Isoniazid (INH) is a cornerstone of antitubercular therapy. Mycobacterium tuberculosis complex bacteria are the only mycobacteria sensitive to clinically relevant concentrations of INH. All other mycobacteria, including M. marinum and M. avium subsp. paratuberculosis are resistant. INH requires activation by bacterial KatG to inhibit mycobacterial growth. We tested the role of the differences between M. tuberculosis KatG and that of other mycobacteria in INH sensitivity.
KEYWORDS: avium, isoniazid, KatG, marinum, mycobacteria
ABSTRACT
Isoniazid (INH) is a cornerstone of antitubercular therapy. Mycobacterium tuberculosis complex bacteria are the only mycobacteria sensitive to clinically relevant concentrations of INH. All other mycobacteria, including M. marinum and M. avium subsp. paratuberculosis are resistant. INH requires activation by bacterial KatG to inhibit mycobacterial growth. We tested the role of the differences between M. tuberculosis KatG and that of other mycobacteria in INH sensitivity. We cloned the M. bovis katG gene into M. marinum and M. avium subsp. paratuberculosis and measured the MIC of INH. We recombinantly expressed KatG of these mycobacteria and tested in vitro binding to, and activation of, INH. Introduction of katG from M. bovis into M. marinum and M. avium subsp. paratuberculosis rendered them 20 to 30 times more sensitive to INH. Analysis of different katG sequences across the genus found KatG evolution diverged from RNA polymerase-defined mycobacterial evolution. Biophysical and biochemical tests of M. bovis and nontuberculous mycobacteria (NTM) KatG proteins showed lower affinity to INH and substantially lower enzymatic capacity for the conversion of INH into the active form in NTM. The KatG proteins of M. marinum and M. avium subsp. paratuberculosis are substantially less effective in INH activation than that of M. tuberculosis, explaining the relative INH insensitivity of these microbes. These data indicate that the M. tuberculosis complex KatG is divergent from the KatG of NTM, with a reciprocal relationship between resistance to host defenses and INH resistance. Studies of bacteria where KatG is functionally active but does not activate INH may aid in understanding M. tuberculosis INH-resistance mechanisms, and suggest paths to overcome them.
INTRODUCTION
Mycobacterium tuberculosis resistance to drugs, and specifically to the first line medication isoniazid (INH), is a major problem in the clinical care of patients. INH has been used since the 1950s for the treatment and prevention of tuberculosis (TB) (1–3), with an MIC against sensitive isolates of M. tuberculosis and M. bovis of ∼0.02 μg/ml (4). Better understanding of the molecular mechanisms of INH resistance holds great value for designing INH derivatives that may overcome resistance, as well as inhibiting nontuberculous mycobacteria (NTM).
INH is a prodrug, activated by the bacterial catalase-peroxidase KatG. Being the only catalase/peroxidase (bifunctional) in M. tuberculosis (5), KatG plays an important role in the physiology and pathogenesis of the bacteria by catabolizing peroxides formed during phagocyte oxidative burst, thus antagonizing the host immune mechanism (6). This oxidative ability of KatG is used against the bacteria to convert INH into an isonicotinoyl acyl radical, capable of binding NAD+/NADH to form an inhibiting adduct (INH-NAD). This adduct inhibits the enoyl-[acyl-carrier-protein] reductase (InhA) protein, an essential enzyme for mycolic acid synthesis, thus impairing mycobacterial cell wall synthesis (7, 8).
Although deletion mutants of katG were shown to be attenuated in laboratory strains of M. tuberculosis (6), numerous clinical isolates of M. tuberculosis have been isolated where katG was either mutated or (more rarely) abolished (by nonsense mutations or genomic deletions). Despite the inactivation of katG and the resulting resistance to INH, these isolates did not manifest loss of virulence (4, 9–11). A recent study showed that the genetic requirement for an active KatG differed among M. tuberculosis clinical isolates in vitro (12), and the degree of the requirement correlated with INH susceptibility. In clinical isolates, some INH resistance-associated mutations were found to impair the enzymatic activity of the catalase reaction, peroxidase reaction, or both, while the mechanism behind other mutations remained elusive (13). Other mutations, such as the most common INH-resistant M. tuberculosis strain Ser315Thr and Asn138Ser, impair INH activation while retaining the catalase and peroxidase activity of KatG (14–16). These mutations impose steric hindrance in the INH binding channel in KatG (17). NTM such as the opportunistic-zoonotic fish pathogen M. marinum, and the possibly zoonotic ruminant pathogen M. avium subsp. paratuberculosis are innately resistant to clinically relevant levels of INH. Yet, these bacteria have KatG and InhA proteins that are postulated to act in a similar manner as the ones in M. tuberculosis. Gaining insight into INH resistance mechanisms of these bacteria could facilitate the development of InhA inhibitors that are activated by their own KatG.
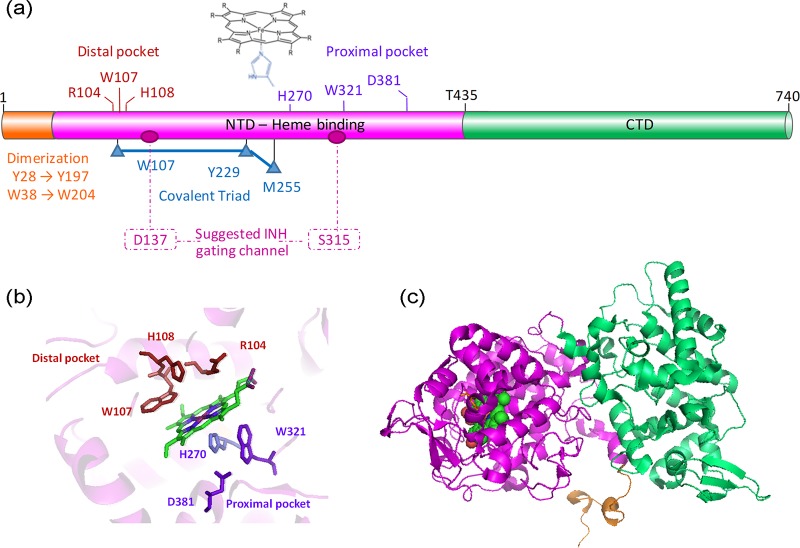
KatG is a heme-binding enzyme that functions as a homodimer. Each monomer contains a catalytic domain at the N-terminal region (residues 100 to 435), which includes the heme-binding sites at the distal and proximal pockets (Fig. 1a) as determined by the first crystal structure of the apo enzyme (PDB ID 1SJ2) (18) (Fig. 1b and c). Further structures of KatG mutants have demonstrated a unique substrate access channel for INH (17). The C-terminal domain (residues 435 to 740) does not bind heme nor have enzymatic capabilities, but its presence is vital as its deletion results in an inactive enzyme, which might point to a regulatory function (5).
