differences between SCA3 and control neurons which were differentiated for 5 to 6 weeks. This suggests that mutant polyQ-ATXN3 does not interfere with the generation of functional neuronal networks. Furthermore, in response to excitatory neurotransmitters, such as l-glutamate, mature neurons increase Ca2+ levels, leading to ATXN3 cleavage in both control and SCA3 conditions [197]. Interestingly, in SCA3 conditions only, ATXN3 cleavage induces formation of SDS-insoluble ATXN3-containing aggregates, a common SCA3 pathologic marker. ATXN3 cleavage is mediated by calpain proteases. This is a consistent finding by Chuang et al. [193], which showed that SCA3 disease modeling is sensitive to glutamate treatment, exacerbating SCA3 pathological phenotypes in terms of cell death and mitochondrial dysfunction. However, neither Hansen et al. [199] nor Ouyang et al. [200] could reproduce SDS-insoluble ATXN3 aggregate formation in their SCA3-iPSC-derived mature neurons, which may be due to differences in neuronal differentiation protocols.
Gene Editing by CRISPR/Cas9 in SCA3
Ouyang et al. [200] demonstrated that CRISPR/Cas9 genetically corrected CAG repeat expansion in the ATXN3 gene in SCA3 patient-derived iPSCs. The authors first illustrated iPSC generation from a SCA3 patient with a Sendai reprogramming vector expressing OCT4, SOX2, KLF4, and c-MYC without affecting the stability of CAG repeats. After CRISPR/Cas9-mediated gene editing of polyQ-ATXN3 in exon 10, a novel stop codon at the start of exon 11 was created, leading to a truncated ATXN3 protein that lacked the toxic polyQ repeat.
iPSC-Based Disease Modeling for SCA6
SCA6 is a progressive cerebellar ataxia characterized by neurodegeneration of the cerebellum with severe loss of Purkinje cells [203]. The thalamus, midbrain, pons, and medulla oblongata are characterized by mild atrophy [204]. SCA6 is caused by unstable CAG repeat expansion in the coding region of the calcium voltage-gated channel subunit alpha1-A (CACNA1A) gene (Table 1). Wild-type polyQ-CACNA1A contains 4 to 18 repeats but the mutant contains 20 to 33 [205, 206]. Notably, CACNA1A is a bicistronic gene encoding 2 proteins: the α1A subunit of P/Q-type voltage-gated calcium channel (Cav2.1) [207] and α1ACT, a newly discovered transcription factor [208]. Interestingly, these 2 structurally distinct proteins both contain the polyQ repeat tract. However, the neuropathogenesis of SCA6 is not fully understood. To date, 2 studies reported iPSC-based disease modeling for SCA6. Ishida et al. [209] interrogated SCA6 neuropathogenesis in an iPSC-derived 3D model of Purkinje cells, and Bavassano et al. [210] used a 2D approach to generate a heterogeneous population of mature neurons.
Characterization of 3D Disease Modeling of SCA6
Ishida et al. [209] generated iPSC lines from 2 SCA6 patients and 2 healthy controls using an episomal reprogramming vector. The authors used an established protocol developed by Muguruma et al. [211] to generate an iPSC-derived 3D culture of Purkinje cells. The culture was based on an understanding of human cerebellum development characterized by the formation of the isthmic, which defines the midbrain–hindbrain boundary and secretes positional cues signals, including FGF8 and WNT1. During the first step of the protocol, iPSC-derived serum-free floating culture of EB-like aggregates (SFEBq) was cultivated in cerebellar differentiation medium supplemented with SB431542, Y-27632, insulin, and FGF2. Notably, iPSCs in culture can self-organize into polarized, neuroepithelial, rosette-like structures, precursors of the neuroectoderm. The presence of SB431542 promotes neuroectodermal differentiation by inhibiting TGF-β, and Y-27632 promotes cellular survival. FGF2 and insulin act as caudalizing factors, promoting isthmic organizer-related functions by FGF8 and WNT1 induction. After 21 days in culture, SFEBq immunohistochemistry illustrated formation of polarized neuroepithelial cells expressing engrailed-2 (EN2) and gastrulation and brain-specific homeobox 2 (GBX2), rostral hindbrain markers. Subsequently, at ~ 35 days, iPSC-derived 3D cerebellar culture started to express Purkinje cell progenitor markers, including LHX5, kirre-like nephrin family adhesion molecule 2 (KIRREL2), pancreas transcription factor 1a (PTF1A), and SKI family transcriptional corepressor 2 (SKOR2). At this stage, FACS was used to purify KIRREL2-positive cells. Purified progenitor cells were cocultured with mouse rhombic lip (RL)-derived granule cells to promote selective differentiation of mature Purkinje cells. The resulting iPSC-derived Purkinje mature culture started to express mature Purkinje cell markers, including L7, calbindin, GABA, and aldolase C. After more than 70 days in culture, L7-positive Purkinje-like cells were characterized by elaborate dendritic branches and spines positive for Purkinje cell-specific glutamate receptor GRID2. Overall, the authors observed no significant differences between SCA6-derived mature Purkinje cell and controls, suggesting that the mutant polyQ-CACNA1A did not interfere with the differentiation protocol.
Ishida et al. showed that mature Purkinje cells derived from 3D cerebellar culture expressed Cav2.1 in the cell body and dendrites of both SCA6 patient and control cells. Moreover, in SCA6-Purkinje cells, Cav2.1 protein levels were increased in the homozygous condition compared with the heterozygous one. The bicistronically translated α1ACT was detected as puncta in the nucleus of mature Purkinje cells but not in the cytoplasm, consistent with a previous study which demonstrated that, under normal conditions, the α1ACT fragment translocates into the nucleus where it acts as a transcription factor for TATA-box binding protein associated factor 1 (TAF1) and BTG antiproliferation factor 1 (BTG1) [208]. However, in SCA6-Purkinje cells, the α1ACT level was lower and negatively correlated with gene dosage. Interestingly, expression of α1ACT-targeted proteins, including TAF1 and BTG1, was lower in levels. Overall, mature Purkinje cells derived from SCA6 patients illustrate that the mutant ATXN6 interferes with α1ACT localization into the nucleus and blocks its function.