FIG 1.
Structural organization of KatGMtb. (a) Schematic representation of KatGMtb domains and important residues in the heme distal pocket (red), heme proximal pocket (purple), covalent triad (blue), dimerization domain (orange), and suggested INH gating channel for heme access channel (pink). (b and c) Structural arrangement of the residues in the heme active site (c) and the structure of a KatG monomer, based on KatGMtb (PDB ID 1SJ2).
In this study, we examined KatG proteins from three mycobacteria and showed that introduction of katG from M. bovis into M. marinum or into M. avium subsp. paratuberculosis increased their sensitivity to INH 20 to 30-fold. The three proteins were recombinantly expressed in E. coli, purified, and analyzed in vitro for their interactions with INH, as well as their catalytic ability to form INH-NAD adduct. Circular dichroism (CD) spectra showed patterns of well folded, mainly helical proteins. Isothermal titration calorimetry (ITC) interaction analysis demonstrated that KatGbov binds INH tighter than KatGavp, while no binding was detected by KatGmar. Enzymatic analysis confirmed that only KatGbov was able to substantially catalyze INH-NAD adduct formation. Multiple sequence alignment of KatG proteins from 24 NTM gave rise to a phylogenetic tree that highlights the unique evolution of KatG of the M. tuberculosis complex. These findings may assist in understanding the molecular events leading to INH resistance in NTM, as well as in MDR-TB, and facilitate development of new antibiotics to target these difficult-to-treat infections.
RESULTS
M. marinum and M. avium subsp. paratuberculosis expressing KatGbov are sensitive to INH.
The katG gene from M. bovis was cloned under the control of the MOP promoter, and placed in an attB-integrating, integrase-negative vector to create pDB246. Removal of the integrase was necessary as it can catalyze excision, producing a background of bacteria that have lost the construct in the face of INH challenge. We then electroporated pDB246 into M. marinum, together with pYUB412 (providing an active integrase), and plated these on zeocin and kanamycin. We examined some of the zeocin/kanamycin resistant transformants by PCR for the presence of the MOP-katG constructs. One of the transformants that was found positive was named mDB85 (M. marinumKatG-bov) and selected for continued work. As a control strain for sensitivity assays, we created mDB72, M. marinum expressing mCherry (M. marinumcherry), which is also resistant to zeocin and kanamycin. This mutant produces bright pink colonies. We also introduced pDB246 into M. avium subsp. paratuberculosis, creating mDB199 (MAPKatG-bov).
To test the sensitivity of M. marinumKatG-bov to INH compared to M. marinumcherry, we grew both mutants in 7H9 medium supplemented with decreasing INH concentrations, as well as plating them on 7H10 solid media with INH (and, in both cases, with zeocin and kanamycin). We found M. marinumKatG-bov was ∼20 times more sensitive to INH than the wild type (WT) M. marinumcherry, with an MIC of 0.1 to 0.2 μg/ml compared to 3.125 to 6.25 μg/ml, respectively (Fig. 2a and b). For MAPKatG-bov (mDB199), we performed a similar experiment in 7H9 medium. WT M. avium subsp. paratuberculosis grew in INH concentrations as high as 25 μg/ml, whereas the MIC of MAPKatG-bov was ∼1.6 μg/ml (Fig. 2c). A percent-inhibition assay was performed and showed MAPKatG-bov to be inhibited by a much lower concentration of INH than that needed to inhibit WT M. avium subsp. paratuberculosis (Fig. 2d).
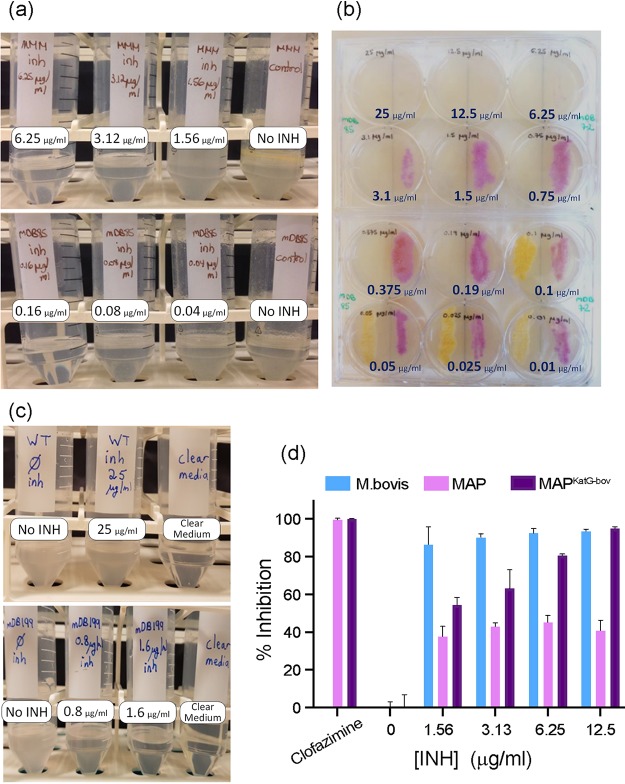
FIG 2.
Expression of KatGbov in M. marinum (a and b) or M. avium subsp. paratuberculosis (c and d) renders them sensitive to isoniazid (INH). (a) The MIC of wild-type M. marinum-moffet (MMM) to INH (top) is 6.25 μg/ml, whereas that of MMM+katGbov (mDB85, bottom) is 0.16 μg/ml. (b) mDB85 (MMM+katGbov, white colonies) and mDB72 (MMM+mCherry, red colonies) were grown on 7H10 plates with INH ranging from 25 to 0.01 μg/ml. (c) Wild-type (K10) M. avium subsp. paratuberculosis is not inhibited by INH at concentrations as high as 25 μg/ml (top), but the MIC for K10+katGbov (bottom) is between 1.6 and 0.8 μg/ml. (d) Percent inhibition of MAPwt versus MAPKatG-bov with increasing concentrations of INH. M. bovis BCG is presented as a control, as well as percent inhibition by the unrelated drug clofazimine.
To ensure the sensitization to INH is a specific result of the enzymatic properties of KatGbov and not simply higher levels of KatG protein due to an additional copy expressed from the mycobacteria optimized promoter (MOP), we cloned katG from M. marinum (KatGmar) using the same method as for katGbov to create plasmid pDB370. pDB370 was introduced into WT M. marinum to create a mutant diploid to katG named mDB202. mDB202 was twice as sensitive to INH (Fig. S2 in the supplemental material) as the parent strain, whereas introduction of KatGbov made the parent strain 20 times more sensitive. This shows the higher expression is responsible for only a small fraction of the effect, whereas most of the effect results from the specific properties of KatGbov.
To conclude, we found that expression of KatGbov in M. marinum and in M. avium subsp. paratuberculosis sensitized both bacteria to INH. These are clinically relevant concentrations, enabling potential experimental treatment of animal models (specifically in murine models of M. avium subsp. paratuberculosis -induced colitis, as mentioned later in the Discussion section).
Sequence alignment of KatG shows conservation across mycobacteria, with divergence in the M. tuberculosis complex.
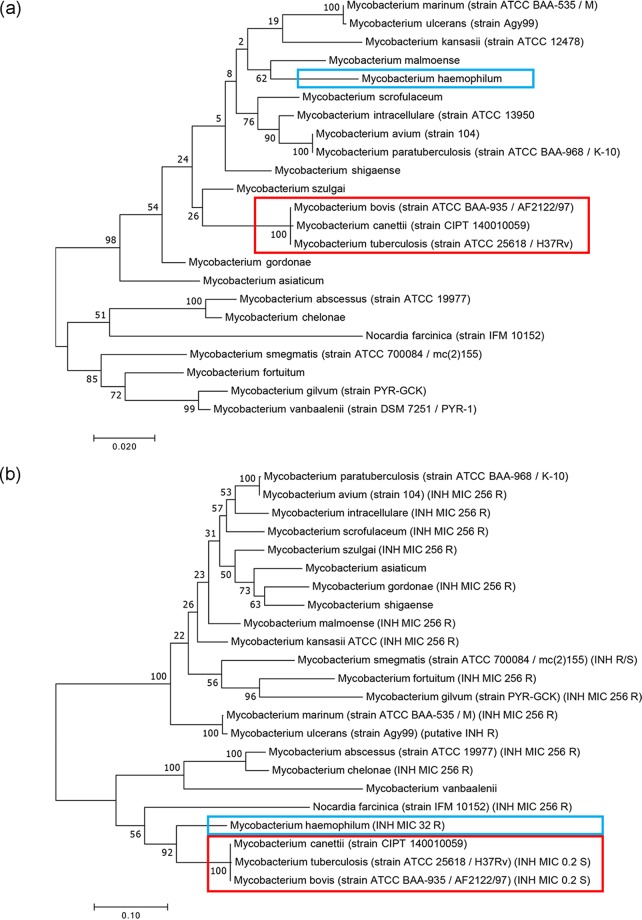
Lowering the MIC to INH by introduction of katGbov suggested that activated INH is an effective InhA inhibitor in both M. marinum and M. avium subsp. paratuberculosis, and that it is the activation of INH by KatG which is the limiting step in INH sensitivity of these bacteria. To examine if INH activation is a unique feature of the M. tuberculosis complex, we first constructed 2 phylogenetic trees of multiple mycobacteria (and the distantly related Nocardia farcinica), one based on the RNA polymerase subunit B (RpoB) protein and the other on KatG (Fig. 3a and b, respectively). The KatG phylogeny diverged from that of RpoB in several surprising aspects. M. haemophilum, usually considered related to M. leprae and unrelated to the M. tuberculosis complex, presented the KatG sequence closest to that of M. tuberculosis (see also Tables S1 and S2). This relatedness is reflected in the reported relative sensitivity of M. haemophilum to INH (MIC to INH is 32 μg/ml), such that INH is marginally effective in management of M. haemophilum infections (19). Interestingly, M. abscessus, usually considered a very distant mycobacterium (up to being sometimes referred to as “Mycobacteroides”), appears quite close to the M. tuberculosis complex when a KatG-based phylogeny is considered, although it is completely resistant to INH.
FIG 3.
Mycobacteria phylogenies analyzed with (a) RNA polymerase subunit B (RpoB) and (b) catalase-peroxidase (KatG) protein sequences. Note the high distance versus close proximity of the M. tuberculosis complex (red) from M. haemophilum (blue) in panel (a) versus (b), reflected in the relative INH sensitivity of the latter.
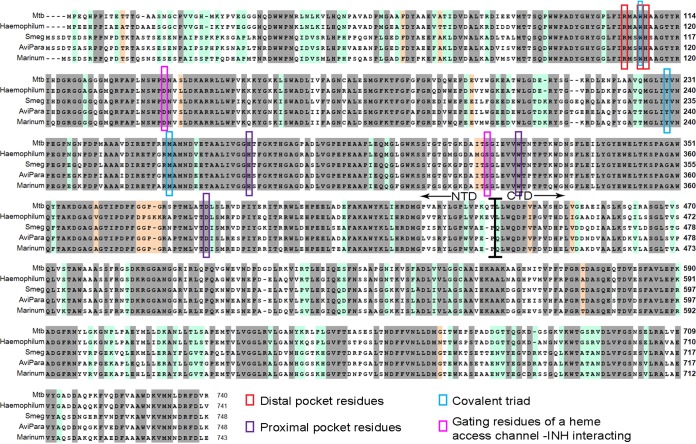
We then sought to compare the amino acid sequences of the three analyzed KatG proteins as well as KatG from M. smegmatis and M. haemophilum. As expected, there is similarity of ∼65% in residues important for the catalytic activity (Fig. 4, gray). Interestingly, an additional 20% similarity was demonstrated in all sequences except KatGbov (Fig. 4, light green). To gain a wider view on sequence conservation, we aligned the amino acid sequences of 24 KatG proteins from pathogenic and nonpathogenic mycobacteria, as well as the sequence of N. farcinica. Again, the conservation of the additional 20% of residues was demonstrated in almost all KatG variants except those of the M. tuberculosis complex (M. tuberculosis, M. bovis, M. africanum, M. microtii, and M. canetii). The full identity matrix is presented in Table S3.