Characterization of 2D Disease Modeling of SCA6
Bavassano et al. generated iPSC lines from 2 SCA6 patients and 2 healthy controls using a polycistronic lentiviral vector containing cDNAs coding for OCT4, SOX2, c-MYC, and KLF4. To generate NSCs, they used 2 different neural induction approaches: (i) an iPSC-derived EB approach and (ii) an iPSC monolayer approach cultured in the presence of dual inhibition of SMAD signaling by LDN-193189 and SB431542. For both protocols, neural rosette-like structures were selected, dissociated, and cultured in neural induction medium containing N2 in presence of FGF2 and EGF. For neuronal differentiation and maturation, NSCs were cultured in the absence of FGF2 and EGF in medium containing N2 and B27 and supplemented with BDNF and cytosine arabinoside. Notably, this protocol generated a heterogeneous population of mature neurons expressing Tau, synapsin, synaptophysin, MAP2, and GABAergic markers.
For functional analysis, Bavassano et al. used a whole-cell patch-clamp and found no functional differences between mature SCA6 and control neurons. These data suggest that the mutant polyQ-CACNA1A did not interfere with the generation of functional neuronal networks. They observed that α1ACT had predominant nuclear localization, as similarly seen in Ishida et al. The authors showed differential expression in α1ACT-targeted genes only for GRN.
iPSC-Based Disease Modeling for SCA7
SCA7 is a progressive cerebellar ataxia characterized by neurodegeneration of the retinal cerebellum. The brainstem, cerebral cortex, basal ganglia, thalamus, and midbrain are also affected. SCA7 is caused by an unstable CAG repeat expansion in the coding region of ATXN7 [204]. The normal poyQ-ATXN7 contains 4 to 19 repeats, whereas the mutant contains 34 to 460 [212]. Although, the ATXN7 protein is involved in transcription and cytoskeleton and microtubule regulation, how polyQ-ATXN7 is associated with SCA7 pathogenesis is not fully understood.
Characterization of NSCs Derived from SCA7 iPSC
Ward et al. [213] used iPSC-disease modeling for SCA7 to address the role of mutant polyQ-ATXN7 in metabolic and mitochondrial dysregulation in SCA7 patients. Because they could not detect disease-specific phenotypes in iPSC-derived NSCs from SCA7 patients with 50, 65, and 70 CAG repeats, they used CRISPR/Cas9 to derive 2 stem cell systems: (i) a knockout rescue model with ATXN7–10Q and (ii) a severe model with ATXN7-113Q. Interestingly, ATXN7-113Q NSC models recapitulated disease-specific phenotypes related to SCA7, including accumulation of mutant polyQ-ATXN7 into aggregates and increased cell death. Furthermore, the ATXN7-113Q NSC model showed a dysregulation of metabolic and mitochondrial phenotypes characterized by decreased expression of enzymes required for nicotinamide adenine dinucleotide (NAD+) salvage in the nucleus and mitochondria.
Summary
The field of polyQ expansion diseases is moving forward using patient-derived iPSCs to model these diseases in many relevant cell types. This includes patient iPSC-derived NSCs, MSNs, cortical neurons, motor neurons, astrocytes, oligodendrocytes, microvascular endothelial, organoids, 3D mature Purkinje cells, and cerebellar cultures. In many cases, the molecular details of polyQ disease can be recapitulated including developmental changes, aggregation, DNA damage, mitochondrial dysfunction, electrophysiological changes in neurons, and cell death. As iPSC-derived modeling of polyQ expansion diseases continues to improve, brain organoids with relevant disease mutations will likely provide further knowledge of developmental mechanisms and selective neuronal vulnerability. As more genetic disease modifiers are identified, the use of genomic engineering will allow us to better understand the molecular details of SNPs that contribute to disease onset and progression. The iPSC models may also help us identify somatic expansions arising from the CAG composition of individual patients and its impact on disease phenotypes.
Another emergent area in this field is the impact of polyQ expansion on peripheral tissue in certain diseases. For example, in HD and SCA1, polyQ proteins in non-neural tissue, such as in the heart, cause metabolic deregulation and other negative effects [162, 215]. Further, HTT lowering was shown to impact the pancreas in HD mouse models [161]. To this end, iPSCs will allow us to model these diseases in non-neural tissue.
Current therapeutic targets for polyQ expansion diseases are focused on decreasing toxic disease protein levels. Tabrizi et al. [214] used antisense oligonucleotide (ASO) designed to reduce the concentration of mHTT in phase I clinical studies with HD patients, demonstrating a significant advancement in the safety and use of ASOs in HD treatment. HD iPSCs and patient-specific design of ASOs will likely contribute to applications in personalized genomic medicine.
As the iPSC technology evolves, we should see the accurate modeling of the polyQ expansion diseases in brain organoids and the development of neurons and other cell types that mimic or capture the processes that are driven by aging in these age-related neurological disorders.