FIG 4.
Multiple sequence alignment of KatG proteins from M. tuberculosis, M. haemophilum, M. marinum, M. avium subsp. paratuberculosis, M. smegmatis and M. avium subsp. avium. Identical residues, highlighted in gray, comprise ∼65% of the residues, and additional ∼20% identity in M. tuberculosis /Haemophilum versus Avp/Mari/Smeg highlighted in light green. Important residues in the heme distal pocket (red), heme proximal pocket (purple), covalent triad (blue) and gating residues of the heme access channel important for the interaction with INH (pink) are all marked. The border between the N- and C-terminal domains is also shown, around residue 450.
Recombinantly expressed KatGavp and KatGmari exhibited well-structured conformation and CD spectra similar to KatGbov.
To better characterize the differences between the KatG proteins, we recombinantly expressed the three enzymes and labeled them with a StrepII tag at the N terminus and a His6 tag at the C terminus to avoid truncated fragments. Purification was done using StrepTactin-based columns. All three proteins eluted from the size exclusion column as a single peak with the corresponding molecular weight of a dimer, relative to a calibration curve of standard molecular weight markers, as was expected.
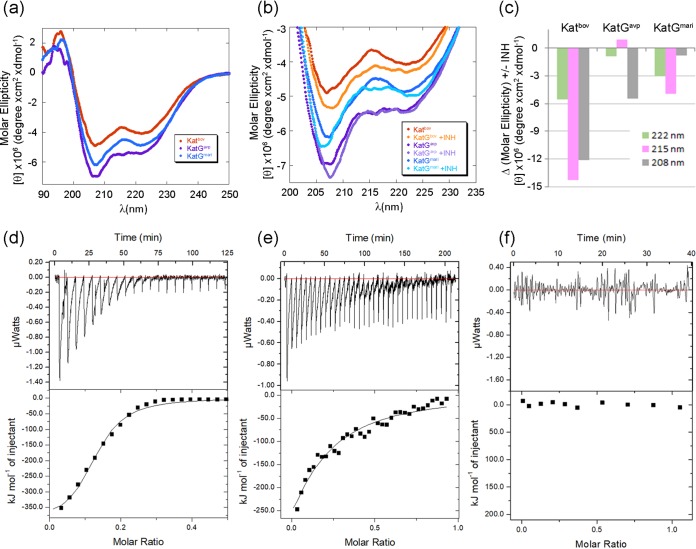
The structural properties of the proteins were studied using circular dichroism (CD). All three proteins exhibited similar far-UV CD spectra in phosphate buffer, pH 7.2, at 25°C, with well-ordered conformations as indicated by the minima at 208 nm and 222 nm. These minima are characteristic of proteins having predominantly α-helical secondary structure (Fig. 5a), as was demonstrated in the KatGMtb crystal structure. A slightly increased signal of KatGavp (purple) at 215 nm compared to KatGbov (red) and KatGmari (blue) might indicate a small increase in the β-sheet content in the fold of the protein. The effect of temperature on the conformation of the proteins was followed by monitoring the changes of the CD spectra with increasing temperature from 5°C to 95°C, in 5°C increments. A set of spectra for KatGbov, KatGavp, and KatGmari is presented in the upper panel (Fig. S1a to c), whereas the changes of the CD signal at 208 nm, 222 nm, and 215 nm as a function of temperature is presented at the lower panel (Fig. S1e to g). The results for all three proteins demonstrate a decrease in signal and a blue shift from ordered to random coil conformation, with a similar melting point between 50 and 55°C, suggesting that the recombinant proteins are folded in a similar manner with similar thermal stabilities.
FIG 5.
Secondary structure and interaction analysis of the free and INH bound KatG proteins. (a) Far-UV CD spectrum of KatGbov (red), KatGavp (purple), and KatGmari (blue), each in 4 μM concentration, in 25 mM phosphate buffer pH 7.2, at 25°C. Strong absorption signals at 222 nm and 208 nm indicates a pattern of well-folded proteins with a high fraction of α-helical secondary structure. (b) Changes in the far-UV CD spectrum of the KatG proteins upon INH addition at a 1:2 molar ratio are more prominent with KatGbov, implying a stronger interaction. (c) Percent changes in the absorption of the signal at 222 and 208 nm (green and pink, respectively, characteristic of α-helix) and at 215 nm (gray, characteristic of β-sheet). KatGbov showed the greater change in absorption upon INH addition. (d to f) KatG-INH interaction analysis studied by isothermal titration calorimetry (ITC). Binding results for titration of INH into KatGbov (d), KatGavp (e), and KatGmari (f) showed that KatGbov binds INH much tighter than KatGavp, whereas no binding was detected to KatGmari. ITC experiments were carried out at 25°C in phosphate buffer (pH 7.2) using 50 to 100 μM KatG and 5 to 10 times INH concentration.
Interaction analysis of INH with KatG proteins using CD and ITC showed differences in binding abilities.
Next, we opted to characterize the direct interaction of INH with each of the KatG enzymes. CD conformational analysis showed a small increase in total signal intensity of the KatGbov CD spectrum, while spectra of KatGavp and KatGmar were essentially unchanged in the presence of INH (Fig. 5b). A slight red shift of the 208 nm minima was detected in the INH bound versus unbound KatGbov and KatGavp, while KatGmar showed a slight blue shift. Signal intensity changes at 208 nm, 222 nm, and 215 nm of INH bound versus unbound KatG proteins are presented in Fig. 5c. These results demonstrate greater conformational changes contributed by the molecular interactions between INH and KatGbov than by KatGavp and KatGmar.
Following CD analysis, we used isothermal titration calorimetry (ITC) to quantitatively and thermodynamically characterize the interaction. Direct binding of KatGMtb with INH by ITC has been previously reported (20, 21). We found that while KatGbov bound INH with a dissociation constant (Kd) of 1 μM, the INH-KatGavp interaction was much weaker, with an affinity of 18 μM (Fig. 5e). No binding was detected for KatGmar (Fig. 5f). Broad isothermal peaks were observed in both INH-KatGbov and INH-KatGavp interactions resulting from considerable heat evolution during binding. This suggests slow binding between INH and KatG, and is a pattern that was previously described (20, 21). The isothermal curves approached apparent saturation at low molar ratios, resulting in a low apparent binding stoichiometry (N ∼0.2 per monomer of KatG) for all interactions. Fractional stoichiometries may arise from a smaller fraction of active KatG (21).
KatGbov is enzymatically more effective than KatGmar or KatGavp in conversion of INH into its active form as INH-NAD adduct.
To further assess the difference between the three KatG proteins, we compared their ability to enzymatically catalyze INH oxidation by monitoring INH-NAD adduct formation. The reaction cell included two binding substrates (INH and NAD+), the enzyme, and a flux of H2O2 generated by the glucose oxidase system as described (22, 23). Adduct formation was monitored spectrophotometrically by recording the absorbance signal at 326 nm, known as the peak formation of INH-NAD complex (Fig. 6a to c). The initial velocities were extracted from the first 5 min of the reaction and plotted against INH concentration to create the Michaelis-Menten plot (Fig. 6d). The corresponding kinetic parameters (Km, Vmax, kcat, and catalytic efficiency) for KatGbov were 4.1 ± 0.5 mM, 0.23 ± 0.01 μM/sec, 0.23 ± 0.015 sec−1, and 55.7, respectively. The KatG proteins from M. marinum and M. avium subsp. paratuberculosis had only negligible activity. We conclude that the enzymatic activity of KatGbov is substantially higher than that of KatGmar and KatGavp in respect to INH oxidation to its active form.
FIG 6.
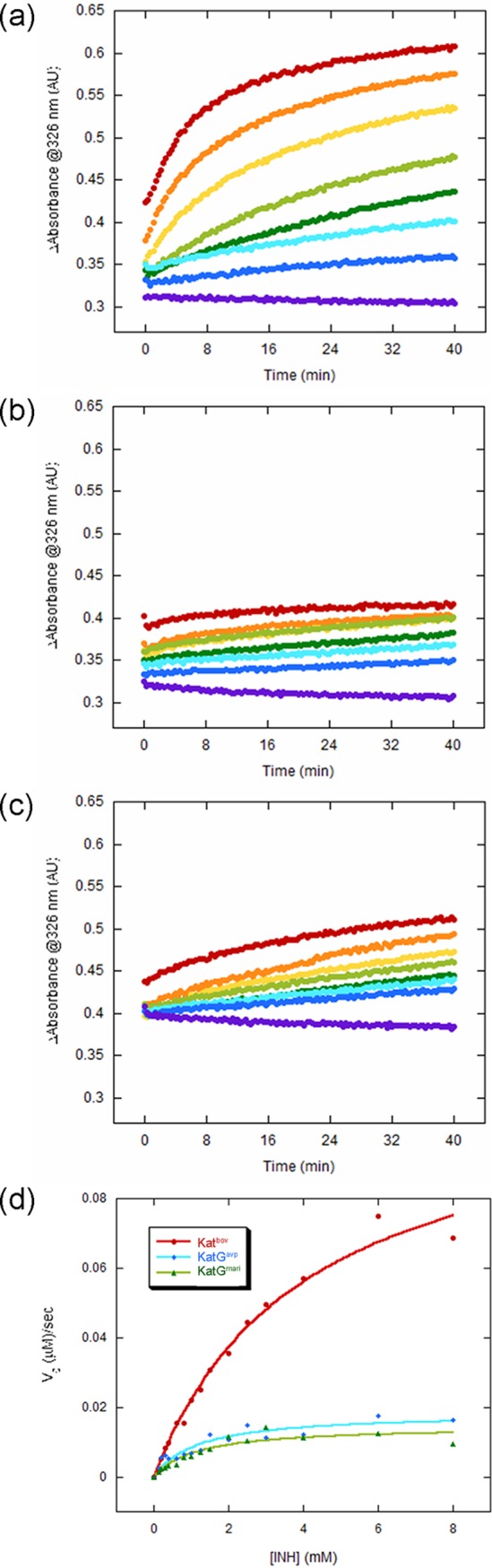
Kinetic analysis of INH-NAD product formation using a spectrophotometric assay. (a to c) IN-NAD adduct formation was followed by monitoring the characteristic signal at 326 nm. The reactions were carried out in triplicate, in 96-well plates with a total volume of 0.2 ml. Each reaction included 0.5 μM KatGbov (a), KatGmari (b), or KatGavp (c) in the presence of 8 mM NAD+, 0.5 units glucose oxidase (Gox), 5 mM glucose, and various concentrations of INH (0.15 to 8 mM). The reaction without NAD+ was carried out as a control and was subtracted from the total reaction. A set of representative results with increasing INH concentrations are shown for simplicity. (d) Michaelis-Menten plot presenting the initial reaction rates of adduct formation as a function of INH concentration, as calculated from a linear fit of the first 5 min of the experiments described in (a) to (c).
DISCUSSION
INH is the backbone and main “workhorse” of anti-tuberculous chemotherapy, thanks to good efficacy, low cost, ease of administration, and a relatively good safety record. However, the continuing emergence of resistant strains threatens to make this valuable drug obsolete. The majority of resistant strains are mutated in their katG gene (such as the S315T mutation), mainly affecting the ability of the protein to oxidize INH to its active form. Additionally, INH has limited utility in the treatment of nontuberculous mycobacteria, as virtually all are resistant to clinically achievable levels of the drug. Here, we show for M. marinum and M. avium subsp. paratuberculosis that this is due to poor drug activation by KatG. Better understanding of the molecular determinants of INH activation, and the differentiation between the physiological activity as a catalase-peroxidase and the nonphysiological activity as an INH activator, could facilitate the design of novel INH analogues that bypass the need for activation. Alternatively, distinct analogues could be envisioned that are activated even by a mutant KatG of M. tuberculosis or the KatG of an NTM that is classically considered to be INH resistant.