Electronic supplementary material
(PDF 498 kb)
Acknowledgments:
This work was supported by T32 training grant AG000266 to S.N. and L.M.E., and NIH R01 NS100529 and NS094422 to L.M.E.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Invited review: Molecular Pathogenesis and Advances in Therapeutics of Repeat Expansion Disorders, guest edited by Dr. Lisa M. Ellerby
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447(7147):932–40. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11(11):786–99. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11(4):247–58. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada M. Dentatorubral-pallidoluysian atrophy (DRPLA): The 50th Anniversary of Japanese Society of Neuropathology. Neuropathology. 2010;30(5):453–7. doi: 10.1111/j.1440-1789.2010.01120.x. [DOI] [PubMed] [Google Scholar]
- 5.Carroll LS, Massey TH, Wardle M, Peall KJ. Dentatorubral-pallidoluysian Atrophy: An Update. Tremor Other Hyperkinet Mov (N Y) 2018;8:577. doi: 10.7916/D81N9HST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971-83. [DOI] [PubMed]
- 7.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90(3):905–81. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 8.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–9. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 9.Cortes CJ, La Spada AR. X-Linked Spinal and Bulbar Muscular Atrophy: From Clinical Genetic Features and Molecular Pathology to Mechanisms Underlying Disease Toxicity. Adv Exp Med Biol. 2018;1049:103–33. doi: 10.1007/978-3-319-71779-1_5. [DOI] [PubMed] [Google Scholar]
- 10.Buijsen RAM, Toonen LJA, Gardiner SL, van Roon-Mom WMC. Genetics, Mechanisms, and Therapeutic Progress in Polyglutamine Spinocerebellar Ataxias. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 11.Ashizawa T, Oz G, Paulson HL. Spinocerebellar ataxias: prospects and challenges for therapy development. Nat Rev Neurol. 2018;14(10):590–605. doi: 10.1038/s41582-018-0051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebert AD, Shelley BC, Hurley AM, Onorati M, Castiglioni V, Patitucci TN, et al. EZ spheres: a stable and expandable culture system for the generation of pre-rosette multipotent stem cells from human ESCs and iPSCs. Stem Cell Res. 2013;10(3):417–27. doi: 10.1016/j.scr.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telezhkin V, Schnell C, Yarova P, Yung S, Cope E, Hughes A, et al. Forced cell cycle exit and modulation of GABAA, CREB, and GSK3beta signaling promote functional maturation of induced pluripotent stem cell-derived neurons. Am J Physiol Cell Physiol. 2016;310(7):C520–41. doi: 10.1152/ajpcell.00166.2015. [DOI] [PubMed] [Google Scholar]
- 14.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480(7378):547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105(43):16707–12. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW, et al. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS One. 2010;5(7):e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Zhang SC. Specification of neuronal and glial subtypes from human pluripotent stem cells. Cell Mol Life Sci. 2011;68(24):3995–4008. doi: 10.1007/s00018-011-0770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12(4):487–96. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci U S A. 2012;109(15):5803–8. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med. 2012;4(5):380–95. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aneichyk T, Hendriks WT, Yadav R, Shin D, Gao D, Vaine CA, et al. Dissecting the Causal Mechanism of X-Linked Dystonia-Parkinsonism by Integrating Genome and Transcriptome Assembly. Cell. 2018;172(5):897–909. doi: 10.1016/j.cell.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoyas CA, La Spada AR. The CAG-polyglutamine repeat diseases: a clinical, molecular, genetic, and pathophysiologic nosology. Handb Clin Neurol. 2018;147:143–70. doi: 10.1016/B978-0-444-63233-3.00011-7. [DOI] [PubMed] [Google Scholar]
- 24.Ross CA. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron. 1997;19(6):1147–50. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 25.Schilling G, Wood JD, Duan K, Slunt HH, Gonzales V, Yamada M, et al. Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA. Neuron. 1999;24(1):275–86. doi: 10.1016/s0896-6273(00)80839-9. [DOI] [PubMed] [Google Scholar]
- 26.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–3. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 27.Paulson HL. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Am J Hum Genet. 1999;64(2):339–45. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parodi S, Pennuto M. Neurotoxic effects of androgens in spinal and bulbar muscular atrophy. Front Neuroendocrinol. 2011;32(4):416–25. doi: 10.1016/j.yfrne.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, et al. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35(5):843–54. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 30.Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN, et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8(5):731–41. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 31.Arnold FJ, Pluciennik A, Merry DE. Impaired Nuclear Export of Polyglutamine-Expanded Androgen Receptor in Spinal and Bulbar Muscular Atrophy. Sci Rep. 2019;9(1):119. doi: 10.1038/s41598-018-36784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matilla-Duenas A, Goold R, Giunti P. Clinical, genetic, molecular, and pathophysiological insights into spinocerebellar ataxia type 1. Cerebellum. 2008;7(2):106–14. doi: 10.1007/s12311-008-0009-0. [DOI] [PubMed] [Google Scholar]
- 33.Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, et al. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95(1):41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 34.Paul S, Dansithong W, Figueroa KP, Scoles DR, Pulst SM. Staufen1 links RNA stress granules and autophagy in a model of neurodegeneration. Nat Commun. 2018;9(1):3648. doi: 10.1038/s41467-018-06041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat Genet. 2000;26(1):44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman AP, Shakkottai VG, Albin RL. Polyglutamine Repeats in Neurodegenerative Diseases. Annu Rev Pathol. 2019;14:1–27. doi: 10.1146/annurev-pathmechdis-012418-012857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6(10):743–55. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 38.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448(7154):704–8. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 39.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447(7146):859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 40.Creus-Muncunill J, Ehrlich ME. Cell-Autonomous and Non-cell-Autonomous Pathogenic Mechanisms in Huntington’s Disease: Insights from In Vitro and In Vivo Models. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 41.Egorova PA, Bezprozvanny IB. Molecular Mechanisms and Therapeutics for Spinocerebellar Ataxia Type 2. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 42.Gottesfeld JM. Molecular Mechanisms and Therapeutics for the GAA.TTC Expansion Disease Friedreich Ataxia. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 43.Liu Q, Pan Y, Li XJ, Li S. Molecular Mechanisms and Therapeutics for SCA17. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 44.Maiuri T, Suart CE, Hung CLK, Graham KJ, Barba Bazan CA, Truant R. DNA Damage Repair in Huntington’s Disease and Other Neurodegenerative Diseases. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 45.Niewiadomska-Cimicka A, Trottier Y. Molecular Targets and Therapeutic Strategies in Spinocerebellar Ataxia Type 7. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 46.Srinivasan SR, Shakkottai VG. Moving Towards Therapy in SCA1: Insights from Molecular Mechanisms, Identification of Novel Targets, and Planning for Human Trials. Neurotherapeutics. 2019. [DOI] [PMC free article] [PubMed]
- 47.Bidollari E, Rotundo G, Altieri F, Amicucci M, Wiquel D, Ferrari D, et al. Generation of induced pluripotent stem cell line CSSi008-A (4698) from a patient affected by advanced stage of Dentato-Rubral-Pallidoluysian atrophy (DRPLA) Stem Cell Res. 2019;40:101551. doi: 10.1016/j.scr.2019.101551. [DOI] [PubMed] [Google Scholar]
- 48.Buijsen RAM, Gardiner SL, Bouma MJ, van der Graaf LM, Boogaard MW, Pepers BA, et al. Generation of 3 spinocerebellar ataxia type 1 (SCA1) patient-derived induced pluripotent stem cell lines LUMCi002-A, B, and C and 2 unaffected sibling control induced pluripotent stem cell lines LUMCi003-A and B. Stem Cell Res. 2018;29:125–8. doi: 10.1016/j.scr.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 49.He L, Ye W, Chen Z, Wang C, Zhao H, Li S, et al. Generation of an induced pluripotent stem cell line (XHCSUi001-A) from urine cells of a patient with spinocerebellar ataxia type 3. Stem Cell Res. 2019;40:101555. doi: 10.1016/j.scr.2019.101555. [DOI] [PubMed] [Google Scholar]
- 50.Kozlowska E, Ciolak A, Olejniczak M, Fiszer A. Generation of human iPS cell line IBCHi001-A from dentatorubral-pallidoluysian atrophy patient's fibroblasts. Stem Cell Res. 2019;39:101512. doi: 10.1016/j.scr.2019.101512. [DOI] [PubMed] [Google Scholar]
- 51.Maguire JA, Gagne AL, Gonzalez-Alegre P, Davidson BL, Shakkottai V, Gadue P, et al. Generation of Spinocerebellar Ataxia Type 2 induced pluripotent stem cell lines, CHOPi002-A and CHOPi003-A, from patients with abnormal CAG repeats in the coding region of the ATXN2 gene. Stem Cell Res. 2019;34:101361. doi: 10.1016/j.scr.2018.101361. [DOI] [PubMed] [Google Scholar]
- 52.van der Graaf LM, Gardiner SL, Tok M, Brands T, Boogaard MW, Pepers BA, et al. Generation of 5 induced pluripotent stem cell lines, LUMCi007-A and B and LUMCi008-A, B and C, from 2 patients with Huntington disease. Stem Cell Res. 2019;39:101498. doi: 10.1016/j.scr.2019.101498. [DOI] [PubMed] [Google Scholar]
- 53.Rawlins MD, Wexler NS, Wexler AR, Tabrizi SJ, Douglas I, Evans SJ, et al. The Prevalence of Huntington’s Disease. Neuroepidemiology. 2016;46(2):144–53. doi: 10.1159/000443738. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh R, Tabrizi SJ. Clinical Features of Huntington’s Disease. Adv Exp Med Biol. 2018;1049:1–28. doi: 10.1007/978-3-319-71779-1_1. [DOI] [PubMed] [Google Scholar]
- 55.Rub U, Seidel K, Heinsen H, Vonsattel JP, den Dunnen WF, Korf HW. Huntington’s disease (HD): the neuropathology of a multisystem neurodegenerative disorder of the human brain. Brain Pathol. 2016;26(6):726–40. doi: 10.1111/bpa.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11(2):253–63. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naphade S, Embusch A, Madushani KL, Ring KL, Ellerby LM. Altered Expression of Matrix Metalloproteinases and Their Endogenous Inhibitors in a Human Isogenic Stem Cell Model of Huntington's Disease. Front Neurosci. 2017;11:736. doi: 10.3389/fnins.2017.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ring KL, An MC, Zhang N, O'Brien RN, Ramos EM, Gao F, et al. Genomic Analysis Reveals Disruption of Striatal Neuronal Development and Therapeutic Targets in Human Huntington’s Disease Neural Stem Cells. Stem Cell Reports. 2015;5(6):1023–38. doi: 10.1016/j.stemcr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Consortium HDi. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci. 2017;20(5):648-60. [DOI] [PMC free article] [PubMed]
- 60.Hunt CPJ, Pouton CW, Haynes JM. Characterising the developmental profile of human embryonic stem cell-derived medium spiny neuron progenitors and assessing mature neuron function using a CRISPR-generated human DARPP-32(WT/eGFP-AMP) reporter line. Neurochem Int. 2017;106:3–13. doi: 10.1016/j.neuint.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Wiatr K, Szlachcic WJ, Trzeciak M, Figlerowicz M, Figiel M. Huntington Disease as a Neurodevelopmental Disorder and Early Signs of the Disease in Stem Cells. Mol Neurobiol. 2018;55(4):3351–71. doi: 10.1007/s12035-017-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 63.Shin E, Palmer MJ, Li M, Fricker RA. GABAergic neurons from mouse embryonic stem cells possess functional properties of striatal neurons in vitro, and develop into striatal neurons in vivo in a mouse model of Huntington’s disease. Stem Cell Rev. 2012;8(2):513–31. doi: 10.1007/s12015-011-9290-2. [DOI] [PubMed] [Google Scholar]
- 64.Lu B, Palacino J. A novel human embryonic stem cell-derived Huntington’s disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 2013;27(5):1820–9. doi: 10.1096/fj.12-219220. [DOI] [PubMed] [Google Scholar]
- 65.Niclis J, Trounson AO, Dottori M, Ellisdon A, Bottomley SP, Verlinsky Y, et al. Human embryonic stem cell models of Huntington disease. Reprod Biomed Online. 2009;19(1):106–13. doi: 10.1016/s1472-6483(10)60053-3. [DOI] [PubMed] [Google Scholar]
- 66.Niclis JC, Pinar A, Haynes JM, Alsanie W, Jenny R, Dottori M, et al. Characterization of forebrain neurons derived from late-onset Huntington’s disease human embryonic stem cell lines. Front Cell Neurosci. 2013;7:37. doi: 10.3389/fncel.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 68.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 69.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 70.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Consortium HDi. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11(2):264-78. [DOI] [PMC free article] [PubMed]
- 73.Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109(31):12538–43. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu X, Tay Y, Sim B, Yoon SI, Huang Y, Ooi J, et al. Reversal of Phenotypic Abnormalities by CRISPR/Cas9-Mediated Gene Correction in Huntington Disease Patient-Derived Induced Pluripotent Stem Cells. Stem Cell Reports. 2017;8(3):619–33. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat Protoc. 2013;8(10):2061–78. doi: 10.1038/nprot.2013.126. [DOI] [PubMed] [Google Scholar]
- 76.Hamilton J, Brustovetsky T, Sridhar A, Pan Y, Cummins TR, Meyer JS, et al. Energy Metabolism and Mitochondrial Superoxide Anion Production in Pre-symptomatic Striatal Neurons Derived from Human-Induced Pluripotent Stem Cells Expressing Mutant Huntingtin. Mol Neurobiol. 2019. [DOI] [PubMed]
- 77.Golas MM. Human cellular models of medium spiny neuron development and Huntington disease. Life Sci. 2018;209:179–96. doi: 10.1016/j.lfs.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen GD, Gokhan S, Molero AE, Mehler MF. Selective roles of normal and mutant huntingtin in neural induction and early neurogenesis. PLoS One. 2013;8(5):e64368. doi: 10.1371/journal.pone.0064368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Golas MM, Sander B. Use of human stem cells in Huntington disease modeling and translational research. Exp Neurol. 2016;278:76–90. doi: 10.1016/j.expneurol.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 80.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4(1):33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanvitale CE, Kerr G, Chaikuad A, Ramel MC, Mohedas AH, Reichert S, et al. A new class of small molecule inhibitor of BMP signaling. PLoS One. 2013;8(4):e62721. doi: 10.1371/journal.pone.0062721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, et al. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18(15):4388–92. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 85.Ericson J, Muhr J, Placzek M, Lints T, Jessell TM, Edlund T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81(5):747–56. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 86.Chamberlain CE, Jeong J, Guo C, Allen BL, McMahon AP. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 2008;135(6):1097–106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- 87.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3(7):683–6. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 88.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391(6665):357–62. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 89.Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136(23):4055–63. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arber C, Precious SV, Cambray S, Risner-Janiczek JR, Kelly C, Noakes Z, et al. Activin A directs striatal projection neuron differentiation of human pluripotent stem cells. Development. 2015;142(7):1375–86. doi: 10.1242/dev.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maira M, Long JE, Lee AY, Rubenstein JL, Stifani S. Role for TGF-beta superfamily signaling in telencephalic GABAergic neuron development. J Neurodev Disord. 2010;2(1):48–60. doi: 10.1007/s11689-009-9035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma L, Hu B, Liu Y, Vermilyea SC, Liu H, Gao L, et al. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10(4):455–64. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nicoleau C, Varela C, Bonnefond C, Maury Y, Bugi A, Aubry L, et al. Embryonic stem cells neural differentiation qualifies the role of Wnt/beta-Catenin signals in human telencephalic specification and regionalization. Stem Cells. 2013;31(9):1763–74. doi: 10.1002/stem.1462. [DOI] [PubMed] [Google Scholar]
- 94.Delli Carri A, Onorati M, Lelos MJ, Castiglioni V, Faedo A, Menon R, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140(2):301–12. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 95.Nekrasov ED, Vigont VA, Klyushnikov SA, Lebedeva OS, Vassina EM, Bogomazova AN, et al. Manifestation of Huntington’s disease pathology in human induced pluripotent stem cell-derived neurons. Mol Neurodegener. 2016;11:27. doi: 10.1186/s13024-016-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Adil MM, Gaj T, Rao AT, Kulkarni RU, Fuentes CM, Ramadoss GN, et al. hPSC-Derived Striatal Cells Generated Using a Scalable 3D Hydrogel Promote Recovery in a Huntington Disease Mouse Model. Stem Cell Reports. 2018;10(5):1481–91. doi: 10.1016/j.stemcr.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kemp PJ, Rushton DJ, Yarova PL, Schnell C, Geater C, Hancock JM, et al. Improving and accelerating the differentiation and functional maturation of human stem cell-derived neurons: role of extracellular calcium and GABA. J Physiol. 2016;594(22):6583–94. doi: 10.1113/JP270655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293(5529):493–8. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 99.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol. 2007;81(5-6):294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 100.Ivkovic S, Ehrlich ME. Expression of the striatal DARPP-32/ARPP-21 phenotype in GABAergic neurons requires neurotrophins in vivo and in vitro. J Neurosci. 1999;19(13):5409–19. doi: 10.1523/JNEUROSCI.19-13-05409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Humpel C, Marksteiner J, Saria A. Glial-cell-line-derived neurotrophic factor enhances biosynthesis of substance P in striatal neurons in vitro. Cell Tissue Res. 1996;286(2):249–55. doi: 10.1007/s004410050694. [DOI] [PubMed] [Google Scholar]
- 102.Kells AP, Fong DM, Dragunow M, During MJ, Young D, Connor B. AAV-mediated gene delivery of BDNF or GDNF is neuroprotective in a model of Huntington disease. Mol Ther. 2004;9(5):682–8. doi: 10.1016/j.ymthe.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 103.Delli Carri A, Onorati M, Castiglioni V, Faedo A, Camnasio S, Toselli M, et al. Human pluripotent stem cell differentiation into authentic striatal projection neurons. Stem Cell Rev. 2013;9(4):461–74. doi: 10.1007/s12015-013-9441-8. [DOI] [PubMed] [Google Scholar]