Here, we show that the basis for INH resistance in the fish and ruminant pathogens M. marinum and M. avium subsp. paratuberculosis is low efficiency of KatG-mediated INH activation, with retained virulence in the respective hosts. Thus, these organisms present as interesting models for investigating INH-KatG interactions, with resulting inhibition (or the lack of it) of InhA, the actual target of the drug. To our knowledge, this is the first time that NTM KatG enzymes were specifically examined for INH activation, providing the first line of evidence for why NTM are INH resistant.
The evolution of KatG between different mycobacteria has been influenced by the positive effect of the catalase and peroxidase activities on the survival of the each species, in its respective environment. Activation of INH was not a driving force, of course, before the introduction of the drug in the 1950s. In this sense, it was a “misfortune” for the organisms of the M. tuberculosis complex to have evolved a KatG that activates INH, allowing us to take advantage of this weakness. As a corollary, it can be argued that it is similarly a misfortune for patients with NTM infections for which KatG does not activate INH, compelling clinicians to use other antibiotics. Whether the M. tuberculosis KatG carries some other advantage for the bacterium over that provided by other KatG enzymes, is not known. It is interesting to see that the evolutionary tree of mycobacteria, when plotted only by KatG evolution, differs from the traditional dendrograms plotted for mycobacteria. When analyzed for KatG, M. tuberculosis appears closer to M. abscessus than to M. kansasii or M. avium, even though the two latter organisms are slow-growing mycobacteria that are phylogenetically closer to M. tuberculosis. Our alignment suggests there are approximately 20% of residues that are quite unique to M. tuberculosis complex but are conserved in other mycobacteria. The introduction of INH treatment, and the evolutionary pressure it imposes, are driving KatG evolution in M. tuberculosis toward that of variants that do not activate INH but participate in virulence, although the exact base-pair changes may differ from those present in other NTM.
These observations also offer therapeutic possibilities, as we showed that INH could be an effective drug in M. marinum and M. avium subsp. paratuberculosis if it were activated. Since the KatG of these bacteria resemble that of M. tuberculosis, it is hypothesized that an INH-like drug that does undergo activation by KatGmar and KatGavp could be developed. Moreover, such a drug could also potentially target INH-resistant clones of M. tuberculosis, since they could represent the evolutionary “destination” of INH-resistant M. tuberculosis bacteria.
The use of INH derivatives to target nontuberculous mycobacteria (NTM) may have therapeutic potential. Although there are good therapeutic options for M. marinum, and the scale of the medical problem it represents is not large, additional drugs could still be of use. M. avium subsp. paratuberculosis is not considered to be a major problem in humans, but if theories on its involvement in Crohn’s disease find more support, this may very well change. Furthermore, on a practical level, our INH-sensitive M. avium subsp. paratuberculosis mutant may have potential utility in a murine model of M. avium subsp. paratuberculosis -induced colitis. As its growth can be inhibited by INH without affecting other members of the bacterial flora, this would enable experiments that investigate the role of M. avium subsp. paratuberculosis in enteric disease through selective depletion of M. avium subsp. paratuberculosis. Finally, in sharp contrast, M. abscessus is recognized as a major, emerging medical problem throughout the world for which current treatments are associated with low cure rates. An INH-like drug against this bacterium could be of great value.
MATERIALS AND METHODS
Mycobacteria and growth conditions.
M. marinum-Moffett (ATCC BAA-535) and M. bovis BCG Pasteur strains were grown in 7H9 liquid media or on 7H10 agar plates, supplemented with 10% oADC (oleic-acid, albumin-dextrose-catalase-NaCl) and glycerol, as widely described. Medium for M. avium subsp. paratuberculosis K-10 was supplemented by mycobactin-J (Allied Monitor, 2 mg/liter). Cultures were kept at 30 to 33°C for M. marinum and 37°C for M. bovis and M. avium subsp. paratuberculosis. The 7H9 medium was supplemented by Tween80. Zeocin (25 μg/ml) and kanamycin (40 μg/ml for M. avium subsp. paratuberculosis, 20 μg/ml for BCG and M. marinum) were added when needed. Isoniazid (INH) (isonicotinic acid hydrazide, CAS number 54- 85-3, Acros) was diluted in water, and added in designated concentrations.
Cloning and expression of KatGbov in mycobacteria.
The katGbov construct was PCR amplified from M. bovis genomic DNA (gDNA). To place it under the mycobacteria optimized promoter (MOP), the PCR was repeated 3 consecutive times with the upstream primers KatGMOP1, KatGMOP2, and KatGMOP3, each reaction adding 25 to 35 bases to rebuild the promoter. The downstream primer was KatGavrnotU in all reactions. The final 2.3 kb product (confirmed by Sanger sequencing) was cloned into an attB positive, integrase negative, kanamycin- and zeocin-selected plasmid, to produce pDB246. This plasmid was electroporated into M. marinum or M. avium subsp. paratuberculosis K-10 (with the integrase supplied in trans) as widely described, and transformants were selected on zeocin and kanamycin. Resulting colonies were checked by PCR using primers katGMOP3 and katGMOPdown, which produces a 0.55KB product only when katG is downstream of the MOP promoter. One of the correct colonies in M. marinum was named mDB85, and in M. avium subsp. paratuberculosis was named mDB199.
MIC determination, cell viability, and percent growth inhibition measurements.
To measure MIC to isoniazid of M. marinum, M. avium subsp. paratuberculosis, and the mutants expressing KatGbov, INH was added to appropriate media (7H9 ± mycobactin-J) to a maximal concentration of 25 μg/ml, and diluted ×2 twelve consecutive times (in 10 ml volume). Log-phase bacteria were diluted to ∼5 × 104 CFU/ml, and 100 μl of that culture (∼5 × 103 CFU were added to each tube. Bacteria were grown in 37°C (M. avium subsp. paratuberculosis) or 32°C (M. marinum), in gentle shaking. Growth was determined 3 days (M. marinum) or 7 days (M. avium subsp. paratuberculosis) after the no-drug control became turbid.