- 104.Lin L, Yuan J, Sander B, Golas MM. In Vitro Differentiation of Human Neural Progenitor Cells Into Striatal GABAergic Neurons. Stem Cells Transl Med. 2015;4(7):775–88. doi: 10.5966/sctm.2014-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Faedo A, Laporta A, Segnali A, Galimberti M, Besusso D, Cesana E, et al. Differentiation of human telencephalic progenitor cells into MSNs by inducible expression of Gsx2 and Ebf1. Proc Natl Acad Sci U S A. 2017;114(7):E1234–E42. doi: 10.1073/pnas.1611473114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, et al. Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat Neurosci. 2018;21(3):341–52. doi: 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, et al. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84(2):311–23. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Richner M, Victor MB, Liu Y, Abernathy D, Yoo AS. MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nat Protoc. 2015;10(10):1543–55. doi: 10.1038/nprot.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu M, Zhang D, Bi C, Mi T, Zhu W, Xia L, et al. A Chemical Recipe for Generation of Clinical-Grade Striatal Neurons from hESCs. Stem Cell Reports. 2018;11(3):635–50. doi: 10.1016/j.stemcr.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, et al. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1(6):703–14. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 111.Kirkeby A, Nelander J, Parmar M. Generating regionalized neuronal cells from pluripotency, a step-by-step protocol. Front Cell Neurosci. 2012;6:64. doi: 10.3389/fncel.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99(22):14071–6. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Danjo T, Eiraku M, Muguruma K, Watanabe K, Kawada M, Yanagawa Y, et al. Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J Neurosci. 2011;31(5):1919–33. doi: 10.1523/JNEUROSCI.5128-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–20. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 115.Roos RA, Bots GT. Nuclear membrane indentations in Huntington’s chorea. J Neurol Sci. 1983;61(1):37–47. doi: 10.1016/0022-510x(83)90053-9. [DOI] [PubMed] [Google Scholar]
- 116.Vigont V, Nekrasov E, Shalygin A, Gusev K, Klushnikov S, Illarioshkin S, et al. Patient-Specific iPSC-Based Models of Huntington’s Disease as a Tool to Study Store-Operated Calcium Entry Drug Targeting. Front Pharmacol. 2018;9:696. doi: 10.3389/fphar.2018.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nekrasov ED, Kiselev SL. Mitochondrial distribution violation and nuclear indentations in neurons differentiated from iPSCs of Huntington’s disease patients. J Stem Cells Regen Med. 2018;14(2):80–5. doi: 10.46582/jsrm.1402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Noakes Z, Keefe F, Tamburini C, Kelly CM, Cruz Santos M, Dunnett SB, et al. Human Pluripotent Stem Cell-Derived Striatal Interneurons: Differentiation and Maturation In Vitro and in the Rat Brain. Stem Cell Reports. 2019;12(2):191–200. doi: 10.1016/j.stemcr.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang C, Ward ME, Chen R, Liu K, Tracy TE, Chen X, et al. Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem Cell Reports. 2017;9(4):1221–33. doi: 10.1016/j.stemcr.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78(5):785–98. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ortiz-Virumbrales M, Moreno CL, Kruglikov I, Marazuela P, Sproul A, Jacob S, et al. CRISPR/Cas9-Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 (N141I) neurons. Acta Neuropathol Commun. 2017;5(1):77. doi: 10.1186/s40478-017-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109(31):12770–5. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mehta SR, Tom CM, Wang Y, Bresee C, Rushton D, Mathkar PP, et al. Human Huntington’s Disease iPSC-Derived Cortical Neurons Display Altered Transcriptomics, Morphology, and Maturation. Cell Rep. 2018;25(4):1081–96. doi: 10.1016/j.celrep.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 124.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7(10):1836–46. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 125.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638–47. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chandrasekaran A, Avci HX, Leist M, Kobolak J, Dinnyes A. Astrocyte Differentiation of Human Pluripotent Stem Cells: New Tools for Neurological Disorder Research. Front Cell Neurosci. 2016;10:215. doi: 10.3389/fncel.2016.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nagata E, Sawa A, Ross CA, Snyder SH. Autophagosome-like vacuole formation in Huntington’s disease lymphoblasts. Neuroreport. 2004;15(8):1325–8. doi: 10.1097/01.wnr.0000127073.66692.8f. [DOI] [PubMed] [Google Scholar]
- 129.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13(5):567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garcia VJ, Rushton DJ, Tom CM, Allen ND, Kemp PJ, Svendsen CN, et al. Huntington’s Disease Patient-Derived Astrocytes Display Electrophysiological Impairments and Reduced Neuronal Support. Front Neurosci. 2019;13:669. doi: 10.3389/fnins.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Drouin-Ouellet J, Sawiak SJ, Cisbani G, Lagace M, Kuan WL, Saint-Pierre M, et al. Cerebrovascular and blood-brain barrier impairments in Huntington’s disease: Potential implications for its pathophysiology. Ann Neurol. 2015;78(2):160–77. doi: 10.1002/ana.24406. [DOI] [PubMed] [Google Scholar]
- 132.Lim RG, Quan C, Reyes-Ortiz AM, Lutz SE, Kedaigle AJ, Gipson TA, et al. Huntington’s Disease iPSC-Derived Brain Microvascular Endothelial Cells Reveal WNT-Mediated Angiogenic and Blood-Brain Barrier Deficits. Cell Rep. 2017;19(7):1365–77. doi: 10.1016/j.celrep.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10(11):1387–94. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 134.Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat Rev Neurosci. 2018;19(10):622–35. doi: 10.1038/s41583-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Crotti A, Benner C, Kerman BE, Gosselin D, Lagier-Tourenne C, Zuccato C, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. 2014;17(4):513–21. doi: 10.1038/nn.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hsiao HY, Chiu FL, Chen CM, Wu YR, Chen HM, Chen YC, et al. Inhibition of soluble tumor necrosis factor is therapeutic in Huntington’s disease. Hum Mol Genet. 2014;23(16):4328–44. doi: 10.1093/hmg/ddu151. [DOI] [PubMed] [Google Scholar]
- 137.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17(1):131–43. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports. 2017;8(6):1727–42. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, et al. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity. 2017;47(1):183–98. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 140.Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci. 2017;20(5):753–9. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94(2):278–93. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22(11):1358–67. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports. 2017;8(6):1516–24. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Haenseler W, Rajendran L. Concise Review: Modeling Neurodegenerative Diseases with Human Pluripotent Stem Cell-Derived Microglia. Stem Cells. 2019. [DOI] [PMC free article] [PubMed]
- 145.Teo RTY, Ferrari Bardile C, Tay YL, Yusof N, Kreidy CA, Tan LJ, et al. Impaired Remyelination in a Mouse Model of Huntington Disease. Mol Neurobiol. 2019. [DOI] [PubMed]
- 146.Jin J, Peng Q, Hou Z, Jiang M, Wang X, Langseth AJ, et al. Early white matter abnormalities, progressive brain pathology and motor deficits in a novel knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2015;24(9):2508–27. doi: 10.1093/hmg/ddv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McCollum MH, Leon RT, Rush DB, Guthrie KM, Wei J. Striatal oligodendrogliogenesis and neuroblast recruitment are increased in the R6/2 mouse model of Huntington’s disease. Brain Res. 2013;1518:91–103. doi: 10.1016/j.brainres.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fennema-Notestine C, Archibald SL, Jacobson MW, Corey-Bloom J, Paulsen JS, Peavy GM, et al. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology. 2004;63(6):989–95. doi: 10.1212/01.wnl.0000138434.68093.67. [DOI] [PubMed] [Google Scholar]
- 149.Myers RH, Vonsattel JP, Paskevich PA, Kiely DK, Stevens TJ, Cupples LA, et al. Decreased neuronal and increased oligodendroglial densities in Huntington’s disease caudate nucleus. J Neuropathol Exp Neurol. 1991;50(6):729–42. doi: 10.1097/00005072-199111000-00005. [DOI] [PubMed] [Google Scholar]
- 150.Gomez-Tortosa E, MacDonald ME, Friend JC, Taylor SA, Weiler LJ, Cupples LA, et al. Quantitative neuropathological changes in presymptomatic Huntington’s disease. Ann Neurol. 2001;49(1):29–34. [PubMed] [Google Scholar]
- 151.Ferrari Bardile C, Garcia-Miralles M, Caron NS, Rayan NA, Langley SR, Harmston N, et al. Intrinsic mutant HTT-mediated defects in oligodendroglia cause myelination deficits and behavioral abnormalities in Huntington disease. Proc Natl Acad Sci U S A. 2019;116(19):9622–7. doi: 10.1073/pnas.1818042116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang B, Wei W, Wang G, Gaertig MA, Feng Y, Wang W, et al. Mutant huntingtin downregulates myelin regulatory factor-mediated myelin gene expression and affects mature oligodendrocytes. Neuron. 2015;85(6):1212–26. doi: 10.1016/j.neuron.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ehrlich M, Mozafari S, Glatza M, Starost L, Velychko S, Hallmann AL, et al. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc Natl Acad Sci U S A. 2017;114(11):E2243–E52. doi: 10.1073/pnas.1614412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Douvaras P, Wang J, Zimmer M, Hanchuk S, O’Bara MA, Sadiq S, et al. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports. 2014;3(2):250–9. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 156.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162(2):375–90. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12(7):671–8. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Conforti P, Besusso D, Bocchi VD, Faedo A, Cesana E, Rossetti G, et al. Faulty neuronal determination and cell polarization are reverted by modulating HD early phenotypes. Proc Natl Acad Sci U S A. 2018;115(4):E762–E71. doi: 10.1073/pnas.1715865115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Coffey SR, Bragg RM, Minnig S, Ament SA, Cantle JP, Glickenhaus A, et al. Peripheral huntingtin silencing does not ameliorate central signs of disease in the B6.HttQ111/+ mouse model of Huntington’s disease. PLoS One. 2017;12(4):e0175968. doi: 10.1371/journal.pone.0175968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang G, Liu X, Gaertig MA, Li S, Li XJ. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc Natl Acad Sci U S A. 2016;113(12):3359–64. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Child DD, Lee JH, Pascua CJ, Chen YH, Mas Monteys A, Davidson BL. Cardiac mTORC1 Dysregulation Impacts Stress Adaptation and Survival in Huntington’s Disease. Cell Rep. 2018;23(4):1020–33. doi: 10.1016/j.celrep.2018.03.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ooi J, Langley SR, Xu X, Utami KH, Sim B, Huang Y, et al. Unbiased Profiling of Isogenic Huntington Disease hPSC-Derived CNS and Peripheral Cells Reveals Strong Cell-Type Specificity of CAG Length Effects. Cell Rep. 2019;26(9):2494–508. doi: 10.1016/j.celrep.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 164.Kedaigle AJ, Fraenkel E, Atwal RS, Wu M, Gusella JF, MacDonald ME, et al. Bioenergetic deficits in Huntington's disease iPSC-derived neural cells and rescue with glycolytic metabolites. Hum Mol Genet. 2019. [DOI] [PMC free article] [PubMed]
- 165.Koyuncu S, Saez I, Lee HJ, Gutierrez-Garcia R, Pokrzywa W, Fatima A, et al. The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington’s disease patients. Nat Commun. 2018;9(1):2886. doi: 10.1038/s41467-018-05320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chae JI, Kim DW, Lee N, Jeon YJ, Jeon I, Kwon J, et al. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington’s disease patient. Biochem J. 2012;446(3):359–71. doi: 10.1042/BJ20111495. [DOI] [PubMed] [Google Scholar]
- 167.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 169.Yang L, Mali P, Kim-Kiselak C, Church G. CRISPR-Cas-mediated targeted genome editing in human cells. Methods Mol Biol. 2014;1114:245–67. doi: 10.1007/978-1-62703-761-7_16. [DOI] [PubMed] [Google Scholar]
- 170.Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics. 2014;41(1):7–19. doi: 10.1016/j.jgg.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 171.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10(10):1028–34. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–8. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 173.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 175.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dabrowska M, Juzwa W, Krzyzosiak WJ, Olejniczak M. Precise Excision of the CAG Tract from the Huntingtin Gene by Cas9 Nickases. Front Neurosci. 2018;12:75. doi: 10.3389/fnins.2018.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.An MC, O'Brien RN, Zhang N, Patra BN, De La Cruz M, Ray A, et al. Polyglutamine Disease Modeling: Epitope Based Screen for Homologous Recombination using CRISPR/Cas9 System. PLoS Curr. 2014;6. [DOI] [PMC free article] [PubMed]
- 178.Shin JW, Kim KH, Chao MJ, Atwal RS, Gillis T, MacDonald ME, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25(20):4566–76. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol Ther. 2017;25(1):12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J Clin Invest. 2017;127(7):2719–24. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nihei Y, Ito D, Okada Y, Akamatsu W, Yagi T, Yoshizaki T, et al. Enhanced aggregation of androgen receptor in induced pluripotent stem cell-derived neurons from spinal and bulbar muscular atrophy. J Biol Chem. 2013;288(12):8043–52. doi: 10.1074/jbc.M112.408211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, Le A, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17(9):1180–9. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grunseich C, Zukosky K, Kats IR, Ghosh L, Harmison GG, Bott LC, et al. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol Dis. 2014;70:12–20. doi: 10.1016/j.nbd.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Amoroso MW, Croft GF, Williams DJ, O'Keeffe S, Carrasco MA, Davis AR, et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci. 2013;33(2):574–86. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jacobi H, Bauer P, Giunti P, Labrum R, Sweeney MG, Charles P, et al. The natural history of spinocerebellar ataxia type 1, 2, 3, and 6: a 2-year follow-up study. Neurology. 2011;77(11):1035–41. doi: 10.1212/WNL.0b013e31822e7ca0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dohlinger S, Hauser TK, Borkert J, Luft AR, Schulz JB. Magnetic resonance imaging in spinocerebellar ataxias. Cerebellum. 2008;7(2):204–14. doi: 10.1007/s12311-008-0025-0. [DOI] [PubMed] [Google Scholar]