Cell viability assays were carried out using the BacTiter-Glo assay (Promega Corporation). Bacteria were grown to an optical density at 600 nm (OD600) of 0.5 and diluted ×103. These cultures were plated in white opaque 96-well plates (in triplicate) with drug dilutions in 100 μl final volume. Plates were incubated at 37°C for 14 days. Wells with no bacteria were used to determine background luminescence. Clofazimine at 1 μg/ml was used as the positive control for M. avium subsp. paratuberculosis K10. An equal volume of BacTiter-Glo reagent was added to each well. Luminescence was measured on day 14 using a NanoQuant infinite M200 Pro plate reader with an integration time of 1,000 ms per well. Growth inhibition was calculated from drug-free conditions (0 μg/ml INH) and defined as: % viability = ([Lumexpt − Lumblank]/[Lumdrugfree average − Lumblank]) × 100. The % inhibition was defined as: 100 − % viability.
The luminescence from experimental conditions is blanked from no bacteria controls and normalized to drug-free controls.
Sequence alignment and analysis.
Multiple sequence alignment was done using CLUSTAL Omega (1.2.4) (13, 24). (https://www.ebi.ac.uk/Tools/msa/clustalo/). Percent identity matrix was created by Clustal2.1 (25). Simple phylogeny tool was generated by the ClustalW2 program (26), using the aligned protein sequences. The accession numbers of the KatG proteins used are reported in Table S1.
Cloning, expression, and purification of recombinant proteins.
The katGbov, katGmari, and katGavp genes were PCR amplified from gDNA of the respective mycobacteria, and cloned into pET29a vectors, enabling introducing of a His6 tag at the C terminus. A StrepII tag was added upstream to each of the katG genes by PCR addition of the DNA sequence. Both N and C termini were tagged to avoid truncated proteins during the purification process. After verification by sequencing, the recombinant KatG proteins were expressed in Escherichia coli BL21(DE3)pLysS cells (Novagen). Cells were grown in LB media at 37°C to an OD600 of 0.6 to 0.8, and induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 21°C for 12 to 16 h. To maximize heme incorporation, hemin chloride (40 mg/liter; Sigma) was exogenously added to the medium, and 50 μg/ml Fe(NH4)2(SO4)2 (Sigma) was supplemented just before induction. Cell pellets were dissolved in lysis buffer that included lysozyme, DNase I, and a protease inhibitor cocktail, in 25 mM phosphate buffer (pH 7.0) and lysed by sonication. The StrepII-tagged proteins were purified using a StrepTrap HP 5 ml column prepacked with StrepTactin (GE Healthcare), equilibrated with binding buffer. Proteins were eluted with binding buffer supplemented with 2.5 mM d-Desthiobiotin (Sigma). The proteins were further purified using size exclusion column (Superdex 200, GE Healthcare) in a 25 mM phosphate buffer (pH 7.2), and eluted as dimers in the gel filtration column, as calculated using a standard curve (not shown). Enzyme concentrations were determined using heme extinction coefficient ε407 = 100 mM−1 cm−1 (23).
Circular dichroism.
Circular dichroism (CD) spectra were recorded using a J-810 spectropolarimeter (Jasco) in 25 mM potassium phosphate buffer, pH 7.2 and 4 μM each protein, with or without 8 μM INH. All samples were measured in a 0.1 cm quartz cuvette for far-UV CD spectroscopy. Far-UV CD spectra were collected over a spectral range of 190 to 260 nm. Data were collected each 1 nm and averaged over 3 acquisitions. Prior to each experiment, the proteins were incubated overnight with or without INH. Wavelength scans were corrected for buffer contributions and converted to molar ellipticity. Changes in the CD spectra were monitored as a function of temperature from 5 to 95°C with 5°C increments, collected each 1 nm and averaged over 3 repeats.
INH-NAD adduct formation kinetics analysis.
INH-NADH adduct-formation assay was adopted from (22, 23). A SpectraMax i3 UV-visible plate reader equipped with 96-well plates was used, and measurements were performed in quadruplets. Adduct formation was measured spectrophotometrically following the increase in absorbance at 326 nm and adduct concentration was calculated using the reported extinction coefficient of ε326 = 9,600 M−1 cm−1 (27). The reaction was carried out at 25°C in a total volume of 200 μl, 25 mM K2HPO4/KH2PO4, pH 7.2, 0.5 μM KatG, and 8 mM NAD+. The reaction proceeded in the presence of an H2O2-generating system glucose/glucose oxidase (G/Gox) (glucose oxidase [0.5 U/ml], glucose [5 mM]). Increasing concentrations of INH (0.15, 0.2, 0.3, 0.4, 0.6, 0.8, 1.0, 1.25, 1.5, 2, 4, 6, and 8 mM) were used and initial velocity was determined from a linear fit of the first 5 min. To correct for background activity, a parallel blank experiment contained all components except NAD+ in each of the INH concentrations tested. Absorbance baseline was subtracted from a mixture that includes NAD+ in the absence of INH. Kinetic parameters (Km, Vmax) were extracted from Michaelis-Menten plots using KALEIDAGRAPH (Synergy Software, Reading, PA).
Isothermal titration calorimetry.
Isothermal titration calorimetry (ITC) experiments were performed using MicroCal iTC200 instrument (Malvern Instruments).
KatG proteins in 50 to 100 μM concentration were stored at 4°C for a week, to enable a higher fraction of the active six-coordinate (6-c) heme iron (21). The proteins were dialyzed against reaction buffer (25 mM K2HPO4 pH 7.2) prior to the experiment. INH solutions were freshly prepared and degassed in a sonication bath before titration. The concentration of INH titrant varied in each experiment, ranging from 5- to 10-fold higher than the concentration of the protein, as the active fraction for each protein changed. Titrations performed at 25°C with 18 to 36 injections of 1 to 2 μl per injection. Slow changes in signals required long intervals between injections, ranging between 300 and 720 s, under continuous stirring. The data were analyzed using Origin 7.0 software. Control titration of buffer into KatG and INH into buffer showed negligible heats of dilutions that were thus ignored in data processing. Binding affinities were calculated from fitting the binding curve to a one set of sites model. The integrated enthalpy changes were best fit to a single-binding site model.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoav Barak of the Bio-Nano unit, Chemical Research Support, Yael Fridmann-Sirkis and Irina Shin of the Department of Life Sciences Core Facilities, Weizmann Institute of Science, Rehovot, Amnon Horovitz of the Weizmann Institute, and Assaf Friedler of Hebrew University for their useful advice, help in data analysis, and technical assistance.