- 187.Lorenzetti D, Bohlega S, Zoghbi HY. The expansion of the CAG repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology. 1997;49(4):1009–13. doi: 10.1212/wnl.49.4.1009. [DOI] [PubMed] [Google Scholar]
- 188.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique. DIRECT. Nat Genet. 1996;14(3):277–84. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 189.Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier JM, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14(3):285–91. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 190.Ralser M, Albrecht M, Nonhoff U, Lengauer T, Lehrach H, Krobitsch S. An integrative approach to gain insights into the cellular function of human ataxin-2. J Mol Biol. 2005;346(1):203–14. doi: 10.1016/j.jmb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 191.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15(16):2523–32. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 192.Xia G, Santostefano K, Hamazaki T, Liu J, Subramony SH, Terada N, et al. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J Mol Neurosci. 2013;51(2):237–48. doi: 10.1007/s12031-012-9930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chuang CY, Yang CC, Soong BW, Yu CY, Chen SH, Huang HP, et al. Modeling spinocerebellar ataxias 2 and 3 with iPSCs reveals a role for glutamate in disease pathology. Sci Rep. 2019;9(1):1166. doi: 10.1038/s41598-018-37774-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Koeppen AH. The Neuropathology of Spinocerebellar Ataxia Type 3/Machado-Joseph Disease. Adv Exp Med Biol. 2018;1049:233–41. doi: 10.1007/978-3-319-71779-1_11. [DOI] [PubMed] [Google Scholar]
- 195.Paulson HL. Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Semin Neurol. 2007;27(2):133–42. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- 196.Chai Y, Berke SS, Cohen RE, Paulson HL. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem. 2004;279(5):3605–11. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- 197.Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480(7378):543–6. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 198.Ou Z, Luo M, Niu X, Chen Y, Xie Y, He W, et al. Autophagy Promoted the Degradation of Mutant ATXN3 in Neurally Differentiated Spinocerebellar Ataxia-3 Human Induced Pluripotent Stem Cells. Biomed Res Int. 2016;2016:6701793. doi: 10.1155/2016/6701793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hansen SK, Stummann TC, Borland H, Hasholt LF, Tumer Z, Nielsen JE, et al. Induced pluripotent stem cell - derived neurons for the study of spinocerebellar ataxia type 3. Stem Cell Res. 2016;17(2):306–17. doi: 10.1016/j.scr.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 200.Ouyang S, Xie Y, Xiong Z, Yang Y, Xian Y, Ou Z, et al. CRISPR/Cas9-Targeted Deletion of Polyglutamine in Spinocerebellar Ataxia Type 3-Derived Induced Pluripotent Stem Cells. Stem Cells Dev. 2018;27(11):756–70. doi: 10.1089/scd.2017.0209. [DOI] [PubMed] [Google Scholar]
- 201.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106(9):3225–30. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yan Y, Shin S, Jha BS, Liu Q, Sheng J, Li F, et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med. 2013;2(11):862–70. doi: 10.5966/sctm.2013-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sasaki H, Kojima H, Yabe I, Tashiro K, Hamada T, Sawa H, et al. Neuropathological and molecular studies of spinocerebellar ataxia type 6 (SCA6) Acta Neuropathol. 1998;95(2):199–204. doi: 10.1007/s004010050787. [DOI] [PubMed] [Google Scholar]
- 204.Seidel K, Siswanto S, Brunt ER, den Dunnen W, Korf HW, Rub U. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124(1):1–21. doi: 10.1007/s00401-012-1000-x. [DOI] [PubMed] [Google Scholar]
- 205.Ishikawa K, Tanaka H, Saito M, Ohkoshi N, Fujita T, Yoshizawa K, et al. Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1-p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am J Hum Genet. 1997;61(2):336–46. doi: 10.1086/514867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shizuka M, Watanabe M, Ikeda Y, Mizushima K, Okamoto K, Shoji M. Molecular analysis of a de novo mutation for spinocerebellar ataxia type 6 and (CAG)n repeat units in normal elder controls. J Neurol Sci. 1998;161(1):85–7. doi: 10.1016/s0022-510x(98)00270-6. [DOI] [PubMed] [Google Scholar]
- 207.Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet. 1997;15(1):62–9. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 208.Du X, Wang J, Zhu H, Rinaldo L, Lamar KM, Palmenberg AC, et al. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell. 2013;154(1):118–33. doi: 10.1016/j.cell.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ishida Y, Kawakami H, Kitajima H, Nishiyama A, Sasai Y, Inoue H, et al. Vulnerability of Purkinje Cells Generated from Spinocerebellar Ataxia Type 6 Patient-Derived iPSCs. Cell Rep. 2017;18(4):1075–6. doi: 10.1016/j.celrep.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 210.Bavassano C, Eigentler A, Stanika R, Obermair GJ, Boesch S, Dechant G, et al. Bicistronic CACNA1A Gene Expression in Neurons Derived from Spinocerebellar Ataxia Type 6 Patient-Induced Pluripotent Stem Cells. Stem Cells Dev. 2017;26(22):1612–25. doi: 10.1089/scd.2017.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10(4):537–50. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 212.David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17(1):65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- 213.Ward JM, Stoyas CA, Switonski PM, Ichou F, Fan W, Collins B, et al. Metabolic and Organelle Morphology Defects in Mice and Human Patients Define Spinocerebellar Ataxia Type 7 as a Mitochondrial Disease. Cell Rep. 2019;26(5):1189–202. doi: 10.1016/j.celrep.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N Engl J Med. 2019;380(24):2307–16. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 215.Lalic NM, Dragasevic N, Stefanova E, Jotic A, Lalic K, Milicic T, et al. Impaired insulin sensitivity and secretion in normoglycemic patients with spinocerebellar ataxia type 1. Mov Disord. 2010;25(12):1976–80. doi: 10.1002/mds.23176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 498 kb)