D.B. and Y.F.C. were supported by research grant IS-4905-16 from BARD, the United States-Israel Binational Agriculture Research and Development fund.
We declare no financial or other conflict of interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fox HH. 1952. The chemical approach to the control of tuberculosis. Science 116:129–134. doi: 10.1126/science.116.3006.129. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein J, Lott WA, Steinberg BA, Yale HL. 1952. Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am Rev Tuberc 65:357–364. doi: 10.1164/art.1952.65.4.357. [DOI] [PubMed] [Google Scholar]
- 3.Pansy F, Stander H, Donovick R. 1952. In vitro studies on isonicotinic acid hydrazide. Am Rev Tuberc 65:761–764. [PubMed] [Google Scholar]
- 4.Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 5.Heym B, Zhang Y, Poulet S, Young D, Cole ST. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol 175:4255–4259. doi: 10.1128/jb.175.13.4255-4259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. 2004. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol 52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR. Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 8.Vilcheze C, Jacobs WR. Jr. 2019. The isoniazid paradigm of killing, resistance, and persistence in Mycobacterium tuberculosis. J Mol Biol 431:3450–3461. doi: 10.1016/j.jmb.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Middlebrook G, Cohn ML, Schaefer WB. 1954. Studies on isoniazid and tubercle bacilli. III. The isolation, drug-susceptibility, and catalase-testing of tubercle bacilli from isoniazid-treated patients. Am Rev Tuberc 70:852–872. doi: 10.1164/art.1954.70.5.852. [DOI] [PubMed] [Google Scholar]
- 10.Nieto RL, Mehaffy C, Creissen E, Troudt J, Troy A, Bielefeldt-Ohmann H, Burgos M, Izzo A, Dobos KM. 2016. Virulence of Mycobacterium tuberculosis after acquisition of isoniazid resistance: individual nature of katG mutants and the possible role of AhpC. PLoS One 11:e0166807. doi: 10.1371/journal.pone.0166807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Garbe T, Young D. 1993. Transformation with katG restores isoniazid-sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol Microbiol 8:521–524. doi: 10.1111/j.1365-2958.1993.tb01596.x. [DOI] [PubMed] [Google Scholar]
- 12.Carey AF, Rock JM, Krieger IV, Chase MR, Fernandez-Suarez M, Gagneux S, Sacchettini JC, Ioerger TR, Fortune SM. 2018. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14:e1006939. doi: 10.1371/journal.ppat.1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marttila HJ, Soini H, Eerola E, Vyshnevskaya E, Vyshnevskiy BI, Otten TF, Vasilyef AV, Viljanen MK. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob Agents Chemother 42:2443–2445. doi: 10.1128/AAC.42.9.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wengenack NL, Rusnak F. 2001. Evidence for isoniazid-dependent free radical generation catalyzed by Mycobacterium tuberculosis KatG and the isoniazid-resistant mutant KatG(S315T). Biochemistry 40:8990–8996. doi: 10.1021/bi002614m. [DOI] [PubMed] [Google Scholar]
- 16.Wengenack NL, Uhl JR, St Amand AL, Tomlinson AJ, Benson LM, Naylor S, Kline BC, Cockerill FR 3rd, Rusnak F. 1997. Recombinant Mycobacterium tuberculosis KatG(S315T) is a competent catalase-peroxidase with reduced activity toward isoniazid. J Infect Dis 176:722–727. doi: 10.1086/514096. [DOI] [PubMed] [Google Scholar]
- 17.Zhao X, Hersleth HP, Zhu J, Andersson KK, Magliozzo RS. 2013. Access channel residues Ser315 and Asp137 in Mycobacterium tuberculosis catalase-peroxidase (KatG) control peroxidatic activation of the pro-drug isoniazid. Chem Commun (Camb) 49:11650–11652. doi: 10.1039/c3cc47022a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertrand T, Eady NA, Jones JN, Jesmin, Nagy JM, Jamart-Gregoire B, Raven EL, Brown KA. 2004. Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991–38999. doi: 10.1074/jbc.M402382200. [DOI] [PubMed] [Google Scholar]
- 19.Bernard EM, Edwards FF, Kiehn TE, Brown ST, Armstrong D. 1993. Activities of antimicrobial agents against clinical isolates of Mycobacterium haemophilum. Antimicrob Agents Chemother 37:2323–2326. doi: 10.1128/aac.37.11.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu S, Girotto S, Lee C, Magliozzo RS. 2003. Reduced affinity for Isoniazid in the S315T mutant of Mycobacterium tuberculosis KatG is a key factor in antibiotic resistance. J Biol Chem 278:14769–14775. doi: 10.1074/jbc.M300326200. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X, Yu S, Magliozzo RS. 2007. Characterization of the binding of isoniazid and analogues to Mycobacterium tuberculosis catalase-peroxidase. Biochemistry 46:3161–3170. doi: 10.1021/bi062218p. [DOI] [PubMed] [Google Scholar]
- 22.Cade CE, Dlouhy AC, Medzihradszky KF, Salas-Castillo SP, Ghiladi RA. 2010. Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: catalase, peroxidase, and INH-NADH adduct formation activities. Protein Sci 19:458–474. doi: 10.1002/pro.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Yu H, Yu S, Wang F, Sacchettini JC, Magliozzo RS. 2006. Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45:4131–4140. doi: 10.1021/bi051967o. [DOI] [PubMed] [Google Scholar]
- 24.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Lei B, Wei CJ, Tu SC. 2000. Action mechanism of antitubercular isoniazid. Activation by Mycobacterium tuberculosis KatG, isolation, and characterization of inha inhibitor. J Biol Chem 275:2520–2526. doi: 10.1074/jbc.275.4.2520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.